Abstract
Streptomyces topoisomerase I (TopA) exhibits exceptionally high processivity. The enzyme, as other actinobacterial topoisomerases I, differs from its bacterial homologs in its C-terminal domain (CTD). Here, bioinformatics analyses established that the presence of lysine repeats is a characteristic feature of actinobacterial TopA CTDs. Streptomyces TopA contains the longest stretch of lysine repeats, which terminate with acidic amino acids. DNA-binding studies revealed that the lysine repeats stabilized the TopA–DNA complex, while single-molecule experiments showed that their elimination impaired enzyme processivity. Streptomyces coelicolor TopA processivity could not be restored by fusion of its N-terminal domain (NTD) with the Escherichia coli TopA CTD. The hybrid protein could not re-establish the distribution of multiple chromosomal copies in Streptomyces hyphae impaired by TopA depletion. We expected that the highest TopA processivity would be required during the growth of multigenomic sporogenic hyphae, and indeed, the elimination of lysine repeats from TopA disturbed sporulation. We speculate that the interaction of the lysine repeats with DNA allows the stabilization of the enzyme–DNA complex, which is additionally enhanced by acidic C-terminal amino acids. The complex stabilization, which may be particularly important for GC-rich chromosomes, enables high enzyme processivity. The high processivity of TopA allows rapid topological changes in multiple chromosomal copies during Streptomyces sporulation.
INTRODUCTION
Only two topoisomerases: topoisomerase I (type I topoisomerase) and gyrase (type II topoisomerase) are sufficient to maintain bacterial chromosome supercoiling (1). Topoisomerase I homologs (named TopA or TopoI) belong to the type IA topoisomerases, which constitute a ubiquitous group of enzymes that remove an excess of negative supercoils using enzyme-bridged DNA strand passage (2). The enzymes cleave one strand of the DNA helix and transfer the second, intact strand through the breakage, which is then religated (3,4). The catalytically active tyrosine within the 5Y-CAP domain forms a transient phosphotyrosine bond with the DNA backbone (5). The active site is located in the toroidal N-terminal fragment of the protein (67 kDa), which also includes a topoisomerase-primase domain (TOPRIM) and DNA-binding sites (6,7). While the N-terminal TopA fragment is highly conserved among various bacteria, the C-terminal domain (CTD) exhibits high diversity (4). In most bacterial species, the CTD contains a varying number of positively charged amino acids and Zn2+ finger motifs (four in Helicobacter pylori TopA, three in Escherichia coli and only one in Thermotoga maritima) (8–11). Removal of the Zn2+ finger motifs from E. coli TopA did not inhibit DNA binding and cleavage but abolished DNA relaxation, while a T. maritima TopA lacking the Zn2+ finger motif still relaxed DNA (4,7,12,13). Thus, Zn2+ fingers were hypothesized to either provide an auxiliary DNA interaction interface or to facilitate strand passage during relaxation. Some bacteria, including E. coli and Bacillus subtilis, possess an additional topoisomerase of type IA, named TopB or TopoIII, that is involved in DNA recombination and decatenation (14,15). The CTD of TopB is shorter than that of TopA (14 kDa instead of 30 kDa) and lacks Zn2+ fingers; however, similar to TopA, the TopB C-terminus is enriched in positively charged amino acids (16). TopA and TopB exhibit different processivities and velocities of supercoil removal, suggesting that the CTD modulates the mechanism of topoisomerase action (17). This idea is reinforced by observations made for actinobacterial homologs of TopA, which stand out as a group of topoisomerases with unique properties (18,19).
Since Actinobacteria represent a phylum that encompasses both clinically (Mycobacterium) and industrially (Streptomyces) important bacteria, the activity of their topoisomerases—enzymes essential for cell survival and involved in the regulation of secondary metabolism—has received considerable attention over the last decade (18–27). Actinobacterial IA topoisomerases differ from other bacterial TopA homologs in their CTD, which, although highly divergent within the class, is elongated, positively charged and lacks Zn2+ fingers (18,19). Instead, the CTDs of actinobacterial TopA homologs include very long stretches of polypeptide chains that are enriched in lysine residues (three such fragments in Mycobacterium and one in Streptomyces) (19,23,25). Biochemical studies of M. smegmatis and S. coelicolor TopAs have demonstrated the indispensability of the CTD for protein activity (19,28). The unique CTD structure was linked to the unusually high processivity that both enzymes exhibit (18,19). Single-molecule studies of S. coelicolor TopA revealed its ability to remove as many as 150 negative supercoils in a single relaxation burst (19). Ahmed and Nagaraja (23) suggested that the high processivity of M. smegmatis TopA may be due to DNA-binding by its CTD, which they suggested to be involved in strand passage. However, it remains unclear whether the participation of the TopA CTD in strand passage may enhance enzyme processivity. Another question that remains unanswered is whether less-processive enzymes would be biologically functional in Actinobacteria.
Streptomyces are soil Actinobacteria that, like all bacteria in this class, possess a GC-rich genome; however, the Streptomyces chromosome is linear and exceptionally long (up to 12 Mb) (29,30). The large genome size of Streptomyces is associated with complex secondary metabolism and regulatory networks, which allow their survival in harsh environmental conditions (30,31). Another adaptation for the soil habitat is the Streptomyces complex life cycle, which involves mycelial vegetative growth and sporulation. Streptomyces grow by forming dense hyphae composed of elongated and branching cells called hyphal compartments (32–34). These hyphal compartments contain multiple copies of chromosomes, which remain unseparated and uncondensed during vegetative growth (35). Sporulation involves the growth of specialized hyphae, which, due to intensive chromosomal replication, contain up to fifty chromosomal copies in a single sporogenic compartment, followed by the transformation of this compartment into a chain of unigenomic spores. At this stage, tens of chromosomes synchronously condense and segregate along the hyphal cell (32,36). The rapid topological changes in chromosomes during sporulation are dependent on a number of proteins, such as segregation proteins (ParA and ParB) and nucleoid-associated proteins (NAPs), and they also require TopA activity (21,37,38). S. coelicolor, one of the model Streptomyces species, possesses only one type I topoisomerase—TopA. While TopA is indispensable, its depletion inhibits growth, blocks sporulation and affects the production of secondary metabolites (such as the easily detectable blue actinorhodin) (21). We hypothesized that impairing S. coelicolor TopA processivity may affect the growth and development of Streptomyces.
Here, we verified the hypothesis that the exceptionally high processivity of S. coelicolor TopA is dependent on basic amino acids in its CTD. We tested the ability of the TopA CTD to interact with DNA, and we used magnetic tweezers to answer the question of how modifications within the CTD influence DNA relaxation at the single-molecule level. Since the CTD of E. coli TopA binds DNA, we tested whether it could restore the DNA binding, activity and processivity of C-terminally truncated S. coelicolor TopA. Finally, we tested whether the C-terminal modifications affect the in vivo functionality of TopA.
MATERIALS AND METHODS
In silico analysis
The amino acid sequences of actinobacterial type IA topoisomerases and the referenced actinobacterial proteomes were obtained from the UniProtKB Protein Knowledgebase (www.uniprot.org). Our dataset contained 4415 proteins belonging to 170 different genera. In this dataset, we searched for a pattern containing one to three lysines or arginines separated by two to nine neutral amino acids. The obtained dataset (3949 proteins with the general pattern) was filtered to contain only unique proteins possessing lysine-rich sequences with a minimum of five repeats and containing a minimum of 10% lysines and 27% alanines. We excluded fragments of proteins and topoisomerases annotated as ATP hydrolyzing. The final dataset contained 1126 proteins belonging to 133 different genera. For each protein, we added an annotation from the GO database indicating its probable function. To search for lysine repeats in different proteins, we downloaded complete proteomes from UniProtKB for 22 different species belonging to the genus Streptomyces. In this dataset, we searched for proteins using the same pattern as described above. We excluded proteins shorter than 120 amino acids and longer than 1500 amino acids (554 proteins in total) and filtered only unique proteins containing at least 3 repeats of a pattern that had a length of at least 17 amino acids and contained a minimum of 10% lysines and 10% alanines. To check whether non-Actinobacteria also contained lysine repeats, we performed a similar search as described for the genus Streptomyces using 20 proteomes from bacteria belonging to different types. Data analyses and searches for lysine repeats were performed using the R program (39). The Basic Local Alignment Search Tool (BLAST) was used for similarity analyses (40).
Bacterial strain preparation and growth conditions
Basic DNA manipulations were performed according to standard protocols (41). The oligonucleotides used for PCR and the construction of vectors are listed in Supplementary Table S1. The strains used in this study are listed in Supplementary Table S2. The culture conditions, antibiotic concentrations, transformation protocols and conjugation methods for S. coelicolor followed typical procedures (42). All S. coelicolor strains were grown on minimal medium that was supplemented with 1% mannitol (MM) or on rich medium (SFM) (42). The construction of the mutant S. coelicolor strains is described in the Supplementary Information. The genetic modifications of the obtained strains were verified by PCR, sequencing and western blot analysis. The growth curves of the S. coelicolor strains were measured in 79 medium (not supplemented with glucose, as previously described (21)) for 96 h at 30°C using Bioscreen C (Automated Growth Curves Analysis System, Growth Curves USA) with five replicates for each strain. The optical density (OD600) of the cultures was measured every 10 minutes.
Purification of mutant proteins
The construction of expression vectors used for the overproduction of modified S. coelicolor TopA proteins (see Figure 2A) in E. coli, and the purification methods are described in the Supplementary Information. All purified proteins were tested for nuclease contamination as follows: 250 ng of protein was incubated for 24 h at 37°C with 200 ng of supercoiled pUC19 plasmid in 50 mM NaH2PO4 (pH 8.1), 300 mM NaCl, 10% glycerol buffer supplemented with 10 mM MgSO4 and 1 mg/ml bovine serum albumin (BSA). Degradation of DNA was assessed by standard 1% agarose gel electrophoresis (data not shown). A circular dichroism (CD) spectra analysis indicated that, for all the analyzed proteins, the incidence of unstructured regions and structural composition was similar (Supplementary Figure S1).
