Abstract
Hypoxia increases both active and repressive histone methylation levels via decreased activity of histone demethylases. However, how such increases coordinately regulate induction or repression of hypoxia-responsive genes is largely unknown. Here, we profiled active and repressive histone tri-methylations (H3K4me3, H3K9me3, and H3K27me3) and analyzed gene expression profiles in human adipocyte-derived stem cells under hypoxia. We identified differentially expressed genes (DEGs) and differentially methylated genes (DMGs) by hypoxia and clustered the DEGs and DMGs into four major groups. We found that each group of DEGs was predominantly associated with alterations in only one type among the three histone tri-methylations. Moreover, the four groups of DEGs were associated with different TFs and localization patterns of their predominant types of H3K4me3, H3K9me3 and H3K27me3. Our results suggest that the association of altered gene expression with prominent single-type histone tri-methylations characterized by different localization patterns and with different sets of TFs contributes to regulation of particular sets of genes, which can serve as a model for coordinated epigenetic regulation of gene expression under hypoxia.
INTRODUCTION
Reduction in oxygen concentration, hypoxia, affects a broad spectrum of physiological cellular processes related to development and growth, as well as pathophyological processes related to cancer, stroke, myocardial infarction, and chronic kidney disease. Hypoxia changes the expression of hypoxia-responsive genes through two main pathways: activation of TFs and inhibition of O2-dependent histone demethylases (1,2). First, hypoxia activates a heterodimeric TF complex of hypoxia-inducible factor-1α (HIF-1α) and HIF-1β (also known as Arnt) (3). Under normoxia, HIF-1α is hydroxylated at 564 and/or 402 proline residues, initiating its ubiquitinylation and subsequent proteasomal degradation. Such hydroxylation is catalyzed by a novel HIF-1α-specific prolyl hydroxylase, which requires O2, α-ketoglutarate (α-KG), vitamin C and Fe2+ (4,5). Under hypoxia, proline hydroxylation ceases and HIF-1α protein accumulates. Second, hypoxia decreases the activity of Jumonji C domain-containing histone demethylases (JMJDs) that require O2 and α-KG as a substrate (6,7). The JMJDs affect gene expression through their regulation of active or repressive histone methylations (8–10).
Histone modifiers including JMJDs are not able to recognize DNA sequences, but are instead recruited on their target chromosome though protein–protein interactions with TFs that bind to specific DNA sequences. Therefore, it should be the TFs to set up the specificity of both histone methylation and gene expression (11,12). These DNA binding abilities of TFs are regulated by several signaling pathways involving in phosphorylation, proteasome degradation, nuclear translocation, and ligand binding. Differently from TFs, histone modifiers are enzymes that use small diffusible metabolites such as O2, α-KG, acetate and S-adenosylmethionine, as substrates. Therefore, with no changes of TF binding, histone methylation of a certain gene can be instantly altered by cellular metabolic status that influences enzyme activities of JMJDs at their target genes.
Many JmjC domain-containing proteins exhibited demethylase activity with different substrate specificities (13): JMJD2A and JMJD2C for H3K9me3, JMJD3 for H3K27me3 and Jumonji/ARID domain-containing protein 1A (JARID1A) and JARID1B for H3K4me3. H3K4 methylation in coding and regulatory regions is associated with active transcription. H3K9 methylation in the coding region is associated with active transcription, but in the regulatory region it is associated with transcriptional repression (14). The fact that JMJDs utilize O2 suggests that the overall active and repressive histone methylation levels would increase under hypoxia. In embryonic stem cells, lineage-specific genes remain in poised states with distinctive bivalent signatures in their promoter regions. The bivalent signature is defined as a histone modification status in which both active and repressive marks are present in their promoters (15–18). Recent studies have shown that hypoxia maintains undifferentiated states of several stem cells (2), suggesting that hypoxia influences pluripotency of stem cell through increasing bivalent histone methylation signatures (19). However, it is not clear how the increased levels of both active and repressive histone methylations coordinately regulate the induction or repression of individual genes under hypoxia.
No global profiling of active and repressive histone methylations under hypoxia has been performed to systematically address this issue. Among five typical types of histones (H1, H2A, H2B, H3 and H4), modification of H3 has been extensively studied (20), and H3 has multiple lysine and arginine residues that can be mono-, di- and tri-methylated to produce many combinatorial patterns of histone methylations. In this study, we examined global profiling of three major tri-methylated histones, H3K4me3, H3K9me3 and H3K27me3, in human adipocyte-derived stem cells (hADSCs) under both normoxia and hypoxia. We then identified differentially methylated genes (DMGs) by hypoxia for each type of histone tri-methylation. Moreover, we performed gene expression profiling of hADSCs and then identified differentially expressed genes (DEGs) by hypoxia. About half of the DEGs showed multiple H3K4me3, H3K9me3 and H3K27me3 under both normoxia and hypoxia, suggesting that alterations in gene expression should involve complex combinatorial changes of H3K4me3, H3K9me3 and H3K27me3. To examine relationships between the DEGs and DMGs, we performed clustering analysis of the DEGs using their changes in H3K4me3, H3K9me3 and H3K27me3. Surprisingly, however, clustering analyses revealed four major clusters and further showed that the DEGs in each cluster involved predominantly the changes in a single type of histone tri-methylations, rather than complex combinatorial changes of multiple types of histone tri-methylations. Furthermore, we identified TFs whose binding motifs significantly enriched in the four groups of DEGs. Therefore, our results suggest the association of altered gene expression with (i) prominent single-type histone tri-methylations characterized by different localization patterns and (ii) TFs as a model for coordinated epigenetic regulation of gene expression under hypoxia.
MATERIALS AND METHODS
Cell culture
Human Adipose-Derived Stem Cells (hADSC) were purchased from Invitrogen (Carlsbad, CA, USA), which are isolated from human adipose tissue collected during liposuction procedures. The medium was supplemented with 10% MSC-qualified FBS. The cells were exposed to hypoxic conditions (21, 2, 1 or <0.5% O2) with the hypoxic gas mixture for 48 h in the In Vivo 200 Hypoxia Workstation (Ruskinn Technology, Leeds, UK) or in an anaerobic incubator (Model 1029; Forma Scientific, Waltham, MA, USA). The level of oxygen in the anaerobic incubator was verified to be <0.5% using the Fyrite Analyzer (Bacharach, New Kensington, PA, USA).
Library construction and ChIP-seq
Each sequenced sample was prepared according to the Illumina's standard protocols. Briefly, chromatin immunoprecipitated DNA (ChIP DNA) was processed with Klenow polymerase and T4 polynucleotide kinase to create 3′-dA. ChIP DNA was then ligated with Illumina adaptors and amplified with Illumina primers for 18 cycles. Following the isolation of 100–500 bp of ChIP DNA from the agarose gel, Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) validated the size, purity, and concentration of DNA, and library quantitation was performed using HiSeq™ 2000 platform to produce 101 bp paired-end samples. Total read numbers obtained for individual samples are listed in Supplementary Table S1. The raw ChIP-seq data were deposited to the GEO database with accession ID: GSE69493.
ChIP-seq data analysis
From the sequencing data, we first removed adapter sequences (TrueSeq universal and index adapters) and then trimmed the reads for which PHRED scores were <20 using the cutadapter software (21). Then, the remaining reads were aligned to the human genome GRCh38 using Bowtie2 software (version 2.0.6) with the default parameters (22). After trimming reads with mapping quality lower than five and duplicated reads, the aligned data were then mapped to 20,654 annotated genes in Ensembl (23) (Supplementary Table S2). After the alignment, we counted the numbers of reads mapped to gene features (GTF file of GRCh38) using Coverage from Bedtools (24) and computed RPKM (reads per kilobase of target per million mapped reads) for the gene features.
TF binding site analysis
To identify TFs whose binding motifs are significantly enriched in the differentially methylated regions (peaks) of the genes in Groups 1–4, we collected 161 TF matrices from the JASPAR CORE database (25). Peak sequences (input sequence) were scanned using the MotifLocator software (26). The second-order custom background model was created by CreateBackgroundModel software in MotifSuite using all peak sequences (26). To assess the statistical significance of MotifLocator scores, we randomized the input sequence and applied MotifLocator to the randomized sequence using the parameters of –b (custom background model) and –t -1. These steps were repeated 1000 times. The resulting scores from random experiments were used to estimate an empirical distribution of MotifLocator scores for each TF by Gaussian kernel density estimation (27). Based on the empirical distribution, we computed false discovery rates (FDRs) for the observed scores using the Storey's method (28). The binding sites for each TF were selected as the ones with FDR < 0.001, and the genes having more than one binding sites were identified as targets of the TF. Next, we identified TFs that have a significant number of target genes in Groups 1–4. To this end, we first randomly shuffled the input sequence 10,000 times, counted the numbers of targets for each TF in individual random experiments, and estimated an empirical distribution for the number of targets using Gaussian kernel density estimation. Using the empirical distribution, we computed FDR for the observed number of targets of the TF in each of Groups 1–4 using the Storey's method (28). Finally, we selected TF candidates with FDR < 0.05 and have at least 5% of group as target genes.
