Abstract
Background and Aims Subfamily Arundinoideae represents one of the last unsolved taxonomic mysteries in the grass family (Poaceae) due to the narrow and remote distributions of many of its 19 morphologically and ecologically heterogeneous genera. Resolving the phylogenetic relationships of these genera could have substantial implications for understanding character evolution in the grasses, for example the twisted geniculate awn – a hygroscopic awn that has been shown to be important in seed germination for some grass species. In this study, the phylogenetic positions of most arundinoid genera were determined using DNA from herbarium specimens, and their placement affects interpretation of this ecologically important trait.
Methods A phylogenetic analysis was conducted on a matrix of full-plastome sequences from 123 species in 107 genera representing all grass subfamilies, with 15 of the 19 genera in subfamily Arundinoideae. Parsimony and maximum likelihood mapping approaches were used to estimate ancestral states for presence of a geniculate lemma awn with a twisted column across Poaceae. Lastly, anatomical characters were examined for former arundinoid taxa using light microscopy and scanning electron microscopy.
Key Results Four genera traditionally included in Arundinoideae fell outside the subfamily in the plastome phylogeny, with the remaining 11 genera forming Arundinoideae sensu stricto. The twisted geniculate awn has originated independently at least five times in the PACMAD grasses, in the subfamilies Panicoideae, Danthonioideae/Chloridoideae and Arundinoideae. Morphological and anatomical characters support the new positions of the misplaced arundinoid genera in the phylogeny, but also highlight convergent and parallel evolution in the grasses.
Conclusions In placing the majority of arundinoid genera in a phylogenetic framework, our study answers one of the last remaining big questions in grass taxonomy while highlighting examples of convergent evolution in an ecologically important trait, the hygroscopic, twisted geniculate awn.
Keywords: Arundinoideae, awns, Crinipes group, Poaceae, plastome phylogenomics, PACMAD grasses
INTRODUCTION
The grass family, Poaceae, has a remarkably stable, phylogenetically based classification as recently summarized by Soreng et al. (2015) and Kellogg (2015). Of the 12 subfamilies, the limits of 11 are largely resolved. Only one subfamily, Arundinoideae, remains an apparent catch-all group, with a history of including heterogeneous and unrelated taxa (summarized in Table 1).
Table 1.
Recent classifications of Arundinoideae showing included genera
| Clayton and Renvoize (1986) | Grass Phylogeny Working Group (2001) | Soreng (2015) | Kellogg (2015) |
|---|---|---|---|
| Alloeochaete | – | Danthonioideae | Alloeochaete |
| Amphipogon | Amphipogon | Amphipogon | Amphipogon |
| Arundo | Arundo | Arundo | Arundo |
| Crinipes | Crinipes | Crinipes | Crinipes |
| Danthonidium | – | Danthonioideae | Danthonidium |
| Dichaetaria | Dichaetaria | Dichaetaria | Dichaetaria |
| Diplopogon | – | – | – |
| Dregeochloa | Dregeochloa | Dregeochloa | Dregeochloa |
| Elytrophorus | Elytrophorus | Elytrophorus | Elytrophorus |
| Chloridoideae | Chloridoideae | ‘Eragrostis’ walteri | ‘Eragrostis’ walteri |
| Hakonechloa | Hakonechloa | Hakonechloa | Hakonechloa |
| Leptagrostis | Leptagrostis | Leptagrostis | Leptagrostis |
| Molinia | Molinia | Molinia | Molinia |
| – | Moliniopsis | Moliniopsis | Moliniopsis |
| Monachather | – | Monachather | Monachather |
| Nematopoa | Nematopoa | Nematopoa | Nematopoa |
| Phaenanthoecium | – | Danthonioideae | Phaenanthoecium |
| Phragmites | Phragmites | Phragmites | Phragmites |
| Piptophyllum | Piptophyllum | Piptophyllum | Piptophyllum |
| Styppeiochloa | Steyppeiochloa | Styppeiochloa | Styppeiochloa |
| Zenkeria | Zenkeria | Zenkeria | Zenkeria |
| Anisopogon | Pooideae | Pooideae | Pooideae |
| Centropodia | Chloridoideae | Chloridoideae | Chloridoideae |
| Chaetobromus | – | Danthonioideae | Danthonioideae |
| Chionochloa | Danthonioideae | Danthonioideae | Danthonioideae |
| Cortaderia | Danthonioideae | Danthonioideae | Danthonioideae |
| Danthonia | Danthonioideae | Danthonioideae | Danthonioideae |
| Gynerium | Incertae sedis | Panicoideae | Panicoideae |
| Lamprothyrsus | Danthonioideae | Danthonioideae | Danthonioideae |
| Notochloe | Danthonioideae | Danthonioideae | Danthonioideae |
| Pentameris | Danthonioideae | Danthonioideae | Danthonioideae |
| Pentaschistis | Danthonioideae | Danthonioideae | Danthonioideae |
| Plinthanthesis | Danthonioideae | Danthonioideae | Danthonioideae |
| Poagrostis | – | Danthonioideae | Danthonioideae |
| Prionanthium | Danthonioideae | Danthonioideae | Danthonioideae |
| Pseudopentameris | Danthonioideae | Danthonioideae | Danthonioideae |
| Pyrrhanthera | – | Danthonioideae | Danthonioideae |
| Rytidosperma | Danthonioideae | Danthonioideae | Danthonioideae |
| Schismus | Danthonioideae | Danthonioideae | Danthonioideae |
| Spartochloa | – | Panicoideae | Panicoideae |
| Tribolium | Danthonioideae | Danthonioideae | Danthonioideae |
| Urochloa | – | Panicoideae | Panicoideae |
| Thysanolaena | Centothecoideae | Panicoideae | Panicoideae |
| Micraira | Micrairoideae | Micrairoideae | Micrairoideae |
| Aristida | Aristidoideae | Aristidoideae | Aristidoideae |
| Sartidia | Aristidoideae | Aristidoideae | Aristidoideae |
| Stipagrostis | Aristidoideae | Aristidoideae | Aristidoideae |
For other earlier classifications, see Barker et al. (1995, 1998).
Most authors were aware that Arundinoideae was an artificial group. Watson and Dallwitz (1992) called the subfamily ‘an unsatisfactory assemblage of convenience, which is not amenable to anything approaching a diagnostic description, and is probably polyphyletic’. Kellogg and Campbell (1987) were the first to identify the Arundinoideae as polyphyletic using an explicitly cladistic approach, and argued that the subfamily ‘should be interpreted as an assemblage of basal groups and evolutionary dead ends’.
Molecular phylogenetic studies confirmed polyphyly of Arundinoideae and removed many disparate elements, notably the subfamily Danthonioideae. These studies included those using sequences of the chloroplast genes rbcL (Barker et al., 1995), ndhF (Clark et al., 1995) and rpoC2 (Barker et al., 1998), the nuclear gene phytochrome B (Mathews et al., 2000) and combined molecular data (Grass Phylogeny Working Group, 2001; Grass Phylogeny Working Group II, 2012; Cotton et al., 2015; Duvall et al., 2017). A few studies (e.g. Linder et al., 1997; Grass Phylogeny Working Group, 2001) included morphological phylogenetics as well. A late addition to the shrinking Arundinoideae was made by Ingram et al. (2011), who added the misnamed ‘Eragrostis’ walteri on the basis of two chloroplast genes and internal transcibed sequence (ITS), solving the mystery of this C3 species in a C4 genus in the mostly C4 subfamily Chloridoideae.
Based on available phylogenetic data, Soreng et al. (2015) and Kellogg (2015) now recognize 17 or 19 genera in Arundinoideae, respectively (Table 1). Soreng et al. (2015) remove Alloeochaete and Phaenanthoecium in Danthonioideae on the basis of ‘well developed, flattened, coiled, geniculate awns diverging between relatively slender lateral lobes, typical of Danthonieae, but not found in Arundinoideae’. These genera were historically associated with the tribe Danthonieae (e.g. Watson and Dallwitz, 1992; Clayton and Renvoize, 1999), which became the basis of subfamily Danthonioideae (Grass Phylogeny Working Group, 2001). However, they are not mentioned in the revision of Danthonioideae by Linder et al. (2010). Soreng et al. (2015) divide their Arundinoideae into tribes, whereas Kellogg (2015) does not.
Following the circumscription of Kellogg (2015; Table 1), Arundinoideae sensu lato (s.l.) is the smallest subfamily in the PACMAD clade (the large clade that includes Panicoideae, Arundinoideae, Chloridoideae, Micrairoideae, Aristidoideae and Danthonioideae), comprising only 50 species in 19 genera. These species are morphologically and ecologically diverse, suggesting that the subfamily may still be polyphyletic. Arundinoideae s.l. thus represents a significant obstacle to inferring character evolution in the PACMAD clade because it contains a heterogeneous group of species of uncertain phylogenetic placement. Some species have characters similar to those found in other distantly related subfamilies, so that their misplacement within Arundinoideae could artificially increase estimates of how many times such characters have evolved independently.