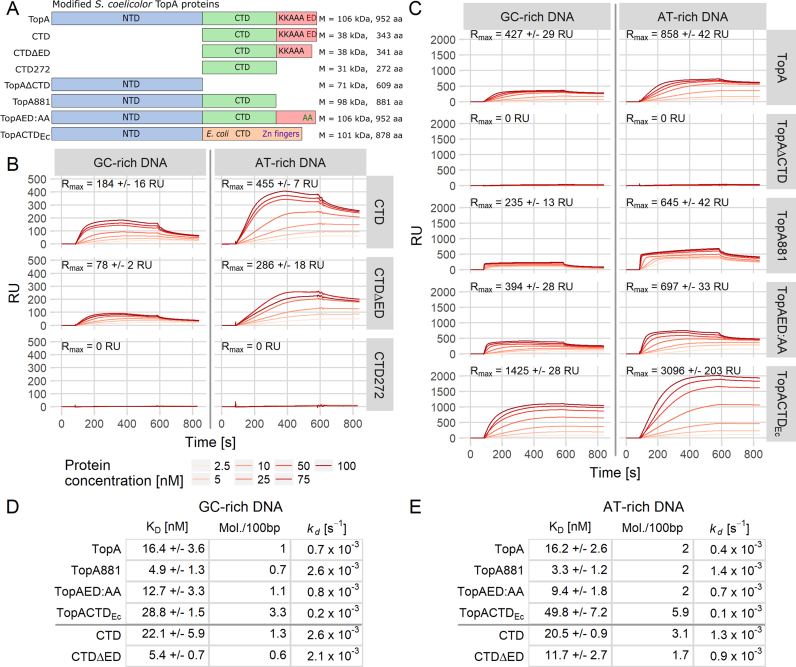
Figure 2.
The C-terminal domain of S. coelicolor TopA binds DNA. (A) Scheme of the modified versions of the TopA protein used in the analyses. Colored bars represent N- (blue) and C-terminal domains (green) with the lysine repeats (red). The E. coli TopA CTD containing Zn2+ fingers (orange) and terminal acidic amino acids (ED) are also shown. The number of amino acids and molecular weight of each protein are indicated. (B) SPR sensorgrams showing the binding of the TopA C-terminal domain (CTD) and its modified versions (CTDΔED and CTD272) at increasing concentrations (2.5–100 nM) to the GC-rich (69% GC, 241 bp) and AT-rich (29% GC, 218 bp) dsDNA. The maximum binding capacity (Rmax) is indicated on each plot. (C) SPR sensorgrams showing the influence of C-terminal truncations (TopAΔCTD, TopA881, TopAED:AA and TopACTDEc) on the binding of TopA to GC-rich (69% GC, 241 bp) and AT-rich (29% GC, 218 bp) DNA. Increasing concentrations (2.5–100 nM) of each protein were used, and the maximum binding capacity (Rmax) is indicated on each plot. (D and E) GC-rich and AT-rich dsDNA-binding parameters calculated for each tested protein: steady-state dissociation constant (KD), number of bound protein molecules/100 bp of DNA fragment and the dissociation rate (kd).
Topoisomerase activity assays
In gel assays of topoisomerase activity, the 200-μl reaction mixture contained 120 ng of negatively supercoiled pUC19 plasmid and 200 nM S. coelicolor TopA, TopA881, TopAΔCTD, TopAED:AA or TopACTDEc in 10 mM MgSO4, 1 mg/ml BSA, 25 mM NaH2PO4 (pH 8.1), 75 mM NaCl and 5% glycerol buffer. The reaction mixture was incubated at 37°C for 0, 0.5, 1, 2, 4, 6, 8 and 12 min and was stopped at that time by the addition of 2 μl of 0.5 M ethylenediaminetetraacetic acid (EDTA). The DNA was subsequently extracted with 1 volume of phenol:chloroform:isoamyl alcohol (25:24:1), and the topoisomers were analyzed in 1% agarose as previously described (19). The calculation of band intensities and the amount of DNA in each band were performed using Fiji software (43). The assay for each protein was repeated three times. To estimate the kinetic parameters Km (Michaelis-Menten constant) and Vmax (maximum velocity), we used 10 ng of TopA (5 nM), 20 ng of TopAED:AA (10 nM), 150 ng of TopA881 (75 nM) or 100 ng of TopACTDEc (50 nM) (the concentration of each protein was determined experimentally, using standard topoisomerase tests, to ensure that the initial velocity of supercoil removal could be precisely measured). The tested proteins were incubated with increasing amounts of negatively supercoiled pUC19 plasmid (100–2500 ng). The reactions were stopped after 30, 60, 90 and 120 s, and the plasmid was resolved and visualized as previously described (19). The calculation of the initial velocity (V0) was performed according to the protocol described by Xu and Leng (44). The obtained data were fitted to the Michaelis–Menten model using R Software and the drc package (45). The Vmax parameters that were calculated for different enzyme values were linearly extrapolated to the same enzyme concentration.
The single-molecule analysis of S. coelicolor TopA, TopA881, TopAED:AA and TopACTDEc protein activity was performed using a 5-kb DNA substrate encompassing a chromosomal fragment from the related S. venezuelae (GC content 72%) in 25 mM NaH2PO4 (pH 8.1), 75 mM NaCl and 10 mM MgCl2 at 34°C. Linear DNA fragments were prepared by cutting the pGEMF08 vector with XbaI and SbfI, purifying them by agarose gel electrophoresis and ligating them with 1-kb DNA labels: SbfI-biotin and XbaI-digoxigenin. Preparation of the 1-kb DNA labels for the magnetic trap experiments was performed according to the protocol described by Szafran et al. (19). In brief, the labeled linear DNA fragments were first attached to streptavidin-coated magnetic beads (Dynal MyOne, Life Technologies) and then to an antidigoxigenin-coated glass flow cell before being placed on a home-built magnetic trap running the PicoTwist software suite. The flow cell was placed on a magnetic trap instrument based on an inverted microscope. To introduce the negative supercoils, the magnetic beads were acted upon by a magnetic field gradient. The experiments were performed with the DNA molecules extended by a force of ∼0.5 pN. This causes DNA instability and the transient, local appearance of single-stranded regions (ssDNA) in the AT-richest parts of the molecule. The fragments were negatively supercoiled by ∼12 turns (leading to the formation of ∼12 plectonemic supercoils; however, after each round of the experiment, unrelaxed supercoils could still be present on the DNA, and the addition of a further 12 negative supercoils to such a DNA molecule would lead to the presence of more than 12 supercoils on the DNA). The removal of the supercoils by topoisomerase was observed as the DNA extension that was measured in real time by analyzing the position of the bead above the surface using nm-resolution particle tracking algorithms. The total time of the experiment amounted to 15 minutes for TopA, TopAED:AA and TopACTDEc and 60 minutes for the TopA881 protein. All the data were analyzed with R Software.
DNA-binding analyses
Surface plasmon resonance (SPR) analyses were performed on a BIAcore T200. For the SPR analysis, the following DNA fragments were obtained: a 241-bp-long fragment of S. coelicolor chromosomal DNA (GC-rich, 69% GC) that was amplified with primers parAB4200 and biotinylated-parS4440rv, as well as a 218-bp-long fragment of Arcobacter butzleri RM4018 chromosomal DNA (AT-rich, 29% GC) that was amplified with primers Abori2PEtv and biotinylated-pOCrv for a comparison of binding to AT-rich and GC-rich DNA. To compare binding to single-stranded and double-stranded DNA, the following DNA fragments were used: a 120-bp-long single-stranded biotinylated oligonucleotide sstopA-biot that was based on the S. coelicolor chromosome (63% GC) and a 120-bp-long fragment of S. coelicolor chromosomal DNA that was amplified with the primers topA_biacore_120 and biotinylated-RV3543RT. All DNA molecules were immobilized on the surface of a Sensor Chip SA (GE Healthcare Life Sciences) with up to 200 response units (RU) in total. An empty flow cell (no DNA bound) was used as a control. DNA not bound to the chip surface was removed, according to the manufacturer's recommendations, by washing the chip first with 50 mM NaOH and 1 M NaCl and subsequently with 50 mM NaOH, 1 M NaCl and 50% isopropanol. The interactions of the proteins with DNA were measured in binding buffer (25 mM NaH2PO4 (pH 8.0) and 150 mM NaCl) for 500 s at flow rate of 15 μl/min, followed by injection of binding buffer for 180 s. After each experiment, the chip surface was regenerated by washing it with the stripping buffer (0.05% SDS, 10 mM HEPES pH 8, 10 mM MgCl2, 1 mM EDTA, 0.05% Tween and 0.9 M NaCl). The interactions were analyzed in a broad range of proteins concentrations (from 2.5 to 100 nM). The results were plotted as SPR sensorgrams after the background signal obtained from empty control flow cell was subtracted. The experimental data were analyzed using the R Software. Since the DNA:protein ratio was not 1:1, all parameters were obtained from the empirical data instead of mechanical models. The stoichiometry analysis was based on the equilibrium phase that was obtained at the highest protein concentration that was used and was calculated using the following equation: Rmax = MwA/MwL * RU * n, where Rmax is the maximal response for protein; MwA and MwL are the molecular mass of the analyte and ligand, respectively; RU is the initial response after the DNA molecule binds to the chip; and n is the number of molecules. The KD (dissociation constant) and maximum binding capacity were obtained from the steady-state equilibrium phase by fitting a plot of equilibrium Req against the protein concentration and were calculated using the following equation: Req = (Rmax * [S])/(KD + [S]), where Req is the response at equilibrium, KD is the dissociation constant, and [S] is the substrate concentration. For each parameter (KD and Rmax), a standard error was calculated and used to compare parameters from different proteins. Differences exceeding standard errors were considered significant. The dissociation rate (kd) was calculated from the dissociation phase in the SPR sensorgrams.
In electrophoretic mobility shift assays (EMSA), up to 250 nM of TopA protein (as well as TopA881 or TopAED:AA mutants) was incubated for 15 min at 37°C in phosphate buffer (25 mM NaH2PO4, pH 8.0, 150 mM NaCl, and 5% glycerol) with 100 ng of supercoiled pUC19 plasmid (2.8 nM). Subsequently, all samples were supplemented with 3.0 μl of 50% glycerol and resolved overnight at low voltage in 0.5% agarose in 0.5× TAE buffer (40 mM Tris, 20 mM acetate and 1 mM EDTA). To visualize the DNA bands, the gel was treated with ethidium bromide solution for 30 min at room temperature.
Fluorescence microscopy and image analysis
Microscopy specimens were prepared by inoculating strains at the acute-angle junction of coverslips inserted at 45° in MM solid medium containing 1% mannitol (42) and cultured for either 24 or 44 h at 30°C. For DNA and cell wall staining, samples were fixed with a 2.8% paraformaldehyde/0.00875% glutaraldehyde mixture for 10 min at room temperature, digested with lysozyme (2 mg/ml in 20 mM Tris–HCl supplemented with 10 mM EDTA and 0.9% glucose) for 2 min, washed with PBS, blocked with 2% BSA in PBS buffer for 10 min and incubated with 0.1–1 μg/ml DAPI (Molecular Probes) and 10 μg/ml WGA-Texas Red (Molecular Probes) for 60 min. Fluorescence microscopy was performed using a Zeiss Axio Imager Z1 fluorescence microscope equipped with a 100× oil immersion objective.
