Abstract
Being able to easily determine the Cu(II) affinity for biomolecules of moderate affinity is important. Such biomolecules include amyloidogenic peptides, such as the well-known amyloid-β peptide involved in Alzheimer’s disease. Here, we report the synthesis of a new water soluble ratiometric Cu(II) dye with a moderate affinity (109 M-1 at pH 7.1) and the characterizations of the Cu(II) corresponding complex by X-ray crystallography, EPR and XAS spectroscopic methods. UV-Vis competition were performed on the Aβ peptide as well as on a wide series of modified peptides, leading to an affinity value of 1.6 109 M-1 at pH 7.1 for the Aβ peptide and to a coordination model for the Cu(II) site within the Aβ peptide that agrees with the one mostly accepted currently.
Copper ions play key biological roles.1 They are essential metal ions that play an important role as catalytic centers in several processes including very fundamental ones like energy production.1 They have also been linked to several diseases. Dyshomeostasis of Cu is very dangerous and is well documented by two lethal genetic diseases, called Wilson's and Menkes' diseases, linked to an overload of Cu and to a depletion of Cu, respectively.2–5 Cu imbalance has also been involved in the etiology of most of neurological disorders6 such as Alzheimer's disease (AD),7–9 Parkinson's disease (PD),10,11 Prion diseases.10,12
At a molecular scale, two parameters are important regarding Cu and the peptides or proteins involved in the above-mentioned diseases: (i) the metal environment, i.e. the nature of the atoms surrounding the metal center and (ii) the affinity of the peptides for the metal center. In the context of AD which is under focus in the present article, there are many reports on the coordination sites of the Cu(II) center to the amyloid-β peptide (Aβ) (for recent reviews, see refs.8,13–15) and on the Cu(II) affinity for Aβ (see refs.16–18 and refs. therein). Some studies also aim at relating the Cu(II) affinity of modified Aβ peptides with the coordination sites.19–21
Determining the affinity of Cu(II) for peptides is thus of interest, first because this is intrinsically an important parameter that mirrors the possibility to have the metal ion-peptide interaction in biological conditions, and second because it give insights into the coordination sphere of the metal center when a series of modified peptides is studied. As a direct consequence, the straightforward and accurate determination of Cu affinity for peptides is an important objective. However, this is an intricate task since it necessitates appropriate analytical tools. The complexity of such studies is well illustrated by the abundant and differing reports on the evaluation of Cu(II) affinities for the Aβ peptide (see refs.16–20 and refs. therein). This is also true for the +I redox state of Cu,18,22–24 which is however not under focus in the present study. While Cu(II) affinity values ranging from 106 M-1 to 1019 M-1 have been proposed,25 a relative consensus has recently been reached around a value of 1010 M-1 at pH 7.417,18 (this value corresponds to the conditional affinity value, i.e. the absolute affinity value at a given pH ; the interaction with buffer is not considered and will lead to a weaker so-called apparent affinity value).25 Hence the objective of the present study is to define a new tool appropriate for the rapid, easy and accurate evaluation of Cu(II) binding affinities to peptides.
There are mainly two ways of determining affinity values.16 The first one relies on potentiometric titrations that lead to absolute constants, from which an apparent affinity value can be calculated at any pH values.16,26 The second one consists in measurements at a given pH either by (i) isothermal calorimetry (ITC), fluorescence when a fluorophore is natively present in the peptide sequence or added for purpose and when the fluorescence is quenched by the addition of the metal ion of interest or (ii) by competition experiments with a ligand of known affinity followed by the appropriate method. Potentiometry is likely the most powerful technique since it gives access to the affinity value at any pH values but could be extremely difficult to implement in case of biomolecules and requires important quantity of the biological material. Results from ITC are far from straightforward although additional data apart from the affinity can be evaluated (enthalpy, entropy…) (reviewed in ref. 16). Competition experiments appear to be a well-suited method. With respect to Cu(II), UV-Vis and fluorescent spectroscopic tools are of interest. In a seminal paper, Wedd and coworkers have designed four peptide-based fluorophores of various Cu(II) affinities ranging from 108 M-1 to 1014 M-1. They were further used to probe the affinity of Cu(II) for different peptides and proteins by competition monitored by fluorescence.27 While Cu(II) quenches the fluorescence of the competitor, the addition of the peptide of interest restores it, removing the Cu(II) from the probe.
In the present paper, we propose an alternative strategy that relies on a UV-visible competition experiment against an easy to synthesize Cu(II) dye with moderate affinity (3.2 109 M-1 at pH 7.1) that is anticipated to be perfectly appropriate for evaluation of Cu(II) affinity to weakly structured peptide, such as Aβ, at physiological pH. By UV-Vis, the addition of the peptide of interest on the colored Cu(II) complex induces the disappearance of the absorption bands.
Using this new dye, we have investigated more than seventeen peptidic sequences derived from the human Aβ peptide, including murine, N-terminally modified peptides and biologically relevant mutants. Their Cu(II) affinities were evaluated and discuss with respect to the coordination models previously proposed in the literature.13,21,28–30
Experimental Section
Chemicals
Reagents were commercially available and were used as received. All the solutions were prepared in milliQ water (resistance: 18.2 MΩ.cm).
The Cu(II) ion source was CuSO4.5H2O, bought from Sigma-Aldrich. A stock solution was prepared at 25 mM.
HEPES buffer (sodium salt of 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid) was bought from Sigma-Aldrich. A stock solution was prepared at 500 mM, pH = 7.3 in order to reach a resulting pH 7.1 and the same buffer stock solution was used over the course of the present study.
Peptides
Aβ16 (DAEFRHDSGYEVHHQK) and the mutants:
Ac-Aβ16, Ac-DAEFRHDSGYEVHHQK; D1N-Aβ16, NAEF-RHDSGYEVHHQK; E3Q-Aβ16, DAQFRHDSGYEVHHQK; D7N-Aβ16, DAEFRHNSGYEVHHQK; E11Q-Aβ16, DAEFRHDSGYQVHHQK; H6R-Aβ16, DAEFRRDSGYEVHHQK; H6A-Aβ16, DAEFRADSGYEVHHQK; H13A-Aβ16, DAEFRHDSGYEVAHQK; H14A-Aβ16, DAEFRHDSGYEVHAQK; mAβ16, DAEFGHDSGFEVRHQK; R5G-Aβ16, DAEFGHD-SGYEVHHQK; Y10F-Aβ16, DAEFRHDSGFEVHHQK; H13R-Aβ16, DAEFRHDSGYEVRHQK; R5G-H13R-Aβ16, DAEFGHDSGYEVRHQK; Aβ28, DAEFRHDSGYEVHHQKLVFFAEDVGSNK; Aβ40, DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAILGLMVGGVV were bought from GeneCust (Dudelange, Luxembourg) with purity grade > 95%.
