Abstract
In the human genome, translation initiation from non-AUG codons plays an important role in various gene regulation programs. However, mechanisms regulating the non-AUG initiation rate remain poorly understood. Here, we show that the non-AUG initiation rate is nearly consistent under a fixed nucleotide context in various human and insect cells. Yet, it ranges from <1% to nearly 100% compared to AUG translation, depending on surrounding sequences, including Kozak, and possibly additional nucleotide contexts. Mechanistically, this range of non-AUG initiation is controlled in part, by the eIF5-mimic protein (5MP). 5MP represses non-AUG translation by competing with eIF5 for the Met-tRNAi-binding factor eIF2. Consistently, eIF5 increases, whereas 5MP decreases translation of NAT1/EIF4G2/DAP5, whose sole start codon is GUG. By modulating eIF5 and 5MP1 expression in combination with ribosome profiling we identified a handful of previously unknown non-AUG initiation sites, some of which serve as the exclusive start codons. If the initiation rate for these codons is low, then an AUG-initiated downstream ORF prevents the generation of shorter, AUG-initiated isoforms. We propose that the homeostasis of the non-AUG translatome is maintained through balanced expression of eIF5 and 5MP.
For eukaryotic translation initiation to proceed, the cap-binding complex eIF4F must bind to m7G-capped mRNAs and recruit them to the 40S small ribosomal subunit (SSU) (1). Prior to this event, the 40S SSU is activated into an open, scanning-competent form through eukaryotic initiation factors bound to Met-tRNAiMet, allowing formation of the 43S ribosome pre-initiation complex (PIC) (2). The 43S PIC attachment to the mRNA 5′-terminal region generates the 48S complex, and the PIC subsequently scans for a start codon, upon recognition of which the large ribosomal subunit joins the 40S SSU thereby forming an elongation-competent ribosome—the 80S initiation complex [for review, see (3–5)]. eIF5 is a crucial component of the multi-initiation factor complex (MFC) involved in the SSU activation, where its GTPase activating protein (GAP) function promotes start codon selection by the 48S complex (6–8).
Immediately after the 43S PIC is loaded onto the 5′-terminal region of capped mRNA, start codon recognition is prevented, except at AUG codons bearing specific cis elements, termed TISU, which are enriched on mRNAs encoding proteins having mitochondrial function (9,10). Subsequently, the efficiency of the PIC recognition of the start codon, AUG, is influenced by a Kozak consensus sequence ([A/G]xxAUGG) in mammals (11) or a similar initiation context (AA[A/G]AUG) in fungi including yeast (12). Despite the stringent AUG selection mechanism, translation of at least 57 genes, including NAT1/EIF4G2/DAP5 encoding an eIF4G-like translational regulatory protein (see below) and the oncogene cMYC, is initiated at non-AUG codons (13) (For review, see (5)). Ribosome profiling studies also suggest that non-AUG initiation occurs more prevalently in mouse embryonic stem cells (mESC) or pre-malignant stage of cancer cells (14,15). Recent work suggests that a non-canonical mechanism involving eIF2A and Leu-tRNAi can specifically enhance CUG initiation (15,16). In contrast, perturbation of the canonical initiation machinery, e.g. by elevation of eIF5 levels above a certain threshold, can also enhance initiation from near cognate start codons, including CUG or GUG codons (17,18). It remains unclear whether these complementary mechanisms cross-talk to modulate the rate of non-AUG initiation.
Translational control can be achieved by the expression or modification of translational regulatory proteins mimicking initiation factors (10,19–21). For example, the NAT1 gene, whose translation is initiated at GUG, encodes a protein related to the eIF4G subunit of eIF4F, but lacks the binding site for the cap-binding subunit eIF4E. Thus, its product is proposed to be involved in cap-independent translation (22). NAT1 is also required for differentiation of human embryonic stem cells and promotes translation of mRNAs coding for mitochondrial proteins and the chromatin modifier HMGN3 (23). Likewise in mESC, it promotes translation of genes encoding proteins important for stem cell differentiation (24). It remains to be determined how its GUG initiation rate is maintained at a high level during these processes and whether it is differentially regulated before and after differentiation.
In this work, we examine another model whereby non-canonical initiation sites are regulated. Here, we study a translation regulatory protein, termed eIF5-mimic protein (5MP), that acts as a translational rheostat, thereby increasing the accuracy of translation initiation by impeding eIF5-dependent translation from non-AUG codons. 5MP bears homology to the C-terminal half of eIF4G and NAT1 and also contains the W2-type HEAT domain. Similar to eIF5, the HEAT domain is capable of binding the Met-tRNAi-binding factor eIF2 and the ribosome-binding factor eIF3 (21), major MFC components (8). However, unlike eIF5, 5MP lacks the GAP function and thus acts as a general inhibitor of translation (21). 5MP is found in most eukaryotes excluding nematodes, yeasts (ascomycetes) and some protozoans (25). Humans encode 5MP1 and a paralog, 5MP2, also known as BZW2 and BZW1, respectively. Importantly, 5MP1 and 5MP2 were reported to promote the tumor growth of salivary mucoepidermoid carcinoma (26) and fibrosarcoma (27), respectively. Their pro-oncogenic properties have been associated with their ability to induce translation of the transcription factor ATF4 through a unique mechansism whereby re-initiation is delayed dependent on upstream ORFs (uORFs) (27) (for a review on uORFs see (28)). Here, we show that non-AUG start codons may be initiated as strongly as canonical AUG codons, depending on specific sequence contexts, including the Kozak consensus. Using eIF5 and 5MP1 as a tool, our ribosome profiling studies identify new non-AUG initiation sites located upstream of the primary start codons. Based on these findings, and cancer genomics databases, we propose a model whereby the interplay between eIF5 and 5MP dictates the levels of non-AUG translation, and discuss the potential impact of this mechanism in neoplasia.
EXPERIMENTAL PROCEDURES
Materials
Construction of plasmids used for yeast, insect, and human cell reporter assays (Supplementary Tables S1 and S2) and preparation of GFP mRNA derivatives (Supplementary Table S3) were described in Supplemental Methods. Yeast translation initiation components used in the reconstitution assays were prepared as described (29). Purification of yeast eIF5 and eIF5-CTD (30) and of human 5MP1 (25) was described previously. Human cell lines HEK293T (ATCC; Rockville, MD, USA), fibrosarcoma HT1080 (ATCC), 293FT (Invitrogen), and iPS cell line 201B7 (a gift of Dr. Makoto Nakagawa, CiRA, Kyoto, Japan) and fly S2 cells (a gift of Dr. Jocelyn MacDonald, Biology, KSU) were grown as described in Supplemental Methods.
Yeast phenotyic and biochemical assays
Yeast phenotypic assays, including histidine auxotrophic assay and β-galactosidase assay, were performed as described previously (31) and described in detail in Supplmental Methods. Measurement of the affinity and kinetics of yeast eIF2-TC binding to yeast 40S was carried out with a native gel assay as described (32). GTP hydrolysis experiments were performed as described (33).
Luciferase assay
Approximately 80–90% confluent HEK293T cells in 75 μl medium loaded on a 96-well assay plate were transfected with 250 ng of plasmid DNA mixture using 0.25 μg PEI (34). For checking the reporter translation under different initiation contexts, 5:1 or 25:1 mixture of each reporter DNA and pSV40 Renilla Kozak AUG (Supplementary Table S1) was employed and the transfection was done in triplicate. To examine the effect of eIF5 or 5MP expression, we transfected cells in duplicate with the 1:5 mixture of the firefly versus Renilla reporter plasmid mixture (5:1) and a pEF1A- derivative plasmid DNA. On the next day (Day 2), firefly and Renilla luciferase activities were measured with the Dual Glo reagents (Promega) using Victor 3 Plate Reader (Perkin Elmer). The ratio of average firefly to Renilla activities was used as a specific ATF4 activity. To check the efficiency of transfection each set of transfection included an experiment with a plasmid expressing GFP. Typically, the efficiency is 50–70% for HEK293T. The luciferase activities from HT1080 were assayed similarly, except using XfectTM for transfection. The typical efficiency of HT1080 transfection measured with a GFP plasmid was 70–80%.