One such trait whose evolutionary interpretation is potentially affected by phylogenetic placement of Arundinoideae s.l. is the presence of a flattened geniculate awn with a twisted basal column at or near the apex of the lemma, which characterizes many members of Danthonioideae (Conert, 1987). This structure is presumed to influence the microsite and orientation in which the grass diaspore is buried through hygroscopic uncoiling and recoiling pushing the diaspore across the ground (Peart, 1979; Garnier and Dajoz, 2001; Johnson and Baruch, 2014). Actively assisted burial by hygroscopic awns has been shown to affect germination rates in grasses in some habitats, suggesting that the trait may be under strong selection in certain lineages (Simpson, 1952; Peart 1979, 1981; Peart and Clifford, 1987).
A hygroscopic awn with a basal twisted column is part of the suite of characters defining the typical ‘danthonioid’ lemma in which the medial awn is flattened and arises from a sinus between two prominent apical lobes (De Wet, 1956; Kabuye and Renvoize, 1975). Humphreys et al. (2010) explored the evolution of awns and other lemma characters in Danthonioideae. Their analysis found that the presence of hygroscopic awns is probably ancestral in Danthonioideae, and only two genera in the subfamily, Tribolium and Schismus, have diversified with an awnless lemma. Losses of awns across the subfamily are typically accompanied by a suite of changes in lemma characters, suggesting that the hygroscopic awn is an important component of a burial syndrome under potentially significant selection pressures.
A twisted geniculate awn is found in several grass clades (Davidse, 1987). The presence of this kind of awn in the Aveneae caused early classifications to include Danthonia and its relatives in the former tribe; analyses of anatomy and cytology showed that the two groups are not closely related (Hubbard, 1948), and thus the twisted geniculate awn must have developed in parallel between them (De Wet, 1956). Similar awns also appear in the Panicoideae, although in most cases the base of the awn is not broadly flattened as it is in Danthonioideae. In addition, members of Panicoideae typically have distinctive two-flowered spikelets, so were not considered closely related to the Danthonieae (Grass Phylogeny Working Group, 2001). Arundinoideae s.l. has four genera with ‘danthonioid’ awns: Alloeochaete, Phaenanthoecium, Dregeochloa and Amphipogon. Only Dregeochloa and Amphipogon have been included in molecular phylogenies (Barker, 1997; Barker et al., 1998; Grass Phylogeny Working Group, 2001; Grass Phylogeny Working Group II, 2012). Placing all four of these genera is necessary to assess the scale of the convergence in this important trait across grasses, and to enable subsequent studies on its ecological and evolutionary significance to be put in a robust phylogenetic framework.
The main historic obstacle to working with the Arundinoideae, namely the distribution of its geographically scattered members in remote areas of the world, remains unchanged. Herbarium specimens are the only sources of morphological, anatomical and genetic information for many genera in the subfamily, and DNA extracted from these specimens is often highly degraded, making PCR amplification and Sanger sequencing difficult or impossible (i.e. Särkinen et al., 2012). In such cases, genome survey sequencing (GSS) can be a powerful tool (e.g. Jankowiak et al., 2005; Besnard et al., 2014). The small fragments used in this type of sequencing (≤500 bp) are potentially well suited to the degraded DNA found in herbarium specimens, and the enormous amount of sequence data generated means that rigorous quality control can be used to remove any contaminants or poor-quality fragments.
In this study, we resolve long-standing questions regarding the phylogenetic placement of 15 genera included in the Arundinoideae s.l. using a new phylogeny of Poaceae based on full chloroplast genomes. In addition to this problematic subfamily, we generate and include several plastomes from the subfamily Micrairoideae, which has been identified as sister to the Arundinoideae (Grass Phylogeny Working Group, 2001; Grass Phylogeny Working Group II, 2012; Cotton et al., 2015; Duvall et al., 2017) and has a taxonomic history almost as complicated (reviewed in Sànchez-Ken et al., 2007). The 32 newly generated sequences include five genera not part of any previous molecular phylogenetic analysis: Alloeochaete, Crinipes, Dichaetaria, Nematopoa and Phaenanthoecium. To test the polyphyly of Arundinoideae s.l., we also sample published plastomes from all other subfamilies in Poaceae. Using this phylogenetic framework, we then reconstruct the evolutionary history of the ‘danthonioid’ awn to see how placement of arundinoid taxa affects interpretations of this distinctive character. Finally, we survey leaf cross-sectional and epidermal anatomy for taxa that are misplaced in the Arundinoideae s.l. to assess compatibility with subfamilial limits based on these traits.
MATERIALS AND METHODS
Taxon sampling
Representatives of 15 of the 19 genera currently assigned to Arundinoideae were sampled, including multiple species within a genus wherever possible (Supplementary Data Table S1). To test possible polyphyly of the subfamily, we also included a broad sample of published plastomes from all other PACMAD subfamilies. We considered the possibility that some ‘arundinoid’ taxa might actually belong in other subfamilies. To test this, we included taxa previously identified as sister to the remaining taxa of each subfamily so we could be confident that placement was not an artefact of limited sampling. Published plastomes for 23 BOP (Bambusoideae, Oryzoideae, Pooideae) clade taxa as well as samples from the basal grade of Anomochlooideae, Pharoideae and Puelioideae were included to test congruence of this larger sample with previously published phylogenies of the family. Sequence data for Eragrostis tef (SRR1463402) were downloaded from the Sequence Read Archive (SRA) to provide another ‘Eragrostis’ species. All other plastomes were taken from GenBank, except for Danthoniopsis dinteri assembled from Washburn et al. (2015), and Chasmanthium laxum, which was assembled from whole-genome shotgun data (Kellogg lab, unpubl. data). In total, 131 full plastomes from 123 species representing all subfamilies in Poaceae were included in the phylogenetic analysis.
DNA isolation and sequencing
Plant material was obtained from either field-dried collections or herbarium specimens, and ground using a mortar and pestle with sterilized sand. Total DNA was extracted using the QIAGEN EasyDNA Plant Mini Kit, a modified cetyltrimethyl-ammonium bromide (CTAB) protocol (Cota-Sánchez et al., 2006), or a combination of the two in which QIAGEN columns were used to clean and isolate the extracted DNA. Sample DNA was sheared using a Covaris S220 sonicator with peak power of 175 and duty factor of 5·0 for 200 cycles for 30 s with a target size of 500 bp. Libraries were prepared using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England BioLabs, Inc.) according to the manufacturer’s instructions. Fragments were size selected to 400–500 bp, purified using Agencourt AMPure XP Beads (Beckman Coulter, Inc.) and sequenced using an Illumina 2 × 250 HiSeq 2500 paired-end Rapid Run at the University of Illinois at Urbana-Champaign Roy J. Carver Biotechnology Center.
Plastome assembly and phylogenetics
Raw paired-end reads were cleaned using Trimmomatic v. 0.32 (Bolger et al., 2014) for TruSeq3-PE adaptors with one mismatch, a palindrome clip threshold of 30 and a simple clip threshold of 10. After adaptor trimming, reads were quality trimmed in Trimmomatic using a sliding window of 10 bp with a minimum average phred score of 20, keeping reads with a minimum length of 40. Plastome-like reads were identified by mapping filtered reads to a set of existing grass chloroplast genomes with bowtie2 v. 2.2.6 under the ‘very-sensitive-local’ parameter set (Langmead and Salzberg, 2012). Mapped reads were assembled with SPAdes v. 3.1.0 using kmers of 55, 87 and 121 and the ‘only-assembler’ option (Bankevich et al., 2012). SPAdes contigs were then meta-assembled in afin (http://github.com/mrmckain/Fast-Plast/tree/master/afin) using the full trimmed read data set under the following parameters: a stop extension value of 0.1, an initial trim of 100 bp, a maximum extension of 100 bp per loop and 50 search loops. afin trims the ends of starting contigs, extends their lengths using matching trimmed reads, attempts to fuse all contigs if the threshold of 10 % mismatch is met for contig overlap and iterates these steps 50 times. Contigs generated by afin were manually assembled into complete plastomes in Sequencher version 5.3 (Gene Codes Corporation) by identifying inverted repeat (IR) boundaries through sequence similarity and, where necessary, searching trimmed reads to connect any remaining fragments through in silico genome walking. Contigs were scaffolded to Schizachyrium scoparium, and gaps in the final assembly were filled with Ns. Some variation was found between the IR regions in some samples, but read lengths were not long enough to phase single nucleotide polymorphisms (SNPs); therefore the inverted repeat B (IRB) region was duplicated and inverted to serve as IRA. A coverage analysis (https://github.com/mrmckain/Fast-Plast) of a 25 bp sliding window was used to check completed plastome assemblies for accuracy, with further modifications to the assemblies made when dips in coverage were identified. Plastome sequences were oriented to start near psbA of the large single copy (LSC) and end with IRA. Annotations and Circos graphs of finished plastomes were made in Verdant (McKain et al., 2017; verdant.iplantcollaborative.org). Full plastome sequences are deposited in both Verdant and GenBank (accession numbers MF035966-MF035997).