Reconstruction of a network model
To construct a network model for the target genes of the six representative TFs in Groups 1–3, we first selected a subset of target genes involved in the GOBPs shown in Figure 6C. We then collected their protein–protein interactions for these genes from five interactome databases: BIND (Biomolecular Interaction Network Database) (29), HPRD (Human Protein Reference Database) (30), BioGRID (Biological General Repository for Interaction Datasets) (31), MINT (Molecular INTeraction Database) (32) and STRING (search tool for recurring instances of neighbouring genes) (33). The initial network model was built with the target genes and their interactors using Cytoscape (34). We then arranged the nodes according to their associated GOBPs and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (35) such that the nodes involved in the same GOBPs and pathways are located closely.
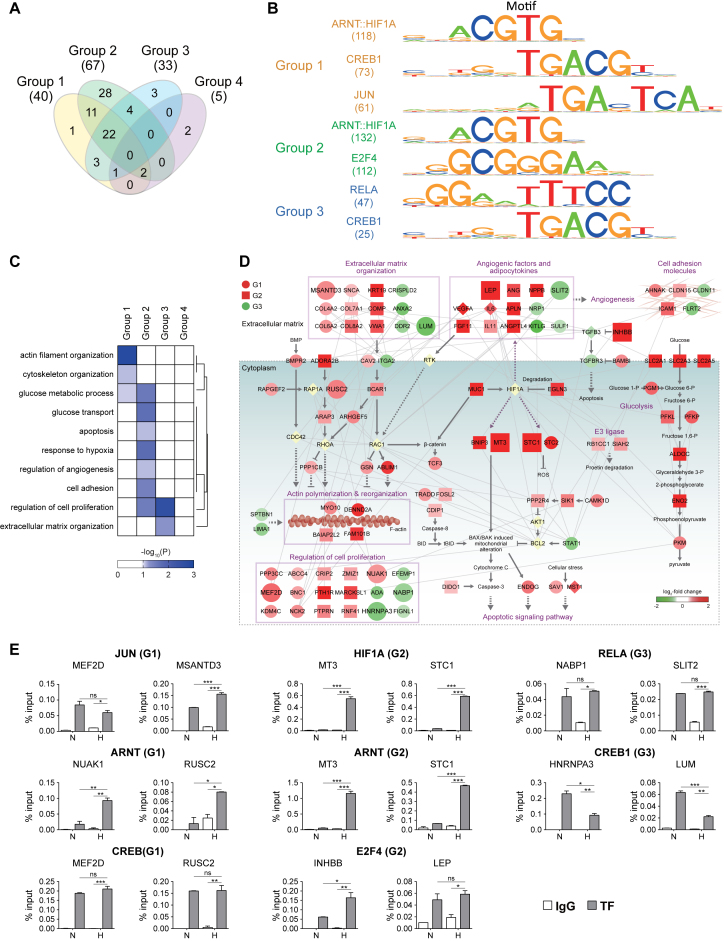
Figure 6.
Differential sets of TFs associated with regulation of mRNA expression under hypoxia. (A) Relationships among the TFs whose binding motifs were significantly enriched in the methylated regions of genes in Groups 1–4. Numbers in parentheses denote the numbers of TFs identified for Groups 1–4. (B) Binding motifs of the TFs for Groups 1–3. HIF1A and ARNT had a shared binding motif. (C) GOBPs represented by the target genes of the six representative TFs in Groups 1–3. A heat map was generated by a hierarchical clustering of –log10(P), where Pij is the enrichment P-values computed for the target genes of the six TFs in Group i and GOBP j by DAVID (Euclidean distance and average linkage method). The color bar represents the gradient of –log10(P). (D) Network model describing interactions among the target genes of the six representative TFs. Node colors represent up- (red) and down-regulation (green) in hypoxia, compared to normoxia, and node shapes denote the groups to which the corresponding genes belonged (red circle, square, and green circle for Groups 1–3, respectively). Large nodes denote the genes for which binding of the six representative TFs to their differentially methylated regions was experimentally confirmed. Arrows and inhibition symbols represent activation and inhibition information obtained from Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. Solid and dotted lines denote direct and indirect activation/inhibition, respectively, and gray lines denote protein–protein interactions. (E) ChIP-PCR analyses of the target genes in Groups 1–3 confirming binding of the six representative TFs to their target genes. The graph shows two or more replicates with SEM. *P < 0.05, **P < 0.01 and ***P < 0.001 by one-way ANOVA with Turkey's correction.
Further information in supplementary materials and methods.
RESULTS
Increased H3K4me3, H3K9me3 and H3K27me3 through decreased catalytic activities of JMJDs under hypoxia
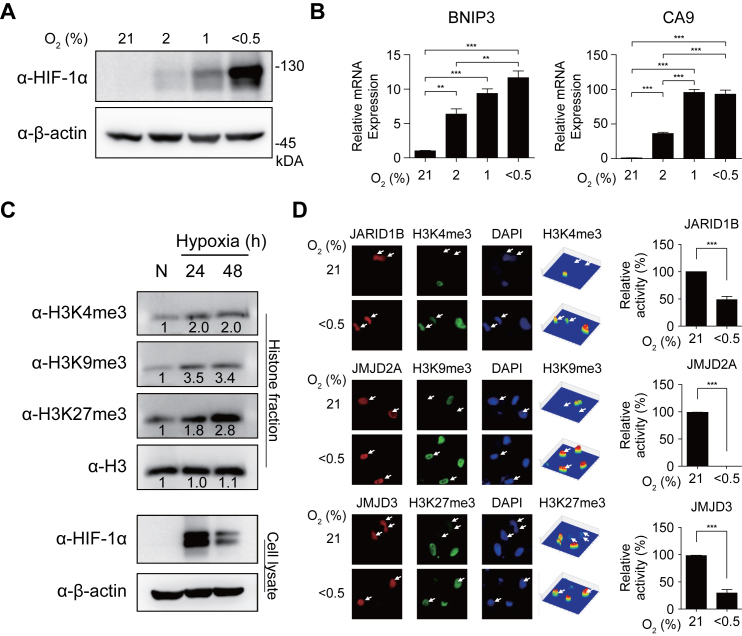
A number of studies have previously shown that hypoxic inhibition of JMJD activities increased total amounts of methylated histones (36,37). Thus, we confirmed that there were increases in the amounts of active (H3K4me3) and repressive (H3K9me3 and H3K27me3) histone methylations in hADSCs as a result of decreased JMJD activity. With decreasing oxygen concentrations (21, 2, 1 and <0.5%), HIF-1α and its target genes, such as BCL2/adenovirus E1B 19kDa interacting protein 3 (BNIP3) and carbonic anhydrase IX (CA9), were induced in hADSCs (Figure 1A and B). Consistent with previous findings, the total amounts of H3K4me3, H3K9me3 and H3K27me3 were found to be increased under hypoxia (Figure 1C). The increased histone methylations can be caused by increased activities of methyltransferases or decreased activities of JMJDs (38). To examine this, we measured the amounts of H3K4me3, H3K9me3 and H3K27me3 after depleting methionine, which is a substrate of methyltransferases. The amounts of the histone methylations were similarly increased under hypoxic conditions, regardless of the presence of methionine (Supplementary Figure S1A), indicating that the histone methylations were increased not by the increased activities of methyltransferases, but mainly by the decreased activities of JMJDs.
Figure 1.
Increased histone tri-methylations through decreased activities of JMJDs in hypoxia. (A) Western blot analyses of HeLa cells treated with hypoxia (21, 2, 1 and <0.5%) for 24 h using indicated antibodies. β-actin was detected as a loading control. (B) Quantitative RT-PCR for two target genes (BNIP3 and CA9) in hypoxia for 16 hours. The data were normalized using levels of 18s rRNA and shown as mean with standard error of means (SEM). *P < 0.05, **P < 0.01 and ***P < 0.001 by one-way ANOVA with Turkey's correction. (C) Increased levels of H3K4me3, H3K9me3 and H3K27me3 at 24 and 48 h after decreasing oxygen concentration (<0.5%) in hADSC. The numbers under the bands represent relative band intensities. The amounts of HIF-1α and histone 3 (H3) were also shown. H3 was used as a loading control for quantification of methylated histones in histone fractions (Supplementary Materials and Methods). β-Actin was used as a loading control for quantification of HIF-1α, a marker protein of hypoxia, in cell lysates. (D) Hypoxic effects on catalytic activities of histone demethylases. HeLa cells were transfected with Myc-tagged JARID1B (H3K4me3), HA-tagged JMJD2A (H3K9me3) and Myc-tagged JMJD3 (H3K27me3) by using calcium phosphate precipitation method as previously described (37). The transfected cells were treated with hypoxia (O2% <0.5) for 16 hours and then immunostained using corresponding tag-antibodies (red) and indicated histone antibodies (green). The nuclei were stained with DAPI (blue). Arrows denote cells transfected with indicated histone demethylases. Using a Pseudo-Color 3D confocal fluorescence microscope, fluorescence intensities of H3K4me3, H3K9me3, and H3K27me3 were estimated as shown in contour maps (right columns). For each histone demethylase, catalytic activity was defined as the difference in estimated fluorescence intensities of its corresponding tri-methylated histones between untransfected and transfected cells. The catalytic activity under hypoxia was normalized by that under normoxia. Data represent the average and standard deviations of the estimated activities of histone demethylases from 6 to 38 transfected cells.