For measurement in Drosophila melanogaster, S2 cells were grown to several layers in 200 μl Schneider's medium on a 48-well plate and transfected with 1 μg of 5:1 mixture of a pAC-firefly derivative and pAc5.1C-RLuc-V5His6, and 2 μg PEI, all of which were pre-mixed in a 40 μl NaCl solution, exactly as described for HEK293T (34). After 2–5 days of transfection, 20–30 μl of the cells in the medium was withdrawn for Dual Glo assay (Promega). Transfection of a GFP-expressing plasmid yielded ∼5–10% of glowing cells under the experimental conditions described here.
GFP mRNA transfection assay
Equal amounts (20 ng) of mRNAs coding for EGFP or iRFP670 were co-transfected by using Stemfect (Stemgent) to cells, which were seeded in a 96-well plate at 2 × 104 cells/well on the day before the day of transfection. Fluorescent images of the transfected cells were captured on the RS100 automated imaging system (Olympus) on 1 day after transfection. Then the cells were washed by PBS once and treated with Accumax (Innovative Cell Technologies) at 37°C for 10 min. The detached cells were passed through a mesh and analyzed by Accuri C6 using FL1 (533/30 nm) and FL4 (675/25 nm) for EGFP and iRFP670, respectively. Flow cytometry data were analyzed using R with flowCore packages (Ellis B, Haaland P, Hahne F, Le Meur N, Gopalakrishnan N, Spidlen J and Jiang M (2017). flowCore: flowCore: Basic structures for flow cytometry data. R package version 1.40.6.). Live and iRFP670 positive cells were gated, then median of ratio of EGFP/iRFP670 of individual cells was calculated and defined as translational efficiency.
Ribosome profiling
HEK293T was transfected with 1:1 mixture of pEF1A-heIF5, pEF1A-h5MP1 (25), and their vector control or with the vector control only. After treatment with lactimidomycin (LTM) or cycloheximide (CHX), cells were subjected to ribosome profiling as described previously (35). The ribosome profiling data was deposited under the accession number GSE102786.
RESULTS
5MP increases the accuracy of translation initiation by counteracting eIF5
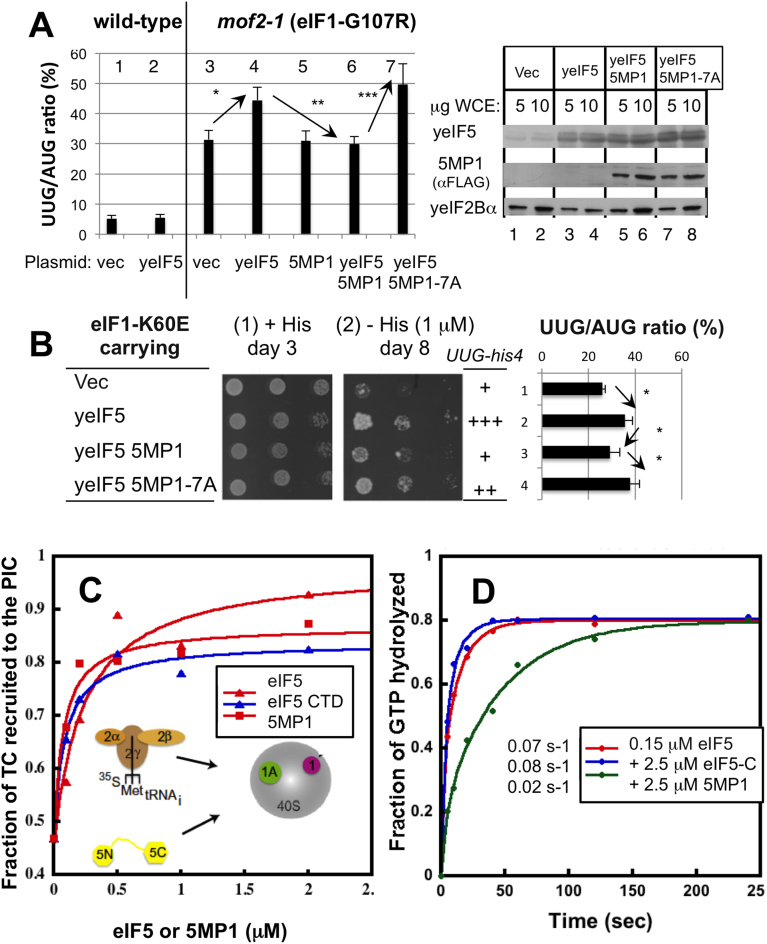
Yeast does not express 5MP and therefore provides a powerful tool to study the functions of 5MP in translation regulation (21,25). eIF5 expression enhances UUG initiation caused by yeast eIF1 mutants (Suppressor of initiation codon mutations, or Sui− phenotype) (36). In addition, 5MP1 and eIF5 compete for eIF2 binding (21). We therefore hypothesized that 5MP1 may increase the accuracy of translation initiation by competing with eIF5 for eIF2 present in the PIC. To test this model, we constructed a vector to co-express yeast eIF5 and human 5MP1, as well as a vector to express eIF5 or 5MP1 alone in high copy (hc), which were used as controls (Figure 1A, right). The yeast mof2-1 mutant altering eIF1-G107 to R (37) was double-transformed with the hc expression vector and a reporter plasmid (AUG-lacZ or UUG-lacZ) and assayed for β-gatactosidase activity. The UUG initiation was estimated by the ratio of β-galactosidase activities from UUG-lacZ versus AUG-lacZ transformants (Figure 1A and B, graphs). hc eIF5 exacerbated the UUG initiation that was elevated by mof2-1 (Figure 1A, columns 1–4), as reported previously (36). Co-expression of eIF5 and 5MP1 decreased the UUG initiation compared to yeast transformed with hc eIF5 alone (Figure 1A; columns 4 and 6). This indicates that human 5MP1 can antagonize eIF5-induced UUG mis-initiation. We next employed a yeast strain harboring eIF1 K60E, which strongly impairs 40S binding in vitro and causes Sui− phenotype in vivo: The strong Sui− phenotype allows the mutant yeast with UUG-his4 allele to grow independent of histidine (38). Thus, the eIF1-K60E transformant carrying vector control grew in the trace amount (1 μM) of histidine (Figure 1B, panel 2, row 1), signifying the occurrence of UUG mis-initiation. Accelerated growth in the trace concentration of histidine was observed in the eIF1-K60E transformant carrying hc eIF5, which was diminished by the co-expression of 5MP1 (Figure 1B, panel 2, rows 2 and 3). Relative to expression of eIF5 alone, 5MP1 co-expression therefore reduces the UUG/AUG initiation ratio (Graph in Figure 1B, rows 2 and 3). Collectively these findings demonstrate that 5MP impedes eIF5-dependent non-AUG initiation independent of genetic background of the yeast or assay used to verify Sui− phenotype. Importantly, the 5MP1 mutant which is defective in eIF2 binding [7A; (21)] failed to suppress eIF5-dependent non-AUG initiation (Figure 1A, left, columns 6 and 7, and 2B, rows 3 and 4). Of note, WT and 7A 5MP1 are expressed at an equal abundance (Figure 1A, right). Therefore, 5MP1 appears to antagonize the effects of eIF5 in non-AUG initiation by competing for eIF2 bound to the PIC.
Figure 1.
