Abstract
A mutation-accumulation (MA) experiment with Caenorhabditis elegans nematodes was conducted in which replicate, independently evolving lines were initiated from a low-fitness mitochondrial electron transport chain mutant, gas-1. The original intent of the study was to assess the effect of electron transport chain dysfunction involving elevated reactive oxygen species production on patterns of spontaneous germline mutation. In contrast to results of standard MA experiments, gas-1 MA lines evolved slightly higher mean fitness alongside reduced among-line genetic variance compared with their ancestor. Likewise, the gas-1 MA lines experienced partial recovery to wildtype reactive oxygen species levels. Whole-genome sequencing and analysis revealed that the molecular spectrum but not the overall rate of nuclear DNA mutation differed from wildtype patterns. Further analysis revealed an enrichment of mutations in loci that occur in a gas-1-centric region of the C. elegans interactome, and could be classified into a small number of functional-genomic categories. Characterization of a backcrossed four-mutation set isolated from one gas-1 MA line revealed this combination to be beneficial on both gas-1 mutant and wildtype genetic backgrounds. Our combined results suggest that selection favoring beneficial mutations can be powerful even under unfavorable population genetic conditions, and agree with fitness landscape theory predicting an inverse relationship between population fitness and the likelihood of adaptation.
Keywords: adaptation, complex I, electron transport chain, mitochondrial dysfunction, mutation accumulation, sign epistasis
Introduction
In agreement with predictions of Fisher’s geometric model of adaptation (Fisher 1930), experimental evidence confirms that new mutations with measurable effects reduce the fitness of well-adapted genotypes (Lynch et al. 1999; Kondrashov and Kondrashov 2010). Original theoretical treatments of the threat to population survival posed by such mutations assumed that they affected fitness independently of genetic or environmental background and that little or no opportunity for reversion or other types of beneficial mutations existed (Lynch et al. 1993, 1995; Lande 1994). Empirical study suggests that both assumptions were wrong: beneficial mutations arise more frequently than originally appreciated (Joseph and Hall 2004; Hall et al. 2008; Achaz et al. 2014) and the most readily harmful mutations—those with moderate fitness effects—may be rare (Estes et al. 2004; Katju et al. 2015). Furthermore, individual mutational effects can be highly dependent upon their environmental and genomic contexts (Lenski and Travisano 1994; Burch and Chao 1999; Elena and Lenski 2001; Maisnier-Patin et al. 2002; Rokyta et al. 2002; Estes and Lynch 2003; Remold and Lenski 2004; Wang et al. 2009, 2013), with a potentially large supply of compensatory epistatic mutations available to partially ameliorate the effects of previously acquired deleterious mutations. Indeed, adaptive landscape theory predicts that the probability of improvement increases with the distance of a population’s mean fitness from a theoretical optimum, and that maladapted populations can access a greater fraction of beneficial mutations than well-adapted ones (Fisher 1930; Whitlock and Otto 1999; Poon and Otto 2000; Tenaillon 2014). In other words, the mutational distribution of fitness effects will depend upon how well adapted a population is (Martin and Lenormand 2006, 2015; Chevin et al. 2010). Another prediction is that small-effect beneficial mutations exist in greater supply than mutations of larger effect (Orr 1998); it follows that adaptive evolution in small populations (with fewer genomes available for mutation) will primarily be driven by fixation of small-effect mutations, whereas larger populations can adapt more quickly by fixing fewer large-effect mutations. These predictions have largely been borne out by data from studies with microbes (Burch and Chao 1999; Sanjuan et al. 2004; Perfeito et al. 2007; Silander et al. 2007; Barrick et al. 2010; Hietpas et al. 2013; Wang et al. 2013), although important exceptions exist (Miller et al. 2011). Studies in complex eukaryotes are still lacking (but see Stearns and Fenster 2016), which is problematic given the potential importance of organismal complexity and pleiotropy in determining real evolutionary outcomes (Orr 2000).
Laboratory mutation-accumulation (MA) experiments allow the impact of mutation to be distinguished from that of other evolutionary forces affecting rates of molecular evolution. Selection is rendered inefficient to the maximum extent possible by maintaining replicate lineages derived from a common ancestral genotype by clonal reproduction, self-fertilization, or extreme inbreeding across many generations, during which time MA lines accumulate and fix independent sets of mutations (Halligan and Keightley 2009; Teotónio et al. 2017). The vast majority of these experiments have initiated MA lines from wildtype ancestral strains that have undergone a period of laboratory domestication and are thus well adapted to laboratory conditions. Although we are aware of no MA experiments using extremely low-fitness ancestors, modifications to the standard MA design have included initiating MA lines from moderately low-fitness ancestors (Sharp and Agrawal 2012) and specific mutant genotypes for the purpose of revealing the effects of deficiencies in DNA repair pathways (Estes et al. 2004; Denver et al. 2006) and mitochondrial functioning (Joyner-Matos et al. 2011). Several MA experiments have now been combined with high-throughput sequencing to provide direct estimates of the average per-generation rate of mutations arising in nuclear genomes along with detailed analyses of their molecular spectra and patterns of bias (Lynch et al. 2008; Denver et al. 2009, 2012; Keightley et al. 2009; Ossowski et al. 2010; Lee et al. 2012; Saxer et al. 2012; Heilbron et al. 2014). When MA line fitness is evaluated, the near universal outcome is one of reduced mean fitness and increased among-line variance for fitness, consistent with mutations having predominantly deleterious effects. A notable exception is the study of Shaw et al. (2000, 2002), which estimated that half of the mutations accumulated in MA lines of Arabidopsis thaliana were beneficial (Shaw et al. 2002)—a result later supported by Rutter et al. (2010, 2012). Shaw et al. (2000) originally suggested that intraindividual selection (e.g., differential cell lineage growth) may have contributed to this result. It would of course be difficult to experimentally minimize or control selection at this level, but by confining our attention to individual-level selection, we may underestimate the power of beneficial mutation to sustain the genetic health and fitness of populations.
The mitochondrial electron transport chain (ETC) offers a compelling framework for studying the endogenous factors that influence mutation. Because the mitochondrial ETC is vital for energy metabolism in all complex life and is thus highly conserved, it is an ideal system for studying the impact of deleterious mutation. Since proper ETC functioning relies on the maintenance of favorable mitonuclear epistatic interactions (Blier et al. 2001; Dowling et al. 2008), it is also an excellent system for addressing the role of epistasis in maintaining population fitness. The mitochondrial ETC also generates reactive oxygen species (ROS) as a byproduct of cellular metabolism (Murphy 2009). ROS are important cell signaling molecules (Mittler et al. 2011), but high ROS levels (e.g., resulting from mitochondrial dysfunction) have long been hypothesized to be mutagenic, especially for mtDNA genomes owing to their proximity to the site of production (Hsie et al. 1986; Demple and Harrison 1994; Cooke et al. 2003). Recent work fails to implicate oxidative stress in somatic mtDNA mutation (Ameur et al. 2011; Yu et al. 2013; Itsara et al. 2014); however, the question of whether ROS are an important contributor to heritable germline mutation remains understudied.
Here, we build upon our previous work to provide a comprehensive, integrative assessment of the impact of mitochondrial dysfunction on nuclear DNA (nDNA) variation and phenotypes related to fitness and physiology. As described in Wernick et al. (2016), which reported on mitochondrial DNA mutation processes, we performed a MA experiment using a mitochondrial ETC-deficient genotype of C. elegans, gas-1(fc21), as the ancestor. We phenotyped replicate MA lines initiated from this mutant and applied Illumina MiSeq technology to analyze the nuclear genomes of a subset of these lines. As before, we compare our findings to those from a previous MA study initiated from a wildtype C. elegans ancestor (Baer et al. 2005). Bioinformatic approaches identified novel, putatively beneficial mutations fixed within MA lines; a subset of these was characterized to determine the contribution to surprising patterns of fitness and phenotypic evolution observed here.
Materials and Methods
Strains
This study utilized MA lines generated from a gas-1 mutant strain of C. elegans. The gas-1(fc21) allele is a single basepair missense mutation (Kayser et al. 1999) associated with deleterious phenotypes including: reduced fecundity, reduced complex I-dependent metabolism (Kayser et al. 2004), hypersensitivity to oxidative stress owing to increased ROS production (Kayser et al. 2001, 2004), and low ATP levels relative to wildtype (Hartman et al. 2001; Kayser et al. 2001, 2004; Lenaz et al. 2006). The gas-1 mutant, derived from ethyl methanesulfonate (EMS) mutagenesis (Kayser et al. 1999), was obtained from the Caenorhabditis Genetics Center (University of Minnesota) and repeatedly backcrossed to N2 in an attempt to create an isogenic mutant strain. As detailed in Wernick et al. (2016), which reported on the mitochondrial DNA mutation processes within the same MA lines, offspring of a single gas-1 (fc21) hermaphrodite were then used to initiate 48 MA lines, maintained for an average of 43 generations (range: 35–47). Failed bottlenecks occasionally resulted from hermaphrodite sterility or death prior to reproduction; in this event, a sibling from the same generation was chosen from a backup plate to initiate the next generation. An MA line was considered extinct when it could not be reconstituted in this way. A subset of five gas-1 MA lines was selected at random for whole-genome sequencing and additional phenotypic analyses. We also utilized data from a previous analysis of MA lines generated from a wildtype N2 strain, which were evolved in the manner described earlier for a maximum of 250 generations (Baer et al. 2005). Namely, gas-1 MA results were compared with those from a subset of five N2 MA lines for which genome sequence (Denver et al. 2009, 2012) and phenotypic data (Denver et al. 2009; Joyner-Matos et al. 2013; Andrew et al. 2015) were available.
Life History Assays and Data Analysis
We assayed nematode life-history and analyzed the resulting data following established methods (Joyner-Matos et al. 2011). Owing to difficulty in working with the poorly performing gas-1 MA lines, life-history assays were conducted in four blocks in which different subsets of gas-1 MA lines were assessed alongside N2 and gas-1 ancestral (hereafter “gas-1 G0” for generation 0) controls. A fifth and sixth block were conducted to increase the sample size of the five gas-1 MA lines selected for whole genome sequencing (below). Specifically, we measured daily offspring production for the gas-1 MA lines, gas-1 G0 and N2 strains; we also recorded the number of unhatched and presumably unviable eggs, easily distinguished from unfertilized eggs under a light microscope. Data were used to calculate mean absolute (total reproductive output, W) and relative fitness (ω) of the gas-1 mutant compared with N2, and of each gas-1 MA line compared with the gas-1 G0 ancestor. Relative fitness of each individual was computed as: ω = Σe−rxl(x)m(x), where l(x) is the number of worms surviving to day x, m(x) is the fecundity at day x, and r is the mean intrinsic population growth rate of the assay-specific N2 or gas-1 G0 control as appropriate. The latter was calculated by solving Euler’s equation for r from the equation ω = Σe−rxl(x)m(x) = 1 using an average value of l(x)m(x) for each block-specific control. We used x = 4.75 on the first reproductive day (c.f., Vassilieva et al. 2000).