Finished plastomes were divided into the IRB, small single copy (SSC) and LSC regions; each region was aligned using MAFFT v. 7.029b with default parameters (Katoh, 2013) and the three alignments were concatenated into a single alignment and trimmed with Gblocks version 0.91b (Castresana, 2000). Three treatments of gaps were used to create edited alignments: (1) all sites with gaps excluded (no gaps): (2) all sites with gaps in less than half of the sampled taxa included (less than half gaps); and (3) all sites included (all gaps). GTR + I + gamma was identified as the best model of base pair substitution based on the Aikake Information Criterion (AIC) using jModelTest2 (Guindon and Gascuel, 2003; Darriba et al., 2012). All four alignments – untrimmed, no gaps, less than half gaps and all gaps – were analysed using maximum likelihood (ML) with RAxML v. 8.0.22 under the GTR + I + gamma model with 500 bootstrap replicates (Stamatakis, 2014). The three early-diverging grass subfamilies – Anomochlooideae, Pharoideae and Puelioideae – were used as outgroups. Alternative topologies were tested using the Shimodaira–Hasegawa (SH) test (Shimodaira and Hasegawa, 1999) in RAxML. The all gaps alignment with Anomochloa as an outgroup was selected for subsequent analyses as a compromise between selecting the least ambiguous alignment while including the maximum amount of data. This alignment was analysed using Bayesian inference in MrBayes v. 3.2.6 (Ronquist et al., 2012). For this analysis, two simultaneous independent MCMCMC runs were conducted with four chains – one cold and three heated – for 1 million generations using a GTR + I + gamma evolutionary model with five rate categories to approximate the continuous gamma distribution. Trees were visualized using FigTree v. 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Morphological character coding and ancestral state reconstruction
The presence or absence of a geniculate lemma awn with a twisted basal column was recorded from herbarium specimens for all genera in Arundinoideae s.l. and combined with data taken from the literature for other taxa (Watson and Dallwitz, 1992; Clayton and Renvoize, 1999; Clayton et al., 2006 onwards) (Supplementary Data Table S2). For the purposes of this study, we did not distinguish between awns in which the column was flattened (‘danthonioid’) and those in which the column was terete. Species with a geniculate awn lacking the twisted column, such as Monachather paradoxus, were coded as lacking this character (0). Other traits associated with the diaspore burial syndrome of Humphreys et al. (2010), such as the presence of a hairy callus and hairs on the lemma body, were not included due to the difficulty of assigning character states across the range of diversity in Poaceae. Additionally, many species in the current study are polymorphic for these characters, and some clades that appear monomorphic in our sample have alternative character states in other unsampled taxa (e.g. Danthonioideae and Amphipogon, both of which have taxa with and without geniculate awns). Thus, a comprehensive study of these important characters is beyond the scope of the current project and would probably be more appropriately addressed at a smaller phylogenetic scale. However, the distribution of a relatively unambiguous character, such as the geniculate and twisted lemma awn, across the phylogeny can help provide context for future evolutionary studies of burial syndromes and suggest taxa on which such studies would be most profitably conducted.
Ancestral state reconstruction was performed on the ML phylogeny with two exceptions: the three outgroup subfamilies were excluded to facilitate visualization of the results, and duplicate species were reduced to a single sample in the phylogeny for trait analysis to prevent bias in the ancestral state reconstruction. Character histories were analysed with parsimony, which ignores branch lengths in the phylogeny, using Mesquite version 3.2 (Maddison and Maddison, 2015) and with ML, which assumes a Brownian motion model, using the R package corHMM (Beaulieu et al., 2013). Two different models of trait evolution were used for likelihood analyses: (1) the equal rates model (ER), which assumes equal rates of change between all character states; and (2) the all rates different model (ARD), which assigns a different rate to each transition, including reversals (Paradis et al., 2004).
Divergence date estimation with BEAST
BEAST v. 1.8.3 (Drummond et al., 2012) was used with the ‘all gaps’ plastome alignment to test the effect of our increased sampling on divergence dates within the PACMAD clade. BEAUti v. 1.8.3 was used to set parameters. The ML tree from RAxML was used as a starting tree after being transformed using the chronopl function in the R package ape with a lambda of 1 and four fossil ages (see below under BEAST priors) used as minimum age constraints (Paradis et al., 2004). Five separate identical runs of 100 million generations each were run on the CIPRES Gateway (Miller et al., 2010), sampling trees every 1000 generations using an uncorrelated relaxed clock model with a lognormal relaxed distribution and with a Yule process model of speciation as a tree prior. A GTR + I + Gamma nucleotide substitution model with four gamma categories was used, with base pair frequencies estimated from the alignment. Four fossil calibrations were specified as stem calibrations with lognormal distributions, with a mean of zero, s.d. of one, an offset from zero equal to the estimated age of the fossil and an initial value of the fossil age. These fossils were assigned positions in the phylogeny according to Vicentini et al. (2008) as follows: 7 million years ago (Mya) for the most recent common ancestor of Setaria and Panicum (Elias, 1942); 19 Mya for stem Chloridoideae (Strömberg, 2005); 35 Mya for the ancestor of BOP + PACMAD (Strömberg, 2005); and 55 Mya for all grass subfamilies excluding Anomochlooideae (Crepet and Feldman, 1991). Each clade involved in the fossil calibrations was also constrained to be monophyletic in the dating analysis to reduce computational effort slightly. LogCombiner v. 1.8.3 was used to combine the last 1000 trees from each of the ten BEAST runs, yielding effective sample sizes (ESSs) >200 for all parameters, and the concatenated tree file was annotated in TreeAnnotator v. 1.8.3.
Alignments, trees, and BEAST control and output files are available in Dryad (http://dx.doi.org/10.5061/dryad.v7m05).
Leaf anatomy
Cross-sections were made from near the middle of mature leaves taken from herbarium specimens, rehydrated in Pohl’s Wetting Solution (Pohl, 1965), fixed in FAA, dehydrated in an ethanol series, infiltrated with paraffin using tert-butanol as an intermediate solvent and embedded in paraffin according to the method in Ruzin (1999). Sections of 10 μm were made using a Microm HM 355 S rotary microtome (Microm International GmbH, Walldorf, Germany) and were stained with the Safranin–Fast Green protocol given in Sass (1951). Sections were photographed using an Olympus BX53 light microscope with a DP25 digital camera attachment (Olympus Corporation, Center Valley, PA, USA).
Leaf fragments were also taken from herbarium specimens to examine epidermal anatomy. These leaf pieces were first rehydrated and dehydrated as above, then transitioned into 100 % xylene solution and sonicated for 10 min each to remove epicuticular wax. The dewaxed fragments were then moved back into 100 % ethanol, dried in a Tousimis Samdri-780a Critical Point Dryer, coated with gold–palladium in a Tousimis Samsputter-2a Sputter Coater (Tousimis, Rockville, MD, USA) and viewed with a Hitachi S-2600H (Hitachi High Technologies America, Inc., Dallas, TX, USA) scanning electron microscope. Critical point drying, sputter coating and scanning electron microscopy (SEM) work was performed at The Research Center for Auditory and Vestibular Studies at Washington University in St. Louis.
Leaf cross-sections and epidermal SEM images were generated for the four misplaced ‘arundinoids’. A full morphological and anatomical analysis of Arundinoideae sensu stricto (s.s.) is in preparation.
RESULTS
Plastome assembly, annotation and alignment
Average coverage for each of the 32 plastomes generated by this study ranged from 31× to 452×, with total lengths of 133 327–141 203 bp (Table S1). Lengths of the LSC, SSC and IR ranged from 79 379 to 82 836 bp, 12 246 to 12 759 bp and 20 048 to 22 762 bp, respectively. Annotations of the plastomes showed conservation of genes in all newly assembled plastomes, with two exceptions. The LSC copy of rpl23 is pseudogenized in sampled Crinipes species, Elytrophorus globularis, ‘Eragrostis’ walteri and both accessions of Monochather paradoxus. The IRA copy of rps19 is pseudogenized in Coelachne africana due to a shift of the IR boundary. Although Hakonechloa macra (GenBank KJ920232.1) was reported to have lost the rpl16 gene and pseudogenized rpl14 (Cotton et al., 2015), the two individuals sampled in this study both have functional versions of these genes.
The alignments range from 77 882 bp when all gaps are excluded to 172 824 bp without trimming, demonstrating the considerable extent of gaps in the full alignment. Part of the reason for this is inclusion of Anomochloa, which differs from the characteristic grass plastome structure in several aspects, such as the absence of an rpoC1 intron and a 39 bp sub-repeat in the rpoC2 insert instead of the 21 bp sub-repeat found in the rest of the grasses (Morris and Duvall, 2010). Use of Pharus as an outgroup reduces the number of ambiguous regions in the alignment because Pharus has the dominant grass plastome architecture, but does not affect inferred phylogenetic relationships.