5MP1 increases the accuracy of translation initiation by competing with eIF5. (A and B) Yeast phenotypic assays. (A) β-Galacosidase assay. Left, Double transformants of WT or mof2-1 strain carrying indicated plasmid and AUG-lacZ or UUG-lacZ reporter plasmid were assayed and the effect of each plasmid treatment on UUG/AUG ratios are presented. Averages from 3 or more indepenent experiments are presented. *P = 0.0002 (n = 11); **P = 0.002 (n = 8); ***P = 0.001 (n = 4). Right, Immnoblot of indicated amounts of whole cell extracts (WCE) showing expression of proteins indicated to the left. (B) His+ phenotype tests with a distinct eIF1 mutant (K60E). Left, indicated yeast eIF1-K60E transformants were assayed for his4-UUG expression based on the growth on a medium limited for histidine (panel 2 with—His plate with a trace amount of histidine at 1 μM). Panel 1, control with + His plate containing a full amount of histidine. Right graph, UUG/AUG initiation ratio was assayed as in panel A. *P < 0.05 (n = 4 or more). (C and D) In vitro reconstitution assays. (C) TC binding assay. 35S-labeled TC was loaded onto 40S/eIF1A/eIF1 (G107R) complex in the presence of different amounts of eIF5 (red triangle), eIF5-CTD (blue triangle) and 5MP1 (red square). The graph shows the percentage of 35S label present in the ribosomal complex. (D) GTPase activation assay. eIF5-dependent GTP hydrolysis for TC/rAUG/40S complex (red) was challenged by indicated amounts of eIF5-CTD (blue) or 5MP1 (green). Numbers to the left of the box indicate GTP hydrolysis rates computed from the graph.
We next examined whether 5MP1 incorporates into the PIC and thereby regulates its activity in vitro. eIF1 is known to oppose Met-tRNAi loading to the 40S in the ternary complex (TC) with eIF2:GTP (36). The eIF1 release in response to AUG recognition is a key step in strong Met-tRNAi binding to the 40S and its accommodation to the P-site (39). We took advantage of the ability of the eIF1 G107R mutant to delay the eIF1 release, thereby destabilizing the TC binding to the 40S. TC binding was monitored by the gel retardation of 35S-Met-tRNAi caused by 40S association (36). Thus, in the presence of eIF1-G107R, only <50% of TC is recruited to the PIC (Figure 1C, [eIF5] = 0). When added in excess, eIF5 promotes TC binding to the ribosome through interaction with eIF2 in the PIC and thereby stimulating eIF1 release (40) (Figure 1C, red triangle). As shown in Figure 1C, red square, 5MP1 promoted TC binding to the ribosome in place of eIF5 or eIF5-CTD (which 5MP1 mimics, red and blue triangles; (30)). Thus, 5MP1 is not only able to interact with the PIC, but also to antagonize eIF1 gatekeeping function, potentially promoting accurate initiation. Furthermore, 5MP1 inhibited eIF5-promoted GTP hydrolysis for eIF2 in the model 48S PIC, even though eIF5-CTD failed to do so (Figure 1D). These results demonstrate that 5MP1 can act on the PIC in order to prevent mis-initiation by eIF5 present in excess of the PIC.
Non-AUG initiation rate is consistent across various cell lines including insect cells and is oppositely regulated by eIF5 and 5MP1
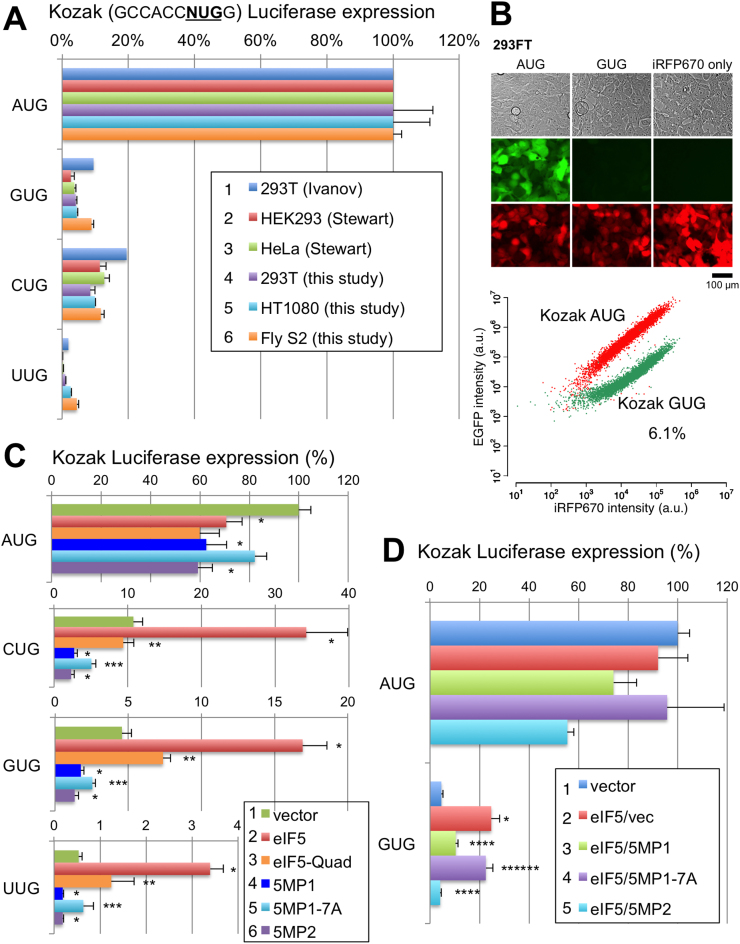
To analyze non-AUG translation in higher eukaryotes including humans, we used the firefly luciferase reporter starting with non-AUG codons under a Kozak context (GCCACCNNNG where NNN is the start codon). As shown in Figure 2A, our assays verified low, but significant, levels of non-AUG initiation, with CUG being the strongest, in human embryonic kidney (HEK) 293T, fibrosarcoma HT1080 and fly S2 cells. A low GUG initiation rate (∼6% compared to AUG) was also confirmed in HEK293FT cells by transfection of capped GFP mRNA with start codons under the same Kozak context (Figure 2B; see Supplementary Figure S1 for RNA used). These results indicate low non-AUG initiation rates, all in the order of CUG >GUG >UUG, in a wide-range of cell types including human immortalized (HEK293), transformed (HEK293T or FT) or cancer (HeLa or HT1080) cell lines as well as insect cells (S2 cells).
Figure 2.
5MP1 and eIF5 conversely regulate non-AUG initiation in human cells. (A) Spectrum of non-AUG initiation rate in various cells. HEK293T, HT1080 (fibrosarcoma), and fly S2 cells were transfected with firefly luciferase reporter plasmids (Supplementary Table S1) initiated by indicated start codons and a control Renilla luciferase plasmid and assayed for both the luciferase activities (see Supplemental method). Firefly/Renilla expression ratio was presented relative to the value from the AUG firefly luciferase reporter. Data was compared to values from different cells determined previously (53,54). (B) mRNA transfection assay. GFP mRNA with an altered initiation site was co-transfected with iRFP670 mRNA in 293FT cells and 10,000 transfected cells were analyzed by FACS. Pictures of cells taken under visible light or green or red fluorescence are shown. Graph, GFP and iRFP signals from each cell were plotted for control GFP mRNA (red) and GFP mRNA with a GUG start codon (green). The value indicates the percentage of GUG initiation compared to control AUG initiation. (C) Effect of eIF5 and 5MP on non-AUG initiation. HEK293T was transfected with the indicated NUG reporter plasmid, Renilla control plasmid, and pEF1A derivatives expressing indicated proteins (5:1 compared to firefly plasmid), assayed for luciferase activities and presented as in panel (B). Asterisks denote statistical significance (P < 0.05) compared to vector control (*), eIF5 (**) or 5MP1 (***) experiments. P values from top of each graph; AUG, 0.01, 0.001 (n = 6), 0.007 (n = 4),; CUG, * 0.02 (n = 4), 0.0001 (n = 8), 0.03 (n = 4), ** 0.016 (n = 4), *** 0.02 (n = 4); GUG, * 0.008 (n = 4), 0.006 (n = 12), 0.018 (n = 6), ** 0.031 (n = 4), *** 0.018 (n = 4); UUG, * 0.00006 (n = 6), 0.01, 0.017 (n = 6), ** 0.007 (n = 6), *** 0.04 (n = 4). (D) Effect of co-expression of eIF5 and 5MP on GUG versus AUG initiation. Assays were done and presented as in panel (C) except using two expression plasmids (1:1) together, as indicated in the inset. P values from top of graph GUG; * 0.0008 (n = 8) compared to row 1, ****0.002 (n = 10), 0.009 (n = 4), compared to row 2; *****0.03 (n = 6) compared to row 3.