Stock solutions of the peptides were prepared by dissolving powder in milliQ water (resulting pH ~ 2), except for the Aβ28 and Aβ40 peptides which were dissolved in NaOH 50 mM. Peptide concentration was determined by UV-Vis absorption of Tyr10 considered as free tyrosine (at pH ~ 2.0, ε276 - ε296 = 1410 cm-1.M-1; at pH ~ 12.5, ε293 – ε360 = 2400 cm-1.M-1).31 For the mutants without Tyr10, the absorption of the two Phe was used (ε258 – ε280 = 390 cm-1.M-1).32
As the Cu(II) coordination sites in the Aβ peptide are localized in the first 16 amino acids residues,8 the Aβ16 and its counterparts were used as a model of the full-length peptides.
ABH and BAH where B corresponds to the β-alanine were bought from Protéogenix (Strasbourg, France) and stock solutions were prepared by solubilizing the peptides in milliQ water. Their concentrations were determined by UV-Vis titration with a titrated Cu(II) solution following the d-d band absorption of the complex,33 according to ref. 34.
Ligand
The 3,4-bis(oxamato)benzoic acid ligand, L (see Scheme S1), was synthetized as described below and used in its sodium salt form. A 8.5 mM stock solution was prepared, increasing the pH until solubilization, determined by UV-Vis titration with a titrated Cu(II) solution following the band absorption at 330 nm of the complex (see text below and Figure 3), according to ref. 34.
Figure 3.
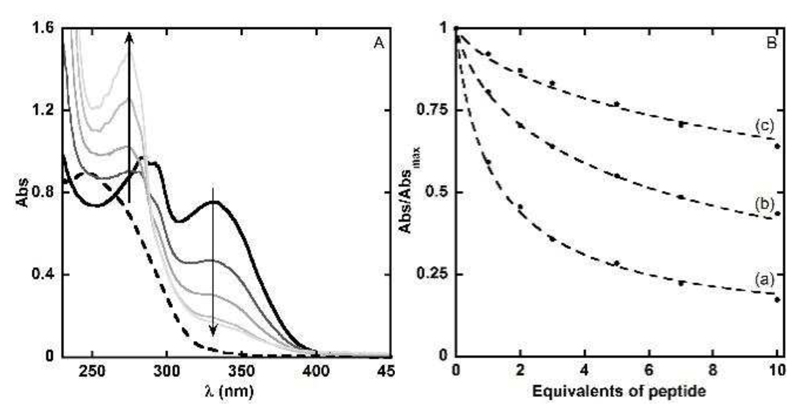
Panel A. UV-Vis spectra of a solution of L (dotted line), in the presence of 0.9 eq. of Cu(II) (solid line) and after addition of increasing amounts (approx.. 1, 3, 7 and 10 eq.) of Aβ16 peptide (grey lines). The arrows indicate the variation of the UV-Vis spectra upon addition of the peptide. The increase at 276 nm is due to Tyr10 absorption. Panel B. Experimental absorbance at 330 nm (dots) and the best fits (dashed lines) of the Cu(L) system upon addition of peptides (Aβ16 (a), H6A-Aβ16 (b) and Ac-Aβ16 (c)). [L] = 50 μM, [Cu(II)] = 45 μM, [peptides] = 0 to 500 μM, [HEPES] = 0.1 M, pH = 7.1, T = 25°C.
Synthesis of H5L
H3Et2L: 3,4-(diethyloxamate)benzoic acid:
The ester form of the L ligand was prepared following the known procedure for oxamate ligands35,36 instead of the previously reported synthesis.37 To 5 g of 3,4-diaminobenzoic acid (32.8 mmol) in THF (200 mL) is added with strong stirring 8.25 mL of ethyl oxalyl chloride (72 mmol, 2.2 eq). The mixture is refluxed for 2 hours, filtered while hot and let to cool down to room temperature. Removal of the solvent leads to a brown precipitate. The solid is abundantly washed with water, filtered, washed with 50 % v. ethanol and then with a minimum of cold 96 % v. ethanol, and dried in air. Yield: 8.9 g (77 %). See Supporting Info for elemental analysis, NMR and IR characterizations.
Na1.5H3.5L·2.2H2O
Aqueous NaOH (2 M, 57 mL) was added dropwise to a suspension of H3Et2L (8 g, 22.7 mmol) in water (400 mL). The solution was stirred for 30 min and filtered. Addition of 4 M HCl (28 mL) to the resulting solution leads to the rapid formation of a brown precipitate. After 10 min the precipitate is collected on a sintered glass filter, washed with cold water, then with 96 % v. EtOH, and finally with ether before being dried for several hours in air and overnight at 45°C in an oven. Yield: 5.4 g (64%). See Supporting Info for elemental analysis, NMR and IR characterizations.
Synthesis of Li3[Cu(L)]
Suitable crystals of the copper dye, Cu(L), were obtained as follow: A suspension of H3Et2L (0.100 g, 0.28 mmol) in water (10 mL) was treated with 0.71 mL of LiOH 2 M (1.42 mmol) and stirred at room temperature until complete dissolution of the ligand. An aqueous solution of Cu(NO3)2·3H2O (0.068 g, 0.28 mmol, 5 mL) was then added dropwise to the oxamate solution resulting in a deep blue solution of the copper complex that was stirred for 30 min at room temperature and filtered. Slow diffusion of acetone yields crystals of Li3[Cu(L)]·5.5H2O in 15 days. Yield: 0.054 g (41 % based on Cu). See Supporting Info for elemental analysis, NMR and IR characterizations.