Phylogenetic analysis
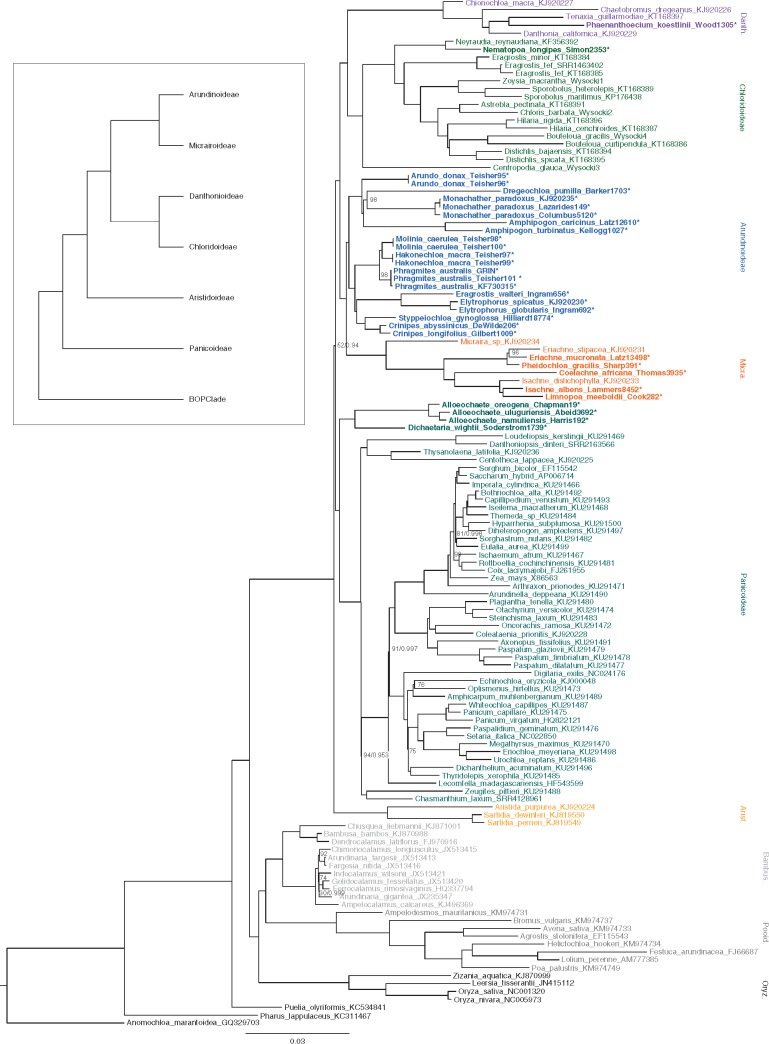
The ML tree in Fig. 2 was the result of analysis of the most inclusive Gblocks-edited alignment (made with the all gaps option) using Anomochloa marantoidea as an outgroup. The tree topology was robust to outgroup sampling and alignment trimming, except that the placement of Aristidoideae changed between two different positions. Bootstrap support for the placement of this subfamily ranged from 51 to 81 %, with alignments with fewer gaps favouring Aristidoideae as sister to the rest of PACMAD and those with more gaps favouring Panicoideae in this position (Table 2). Despite higher bootstrap values in phylogenetic estimates of the alignments with more gaps, the two topologies were not significantly different according to S–H tests (Shimodaira and Hasegawa, 1999). The Bayesian analysis of the all-gaps alignment yielded an identical topology to the ML tree, with a posterior probability of 0·94 for the placement of Aristidoideae sister to the rest of PACMAD.
Fig. 2.
Maximum likelihood phylogeny of 131 full plastomes across the grass family, with Anomochloa used as an outgroup. Bootstrap values <100 are shown above nodes, followed by posterior probabilities <1·0 from Bayesian analysis. Subfamilies in PACMAD are grouped by colour, while those in BOP are shades of grey. Samples in bold with asterisks were generated for the current study. The insert shows alternative topology for PACMAD relationships in which Panicoideae is sister to the rest of the clade.
Table 2.
Alternative phylogenetic analyses and their effects on the placement of Aristidoideae (A*) within PACMAD as well as the bootstrap support for that placement
| Alignment | Topology supported | Bootstrap |
|---|---|---|
| Untrimmed Anomochloa Out | A*(P(CMAD)) | 51 |
| AllGaps Anomochloa Out | A*(P(CMAD)) | 52 |
| HalfGaps Anomochloa Out | P(A*(CMAD)) | 57 |
| NoGaps Anomochloa Out | P(A*(CMAD)) | 81 |
Monophyly of Arundinoideae s.l. was not supported in any tree, with four genera consistently falling into other subfamilies in all analyses. The Zimbabwean monospecific genus Nematopoa groups with members of Chloridoideae, the Ethiopian monospecific genus Phaenanthoecium groups with Danthonioideae, and Dichaetaria and Alloeochaete form a clade sister to Panicoideae. These placements have 100 % bootstrap support and posterior probabilities of 1·0 under all variations of tree estimation, as does monophyly of the remaining Arundinoideae. The latter clade, referred to hereafter as Arundinoideae s.s., includes the cosmopolitan reeds Arundo and Phragmites; the temperate genera Molinia and Hakonechloa; the African Crinipes, Styppeiochloa, Dregeochloa plus the misnamed ‘Eragrostis’ walteri; the Australian genera Amphipogon and Monachather; and the African–Australian–Asian genus Elytrophorus. Relationships among genera in this subfamily are strongly supported, with all nodes receiving >95 % bootstrap support.
Molecular dating analysis
BEAST recovered a maximum clade credibility tree (Supplementary Data Fig. S1) with identical topology and similar support values to the ML tree in Fig. 2. Aristidoideae is sister to the rest of PACMAD with a posterior probability of 0·91. Despite the large number of generations in the combined analysis, ESS values for many parameters were well below the recommended 200. For example, the common ancestor of Arundinoideae and Micrairoideae is estimated to have lived between approx. 10 and 26 Mya based on the 95 % highest probability density (HPD). Similarly, the BOP and PACMAD clades are estimated to share a common ancestor that lived between approx. 28 and 47 Mya.
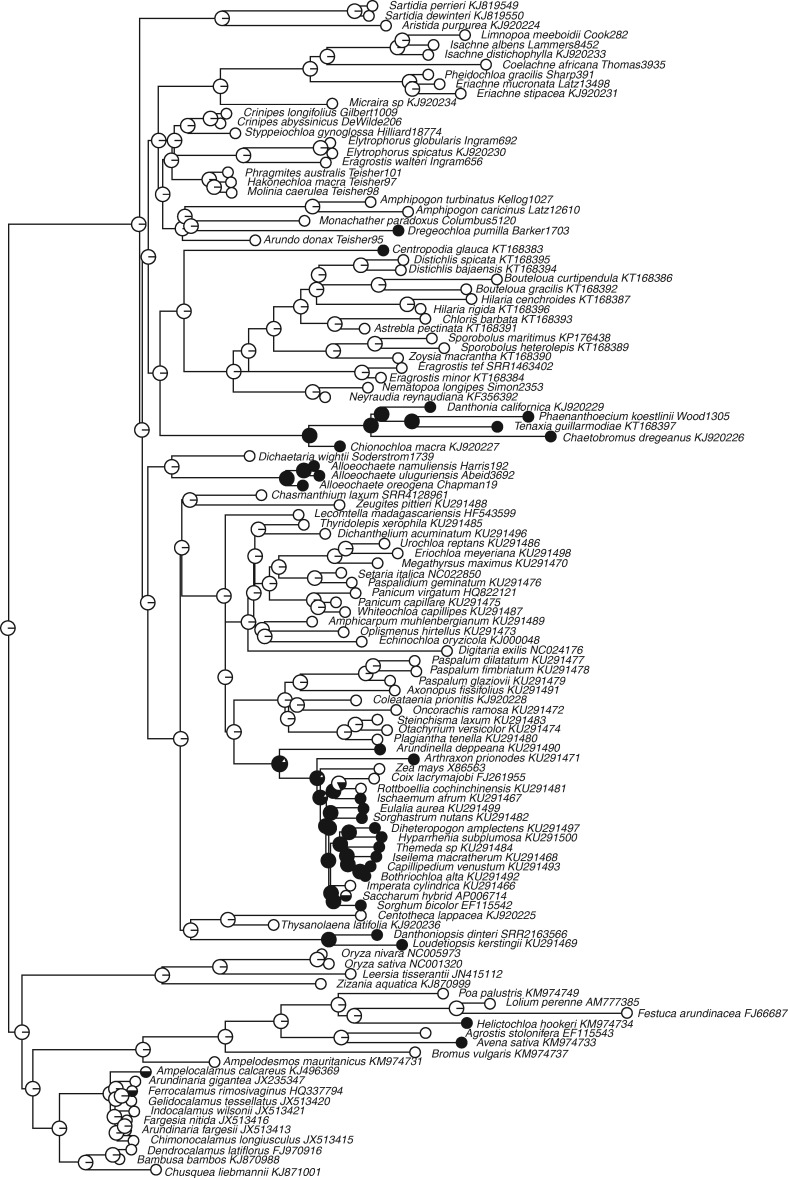
Phylogenetic distribution of the twisted geniculate lemma awn
A geniculate lemma awn with a twisted basal column is estimated to have originated at least five times independently across the PACMAD grasses according to both ML (Fig. 3) and parsimony analyses (Supplementary Data Fig. S2), with at least two separate origins in the BOP clade. The best fit model of evolution under the ML ‘all rates different’ scenario estimates gains of twisted geniculate awns to be almost three times as frequent as losses (AIC = 80·90), but this fit is not a significant improvement over the simpler ‘equal rates’ model (AIC = 80·62). With the current taxonomic sampling, the presence of twisted geniculate awns is estimated to be ancestral in the Danthonioideae as well as the Andropogoneae + Arundinelleae in subfamily Panicoideae. All members of Danthonioideae in our sample have a flattened geniculate awn with a twisted column, though many members of that subfamily have lost the trait (Humphreys et al., 2010). Based on our sampling in the Panicoideae, geniculate awns with twisted columns have evolved at least three times independently in the subfamily, with members of the Andropogoneae experiencing at least three separate subsequent losses of awns.
Fig. 3.
Maximum likelihood ancestral states for a geniculate awn with a twisted basal column across the BOP and PACMAD clades under the equal rates model of trait evolution using the R package corHMM. Filled circles, presence of the trait; open circles, absence of the trait.