To understand how JMJDs contributed to the hypoxia-induced increase in histone tri-methylation, we examined hypoxia-induced changes in both abundances and catalytic activities of JMJDs that have been shown to act on H3K4me3 (JARID1A-D), H3K9me3 (JMJD2A-D) and H3K27me3 (JMJD3) because enzymatic activities of JMJDs are determined not only by their abundances but also by their catalytic activities. First, to examine hypoxic changes of JMJD abundances, we measured mRNA expression levels of the above JMJDs under normoxia (%O2 = 21) and hypoxia (%O2 < 0.5) using quantitative real time-polymerase chain reaction (qRT-PCR). JARID1B, JMJD2B, and JMJD2C increased in their expression levels under hypoxia (%O2 < 0.5), compared to under normoxia, although the other JMJDs showed no expression changes (Supplementary Figure S1B). Such hypoxic changes of JMJDs were consistent with those detected from microarray analyses (Supplementary Figure S1C). These data indicate that variable expression changes of JMJD (up-regulation or no change) showed no correlation with those of their corresponding histone tri-methylation levels under hypoxia, implying that the mRNA expression of the JMJDs has no contribution to the increase in histone tri-methylation. Second, to examine hypoxic changes of catalytic activities of JMJDs, we performed an assay to measure demethylation catalytic activities of JMJDs under nomoxia and hypoxia as previously described (37). In this assay, we overexpressed JMJDs by cDNA transfection and measured levels of H3K4me3, H3K9me3 and H3K27me3 in cells overexpressing JMJDs under both normoxia and hypoxia by immunostaining. H3K4me3, H3K9me3, and H3K27me3 in JMJD-overexpressing cells disappeared under normoxia, but were still present under hypoxia (%O2 < 0.5), despite overexpression of JMJDs (Figure 1D and Supplementary Figure S1D), indicating that overexpression of JMJDs cannot compensate hypoxic inhibition of their catalytic activities (Supplementary Discussion). Taken together, these data suggest that increased histone tri-methylation was mainly due to decreased demethylation catalytic activities of JMJDs, not to altered abundances of JMJDs.
Distributions of multiple histone tri-methylations within gene structures
To analyze the change in the multi-dimensional landscape of histone methylations under hypoxia, we performed chromatin immunoprecipitation-sequencing (ChIP-seq) to generate genome-wide maps of H3K4me3, H3K9me3 and H3K27me3 in hADSCs under both normoxia (%O2 = 21) and hypoxia (%O2 < 0.5) (Materials and Methods; Supplementary Table S1). ChIP-seq yielded a total of 199.1 and 194.4 million reads for the three types of histone tri-methylations under hypoxia and normoxia, respectively (Supplementary Table S2). These reads were aligned to the human genome (UCSC GRCh38) using BOWTIE2 (Materials and Methods), resulting in a total of 5.54 Gbp of mapped sequences for all three types of histone tri-methylations under both normoxia and hypoxia, which corresponded to 3.7-fold coverages of the human genome on average.
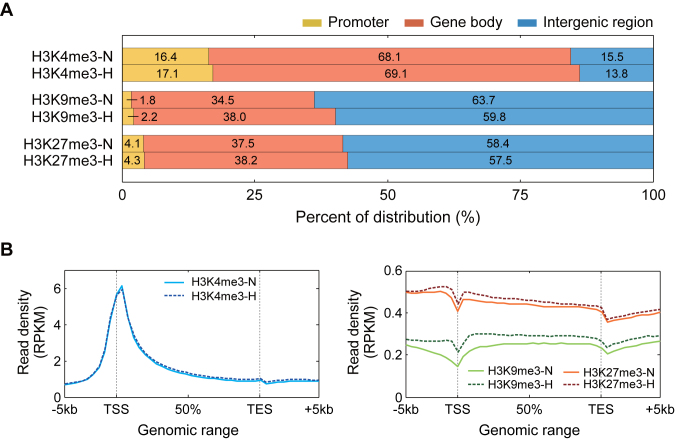
To examine the characteristics of the three types of histone methylations, we first compared their respective distributions within the chromosomes (Figure 2A). The comparison revealed that H3K4me3 was preferentially enriched in gene bodies, while H3K9me3 and H3K27me3 were most significantly enriched in intergenic regions, followed by gene bodies. Interestingly, these enrichment patterns were shared in both normoxia and hypoxia. We further analyzed how the histone tri-methylations were distributed within the gene structures (Materials and Methods). H3K4me3 was preferentially enriched near transcription start sites (TSS) (Figure 2B, left). The enrichment patterns of H3K4me3 were shared between normoxia and hypoxia. On the other hand, both H3K9me3 and H3K27me3 were enriched in both distal promoters (<−1 Kb) and gene bodies, but not in TSS (Figure 2B, right). Although H3K9me3 was equally enriched in both promoters and gene bodies, H3K27me3 was more preferentially enriched in promoters than in gene bodies, which is consistent with previous findings (39). The enrichment patterns of H3K9me3 and H3K27me3 were shared between normoxia and hypoxia, but the extent of the enrichment increased under hypoxia clearly in proximal promoter (≥−1 Kb) and gene bodies. These data showed that different types of histone tri-methylations had distinct distributions within gene structures, thus suggesting that the differential distributions may serve as an important regulatory dimension in transcriptional regulation based on the multi-dimensional landscape of histone tri-methylations.
Figure 2.
Localization of histone modifications across genome and gene structures. (A) Distribution of H3K4me3, H3K9me3, and H3K27me3 at the chromosomal level. The stacked bar graphs show proportions of the aligned reads for the three types of histone methylations (see the percentages shown) under normoxia and hypoxia in the promoters, gene bodies, and intergenic regions. (B) Distributions of the histone methylations within the gene structures. For each gene, the gene structure (-5 kb-TSS-TES-+5 kb) was parsed into 45 bins (Materials and Methods). The average RPKM read density profiles (y-axis) of H3K4me3 (blue), H3K9me3 (green) and H3K27me3 (orange) for all the genes in individual bins (x-axis) were computed and plotted. Solid and dotted lines represent profiles of read densities under normoxia and hypoxia, respectively.
Correlations of multiple histone methylations with mRNA expression
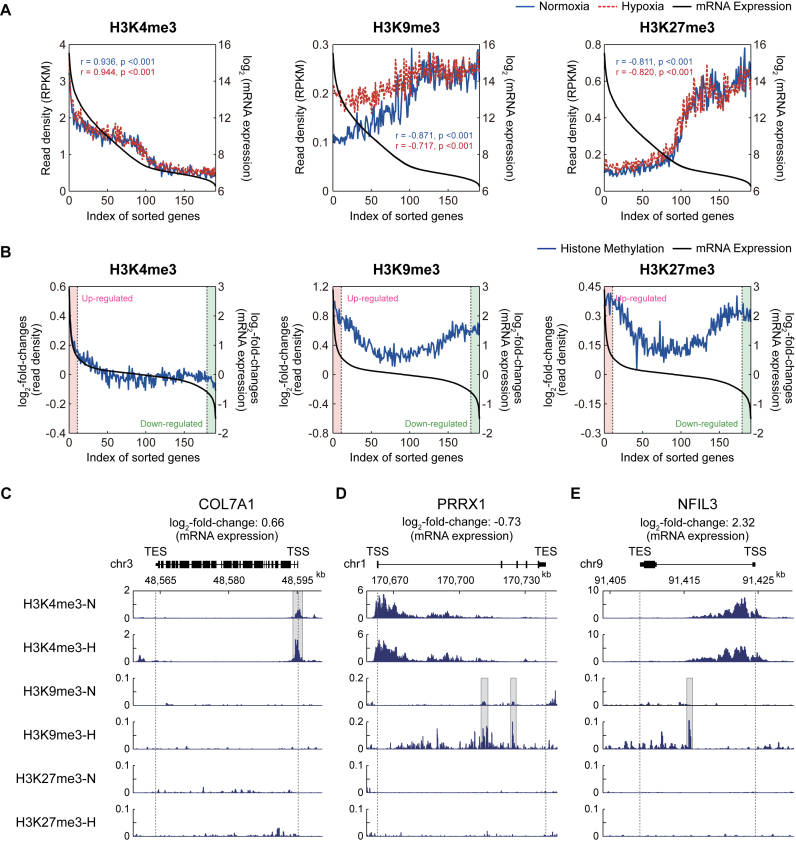
To explore the link of the changes in the multi-dimensional histone methylation to the expression changes of hypoxia target genes, we then performed gene expression profiling of hADSCs under the same normoxic and hypoxic conditions (Materials and Methods, Supplementary Discussion). We first analyzed the correlations between the levels of histone methylation and mRNA expression using the correlation analysis previously reported (Materials and Methods). The correlation analysis revealed that H3K4me3 levels showed a significant (P < 0.01; Pearson correlation) positive correlation with mRNA expression levels (Figure 3A, left), whereas the levels of both H3K9me3 and H3K27me3 showed significant (P < 0.01) negative correlations (Figure 3A, middle and right). These observations are consistent with the conventional roles of H3K4me3 as an active mark for gene expression and of H3K9me3 and H3K27me3 as repressive marks. Interestingly, both the positive and negative correlations were apparent for the genes with relatively high mRNA expression levels, whereas the correlations become less significant (H3K4me3 and H3K27me3) or even disappeared (H3K9me3) for the genes with low mRNA expression levels, consistent to the findings previously reported by Cui et al. (40). These correlation patterns were shared between under normoxia and hypoxia. However, the negative correlation of H3K9me3 with the gene expression was relatively stronger under normoxia (black versus blue lines in Figure 3A, middle), compared to hypoxia (black versus red lines in Figure 3A, middle), suggesting that there could be some factors to cause the reduced negative correlation under hypoxia.