Methods
X-Ray structure
Crystallographic data were collected on a Bruker Kappa-APEX II CCD diffractometer (CuKα, λ = 1,54178). Crystal data: purple plates, monoclinic C2/c, a = 26.3214(15), b = 6.7217(4), c = 20.4183(11) Å, β = 112.590(3)°, V = 3335.3(3) Å3, Z = 8, T = 200(1) K, ρ = 1.567 g.cm-3, F(000) = 1560, μCuKα= 2.293 mm-1. Crystals were mounted on a Hamilton cryoloop using Paratone-N oil and placed in the cold flow produced with an Oxford Cryocooling device. Partial hemispheres of data –preselected with the APEX II software38 were collected using φ and ω scans. Integrated intensities were obtained with SAINT+ and were corrected for absorption with SADABS;38,39 the structure was solved with SIR9240 and refined with SHELXL-2014/741 (WinGX software package).42 Data refinement gives (using 271 parameters and 6 restraints) wR2 = 0.2432 (2923 unique reflections), R1 = 0.0720 (2514 reflections with I>2σ(I)), GOF = 1.128. Crystallographic details are available in CIF format, free of charge via www.ccdc.cam.ac.uk/conts/retrieving.html (or from the Cambridge Crystallographic Data Centre, 12, Union Road, Cambridge CB2 1EZ, UK; fax: (+44) 1223-336-033; or deposit@ccdc.cam.ac.uk). CCDC number 1516020.
NMR, IR and Elemental Analysis
1H and 13C NMR spectra were collected on a 400 MHz Bruker Avance spectrometer at 298 K.
ATR/FT-IR spectra were collected on a Bruker TENSOR 27 equipped with a simple reflexion ATR diamond plate of the Harrick MPV2 series.
Elemental analysis was performed at the ICSN, CNRS UPR 2301, in Gif-sur-Yvette, France.
Electron Paramagnetic Resonance
Electron Paramagnetic Resonance (EPR) data were recorded using an Elexsys E 500 Bruker spectrometer, operating at a microwave frequency of approximately 9.5 GHz. Spectra were recorded using a microwave power of 20 mW across a sweep width of 150 mT (centred at 310 mT) with modulation amplitude of 0.5 mT. Experiments were carried out at 110 K using a liquid nitrogen cryostat.
EPR samples were prepared from stock solution of ligand diluted down to 0.2 mM in H2O. 0.9 eq. of 65Cu(II) was added from 25 mM 65Cu(NO3)2 stock solution home-made from a 65Cu foil. Samples were frozen in quartz tube after addition of 10% glycerol as a cryoprotectant and stored in liquid nitrogen until used.
X-ray Absorption Spectroscopy (XAS)
Cu(II) K-edge EXAFS (Extended X-Ray Absorption Fine Structure) and XANES (X-ray Absorption Near–Edge Structure) spectra were recorded at the BM30B (FAME) beamline at the European Synchrotron Radiation Facility (ESRF, Grenoble, France).43 The storage ring was operated in 7/8+1 mode at 6 GeV with a 200 mA current. The beam energy was selected using a Si(220) N2 cryo-cooled double-crystal monochromator with an experimental resolution close to that theoretically predicted (namely ~ 0.5 eV FWHM at the Cu energy). The beam spot on the sample was approximately 300 x 100 µm2 (H x V, FWHM). Because of the low Cu(II) concentration, spectra were recorded in fluorescence mode with a 30-element solid state Ge detector (Canberra) in frozen liquid cells in a He cryostat. The temperature was kept at 20 K during data collection. The energy was calibrated with Cu metallic foil, such that the maximum of the first derivative was set at 8979 eV. EXAFS Cu(II) data were collected from 8840 to 8960 eV using 5 eV step of 2 s, from 8960 to 9020 eV using 0.5 eV step of 3 s, and from 9020 to 9300 eV with a k-step of 0.05 Å-1 and 3 s per step. At least six scans recorded on different spots were averaged. XAS samples were prepared from stock solutions of ligand and Cu(II) diluted down to approx. 1.0 mM in buffered solution. Samples were frozen in the sample holder after addition of 10% glycerol as a cryoprotectant and stored in liquid nitrogen until used. Cu(II) photoreduction was controlled by recording successive scans at the same spot. It was considered that during the first 20 minutes of recording the photoreduction is insignificant.
The data analysis was performed using the “Multi-Plateform Applications for X-ray Absorption” package, including Cherokee and Roundmidnight programs,44 according to the standard and previously reported data analysis procedures.45,46 Spectra were background-corrected by a linear regression through the pre-edge region and a polynomial through the post-edge region. The backscattering phase, Φi(k, Ri), and amplitude, Ai(k, Ri), functions were obtained using the ab initio FEFF7 code.47 Since theoretical phase shifts were used, it is necessary to fit the energy threshold E0 by adding an extra fitting parameter, ΔE0. Moreover, the FEFF7 code was used to check if the multiple scattering of our reference compounds of known crystallographic structure is negligible in the 0–3 Å range. The estimated errors for distances and coordination numbers are ± 0.02 Å and ± 20%, respectively.
UV-Visible spectrophotometry
UV-vis spectra were recorded on a spectrophotometer SPECORD S600 Analytik Jena at 25°C in 1 cm path length quartz cuvette.
Competition experiments between the ligand and the peptides
The experiments have been monitored by UV-Vis in HEPES buffer 0.1 M (at a resulting pH = 7.1). The ligand (50 µM in theory) and Cu(II) (45 µM in theory) were mixed and successive additions of the different peptides (0 to 500 µM in total) were added. Each addition of the peptide is made once the thermodynamic equilibrium of the previous reaction is reached (this takes about 20 min). To be reproducible with respect to pH condition, the same stock buffer solution (pH = 7.3 to reach a resulting pH of 7.1) was used for all the experiments. The competition experiments were performed at 25°C in triplicate.
Analysis of the data
The data analysis was performed with a 2-step procedure for each experiment (corresponding to a given peptide).
Step 1. The theoretical concentrations of the ligand L and the Cu(II) were adjusted using the absorbance of ligand at 246 nm (ε246 - ε450 = 18000 cm-1.M-1) and the absorbance of the Cu(L) complex at 330 nm (ε330 - ε450 = 18000 cm-1.M-1) respectively.
Step 2. The absorbance of the competition experiments measured at 330 nm after completion of the reaction was plotted as a function of the number of peptides equivalents recalculated with respect to the real ligand concentration (see step 1). Then these data were fitted with an in-house procedure using the adjusted concentrations as starting parameters.
Absorbance was calculated according to:
where P stands for peptide, L for the ligand, and CuP and CuL for the complex between Cu(II) and peptide and the ligand, respectively.