To determine the impact of MA on evolution of the above phenotypes, the per generation change in the mean, ΔM, of each trait was estimated as the generalized linear regression coefficient of trait value scaled as a fraction of the block-specific gas-1 G0 mean on line-specific MA generation number. For the MA lines, within- and among-line components of variance were calculated using restricted maximum likelihood (REML) with variance estimates constrained to be nonnegative (except for when calculating confidence intervals). We tested the model trait mean = μ + block + MA line(block) + parent[MA line(block)] + ɛ, wherein all effects are random—and MA line(block) represents among-line (genetic) variance and ɛ represents the within-line (environmental) component of variance, VE. A frequency distribution of gas-1 MA line fitness relative to gas-1 G0 was created using least squares means for the gas-1 MA lines derived from the MA line(block) term using the expected mean squares (EMS) method as applied in JMP12 (SAS). For N2 and gas-1 G0 control lines, we tested the model trait mean = μ + block + parent(block) + ɛ, wherein all effects are random and parent(block) represents among-line (or pseudoline) variance. Second, the per-generation change in genetic variance owing to MA, the mutational variance VM, was calculated for each trait as the difference in the among-line components of variance of the gas-1 G0 control and the MA lines divided by 2t, where t = the maximal number of MA generations. The per generation change in among-line variance, Vb, was calculated for the gas-1 MA lines by dividing the among line variance for each line by the average number of generations, 42.8.
Steady-State ROS and ATP Levels
Prior to all experiments, strains were allowed to recover from freezing for two generations at 20 °C on Nematode Growth Medium-Lite (NGML) plates containing 20 µg/ml streptomycin and seeded with Escherichia coli OP50-1. Lines were age synchronized by standard bleach treatment prior to each assay. We quantified in vivo steady-state ROS levels of the five gas-1 MA lines compared with N2 and gas-1 G0 strains using established fluorescence microscopy methods (Hicks et al. 2012; Joyner-Matos et al. 2013; Smith et al. 2014). ROS levels of the five sequenced N2 MA lines were previously reported (Joyner-Matos et al. 2013). We quantified ATP content for four to five independent samples from each of the five N2 and gas-1 MA lines compared with N2 and gas-1 G0 strains following methods adapted from Van Raamsdonk et al. (2010) and Yang and Hekimi (2010). For each sample, roughly 200 age-synchronized young adult worms were washed and worm cuticles were broken via successive freeze-thaw cycles. Samples were boiled for 15 min, sonified (20% amplitude, twice for 12 s), and centrifuged at 10,000×g for 12 min. About 90 µl of supernatant was used to quantify ATP levels with a luminescence ATP detection kit (Invitrogen, Carlsbad, CA) and a TECAN Infinite M200 Pro plate reader (Tecan, Männedorf, Switzerland). ATP measurements were normalized by protein content measured with a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA).
Illumina MiSeq Read Mappings and Analyses
Whole-genome sequencing was conducted for five randomly chosen gas-1 MA lines, N2 and the backcrossed gas-1 G0. Samples were prepared from first larval (L1) stage nematodes as described in (Wernick et al. 2016). Following each Illumina MiSeq run, reads were aligned to the C. elegans genome (version WS242) using CLC Genomics Workbench (CLC Bio-Qiagen, Aarhus, Denmark). All reads were paired-end (2×150 bp) and mapped using the following parameters: no masking, mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.98, read fraction = 0.98, global alignment = no, nonspecific match handling = map randomly. Illumina data were deposited in the SRA database under accession number SRP069774.
Characterization of gas-1 G0 Genetic Background
Two potential sources of genetic difference between the gas-1 G0 and N2 progenitor strains are: (1) mutations resulting from the EMS mutagenesis employed to create the original gas-1 mutant, and (2) spontaneous mutations fixed during the ten generations of backcrossing. The majority of mutations from the first source should have been eliminated during the ten generations of backcrossing gas-1 to N2 males, but some EMS-induced mutations, particularly those closely linked to the gas-1 locus, may have remained. C. elegans experiences base substitutions at the rate of approximately two per generation (Denver et al. 2004), meaning that roughly 20 mutations were expected to have accumulated during the backcrossing process. All single-basepair mutations in gas-1 G0 were identified as sites that differed from the C. elegans reference genome (WS242) and were not present in our N2 strain. To eliminate false positives resulting from sequencing artifacts, candidate mutations for which coverage was less than the average coverage of gas-1 candidate SNPs (18×) were omitted. Additionally, the following criteria were applied: (1) 100% of reads indicated a single nonreference base, (2) at least one read was present from both the reverse and forward strand, and (3) reads in a given direction varied upon start/end positions. Mutations of the six possible substitution types were counted in a chromosome-specific manner. Characteristics of gas-1 G0 mutations meeting the above criteria were compared with those published for N2 (Denver et al. 2012). χ2 tests were applied to test for significance.
Identification and Characterization of gas-1 MA Line Mutations
Candidate SNPs in gas-1 MA lines were identified as variants from the C. elegans reference genome (WS242) and our N2 lab strain. Each MA line was also surveyed for retention or loss of SNPs identified in gas-1 G0. To eliminate false positives, we applied the aforementioned criteria and required at least 5-fold coverage. Lastly, we only considered mutations found in one line to eliminate false positives associated with cryptic heterozygosity or paralogy following our previous approaches (Denver et al. 2009, 2012).
Mutation Rate Analysis
The nuclear mutation rate was calculated as previously described (Denver et al. 2009) from pooled gas-1 MA lines using the equation μbs= m/(LnT) where μbs is the base substitution rate (per nucleotide site per generation), m is the number of observed mutations, L is the number of MA lines, n is the number of nucleotide sites, and T is the time in generations. Values for n reflect the total number of base pairs surveyed that met the criteria for consideration of a possible mutation site. Conditional rate estimates for the six possible mutation types (A: T → G: C, G: C → A: T, A: T → C: G, G: C → T: A, A: T →T: A, and G: C → C: G) were also determined in a nonstrand specific manner using the pooled gas-1 MA line mutations. Standard errors for these estimates were approximated as [μbs/(nT)]1/2. We compared observed conditional rate estimates in the gas-1 MA lines to published values for N2 MA lines (Denver et al. 2012).
Gene Ontology Enrichments
We used GoMiner (Zeeberg et al. 2003, application build 457) and the GO MySQL Database (MySQL 3.7.13, Oracle Corporation, Cupertino, CA; GO database build 2016-06-07, geneontology.org) to calculate gene ontology (GO) enrichments for genes in the gas-1 interactome and all nDNA SNPS that arose within protein coding regions of gas-1 MA and the N2 MA lines within three functional domains—biological process, cellular component, and molecular function. GoMiner reports a two-sided Fisher’s exact P value based upon the number of genes in a category and a false discovery rate (FDR)-corrected P value for multiple testing (Zeeberg et al. 2005); however, the FDR can be overly conservative when the sample size or number of discoveries is small; that is, <100 (Zeeberg et al. 2005; Tong and Zhao 2008), so we based interpretations on results of the Fisher’s exact tests with α = 0.05. We then determined which broader-level functional “GO slim” categories, as defined by the GO Consortium (Version 1.2, 2012-09-21, geneontology.org), were enriched using CateGOrizer (Hu et al. 2008), which classifies enriched GO terms into their respective GO slim categories, giving a coarser-scale overview of enrichment patterns. GO slim categories were then checked against the GoMiner output to assess statistical significance.
Interactome Analysis of gas-1 and N2 MA Line Mutations
Following our previous approach (Denver et al. 2010), GeneOrienteer version 2.25 (Zhong and Sternberg 2006) was used to construct an interactome, a list of genes predicted to interact with a gene of interest (gas-1), and determine the extent to which mutated MA line genes showed membership within this network. We first queried the entire C. elegans interactome to determine the interactions involving gas-1. The query returned 181 genes predicted to “directly” interact with gas-1 within one degree. We then queried this smaller interactome to identify genes predicted to interact within two degrees of gas-1. We refer to the comprehensive list of first- and second-degree gas-1 interacting genes, totaling 4,189, as the “gas-1 centric interactome.” We next identified all genic mutations within the gas-1 and N2 MA line sets that were members of the gas-1-centric interactome and used Monte Carlo Maximum Likelihood simulations to assess the probability of observing the results by chance. Twenty-three mutations (the number of genic mutations discovered in gas-1 MA lines) were randomly generated using a simulation written in Python. This simulation was repeated for 1,000 iterations, and the average and standard deviation of the number of mutations occurring within the gas-1 centric interactome were calculated. This simulation was repeated for N2 with the exception that 59 mutations, the number of genic mutations discovered within N2 MA lines (Denver et al. 2012), were randomly generated.
Isolation and Characterization of Candidate Beneficial Mutations from MA431
Because results of the above analyses indicated that some degree of adaptive evolution had occurred in the gas-1 MA lines, we aimed to characterize candidate beneficial or compensatory SNPs that arose within one particular line. Line MA341 was chosen for analysis as it was found to contain SNPs affecting genes identified by the GO and interactome analyses and/or with known function—namely, rheb-1, daf-2, and sel-2, all of which reside on chromosome III. Toward this goal, MA431 was backcrossed to N2 following the same approach used to generate the gas-1 G0 strain. At each generation, DNA was extracted from the parental MA431 hermaphrodite (n = 30–44) for PCR and Sanger DNA sequencing at three SNP locations within the rheb-1, daf-2, and sel-2 genes. Offspring from hermaphrodites retaining at least one of these SNPs were used in the next round of crossing. Although our aim was to isolate each chromosome III SNP individually, we were unsuccessful at maintaining strains that did not contain all three (see Results for MA431 SNP linkage). After seven rounds of backcrossing, hermaphrodites containing MA431 mutations were allowed to self and tested to find homozygotes, resulting in a strain referred to hereafter as N2MA431. Finally, the MA431 chromosome III homozygous mutant lines were crossed with our near-isogenic gas-1 G0 strain and offspring allowed to self to produce a strain that contained only the chromosome III SNP mutations on the gas-1 background, gas-1MA431.