In human cells, an eIF1 mutation is not required for eIF5 in excess to cause mis-initiation from non-AUG codons (18). Accordingly, in HEK293T cells, introduction of the eIF5-expressing plasmid alone increased luciferease reporter expression from CUG, GUG or UUG codon by 2- to 3-fold (Figure 2C, red columns). This effect was alleviated by eIF5-CTD Quad mutation (H305D N306D E347K E348K) (40), which is defective in eIF1 and eIF2 binding (Figure 2C, orange columns). This indicates that the observed effects depend on eIF5 binding to the PIC through eIF1 and eIF2. 5MP1 co-expression counteracted the effect of eIF5 in a manner depending on the seven residues in the 5MP1-CTD which are esential for eIF2 binidng (21) (Figure 2D, columns 2–4). Comparable results were observed using 5MP2 (Figure 2D, column 5). Therefore, both 5MP1 and 5MP2 increase the accuracy of selection of initiation codon, likely via competing with eIF5 for eIF2 bound to the PIC. Curiously, expression of 5MP1 or 5MP2 alone was sufficient to suppress luciferase reporter expression from GUG, UUG or CUG codons more strongly than their effect on AUG initiation in general translation (Figure 2C). This suggests that the low but sigificant level of non-AUG initiation (∼3–10% of AUG initiation) is due to the slight excess of eIF5 present in the PIC, which can be antagonized by forced 5MP expression.
An unusually high rate of initiation from NAT1 GUG codon depends on a specific nucleotide context including the Kozak consensus
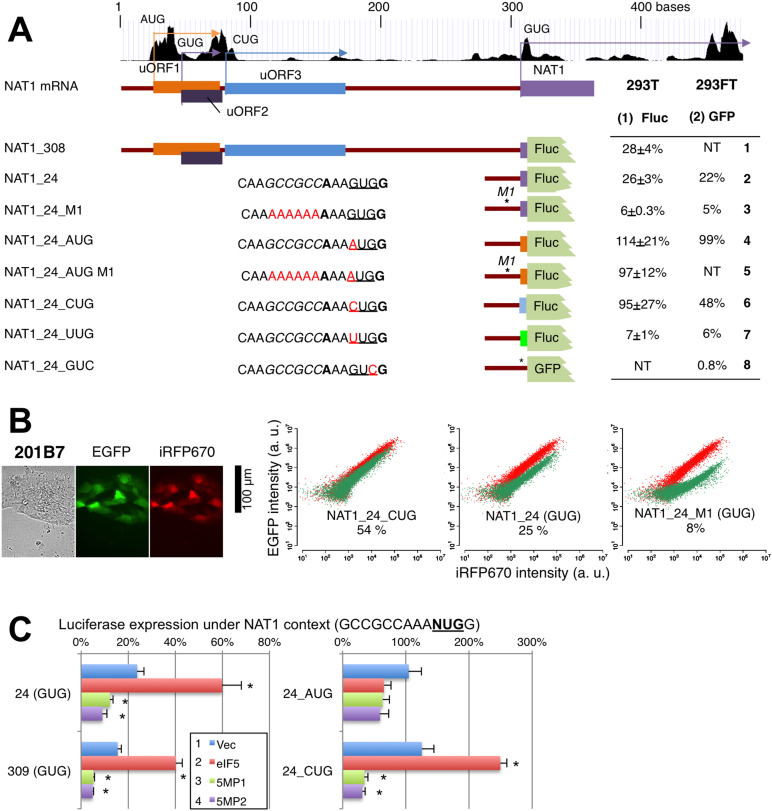
The GUG-initiation rate for NAT1 is unusually high, ∼30% compared to an AUG-mutant version of the same NAT1 expression plasmid (41). Here we confirmed this finding by generating firefly luciferase plasmids whose translation starts from 309-base-long NAT1 5′UTR or from its 24-nt-long UTR (Figure 3A, column 1, rows 1 and 2). Alteration of the GUG codon to AUG elevated the reporter expression to a similar level of AUG initiation seen with a Kozak context (row 4), in agreement with a strong Kozak context (AxxGUGG) around the start codon. Mutational analysis of the NAT1 24-nt construct demonstrated that the strong GUG initiation, but not initiation from its AUG version, depends on the rGCCGCC sequence located 4- to 9-nt upstream of the start codon (Figure 3A, column 1, rows 2–5). Considering that the level of GUG initiation from NAT1_24_M1 is equivalent to GUG initiation under a typical Kozak context ‘GCCACCNUGG’ (Figure 3A, GUG, row 4), it is plausible that the rGCCGCC sequence contains a cis element responsible for the enhanced GUG initiation. Because the typical Kozak sequence used as the reference here overlaps with the rGCCGCC sequence, it remains to be determined whether the rGCCGCC sequence enhances the initiation rate as a part of the Kozak consensus or beyond the effect of the Kozak sequence. Here we define this specific context enhancing non-AUG initiation as the NAT1 context.
Figure 3.
5MP and eIF5 conversely control strong GUG-initiation of NAT1/EIF4G2. (A) Analysis of the NAT1 GUG initiation signal. Schematics on top depict the structure of NAT1 mRNA. uORFs and NAT1 main ORF were color-coded by start codons (purple, GUG; orange/red, AUG; blue, CUG). Graph on top shows the ribosome profile from the UCSC genome browser. Below the schematics is described the initiation frequency from reporter constructs listed to the left and depicted as schematics in the middle (color-coded as the schematics on top except green for UUG). Rows 2–7 list the last 12-nt RNA sequence before the start codon of the 24-nt 5′UTR used for the assay. Altered bases are shown in red, while the rGCCGCC sequence is italicized. Asterisks in the schematics indicate the location of mutations introduced. Columns 1 and 2 list the results of experiments with the firefly luciferase reporter plasmid (with SE) and the GFP mRNA derivative, respectively. SD for the latter is listed in Supplementary Figure S5. (B) GFP mRNA assays in human iPS cells (201B7). Pictures show 201B7 cells transfected with control GFP mRNA (Kozak AUG, green) and iRFP mRNA (red). Graphs, FACS analysis of the cells transfected with GFP mRNA listed. Values indicate % compared to control GFP mRNA. (C) Effect of eIF5 and 5MP1/2 expression was tested with indicated NAT1 reporter plasmid and presented as in Figure 2C. Asterisks denote statistical significance (P < 0.05) compared to vector control (*). P values from top of each graph; 24(GUG), 0.03, 0.02, 0.008 (n = 4); 309 (GUG), 0.0001, 0.0005, 0.0006 (n = 8); 24_CUG, 0.0005 (n = 4), 0.003, 0.002 (n = 6).
Surprisingly, the replacement of the start codon with CUG dramatically increased (Figure 3A, column 1, row 6), whereas replacement with UUG substantially decreased the reporter expression relative to the original NAT1_GUG initiation (Figure 3A, column 1, row 7). Yet, all of these non-AUG initiation rates were higher than corresponding non-AUG initiation rates from the typical Kozak context (Figure 2A). This suggests that the NAT1 initiation context generally enhances non-AUG initiation rates, but not that of AUG. To verify these findings, we generated capped GFP mRNA with 24-nt NAT1 5′-UTR containing the same nucleotide alterations (Supplementary Figure S1). When co-transfected with a control iRFP mRNA, NAT1 GUG initiation was ∼25% compared to its AUG version or Kozak AUG control and depended on the rGCCGCC sequence, whereas its CUG mutant version expressed a much higher level of GFP (Figure 3A, column 2, and Supplementary Figure S2). We also confirmed that a start codon mutation diminishes luciferase expression to <1% compared to the AUG construct expressed under the NAT1 or Kozak context (Figure 3A, row 8).