Figure 3.
Correlation between hypoxia-induced changes of histone modification and gene expression. (A) Correlation between the levels of histone modification and gene expression under normoxia and hypoxia. The expressed genes were sorted in a descending manner by their expression levels and then binned such that each bin includes 100 genes. For each of the three types of histone methylations, the averaged RPKMs (read density; y1-axis) and the averaged expression levels (log2-intensities; y2-axis) in individual bins (x-axis) were plotted. Blue solid, red dotted, and black solid lines represent the averaged RPKMs under normoxia and hypoxia and the averaged expression levels, respectively. (B) Correlation of log2-fold-changes of histone modifications between hypoxia and normoxia with those of gene expression. The expressed genes were sorted using their log2-fold-changes and binned as described above. For each of the three types of histone methylations, the averaged log2-fold-changes of RPKMs (y1-axis) and the averaged log2-fold-changes of gene expression levels (y2-axis) in individual bins (x-axis) were plotted. Black and blue lines represent the averaged log2-fold-changes of gene expression and histone methylation levels, respectively. Up- and down-regulated genes (red and green backgrounds, respectively) were defined as the ones with fold-changes larger than 1.5. (C–E) The aligned reads of three representative genes for the three types of histone methylations: COL7A1 (C), NFIL3 (D), and PRRX1 (E). The regions that include the differentially methylated peaks for H3K4me3 (C) and H3K9me3 (D and E) were denoted with gray backgrounds.
Furthermore, we also examined the correlation of the log2-fold-changes of histone methylation levels between normoxia and hypoxia with those of mRNA expression levels. For up-regulated genes under hypoxia (Figure 3B, red regions), there was a positive correlation (r = 0.99; P = 3.95 × 10−10) between the log2-fold-changes of H3K4me3 and mRNA expression levels (Figure 3B, left). A representative up-regulated gene with the increased levels of H3K4me3 under hypoxia, compared to normoxia, was shown in Figure 3C. On the other hand, the down-regulated genes under hypoxia (Figure 3B, green regions) showed increased levels (i.e. positive log2-fold-changes) of H3K9me3 (Figure 3B, middle; Figure 3D) or H3K27me3 (Figure 3B, right). Unexpectedly, however, for the up-regulated genes, the log2-fold-changes of H3K9me3 levels showed a positive correlation (r = 0.69; P = 1.26 × 10−2) with those of mRNA expression levels, indicating the possibility that the up-regulated genes can have increased levels of H3K9me3 (Figure 3B, middle; Figure 3E). These up-regulated genes with increased H3K9me3 levels can account for the reduced negative correlation between H3K9me3 and the gene expression (Figure 3A, middle). Unlike H3K9me3, H3K27me3 showed no significant correlation (r = –0.22; P = 0.50), despite its increased level, for the up-regulated genes (Figure 3B, right, red regions). These data collectively showed that the three types of histone tri-methylations had the distinct alteration patterns under hypoxia. This suggests that in addition to their differential localization (Figure 2B), the differential alteration patterns of H3K4me3, H3K9me3 and H3K27me3 also contribute to the regulation of the mRNA expression under hypoxic conditions.
Relationships between changes of histone tri-methylation and mRNA expression under hypoxia
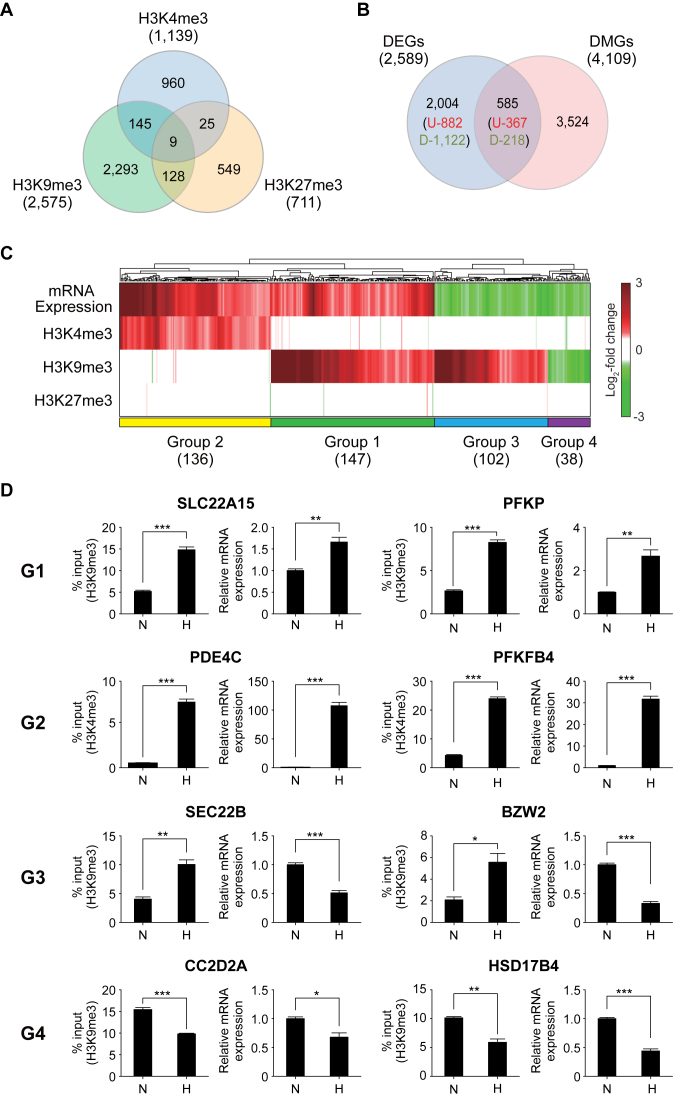
We showed above that there was the correlation between hypoxia-induced changes of histone tri-methylation and gene expression levels at the global level. Next, we examined the correlation between histone tri-methylation and mRNA expression changes at an individual gene level. To this end, we first identified H3K4me3, H3K9me3 and H3K27me3 peaks in all annotated genes by applying MACS to the ChIP-seq data (Supplementary Materials and Methods). H3K4me3, H3K9me3 and H3K27me3 peaks were found in 14,208 genes under normoxia and in 14,072 genes under hypoxia (Supplementary Figure S2A). Of these genes, we then identified 4,109 DMGs (Supplementary Table S4) that showed differences in the levels for any of the three histone tri-methylations between normoxia and hypoxia (Figure 4A; Supplementary Materials and Methods). Second, we identified 2,589 DEGs (1,149 up-regulated and 1,340 down-regulated genes) in hypoxia, compared to normoxia, using an integrative statistical method previously reported (Supplementary Table S5; Materials and Methods; Supplementary Discussion). These DEGs included three representative hypoxia marker genes (BNIP3, CA9 and VEGF) showing significant (7.1-, 28.5- and 6.4-fold, respectively) up-regulation under hypoxia compared to under normoxia (Supplementary Figure S2B). The comparison of the DMGs with the DEGs revealed that the 585 DEGs (367 up-regulated and 218 down-regulated genes) had the peaks with differential levels of H3K4me3, H3K9me3 or H3K27me3 between normoxia and hypoxia (Figure 4B; Supplementary Discussion).
Figure 4.