We performed a life-history assay as previously described for the N2MA431 and gas-1MA431 strains alongside N2 and gas-1 G0 controls, which included 40 replicates of each strain. Selection coefficients of the MA431 SNPs on each genetic background were calculated as the difference in relative fitness between N2MA431 and N2 and between gas-1MA431 and gas-1 G0. A Cox proportional hazards method was used to compare age-specific mortality of the N2MA431 and gas-1MA431 strains and their N2 and gas-1 G0 controls, with strain and parent(strain) as main effects. Lastly, we quantified in vivo steady-state ROS and ATP levels of the N2MA431 and gas-1MA431 lines compared with N2 and gas-1 G0 strains as previously described.
Results
Characterization of gas-1 G0 Genetic Background
Bioinformatic analysis of the gas-1 G0 strain identified 76 genetic differences relative to our N2 strain (supplementary fig. S1 and table S1, Supplementary Material online). The X-chromosome (where gas-1 is located) harbored the majority, ∼62%, of these mutations compared with 15.7% of the total mutations identified in the N2 MA lines (Denver et al. 2012)—a highly significant difference (P < 0.00001, χ2 test). The gas-1 G0 X-chromosome also experienced an increased proportion of G: C→A: T mutations compared with published N2 MA line results (80.9% vs. 23.5%; P < 0.00001, χ2 test). Genome-wide, the gas-1 G0 strain also showed a much higher proportion of G: C→A: T mutations compared with the published N2 value (61.8% vs. 33.3%; P < 0.00001, χ2 test). All 76 background mutations present within gas-1 G0 were retained within the sequenced MA lines (below).
gas-1 G0 Phenotypes
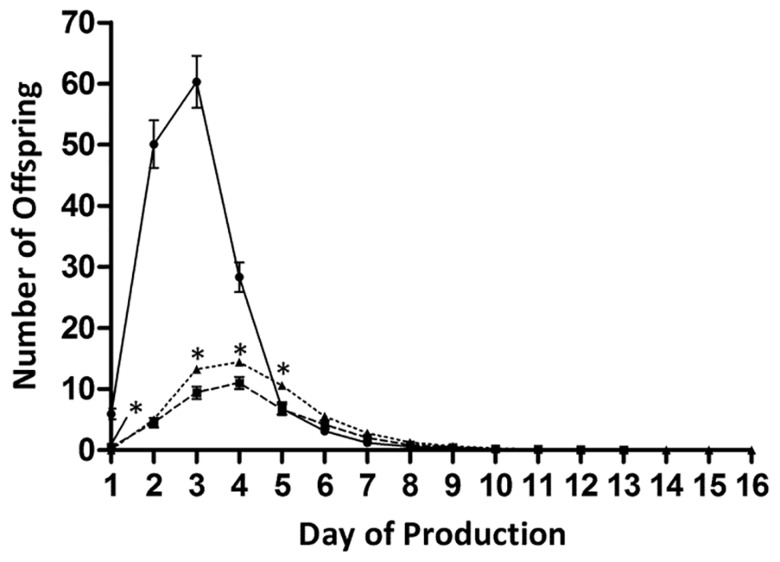
There were no significant effects of assay block on any life-history trait (not shown). As expected based on previous studies of the CGC gas-1 strain (Morgan and Sedensky 1994; Kayser et al. 1999), our backcrossed gas-1 G0 strain produced far fewer offspring than N2 (fig. 1 and table 1;supplementary table S2, Supplementary Material online), resulting in lower relative fitness (t(148) = 9.954, P < 0.0001; table 1). gas-1 G0 reproductive maturity and peak reproductive output were also delayed by a full day compared with N2 (fig. 1).
Fig. 1.—
Reproductive schedules. Daily offspring production of N2 (solid line), gas-1 G0 (dashed line, squares), and the combined gas-1 MA lines (dashed line, triangles). Error bars are 1 SEM. Summary statistics are reported in supplementary table S2, Supplementary Material online.
Table 1.
Summary Statistics for Fitness Traits
| W | ω | |
|---|---|---|
| N2 | 151.6±10.03 | 1.000±0.053 |
| gas-1 G0 | 38.40±3.277 | 0.225±0.057 |
| gas-1 G0a | 38.40±3.277 | 1.000±0.085 |
| gas-1 MAa | 50.58±1.647 | 1.260±0.058 |
| ΔM (x 10−3) | 127.9±38.20 | 5.757±1.317 |
| Vb gas-1 G0 | 76.74±84.81 | 0.054±0.059 |
| Vb gas-1 MA | 5.191±1.824 | 0.010±0.004 |
| VE gas-1 G0 | 628.6±105.7 | 0.014±0.002 |
| VE gas-1 MA | 20.58±1.439 | 0.017±0.001 |
| VM | −1.547 | −0.004 |
Note.—All values are means ± 1 SEM.
Fitness relative to gas-1 G0; fitness is otherwise reported relative to N2.
W, absolute fitness; ω, relative fitness.
Potential for Selection during gas-1 MA
Eight of the original 48 gas-1 MA lines (16.7%) went extinct prior to the 50 generation cut-off, as compared with only one extinction among 48 MA lines initiated from N2 (2%) during 125 generations of evolution (Joyner-Matos et al. 2011). The frequency of failed bottlenecks occurring during the gas-1 MA experiment, 14.2%, was twice that reported for the N2 MA lines, 7.1% (Joyner-Matos et al. 2011). Additionally, gas-1 MA lines laid an increased number of fertilized but ultimately unhatched eggs compared with ancestral controls. The difference in unhatched egg counts among N2, gas-1 G0 and MA groups (F2, 677 = 8.9345, P < 0.0001) was due to the gas-1 MA line group having a small but significantly greater number of unhatched eggs (3.68 ± 0.31) than either gas-1 G0 (1.51 ± 0.41) or N2 (1.09 ± 0.18) (Tukey HSD; α = 0.05, P < 0.01). In contrast, the prevalence of “bagging” (where internal hatching of eggs kill the parent) in the MA lines (8.2%) was indistinguishable from that in N2 (12.4%) or gas-1 G0 (10.2%) (χ2 test), and thus did not significantly impact MA line fitness.
gas-1 MA Life-History Evolution
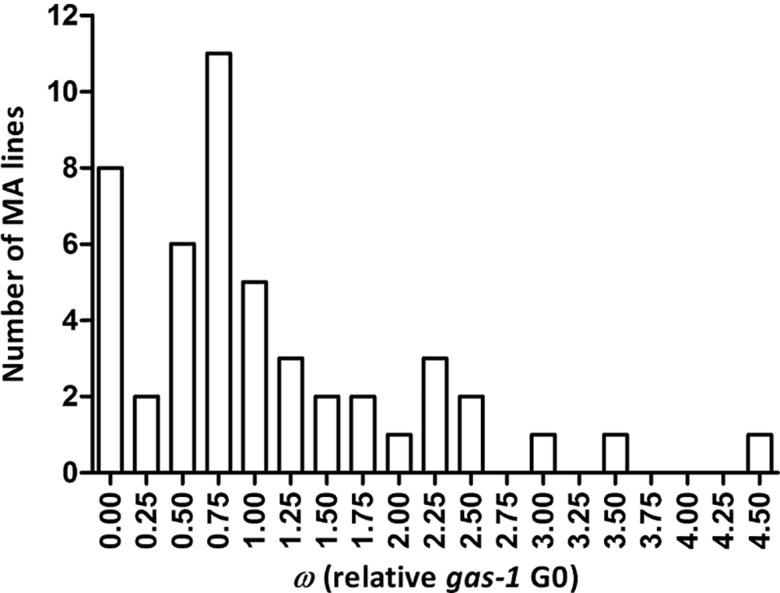
gas-1 lines evolved higher average absolute and relative fitness during the MA process, leading to positive values for ΔM, the per generation change in mean phenotype (table 1). These differences were due to MA lines producing, on an average, significantly more offspring on days 1, 3, 4, and 5 of their reproductive period (fig. 1 and supplementary table S2, Supplementary Material online). Eighteen MA lines were the major contributors to this pattern; of the remaining 30 MA lines, 8 went extinct (above) and 22 exhibited unchanged or slightly reduced fitness relative to gas-1 G0 (fig. 2).
Fig. 2.—
Frequency histogram of gas-1 MA relative fitness. Histogram of the 48 gas-1 MA lines’ fitness (w) relative to gas-1 G0 (equal to 1.0) using least squares means including the 8 gas-1 MA lines that went extinct.
As expected, phenotypic variance among ancestral N2 or gas-1 G0 control lines did not differ from zero for absolute or relative fitness (table 1); within-line (environmental) variance estimates are shown in supplementary table S3, Supplementary Material online. Significant per-generation changes in among-line (genetic) variance, Vb, were detected for both fitness metrics in gas-1 MA lines (table 1). However, point estimates for Vb were uniformly lower for gas-1 MA lines compared with those for the gas-1 G0 ancestor for both traits (table 1). If we had corrected for the (nonsignificant) among-(pseudo)line variance observed for the gas-1 G0 control (c.f., Joyner-Matos et al. 2011), we would have detected no genetic variance for the MA lines. Accordingly, mutational variance, VM, estimates are low and slightly negative for both traits (table 1). Finally, a roughly similar pattern of reduced variance was observed for VE, the per-generation within-line variance, meaning that average MA line performance became more uniform compared with that of its ancestor.
gas-1 MA ROS and ATP
No significant block or interaction effects for either ROS or ATP were found when N2 and gas-1 G0 control performance was modeled as a two-way ANOVA: y = μ + strain + block + strain*block + ɛ, where block and strain* block are random effects (not shown). gas-1 G0 exhibited significantly higher ROS (supplementary fig. S2A, Supplementary Material online) and lower ATP (supplementary fig. S2B, Supplementary Material online) than N2. gas-1 MA line ROS levels were significantly reduced compared with gas-1 G0 and statistically indistinguishable (although higher) than wildtype levels (supplementary fig. S2A, Supplementary Material online). gas-1 MA lines also exhibited lower amounts of within-line variance than gas-1 G0, approaching levels observed for N2 (not shown). Average steady-state ATP levels in the gas-1 MA lines were unchanged compared with gas-1 G0; both gas-1 G0 and its MA lines had significantly reduced ATP levels compared with wildtype N2 (supplementary fig. S2B, Supplementary Material online).