RESULTS
C-terminal lysine repeats are a characteristic feature of actinobacterial TopA homologs
Actinobacterial type IA topoisomerases (TopA) differ from their other bacterial homologs at their CTD, which is longer and lacks Zn2+ finger motifs. Instead, the C-terminal fragment of S. coelicolor TopA encompasses a lysine-rich stretch of polypeptide chain (71 aa) (Figure 1A). The lysine residues alternate with three to five neutral amino acids (usually alanines or threonines), constituting a K1–2(A/T/N)3–5 motif named ‘the lysine repeat’ (Figure 1B). To determine whether the lysine repeats are a unique characteristic of Streptomyces TopA homologs, we searched 2212 actinobacterial type IA topoisomerase sequences for similar patterns.
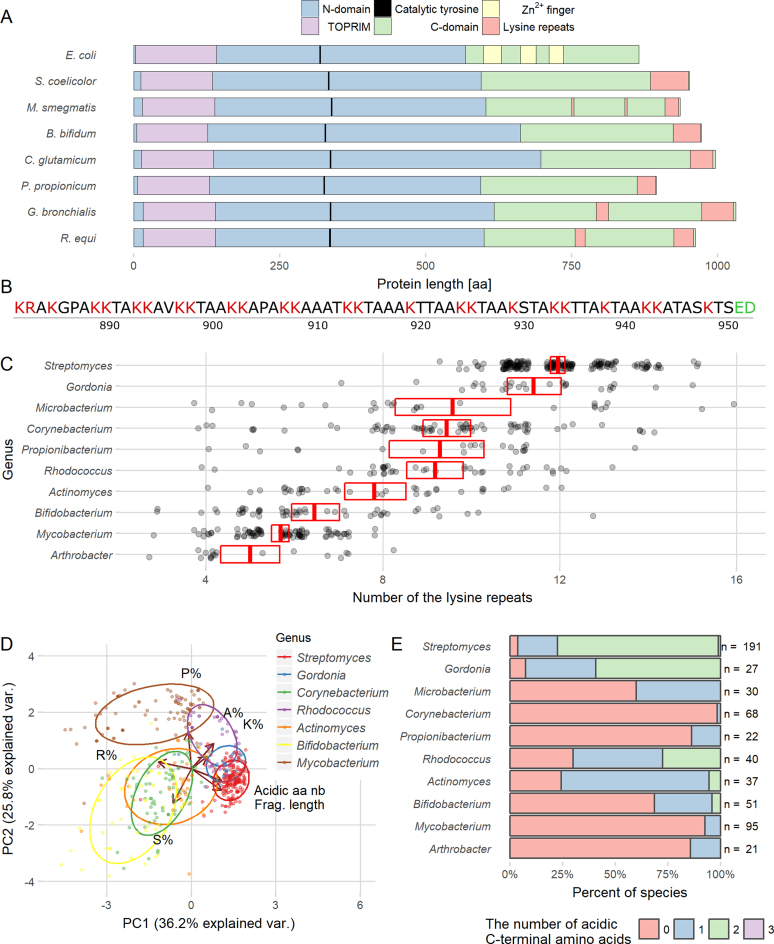
Figure 1.
Lysine repeats are characteristic of the actinobacterial TopA C-terminal domain. (A) Comparison of TopA sequences from various bacterial species. Blue and green color bars represent N- and C-terminal domains, respectively. Positions of the TOPRIM domain (purple), the catalytical tyrosine residue (black), Zn2+ fingers (yellow) and the lysine repeats (red) are indicated. (B) Sequence of S. coelicolor TopA 71 terminal amino acids encompassing the lysine repeats (K1–2(A/T/N)3–5). Basic amino acids are marked in red, and acidic in green. (C) The number of lysine repeats present in the TopA C-terminal domains of various genera belonging to Actinobacteria. Only data for the genera that had at least 20 identified proteins (699 proteins in total) are presented. The red box with a crossbar indicates the mean with 95% confidence interval, and all observations are marked by semitransparent points. (D) Principal component analysis (PCA) of the lysine repeats found in TopA C-terminal domains of Actinobacteria showing the variation in length and amino acid content. PCA was performed using the amino acid composition shown as the percentage of each amino acid: proline (P%), lysine (K%), alanine (A%), arginine (R%) and serine (S%), the length of the lysine repeats (Frag. length) and the number of acidic amino acids present among the last three amino acids of the protein (Acidic aa nb). On the plot, the first two principal components that have the largest variance (indicated on the plot) are shown. Color points indicate individual species belonging to different genera with normal data ellipse for each group (probability = 0.68). The analyzed genera are indicated. (E) Percentage of Actinobacteria TopA homologs containing acidic amino acids at the C-terminus. The analyzed genera are indicated.
Lysine repeats were identified in the C-termini of almost all the analyzed actinobacterial TopA homologs. While the number of repeats differed among and within genera, the Streptomyces TopA homologs contained the most numerous motifs—12 on average, but even up to 15 (Figure 1C). Different genera could be distinguished by the presence of additional amino acids, such as prolines or serines, in the lysine repeats, as shown by a principal component analysis (PCA) (Figure 1D). Interestingly, in 80% of the analyzed Streptomyces species (191 species), TopA homologs terminated with two acidic amino acids (glutamate, aspartate or both) (Figure 1E). This feature was found to be highly conserved in almost all analyzed Streptomyces TopA-like sequences but not in the sequences from other Actinobacteria.
To determine whether lysine repeats could be identified in proteins other than TopA, we searched for them in 22 complete Streptomyces proteomes. The search identified 554 proteins that contained lysine repeats similar to those present in S. coelicolor TopA. Among all the identified proteins, 175 were annotated as DNA-binding proteins (such as FtsK, HupS, Ku and HrdB), suggesting their overrepresentation in the group, which was confirmed by a statistical analysis using a hypergeometric test (p-values < 0.001). This result indicated that the lysine repeats may be important for interactions with DNA.
Interestingly, when we searched for lysine repeats in 20 proteomes of bacterial species from unrelated genera, we identified only a few proteins, including Caulobacter crescentus TopA and Bordetella pertussis sigma factor RpoD (Supplementary Figure S2). This finding may indicate that lysine repeats are characteristic of bacteria with GC-rich genomes.
In summary, our analyses showed that the multiple lysine repeats present in the CTD are characteristic of actinobacterial TopA homologs. In Streptomyces TopAs, the number of repeats is higher than the TopA-like proteins of other analyzed bacteria, and two C-terminal amino acids are acidic. Interestingly, among the proteins containing lysine repeats, the DNA-binding proteins are overrepresented, suggesting that they play a role in protein–DNA interactions.
The C-terminal domain of S. coelicolor TopA requires lysine repeats to bind DNA
We then examined whether the lysine repeats in the S. coelicolor TopA CTD are indeed engaged in DNA binding. To this end, we used SPR to measure the DNA binding of the intact TopA CTD as well as its truncated versions that lack the two terminal acidic amino acids, ED (CTDΔED), or the lysine repeats (CTD272, lacking 71 amino acids at the C-terminus) (Figure 2A).
The intact CTD efficiently bound double-stranded linear DNA (dsDNA) fragments (Figure 2B). The binding curves showed that the CTD had a higher binding capacity (Rmax) for AT-rich DNA than for GC-rich DNA. The calculated number of protein molecules bound to the DNA fragment was 1.3 molecules per 100 bp GC-rich DNA and 3.1 molecules per 100 bp AT-rich DNA fragment (Figure 2D and E). The CTD also bound efficiently to ssDNA (GC-rich); however, due to the high efficiency of protein binding and equipment limitations, we could not use sufficiently high concentrations of the tested protein to allow a reliable calculation of KD (Supplementary Figure S3). The affinity of CTD for GC-rich and AT-rich DNA fragments was similar, based on the steady-state calculation of the dissociation constant, while the dissociation rate for GC-rich DNA was two-fold higher (kd = 2.64 × 10−3 s−1) than that for AT-rich DNA (kd = 1.26 × 10−3 s−1) (Figure 2D and E).
The removal of the lysine motif from the TopA CTD (CTD272) completely abolished its interaction with DNA, while the terminal ED-truncation (CTDΔED) affected the binding properties of the CTD (Figure 2B). The binding capacity of CTDΔED was less than half that of the intact CTD protein fragment; the stoichiometry of the complex was similarly decreased (0.6 molecules/100 bp instead of GC-rich DNA compared to 1.3 for CTD), while the dissociation rate, kd, was only slightly affected (Figure 2D). Similar to the CTD, CTDΔED showed more than two-fold higher binding capacity to AT-rich DNA or to ssDNA than to GC-rich dsDNA (Figure 2D and E, Supplementary Figure S3B). Surprisingly, the steady-state dissociation constant of CTDΔED was 4-fold lower than that calculated for the intact CTD protein fragment (KD 5.4 ± 0.7 nM for CTDΔED compared to 22.1.5 ± 5.9 nM for CTD), indicating its increased affinity for DNA (Figure 2D).
Thus, the S. coelicolor TopA CTD binds linear DNA fragments, exhibits higher binding capacity and forms a more stable complex with AT-rich or ssDNA than with GC-rich dsDNA. The binding affinity was increased by the removal of the two C-terminal acidic amino acids, possibly due to the lack of charge repulsion. Removal of the lysine repeats completely abolished the interaction of the TopA CTD with the DNA.
Truncation of the C-terminal domain destabilizes the TopA–DNA complex
Having confirmed that the lysine repeats in S. coelicolor TopA CTD are required for interaction with DNA, we investigated whether CTD modifications affect the DNA binding by the enzyme. To this end, we studied the DNA-binding behavior of the TopA N-terminal domain alone (TopAΔCTD), the TopA variant that lacks the lysine repeats in its CTD (TopA881, truncated as in CTD272) and the TopA variant with the two acidic C-terminal amino acids (ED) replaced with AA (TopAED:AA) (Figure 2A).