The leader region of NAT1 mRNA contains three uORFs, the AUG-initiated uORF1 and GUG- or CUG initiated uORF2 or uORF3 (Figure 3A, top). The equivalent levels of GUG or AUG initiation from either NAT1_308 or _24 constructs (Figure 3A, rows 1, 2 and 4) suggests that these uORFs do not inhibit translation of NAT1. This idea was verified by examining mutations altering the start codons of all the three uORFs (Supplementary Figure S3A, rows 6–9). We confirmed initiation from these start codons and showed that uORF1 suppresses translation of uORF2 and uORF3 (Supplementary Figure S3A, see Supplemental text). Furthermore, GFP mRNA assays demonstrate that the strong GUG initiation under the NAT1 context is cap-dependent (Supplementary Figure S3B). Therefore, we suggest that the NAT1 mRNA is translated by a cap-dependent mechanism involving re-initiation, whereby sufficient distance between uORF1 (the most strongly translated uORF) and the main ORF allows downstream re-initiation by ribosomes that have finished uORF1 translation.
Based on the recent reports that NAT1/DAP5 plays an important role in stem cell differentiation (23,24), we next examined whether the strong NAT1 GUG initiation rate is altered in human stem-like cells. The capped GFP mRNAs used in Figure 3A were transfected into a human induced pluripotent stem (iPS) cell line 201B7 and assayed for GFP expression (Supplementary Figure S4). However, compared to HEK293FT, the stem cell background did not alter GFP expression from all the mRNAs tested (Supplementary Figure S5), including those with GUG or CUG codon enhanced under the NAT1 context (NAT1_24, -_24_CUG) or with normally low GUG initiation signal (NAT1_24_M1) (Figure 3B). This last result also argues against the possibility that the stringency of start codon selection is generally lower in stem cells.
5MP suppresses and eIF5 enhances the strong GUG or CUG initiation under the NAT1 context
Similar to non-AUG initiation from a regular Kozak context, eIF5 expression increased and 5MP1 or 5MP2 expression decreased GUG initiation from the luciferase reporter with the 309-nt or 24-nt-long NAT1 5′UTR (Figure 3C). The effect of 5MP on the NAT1 GUG initiation was stronger than its general effect on the AUG variant of 24-nt-long construct (Figure 3C, AUG). Moreover, 5MP expression decreased upstream AUG (uORF1) initiation under the suboptimal Kozak context (UxxAUGG) more strongly (Supplementary Figure S3C, see Supplementary Figure S3A also) than AUG initiation under the strong Kozak context (Figure 2C, AUG and 3C, 24_AUG). This is in agreement with the idea that initiation from the weaker Kozak sequence can be affected by altering the canonical stringent initiation mechanism (12). However, eIF5 expression did not increase the uAUG initiation (Figure 3D), suggesting that the effect of eIF5 is specific to non-AUG initiation. Finally, eIF5 expression increased, and 5MP expression decreased the CUG initiation from the CUG variant of 24-nt NAT1 construct (Figure 3C, GUG). Thus, in the context of the NAT1 5′-UTR, the CUG codon initiates translation as strongly as the AUG codon does and, unlike the AUG codon, is regulated by altered levels of eIF5 or 5MP.
GTI-seq confirms opposite regulation of non-AUG translation by eIF5 and 5MP1 on a transcriptome-wide scale
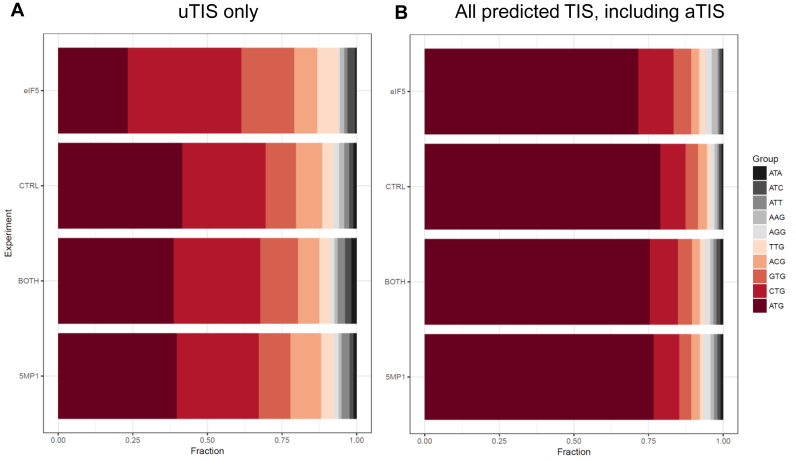
In order to determine whether eIF5 and 5MP1 conversely regulate non-AUG translation genome-wide, we transfected HEK293T with 1:1 mixture of eIF5- or 5MP1-expressing plasmid and the vector control (eIF5 or 5MP1, respectively), both eIF5- and 5MP1-expressing plasmids (both), or the vector control only (Ctrl), and performed global translation initiation sequencing (GTI-seq), using two related but distinct translation inhibitors, LTM and CHX. As shown in Figure 4A, eIF5 expression increases GUG or CUG initiation and decreases AUG initiation from 5′UTR (uTIS) and this trend was reversed by co-expression of eIF5 and 5MP1. Likewise, eIF5 expression increases GUG or CUG initiation and decreases AUG initiation from all the predicted start sites including those initiating protein coding regions (Figure 4B, aTIS). This further confirms that the competition between eIF5 and 5MP1 determines non-AUG initiation genome-wide. However, we did not observe that 5MP1 expression per se decreases GUG or CUG initiation or increases AUG initiation in these assays. We do not believe that this method allows us to judge the decrease in the intensity of each TIS.
Figure 4.
GTI-seq verifies converse regulation of non-AUG translation by eIF5 and 5MP1 genome-wide. The proportion of translation initiation sites (TIS) with indicated start codons (color-code to the right) is shown for HEK293T transfected with eIF5 or 5MP1-expression plasmid, both (BOTH), or with the vector control (Ctrl). (A) uTIS, TIS located upstream of authentic TIS. (B) All predicted TIS (aTIS) including uTIS.
Ribosome profiling identifies new non-AUG start codons regulated conversely by eIF5 and 5MP, similar to the NAT1 GUG codon
Our analysis of ribosome profiling with CHX-treated cells (Ribo-seq) also identified a handful of genes whose ribosome density is conversely regulated by eIF5 and 5MP1 (Table 1 and Supplementary Table S4). Of these, translation of ARL6IP1, UBEC2 and LSM8 is initiated by upstream in-frame GUG, CUG and CUG start codons, respectively, indicating that translation from the non-AUG codons greatly contributes to expression of these proteins (Supplementary Figure S8A). RPS27L belonging to this group is initiated by AUG, but the main ORF is preceded by a UUG-initiated uORF, which is also regulated similarly by eIF5 and 5MP1 (Table 1, row 7). The ribosome profile of RPS27L mRNA suggests that translation of the uORF is permissive for RPS27L translation, and that RPS27L is regulated through the UUG-initiated uORF (Supplementary Figure S8A).
Table 1. Ribosome density (RKPM) of genes controlled by eIF5 and 5MP1.
| Gene | Acc # | Start codon | eIF5 | Vec | 5MP1 | Feature |
|---|---|---|---|---|---|---|
| Increased by eIF5, decreased by 5MP1 | ||||||
| ARL6IP1 | NM_015161 | GUG | 272 | 135 | 19 | uGUG in frame |
| UBE2C | NM_007019 | CUG | 462 | 223 | 50 | uCUG in frame |
| LSM8 (NIAA38) | NM_016200 | CUG | 252 | 106 | 42 | uCUG in frame |
| RPS27L | NM_015920 | AUG | 448 | 223 | 107 | Re-initiation, uORF |
| RPS27L (uORF) | NM_015920 | UUG | 559 | 298 | 233 | |
| GTF3A | NM_002097 | CUG | 45 (1.5) | 32 (1) | 18 (0.4) | CUG initiation (13) |
| R3HCC1 | NM_001136108 | CUG | 30 (1.6) | 21 (1) | 0 (0.3) | CUG initiation (13) |
| BAG1 | NM_001172415 | CUG | 39 (2.9) | 33 (1) | 10 (0.6) | CUG initiation (13) |
| PTEN | NM_000314 | CUG/AUU | 11 | 7 | 3 | CUG/AUU initiation (55) |
| Decreased by eIF5, increased by 5MP1 | ||||||
| CEBPA | NM_004364 | AUG | 31 | 85 | 230 | Re-initiation control? |
| AIF1L | NM_001185095 | AUG | 30 | 72 | 156 | uCUG out of frame regulated? |
| Decreased by both eIF5 and 5MP1 | ||||||
| H1F0 | NM_005318 | CUG | 67 | 158 | 76 | uCUG in frame |
| TMSB4X | NM_021109 | AUG | 46 | 157 | 0 | uGUG out of frame regulated? |
| cMyc | NM_002467 | CUG | 52 (0.6) | 66 (1) | 20 (0.3) | uCUG in frame inhibits main AUG |
| DDX17 | NM_001098504 | CUG/GUG | 92 | 129 | 91 | Similar to cMyc? |
Data in parenthesis are fold increase measured by luciferase assays compared to vector control.