Rates and Spectra of gas-1 MA Line Mutations
Forty novel mutations were detected across the five sequenced gas-1 MA lines (table 2); additionally, all 76 background mutations detected within gas-1 G0 were retained by each MA line. Considering only the novel mutations that arose during MA, MA-line specific mutation numbers ranged from 5 to 11. The per-generation base-substitution mutation rate, 2.13 (±0.34)×10−9, was statistically indistinguishable (within two SEM) from that previously reported for N2, 2.7 (±0.40)×10−9 (Denver et al. 2009). The largest mutation class (16/40) was composed of G: C → A: T transition events. Compared with estimates published for N2 MA lines (Denver et al. 2012), these events alone were substantially elevated in the gas-1 MA lines (SEM values for N2 and gas-1 MA G: C → A: T conditional rates did not overlap; supplementary fig. S3, Supplementary Material online). There was no significant difference between the proportion of synonymous to nonsynonymous mutations between the gas-1 MA and N2 MA values (P = 0.92, χ2 test).
Table 2.
gas-1 MA Line Mutations
| gas-1 MA Line | Position | Type Conversion | Mutation Type | Gene | Classification | Ref Codon | Ref AA | Var Codon | Var AA | Syn or Non |
|---|---|---|---|---|---|---|---|---|---|---|
| MA412 | II: 11,182,784 | A: T → C: G | A → C | F37H8.3 | Exon | CCA | Pro | CCC | Pro | Syn |
| MA412 | II: 13,912,231 | G: C → A: T | G → A | W02B8.2 | Exon | GAG | Glu | AAG | Lys | Non |
| MA412 | V: 11,394,089 | A: T → G: C | A → G | Intergenic | ||||||
| MA412 | V: 12,318,251 | A: T → C: G | T → G | Intergenic | ||||||
| MA412 | V: 19,920,250 | A: T → G: C | A → G | Intergenic | ||||||
| MA412 | V: 20,674,727 | G: C → T: A | G → T | mlt-11 | Exon | TCG | Ser | TCT | Ser | Non |
| MA412 | X: 5,107,789 | A: T → G: C | T → C | Intergenic | ||||||
| MA412 | X: 6,529,937 | A: T → G: C | T → C | Intergenic | ||||||
| MA412 | X: 10,265,783 | G: C → C: G | G → C | Intergenic | ||||||
| MA419 | III: 896,522 | G: C → T: A | G → T | F23H11.2 | Exon | TGC | Cys | TTC | Phe | Non |
| MA419 | V: 3,152,259 | A: T → T: A | T → A | Intergenic | ||||||
| MA419 | X: 10,274,961 | G: C → A: T | C → T | F41E7.2 | Intron | |||||
| MA419 | X: 4,975,182 | G: C → C: G | G → C | ZC8.6 | Exon | TTC | Phe | TTG | Phe | Syn |
| MA419 | X: 10,368,359 | G: C → T: A | G → T | Intergenic | ||||||
| MA419 | X: 12,384,471 | G: C → A: T | C → T | Intergenic | ||||||
| MA429 | I: 12,870,206 | G: C → A: T | C → T | Intergenic | ||||||
| MA429 | II: 4,663,692 | G: C → A: T | C → T | T05A7.11 | Exon | GCA | Ala | ACA | Thr | Non |
| MA429 | IV: 3,470,137 | G: C → A: T | G → A | F58E2.2 | Exon | AAG | Lys | AAA | Lys | Syn |
| MA429 | X: 17,133,019 | G: C → A: T | C → T | K09E3.7 | Exon | GTG | Vak | ATG | Met | Non |
| MA429 | X: 17,132,998 | A: T → G: C | T → C | K09E3.7 | Exon | ACA | Thr | GCA | Ala | Non |
| MA431 | I: 10,724,831 | A: T → G: C | A → G | srz-85 | Exon | CAT | His | CAC | His | Syn |
| MA431 | III: 9,455,062 | A: T → T: A | A → T | rheb-1 | Intron | |||||
| MA431 | III: 3,002,544 | G: C → T: A | G → T | daf-2 | Intron | |||||
| MA431 | III: 4,598,014 | G: C → A: T | C → T | sel-2 | Exon | CGT | Arg | CAT | His | Non |
| MA431 | IV: 12,986,492 | G: C → A: T | C → T | Intergenic | ||||||
| MA431 | IV: 14,058,054 | A: T → T: A | T → A | Intergenic | ||||||
| MA431 | IV: 17,181,972 | G: C → A: T | G → A | C52D10.3 | Intron | |||||
| MA431 | V: 3,379,498 | G: C → C: G | G → C | C04E12.10 | Exon | GAT | Asp | CAT | His | Non |
| MA431 | X: 5,964,400 | A: T → T: A | T → A | R07E4.1 | Intron | |||||
| MA438 | I: 8,987,554 | G: C → A: T | G → A | Intergenic | ||||||
| MA438 | II: 13,977,894 | G: C → A: T | G → A | sre-48 | Intron | |||||
| MA438 | III: 2,792,754 | G: C → T: A | G → T | Y71H2AM.24 | Intron | |||||
| MA438 | III: 11,412,255 | A: T → C: G | A → C | twk-31 | Intron | |||||
| MA438 | IV: 2,620,955 | G: C → A: T | G → A | smf-3 | Intron | |||||
| MA438 | IV: 7,452,889 | G: C → A: T | G → A | Intergenic | ||||||
| MA438 | V: 14,423,155 | G: C → A: T | C → T | Intergenic | ||||||
| MA438 | V: 1,646,965 | A: T → C: G | A → C | alh-2 | Exon | TGA | Trp | TGC | Cys | Non |
| MA438 | V: 18,784,124 | A: T → G: C | T → C | Y17D7B.10 | Intron | |||||
| MA438 | X: 9,580,008 | A: T → T: A | A → T | Intergenic | ||||||
| MA438 | X: 17,456,803 | G: C → A: T | G → A | Intergenic |
Ref Allele, reference allele; Var allele, variant allele; Ref Codon, Reference Codon; Ref AA, reference amino acid; Var Codon, variant codon; Var AA, variant amino acid; Syn or Non, synonymous or nonsynonymous mutation.
GO Term Enrichment
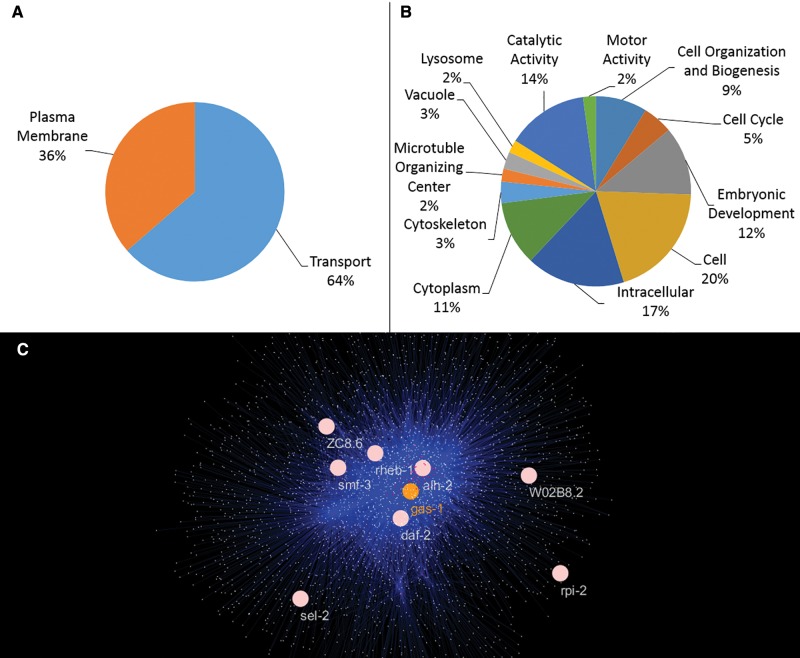
Supplementary file S1, Supplementary Material online, gives a complete accounting of GO enrichment results. No significantly enriched GO slims were shared between the gas-1 MA and N2 MA line groups (supplementary file S1, Supplementary Material online and fig. 3A and B). For gas-1 MA lines, two categories were significantly enriched: the biological process of transport and the cellular component of plasma membrane (fig. 3A). Four gas-1 MA line genic mutations were classified under both transport and plasma membrane; those in: smf-3, daf-2, twk-3, and rheb-1 genes. In contrast, 12 GO slims were enriched in the N2 MA lines (fig. 3B). In addition to the lack of overlap between enriched gas-1 and N2 MA line GO slims (fig. 3A and B), we found that all 12 GO slim categories enriched among N2 MA lines were also enriched in the entire gas-1 interactome set (see below), which contained 80 significantly enriched GO slim categories. Conversely, transport was the only term enriched among both the gas-1 MA line and gas-1 interactome gene sets.
Fig. 3.—
GO slim enrichments and gas-1-centric interactome.Nonoverlapping sets of GO slims were in enriched in gas-1 (A) and N2 lines (B), which experienced maxima of 47 and 250 generations of mutation accumulation, respectively. (C) Depiction of genes (blue points) predicted to interact within 2-degrees of the gas-1 gene (orange); genes within this network that acquired mutations in one of the five sequenced gas-1 MA lines are shown in pink.
gas-1 Interactions
Of the 23 genic mutations (i.e., those discovered within exons or introns; table 2) found within sequenced gas-1 MA lines, 8 were predicted to interact within 2-degrees of the gas-1 gene (fig. 3C); 1 of these, alh-2—encoding a mitochondrial aldehyde dehydrogenase—was a direct interactor with gas-1. Simulations revealed that this number (eight) was higher than that expected by chance, 3.3 (P = 0.005, χ2 test; supplementary fig. S4, Supplementary Material online), and in only 6 of the 1,000 iterations did eight or more mutations reside within this network. None of the 76 gas-1 G0 background mutations was found within the interactome. Of the 59 genic mutations found within sequenced N2 MA lines, 12 were located within the gas-1-centric interactome. In contrast to the gas-1 MA line result, this number was no different than that expected by chance, 8.4 (P = 0.177, χ2 test; supplementary fig. S5, Supplementary Material online); 131/1,000 iterations saw at least 12 mutations located in genes predicted to interact within 2-degrees of gas-1.