The same equation can be expressed as:
where α stands for the progression of the following reaction : peptide + Cu(II) → peptide-Cu(II)
and the [ ]0 correspond to the starting values in which the dilution due to addition of the peptide has been taken into account when required.
Cu(II) is supposed to be chelated by the peptide or by L, there is no free Cu(II).
With the starting concentrations:
This is a quadratic equation having the form
Then, the value was adjusted to obtain the best reproduction of the experimental data.
NB: Although Kd / Ka are dimensionless values, they are given here in M or M-1, respectively, for convenience. Indeed for 1:1 system, the Kd value in M gives a direct idea at which concentration the complex is predominantly formed.
NB: in a preliminary experiment, was evaluated by competition with ABH and BAH knowing and 33 with the similar procedure.
Results
The competitor used in the present work to evaluate the Cu(II) affinity of a wide series of Aβ derived peptides by UV-Vis competition experiments, is an oxamate ligand (H5L, Scheme S1, for the sake of clarity L5- will be written L. Similarly, the [Cu(L)]3- will be noted Cu(L)). Oxamate ligands have largely proved their appeal for the design of metalloligands and the subsequent preparation of multidimensional compounds with controlled architectures and properties.48 In particular, the use of oxamate ligands and the multi-step assembly of oxamate-based complexes have successfully led to relevant objects in molecular magnetism: switchable molecules, high nuclearity coordination complexes, single-chain magnets, porous magnets.49,50 Recently oxamate-based systems have also shown attractiveness in broader coordination chemistry fields such as catalysis.51,52 Here, we will take advantage of the mild Cu(II) affinity and of the presence of an intense UV-Visible band for the complex with Cu(II) (λ = 330 nm, ε = 18000 cm-1.M-1).
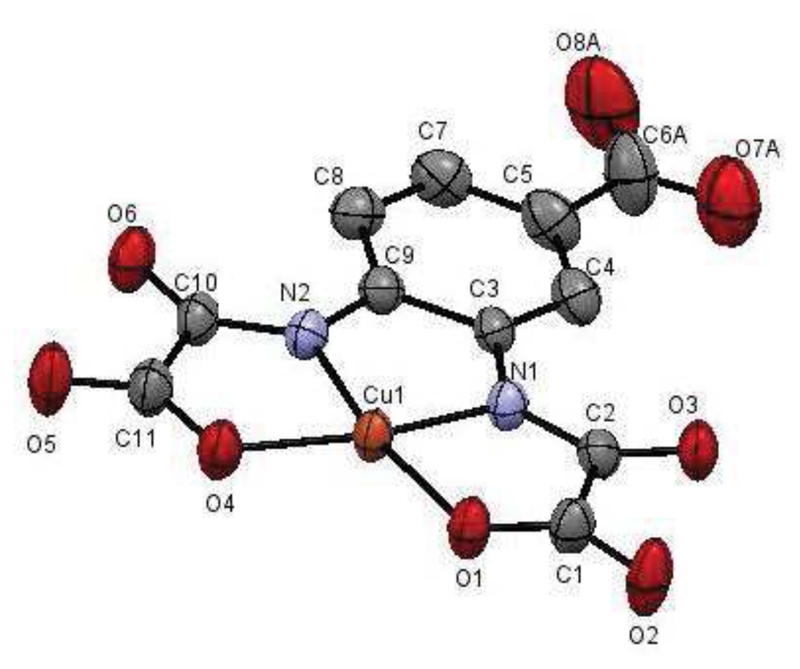
The X-ray structure of Cu(L) is shown in Figure 1 and the crystallographic parameters are given in Table 1, top, and in the Supporting Info. The complex adopts a square-planar geometry where the L ligand coordinates the Cu(II) ion via its two oxamate groups each binding the metal center through one nitrogen and one oxygen atom. The square-planar geometry is fairly regular with a 2.4° dihedral angle between the oxamate groups and a 3.1° dihedral angle between the Cu(II) coordination plane and the phenyl plane of the ligand. The carboxylate group of the ligand remains uncoordinated. Supramolecular arrangements of the complexes are described in the Supporting Information (Figure S1).
Figure 1.
Ellipsoid plot (H and disorders atoms are omitted for clarity) of Cu(L).
Table 1. Crystallography distances between Cu(II) and their neighbours (top) and EXAFS parameters (bottom): N = number of neighbours, R = absorber-neighbour distance, σ = Debye-Waller factor.
| N | Distance (Å) |
|---|---|
| Cu-N1 | 1.9061(55) |
| Cu-N2 | 1.9127(37) |
| Cu-O1 | 1.9588(34) |
| Cu-O4 | 1.9642(51) |
| N | Distance (A) |
| 2 Cu-N | 1.92 |
| 2 Cu-O | 1.97 |
| σ2 (Å-2) | R factor |
| 0.00471 | 0.18 % |
The persistence of the structure in aqueous solution has been probed by several complementary techniques. The most appropriate one is EXAFS by which the Cu-N and Cu-O distances have been determined in solution and are in perfect agreement with those obtained for the crystal structure (for crystallography, see Table 1, bottom and Figure S1, for EXAFS fitting, see Figure S2).
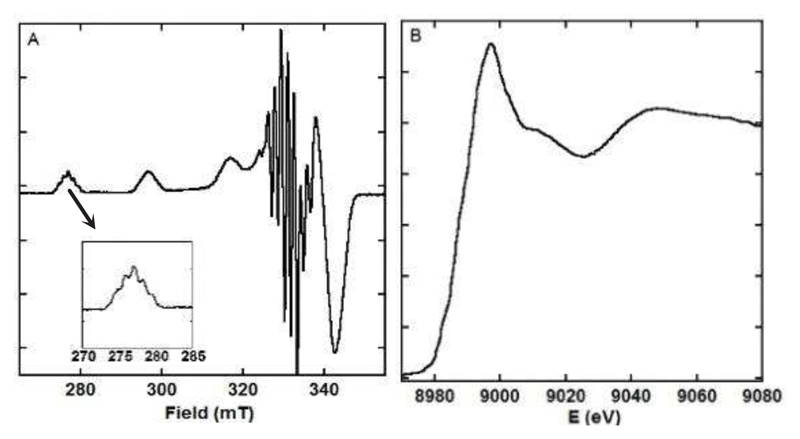
The parameters estimated from the EPR signature (Figure 2A) are also in line with the Cu(L) solid-state structure. Indeed, the g// and A//(65Cu) values equal 2.22 and 212 104 cm-1, respectively and are thus in line with a 2N2O equatorial binding mode according to the Peisach and Blumberg correlation.53 A weak rhombicity linked to the presence of four non equivalent equatorial ligands is at the origin of the complex features observed in the g⊥ region (coupling between ICu = 3/2 with SCu = 1/2 for the two gx and gy transitions). The presence of two equivalent nitrogen atoms is consistent with the five super-hyperfine lines observed on the first parallel transition, arising from the coupling between IN = 1 with SCu = 1/2. The XANES spectrum of Cu(L) is given in Figure 2B. It is in line with Cu(II) complexes where the Cu(II) center lies in a square planar environment.1
Figure 2.