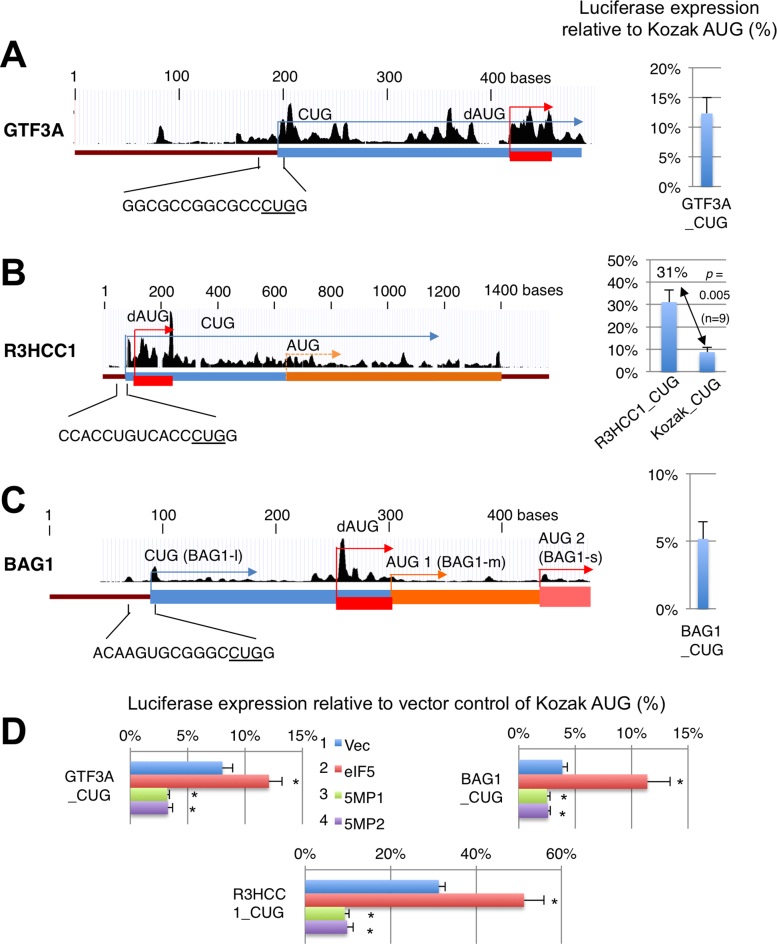
We also found that proposed CUG-initiated translation of GTF3A, R3HCC1, BAG1 and PTEN (13) is conversely regulated by eIF5 and 5MP1, similar to NAT1 (Table 1). UCSC ribosome profiling data supports (almost) sole CUG initiation of GTF3A, R3HCC1 and BAG1 (Figure 5A–C top). GTF3A encodes the general transcription factor 3A, responsible for RNA polymerase III transcription (42). Intriguingly, its translation starts from a CUG codon in mammals, but in birds, frogs, fish, insects or lower eukaryotes, initiation occurs from the AUG codon located at the position equivalent to the CUG codon (Supplementary Figure S6). R3HCC1 is a possible RNA-binding protein with the R3H and RNA-recognition motif (RRM) domains, which is conserved in vertebrates (35). The combination of R3H and RRM domains is found in the C-terminal half of poly(A)-specific ribonuclease (PARN) (43). BAG1 is an oncogene encoding a co-chaperone and has three isoforms sharing the same C-terminus, BAG1L initiated with CUG, and BAG1M and BAG1S initiated with AUG (44). BAG1L and BAG1S are the major products of this gene (45) (see below Figure 5C for how BAG1M AUG might be skipped).
Figure 5.
5MP and eIF5 conversely control CUG initiation of GTF3A (A), R3HCC1 (B) and BAG1 (C). (A–C) The mRNA structures were depicted with UCSC ribosome profiles and ORFs color-coded by start codons as in Figure 3A. RNA sequences near the CUG start codons are listed below the schematics. Graphs indicate firefly luciferase activities from the reporters bearing 24-nt 5′UTR relative to activity from Kozak AUG control. In (B), the activity was compared to CUG under Kozak context. (D) Effect of eIF5 and 5MP1/2 expression was tested with indicated CUG reporter plasmids and presented as in Figure 2C. Asterisks denote statistical significance (P < 0.05) compared to vector control (*). P values from top of each graph; GTF3A, 0.003, 0.006, 0.01 (n = 6); R3HCC1, 0.01 (n = 6), 0.00006, 0.0003 (n = 5); BAG1, 0.03, 0.009, 0.03 (n = 4).
Using luciferase reporter bearing 24-nt 5′UTR, we found that the CUG initiation rate is very low for GTF3A and BAG1—only 5–10% compared to Kozak AUG, whereas it is reasonably high for R3HCC1—∼30% compared to Kozak AUG, significantly higher than CUG initiation under the Kozak context (Figure 5A–C, Graph). As shown in Figure 5D, the reporter translation from the CUG codon of each gene was conversely regulated by eIF5 and 5MP1/2, verifying the results of our ribosome profiling. The low CUG initiation rate for GTF3A or BAG1 was surprising as it would allow for leaky-scanned ribosomes to initiate at downstream in-frame AUG codons. To assess how the downstream AUG initiation is prevented, we examined the nucleotide sequences of the genes. First, the GTF3A-coding region does not possess AUG codons for the first two thirds of the protein, and this feature is conserved throughout mammals (which utilize CUG initiation for GTF3A), but not in frogs, fish, insects or yeasts (which do not) (Supplementary Figure S6). Thus, the absence of downstream AUG codons in the reading frame prevents generation of nonfunctional isoforms with the same C-terminus. More importantly, ribosome profiling data indicate that all the three mRNAs possess a short ORF initiated with an AUG codon located downstream of and out-of-frame to the CUG codon (dAUG) (red square and arrows in Figure 5A–C, top). We propose that translation of such downstream ORFs (dORFs) prevents re-initiation of AUG codons located immediately downstream, making the CUG codon the sole start codon [or, in the case of BAG1, preventing BAG1M translation from upstream in-frame AUG (45)]. In agreement with this idea, the ribosome profiling data show that 5MP1 expression suppresses CUG-initiated translation of GTF3A and R3HCC1, yet allowing translation of the dORFs (Supplementary Figure S7).
eIF5 and 5MP1 regulate non-AUG initiation differentially
The Ribo-seq study also identified non-AUG or AUG-initiated genes regulated differently by eIF5 and 5MP1, compared to regulation of NAT1. As shown in Tables 1 and Supplementary Table S4, translation of a subset of genes, including TMSB4X and H1F0, is decreased by both eIF5 and 5MP1. TMSB4X has an inhibitory, overlapping uORF initiated by a GUG codon out of frame by +2 (Supplementary Figure S8B). The ribosome profile of its mRNA suggests that eIF5 increases translation of this uORF, thereby repressing TMSB4X translation (Supplementary Figure S8B). In the case of H1F0, its mRNA possesses an in-frame uCUG that appears to work as a start codon and a CUG-initiated uORF out of frame by +1 (Supplementary Figure S8B). The ribosome profile suggests that eIF5 increases translation from both the CUG codons, yet resulting in repression of translation of the main ORF (Supplementary Figure S8B). As shown below in Figure 6, a similar trend is observed for translation of cMYC mRNA possessing an in-frame uCUG codon.