MA431 SNP Linkage and Characterization
The above analyses revealed several notable SNPs to be contained within one gas-1 MA line, MA431; we therefore sought to isolate and individually characterize these mutations on both gas-1 G0 and wildtype N2 backgrounds. Among the nine total SNPs discovered within MA431, three affected genes within the gas-1-centric interactome: rheb-1, daf-2, and sel-2. All reside on chromosome III separated by approximately: 8.5 cM (rheb-1 and daf-2), 3.5 cM (rheb-1 and sel-2), and 11.0 cM (daf-2 and sel-2) (Wormbase, version WS254), meaning we could expect ∼8.5%, 3.5%, and 11% of chromatids from each cross to be recombinant for each pair, respectively. Contrary to this expectation, we observed no recombinant genotypes over the course of the seven-generation backcrossing experiment, but our power to detect such genotypes was limited by the small number of offspring (n = 30–44) we could practicably sample and genotype at each generation. We were, however, successful in separating this group of SNPs from all others except one; a fourth SNP on chromosome V remained linked to the chromosome III set. This SNP resided within C04E12.10 (Wormbase, version WS254), an orthologue of human NGLY1 (N-glycanase 1) (Camon 2003; Mulder 2003), resulting in replacement of an asparagine with a histidine.
We found no evidence that the latter observation resulted from a translocation event occurring within either gas-1 G0 or MA431. To investigate this possibility, de novo assemblies of both the gas-1 progenitor strain and gas-1 MA431 were conducted using CLC Genomics Workbench and used to create custom BLAST databases. A 500-bp section of the MA431 chromosome V containing the SNP (V: 3,379,000–3,379,499 bp) was then used as the BLAST query against both custom BLAST databases. Finally, the top contigs—with 6-kb flanking sequences—returned from each BLAST search were then used as a BLAST query against the N2 reference genome (WS242) with the expectation that, if the chromosome V SNP was both specific to MA431 and not the result of a translocation, the BLAST searches would return only chromosome V sequences. A caveat is that if a segment of highly repetitive DNA joined the translocated region, the de novo assembly would be unable to construct a contig across that region.
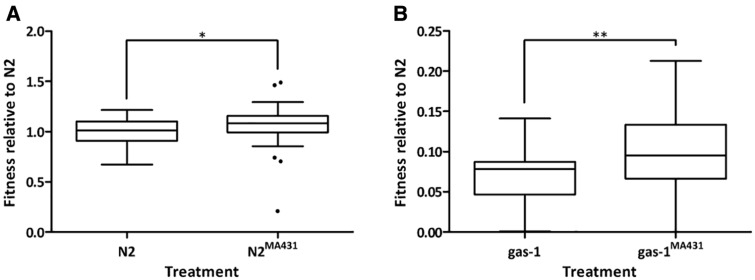
The four isolated MA431 SNPs increased relative fitness on both gas-1 G0 and wildtype N2 backgrounds (fig. 4), exhibiting a slightly greater selective benefit on the N2 (s = 0.065; fig. 4A) compared with the gas-1 G0 background (s = 0.029; fig. 4B). Fitness of the gas-1 G0 and gas-1MA431 pair of strains was dramatically reduced compared with the N2 and N2MA431 pair (Tukey HSD, α = 0.05). This between-pair difference overshadowed the small but significant differences within each pair (fig. 4). The improved fitness of MA431-SNP containing strains was mainly due to increased early life (but not total) reproduction compared with gas-1 G0 or N2 (supplementary fig. S6 and table S4, Supplementary Material online). Note that N2 “catches up” on the second reproductive day, producing significantly more offspring than N2MA431. Strains also varied with respect to age-specific survival (supplementary fig. S7, Supplementary Material online) and mortality risk (log-rank χ23 = 19.29, P < 0.0001) such that gas-1MA431 mortality was reduced compared with gas-1 G0 (risk ratio: 0.546, 95% C.I. = 0.347–0.856, P < 0.008), but N2MA431 mortality was indistinguishable from that of N2. No differences in average lifespan (mean number of days ± SEM) were detected among gas-1MA431 (14.67 ± 0.773), gas-1 G0 (12.29 ± 0.613), N2MA431 (15.80 ± 0.673), or N2 (17.53 ± 0.586).
Fig. 4.—
Relative fitnesses for MA431 SNP-containing lines and ancestral controls. Fitness of MA431 backcrossed lines relative to N2 reported (mean ± 1 SEM); note the different y-axis scales in each panel: Wilcoxan rank sums tests showed that N2MA431 had improved fitness compared with N2 (χ2(1) = 5.16, P < 0.02) and that gas-1MA431 had improved fitness compared with gas-1 G0 (χ2(1) = 7.34, P < 0.01). (A) Wildtype N2 (1.000 ± 0.021) and N2MA431 (1.065 ± 0.033) and (B) gas-1 G0 (0.068 ± 0.005) and gas-1MA431 (0.097 ± 0.008). * and ** denote statistical significance at P < 0.01 and < 0.05 levels, respectively.
ROS levels differed among the four experimental strains (supplementary fig. S8, Supplementary Material online; F3, 166 = 20.95, P < 0.0001), such that gas-1MA431 ROS was reduced compared with gas-1 G0, and both gas-1 (fc21)-containing strains exhibited higher ROS levels than N2 and N2MA431 at P < 0.01 (Tukey’s HSD, α = 0.05). Strains also varied in their ATP content (supplementary fig. S9, Supplementary Material online; F3, 16 = 9.781, P = 0.0007), such that N2 had significantly higher ATP levels than either of the gas-1 (fc21)-containing strains at P < 0.01, and marginally higher levels than N2MA431 at P = 0.0695 (Tukey’s HSD, α = 0.05). N2MA431, gas-1 G0 and gas-1MA431 ATP levels were statistically indistinguishable.
Discussion
We reported on phenotypic and nuclear genome evolution of C. elegans MA lines initiated from a mitochondrial ETC mutant (gas-1) compared with previously studied lines initiated from a wildtype (N2) ancestor (Baer et al. 2005). We also functionally characterized a set of mutations that arose during controlled laboratory evolution—the first C. elegans experiment to do so.
Genetic Analysis of gas-1 G0 Reveals Residual Mutations Consistent with EMS Generation
Ideally, MA lines initiated from isogenic strains of gas-1 (fc21) and N2 would be compared with determine how gas-1 (fc21) affects evolution under drift. Because the gas-1 strain resulted from EMS mutagenesis, we backcrossed it to our laboratory N2 to eliminate extraneous mutations resulting from this treatment, ∼99% of which were expected to be G: C → A: T transitions (Greene et al. 2003; Kim et al. 2006). Our results were consistent with a swath of EMS-generated mutations having been maintained even after backcrossing due to linkage with the selected gas-1 (fc21) mutation. The original gas-1 (fc21) strain was not sequenced, but we speculate that it contained vastly more EMS-induced mutations than our backcrossed strain. These findings are concerning considering the large number of previous studies employing the CGC gas-1 (fc21)-containing strain and others derived from chemical mutagenesis. In any case, whereas we can safely compare gas-1 G0 with its resulting MA lines, comparisons of gas-1 G0 with N2 and between MA lines generated from each strain must be viewed with some caution because we failed to eliminate all genetic differences between the two ancestral strains. Future work targeting single SNPs using CRISPR-Cas9 gene editing can avoid these potential problems.
gas-1 G0 Exhibits Elevated ROS and a “Slow-Living” Phenotype
Keeping the above caveat in mind, gas-1 G0 was substantially less fit than N2 owing to reduced reproductive output alongside delayed reproductive maturity. As in previous studies (Kayser et al. 2001, 2004), gas-1 G0 exhibited increased ROS and depressed ATP levels relative to wildtype. We also previously found that pharyngeal pumping rates, a biomarker of age in C. elegans (Collins et al. 2008), were much lower in gas-1 G0 than in N2 and failed to show the typical pattern of age-correlated decline (Lue 2015). These phenotypes make sense in light of previous work showing that, whereas the CGC gas-1 (fc21) mutant exhibits elevated ROS and oxidative stress, it also upregulates expression of numerous metabolic and cellular defence pathways (Falk et al. 2008). The latter is consistent with the fact that DAF-16, the single FOXO transcription factor present in C. elegans, is constitutively translocated to nuclei in this mutant, an event that normally occurs only under conditions of starvation, oxidative, and other forms of stress in wildtype nematodes (Kondo et al. 2005). daf-16 is negatively regulated by the insulin/insulin-like growth factor (IGF) signaling pathway and, in response to environmental stimuli, regulates ∼100 genes encoding protective products such as heat shock proteins and antioxidants (Hu 2007). The presence of DAF-16 in the nuclei of gas-1 mutants could result from either or both its high intracellular ROS levels or reduced rates of pharyngeal pumping if the latter leads to dietary restriction, and account for its “slow-living” phenotype (c.f. Feng et al. 2001). The former scenario seems more likely since gas-1 worms did not have noticeable frequencies of Dauer larva (personal observation), which might be expected under diet restriction (Hu 2007).
gas-1 nDNA Mutation Spectrum but Not Per-Generation Rate Differs from Wildtype
Oxidative stress is associated with two major base-substitution types; G: C → A: T transitions and G: C → T: A transversions, caused by 5-hydroxyuracil and 8-oxo-dG lesions, respectively (Wiseman and Halliwell 1996; Cooke et al. 2003; Cooke and Evans 2007). Both types were found to predominate the nDNA mutational spectrum in N2 MA lines (Denver et al. 2009); conversely, the gas-1 mutational spectrum was dominated by G: C → A: T transitions alone. Although this difference could indicate different underlying mechanisms of mutation, considerable evidence from this study indicates that gas-1 MA line mutations had been filtered by selection. G: C → T: A transversions may therefore have been selectively removed in gas-1 MA lines. Similarly, our previous analysis of mtDNA mutation within the same lines found that, whereas rates of heritable mtDNA mutation in gas-1 MA lines were indistinguishable from wildtype, the molecular spectrum of mutations was skewed—in that case toward an increased frequency of single nucleotide substitutions and a reduced frequency of indels compared with wildtype (Wernick et al. 2016). The potentially biased sample of mutations, combined with the downward evolution of ROS levels during gas-1 MA, makes it impossible to test for a relationship between endogenous ROS and germline mutation rate in this system. Our results are also in general agreement with those of Joyner-Matos et al. (2011) who found that MA lines initiated from a mev-1 (kn-1) ETC complex II mutant, known to experience high endogenous ROS levels (Senoo-Matsuda et al. 2001), exhibited wildtype rates of fitness decay. It is possible that ROS levels decreased during the MA phase in this experiment as well.