Relationships between changes of histone methylation and mRNA expression in hypoxia. (A) Relationships of the DMGs identified for H3K4me3, H3K9me3 and H3K27me3. The numbers in parentheses denote total numbers of DMGs identified for individual histone methylations. (B) Comparison of the DMGs and DEGs. The numbers of up- and down-regulated genes were also shown in parenthesis. (C) Four major groups (Groups 1–4) of the shared genes between the DMGs and DEGs. A heat map was generated by a hierarchical clustering of log2-fold-changes of gene expression and histone methylation levels (RPKMs) using Euclidean distance and the average linkage method. Colors in the heat map represent up- (red) and down-regulation (green) in hypoxia, compared to normoxia. Color bar shows the gradient of log2-fold-changes of gene expression and histone methylation levels. The number of the genes in each group was shown in parenthesis. (D) ChIP-PCR and RT-PCR analyses of the representative genes in Groups 1–4 confirming the differential methylation and expression in the groups. The graph shows two or more replicates with SEM. *P < 0.05, **P<0.01 and ***P < 0.001 by Students’ t-test.
To systematically explore the relationships between the differential tri-methylation and expression, we clustered the 585 shared genes between the DEGs and DMGs into 27 groups based on their differential expression and methylation patterns (Supplementary Figure S2C). Of the 27 groups, we focused on the top four major groups, Groups 1 to 4, each of which included >5% of the 585 shared genes, and Groups 1–4 included 72.3% of the 585 shared genes (Figure 4C; Supplementary Tables S4 and S5). Group 2 included the genes with increased mRNA expression and H3K4me3 levels under hypoxia, compared to normoxia (Figure 4C; Supplementary Figure S3B). Group 3 included the genes with decreased mRNA expression levels and increased H3K9me3 levels under hypoxia (Figure 4C; Supplementary Figure S3C). These two alteration patterns were consistent with archetypal roles of H3K4me3 and H3K9me3 as active and repressive marks, respectively. On the other hand, Group 1 included the genes with increased mRNA expression and H3K9me3 levels in hypoxia (Figure 4C; Supplementary Figure S3A), which is consistent with the positive correlation of log2-fold-changes between normoxia and hypoxia for up-regulated genes (Figure 3B, middle). Group 4 included the genes with decreased mRNA expression and H3K9me3 levels (Figure 4C; Supplementary Figure S3D). These two alteration patterns were contradictory to the archetypal role of H3K9me3. Interestingly, of the genes with any of the three types of histone tri-methylations, 5,767 (40.6% of 14,207) and 5,757 (40.9% of 14,071) genes had multiple types of histone tri-methylations under normoxia and hypoxia, respectively (Supplementary Figure S2A). However, the clustering analysis (Figure 4C) revealed that the DEGs in each of the major Groups 1–4 showed the changes in only one type of histone tri-methylations. Taken together, all these data suggest that the single types of the histone tri-methylations predominantly contribute to differential regulation of the gene expression in Groups 1–4 under hypoxia. Next, we confirmed the hypoxia-induced alterations of mRNA expression and histone tri-methylation (H3K4me3 and H3K9me3) for two representative genes in Groups 1–4 by using qRT-PCR and ChIP-PCR analyses (Figure 4D).
In these analyses, we hypothesized that reduced activities of JMJDs by hypoxia would be one of the main causes for the changes in histone methylation and mRNA expression. Finally, to test this hypothesis, we treated hADSCs under normoxia with JIB-04, a pan-inhibitor of the Jumonji demethylase superfamily (41–43), and then analyzed whether both histone tri-methylation and expression levels of the representative genes (Figure 4D) were changed by JIB-04 treatment, as they were by hypoxia. For this analysis, JIB-04 can serve as an appropriate JMJD inhibitor because it is known to primarily act on H3K4me3 and H3K9me3 demethylases (44). Consistently, we found that JIB-04 treatment increased total amounts of H3K4me3 and H3K9me3 (Supplementary Figure S4A; Supplementary Discussion). Both qRT-PCR and ChIP-PCR analyses showed that JIB-04 treatment changed H3K4me3 or H3K9me3 levels and expression levels of the selected genes from each group in the same direction as hypoxia, but with relatively less extents of changes to hypoxia (Supplementary Figure S4B). For example, for SLC22A15 and PFKP in G1, JIB-04 treatment increased their H3K9me3 and gene expression levels, similar to hypoxia, but with relatively smaller fold-changes than hypoxia in up-regulation of H3K9me3 and gene expression. Hypoxia showed higher increases in total amounts of H3K4me3 and H3K9me3 than JIB-4 treatment (Supplementary Figure S4A), which can explain the less changes in H3K4me3 or H3K9me3 levels and expression levels of the selected genes by JIB-04 treatment than those by hypoxia (Supplementary Figure S4B).
Differential histone tri-methylation programs for regulation of mRNA expression under hypoxia
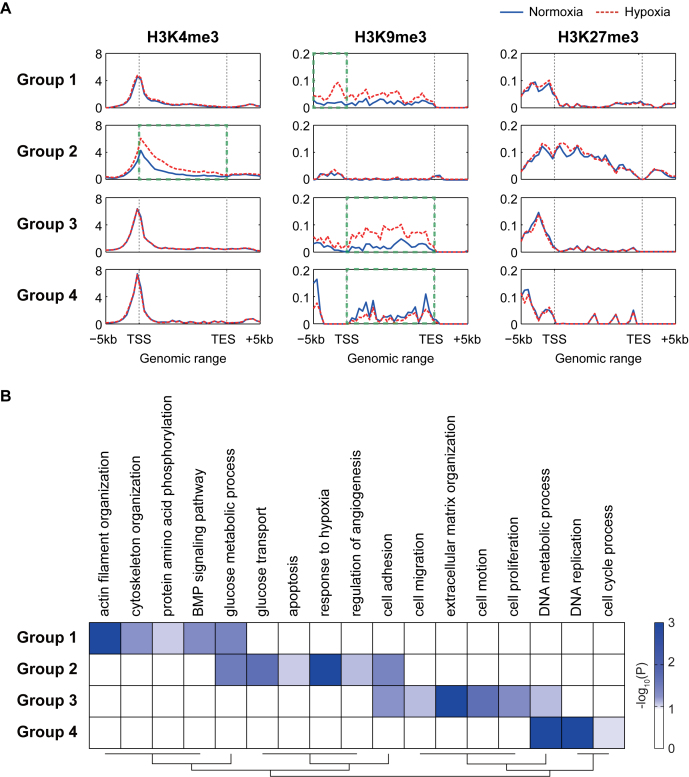
We next compared the characteristics of the four major groups (Groups 1–4) of the DEGs and DMGs. To this end, we examined whether Groups 1–4 showed different localizations of the histone methylation peaks within gene structures (Figure 5A). In Group 2, H3K4me3 was increased under hypoxia near the TSS, which is its typical location (Figure 2B), and further increased additionally across gene bodies, compared to normoxia (dotted box for H3K4me3 in Figure 5A). In the other groups, H3K4me3 was enriched near the TSS, but showed no change under hypoxia (Figure 5A). In Groups 1 and 3, H3K9me3 was increased under hypoxia. However, in Group 1, H3K9me3 was significantly increased in the promoters where H3K4me3 was relatively less enriched, whereas in Group 3, H3K9me3 was predominantly increased in gene bodies (dotted box for H3K9me3 in Figure 5A). In Group 4, H3K9me3 was decreased preferentially in gene bodies (dotted box for H3K9me3 in Figure 5A). These data indicate that in addition to the differential alteration patterns of histone tri-methylations shown in Figure 4C, Groups 1–4 were further characterized by differential localization patterns of the predominant histone tri-methylations for the groups.
Figure 5.
Differential histone methylation for the regulation of mRNA expression in hypoxia. (A) Differential locational distributions of H3K4me3, H3K9me3, and H3K27me3 for the genes in Groups 1–4 across the gene structures. For each gene, the gene structure (–5 kb-TSS-TES-+5 kb) was binned. The averaged RPKMs (read density; y-axis) for all the genes in individual bins (x-axis) were plotted. Solid and dotted lines represent profiles of read densities under normoxia and hypoxia, respectively. (B) GOBPs represented by the genes in Groups 1–4. A heat map was generated by a hierarchical clustering of –log10(P), where Pijis the enrichment P-values computed for Group i and GOBP j by DAVID (Euclidean distance and average linkage method). The color bar represents the gradient of –log10(P).
Next, we examined whether the genes in the four major groups were involved in different cellular processes. To this end, we performed the enrichment analysis of gene ontology biological processes (GOBPs) for Groups 1–4 using the DAVID software (Figure 5B and Supplementary Table S6). First, the responses to hypoxia (‘response to hypoxia’, ‘glucose transport’ and ‘regulation of angiogenesis’) were mainly represented by Group 2. Second, the processes related to cytoskeleton organization (‘actin filament organization’ and ‘cytoskeleton organization’) and cellular signaling (‘BMP signaling pathway’ and ‘protein amino acid phosphorylation’) were uniquely represented by Group 1. Third, cell migration-related processes (‘cell motion’, ‘cell migration’ and ‘ECM reorganization’) were represented mainly by Group 3. Finally, the processes related to cell cycles (‘cell cycle process’ and ‘DNA replication’) were uniquely represented by Group 4. Taken together, these data suggest that the four major groups can represent different modes of regulation of mRNA expression by histone methylation, thereby controlling different target cellular processes.