Figure 6.
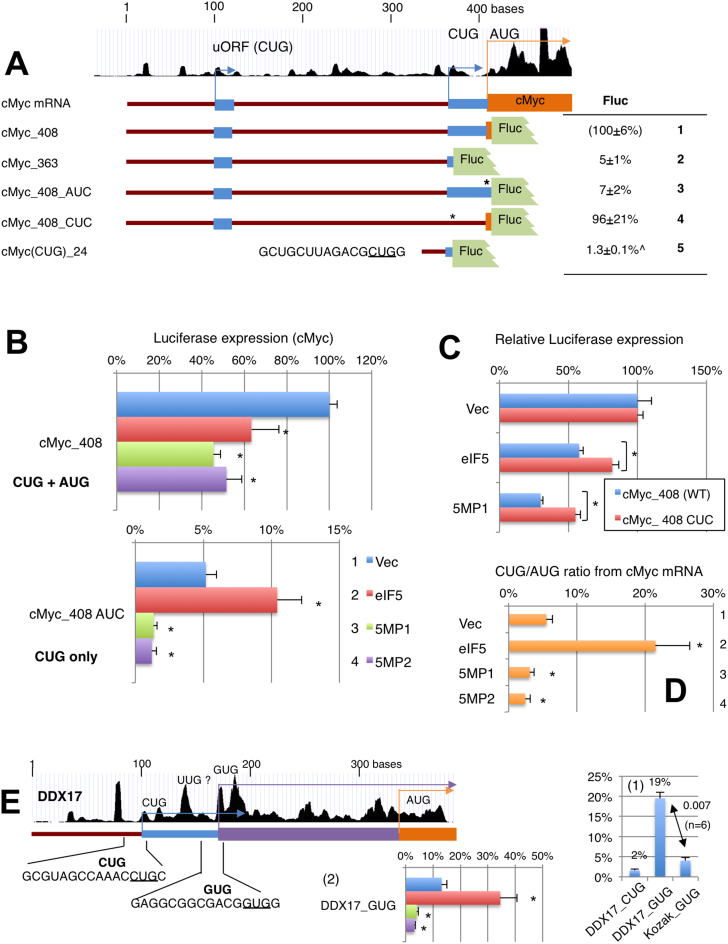
Translational control of cMYC and DDX17 by eIF5 and 5MP. (A) Analysis of cMYC start codons. Top, schematics depict the cMYC leader region with UCSC ribosome profile and boxes representing reading frames color-coded as in Figure 3A. The table describes firefly luciferase activities from constructs listed to the left and depicted in the middle, relative to the activity from cMyc_408 (row 1). ∧, activity for cMyc(CUG)_24 is presented relative to that from Kozak AUG. Asterisks, start codon mutations. (B) Effect of eIF5 and 5MP1/2 expression was tested with indicated cMyc reporter plasmids, and presented as values relative to cMyc_408 vector control, as in Figure 2C. P values of significance from top; cMyc_408, 0.04, 0.00004 (n = 8), 0.01 (n = 4); cMyc_408 AUC, 0.01, 0.0007, 0.0005 (n = 10) compared to vector control. (C) Effect of eIF5 and 5MP1 on cMyc_408 and cMyc_408_CUC was compared to the values from vector control for each, and statistical significance was computed using students’ Ttest. *P values of significance; from top, 0.04, 0.02 (n = 4). (D) CUG/AUG ratio (R) in cMyc_408 (WT) was computed based on the following formula and presented with asterisks denoting statistical significance (P < 0.05) compared to the value with vector control. R = a/(a – b), where a is activity from cMyc_408 AUC and b is activity from cMyc_408 (WT). *P values of significance; from top, 0.01, 0.02, 0.009 (n = 10) compared to vector control. (E) DDX17 mRNA structure was depicted as in Figure 5A–C. Graph 1 indicates firefly luciferase activities from the reporter with DDX17 CUG and GUG codons carrying the 24-nt mRNA sequences preceding each (columns 1 and 2) and compared to activity from Kozak GUG (column 3). Graph 2, the effect of eIF5 and 5MP1/2 on the DDX17 GUG reporter translation. p values from top; *0.02, 0.003, 0.003 (n = 6) compared to vector control.
Interestingly, translation of even a smaller subset of genes, including CEBPA and AIF1L, is decreased by eIF5 and increased by 5MP1 (Table 1 and Supplementary Table S4). The mechanism of this interesting observation requires further investigation, but the list includes CEBPA that encodes two isoforms of C/EBPα sharing the same C-terminus. Under mTOR signaling, re-initiation after uORF translation occurs past the first AUG codon of CEBPA, generating a shorter isoform initiated at the second AUG (46). It would be intriguing to investigate whether the differential regulation by eIF5 and 5MP1 is related to the differential re-initiation of CEBPA start codons. In the case of AIF1L, an out-of-frame uCUG generates an uORF overlapping with the AIF1L start codon (UCSC genome browser). eIF5-mediated up-regulation of this reading frame might result in repression of the main AIF1L ORF, similar to TMSB4X.
5MP diminishes CUG initiation from cMYC mRNA, while eIF5 increases CUG initiation but represses overall cMYC mRNA translation
Translation of cMYC, which has CUG followed by AUG start codons, was repressed by both eIF5 and 5MP1 (Table 1). Because CUG-initiation of this oncogene was highlighted recently (16), this observation deserves further attention. The UCSC ribosome profile indicates a minor peak corresponding to the in-frame uCUG and a major peak following the location of the main AUG codon (Figure 6A, top), suggesting a weak CUG initiation from the uCUG. We generated a luciferase reporter plasmid with a 5′UTR whose 5′ end is located 363-nt upstream of this CUG (cMyc_363), and a construct with a 5′UTR bearing the same 5′ end, but additionally carrying the 48-nt cMYC-coding region downstream including the cMYC AUG codon (cMyc_408). As a control, we also designed reporters bearing a mutation altering the CUG or AUG start codons (Figure 6A).
The luciferase assay confirmed that both eIF5 and 5MP1 repress the reporter translation from cMyc_408 (WT) (Figure 6B and Table 1). To examine the contribution of the CUG initiation, we measured luciferase expression from cMyc-363 and cMyc_408 AUC, the AUG-to-AUC mutant version of cMyc_408. The result confirms ∼5-7% uCUG initiation compared to AUG initiation rate, which was determined using cMyc_408 CUC, the CUG-CUC mutant version of cMyc_408 (Figure 6A, rows 2 and 3). Thus, overall, cMYC expression from the longer construct cMyc_408 (WT) is only marginally higher than expression from the cMyc_408 CUC construct (Figure 6A, rows 1 and 4). The minor contribution of uCUG initiation is at least partially attributable to a weak CUG initiation rate, as measured with the cMYC CUG construct bearing 24-nt UTR in both the luciferase assay and the assay using purified GFP mRNA (Figure 6A, row 5, and Supplementary Figure S3D, row 1). Surprisingly, while ribosome protection of a CUG-initiated uORF was noted in mESC (14) and we found a ribosome protection in cMyc mRNA (Figure 6A, top), we did not find a high rate of CUG initiation from that site (Supplementary Figure S3D, row 3).