The intact TopA protein, similar to the CTD, bound to AT-rich dsDNA or ssDNA more efficiently than to GC-rich dsDNA (two TopA molecules/100 bp to AT-rich or ssDNA and 1 molecule/100 bp to GC-rich DNA) (Figure 2C, D and E, Supplementary Figure S3A and C). Although the calculated steady-state dissociation constant for the TopA–DNA interaction was similar to that calculated for the CTD-DNA binding (Figure 2D), the DNA–TopA complex was very stable and dissociated inefficiently only at the highest protein concentrations used in the experiment (kd = 0.7 × 10-3 s−1; 80% of protein molecules remained bound to DNA, Figure 2C, D, and Supplementary Figure S3D).
The C-terminal truncation of TopA (TopAΔCTD) completely abolished binding to any of the tested DNA fragments (Figure 2C). The binding profile of TopA881 was also strikingly different from that of the wild-type TopA (Figure 2C). Sensorgrams showed that TopA881 rapidly bound to the DNA and rapidly dissociated from GC-rich DNA (kd = 2.56 × 10−3 s−1, compared to kd = 0.71 × 10−3 s−1 for the wild-type TopA) and AT-rich DNA (kd = 1.39 × 10−3 s-1, compared to kd = 0.44 × 10−3 s−1 for the wild-type TopA); in addition, only 40% of the molecules remained bound to the DNA (Figure 2D and Supplementary Figure S3D). TopA881, similar to wild-type TopA, showed a lower dissociation rate and higher binding capacity for AT-rich DNA (and ssDNA) than for GC-rich DNA (Figure 2D and E and Supplementary Figure S3A and C). Surprisingly, the TopA881 affinity for DNA was higher than that of the wild-type TopA (KD = 4.9 ± 1.3 nM, compared to KD = 16.4 ± 3.6 nM calculated for wild-type TopA), but the stoichiometry of TopA881 binding was similar to the wild-type TopA binding stoichiometry (Figure 2D). The increased affinity of TopA881 for DNA was additionally confirmed by a gel retardation experiment using supercoiled plasmid DNA (Supplementary Figure S4). The TopAED:AA binding (KD) was similar to that of wild-type TopA, and its slow dissociation rate (70% of molecules remained bound to DNA, Supplementary Figure S3D) and binding stoichiometry (Figure 2D) were also similar; however, the binding capacity of the modified protein was marginally lower.
To recapitulate, we showed that the elimination of the CTD abolished DNA binding by S. coelicolor TopA. Unexpectedly, the removal of the lysine repeats increased the enzyme's affinity for DNA and destabilized the protein–DNA complex. We speculate that the higher affinity toward DNA could be the result of the exposure of the binding site in the N-terminal domain by the removal of the CTD fragment. Replacement of the terminal acidic amino acids with alanines only marginally affected DNA binding by TopA.
Fusion of the E. coli TopA CTD to S. coelicolor TopAΔCTD restores its enzymatic activity but not its processivity
Previously, we demonstrated that a truncated S. coelicolor TopA lacking the CTD (TopAΔCTD) was unable to relax DNA (19). Having shown here that the CTD binds DNA, we expected that an E. coli TopA CTD that contains Zn2+ fingers and interacts with DNA could restore topoisomerase activity when fused to TopAΔCTD. To test this hypothesis, we constructed a hybrid protein (TopACTDEc) that was a fusion of the E. coli TopA CTD (269 aa) with the S. coelicolor TopAΔCTD construct (609 aa) (Figure 2A).
First, we tested whether the hybrid protein TopACTDEc could bind DNA. The SPR analysis showed that, although the binding capacity (Rmax) of TopACTDEc was higher than that of the wild-type TopA and the number of TopACTDEc molecules that bound to both GC-rich and AT-rich DNA fragments increased 3-fold, the dissociation constant increased compared to wild-type TopA (KD = 28.8 ± 1.5 nM for hybrid protein versus 16.4 ± 3.6 nM for wild-type TopA) (Figure 2C–E and Supplementary Figure S3). Thus, the hybrid protein exhibited lower affinity for DNA than wild-type S. coelicolor TopA.
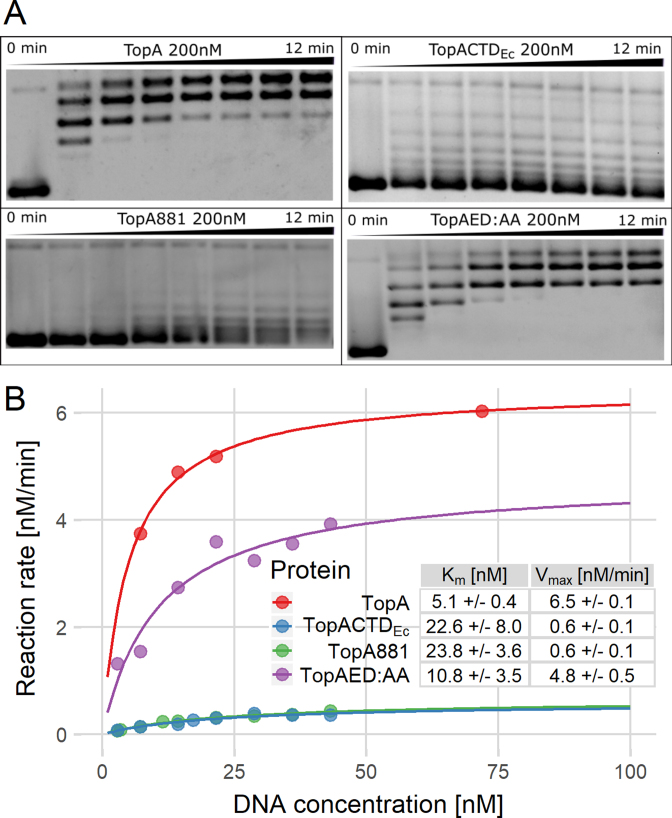
Next, we tested the enzymatic activity of the hybrid protein. While the DNA relaxation activity of TopAΔCTD was completely abolished (19), TopACTDEc relaxation activity was restored; however, the relaxation pattern was more distributive than observed for the wild-type S. coelicolor TopA (Figure 3A). Using the substrate concentration range from 0 to 70 nM, we measured the reaction velocities (Vmax) within their linear range and fitted the reaction kinetics to the Michaelis–Menten model. This analysis showed decrease of the maximum reaction velocity of the hybrid protein (Vmax = 0.6 ± 0.1 nM/min) and its substrate affinity (Km = 22.6 ± 8 nM) compared to both parameters (Vmax = 6.5 ± 0.12 nM/min and Km = 5.1 ± 0.4 nM) of the wild-type TopA (Figure 3B).
Figure 3.
Bulk topoisomerase assays show that the activity of TopA depends on the presence of the lysine repeats in the C-terminal domain. (A) Images of agarose gel-based assays demonstrating the activity of TopA, TopACTDEc, TopA881 and TopAED:AA (200 nM); the reaction mix was sampled at time points from 0 to 12 min. (B) Estimation of kinetic parameters based on the agarose gel assays. Vmax and Km were calculated by fitting the data for TopA (red), TopACTDEc (blue), TopA881 (green) and TopAED:AA (purple) to the Michaelis–Menten equation.
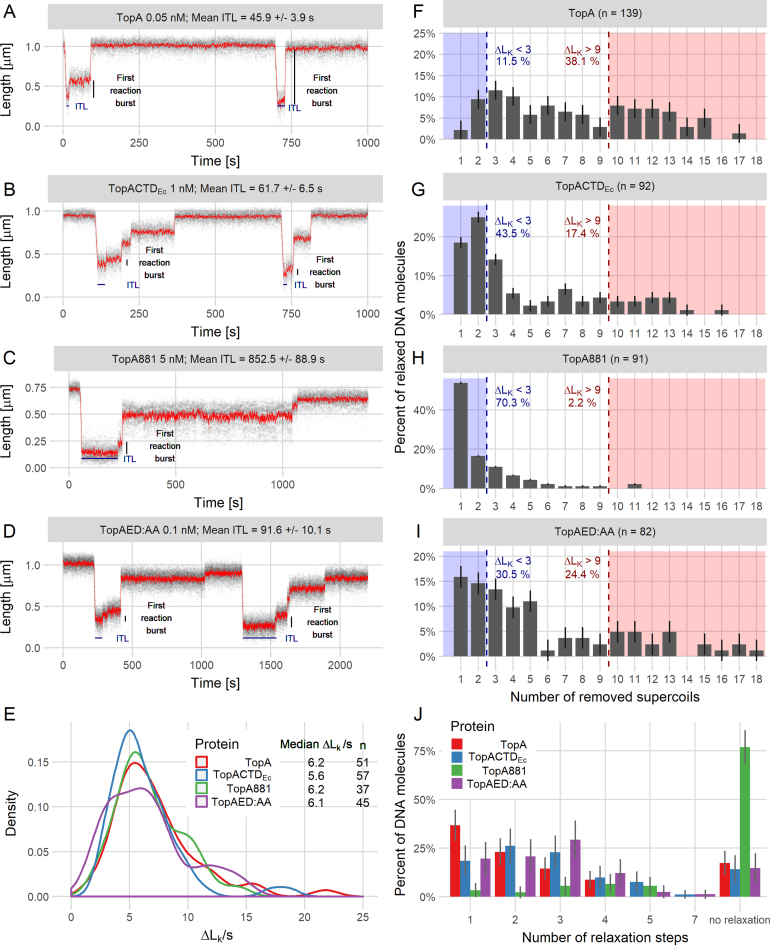
Our previous report showed that S. coelicolor TopA is substantially more processive than the E. coli enzyme; this enhanced processivity was associated with the differences in the CTDs (19). Thus, we expected that the hybrid enzyme TopACTDEc, although active, would exhibit lower processivity than the wild-type S. coelicolor TopA. This behavior was already suggested by the bulk assay of topoisomerase activity. The magnetic tweezers experiment allowed us to measure the number of negative supercoils (Lk—linking number) that were removed in a single enzymatic burst from a 5-kb DNA molecule in which 12 supercoils were initially introduced (Figure 4A and B). The analysis revealed that although TopACTDEc exhibited the same velocity of supercoil removal as wild-type TopA (Figure 4E), the initial time lag (ITL—the time between the addition of the enzyme and the first observed DNA relaxation event) was significantly extended (the concentration of TopACTDEc was 200-fold higher than that of wild-type TopA, Figure 4A and B). Moreover, while the wild-type S. coelicolor TopA removed at least 10 supercoils from 38% of DNA molecules in the first reaction burst (Figure 4F), TopACTDEc removed 10 or more supercoils from only 17% of DNA molecules and usually (44% of molecules) removed fewer than three supercoils (Figure 4G and J).
Figure 4.