Different sets of TFs associated with regulation of mRNA expression under hypoxia.
We showed above that histone tri-methylations under hypoxia were mainly determined by activities of histone demethylases (JMJDs) (Figure 1D and Supplementary Figure S1A). However, these JMJDs cannot recognize directly DNA sequences, but are commonly recruited to nucleosomes through interactions with TFs (11,45). Thus, we examined what TFs could be involved in the recruitment of the JMJDs to the methylated sites of the genes, which could account for altered histone tri-methylations in the four major groups (Groups 1–4). To this end, we first searched for the TFs that could bind to differentially methylated regions of the genes, (i.e., DNA sequence regions covering differential H3K4me3 and H3K9me3 peaks) in Groups 1–4 using MotifLocator software based on position weight matrixes for 133 TFs (161 motifs) in JASPAR database (46). We then identified a total of 77 TFs (40, 67, 33 and 5 TFs for Groups 1–4, respectively) whose binding motifs were significantly (P<0.05) enriched in the methylated regions of the genes in Groups 1–4 (Supplementary Table S7; Materials and Methods). Of them, 43 TFs were shared in at least more than two of Groups 1–4 while 34 were uniquely identified in individual groups, suggesting that different sets of TFs could be associated with recruitment of JMJDs corresponding to differential tri-methylations in Groups 1–4 (Figure 6A).
Of these enriched TFs, we focused on 75 TF for Groups 1–3 in the following analyses because Group 4 was the smallest group with a smaller number of the enriched TFs than Groups 1–3. Among the 75 TFs, we selected the following six representative TFs based on the criteria described in Supplementary Discussion: ARNT/HIF1A, CREB1, and JUN for Group 1; ARNT/HIF1A, and E2F4 for Group 2; and RELA and CREB1 for Group 3. Enriched binding motifs of the six representative TFs for Groups 1–3 were shown in Figure 6B. To understand functional roles of the six representative TFs, we performed the GOBP enrichment analysis for their target genes in Groups 1–4 (Figure 6C) and found that these target genes also represented a majority of the cellular processes that were represented by Groups 1–4 (Figure 5B): cytoskeleton and extracellular matrix organization, glucose metabolic process, apoptosis, response to hypoxia, regulation of angiogenesis, and cell adhesion. To examine collective associations of the six TFs with these processes, we constructed a network model describing interactions among their target genes (Figure 6D). The network model showed that the six representative TFs mainly contributed to regulation of actin cytoskeleton (ARHGEF5, RAPGEF2, BCAR1, GSN, RUSC2 and ARAP3) together with its upstream (COL4A2/6A2/7A1/8A2, MSANTD3, VWA1, COMP1, CAV2, ADORA2B and LUM in extracellular matrix organization) and downstream molecules (TRADD, CDIP1, DIDO1, and INHBB in apoptotic signaling pathway and APLN, MARCKSL1, PTHIR, NUAK1, and HNRNPA3 in cell proliferation) (Figure 6D, left). Moreover, the network model also showed that the six TFs contributed to HIF-1α pathway (MUC1, EGLN3, BNIP3, MT3 and STC1/2 in HIF-1α pathway and ANG, ANGPTL4, FGF11, VEGFA and IL6/11 in angiogenic factors/adipocytokines), as well as regulation of glucose transport (SLC2A1/3/5) and metabolism (PGM1, PFKL, PFKP, ALDOC, ENO2 and PKM) (Figure 6D, right). These data suggest that different sets of the TFs associated with Groups 1–3 could collectively contributed with the key cellular processes (actin cytoskeleton, HIF-1α pathway, and glucose metabolism) altered by hypoxia.
We then experimentally tested whether the six representative TFs bound to a set of the target genes in the network model. It is well-known that hypoxia up-regulates the genes in glucose metabolism (47). Thus, we focused on the following target genes involved in regulation of actin cytoskeleton and HIF-1α pathway in Groups 1–3 using chromatin immunoprecipitation (ChIP)-PCR experiments (Figure 6E; Supplementary Figure S5): MSANTD3, RUSC2 and NUAK (Group 1), INHBB (Group 2), and LUM, NABP1 and HNRNPA3 (Group 3) in actin cytoskeleton regulation; and MEF2D (Group 1), LEP, MT3, and STC1 (Group 2), and SLIT2 (Group 3) in HIF-1α pathway. ChIP-PCR data confirmed that the six representative TFs bound to the differentially methylated regions of these target genes in Groups 1–3 (Figure 6E). Interestingly, however, several of the six representative TFs showed different binding strengths to the target genes between normoxia and hypoxia. ARNT and HIF1A showed higher binding strengths in hypoxia, compared to normoxia, whereas CREB1 showed weaker binding strengths to Group 3 target genes in hypoxia. On the other hand, CREB1, E2F4 and RELA showed no significant difference in binding strength to their target genes in Groups 1–3, respectively, while JUN showed a mixed pattern for MEF2D and MSANTD3 with no significant difference and higher binding strength in hypoxia, respectively (see DISCUSSION).
Next, to examine the effects of these TF bindings on gene expression and histone tri-methylation of the selected target genes, we first generated hADSCs with knockdown of HIF1A or CREB1 (Supplementary Discussion) by infecting hADSCs with a lentivirus encoding shRNAs against HIF1A or CREB1. We then examined whether changes in H3K4me3 or H3K9me3 levels and expression levels of the selected target genes were affected by knockdown of HIF1A or CREB1. Knockdowns decreased mRNA expression levels of HIF1A or CREB1 by 91.22% and 84.8% in hADSCs (Supplementary Figure S6A). Knockdowns significantly (P < 0.001) reduced hypoxia-induced changes of the selected target genes of HIF1A or CREB1 in their H3K4me3 or H3K9me3 levels and expression levels (Supplementary Figure S6B). For example, HIF1A knockdown significantly (P < 0.01) reduced hypoxia-induced increases of both H3K4me3 and mRNA expression levels for MT3 and STC1, target genes of HIF1A in Group 2. CREB1 knockdowns led to similar reductions for MEF2D and RUSC2 in Group 1 and HNRNPA3 and LUM in Group 3. These data suggest that the representative TFs could be involved in the recruitments of the JMJDs to differentially methylated regions of the target genes, contributing to regulation of histone methylation and gene expression of target genes in Groups 1–3 and their associated cellular processes (actin cytoskeleton and HIF-1α pathway). In summary, together with different types of histone methylations (Figure 4C) and differential localization patterns of histone methylations (Figure 5A), different sets of TFs involved in recruitment of JMJDs, can collectively contribute to regulation of the differential cellular processes associated with Groups 1–4 (Figure 5B).
DISCUSSION
How the multi-dimensional histone methylation contributes to the regulation of the gene expression remains largely elusive. In this study, we investigated how the three types of histone tri-methylations (H3K4me3, H3K9me3, and H3K27me3) were linked to the alteration patterns in mRNA expression under hypoxia. Previous studies have shown the coexistence of active H3K4me3 and repressive H3K27me3 in the genes involved in the development of mammalian embryonic stem cells (48–50) and adult stem/progenitor cells (40,51). These multiple histone modifications have been suggested to poise genes for fast induction during the differentiation into different cell lineages. In our study, many of the genes were also shown to have multiple histone modifications under normoxia and hypoxia (Supplementary Figure S2A). Thus, combinatorial alterations of the multiple histone methylations were expected to be associated with the DEGs under hypoxia. However, our integrative analysis of the ChIP-seq and the gene expression data revealed that the DEGs were categorized into the four major groups based on changes in three types of histone tri-methylations, H3K4me3, H3K9me3, H3K27me3, and further showed that the DEGs in each group were associated with changes in one prominent type of the histone tri-methylations (Figure 4C), although many of these genes had multiple histone methylations under hypoxic conditions (Supplementary Figure S2A).
A number of previous studies have examined the relationships between the levels of histone methylation and the gene expression (40,52). However, only a few studies have investigated the relationships between the changes in histone methylation and gene expression levels between two different conditions (e.g. normoxia and hypoxia in this study). For example, Papait et al. (53) analyzed genome-wide changes in seven types of histone modifications (H3K4/9/27me3, H3K9/72me2 and H3K9/27ac) for the genes regulated during cardiac hypertrophy. They clustered up- and down-regulated genes during cardiac hypertrophy based on alteration patterns of the histone modifications. Consistent to our findings, they also found that the up- or down-regulated genes could be further clustered into several subgroups, and each subgroup showed the changes predominantly in only one type of histone modifications.