Life History and Bioinformatic Results Signal Positive and Purifying Selection in gas-1 MA Lines
Contrary to results of standard MA studies, after fewer than 50 generations of single-individual bottlenecking, mean absolute, and relative fitness of gas-1 MA lines improved slightly beyond that of their ancestral gas-1 G0 control, whereas among-line variance in these traits declined. For comparison, the mutational variance, VM, for absolute fitness (W) was 5.77 for the N2 MA lines (Baer et al. 2005) compared with a negative estimate, −1.547, for the gas-1 MA lines. Improved gas-1 MA line fitness was entirely due to improvements in early life reproduction beyond that of gas-1 G0. Because gas-1 MA lines were initiated from a genetically homogeneous ancestral population and evolved by strict selfing, and since neither reversion of the gas-1 mutant allele nor loss of the gas-1 G0 background mutations was observed in any MA line, their fitness gains were necessarily fuelled by fixation of de novo beneficial or compensatory epistatic mutations. The most likely fate of such mutations arising in this unfavorable population genetic environment would be loss by drift, but the high numbers of failed bottlenecks and gas-1 MA line extinctions is consistent with selection operating at individual and/or intraindividual (e.g., gametic) levels. Additionally, the increased numbers of nonviable eggs is indicative of purifying selection (e.g., at zygotic or postzygotic levels) against new deleterious mutations, and suggests that an increased fraction of mutations was lethal on the gas-1 genetic background compared with N2. More work would be required to identify the mechanism(s) of selection in the gas-1 MA lines, but our findings recall those of the Arabidopsis thaliana MA experiments of Shaw and colleagues (Shaw et al. 2000, 2002; Rutter et al. 2010) where mean fitness of wildtype MA lines remained unchanged over generations of inbreeding, a pattern that may have resulted from the action of somatic cell lineage selection occurring in the meristem prior to formation of germ cells (Otto and Orive 1995). Our gas-1 MA lines may have benefitted from a different form of intraindividual selection that yielded a similar outcome.
Results of our bioinformatics analyses also bore the signature of selection and were consistent with the idea that some gas-1 MA line mutations conferred beneficial or compensatory effects. First, fewer GO slim categories were significantly enriched in the gas-1 MA line mutated gene set compared with that of the N2 MA lines. The GoMiner enrichment algorithm takes the total number of altered genes into account when calculating enrichment scores, so this pattern is not a consequence of N2 MA lines having more mutations and is instead consistent with a weaker role for genetic drift in gas-1 MA versus N2 MA line molecular evolution. Second, four genic mutations in the gas-1 MA lines—daf-2, rheb-1, twk-31, and smf-3—had membership in the same GO slim categories—transport, plasma membrane, and cell (the latter not significantly enriched)—suggesting or confirming known overlap in their function. For instance, daf-2 (the C. elegans insulin/IGF-1 receptor) and rheb-1 (a GTPase) are known members of the nutrient-sensing target of rapamycin, TOR, pathway that affects growth and longevity; reviewed in Lapierre and Hansen (2012). It may be that altering the transport of nutrients, signaling molecules, or other proteins and ions across membranes counterbalances gas-1-associated mitochondrial dysfunction. Given these genes’ membership in important metabolic pathways (insulin signaling; mitochondrial unfolded protein response; K+ transport), it is possible that any ameliorating effects act by altering flux in these pathways at plasma membrane junctions. Third, the number of gas-1 MA line mutations occurring within gas-1 interactome genes was higher than expected by chance, suggesting that some gas-1 MA line mutations were fixed in response to the gas-1 mutation rather than resulting from drift. Finally, four of the mutated genes—daf-2, rheb-1, smf-3, and sel-2—with membership in the enriched gas-1 MA GO slim categories (plasma membrane and transport) were also members of the gas-1 interactome. Interestingly, the plasma membrane category was not significantly enriched among gas-1 interactome genes, yet they may have been an important target for compensatory or beneficial mutation in the gas-1 MA lines.
Potential for Protective Metabolic Shift in gas-1 MA Lines
The evolution of gas-1 MA ROS levels toward lower, wildtype levels contrasts with the elevated ROS levels we previously observed among N2 MA lines (Joyner-Matos et al. 2013). Conversely, the MA process had little impact on steady-state ATP levels in gas-1 MA lines—the same was true for pharyngeal pumping (Lue 2015). A potential explanation for these combined results is that the gas-1 MA lines benefitted from a metabolic shift toward increased reliance upon fermentation and away from oxidative phosphorylation—a phenomenon known to occur in certain cancer cells (the Warburg effect, Warburg 1956). By utilizing one or more of their diverse fermentation pathways (Holt and Riddle 2003; Hulme and Whitesides 2011), C. elegans could reduce mitochondrial ROS production, perhaps offsetting the cost of the strain’s already low ATP (Tielens et al. 2002), preventing apoptosis (Ruckenstuhl et al. 2009), or increasing mtDNA genome stability (Ericson et al. 2012). In the absence of gene expression or other supporting data, we do not know whether such a shift, if one occurred, resulted from fixation of beneficial mutations and/or a stress response to deleterious MA in gas-1 MA lines.
Beneficial Haplotype Characterization Indicates Role for Epistasis
Although we were unable to isolate single, candidate beneficial gas-1 MA431 SNPs to ascertain their individual contributions to the line’s slight fitness recovery, this failure revealed a potentially large role for epistasis in gas-1 MA line fitness evolution. Our inability to break up the three chromosome III SNPs may have been due to their strong linkage and our limited power to detect recombinant genotypes. But failure to isolate the chromosome V SNP from the chromosome III group is consistent with some form of epistasis operating between this SNP and one or more on chromosome III. Interestingly, the four-SNP combination proved to be slightly more advantageous on its nonnative N2 background, and thus did not appear to specifically compensate for the gas-1 (fc21) mutation. This result was somewhat surprising given the interactome results and the reasonable expectation that N2 had achieved maximal laboratory adaptation, and is consistent with the existence of a rugged genetic landscape in which a new local fitness peak was opened to N2 by addition of the MA431 SNP set (c.f., Poelwijk et al. 2007). Two genetic explanations for this outcome include one of strong positive epistasis operating between some combination of MA431 SNPs, which would be neutral or beneficial on their own but advantageous in combination (Phillips 2008); in this scenario, the MA431 SNPs could have conferred a greater advantage on the N2 background simply because they did not have to contend with the 76 background mutations present within gas-1 G0. A second possibility is one of reciprocal sign epistasis (Kvitek and Sherlock 2011; Poelwijk et al. 2011), in this case involving individually deleterious mutations conferring a beneficial effect in combination. As previously noted, when any one of four MA431 SNPs was lost during the backcrossing phase, all others would be lost in the following generation—an outcome consistent with this form of epistasis operating between some combination of SNPs. If true, the mutational route to this beneficial SNP combination may have been complex given the likely existence of low-fitness intermediate genotypes; a transgenerational analysis of gas-1 MA431 mutational dynamics would be necessary to distinguish among these possibilities.
In contrast to the similar fitness effects of the MA431 SNPs in the gas-1 GO versus N2 strains, their physiological effects were strongly strain-dependent. The four SNPs’ gas-1 G0-specific consequences (reduced ROS levels with no effect on ATP levels) are consistent with our metabolic shift hypothesis (above), whereas the consequences for N2 (reduced ATP levels with no effect on ROS levels) are more mysterious. Clearly, more work would be required to fully characterize the MA431 SNP set, but these results reveal strong context-dependency in its effects at the physiological level. Finally, we note that the above interpretations ignore the possibility of epistatic interactions between the MA431 SNP set and mtDNA variants and/or nDNA mutation types not considered here. The former at least seems unlikely since, although we previously identified four heteroplasmic mtDNA variants segregating within the MA431 line, none were detected at levels exceeding ∼4% in the sampled population (table 4 in Wernick et al. 2016).
Conclusion
Our study indicates that, even under conditions of extreme genetic drift, low-fitness populations can maintain or slightly improve their fitness through both selective elimination of deleterious mutations and fixation of novel beneficial mutations or allelic combinations. These unusual results are consistent with predictions of adaptive landscape theory—namely, with the existence of an inverse relationship between a population’s fitness and its likelihood of adaptation, and considerable context dependency of the mutational distribution of fitness effects. A greater fraction of new mutations may confer beneficial and lethal effects in populations existing at low-fitness extremes, simultaneously opening more mutational routes to both fitness maintenance/recovery and extinction. In addition, the fact that we observed only minor fitness improvements—nothing approaching complete recovery of ancestral fitness levels—in gas-1 MA lines is consistent with the prediction that most advantageous mutations confer only small fitness benefits; however, the individual effects of any medium- or large-effect beneficial mutations acquired by the gas-1 MA lines were likely muted by deleterious mutations present in the same genomes. The above interpretation was supported by our bioinformatic analyses, which revealed highly nonrandom patterns of genetic network location and functional annotation among gas-1 versus N2 MA line mutations. Interestingly, these analyses implicated some of the same pathways known to be upregulated in the gas-1 mutant ancestor (Falk et al. 2008), suggesting that both genetic and nongenetic responses may tend to occur via similar or overlapping pathways. This study also revealed the capacity of ETC-deficient populations experiencing relaxed selection to rapidly recover near-wildtype levels of ROS—a pattern that may have been at least partly due to nongenetic mechanisms (i.e., compensatory transcriptional upregulation of metabolic and damage response pathways, Falk et al. 2008), which were further activated in response to deleterious MA in the gas-1 background. The downward evolution of ROS levels was accompanied by maintenance of a wildtype mutation rate but an altered mutational spectrum—a result similar to that of our previous analysis of mtDNA mutation processes in the same lines (Wernick et al. 2016). Understanding the extent to which this pattern may be due to selection favoring or eradicating particular base-substitution types would require further study. Lastly, our characterization of a SNP combination fixed within one gas-1 MA line was consistent with a role for reciprocal sign epistasis in fitness evolution in our system, and adds to the growing body of evidence that fitness landscapes are rugged and, whereas possibly dominated by genetic constraint, can provide routes to adaptation and “accessible innovation” (c.f., fig. 3 in Barrick and Lenski 2013).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank B. Taylor, J. Podrabsky and T. Rosenstiel for helpful discussion; and A. Basler, A. Coleman-Hulbert, L. Munoz-Tremblay, A. Mustain, and J. Sullins for laboratory support; and anonymous reviewers for thoughtful comments on the manuscript. This work was supported the National Science Foundation [MCB-1330427 to S.E. and D.R.D., and HRD-140465, which supported the undergraduate research of G.V.]; and the PSU Biology Department [Forbes-Lea grant to S.F.C.]. We also thank the OSU Center for Genome Research and Biocomputing for DNA sequencing and bioinformatics support.