Single-molecule analysis shows that modifications of the C-terminal domain decrease the processivity of TopA. (A–D) Example trace lines from single-molecule experiments for TopA (A) and the modified TopA versions TopACTDEc (B), TopA881 (C) and TopAED:AA (D) with the ITL and first reaction burst marked. (E) The supercoil relaxation rates (ΔLk/s) for TopA (red), TopACTDEc (blue), TopA881 (green) and TopAED:AA (purple). The plot shows the distribution as a probability density function. n is the number of analyzed events, in which between 6 and 12 supercoils were removed from the DNA molecule. (F–I) Decreased processivity of the modified proteins TopACTDEc (G), TopA881 (H), and TopAED:AA (I) compared to the wild-type TopA (F), calculated as the number of supercoils removed—ΔLk (change in the linking number) during the first relaxation burst. Dashed lines indicate the fraction of the molecules in which fewer than three (blue) or more than nine supercoils (red) were removed. ‘n’ is the total number of relaxation events analyzed for each protein. (J) The increased number of steps required by TopA (red), TopACTDEc (blue), TopA881 (green) and TopAED:AA (purple) for the full relaxation of 5-kb DNA molecules. The histogram shows the percentage of the non-relaxed DNA molecules and the DNA molecules that were relaxed in the specified number of steps (1–7) for each tested protein.
To recapitulate, our experiments showed that the E. coli CTD restored DNA-binding and relaxation activity in the truncated S. coelicolor TopAΔCTD protein. The hybrid protein, however, exhibited lower DNA affinity (higher KD and higher Km) and lower Vmax than the wild-type protein. The E. coli CTD fusion could not re-establish the high processivity of S. coelicolor TopA, confirming that it was dependent on its CTD.
Modifications of TopA C-terminus affect enzyme processivity
Since we showed that the lysine repeats confer the DNA-binding activity of the S. coelicolor CTD and its substitution with the E. coli TopA CTD restored DNA binding but not enzyme processivity, we expected that the lysine repeats are a prerequisite for high processivity. Thus, we examined how the removal of the lysine repeats (71 aa; TopA881) or substitution of acidic C-terminal glutamate and aspartate with alanines (TopAED:AA) would affect the enzyme's activity.
First, using the bulk topoisomerase activity assay, we compared the activities of the wild-type TopA, TopA881 and TopAED:AA (Figure 3A). As previously observed (19), the wild-type TopA relaxed DNA efficiently (Figure 3A) and removal of CTD (TopAΔCTD) abolished TopA activity, however mixing TopAΔCTD with the CTD restored DNA relaxation (Supplementary Figure S5) (as observed earlier for M. smegmatis TopA (28)), indicating the proper folding of TopAΔCTD. Markedly, removal of the lysine motif changed the DNA relaxation pattern from processive to distributive (Figure 3A). Interestingly, the replacement of the terminal Glu-Asp with Ala-Ala also slightly lowered the TopA activity (Figure 3A). At the end of the experiment, the wild-type TopA completely relaxed approximately 45% of the plasmid molecules, while TopAED:AA completely relaxed only 20% of plasmids. The maximum reaction velocity of TopA881 and TopAED:AA mutants was decreased (Vmax = 0.6 ± 0.1 nM/min and 4.8 ± 0.5 nM/min, respectively), while their Michaelis–Menten constants increased (TopA881 Km = 23.8 ± 3.6 nM and TopAED:AA Km = 10.8 ± 3.5 nM) compared to the wild-type enzyme TopA (Vmax = 6.5 ± 0.1 nM/min and Km = 5.1 ± 0.4 nM) (Figure 3B). These results suggest that both modifications notably influenced the efficiency of DNA relaxation.
Having confirmed that modifications of the TopA CTD affected the enzymatic activity parameters, we used magnetic tweezers to examine their influence on the enzyme's processivity (Figure 4C and D). Whereas the wild-type TopA removed all supercoils (ΔLK > 9) from 38% of molecules in a first relaxation burst, TopAED:AA fully relaxed 24% and TopA881 only 2% of DNA molecules (Figure 4F, H and I). Both modified enzymes more often removed only one or two supercoils (ΔLK < 3 for 70% of DNA molecules relaxed by TopA881 and 31% of molecules relaxed by TopAED:AA) (Figure 4H and I). Thus, the number of steps required to fully relax DNA molecules increased significantly for both modified proteins, and TopA881 was unable to fully relax 80% of the DNA molecules during the experiment time (Figure 4J). These analyses showed that TopAED:AA removed supercoils less effectively than the wild-type TopA, while the processivity of TopA881 was completely abolished.
The initial time lag for TopA881 was increased >20-fold compared to TopA, although the protein concentration was 100-fold higher (5 nM TopA881, ITL = 852.5 s ± 72.3 versus 0.05 nM TopA, 45.9 s ± 3.9), while the ITL of the TopAED:AA protein was 2-fold longer than that of wild-type TopA (0.1 nM, 91.6 ± 7.8) (Figure 4C and D). Histograms of individual ITLs that were measured for all tested proteins showed that the ITLs were distributed according to single-exponential statistics (Supplementary Figure S6), which suggests that the binding was not cooperative. Interestingly, the relaxation velocities measured for reaction steps that removed more than six supercoils remained similar in all tested proteins (6–7 Lk/s) (Figure 4E). This result also confirmed that the analyzed proteins were properly folded.
Thus, our results establish that the CTD is essential for high TopA processivity. The removal of the lysine repeats and the mutation of the terminal acidic amino acids lowers protein processivity and increases the time before reaction initiation. These observations are in agreement with the observation that modification of the CTD destabilizes the TopA:DNA complex.
Growth of the TopA-depleted strain cannot be restored by TopACTDEc and is further impaired by the presence of TopAΔCTD
The CTD truncation of S. coelicolor TopA abolished its activity, while its substitution with the E. coli CTD restored enzyme activity but not its high processivity. Since TopA depletion inhibited the growth and development of S. coelicolor, we expected that C-terminal TopA modifications (topAΔCTD and topACTDEc) may also delay the growth and development of mutant strains. To test this hypothesis, we constructed S. coelicolor strains that produced modified TopA proteins and analyzed their growth.
As expected, the deletion of the topA gene fragment encoding the CTD was unsuccessful, which may be explained by impaired DNA binding of the TopAΔCTD protein. We were also unable to complement the S. coelicolor topA deletion with the topACTDEc gene. Thus, we used a merodiploidic strain (PS07 strain with a second copy of the topA gene under the control of the ptipA inducible promoter (21)) to delete the CTD-encoding fragment of the topA gene in its native locus or to replace it with the CTD-encoding fragment of E. coli topA (with the sequence optimized for Streptomyces codon usage) (strains AS02 and AS09, respectively, Figure 5A). Due to topA promoter autoregulation (46), the production of the modified TopAs (under the control of the native promoter) was detected only in the absence of the ptipA inducer and not at the induction of wild-type topA gene expression (Supplementary Figure S7). The growth of the obtained strains was compared to the growth of the wild-type and PS04 strains (with a single copy of topA under control of the ptipA promoter (21)), in which the TopA protein level was depleted 20-fold compared to the wild-type strain when cultured without inducer.
Figure 5.
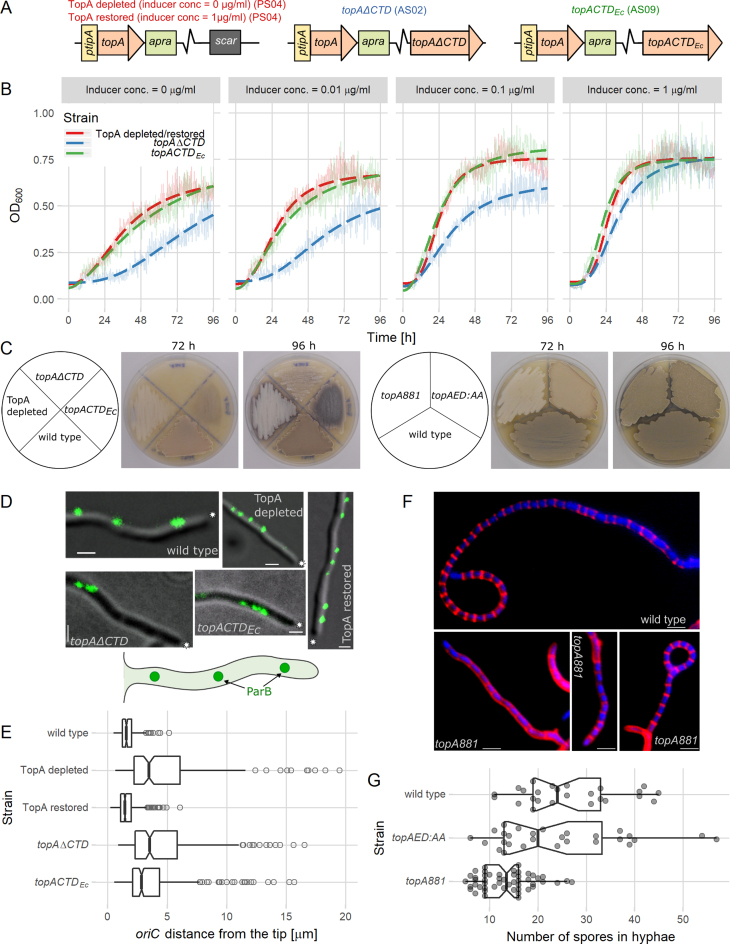
C-terminal TopA domain modifications impair S. coelicolor growth and development. (A) Scheme of the strains used in the in vivo experiments: PS04 (TopA depleted), AS02 (topAΔCTD) and AS09 (topACTDEc). (B) Inhibited growth of TopA-depleted (PS04, red), topAΔCTD (AS02, blue) and topACTDEc (AS09, green) strains cultured with different concentrations of wild-type topA expression inducer (0–1 μg/ml) compared to TopA-restored strain (PS04, inducer concentration 1 μg/ml). Semitransparent lines show the mean absorbance values obtained from five replicates, and the dashed lines correspond to the fit to a log-logistic growth model. (C) Slower development TopA-depleted (PS04), topAΔCTD (AS02), topACTDEc (AS09), topA881 (AS03) and topAED:AA (AS05) strains compared to the wild-type (M145) at 72 and 96 h of growth on SFM medium. Gray pigment is characteristic of fully developed aerial hyphae and white color indicates the inhibition of S. coelicolor sporulation (the dark appearance of AS09 results from the production of blue actinorhodin). (D) Disturbed positions of chromosomal oriC regions in hyphae of wild-type (AK101), TopA-depleted (AS11, with and without the presence of the inducer of TopA expression), topAΔCTD (AS12) and topACTDEc (AS19) strains. Images show ParB-EGFP fluorescence (green) overlaid with a DIC image (gray). Scale bar 1 μm, asterisks indicate the tips of hyphae, and the cartoon represents the imaged hyphae. (E) Increased distance between the hyphal tip and the apical chromosome oriC in the TopA-depleted (AS11, 202 hyphae analyzed), TopA-restored (AS11 cultured in the presence of inducer, 578 hyphae analyzed), topAΔCTD (AS12, 296 hyphae analyzed) and topACTDEc (AS19, 312 hyphae analyzed)) strains compared to the wild-type (AK101, 202 hyphae analyzed). Boxplots show the median of distribution (line), the first and third quartiles (lower and upper ‘hinges’), and the 95% confidence interval (notches around the median). (F) Prespore chains of normal length in wild-type (M145) and examples of shortened prespore chains in topA881 (AS03) strains. Images show hyphae (after 44 h of culture); cell walls were stained with Texas Red (red), and the DNA was stained with DAPI (blue). Scale bar 2 μm. (G) The number of compartments in prespore chains of the wild-type (M145, 25 hyphae analyzed), topAED:AA (AS05, 28 hyphae analyzed) and topA881 (AS03, 50 hyphae analyzed) strains. Boxplots show the median of distribution (line), first and third quartiles (lower and upper ‘hinges’), and the 95% confidence interval (notches around the median). Semitransparent points indicate all observations.