Moreover, Cui et al. (40) analyzed genome-wide changes in eight types of histone methylations (H3K4/9/20/27me1 and H3K4/9/27/36me3) for the genes regulated during differentiation of multipotent human primary hematopoietic stem cells/progenitor cells into erythrocyte precursors. They performed no clustering analysis of the up- and down-regulated genes, but they examined the correlations between the changes in histone modification and gene expression levels. Consistent to our findings, they also found that the positive and negative correlations of active and repressive methylations between histone methylation and gene expression levels were more apparent for the genes with high mRNA expression levels, as described in Figure 3A. They also found that there was also a positive correlation of H3K9me3 between the changes of histone methylation and gene expression levels during the differentiation for up-regulated genes in differentiated cells, as described in Figure 3B (middle).
The four major groups of the DEGs showed distinct alteration patterns in histone methylation and mRNA expression (Figure 4C). Groups 2 and 3 corresponded to the archetypal relationships between histone methylation and transcriptional induction/repression (i.e. increased H3K4me3 and H3K9me3 to up- and down-regulation of mRNA expression, respectively). On the contrary, Groups 1 and 4 were not consistent with the archetypal relationships. However, our comparative analysis of Groups 1 and 3 (Figure 5A) suggested that the localization of H3K9me3 could determine the up- or down-regulation of the target genes, depending on whether H3K9me3 was enriched in the promoter for up-regulation or the gene body for down-regulation. Previously, Wiencke et al. (54) compared the genes with and without H3K9me3 in their promoters and found that H3K9me3 could be associated with active genes, rather than being a repressive mark, consistent to our findings. Therefore, our findings of the location-dependent associations of H3K9me3 with mRNA expression provide a model that can explain the non-archetypal relationship between H3K9me3 and mRNA expression.
The four major groups of the DEGs showed that H3K4me3 and H3K9me3 were exclusively associated with different sets of the DEGs (Figure 4C): for example, Group 2 predominantly with H3K4me3, but minimally with H3K9me3; and Groups 1 and 3 predominantly with H3K9me3, but insignificantly with H3K4me3. In support of this finding, previous studies showed that H3K9me3 demethylases formed complexes with H3K4 methyltransferases and vice versa. JMJD2B, an H3K9me3 demethylase, interacted with MLL2, a H3K4 methyltransferase in MCF-7 cells, but with no other histone demethylases (55). Also, JARID1A, an H3K4me3 demethylase, formed a complex with G9A, an H3K9 methyltransferase, in murine erythroid cells (56,57). Both G9A and JARID1A repressed Eygene by increasing simultaneously both methylation of H3K9me3 and demethylation of H3K4me3 (57). Knockdown of JARID1A led to derepression of Ey gene by increasing H3K4me3 with no changes of H3K9me3. Hypoxia would decrease the activity of JARID1A in the G9A-JARID1A, similar to knockdown of JARID1A, eventually increasing H3K4me3. This observation is consistent to the mutually exclusive association of H3K4me3 and H3K9me3 and further to the predominant association of the DEGs in Group 2 with H3K4me3 (Figure 4C). Similarly, for other complexes of H3K9me3 demethylase-H3K4 methyltransferase, such as JMJD2B-MLL2, hypoxia can result in the predominant association of the DEGs in Groups 1 and 3 with H3K9me3 (Figure 4C). Thus, these data may explain our finding for the association between gene expression and predominant single-type histone methylation (55).
Several pieces of evidence showed that histone demethylases are recruited by TFs to their target genes. By co-IP experiments, Jin et al. (2014) showed that RELA physically interacted with JMJD2A that is important to engagement of JMJD2A to the promoter of IFNB1, a RELA target gene, in bone marrow-derived macrophages (58). Drosophila c-Jun homolog, Jra recruited JMJD2A/heterochromatin protein1 (HP-1a) complex to gene body of target genes of Jra through its interaction with HP-1a (59). In VHL-deficient clear-cell renal cell carcinoma, a subset of HIF1 target genes such as IGFBP3, DNAJC12, COL6A1, GDF15 are repressed because in this carcinoma, constitutive HIF1A/Arnt heterodimer recruited JARID1C on these target genes, leading to reduction of H3K4me3. (60). Similarly, ChIP-reChIP analysis of squamous cell carcinoma revealed that JMJD2A was recruited to several target genes of JUN/FOS (61). Several studies showed that JARID1A and B were recruited to the E2F target genes to repress their expression in senescent cells through their interactions with E2F/RB1 complex (62). Our TF binding assays showed similar and different binding strengths of the TFs to their target genes between normoxia and hypoxia (Figure 6E). Hypoxia activates signaling cascades that activate downstream TFs. Such TFs can be involved in the recruitment of histone demethylase (signaling control). On the other hand, hypoxia also could decrease activity of histone demethylases due to the decreased amount of O2 substrate, thereby affecting histone methylation (metabolic control). These data suggest that the TFs showing similar binding strengths to their target genes under hypoxia might be under the metabolic control, whereas the TFs showing different binding strengths might be under the signaling control. Based on this notion, the types of TFs, together with the association between gene expression and prominent single-type histone methylation, can further characterize whether the single-type histone methylation can be determined by environmental signaling or metabolic states associated with amounts of metabolites (O2, α-KG, and/or vitamin C), which can affect activities of histone demethylases.
Many pieces of evidence (63–65) suggest that aberrant gene expression and histone methylation patterns are associated with hypoxia-related diseases, such as cancer and cardiomyopathy. Our model for the association between gene expression and prominent single-type histone tri-methylation can be used to understand the relationships between altered gene expression and histone methylation patterns in the hypoxia-related diseases. Beside of gene expression, histone methylations also involve in regulating DNA replication. Recently, Wu et al. proposed a novel function of JMJD2D to facilitate DNA replication by reducing H3K9me3 (66), suggesting that hypoxic increase of H3K9me3 can influence DNA replication. Histone demethylases that play important roles in association with gene expression as well as with DNA replication can contribute to pathogenesis of hypoxia-related disease (67,68). Thus, our model can help facilitate the identification of such important histone demethylases as potential therapeutic targets in the hypoxia-related diseases.
DATA AVAILABILITY
The data from this study is provided in Additional files. In addition, the microarray datasets are available from GEO under listed accession numbers [GSE69495]. The raw ChIP-seq data were deposited to the GEO database under listed accession numbers [GSE69493].
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea [NRF-2012M3A9B6055343, 2016R1A2B4012840, 2013M3A9D3046248, 2014M3C7A1046047]; Institute for Basic Science [IBS-R013-G1-2016-a00] funded by the Korean Ministry of Education, Science and Technology. Funding for open access charge: National Research Foundation of Korea [NRF-2012M3A9B6055343, 2016R1A2B4012840, 2013M3A9D3046248, 2014M3C7A1046047]; Institute for Basic Science [IBS-R013-G1-2016-a00].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gunaratnam L., Bonventre J.V.. HIF in kidney disease and development. J. Am. Soc. Nephrol.: JASN. 2009; 20:1877–1887. [DOI] [PubMed] [Google Scholar]
- 2. Mohyeldin A., Garzon-Muvdi T., Quinones-Hinojosa A.. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010; 7:150–161. [DOI] [PubMed] [Google Scholar]
- 3. Semenza G.L. HIF-1, O(2), and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell. 2001; 107:1–3. [DOI] [PubMed] [Google Scholar]
- 4. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J.M., Lane W.S., Kaelin W.G. Jr. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001; 292:464–468. [DOI] [PubMed] [Google Scholar]
- 5. Jaakkola P., Mole D.R., Tian Y.M., Wilson M.I., Gielbert J., Gaskell S.J., Kriegsheim A., Hebestreit H.F., Mukherji M., Schofield C.J. et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001; 292:468–472. [DOI] [PubMed] [Google Scholar]
- 6. Tausendschon M., Dehne N., Brune B.. Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity. Cytokine. 2011; 53:256–262. [DOI] [PubMed] [Google Scholar]
- 7. Zhou X., Sun H., Chen H., Zavadil J., Kluz T., Arita A., Costa M.. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010; 70:4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seward D.J., Cubberley G., Kim S., Schonewald M., Zhang L., Tripet B., Bentley D.L.. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat. Struct. Mol. Biol. 2007; 14:240–242. [DOI] [PubMed] [Google Scholar]
- 9. Whetstine J.R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M. et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006; 125:467–481. [DOI] [PubMed] [Google Scholar]
- 10. Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K.. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007; 449:731–734. [DOI] [PubMed] [Google Scholar]
- 11. Ptashne M. Epigenetics: core misconcept. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7101–7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Slattery M., Zhou T., Yang L., Dantas Machado A.C., Gordan R., Rohs R.. Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci. 2014; 39:381–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi Y., Whetstine J.R.. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell. 2007; 25:1–14. [DOI] [PubMed] [Google Scholar]
- 14. Li B., Carey M., Workman J.L.. The role of chromatin during transcription. Cell. 2007; 128:707–719. [DOI] [PubMed] [Google Scholar]