Panel A. EPR signature of Cu(L). [L] = 200 μM, [65Cu(II)] = 180 μM, [HEPES] = 50 mM, pH = 7.1, T = 120 K. Inset: superhyperfine structure on the first parallel line. Panel B. Normalized Cu K-edge X-ray Absorption Near –Edge Structure (XANES) spectrum of Cu(L). [L] = 1 mM, [Cu(II)] = 0.9 mM, [HEPES] = 0.1 M, pH = 7.0, T = 20K.
The other important parameter of this ligand is its affinity constant for Cu(II). This affinity was evaluated by the same type of competition experiments than those detailed below, performed first between L and two short peptides, namely ABH and BAH (B stands for β-alanine).33 These peptides were studied by potentiometry to determine the impact of the 5-membered versus 6-membered metallacycles with regard to the Cu(II) affinity in an ATCUN-like (Amino Terminal CU and Nickel) sites.54 Their conditional Cu(II) affinity values calculated at pH 7.1 from the potentiometric data equal: 3.0 108 and 3.7 109 M-1, respectively. Competition experiments with the ABH (resp. BAH) lead to a Cu(II) affinity value of 2.2 109 M-1 (resp. 4.1 109 M-1) for L. The average value of = 3.2 ± 1.0 109 M-1 was used in the rest of the study.
The competition experiment between L and the Aβ peptides for Cu(II) binding were performed by using the peptides to remove the Cu(II) from the colored Cu(L) complex leading to a disappearance of the signature at 330 nm characteristic of the Cu(L) complex. Figure 3A shows the typical series of spectra obtained for a given peptide (here Aβ16). L was first added (dotted line), then Cu(II) (solid line) and then successive additions of the peptide were performed (grey lines, See Experimental Section for more details). Note that the increase at 276 nm is due to Tyr10 absorbance and perfectly matches the amount of peptide added.
The absorbance at λ = 330 nm of the mixture was plotted as a function of the equivalent numbers of peptide added (dots in Figure 3B). Then, these data were fitted with an in-house procedure (dashed lines, for more details see Experimental section).
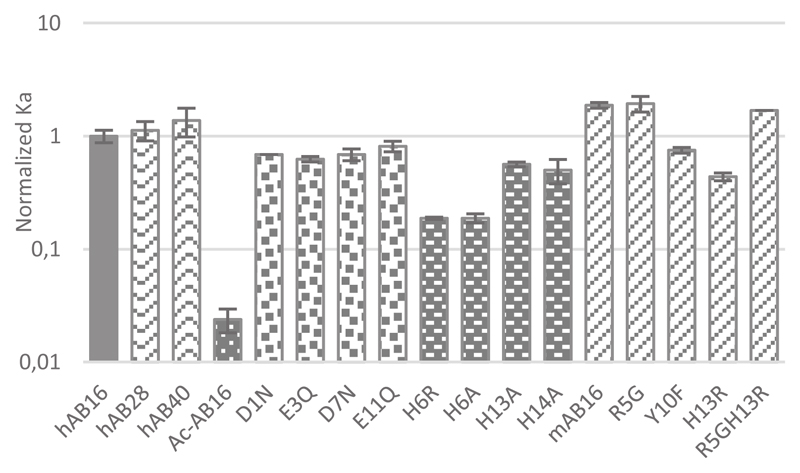
Experimental absorbance values and their fits are shown in Figure 3B as a matter of illustration for Aβ16, H6A-Aβ16 and Ac-Aβ16, (curves a, b, c, respectively). Qualitative analysis indicates that the Cu(II) affinity of the three peptides follow the trend: Aβ16 > H6A-Aβ16 > Ac-Aβ16. All the Cu(II) affinity values obtained in the present study are gathered in Table 2 while the corresponding reproductions of the experimental data are given in the Supporting Info (Figure S3). The accuracy of the fitting procedure is shown in the Figure S4. All the calculated Cu(II) affinity values were normalized with respect to the value of Aβ16 for an easier comparison, as shown in Table 2 and in Figure 4.
Table 2. Cu(II) affinity values of the Aβ peptides studied here. The reference for the normalization is the Aβ16. A Cu(II) affinity value of 3.2 ± 1.0 109 M-1 for the competing ligand L was taken.
| Peptide | Ka (109 M-1) a | Ka(peptide)/Ka(hAβ16) |
|---|---|---|
| Aβ16 | 1.6 | 1.0 |
| Aβ28 | 1.8 | 1.1 |
| Aβ40 | 2.2 | 1.4 |
| Ac-Aβ16b | < 0.1 | < 0.1 |
| D1N-Aβ16 | 1.1 | 0.7 |
| E3Q-Aβ16 | 1.0 | 0.6 |
| D7N-Aβ16 | 1.1 | 0.7 |
| E11Q-Aβ16 | 1.3 | 0.8 |
| H6R-Aβ16 | 0.3 | 0.2 |
| H6A-Aβ16 | 0.3 | 0.2 |
| H13A-Aβ16 | 0.9 | 0.6 |
| H14A-Aβ16 | 0.8 | 0.5 |
| mAβ16 | 3.0 | 1.9 |
| R5G-Aβ16 | 3.1 | 1.9 |
| Y10F-Aβ16 | 1.2 | 0.8 |
| H13R-Aβ16 | 0.7 | 0.4 |
| R5G-H13R-Aβ16 | 2.7 | 1.7 |
The error bars are evaluated in the S.I. (see Figure S4).
The Ka value for Ac-Aβ16 is too weak to be thoroughly determined by this method. Only a higher value can be given.