The vegetative growth (liquid culture) of all modified strains was retarded at the low level of full-length topA gene expression in the absence of the inducer. The growth of the strain that produced the C-terminal-truncated variant, TopAΔCTD (AS02), was significantly slower than the growth of TopA-depleted strain PS04 at all tested inducer concentrations. Even at the inducer concentration of 1 μg/ml (the TopA level restored to the wild-type level and decreased topAΔCTD expression due to autoregulation (Supplementary Figure S7) (21,46)), the growth of the topAΔCTD (AS02) strain was still somewhat slower than that of the TopA-restored strain (PS04), suggesting a dominant-negative effect of the production of TopAΔCTD. The growth of the topACTDEc (AS09) strain was similar to the growth of the TopA-depleted strain (Figure 5B).
The depletion of the TopA level in S. coelicolor inhibits sporulation, and as the gray spore chains are not formed, the colonies remain white (21). If the production of the wild-type TopA was not induced, neither the topACTDEc (AS09) strain nor the topAΔCTD (AS02) strain was able to generate spores (indicated by the presence of gray pigment), even after prolonged incubation, similar to the TopA-depleted PS04 (the dark AS09 appearance resulted from enhanced blue actinorhodin production, which was also associated with increased chromosome supercoiling (21)) (Figure 5C, left panel). Moreover, the topAΔCTD strain (AS02) required 96 h to produce the white aerial hyphae, while the TopA-depleted strain took 72 h. Thus, the expression of topAΔCTD impaired AS02 strain development to an even greater extent than TopA depletion by itself.
To determine how the expression of the modified TopA proteins affects the distribution of multiple chromosomes in vegetative hyphae, we marked the chromosomal oriC (origin of chromosome replication) region using a ParB-EGFP protein (35). Our earlier studies showed that chromosomes in multigenomic hyphae follow the extending hyphal tip and that the apical chromosome oriC complexed with ParB maintains a constant distance to the hyphal tip (35). In the vegetative hyphae of the TopA-depleted strain (AS11), the distance between the first ParB complex and the tip was significantly increased compared to the wild-type strain (mean distance: 4.6 and 1.7 μm, respectively); the apical anchorage of oriC was restored upon induction of topA expression. In the topAΔCTD (AS12) strain, the position of the apical chromosome was similar to that in the TopA-depleted strain (mean distance: 4.8 μm) (Figure 5D and E), suggesting substantial problems with the chromosome distribution in the vegetative hyphae. In the hyphae of the topACTDEc strain (AS19), the position of the apical chromosome was partially restored (mean distance: 3.6 μm) (Figure 5D and E), indicating that the processivity of the hybrid protein was sufficient to maintain the chromosome distribution in vegetative hyphae.
In summary, these results indicate that the hybrid TopA protein that contains the E. coli CTD is able to complement the TopA depletion to some extent, but only during vegetative growth, and it does not restore sporulation. Markedly, the production of truncated TopAΔCTD apart from the wild-type TopA protein severely impaired the growth of S. coelicolor. The growth inhibition of strains producing modified TopA proteins was associated with impaired chromosome distribution in the hyphae.
C-terminal modifications of TopA affect the sporulation stage of S. coelicolor growth
The depletion of the TopA level completely blocks sporulation. During the growth of sporogenic hyphae and their conversion to spore chains, multiple copies of chromosomes in the hyphal cell undergo intensive replication and rapid topological changes, suggesting the highest demand for TopA processivity. Having demonstrated that modification of the lysine repeats in the CTD of TopA and substitution of the last two acidic amino acids affected DNA binding and enzyme processivity, we expected that the biological consequences of the CTD modifications should be pronounced at the sporulation stage. To verify this hypothesis, we constructed S. coelicolor strains in which the topA gene in its native locus was replaced with topA881 (AS03) or topAED:AA (AS05) (expressed from the native promoter) and analyzed their development.
The vegetative growth (liquid culture) of strains topA881 (AS03) and topAED:AA (AS05) was the same as the growth of the wild-type strain (Supplementary Figure S8); however, the development of the mutant strains was affected. In the culture of the topA881 strain, the gray pigment, indicating spores, was detected only after 96 h, compared to the 72 h required for the wild-type strain (Figure 5C, right panel), indicating a delay in sporulation. After 72 h of growth, the lighter color of the topAED:AA strain (AS05) suggested that its development of gray spores was less efficient than in the wild-type strain (Figure 5C, right panel).
Having established that the topA881 strain (AS03) exhibited severely delayed sporulation, we further examined its aerial hyphae microscopically. The extension of Streptomyces aerial hyphae is inhibited before synchronized septation and transformation into the prespore chain. The prespore chains of the wild-type strain had an average length of 30–50 μm and were usually divided into 20–30 prespores (Figure 5F and G). The analysis of the topAED:AA (AS05) prespore chains did not show clear differences in the number of prespore compartments or chromosome distribution (Figure 5G). In contrast, the topA881 strain (AS03) produced much shorter spore chains, containing 2-fold fewer prespores than the wild-type strain (Figure 5F and G). The average size of the prespores was not disturbed in this strain, but 5% of the spores lacked the chromosome (compared to 1% in the wild-type strain).
To conclude, our analysis showed that the high processivity of topoisomerase I conferred by the lysine repeats is not essential for vegetative growth but is required during Streptomyces sporulation for efficient production of spore chains.
DISCUSSION
Our in silico analyses revealed that the presence of lysine repeats (K1–2(A/T/N)3–5), similar to the lack of Zn2+ fingers, is a characteristic feature of actinobacterial TopA CTDs. Streptomyces TopA homologs contain a single stretch of lysine repeats, the longest among actinobacteria, that terminates with two negatively charged amino acids. The S. coelicolor TopA structure prediction indicated that the lysine repeats may form an α-helix. Indeed, the analysis of the CD spectra indicated a lowered incidence of α-helixes for the truncated proteins and a higher incidence of β-structures for the hybrid protein TopACTDEc, which is consistent with the higher incidence of β-structures in the E. coli TopA CTD (32%, based on crystallographic data) compared to the S. coelicolor TopA CTD (16%, based on structure prediction using DeepCNF (Deep Convolutional Neural Fields) (47)). The recent sequence and structure analysis of M. tuberculosis TopA revealed that its CTD encompasses four repeated subdomains and ends with a lysine-rich stretch of peptide chain (25). Each subdomain is formed by four-stranded antiparallel β-sheets that are stabilized by crossing over an α-helix, constituting a novel protein fold. However, this study did not elucidate the structure of the whole CTD and the role of the lysine stretch.
We demonstrated here that the lysine repeats in the CTD of S. coelicolor TopA interact with DNA. The removal of the lysine repeats from the CTD (CTD272) completely abolished its interaction with DNA, while the truncation of the lysine repeats from the full-length S. coelicolor TopA (TopA881) significantly changed the enzyme's binding properties. The impaired DNA binding of the CTD272 protein fragment could result from its partial misfolding or the removal of the DNA-binding domain. Removal of lysine repeats from the whole enzyme also affected its DNA binding—TopA881 exhibited an affinity for DNA that was higher than that of wild-type TopA, and it rapidly associated but also readily dissociated from DNA. The dissociation of the wild-type TopA from DNA in the absence of Mg2+ is very limited, presumably due to impaired religation of the cleaved DNA strand and the inability of TopA to finish the catalytical cycle (19). The rapid dissociation of TopA881 may indicate that, in the absence of lysine motifs, the formation of the complex does not lead to DNA cleavage. The formation of the unstable complex by TopA881 corroborates its decreased enzymatic activity (increased Km), significantly decreased processivity of supercoil removal and significantly increased initial time lag (ITL). Markedly, the modification of the characteristic Streptomyces TopA homologs’ C-terminal acidic amino acids (TopAED:AA) also led to the modification of the enzyme's affinity for substrate (Km), processivity and initial time lag; however, as expected, these changes occurred to a much lesser extent than in the case of the removal of the lysine repeats. Strikingly, the velocity of supercoil removal by TopA881, TopAED:AA and the wild-type TopA were identical, which indicates that C-terminal truncation affects only the stability of the complex, not the reaction mechanism.
Thus, DNA binding by the CTD is a conserved property of TopA homologs, which is conferred by Zn2+ fingers in E. coli TopA and by the positively charged amino acids in TopB (Topo III), Mycobacterium and Streptomyces TopA (11,23). It has been suggested that Mycobacterium species eliminated the Zn2+ fingers from TopA to increase the enzyme's pH and oxidative stress tolerance or to endure the intracellular paucity of Zn2+ ions (maintained due to their toxicity) (23). It is difficult to determine whether similar reasons could account for the lack of Zn2+ fingers in Streptomyces TopA. Nevertheless, actinobacterial TopA homologs apparently use the lysine repeats as the DNA-binding interface in their CTDs. The results from this work and those described previously indicate that S. coelicolor TopA, similar to most topoisomerase IA homologs, preferably interacts with single-stranded DNA (19). This preference was conferred by both the CTD and the truncated TopA881 lacking lysine repeats. Notably, the occurrence of single-stranded regions is substantially diminished in GC-rich chromosomes, thus limiting the number of interaction sites for TopA. The DNA-binding site based on the lysine repeats may play a significant role in the additional stabilization of the complex in the shortage of single-stranded DNA regions. Interestingly, lysine repeats were identified in a number of Streptomyces DNA-binding proteins, such as HupS (HU homolog), FtsK and the HrdB sigma factor as well as proteins from bacteria with GC-rich genomes that interact with DNA (e.g. B. pertussis sigma factor RpoD and C. crescentus TopA). Lysine repeats were also shown to be required for DNA interaction of M. smegmatis HupB (Holowka et al., submitted). Markedly, lysine repeats were not recognized in the homologs of these proteins in other species.