Literature Cited
- Achaz G, et al. editors. 2014. Ecological genomics: ecology and the evolution of genes and genomes. Netherlands: Springer; p. 211–231. [Google Scholar]
- Ameur A, et al. 2011. Ultra-deep sequencing of mouse mitochondrial DNA: mutational patterns and their origins. PLoS Genet. 7(3):e1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew JR, et al. 2015. Abiotic stress does not magnify the deleterious effects of spontaneous mutations. Heredity 115:503–508.http://dx.doi.org/10.1038/hdy.2015.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer CF, et al. 2005. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc Natl Acad Sci U S A. 102:5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Kauth MR, Strelioff CC, Lenski RE.. 2010. Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol Biol Evol. 27(6):1338–1347.http://dx.doi.org/10.1093/molbev/msq024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE.. 2013. Genome dynamics during experimental evolution. Nat Rev Genet. 14(12):827–839.http://dx.doi.org/10.1038/nrg3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blier PU, Dufresne F, Burton RS.. 2001. Natural selection and the evolution of mtDNA-encoded peptides: evidence for intergenomic co-adaptation. Trends Genet. 17(7):400–406.http://dx.doi.org/10.1016/S0168-9525(01)02338-1 [DOI] [PubMed] [Google Scholar]
- Burch CL, Chao L.. 1999. Evolution by small steps and rugged landscapes in the RNA virus phi6. Genetics 151(3):921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camon E. 2003. The gene ontology annotation (GOA) project: Implementation of GO in SWISS-PROT, TrEMBL, and InterPro. Genome Res. 13:662–672.http://dx.doi.org/10.1101/gr.461403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevin L-M, Martin G, Lenormand T.. 2010. Fisher’s model and the genomics of adaptation: restricted pleiotropy, heterogenous mutation, and parallel evolution. Evolution 64(11):3213–3231. [DOI] [PubMed] [Google Scholar]
- Collins JJ, Huang C, Hughes S, Kornfeld K.. 2008. The measurement and analysis of age-related changes in Caenorhabditis elegans. WormBook. p. 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke MS, Evans MD.. 2007. 8-oxo-deoxyguanosine: reduce, reuse, recycle? Proc Natl Acad Sci U S A. 104(34):13535–13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J.. 2003. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 17(10):1195–1214.http://dx.doi.org/10.1096/fj.02-0752rev [DOI] [PubMed] [Google Scholar]
- Demple B, Harrison L.. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 63:915–948.http://dx.doi.org/10.1146/annurev.bi.63.070194.004411 [DOI] [PubMed] [Google Scholar]
- Denver DR, et al. 2009. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc Natl Acad Sci U S A. 106(38):16310–16314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, et al. 2010. Selective sweeps and parallel mutation in the adaptive recovery from deleterious mutation in Caenorhabditis elegans. Genome Res. 20(12):1663–1671.http://dx.doi.org/10.1101/gr.108191.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, et al. 2012. Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biol Evol. 4(4):513–522.http://dx.doi.org/10.1093/gbe/evs028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Feinberg S, Steding C, Durbin M, Lynch M.. 2006. The relative roles of three DNA repair pathways in preventing Caenorhabditis elegans mutation accumulation. Genetics 174(1):57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Thomas WK.. 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430:679-682.http://dx.doi.org/10.1038/nature02697 [DOI] [PubMed] [Google Scholar]
- Dowling DK, Friberg U, Lindell J.. 2008. Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol. Evol 23(10):546–554. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE.. 2001. Epistasis between new mutations and genetic background and a test of genetic canalization. Evolution 55:1746–1752.http://dx.doi.org/10.1111/j.0014-3820.2001.tb00824.x [DOI] [PubMed] [Google Scholar]
- Ericson NG, et al. 2012. Decreased mitochondrial DNA mutagenesis in human colorectal cancer. PLoS Genet. 8(6):e1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes S, Lynch M.. 2003. Rapid fitness recovery in mutationally degraded lines of Caenorhabditis elegans rapid fitness recovery in mutationally degraded lines of Caenorhabditis elegans. Evolution 57(5):1022–1030.http://dx.doi.org/10.1111/j.0014-3820.2003.tb00313.x [DOI] [PubMed] [Google Scholar]
- Estes S, Phillips PC, Denver DR, Thomas WK, Lynch M.. 2004. Mutation accumulation in populations of varying size: the distribution of mutational effects for fitness correlates in Caenorhabditis elegans. Genetics 166(3):1269–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk MJ, et al. 2008. Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Mol Genet Metab. 93(4):388–397.http://dx.doi.org/10.1016/j.ymgme.2007.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussière F, Hekimi S.. 2001. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell 1(5):633–644. [DOI] [PubMed] [Google Scholar]
- Fisher R. 1930. On the genetical theory of natural selection. Oxford (United Kingdom: ): Oxford University Press. [Google Scholar]
- Greene EA, et al. 2003. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164(2):731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DW, Mahmoudizad R, Hurd AW, Joseph SB.. 2008. Spontaneous mutations in diploid Saccharomyces cerevisiae: another thousand cell generations. Genet Res. 90(3):229–241. [DOI] [PubMed] [Google Scholar]
- Halligan DL, Keightley PD.. 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu Rev Ecol Evol Syst. 40(1):151–172.http://dx.doi.org/10.1146/annurev.ecolsys.39.110707.173437 [Google Scholar]
- Hartman PS, Ishii N, Kayser E-B, Morgan PG, Sedensky MM.. 2001. Mitochondrial mutations differentially affect aging, mutability and anesthetic sensitivity in Caenorhabditis elegans. Mech Ageing Dev. 122(11):1187–1201. [DOI] [PubMed] [Google Scholar]
- Heilbron K, Toll-Riera M, Kojadinovic M, MacLean RC.. 2014. Fitness is strongly influenced by rare mutations of large effect in a microbial mutation accumulation experiment. Genetics 197(3):981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Howe DK, Leung A, Denver DR, Estes S.. 2012. In vivo quantification reveals extensive natural variation in mitochondrial form and function in Caenorhabditis briggsae. PLoS ONE 7(8):e43837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas RT, Bank C, Jensen JD, Bolon DNA.. 2013. Shifting fitness landscapes in response to altered environments. Evolution 67(12):3512–3522.http://dx.doi.org/10.1111/evo.12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SJ, Riddle DL.. 2003. Sage surveys C. elegans carbohydrate metabolism: evidence for an anaerobic shift in the long-lived dauer larva. Mech Ageing Dev. 124(7):779–800. [DOI] [PubMed] [Google Scholar]
- Hsie AW, et al. 1986. Evidence for reactive oxygen species inducing mutations in mammalian cells. Proc Natl Acad Sci U S A. 83(24):9616–9620.http://dx.doi.org/10.1073/pnas.83.24.9616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu PJ. 2007. Dauer. WormBook; p. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z-L, Bao J, Reecy JM.. 2008. CateGOrizer: a web-based program to batch analyze gene on-tology classification categories. Online J Bioinform. 9:108–112. [Google Scholar]
- Hulme SE, Whitesides GM.. 2011. Chemistry and the worm: Caenorhabditis elegans as a platform for integrating chemical and biological research. Angew Chem Int Ed. 42(35):4774–4807. [DOI] [PubMed] [Google Scholar]
- Itsara LS, et al. 2014. Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet. 10(2):e1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SB, Hall DW.. 2004. Spontaneous mutations in diploid Saccharomyces cerevisiae: more beneficial than expected. Genetics 168(4):1817–1825.http://dx.doi.org/10.1534/genetics.104.033761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner-Matos J, Bean LC, Richardson HL, Sammeli T, Baer CF.. 2011. No evidence of elevated germline mutation accumulation under oxidative stress in Caenorhabditis elegans. Genetics 189(4):1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner-Matos J, et al. 2013. Evolution of a higher intracellular oxidizing environment in Caenorhabditis elegans under relaxed selection. PLoS One 8(6):4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V, Packard LB, Bu L, Keightley PD, Bergthorsson U.. 2015. Fitness decline in spontaneous mutation accumulation lines of Caenorhabditis elegans with varying effective population sizes. Evolution 69(1):104–116. [DOI] [PubMed] [Google Scholar]
- Kayser EB, Morgan PG, Hoppel CL, Sedensky MM.. 2001. Mitochondrial expression and function of GAS-1 in Caenorhabditis elegans. J Biol Chem. 276(23):20551–20558. [DOI] [PubMed] [Google Scholar]
- Kayser EB, Morgan PG, Sedensky MM.. 1999. GAS-1: a mitochondrial protein controls sensitivity to volatile anesthetics in the nematode Caenorhabditis elegans. Anesthesiology 90(2):545–554.http://dx.doi.org/10.1097/00000542-199902000-00031 [DOI] [PubMed] [Google Scholar]
- Kayser EB, Sedensky MM, Morgan PG.. 2004. The effects of complex I function and oxidative damage on lifespan and anesthetic sensitivity in Caenorhabditis elegans. Mech Ageing Dev. 125(6):455–464. [DOI] [PubMed] [Google Scholar]
- Keightley PD, et al. 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19(7):1195–1201.http://dx.doi.org/10.1101/gr.091231.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Schumaker K, Zhu J-K.. 2006. EMS mutagenesis of Arabidopsis. Methods Mol Biol. 323:101–103. [DOI] [PubMed] [Google Scholar]
- Kondo M, et al. 2005. Effect of oxidative stress on translocation of daf-16 in oxygen-sensitive mutants, mev-1 and gas-1 of Caenorhabditis elegans. Mech Ageing Dev. 126(6–7):637–641. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Kondrashov AS.. 2010. Measurements of spontaneous rates of mutations in the recent past and the near future. Philos Trans R Soc B Biol Sci. 365(1544):1169–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek DJ, Sherlock G.. 2011. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genetics 7(4):e1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. 1994. Risk of population extinction from fixation of new deleterious mutations. Evolution 48(5):1460–1469.http://dx.doi.org/10.1111/j.1558-5646.1994.tb02188.x [DOI] [PubMed] [Google Scholar]
- Lapierre LR, Hansen M.. 2012. Lessons from C. elegans: signaling pathways for longevity. Trends Endocrinol Metab. 23(12):637–644.http://dx.doi.org/10.1016/j.tem.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Popodi E, Tang H, Foster PL.. 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A. 109(41):E2774–E2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenaz G, et al. 2006. Mitochondrial complex I: structural and functional aspects. Biochim Biophys Acta 1757(9–10):1406–1420.http://dx.doi.org/10.1016/j.bbabio.2006.05.007 [DOI] [PubMed] [Google Scholar]