Leaf anatomy
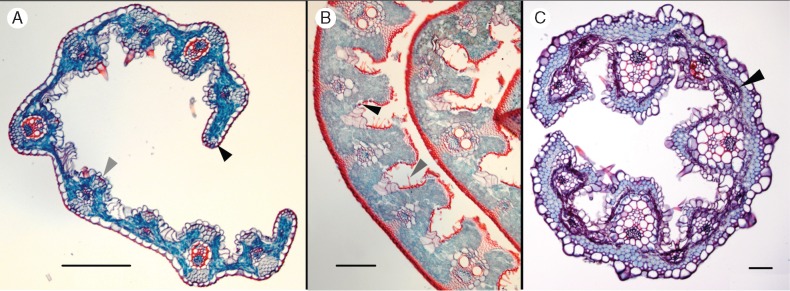
Leaf cross-sections were generated for three of the four genera falling outside Arundinoideae s.s. Dichaetaria specimens did not yield satisfactory sections. Both Phaenanthoecium and Alloeochaete exhibit C3 leaf anatomy, with the veins separated by more than two mesophyll cells (Fig. 4A, B). In contrast, Nematopoa longipes has Kranz anatomy (Fig. 4C), consistent with its placement in the largely C4 Chloridoideae in the plastome phylogeny. Nematopoa also has an unusual distribution of sclerenchyma, which extends the entire width of the leaf just inside the abaxial epidermis and forms a layer 2–4 cells thick. Leaves of all three genera are curled adaxially and exhibit prominent ribs on the adaxial side. Phaenanthoecium and Alloeochaete have large bulliform cells, and Alloeochaete and Nematopoa exhibit macrohairs.
Fig. 4.
Cross-sections of species from three of the four genera misplaced in Arundinoideae s.l. Black scale bars are approx. 500 μm. (A) Phaenanthoecium koestlinii (Wood 1305) showing schlerenchyma around the vascular bundles (grey arrow) and forming a cap at the leaf margins (black arrow). (B) Alloeochaete andongensis (Gossweiler 11810) has well-defined ribs around vascular bundles that form deep pits containing stomata (black arrow) and macrohair-like prickles (grey arrow, also in Fig. 5B). (C) Nematopoa longipes (Simon 2353) appears to be C4 with small tertiary vascular bundles (black arrow) limiting the distance between bundle sheath cells.
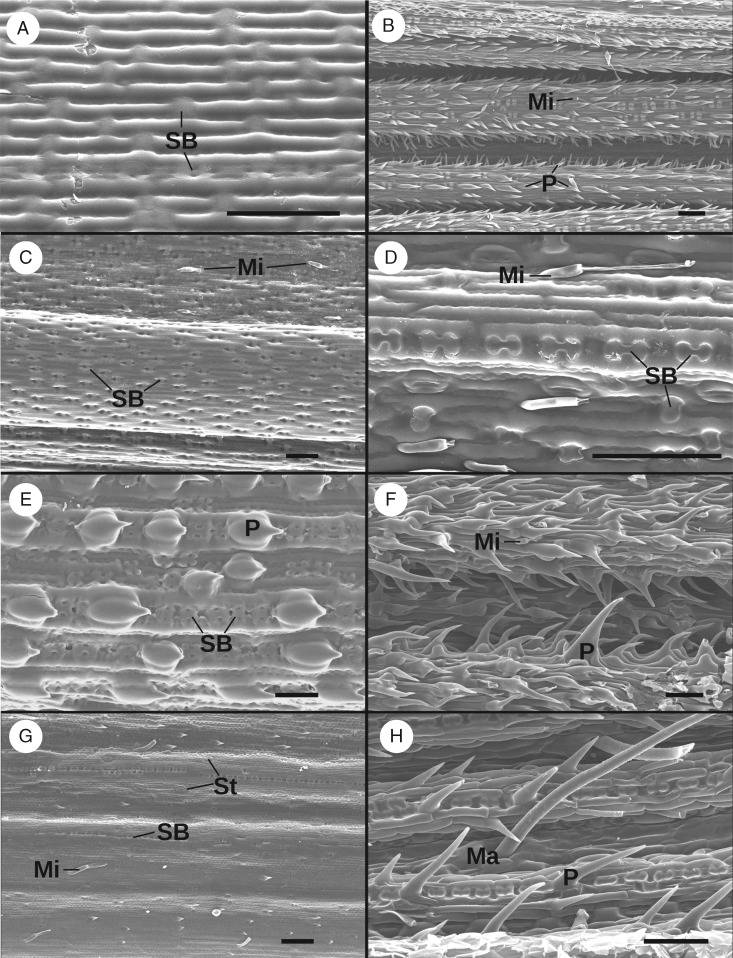
Adaxial and abaxial epidermal SEM images for representatives of all four misplaced genera are shown in Fig. 5. All sections are oriented with the long axis of the leaf horizontal and the apex to the left. Some features used in previous grass classifications, such as microhairs, macrohairs, prickles and silica bodies, are labelled and discussed below.
Fig. 5.
Epidermal SEM images from the abaxial (A, C, E, G) and adaxial (B, D, F, H) sides of the middle sections of mature leaves from herbarium specimens. (A, B) Alloeochaete andongensis (Gossweiler 11810), (C, D) Dichaetaria wightii (Soderstrom 1739), (E, F) Nematopoa longipes (Simon 2353) and (G, H) Phaenanthoecium koestlinii (Wood 1305). Selected features are marked as follows: Mi, microhair; Ma, macrohair; P, prickle; SB, silica body; St, stomate. Black and white scale bars are approx. 50 μm.
DISCUSSION
Our analyses show that Arundinoideae s.l. is polyphyletic. This finding has significant consequences for evolutionary inferences of morphology and anatomy across the PACMAD clade. In particular, the geniculate awn with a twisted column appears to be more homoplasious than previously thought, with two additional independent origins of the trait inferred by parsimony and ML reconstruction on the chloroplast tree. The new placements of the four pseudo-arundinoid genera are surprising in the case of Alloeochaete and Dichaetaria, anatomically sensible in the case of Nematopoa, and morphologically corroborated in the case of Phaenanthoecium.
The twisted geniculate awn
Arundinoideae s.s. contains two taxa, Dregeochloa and Amphipogon, with a geniculate awn with a twisted basal column. Within Amphipogon, only A. setaceus has such an awn; the species is not sampled here but morphologically otherwise is a ‘good’ Amphipogon. The two taxa fall in a clade with Monachather, which has spikelets that are similar to those of Dregeochloa and awns that are geniculate, but not twisted into a column. Dregeochloa was segregated from Danthonia (Conert, 1966) and was previously considered to be a member of Danthonieae on the basis of its lemmas (e.g. Renvoize, 1981; Clayton and Renvoize, 1986; Watson and Dallwitz, 1992), though Barker et al. (1998) found some moderate support for its placement in the Arundineae based on rpoC2 sequence data. Monachather was similarly thought to be danthonioid until the analysis of rbcL sequence data by Linder et al. (1997) placed it with Arundo.
Like Dregeochloa, Phaenanthoecium was removed from Danthonia (Hubbard et al., 1936), but retained in the tribe Danthonieae by most authors (e.g. Kabuye and Renvoize, 1975; Watson and Dallwitz, 1992). In this case, morphology of the spikelet is concordant with the phylogenetic history of the plastome. Phaenanthoecium is placed sister to Tenaxia guillarmodiae in our phylogeny, but our sample of Danthonioideae is too sparse to assess the implications of this result for the classification of this subfamily.
Alloeochaete was also based on a former Danthonia, Danthonia andongensis Rendle (Hubbard, 1940). Kabuye and Renvoize (1975) considered whether the genus belonged in tribe Danthonieae, concluding that despite being unique among the danthonioid genera in several morphological attributes (i.e. the male lowest floret, five-nerved lemmas and florets longer than glumes), ‘the similarity in lemma morphology clearly demonstrates that Alloeochaete is closely related to Danthonia sensu lato’. The placement of this genus as sister to the Panicoideae (Fig. 2) provides another example of parallel development in lemma characters in grasses (Fig. 3).
In this study, we have not incorporated development in coding character states for the awn, largely because little is known about awn development and genetics (Kellogg, 2015). The twisted geniculate awns in various subfamilies may arise from different developmental pathways using different gene networks. This seems likely to be the case in the Aveneae and Danthonieae given that awns arise from the back of the lemma in the former and from a sinus at the apex of the lemma in the latter (De Wet, 1956). Dregeochloa and Monachather are closely related according to the chloroplast tree, and both have geniculate awns, though only the former has a twisted column. The developmental changes involved in producing this difference are unknown, as is the ecological significance of a coil as opposed to a hygroscopic kink.
The multiple independent origins of the twisted geniculate awn suggest that it is not a particularly difficult structure to evolve. Furthermore, Humphreys et al. (2010) showed that lineages in Danthonioideae lose these awns less frequently than might be expected by chance, and in only two cases of loss did the awnless state persist through subsequent diversification, suggesting selection for the awned lemma in most of the subfamily. A similarly interesting evolutionary situation may be present in the Andropogoneae (Panicoideae), which appear to be characterized by multiple losses of the twisted geniculate awn. Expanded sampling in this clade would potentially provide a valuable parallel to the study by Humphreys et al. (2010) to understand the evolutionary role of hygroscopic awns in grass adaptation and diversification.