Previously, Ahmed and co-workers noted that the removal of one or two lysine-rich stretches impaired M. smegmatis TopA processivity and suggested that the basic amino acids in the mycobacterial TopA CTD are involved in strand passage (23). That suggestion is in agreement with the earlier notion that in E. coli TopA, Zn2+ fingers interact with the transferred DNA strand during the reaction. However, it was also observed that modifications of the CTD of T. maritima TopA did not affect strand passage, suggesting that this protein fragment instead provides an accessory platform for DNA binding (4,48). Our studies of S. coelicolor TopA cannot confirm the engagement of lysine repeats in strand passage, although the single-stranded DNA binding preference could support this hypothesis. However, the single-molecule experiments revealed the identical velocity of supercoil removal by the truncated (TopA881) and the wild-type TopA proteins. This result strongly questions the involvement of the lysine repeats in the relaxation reaction itself. Instead, our observations indicate that the interaction of the lysine repeats with DNA stabilizes the complex before the initiation of the reaction and during supercoil removal. Thus, our results provide new insight into the mechanism of TopA activity.
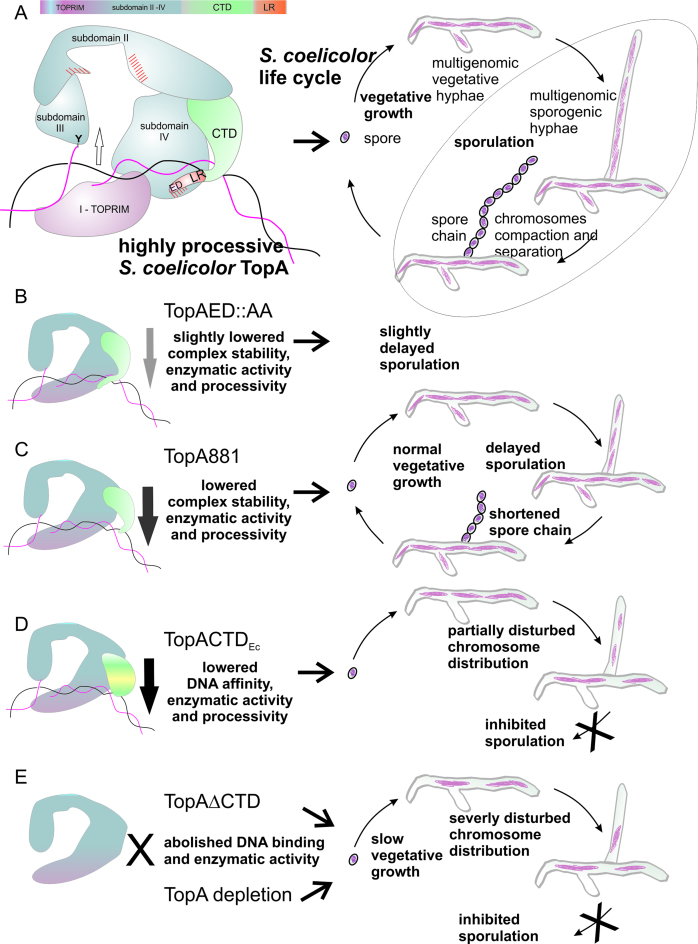
On the basis of our observations, we propose a speculative model (Figure 6A) that explains the obtained results: the differences in binding affinity and complex stability resulting from TopA modifications. According to our model, the CTD not only may bind DNA but also possibly interacts with the NTD, forming a clamp that stabilizes the complex. The sequence analysis indicated that the S. coelicolor TopA NTD in the last helix of the D4 domain contains an unusual (not present in E. coli TopA homologs) insertion of 23 amino acids (including 5 acidic amino acids) flanked by basic amino acids that are not conserved (Supplementary Figure S9). A similar but shorter insertion was noted in M. tuberculosis TopA (25). This insertion (or another unique fragment of the NTD sequence, Figure 6, red dashed area) could provide the interaction interface for C-terminal amino acids. The fact that the insertion is located in proximity to the DNA-binding groove in subdomain IV of the NTD may suggest that its interaction with the terminal fragment of the CTD may hinder the DNA-binding site in subdomain IV. That could explain the higher affinity of TopA881 for DNA as resulting from the increased accessibility of the DNA-binding groove in the NTD. The observation that the elimination of the ED amino acid pair from the whole-length protein (TopAED:AA) affected the protein's interaction with DNA to a much lesser extent than their mutation in the CTD protein fragment could be explained by the interaction of these acidic amino acids with basic amino acids in the NTD. Because of such an interaction in the wild-type TopA, the terminal ED would not be as exposed as in the CTD, where they could lower the affinity for DNA via charge repulsion. Thus, we speculate that the interaction of the C-terminal ED with basic amino acids in the NTD could help to maintain the clamp closed on DNA, additionally increasing complex stability and enzyme processivity (Figure 6). It must be noted that the removal of the C-terminal domain completely abolished S. coelicolor TopA activity (19) and the DNA binding of the NTD (Figure 6). The impaired DNA binding of S. coelicolor TopAΔCTD may result from its partial misfolding, however this explanation seems unlikely since mixing TopAΔCTD with CTD restores enzyme relaxation activity. Thus, the presence in CTD of additional (other than lysine repeats) subdomain required for the formation of the TopA–DNA complex could explain the obtained results. Interestingly, the M. smegmatis TopA NTD and CTD interacted with DNA independently; moreover, they could catalyze the reaction when mixed together (28). This observation and the finding that also S. coelicolor TopAΔCTD and CTD when mixed relax DNA support our hypothesis that an interaction between the CTD and NTD stabilizes the complex on DNA. Thus, the clamp formation that is suggested here may be a conserved feature of actinobacterial TopA that stabilizes enzymes on GC-rich chromosomes.
Figure 6.
Model of DNA complex formation by the S. coelicolor TopA. (A) A speculative model of the TopA–DNA complex that involves the formation of the DNA-stabilizing clamp by the CTD, fastened due to the interaction between the lysine repeats (LR, red) with DNA and actinobacterial unique sequence (red hatched area) in subdomain IV. (B–E) The influence of TopA modifications—exchange of C-terminal Glu and Asp for Ala (TopAED:AA) (B); truncation of the lysine repeats (TopA881) (C); the fusion of E. coli TopA C-terminal domain (TopACTDEc) (D) and the CTD truncation (TopAΔCTD) (E)—on DNA binding, enzymatic activity, and processivity is reflected in in vivo functionality either during sporulation alone or also during vegetative growth.
The fusion of the S. coelicolor TopA NTD to the DNA-binding E. coli TopA CTD was unable to restore the high processivity of S. coelicolor TopA. The hybrid TopACTDEc enzyme was unable to reestablish the growth and sporulation of the S. coelicolor strain that was depleted of TopA (Figure 6D). The hybrid protein exhibited lowered DNA affinity, consistent with its elongated initial time lag and lower processivity. All of these observations most likely explain the lack of complementation of TopA-depleted strain by the hybrid protein. The other possible explanation is the lower stability of the hybrid protein (not confirmed by the Western blotting) or an essential interaction occurring between the S. coelicolor CTD and an unknown factor that is abolished in the hybrid protein. At this stage, we cannot suggest which explanation is more likely. The presence of the truncated TopAΔCTD had an even more severe influence on growth than the production of the hybrid protein TopACTDEc and TopA depletion. A similar reduction in growth was observed in an M. smegmatis strain overproducing the truncated TopA that lacked the lysine-rich amino acid stretches (23). The authors suggested that the wild-type protein was outcompeted by the truncated form, which was unable to perform the full catalytical cycle. The extremely slow growth of the S. coelicolor that produced the TopAΔCTD protein apart from the wild-type protein may presumably be similarly explained by the outcompeting of a possible interaction partner or regulator.
During the formation of Streptomyces sporogenic hyphae, multiple chromosomal copies undergo intensive replication (providing up to 50 genome copies in one compartment), followed by the synchronized separation of nucleoids. Having multiple copies of the genome that must be rapidly processed during sporulation creates an extra challenge for TopA processivity. The slower vegetative growth of S. coelicolor strains with impaired TopA processivity may be explained, at least partially, by the diminished distribution of the multiple chromosomes in the hyphae. We noted that a proper level of S. coelicolor TopA is required to maintain the constant distance between apical chromosomes and the tip of extending hyphal branches. This result is the first indication that the chromosome topology affects chromosome distribution during hyphal growth. At the sporulation stage, the multiple copies of chromosomes intensely replicate and need to be uniformly distributed throughout the hyphal cell. The diminished processivity of the S. coelicolor strain producing TopA881 compared to the wild-type protein, although it did not affect the vegetative growth, impaired sporogenic hyphae formation (Figure 6C). The lower number of chromosomal copies in the sporogenic hyphae indicates that the reduced processivity of the truncated TopA was not sufficient to remove the topological constraints during the simultaneous replication and/or segregation of multiple chromosomes. Even the elimination of the terminal ED, which led to a modest inhibition of TopA processivity, delayed sporulation (Figure 6B). Thus, the S. coelicolor TopA processivity that is required for the topological maintenance of multiple copies of GC-rich chromosomes, particularly during the development of sporogenic hyphae, is conferred by lysine repeats and may be additionally enhanced by the fastening of the stabilizing clamp caused by the interaction of the terminal acidic amino acids.