- Lenski RE, Travisano M.. 1994. Dynamics of adaptation and diversification: a 10, 000-generation experiment with bacterial populations. Proc Natl Acad Sci U S A. 91(15):6808–6814.http://dx.doi.org/10.1073/pnas.91.15.6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue M. 2015. Phenotypic and mutational consequences of mitochondrial ETC genetic damage [thesis]. [Portland (OR)]: Portland State University. [Google Scholar]
- Lynch M, Bürger R, Butcher D, Gabriel W.. 1993. The mutational meltdown in asexual populations. J Hered. 84(5):339–344. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery J, Burger R.. 1995. Mutation accumulation and the extinction of small populations. Am Nat. 146(4):489–518. [Google Scholar]
- Lynch M, et al. 1999. Perspective: spontaneous deleterious mutations. Evolution 53(3):645–663.http://dx.doi.org/10.1111/j.1558-5646.1999.tb05361.x [DOI] [PubMed] [Google Scholar]
- Lynch M, et al. 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci U S A. 105(27):9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisnier-Patin S, Berg OG, Liljas L, Andersson DI.. 2002. Compensatory adaptation to the deleterious effect of antibiotic resistance in Salmonella typhimurium. Mol Microbiol. 46(2):355–366. [DOI] [PubMed] [Google Scholar]
- Martin G, Lenormand T.. 2006. A general multivariate extension of Fisher’s geometrical model and the distribution of mutation fitness effects across species. Evolution 60(5):893–907. [PubMed] [Google Scholar]
- Martin G, Lenormand T.. 2015. The fitness effect of mutations across environments: Fisher’s geometrical model with multiple optima. Evolution 69(6):1433–1447. [DOI] [PubMed] [Google Scholar]
- Miller CR, Joyce P, Wichman HA.. 2011. Mutational effects and population dynamics during viral adaptation challenge current models. Genetics 187(1):185–202.http://dx.doi.org/10.1534/genetics.110.121400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, et al. 2011. ROS signaling: the new wave? Trends Plant Sci. 16(6):300–309. [DOI] [PubMed] [Google Scholar]
- Morgan PG, Sedensky MM.. 1994. Mutations conferring new patterns of sensitivity to volatile anesthetics in Caenorhabditis elegans. Anesthesiology 81(4):888-898.http://dx.doi.org/10.1097/00000542-199410000-00016 [DOI] [PubMed] [Google Scholar]
- Mulder NJ. 2003. The InterPro database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31:315–318.http://dx.doi.org/10.1093/nar/gkg046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP. 2009. How mitochondria produce reactive oxygen species. Biochem J. 417(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52(4):935–949.http://dx.doi.org/10.1111/j.1558-5646.1998.tb01823.x [DOI] [PubMed] [Google Scholar]
- Orr HA. 2000. Adaptation and the cost of complexity. Evolution 54:13–20. [DOI] [PubMed] [Google Scholar]
- Ossowski S, et al. 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327(5961):92–94.http://dx.doi.org/10.1126/science.1180677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Orive ME.. 1995. Evolutionary consequences of mutation and selection within an individual. Genetics 141(3):1173–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeito L, Fernandes L, Mota C, Gordo I.. 2007. Adaptive mutations in bacteria: high rate and small effects. Science 317(5839):813–815.http://dx.doi.org/10.1126/science.1142284 [DOI] [PubMed] [Google Scholar]
- Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ.. 2007. Empirical fitness landscapes reveal accessible evolutionary paths. Nature 445(7126):383–386.http://dx.doi.org/10.1038/nature05451 [DOI] [PubMed] [Google Scholar]
- Poelwijk FJ, Tănase-Nicola S, Kiviet DJ, Tans SJ.. 2011. Reciprocal sign epistasis is a necessary condition for multi-peaked fitness landscapes. Journal of Theoretical Biology 272(1):141-144. [DOI] [PubMed] [Google Scholar]
- Poon A, Otto SP.. 2000. Compensating for our load of mutations: freezing the meltdown of small populations. Evolution 54(5):1467–1479.http://dx.doi.org/10.1111/j.0014-3820.2000.tb00693.x [DOI] [PubMed] [Google Scholar]
- Phillips PC. 2008. Epistasis — the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 9:855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remold SK, Lenski RE.. 2004. Pervasive joint influence of epistasis and plasticity on mutational effects in Escherichia coli. Nat Genet. 36(4):423–426.http://dx.doi.org/10.1038/ng1324 [DOI] [PubMed] [Google Scholar]
- Rokyta D, Badgett MR, Molineux IJ, Bull JJ.. 2002. Experimental genomic evolution: extensive compensation for loss of DNA ligase activity in a virus. Mol Biol Evol. 19(3):230–238.http://dx.doi.org/10.1093/oxfordjournals.molbev.a004076 [DOI] [PubMed] [Google Scholar]
- Ruckenstuhl C, et al. 2009. The Warburg effect suppresses oxidative stress induced apoptosis in a yeast model for cancer. PLoS One 4(2):e4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter MT, et al. 2012. Fitness of Arabidopsis thaliana mutation accumulation lines whose spontaneous mutations are known. Evolution 66(7):2335–2339.http://dx.doi.org/10.1111/j.1558-5646.2012.01583.x [DOI] [PubMed] [Google Scholar]
- Rutter MT, Shaw FH, Fenster CB.. 2010. Spontaneous mutation parameters for Arabidopsis thaliana measured in the wild. Evolution 64(6):1825–1835.http://dx.doi.org/10.1111/j.1558-5646.2009.00928.x [DOI] [PubMed] [Google Scholar]
- Sanjuan R, Moya A, Elena SF.. 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci U S A. 101(22):8396–8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxer G, et al. 2012. Whole genome sequencing of mutation accumulation lines reveals a low mutation rate in the social amoeba Dictyostelium discoideum. PLoS One 7(10):e46759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo-Matsuda N, et al. 2001. A defect in the cytochrome b large subunit in complex ii causes both superoxide anion overproduction and abnormal energy metabolism in Caenorhabditis elegans. J Biol Chem. 276:41553–41558.http://dx.doi.org/10.1074/jbc.M104718200 [DOI] [PubMed] [Google Scholar]
- Sharp NP, Agrawal AF.. 2012. Evidence for elevated mutation rates in low-quality genotypes. Proc Natl Acad Sci U S A. 109:6142–6146.http://dx.doi.org/10.1073/pnas.1118918109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw FH, Geyer CJ, Shaw RG.. 2002. A comprehensive model of mutations affecting fitness and inferences for Arabidopsis thaliana. Evolution 56(3):453–463.http://dx.doi.org/10.1111/j.0014-3820.2002.tb01358.x [DOI] [PubMed] [Google Scholar]
- Shaw RG, Byers DL, Darmo E.. 2000. Spontaneous mutational effects on reproductive traits of Arabidopsis thaliana. Genetics 155(1):369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander OK, Tenaillon O, Chao L.. 2007. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 5(4):e94.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SW, Latta LC, Denver DR, Estes S.. 2014. Endogenous ROS levels in C. elegans under exogenous stress support revision of oxidative stress theory of life-history tradeoffs. BMC Evol Biol. 14:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns FW, Fenster CB.. 2016. The effect of induced mutations on quantitative traits in Arabidopsis thaliana: natural versus artificial conditions. Ecol Evol. 6(23):8366–8374.http://dx.doi.org/10.1002/ece3.2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O. 2014. The utility of Fisher’s geometric model in evolutionary genetics. Annu Rev Ecol Evol Syst. 45:179–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teotónio H, Estes S, Phillips PC, Baer CF.. 2017. Experimental evolution with Caenorhabditis nematodes. Genetics 206(2):691–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tielens AGM, Rotte C, Van Hellemond JJ, Martin W.. 2002. Mitochondria as we don’t know them. Trends Biochem Sci. 27(11):564–572. [DOI] [PubMed] [Google Scholar]
- Tong T, Zhao H.. 2008. Practical guidelines for assessing power and false discovery rate for a fixed sample size in microarray experiments. Stat Med. 27(11):1960–1972.http://dx.doi.org/10.1002/sim.3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, et al. 2010. Decreased energy metabolism extends life span in Caenorhabditis elegans without reducing oxidative damage. Genetics 185(2):559-571.http://dx.doi.org/10.1534/genetics.110.115378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilieva LL, Hook AM, Lynch M.. 2000. The fitness effects of spontaneous mutations in Caenorhabitis elegans. Evolution 54(4):1234-1246.http://dx.doi.org/10.1111/j.0014-3820.2000.tb00557.x [DOI] [PubMed] [Google Scholar]
- Wang AD, et al. 2009. Selection, epistasis, and parent‐of‐origin effects on deleterious mutations across environments in Drosophila melanogaster. Am Nat. 174(6):863–874. [DOI] [PubMed] [Google Scholar]
- Wang Y, Arenas CD, Stoebel DM, Cooper TF.. 2013. Genetic background affects epistatic interactions between two beneficial mutations. Biol Lett. 9(1):20120328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. 1956. On the origin of cancer cells. Science 123(3191):309–314.http://dx.doi.org/10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- Wernick RI, Estes S, Howe DK, Denver DR.. 2016. Paths of heritable mitochondrial DNA mutation and heteroplasmy in reference and gas-1 strains of Caenorhabditis elegans. Front Genet. 7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, Otto SP.. 1999. The panda and the phage: compensatory mutations and the persistence of small populations. Trends Ecol Evol. 14(8):295–296.http://dx.doi.org/10.1016/S0169-5347(99)01662-6 [DOI] [PubMed] [Google Scholar]
- Wiseman H, Halliwell B.. 1996. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem J. 313(1):17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Hekimi S.. 2010. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biology 8(2):e1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu E, et al. 2013. Mitochondrial DNA damage can promote atherosclerosis independently of reactive oxygen species through effects on smooth muscle cells and monocytes and correlates with higher-risk plaques in humans. Circulation 128(7):702–712.http://dx.doi.org/10.1161/CIRCULATIONAHA.113.002271 [DOI] [PubMed] [Google Scholar]
- Zeeberg BR, et al. 2003. Gominer: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 4(4):R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg BR, et al. 2005. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID). BMC Bioinformatics 6:168.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Sternberg P.. 2006. Genome-wide prediction of c. elegans genetic interactions. Science 311(5766):1481–1484.http://dx.doi.org/10.1126/science.1123287 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.