A pointed, hairy callus and tufts of hairs frequently in rows across the back of the lemma together with the twisted geniculate awn form the ‘active burial syndrome’ characterizing most Danthonoideae (Humphreys et al., 2010). These traits also appear together in non-danthonioids such as Alloeochaete and Dregeochloa, suggesting either strong selection for all three operating in unison in distantly related taxa or a shared developmental pathway, or both. Ecological experiments on germination, such as those done by Peart (1979, 1981, 1984) and Peart and Clifford (1987) in Australian grasses, and developmental genetic investigations into the genetic architecture underlying lemma formation will be necessary to understand fully the evolution of grass awns. Further study is needed to analyse awn states, as described here, with other ‘active burial syndrome’ lemma features.
Anatomy of arundinoid imposters
Nematopoa longipes belongs in the Chloridoideae based on plastome sequence data and leaf anatomy. The monotypic genus was separated from the chloridoid Triraphis by Hubbard (1957a), but the similarity between the two seems likely to be due to shared ancestry. Leaf cross-sectional anatomy (Fig. 4C) shows Nematopoa to be C4, with close vein spacing and double bundle sheaths with chlorenchyma arranged radially in the outer sheath, supporting placement of the genus in the Chloridoideae. Curiously, both Renvoize (1981) and Linder et al. (1997; Fig. 5C) concluded that this genus is C3 on the basis of leaf anatomy. The specimen used in the current study was identified and examined by Linder et al. for their morphological analysis, so we do not think misidentification is the cause of the discrepancy. More probably, the difference in interpretation may be because the tertiary vascular bundles shown in Fig. 4C (black arrow) are quite small. Depending on the stain used and the orientation and quality of the section, they could easily be missed, causing the leaf to appear to have widely spaced vascular bundles consistent with the C3 pathway. In addition, the extensive sclerenchyma just inside the abaxial epidermis is in the position occupied by chlorenchyma in many C3 species, so could easily be misinterpreted. Sclerenchyma is more commonly associated with veins, as observed here in Phaenanthoecium and Alloeochaete, and the distribution in Nematopoa is unusual.
Our specimen of Nematopoa also has short and relatively squat microhairs with the apical cell approximately the same length as the basal cell like those found in Chloridoideae (Fig. 5F), though the apical cell does not appear obviously spherical as in the rest of this subfamily (e.g. Palmer et al., 1985). The longitudinally oriented dumb-bell and cruciform silica bodies found on the adaxial (not shown) and abaxial leaf surface in this specimen are similarly consistent with structures found in Chloridoideae; however, these characters are also found among other subfamilies (Metcalfe, 1960; Renvoize, 1981, Reimer and Cota-Sànchez, 2007).
Phaenanthoecium koestlinii is the only member of its genus (Watson and Dallwitz, 1992), and has a lemma similar to those found in Danthonioideae. Anatomical justification for the placement of Phaenanthoecium in this subfamily is difficult, since there are no obvious leaf anatomical synapomorphies for Danthonioideae or any substantial sub-set thereof. However, the anatomical features found in Phaenanthoecium can at least be shown to be compatible with those found across the subfamily. In cross-section, Phaenanthoecium has non-Kranz anatomy, with midveins indistinguishable from other primary vascular bundles, evenly distributed chlorenchyma cells, sclerenchyma bands on the abaxial and adaxial sides of the vascular bundles and small sclerenchyma caps along the margins of the leaf. Similar traits are found in the danthonioid genus Chaetobromus (C. involucratus; Ellis, 1988b) and some species of Pentameris (P. thuarii and P. dregeana; Ellis, 1985a, 1986). In addition, the leaf margins are distinctly rounded rather than reaching a point, a character that is similar to that seen in Pentaschistis dentata and P. ecklonii (formerly placed in Prionanthium; Ellis, 1989). The microhairs in Fig. 5G and macrohair and macrohair-like prickles in Fig. 5H resemble those found in species of Danthonia (Reimer and Cota-Sànchez, 2007) and Urochlaena (Ellis, 1988a), though similar traits can be found in other subfamilies (e.g. Palmer and Tucker, 1981, 1983; Palmer et al., 1985; Palmer and Gerbeth-Jones, 1986, 1988). Verboom et al. (1994) found that haustorial synergids in the embryo sac may be a synapomorphy for the Danthonioideae, but this trait requires ample living material to assess and has thus been sampled for a relatively small sub-set of the subfamily.
Dichaetaria and Alloeochaete form a clade sister to the Panicoideae, despite morphological similarity to members of Arundinoideae and Danthonioideae. The other early-diverging members of Panicoideae are highly heterogeneous, explaining in part why this affiliation was not previously identified. Anatomically, the samples from these two genera in our analysis share a few epidermal features. Both have microhairs on the costal zones that are narrow and have apical cells twice as long as the basal cells (Fig. 5B, D) and a mix of dumb-bell- and saddle-shaped silica bodies (Fig. 5A, D). In cross-section, Alloeochaete andongensis (Fig. 4B) strongly resembles A. gracillima and A. uluguruensis (Linder et al., 1997). Alloeochaete has broad adaxial ribs that are nearly square on the adaxial side. The major veins have scelerenchyma girders on both sides, whereas minor veins lack adaxial girders. The overall shape of the ribs is reminiscent of that in the danthonioid Pseudopentameris brachyphylla, although P. brachyphylla has adaxial and abaxial sclerenchyma girders associated with all veins (Ellis, 1985b). Anatomical evidence is thus apparently at odds with the phylogenetic placement of Alloeochaete.
Arundinoideae sensu stricto
Resolution of relationships within Arundinoideae s.s. in our study is significantly improved over phylogenies based on individual chloroplast genes (e.g. Grass Phylogeny Working Group II, 2012) and those based on full plastome sequences but with only a few arundinoid genera (e.g. Cotton et al., 2015; Duvall et al., 2017). The genera of Arundinoideae s.s. still form a morphologically and ecologically heterogeneous assemblage. The genera form two clades, one with glumes shorter than the spikelet and the other with glumes as long as or longer than the spikelet. The ‘short glumes’ group consists of Phragmites, Hakonechloa and Molinia, as well as another clade of mostly African genera. ‘Eragrostis’ walteri, formerly thought to be a unique example of reversion from C4 to C3 photosynthesis (Ingram et al., 2011), falls in this clade and is sufficiently distinct from its sister taxon Elytrophorus that it should be assigned its own genus. The other two taxa in the ‘short glumes’ clade, Styppeiochloa and Crinipes, are sister taxa, as suggested by their taxonomic history (the type species of Styppeiochloa was segregated from Crinipes) and supported by their similar preference for seasonally wet, rocky habitats, their one-nerved glumes and their often two-flowered spikelets. The ‘long glumes’ clade of Arundinoideae contains, in addition to Arundo, the Australian genera Amphipogon and Monachather, and the South African genus Dregeochloa.
Four genera possibly belonging in the Arundinoideae are not included in this study. The monotypic African genera Leptagrostis and Piptophyllum have insufficient collected material to conduct destructive sampling. Herbarium samples of the Indian genera Danthonidium and Zenkeria yielded DNA that was too degraded to be sequenced. None of these taxa has unambiguous synapomorphies to support their placement in the current phylogeny. Linder et al. (1997) placed Leptagrostis, Piptophyllum and Zenkeria in the ‘crinipoid’ group, whose other members are placed in the plastome tree in the Arundinoideae, Chloridoideae and at the base of the Panicoideae (see above). Like Nematopoa, Piptophyllum was considered intermediate between Triraphis and Crinipes (Hubbard, 1957b), but leaf anatomical data for this monotypic genus are lacking. Interestingly, and unlike Nematopoa, Triraphis or Crinipes, the lemma awn in Piptophyllum is slightly twisted at the base, suggesting a possible additional independent origin of this trait. The spikelets of Danthonidium have lemmas similar to Dregeochloa and Monachather, but also to several taxa in subfamily Danthonioideae and Panicoideae. Soreng et al. (2015) treat Danthonidium as incertae sedis in the Danthonioideae along with Alloeochaete and Phaenanthoecium, which are recovered in very different clades in our analysis.
Micrairoideae
Our analysis places Limnopoa within the large genus Isachne, and Duvall et al. (2017) found that Hubbardia is sister to Limnopoa, suggesting that generic limits in these taxa need to be revised. Isachne is reported to have almost 100 species (Kellogg, 2015), but the most comprehensive revision of its members included only the 23 species from Malesia (Iskandar and Veldkamp, 2003). Limnopoa meeboldii is the sole species of Limnopoa, and Hubbardia heptaneuron is the only species of Hubbardia, so transferring these names to Isachne would not be difficult.
Two monotypic genera in Micrairoideae, Heteranthoecia and Sphaerocaryum, have not been sampled for molecular sequence data, and only one species of Coelachne is included in our phylogeny, so relationships within the tribes Hubbardieae and Isachneae sensuSoreng et al. (2015) are difficult to assess. Given the size, morphological heterogeneity and geographic distribution of Isachne, especially if Hubbardia and Limnopoa are included, it may be best to assign these genera to a single tribe, Isachneae, pending further phylogenetic sampling. Alternatively, Kellogg (2015) suggests that segregation of genera into tribes in this relatively small subfamily is unnecessary.