In summary, our findings elucidate the role of actinobacterial TopA CTDs in DNA relaxation. We suggest that the lysine repeats stabilize the TopA–DNA complex, and we hypothesize that this may be possibly due to formation of the clamp, closed by the interaction between terminal acidic amino acids and the NTD. The lysine repeat structure confers unusually high enzyme processivity, which in Streptomyces is particularly required for the replication and segregation of multiple chromosomes in sporogenic hyphae. However, severe modification of TopA activity also affects the chromosome distribution in vegetative hyphae. Thus, the lysine repeats represent a DNA-interaction interface specific to GC-rich bacteria.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Mr. Paweł Jaworski for help with the BIAcore measurements, to Dr. Jerzy Majka for the discussion on BIAcore analyses and to Dr. Daniel Krowarsch for the help with CD measurements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Foundation for Polish Science under the Parent Bridge program [POMOST/2010-2/5]; National Science Center of Poland [HARMONIA 2016/22/M/NZ1/00122 to M.S. and OPUS 2014/15/B/NZ2/01067 to D.J.]; AS’s visit to Institut Jacques Monod and the publications cost were financed by the Wroclaw Centre of Biotechnology Program of the Leading National Research Centre (KNOW) for the years 2014–2018. Funding for open access charge: Wroclaw Centre of Biotechnology Program of the Leading National Research Centre (KNOW) [2014/15/B/NZ2/01067].
Conflict of interest statement. None declared.
REFERENCES
- 1. Forterre P., Gadelle D.. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009; 37:679–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Perry K., Mondragón A.. Structure of a complex between E. coli DNA topoisomerase I and single-stranded DNA. Structure. 2003; 11:1349–1358. [DOI] [PubMed] [Google Scholar]
- 3. Lima C.D., Wang J.C., Mondragon A.. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994; 367:138–146. [DOI] [PubMed] [Google Scholar]
- 4. Viard T., de La Tour C.B.. Type IA topoisomerases: a simple puzzle?. Biochimie. 2007; 89:456–467. [DOI] [PubMed] [Google Scholar]
- 5. Zhang Z., Cheng B., Tse-Dinh Y.-C.. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:6939–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feinberg H., Changela A., Mondragón A.. Protein-nucleotide interactions in E. coli DNA topoisomerase I. Nat. Struct. Mol. Biol. 1999; 6:961–968. [DOI] [PubMed] [Google Scholar]
- 7. Bugreev D.V., Nevinsky G.A.. Structure and Mechanism of Action of Type IA DNA Topoisomerases. Biochemistry. 2009; 74:1467–1481. [DOI] [PubMed] [Google Scholar]
- 8. Tse-Dinh Y.-C., Beran-Steed R.K.. Escherichia coli DNA topoisomerase I is a zinc metalloprotein with three repetitive zinc-binding domains. J. Biol. Chem. 1988; 263:15857–15859. [PubMed] [Google Scholar]
- 9. Suerbauma S., Brauer-Steppkesa T., Labigneb A., Cameronc B., Drlica K.. Topoisomerase I of Helicobacter pylori: juxtaposition with a flagellin gene (flaB) and functional requirement of a fourth zinc finger motif. Gene. 1998; 210:151–161. [DOI] [PubMed] [Google Scholar]
- 10. de la Tour B.C., Kaltoum H., Portemer C., Confalonieri F., Huber R., Duguet M.. Cloning and sequencing of the gene coding for topoisomerase I from the extremely thermophilic eubacterium, Thermotoga maritima. Biochim. Biophys. Acta. 1995; 1264:279–283. [DOI] [PubMed] [Google Scholar]
- 11. Ahumada A., Tse-Dinh Y.-C.. The role of the Zn (II) binding domain in the mechanism of E. coli DNA topoisomerase I. BMC Biochem. 2002; 13:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tse-Dinh Y.-C. Zinc (II) coordination in Escherichia coli DNA topoisomerase I is required for cleavable complex formation with DNA. J. Biol. Chem. 1991; 266:14317–14320. [PubMed] [Google Scholar]
- 13. Zumstein L., Wang J.C.. Probing the structural domains and function in vivo of Escherichia coli DNA topoisomerase I by mutagenesis. J. Mol. Biol. 1986; 191:333–340. [DOI] [PubMed] [Google Scholar]
- 14. Terekhova K., Marko J.F., Mondragón A.. Studies of bacterial topoisomerases I and III at the single molecule level. Biochem. Soc. Trans. 2013; 41:571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Changela A., Digate R.J., Mondragon A.. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001; 411:1077–1081. [DOI] [PubMed] [Google Scholar]
- 16. Zhang H.L., Malpure S., Li Z., Hiasa H., DiGate R.J.. The role of the carboxyl-terminal amino acid residues in Escherichia coli DNA topoisomerase III-mediated catalysis. J. Biol. Chem. 1996; 271:9039–9045. [DOI] [PubMed] [Google Scholar]
- 17. Terekhova K., Gunn K.H., Marko J.F., Mondragón A.. Bacterial topoisomerase I and topoisomerase III relax supercoiled DNA via distinct pathways. Nucleic Acids Res. 2012; 40:10432–10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bhaduri T., Bagui T.K., Sikder D., Nagaraja V.. DNA Topoisomerase I from Mycobacterium smegmatis. J. Biol. Chem. 1998; 273:13925–13932. [DOI] [PubMed] [Google Scholar]
- 19. Szafran M.J., Strick T., Strzałka A., Zakrzewska-Czerwińska J., Jakimowicz D.. A highly processive topoisomerase. I: Studies at the single-molecule level. Nucleic Acids Res. 2014; 42:7935–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Narula G., Becker J., Cheng B., Dani N., Abrenica M.V., Tse-Dinh Y.-C.. The DNA relaxation activity and covalent complex accumulation of Mycobacterium tuberculosis topoisomerase I can be assayed in Escherichia coli: application for identification of potential FRET-dye labeling sites. BMC Biochem. 2010; 11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szafran M., Skut P., Ditkowski B., Ginda K., Chandra G., Zakrzewska-Czerwińska J., Jakimowicz D.. Topoisomerase I (TopA) is recruited to ParB complexes and is required for proper chromosome organization during Streptomyces coelicolor sporulation. J. Bacteriol. 2013; 195:4445–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhat A.G., Leelaram M.N., Hegde S.M., Nagaraja V.. Deciphering the distinct role for the metal coordination motif in the catalytic activity of Mycobacterium smegmatis topoisomerase I. J. Mol. Biol. 2009; 393:788–802. [DOI] [PubMed] [Google Scholar]
- 23. Ahmed W., Bhat A.G., Leelaram M.N., Menon S., Nagaraja V.. Carboxyl terminal domain basic amino acids of mycobacterial topoisomerase I bind DNA to promote strand passage. Nucleic Acids Res. 2013; 41:7462–7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Godbole A.A., Leelaram M.N., Bhat A.G., Jain P., Nagaraja V.. Characterization of DNA topoisomerase I from Mycobacterium tuberculosis: DNA cleavage and religation properties and inhibition of its activity. Arch. Biochem. Biophys. 2012; 528:197–203. [DOI] [PubMed] [Google Scholar]
- 25. Tan K., Cao N., Cheng B., Joachimiak A.. Insights from the structure of Mycobacterium tuberculosis topoisomerase I with a novel protein fold. J. Mol. Biol. 2015; doi:10.1016/j.jmb.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chopra S., Matsuyama K., Tran T., Malerich J.P., Wan B., Franzblau S.G., Lun S., Guo H., Maiga M.C., Bishai W.R. et al. Evaluation of gyrase B as a drug target in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2012; 67:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahmed W., Menon S., Karthik P.V., Nagaraja V.. Reduction in DNA topoisomerase I level affects growth, phenotype and nucleoid architecture of Mycobacterium smegmatis. Microbiology. 2015; 161:341–353. [DOI] [PubMed] [Google Scholar]
- 28. Jain P., Nagaraja V.. Indispensable, functionally complementing N and C-terminal domains constitute site-specific topoisomerase I. J. Mol. Biol. 2006; 357:1409–1421. [DOI] [PubMed] [Google Scholar]
- 29. Harrison J., Studholme D.J.. Genomics update Recently published Streptomyces genome sequences. Microb. Biotechnol. 2014; 7:373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bentley S.D., Chater K.F., Cerdeño-Tárraga A.M., Challis G., Thomson N., James K., Harris D., Quail M., Kieser H., Harper D., Bateman A. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2004; 417:141–147. [DOI] [PubMed] [Google Scholar]
- 31. Liu G., Chater K.F., Chandra G., Niu G., Tan H., Liu G., Chater K.F., Chandra G., Niu G.. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013; 77:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Flärdh K., Buttner M.J.. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 2009; 7:36–49. [DOI] [PubMed] [Google Scholar]
- 33. Chater K.F. Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2006; 361:761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCormick J.R., Flardh K.. Signals and regulators that govern Streptomyces development ¨. FEMS Microbiol. Lett. 2012; 36:206–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kois-Ostrowska A., Strzałka A., Lipietta N., Tilley E., Zakrzewska-Czerwińska J., Herron P.R., Jakimowicz D.. Unique function of the bacterial chromosome segregation machinery in apically growing Streptomyces—targeting the chromosome to new hyphal tubes and its anchorage at the tips. PLoS Genet. 2016; 12:e1006488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jakimowicz D., van Wezel G.P.. Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere?. Mol. Microbiol. 2012; 85:393–404. [DOI] [PubMed] [Google Scholar]
- 37. Jakimowicz D., Zydek P., Kois A., Zakrzewska-Czerwińska J., Chater K.F.. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 2007; 65:625–641. [DOI] [PubMed] [Google Scholar]
- 38. Swiercz J.P., Nanji T., Gloyd M., Guarné A., Elliot M.A.. A novel nucleoid-associated protein specific to the actinobacteria. Nucleic Acids Res. 2013; 41:4171–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. R Core Team R: A language and environment for statistical computing. 2016. [Google Scholar]
- 40. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 41. Russell D.W., Sambrook J.. Molecular cloning: a laboratory manual. 2001; NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 42. Kieser T., Bibb M.J., Buttner M.J., Chater K.F., Hopwood D.A.. Practical Streptomyces Genetics. John Innes Cent. Ltd. 2000; doi:10.4016/28481.01. [Google Scholar]
- 43. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu X., Leng F.. A rapid procedure to purify Escherichia coli DNA topoisomerase I. Protein Expr. Purif. 2011; 77:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ritz C., Baty F., Streibig J.C., Gerhard D.. Dose-response analysis using R. PLoS One. 2015; 10:e0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Szafran M.J., Gongerowska M., Gutkowski P., Zakrzewska-Czerwinska J.. The coordinated positive regulation of topoisomerase genes maintains topological homeostasis in Streptomyces coelicolor. J. Bacteriol. 2016; 198:3016–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang S., Li W., Liu S., Xu J.. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res. 2016; 44:W430–W435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Viard T., Cossard R., Duguet M., de la Tour C.B.. Thermotoga maritima-Escherichia coli chimeric topoisomerases. Answers about involvement of the carboxyl-terminal domain in DNA topoisomerase I-mediated catalysis. J. Biol. Chem. 2004; 279:30073–30080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.