Figure 4.
Bar graph of the normalized affinity constants in log units of the Aβ peptides and some of their mutants for Cu(II). The reference for the normalization is the hAβ16. The error bars indicate the standard deviation. [L] = 50 μM, [Cu(II)] = 45 μM, [peptides] = 0 to 500 μM, [HEPES] = 0.1 M, pH = 7.1, T = 25°C. The affinity constant of the ligand is Ka = 3.2 ± 1.0 109 M-1.
Discussion
The Aβ peptides used in this study were modified with respect to the human sequence on any potential binding amino-acids, namely the carboxylate containing amino-acid residues, the His residues and the N-terminal amine. The mutation involved between the human and murine sequences were also studied.
Human-derived peptides
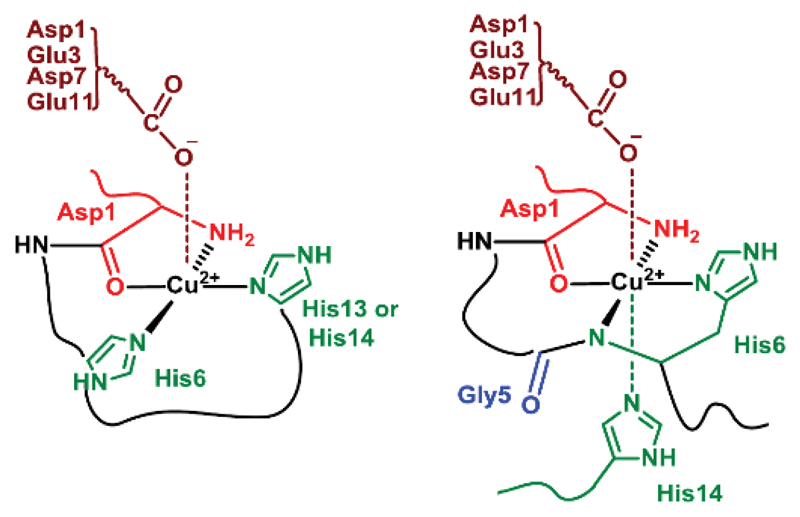
The first modification is the acetylation of the N-terminal amine (Asp1) precluding the possibility of Cu(II) binding. The affinity constant of Cu(II) for this modified peptide is at least 10 times weaker than that of the Aβ16 peptide, in line with an important contribution of the N-terminal amine in Cu(II) binding to Aβ16, as discussed in the literature.28,55,56 In the second group of modified peptides, the impact of the carboxylate function is evaluated. The carboxylate group of each amino acid residues Asp1, Glu3, Asp7 and Glu11 is mutated with an amide function. Only a very weak impact is observed with a decrease in the affinity upon modification that doesn’t exceed a factor of 2. In the Aβ16 there are 4 carboxylate groups available for the Cu(II) coordination. Therefore, if one carboxylate is mutated, 3 other ones remain to complete the coordination sphere. This is in perfect agreement with the possibility of having an equilibrium between the four carboxylate groups for one binding position.57 The high similarity of affinity values for the four carboxylate-modified peptides also suggest that there is no preference for one given carboxylate group. The last group of modified peptides consist in the His mutants (His6, His13 and His14). While His6 mutation (H6A or H6R) has a significant impact (a 5-fold decrease of the affinity value), His13 or His14 mutation has a lesser effect (with a maximal 2-fold decrease of the affinity value). This indicates that (i) none of the His is essential but (ii) the His13 and His14 can more easily exchange for the binding position on the peptide than His6 and His13 or than His6 and His14 do. This is in line with the proposition that in the main coordination site His13 and His14 exchange for one binding position while His6 is constantly bound.13,30,58 It is worth mentioning that similar differences have been previously determined by ITC.19,20 All together these data fit very well with the coordination site of Cu(II) proposed based on spectroscopic data (reviewed in refs.13,30) and reminded in Scheme 1. In such proposition, the Cu(II) ion is equatorially bound to the N-terminal amine, the adjacent carbonyl group from the Asp1-Ala2 bond, the imidazole group form His 6 and from His13 or 14 in equilibrium (see Scheme S2, top).8 A carboxylate-containing residue has been proposed to occupy the apical position.57 Last, the Cu(II) affinity seems to weakly increase with the length of the peptide, in line with previous reports,17,59 but this result has to be taken with cautious due to the increase in the error bar when working with the Aβ40 peptide. This confirms that the Aβ16 moiety is a good model with respect to Cu(II) binding site within the full-length peptide.
Scheme 1. Predominant forms at pH 7.1 of the Cu(II) coordination with human (left) and murine (right) Aβ peptides.
Human versus murine peptides
The murine sequence differs from the human one by 3 mutations: R5G, Y10F and H13R. With respect to Cu(II) affinity values, the murine peptide is twice stronger than the human one, in line with previous measurements by direct competition,17,29 ITC,19 and potentiometry.55 The main mutation responsible for such a difference is the R5G mutation. This has been previously related to the formation of a 6-membered metallacycle between the side chain of His6 and the deprotonated Gly5-His6 peptide function for the murine peptide that is not observed in the human case (see Schemes 1 and S2 bottom).29,30
Conclusions
In the present study, a new ratiometric dye was synthetized and fully characterized. It was further used to determine the affinity constant of Cu(II) for several Aβ peptides by UV-Vis competition experiments. This was made possible by the intense (18000 cm-1.M-1) LMCT band displayed at λ = 330 nm by the Cu(L) complex. The affinity values obtained in this study for the various modified peptides agree perfectly with published coordination sites, thus validating the proposed strategy that relies on the determination of affinity constants of a wide series of modified peptides to extrapolate a coordination model for a given peptide. In addition, the designed dye could be further used to straightforwardly determine Cu(II) affinity for other flexible peptides such as alpha-synuclein involved in Parkinson’s disease11 or Prion protein,12 for which the Cu(II) affinities are proposed to lie in the 108-1010 M-1 range. However, outside this range, competition with L will not give reliable values. In addition, the molar extinction coefficient is relatively low implying the use of quite large biomolecule concentration. As a consequence, to generalize this approach to the evaluation of Cu(II) affinity for any other (bio)molecules relies on the ability to modulate the Cu(II) affinities and to increase the molar extinction coefficient of oxaloamino-benzoic acid derivatives by playing with the scaffold.