- 15. Mendenhall E.M., Bernstein B.E.. Chromatin state maps: new technologies, new insights. Curr. Opin. Genet. Dev. 2008; 18:109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M. et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006; 8:532–538. [DOI] [PubMed] [Google Scholar]
- 17. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou Y., Kim J., Yuan X., Braun T.. Epigenetic modifications of stem cells: a paradigm for the control of cardiac progenitor cells. Circ. Res. 2011; 109:1067–1081. [DOI] [PubMed] [Google Scholar]
- 19. Su X., Wellen K.E., Rabinowitz J.D.. Metabolic control of methylation and acetylation. Curr. Opin. Chem. Biol. 2016; 30:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hancock R.L., Dunne K., Walport L.J., Flashman E., Kawamura A.. Epigenetic regulation by histone demethylases in hypoxia. Epigenomics. 2015; 7:791–811. [DOI] [PubMed] [Google Scholar]
- 21. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011; 17:10–12. [Google Scholar]
- 22. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S. et al. Ensembl 2013. Nucleic Acids Res. 2013; 41:D48–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H. et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014; 42:D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thijs G., Moreau Y., De Smet F., Mathys J., Lescot M., Rombauts S., Rouze P., De Moor B., Marchal K.. INCLUSive: integrated clustering, upstream sequence retrieval and motif sampling. Bioinformatics. 2002; 18:331–332. [DOI] [PubMed] [Google Scholar]
- 27. Bowman A.W., Azzalini A.. Applied Smoothing Techniques for Data Analysis: The Kernel Approach with S-Plus Illustrations. 1997; Oxford; NY: Clarendon Press; Oxford University Press. [Google Scholar]
- 28. Storey J.D., Tibshirani R.. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bader G.D., Betel D., Hogue C.W.. BIND: the biomolecular interaction network database. Nucleic Acids Res. 2003; 31:248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peri S., Navarro J.D., Kristiansen T.Z., Amanchy R., Surendranath V., Muthusamy B., Gandhi T.K.B., Chandrika K.N., Deshpande N., Suresh S. et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004; 32:D497–D501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M.. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006; 34:D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chatr-Aryamontri A., Ceol A., Palazzi L.M., Nardelli G., Schneider M.V., Castagnoli L., Cesareni G.. MINT: the molecular INTeraction database. Nucleic Acids Res. 2007; 35:D572–D574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cline M.S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B. et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007; 2:2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M.. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012; 40:D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beyer S., Kristensen M.M., Jensen K.S., Johansen J.V., Staller P.. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 2008; 283:36542–36552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee H.Y., Yang E.G., Park H.. Hypoxia enhances the expression of prostate-specific antigen by modifying the quantity and catalytic activity of Jumonji C domain-containing histone demethylases. Carcinogenesis. 2013; 34:2706–2715. [DOI] [PubMed] [Google Scholar]
- 38. Kaelin W.G. Jr, McKnight S.L.. Influence of metabolism on epigenetics and disease. Cell. 2013; 153:56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kooistra S.M., Helin K.. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012; 13:297–311. [DOI] [PubMed] [Google Scholar]
- 40. Cui K., Zang C., Roh T.Y., Schones D.E., Childs R.W., Peng W., Zhao K.. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009; 4:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Woon E.C., Tumber A., Kawamura A., Hillringhaus L., Ge W., Rose N.R., Ma J.H., Chan M.C., Walport L.J., Che K.H. et al. Linking of 2-oxoglutarate and substrate binding sites enables potent and highly selective inhibition of JmjC histone demethylases. Angew. Chem. Int. Ed. Engl. 2012; 51:1631–1634. [DOI] [PubMed] [Google Scholar]
- 42. Cascella B., Lee S.G., Singh S., Jez J.M., Mirica L.M.. The small molecule JIB-04 disrupts O2 binding in the Fe-dependent histone demethylase KDM4A/JMJD2A. Chem. Commun. (Camb.). 2017; 53:2174–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leurs U., Lohse B., Rand K.D., Ming S., Riise E.S., Cole P.A., Kristensen J.L., Clausen R.P.. Substrate- and cofactor-independent inhibition of histone demethylase KDM4C. ACS Chem. Biol. 2014; 9:2131–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang L., Chang J., Varghese D., Dellinger M., Kumar S., Best A.M., Ruiz J., Bruick R., Pena-Llopis S., Xu J. et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat. Commun. 2013; 4:2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Benveniste D., Sonntag H.J., Sanguinetti G., Sproul D.. Transcription factor binding predicts histone modifications in human cell lines. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:13367–13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakazawa M.S., Keith B., Simon M.C.. Oxygen availability and metabolic adaptations. Nat. Rev. Cancer. 2016; 16:663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 49. Pan G., Tian S., Nie J., Yang C., Ruotti V., Wei H., Jonsdottir G.A., Stewart R., Thomson J.A.. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007; 1:299–312. [DOI] [PubMed] [Google Scholar]
- 50. Zhao X.D., Han X., Chew J.L., Liu J., Chiu K.P., Choo A., Orlov Y.L., Sung W.K., Shahab A., Kuznetsov V.A. et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007; 1:286–298. [DOI] [PubMed] [Google Scholar]
- 51. Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., Cui K., Kanno Y., Roh T.Y., Watford W.T. et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009; 30:155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee B.M., Mahadevan L.C.. Stability of histone modifications across mammalian genomes: implications for ‘epigenetic’ marking. J. Cell. Biochem. 2009; 108:22–34. [DOI] [PubMed] [Google Scholar]
- 53. Papait R., Cattaneo P., Kunderfranco P., Greco C., Carullo P., Guffanti A., Vigano V., Stirparo G.G., Latronico M.V., Hasenfuss G. et al. Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:20164–20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wiencke J.K., Zheng S., Morrison Z., Yeh R.F.. Differentially expressed genes are marked by histone 3 lysine 9 trimethylation in human cancer cells. Oncogene. 2008; 27:2412–2421. [DOI] [PubMed] [Google Scholar]
- 55. Shi L., Sun L.Y., Li Q., Liang J., Yu W.H., Yi X., Yang X.H., Li Y.Y., Han X., Zhang Y. et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:7541–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li F., Huarte M., Zaratiegui M., Vaughn M.W., Shi Y., Martienssen R., Cande W.Z.. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008; 135:272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chaturvedi C.P., Somasundaram B., Singh K., Carpenedo R.L., Stanford W.L., Dilworth F.J., Brand M.. Maintenance of gene silencing by the coordinate action of the H3K9 methyltransferase G9a/KMT1C and the H3K4 demethylase Jarid1a/KDM5A. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:18845–18850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jin J., Hu H., Li H.S., Yu J., Xiao Y., Brittain G.C., Zou Q., Cheng X., Mallette F.A., Watowich S.S. et al. Noncanonical NF-kappaB pathway controls the production of type I interferons in antiviral innate immunity. Immunity. 2014; 40:342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Y., Zhang D.. HP1a/KDM4A is involved in the autoregulatory loop of the oncogene gene c-Jun. Epigenetics. 2015; 10:453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Niu X., Zhang T., Liao L., Zhou L., Lindner D.J., Zhou M., Rini B., Yan Q., Yang H.. The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C. Oncogene. 2012; 31:776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ding X., Pan H., Li J., Zhong Q., Chen X., Dry S.M., Wang C.Y.. Epigenetic activation of AP1 promotes squamous cell carcinoma metastasis. Sci. Signal. 2013; 6, doi:10.1126/scisignal.2003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chicas A., Kapoor A., Wang X., Aksoy O., Evertts A.G., Zhang M.Q., Garcia B.A., Bernstein E., Lowe S.W.. H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:8971–8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cascella B., Mirica L.M.. Kinetic analysis of iron-dependent histone demethylases: alpha-ketoglutarate substrate inhibition and potential relevance to the regulation of histone demethylation in cancer cells. Biochemistry. 2012; 51:8699–8701. [DOI] [PubMed] [Google Scholar]
- 64. Dunwoodie S.L. The role of hypoxia in development of the mammalian embryo. Dev. Cell. 2009; 17:755–773. [DOI] [PubMed] [Google Scholar]
- 65. Luo W.B., Chang R., Zhong J., Pandey A., Semenza G.L.. Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E3367–E3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu R., Wang Z., Zhang H., Gan H., Zhang Z.. H3K9me3 demethylase Kdm4d facilitates the formation of pre-initiative complex and regulates DNA replication. Nucleic Acids Res. 2017; 45:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hohl M., Wagner M., Reil J.C., Muller S.A., Tauchnitz M., Zimmer A.M., Lehmann L.H., Thiel G., Bohm M., Backs J. et al. HDAC4 controls histone methylation in response to elevated cardiac load. J. Clin. Invest. 2013; 123:1359–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hojfeldt J.W., Agger K., Helin K.. Histone lysine demethylases as targets for anticancer therapy. Nat. Rev. Drug Discov. 2013; 12:917–930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data from this study is provided in Additional files. In addition, the microarray datasets are available from GEO under listed accession numbers [GSE69495]. The raw ChIP-seq data were deposited to the GEO database under listed accession numbers [GSE69493].