Phylogenetic position of Aristidoideae
Many previous phylogenies have placed Aristidoideae sister to the remainder of the PACMAD clade (e.g. Clark et al., 1995; Grass Phylogeny Working Group II, 2012). In contrast, some more recent studies using whole plastomes have put Panicoideae in that position, with Aristidoideae sister to the CMAD group (Cotton et al. 2015; Burke et al., 2016; Fig. 2, inset), although data from these studies could not reject the possibility of Aristidoideae being sister to a clade comprising the remainder of PACMAD. Here we find that placement of Aristidoideae is sensitive to inclusion of gaps in the alignment. This sensitivity means that either regions with high insertion/deletion rates contain phylogenetic signal and thus the more gap-inclusive alignments more closely approximate the ‘true’ plastome phylogenetic history, or these gap-filled regions introduce more phylogenetic noise into the data. Given the large number of taxa sampled and the fact that full chloroplast genomes were used in the analysis, it seems unlikely that the question of the correct placement of Aristidoideae will be resolved using plastome sequences.
Divergence date estimates
Ages inferred in the current analysis are younger than previously published estimates. Vicentini et al. (2008) reported age estimates for the ancestor of BOP and PACMAD ranging from 48 to 85 Mya, while Christin et al. (2014) reported ages for the same divergence of 20–62 Mya from plastid and 51–63 Mya from nuclear sequence data across four different dating analyses. Our analysis yielded a median age of approx. 33 Mya for this clade, with a 95 % HPD range of approx. 28–47 Mya, falling within the range estimated by Christin et al. (2014). Accurate absolute dating for events in grasses may not be possible given the family’s sparse fossil record and heterogeneous molecular evolutionary rates.
Conclusion
This study represents the largest plastome phylogeny of the grass family to date as well as the most complete sampling of genera in the taxonomically difficult subfamily Arundinoideae. Resolving the polyphyly of this poorly studied group has substantial implications for ancestral trait estimations across the PACMAD clade as shown by our new understanding of the phylogenetic distribution of the ‘danthonioid’ awn. Four genera are removed from the Arundinoideae s.s. on the basis of the plastome phylogeny: Alloeochaete and Dichaetaria are sister to Panicoideae; Phaenanthoecium is placed in Danthonioideae; and Nematopoa appears to belong in Chloridoideae. These new placements have some support from morphological and anatomical traits, but are equally representative of the notorious tendency for parallel evolution in the Poaceae. Herbarium specimens were vital for resolving these long-standing issues in grass classification and will continue to be an essential resource for phylogenetics of taxa for which field collections are not practical.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: concatenated tree file from 1 million trees taken from BEAST analysis. Figure S2: parsimony ancestral state reconstruction of twisted geniculate awns on the phylogeny from Fig. 2 using Mesquite. Table S1: plastomes included in the phylogenetic analysis, with assembly statistics for newly generated sequences. Table S2: character states for presence/absence of a twisted geniculate lemma awn in species sampled in the plastome phylogeny.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Maria Vorontsova at Kew for assistance in sampling specimens, Gerrit Davidse for help with character coding, Jim Solomon for co-ordinating herbarium loans, and Paul Calloman for help with Fig. 1, and the editors and two anonymous reviewers for helpful suggestions on an earlier version of the manuscript. This work was supported by the National Science Foundation grant DEB-1457748 to E.A.K., Washington University in St. Louis, and the Research Center for Auditory and Vestibular Studies is supported by the National Institutes of Health NIDCD Grant P30DC04665. J.K.T., E.A.K. and M.R.M. contributed to the conception and design of the study. J.K.T. and M.R.M. performed the research, and acquired and analysed the data. All authors contributed to interpretation of the data and writing of the manuscript.
Fig. 1.

Sample of the diversity of floret form in grasses. From left to right: awnless Isachne arundinacea (Harris 12487); straight-awned Eriachne pallescens (Beaman 10813); and twisted and geniculately awned Tenaxia californica (Schlechter 9499). Florets were photographed individually with focus stacking using a Nikon D90 camera on an Infinity K2/SC™ Long-Distance Microscope with either a CF4 or CF3 objective. Stacked images were assembled using Helicon Focus, and the resulting compiled images were edited in GIMP (http://gimp.org/). Floret images were extracted individually, size-adjusted to be at the same scale and combined onto a solid black background. No other adjustments were made. Abbreviations: A, awn; L, lemma; C, callus; H, hair on lemma body. Scale bar = 1 mm.
LITERATURE CITED
- Bankevich A, Nurk S, Antipov D, et al. 2012. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. Journal of Computational Biology 19: 455–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker NP. 1997. The relationships of Amphipogon, Elytrophorus and Cyperochloa (Poaceae) as suggested by rbcL sequence data. Telopea 7: 205–213. [Google Scholar]
- Barker NP, Linder HP, Harley EH.. 1995. Polyphyly of Arundinoideae (Poaceae): evidence from rbcL sequence data. Systematic Botany 20: 423–435. [Google Scholar]
- Barker NP, Linder HP, Harley EH, Town C, Bag P, Lavin M.. 1998. Sequences of the grass-specific insert in the chloroplast rpoC2 gene elucidate generic relationships of the Arundinoideae (Poaceae). Systematic Botany 23: 327–350. [Google Scholar]
- Beaulieu JM, O’Meara BC, Donoghue MJ.. 2013. Identifying hidden rate changes in the evolution of a binary morphological character: the evolution of plant habit in campanulid angiosperms. Systematic Biology 62: 725–737. [DOI] [PubMed] [Google Scholar]
- Besnard G, Christin P-A, Malé P-JG, et al. 2014. From museums to genomics: old herbarium specimens shed light on a C3 to C4 transition. Journal of Experimental Botany 65: 6711–6721. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke SV, Wysocki WP, Zuloaga FO, et al.2016. Evolutionary relationships in Panicoid grasses based on plastome phylogenomics (Panicoideae; Poaceae). BMC Plant Biology 16: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Christin P-A, Spriggs E, Osborne CP, Strömberg CAE, Salamin N, Edwards EJ.. 2014. Molecular dating, evolutionary rates, and the age of the grasses. Systematic Biology 63: 153–165. [DOI] [PubMed] [Google Scholar]
- Clark LG, Zhang W, Wendel JF.. 1995. A phylogeny of the grass family (Poaceae) based on ndhF sequence data. Systematic Botany 20: 436–460. [Google Scholar]
- Clayton WD, Renvoize SA.. 1999. Genera Graminum. Grasses of the world. London: Royal Botanic Gardens, Kew. [Google Scholar]
- Clayton WD, Vorontsova MS, Harman KT, Williamson H.. 2006. onwards. GrassBase The Online World Grass Flora http://www.kew.org/data/grasses-db.html (accessed December 2016 January 2017).
- Conert HJ. 1966. Dregeochloa, eine neue Gattung der Gramineen. Senckenbergiana Biologica 47: 335–345. [Google Scholar]
- Conert HJ. 1987. Current concepts in the systematics of the Arundinoideae In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME, eds. Grass systematics and evolution. Washington, DC: Smithsonian Institution Press, 239–250. [Google Scholar]
- Cota-Sánchez JH, Remarchuk K, Ubayasena K.. 2006. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Molecular Biology Reporter 24: 161–67. [Google Scholar]
- Cotton JL, Wysocki WP, Clark LG, et al. 2015. Resolving deep relationships of PACMAD grasses: a phylogenomic approach. BMC Plant Biology 15: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepet WL, Feldman GD.. 1991. The earliest remains of grasses in the fossil record. American Journal of Botany 78: 1010‒1014. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidse G. 1987. Fruit dispersal in the Poaceae In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME eds. Grass systematics and evolution. Washington, DC: Smithsonian Institution Press, 143–155. [Google Scholar]
- De Wet JMJ. 1956. Leaf anatomy and phylogeny in the tribe Danthonieae. American Journal of Botany 43: 175–182. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall MR, Yadav SR, Burke SV, Wysocki WP.. 2017. Grass plastomes reveal unexpected paraphyly with endemic species of Micrairoideae from India and new haplotype markers in Arundinoideae. American Journal of Botany 104: 1–10. [DOI] [PubMed] [Google Scholar]
- Elias MK. 1942. Tertiary prairie grasses and other herbs from the High Plains. Geological Society of America Special Paper (Regular Studies) 41: 1–176. [Google Scholar]
- Ellis RP. 1985a. Leaf anatomy of the South African Danthonieae (Poaceae). XII. Pentameris thuarii. Bothalia 15: 573–578. [Google Scholar]
- Ellis RP. 1985b. Leaf anatomy of the South African Danthonieae (Poaceae). X. Pseudopentameris. Bothalia 15: 561–566. [Google Scholar]
- Ellis RP. 1986. Leaf anatomy of the South African Danthonieae (Poaceae). XIV. Pentameris dregeana. Bothalia 16: 235–241. [Google Scholar]
- Ellis RP. 1988a. Leaf anatomy of the South African Danthonieae (Poaceae). XVI. The genus Urochlaena. Bothalia 18: 101–104. [Google Scholar]
- Ellis RP. 1988b. Leaf anatomy of the South African Danthonieae (Poaceae). XVII. The genus Chaetobromus. Bothalia 18: 195–209. [Google Scholar]
- Ellis RP. 1989. Leaf anatomy of the South African Danthonieae (Poaceae): XIX. The genus Prionanthium. Bothalia 19: 217–223. [Google Scholar]
- Garnier LKM, Dajoz I.. 2001. Evolutionary significance of awn length variation in a clonal grass of fire-prone savannas. Ecology 82: 1720–1733. [Google Scholar]
- Grass Phylogeny Working Group. 2001. Phylogeny and subfamilial classification of the grasses (Poaceae). Annals of the Missouri Botanical Garden 88: 373–457. [Google Scholar]
- Grass Phylogeny Working Group II. 2012. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytologist 193: 304–312. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O.. 2003. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Hubbard CE. 1940. Alloeochaete andongensis (Rendle) C. E. Hubbard. In: Hooker’s Icones Plantarum, Ser. 5, 34, tab. 3418.