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website. Characterization of the ligand and of the Cu(L) complex, EXAFS parameters, Competition data, Error bar, Cu coordination model.
Acknowledgements
CH thanks the ERC aLzINK - Contract n° 638712 for financial support. The authors thank the European Synchrotron Radiation Facility for provision of beamtime (experiment 30-02-1060), the FAME staff for their support, F. Collin, and C. Cheignon for their help in recording the XANES and EXAFS data. Prof. P. Faller is acknowledged for interesting discussions.
References
- (1).Solomon EI, Heppner DE, Johnston EM, Ginsbach JW, Cirera J, Qayyum M, Kieber-Emmons MT, Kjaergaard CH, Hadt RG, Tian L. Chem Rev. 2014;114:3659–3653. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Sarkar B. Chem Rev. 1999;99:2535–2544. doi: 10.1021/cr980446m. [DOI] [PubMed] [Google Scholar]
- (3).Delangle P, Mintz E. Dalton Trans. 2012;41 doi: 10.1039/c2dt12188c. [DOI] [PubMed] [Google Scholar]
- (4).Lalioti V, Muruais G, Tsuchiya Y, Pulido D, Sandoval IV. Frontiers Biosciences. 2009;14:4878–4903. doi: 10.2741/3575. [DOI] [PubMed] [Google Scholar]
- (5).Scheiber IF, Mercer JFB, Dringen R. Prog Neurobiol. 2014;116:33–57. doi: 10.1016/j.pneurobio.2014.01.002. [DOI] [PubMed] [Google Scholar]
- (6).Kozlowski H, Luczkowski M, Remelli M, Valensin D. Coord Chem Rev. 2012;256:2129–2141. [Google Scholar]
- (7).Ayton S, Lei P, Bush AI. Free Radic Biol Med. 2013;62:76–89. doi: 10.1016/j.freeradbiomed.2012.10.558. [DOI] [PubMed] [Google Scholar]
- (8).Hureau C. Coord Chem Rev. 2012;256:2164–2174. [Google Scholar]
- (9).Barnham KJ, Bush AI. Chem Soc Rev. 2014;43:6727–6749. doi: 10.1039/c4cs00138a. [DOI] [PubMed] [Google Scholar]
- (10).D'Ambrosi N, Rossi L. Neurochem Int. 2015;90:36–45. doi: 10.1016/j.neuint.2015.07.006. [DOI] [PubMed] [Google Scholar]
- (11).Binolfi A, Quintanar L, Bertoncini CW, Griesinger C, Fernández CO. Coord Chem Rev. 2012;256:2188–2201. [Google Scholar]
- (12).Arena G, La Mendola D, Pappalardo G, Sóvágó I, Rizzarelli E. Coord Chem Rev. 2012;256:2202–2218. [Google Scholar]
- (13).Drew SC, Barnham KJ. Acc Chem Res. 2011;44:1146–1155. doi: 10.1021/ar200014u. [DOI] [PubMed] [Google Scholar]
- (14).Faller P, Hureau C, Berthoumieu O. Inorg Chem. 2013;52:12193–12206. doi: 10.1021/ic4003059. [DOI] [PubMed] [Google Scholar]
- (15).Telpoukhovskaia MA, Orvig C. Chem Soc Rev. 2013;42:1836–1846. doi: 10.1039/c2cs35236b. [DOI] [PubMed] [Google Scholar]
- (16).Zawisza I, Rozga M, Bal W. Coord Chem Rev. 2012;256:2297–2307. [Google Scholar]
- (17).Alies B, Renaglia E, Rozga M, Bal W, Faller P, Hureau C. Anal Chem. 2013;85:1501–1508. doi: 10.1021/ac302629u. [DOI] [PubMed] [Google Scholar]
- (18).Young TR, Kirchner A, Wedd AG, Xiao Z. Metallomics. 2014;6:505–517. doi: 10.1039/c4mt00001c. [DOI] [PubMed] [Google Scholar]
- (19).Hong L, Carducci TM, Bush WD, Dudzik CG, Millhauser GL, Simon JD. J Phys Chem B. 2010;114:11261–11271. doi: 10.1021/jp103272v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hong L, Simon JD. Metallomics. 2011;3:262–266. doi: 10.1039/c0mt00052c. [DOI] [PubMed] [Google Scholar]
- (21).Alies B, Bijani C, Sayen S, Guillon E, Faller P, Hureau C. Inorg Chem. 2012;51:12988–13000. doi: 10.1021/ic302097d. [DOI] [PubMed] [Google Scholar]
- (22).Alies B, Badei B, Faller P, Hureau C. Chem Eur J. 2012;18:1161–1167. doi: 10.1002/chem.201102746. [DOI] [PubMed] [Google Scholar]
- (23).Feaga HA, Maduka RC, Foster MN, Szalai VA. Inorg Chem. 2011;50:1614–1618. doi: 10.1021/ic100967s. [DOI] [PubMed] [Google Scholar]
- (24).Xiao Z, Gottschlich L, van der Meulen R, Udagedara SR, Wedd AG. Metallomics. 2013;5:501–513. doi: 10.1039/c3mt00032j. [DOI] [PubMed] [Google Scholar]
- (25).Faller P, Hureau C. Dalton Trans. 2009:1080–1094. doi: 10.1039/b813398k. [DOI] [PubMed] [Google Scholar]
- (26).Arena G, Papppalardo G, Sovago I, Rizzarelli E. Coord Chem Rev. 2012;256:3–12. [Google Scholar]
- (27).Young TR, Wijekoon CJK, Spyrou B, Donnelly PS, Wedd AG, Xiao Z. Metallomics. 2015;7:567–578. doi: 10.1039/c4mt00301b. [DOI] [PubMed] [Google Scholar]
- (28).Alies B, Eury H, Bijani C, Rechignat L, Faller P, Hureau C. Inorg Chem. 2011;50:11192–11201. doi: 10.1021/ic201739n. [DOI] [PubMed] [Google Scholar]
- (29).Eury H, Bijani C, Faller P, Hureau C. Angew Chem Int Ed Engl. 2011;50:901–905. doi: 10.1002/anie.201005838. [DOI] [PubMed] [Google Scholar]
- (30).Hureau C, Dorlet P. Coord Chem Rev. 2012;256:2175–2187. [Google Scholar]
- (31).Faller P, Hureau C, Dorlet P, Hellwig P, Coppel Y, Collin F, Alies B. Coord Chem Rev. 2012;256:2381–2396. [Google Scholar]
- (32).Fasman GD. HoBaMB. 3rd ed. CRC; 1976. Proteins, I; p. 183. [Google Scholar]
- (33).Nagaj J, Stokowa-Sołtys K, Zawisza I, Jeżowska-Bojczuk M, Bonna A, Bal W. J Inorg Biochem. 2013;119:85–89. doi: 10.1016/j.jinorgbio.2012.11.002. [DOI] [PubMed] [Google Scholar]
- (34).Hureau C, Eury H, Guillot R, Bijani C, Sayen S, Solari PL, Guillon E, Faller P, Dorlet P. Chem Eur J. 2011;17:10151–10160. doi: 10.1002/chem.201100751. [DOI] [PubMed] [Google Scholar]
- (35).Stumpf HO, Pei Y, Kahn O, Sletten J, Renard JP. J Am Chem Soc. 1993;115:6738–6745. [Google Scholar]
- (36).Cervera B, Sanz JL, Ibanez MJ, Vila G, Lloret F, Julve M, Ruiz R, Ottenwaelder X, Aukauloo A, Poussereau S, Journaux Y, et al. J Chem Soc Dalton Trans. 1998;5:781–790. [Google Scholar]
- (37).Paul-Roth CO. C R Chimie. 2005;8:1232–1236. [Google Scholar]
- (38).Bruker AXS Inc. B. Madison; Wisconsin USA: 1998. [Google Scholar]
- (39).Blessing RH. Acta Crystallogr, Sect A: Found Crystallogr. 1995;51:33. doi: 10.1107/s0108767394005726. [DOI] [PubMed] [Google Scholar]
- (40).Altomare A, Cascarano G, Giacovazzo C, Guagliardi A, Burla MC, Polidori G, Camalli M. J Appl Crystallogr. 1994;27:435. [Google Scholar]
- (41).Sheldrick GM. Acta Crystallogr, Sect C: Cryst Struct Commun. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Farrugia LJ. J Appl Crystallogr. 2012;45:849–854. [Google Scholar]
- (43).Proux O, Biquard X, Lahera E, Menthonnex JJ, Prat A, Ulrich O, Soldo Y, Trévisson P, Kapoujvan G, Perroux G, Taunier P, et al. Phys Scr. 2005;115:970–973. [Google Scholar]
- (44).Michalowicz A, Moscovici J, Muller-Bouvet D, Provost K. J Phys: Conf Ser. 2009;190:012034–012035. [Google Scholar]
- (45).Lengeler B, Eisenberg P. Phys Rev B: Condens Matter Mater Phys. 1980:4507–4520. [Google Scholar]
- (46).Guillon E, Merdy P, Aplincourt M. Chemistry - A European Journal. 2003;9:4479–4484. doi: 10.1002/chem.200304920. [DOI] [PubMed] [Google Scholar]
- (47).Zabinsky SI, Rehr JJ, Ankudinov AL, Albers RC, Eller M. J Phys Rev B: Condens Matter Mater Phys. 1995;52:2995–3009. doi: 10.1103/physrevb.52.2995. [DOI] [PubMed] [Google Scholar]
- (48).Pardo E, Ruiz-García R, Cano J, Ottenwaelder X, Lescouëzec R, Journaux Y, Lloret F, Julve M. Dalton Trans. 2008;21:2780. doi: 10.1039/b801222a. [DOI] [PubMed] [Google Scholar]
- (49).Dul M-C, Pardo E, Lescouëzec R, Journaux Y, Ferrando-Soria J, Ruiz-Garcia R, Cano J, Julve M, Lloret F, Cangussu D, Pereira CLM, et al. Coord Chem Rev. 2010;254:2281–2296. [Google Scholar]
- (50).Grancha T, Ferrando-Soria J, Castellano M, Julve M, Pasán J, Armentano D, Pardo E. Chem Commun. 2014;50:7569–7585. doi: 10.1039/c4cc01734j. [DOI] [PubMed] [Google Scholar]
- (51).Fortea-Pérez FR, Schlegel I, Julve M, Armentano D, De Munno G, Stiriba S-E. J Organomet Chem. 2013;743:102–108. [Google Scholar]
- (52).Pim WDd, Oliveira WXC, Ribeiro MA, Faria ÉNd, Teixeira IF, Stumpf HO, Lago RM, Pereira CLM, Pinheiro CB, Figueiredo-Júnior JCD, Nunes WC, et al. Chem Commun. 2013;49:10778–10780. doi: 10.1039/c3cc46242k. [DOI] [PubMed] [Google Scholar]
- (53).Peisach J, Blumberg WE. Arch Biochem Biophys. 1974;165:691–708. doi: 10.1016/0003-9861(74)90298-7. [DOI] [PubMed] [Google Scholar]
- (54).Bal W, Sokołowska M, Kurowska E, Faller P. Biochim Biophys Acta. 2013;1830:5444–5455. doi: 10.1016/j.bbagen.2013.06.018. [DOI] [PubMed] [Google Scholar]
- (55).Kowalik-Jankowska T, Ruta M, Wisniewska K, Lankiewicz L. J Inorg Biochem. 2003;95:270–282. doi: 10.1016/s0162-0134(03)00128-4. [DOI] [PubMed] [Google Scholar]
- (56).Syme CD, Nadal RC, Rigby SE, Viles JH. J Biol Chem. 2004;279:18169–18177. doi: 10.1074/jbc.M313572200. [DOI] [PubMed] [Google Scholar]
- (57).Hureau C, Coppel Y, Dorlet P, Solari PL, Sayen S, Guillon E, Sabater L, Faller P. Angew Chem Int Ed Engl. 2009;48:9522–9525. doi: 10.1002/anie.200904512. [DOI] [PubMed] [Google Scholar]
- (58).Dorlet P, Gambarelli S, Faller P, Hureau C. Angew Chem Int Ed Engl. 2009;48:9273–9276. doi: 10.1002/anie.200904567. [DOI] [PubMed] [Google Scholar]
- (59).Kowalik-Jankowska T, Ruta-Dolejsz M, Wisniewska K, Lankiewicz L. J Inorg Biochem. 2001;86:535–545. doi: 10.1016/s0162-0134(01)00226-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.