- Hubbard CE. 1948. The genera of British grasses In: British flowering plants. London: P.R. Gawthorn Ltd, 284–348. [Google Scholar]
- Hubbard CE. 1957a. Notes on African grasses: XXV. Nematopoa, a new genus from Southern Rhodesia. Kew Bulletin 12: 51–52. [Google Scholar]
- Hubbard CE. 1957b. Notes on African grasses: XXVI. Piptophyllum, a new genus from Angola. Kew Bulletin 12: 52–53. [Google Scholar]
- Hubbard CE, Schweickerdt HG, Snowden JD.. 1936. Notes on African grasses: XIX. Miscellaneous notes and new species. Bulletin of Miscellaneous Information (Royal Botanic Gardens, Kew) 1936: 293–335. [Google Scholar]
- Humphreys AM, Antonelli A, Pirie MD, Linder HP.. 2010. Ecology and evolution of the diaspore burial syndrome. Evolution 65: 1163–1180. [DOI] [PubMed] [Google Scholar]
- Ingram AL, Christin P-A, Osborne CP.. 2011. Molecular phylogenies disprove a hypothesized C4 reversion in Eragrostis walteri (Poaceae). Annals of Botany 107: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskandar EAP, Veldkamp JF.. 2004. A revision of Malesian Isachne sect. Isachne (Gramineae, Panicoideae). Reinwardtia 12: 159–179. [Google Scholar]
- Jankowiak K, Buczkowska K, Szweykowska-Kulinska Z.. 2005. Successful extraction of DNA from 100-year-old herbarium specimens of the liverwort Bazzania trilobata. Taxon 54: 335–336. [Google Scholar]
- Johnson EE, Baruch Z.. 2014. Awn length variation and its effect on dispersal unit burial of Trachypogon spicatus (Poaceae). Revista de Biología Tropical/International Journal of Tropical Biology and Conservation 62: 321–326. [PubMed] [Google Scholar]
- Kabuye CHS, Renvoize SA.. 1975. The genus Alloeochaete, tribe Danthonieae (Gramineae). Kew Bulletin 30: 569–577. [Google Scholar]
- Katoh S. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA. 2015. Flowering plants. Vol. 13. Monocots: Poaceae. Heidelberg: Springer. [Google Scholar]
- Kellogg EA, Campbell CS.. 1987. Phylogenetic analyses of the Gramineae In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME eds. Grass systematics and evolution. Washington, DC: Smithsonian Institution Press, 310–322. [Google Scholar]
- Langmead B, Salzberg S.. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder HP, Verboom GA, Barker NP.. 1997. Phylogeny and evolution in the Crinipes group of grasses (Arundinoideae: Poaceae). Kew Bulletin 52: 91–110. [Google Scholar]
- Linder HP, Baeza M, Barker NP, et al. 2010. A generic classification of the Danthonioideae (Poaceae). Annals of the Missouri Botanical Garden 97: 306–364. [Google Scholar]
- Maddison WP, Maddison DR.. 2015. Mesquite: a modular system for evolutionary analysis. Version 3.04 http://mesquiteproject.org
- Mathews S, Tsai RC, Kellogg EA.. 2000. Phylogenetic structure in the grass family (Poaceae): evidence from the nuclear gene phytochrome B. American Journal of Botany 87: 96–107. [PubMed] [Google Scholar]
- McKain MR, Hartsock RH, Wohl MM, Kellogg EA.. 2017. Verdant: automated annotation, alignment, and phylogenetic analysis of whole chloroplast genomes. Bioinformatics 33: 130–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe CE. 1960. Anatomy of the monocotyledons, I: Gramineae. London: Oxford University Press. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: SC10 Workshop on Gateway Computing Environments (GCE10). [Google Scholar]
- Morris LM, Duvall MR.. 2010. The chloroplast genome of Anomochloa marantoidea (Anomochlooideae; Poaceae) comprises a mixture of grass-like and unique features. American Journal of Botany 97: 620–627. [DOI] [PubMed] [Google Scholar]
- Palmer PG, Gerbeth-Jones S.. 1986. A scanning electron microscope survey of the epidermis of East African grasses, IV. Smithsonian Contributions to Botany 62: 1–120. [Google Scholar]
- Palmer PG, Gerbeth-Jones S.. 1988. A scanning electron microscope survey of the epidermis of East African grasses, V, and West African supplement. Smithsonian Contributions to Botany 67: 1–157. [Google Scholar]
- Palmer PG, Tucker AE.. 1981. A scanning electron microscope survey of the epidermis of East African grasses, I. Smithsonian Contributions to Botany 49: 1–84. [Google Scholar]
- Palmer PG, Tucker AE.. 1983. A scanning electron microscope survey of the epidermis of East African grasses, II. Smithsonian Contributions to Botany 53: 1–72. [Google Scholar]
- Palmer PG, Gerbeth-Jones S, Hutchinson S.. 1985. A scanning electron microscope survey of the epidermis of East African grasses, III. Smithsonian Contributions to Botany 55: 1–136. [Google Scholar]
- Paradis E, Claude J, Strimmer K.. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Peart MH. 1979. Experiments on the biological significance of the morphology of seed dispersal units in grasses. Journal of Ecology 67: 843–863. [Google Scholar]
- Peart MH. 1981. Further experiments on the biological significance of the morphology of seed-dispersal units in grasses. Journal of Ecology 69: 425–436. [Google Scholar]
- Peart MH. 1984. The effects of morphology, orientation and position of grass diaspores on seedling survival. Journal of Ecology 72: 437–453. [Google Scholar]
- Peart MH, Clifford HT.. 1987. The influence of diaspore morphology and soil-surface properties on the distribution of grasses. Journal of Ecology 75:569–576. [Google Scholar]
- Pohl RW. 1965. Dissecting equipment and materials for the study of minute plant structures. Rhodora 67: 95–96. [Google Scholar]
- Renvoize SA. 1981. The sub-family Arundinoideae and its position in relation to a general classification of the Gramineae. Kew Bulletin 36: 85–102. [Google Scholar]
- Reimer E, Cota-Sànchez JH.. 2007. An SEM survey of the leaf epidermis in danthonioid grasses (Poaceae: Danthonioideae). Systematic Botany 32: 60–70. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, et al. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin SE. 1999. Plant microtechnique and microscopy. New York: Oxford University Press. [Google Scholar]
- Sánchez-Ken JG, Clark LG, Kellogg EA, Kay EE.. 2007. Reinstatement and emendation of subfamily Micrairoideae (Poaceae). Systematic Botany 32: 71–80. [Google Scholar]
- Särkinen T, Staats M, Richardson JE, Cowan RS, Bakker FT, Caramelli D.. 2012. How to open the treasure chest? Optimising DNA extraction from herbarium specimens. PLoS One 7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass JE. 1951. Botanical microtechnique. Ames, IA: The Iowa State College Press. [Google Scholar]
- Shimodaira H, Hasegawa M.. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution 16: 1114. [Google Scholar]
- Simpson M. 1952. Value of the awn in establishing seed of Danthonia penicillata (Labill.) Palisot. New Zealand Journal of Science and Technology 34: 360–364. [Google Scholar]
- Soreng RJ, Peterson PM, Romaschenko K, et al. 2015. A worldwide phylogenetic classification of the Poaceae (Gramineae): phylogenetic classification of the grasses. Journal of Systematics and Evolution 53: 117–137. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strömberg CAE. 2005. Decoupled taxonomic radiation and ecological expansion of open-habitat grasses in the Cenozoic of North America. Proceedings of the National Academy of Sciences, USA 102: 11980–11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboom GA, Linder HP, Barker NP.. 1994. Haustorial synergids: an important character in the systematics of danthonioid grasses (Arundinoideae: Poaceae)? American Journal of Botany 81: 1601–1610. [Google Scholar]
- Vicentini A, Barber JC, Aliscioni SS, Giussani LM, Kellogg EA.. 2008. The age of the grasses and clusters of origins of C4 photosynthesis. Global Change Biology 14: 2963–2977. [Google Scholar]
- Washburn JD, Schnable JC, Davidse G, Pires JC.. 2015. Phylogeny and photosynthesis of the grass tribe Paniceae. American Journal of Botany 102: 1493–1505. [DOI] [PubMed] [Google Scholar]
- Watson L, Dallwitz MJ.. 1992. onward. The grass genera of the world: descriptions, illustrations, identification, and information retrieval; including synonyms, morphology, anatomy, physiology, phytochemistry, cytology, classification, pathogens, world and local distribution, and references. Version: 7 December 2015. http://delta-intkey.com.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.