Similar to other non-AUG codons examined, eIF5 expression increases and 5MP1/2 expression decreases the CUG initiation from cMyc_408 AUC construct altering the cMYC AUG codon (Figure 6B, cMyc_408 AUC). We also observed a relatively large decrease in cMyc_408 expression by 5MP1/2 (Figure 6B, cMyc_408), which may be attributed to the suboptimal Kozak context of its AUG start codon (AxxAUGC). This is reminiscent of the 5MP1/2 effect on NAT1_uAUG initiation from a suboptimal Kozak context (UxxAUGG) (Supplementary Figure S3C). A comparable decrease in overall cMYC expression (in cMyc_408) by eIF5 is surprising (Figure 6B, also see Table 1) and instead, suggests that the increase in CUG initiation inhibits downstream AUG initiation in this particular case. In agreement with this idea, eIF5 represses expression from cMyc_408 CUC devoid of CUG initiation less strongly than expression from cMyc_408_WT (Figure 6C). Together, these results indicate that, overall, eIF5 or 5MP expression decreases cMYC translation, in agreement with ribosome profiling (Table 1). However, eIF5 and 5MP conversely alters the proportion of cMyc isoforms with or without the CUG-initiated polypeptide, as shown in Figure 6D.
Similar to cMYC and H1F0, Ribo-seq data suggest that non-AUG translation of DDX17 encoding a DEAD-box RNA helicase (47) was repressed by both eIF5 and 5MP1 (Table 1). We found that the in-frame uGUG codon, as more recently proposed (13), rather than the CUG start codon as originally proposed (47), is a stronger initiation site (Figure 6E, graph 1, columns 1–2), even stronger than a typical Kozak GUG (column 3). Since eIF5 increases and 5MP1/2 decreases DDX17 GUG initiation (Figure 6E, graph 2), the increased GUG initiation might contribute to overall inhibition of AUG initiation from the suboptimal context (ACCAUGC), similarly to the case with cMYC.
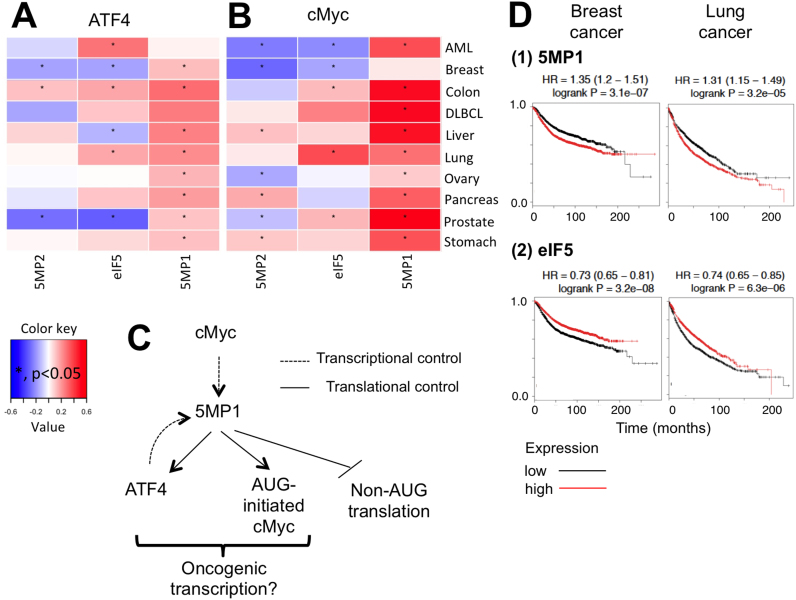
Cancer genomics databases suggest feedback control of 5MP1 transcription
Given the consistent non-AUG initiation rate across different cell types (Figures 2A and 3B, and Supplementary Figure S5), we pondered whether eIF5 or 5MP are significantly controled at the level of transcription. To address this, we took advantage of the TCGA database including ∼10 000 sets of patient data. We examined corelations with ATF4 and cMYC, which 5MP regulates on a translational level. As shown in Figure 7A and B, 5MP1, but not 5MP2 or eIF5, positively correlates with ATF4 and cMYC expression in most cancer types investigated. Furthermore, 5MP1 expression correlates with levels of several ATF4 target genes in liver, lung and stomach cancers (Supplementary Figure S9). Collectively, these results strongly suggest the existance of a positive feedback loop consisting of cMyc, 5MP1 and ATF4 (Figure 7C). Additional co-expression analysis supports this view (Supplementary Figure S10; see Supplemental text for details). In agreement with the role of 5MP1 in promoting fibrosarcoma tumorigenesis (27), 5MP1 expression correlates with a poor prognosis in breast and lung cancer patients (Figure 7D). Overall, this data supports a transcriptional program to maintain non-AUG translatome homeostais and warrants future investigation of 5MP1 as a potential oncogene.
Figure 7.
Possible feedback control of 5MP1 transcription. Pearson correlation between ATF4 mRNA levels (A) or cMYC mRNA (B) and 5MP1, 5MP2 and eIF5 mRNA levels. Correlation coefficients were calculated in 10 different cancers. Significant correlations are demarcated with an asterisk (P < 0.05). (C) A model of cMyc-5MP1-ATF4 pathway involved in oncogenic transcription. Dotted arrows indicate transcriptional control, whereas straight arrows and stop bar denote translational control. (D) Correlation between 5MP1 or eIF5 expression with the prognosis of breast or lung cancer patients (see Supplemental text for details).
DISCUSSION
5MP contributes to accurate initiation in eukaryotes
eIF5′s GAP function is a prerequisite for accurate initiation (7). Yet, eIF5 expression above a certain threshold leads to an increased frequency of non-AUG initiation (18,36,40). In this work, we showed that 5MP increases the initiation accuracy through competition with eIF5 (Figures 1 and 2C and D). eIF5 is thought to be responsibe for stabilizing PIC conformations favoring accurate initiation (2,40,48). We propose that high eIF5 expression permits a second eIF5 copy to bind the PIC during mRNA scanning. This shifts the PIC conformation to favor non-AUG initiation. Consistent with this idea, eIF5 overexpression in yeast doubles the amount of eIF5 bound to eIF3 (49). Upon elevation of 5MP expression, we predict that 5MP binding to the PIC will preclude binding of the second eIF5 copy to the scanning PIC, thereby preventing non-AUG initiation. Detailed mapping studies identified the β-subunit of eIF2 as a crucial eIF5-binding partner when the PIC changes its conformation in response to AUG recognition (40). Prior to this event, eIF2β would be available as a competitive binding site for the second eIF5 copy or 5MP during mRNA scanning (see the model in (2)). An alternative explanation may be that excess of eIF5, in respect to PIC, may sequester a factor that facilitates scanning. This would result in scanning arrest at non-AUG start codons. Notably however, we did not identify such a factor bound to FLAG-eIF5 in our previous immunoprecipitation/MS studies (27).
Non-AUG initiation in mammals
Herein, we showed that the non-AUG initiation rate strongly depends on the surrounding nucloetide contexts, leading to expression levels ranging from <1% to almost equivalent to the AUG-mediated initiation rate (Figures 1–3). One of the best contexts for non-AUG initiation was found within the 24-nt region immediately upstream of NAT1 GUG codon, specifically the Kozak context and the rGCCGCC sequence, which partially overlaps with the Kozak. Surprisingly, the CUG codon placed under the same NAT1 context displayed expression nearly equivalent to AUG under the Kozak context. The minimum requirement for appreciable GUG or CUG initiation is the Kozak context of (G/A)xxxNUGG; any CUG or GUG codon that falls outside of this context was translated very poorly (Figures 3 and 5 and Supplementary Figure S3), as reported previously (13). In addition to the GUG of NAT1, the GUG of DDX17 and the R3HCC1 CUG codons initiated translation more strongly than their counterparts when placed under the Kozak context (Figure 5A). The nucleotide contexts that allow higher rate of translation from these non-AUG codons remains to be investigated. Finally, we noted that the coding regions initiated by non-AUG codons are often followed by an AUG-initiated downstream ORF, which prevents leaky scanned ribosomes to initiate downstream in-frame AUG codons (Figure 5 and Supplementary Figure S7).
Possible role of non-AUG translatome in regulating cell fate
The non-AUG initiation rates are consistent across different cell-types, whether normal, cancer, non-vertebrate, or pluripotent stem cells (Figures 2A and 4B and Supplementary Figure S2-4). Yet, the non-AUG initiation rates can be altered by elevated levels of eIF5 and 5MP (Figures 2–5). We therefore propose that the maintenance of consistent non-AUG initiaion rate is important for optimal cell function which is actively achieved via the balance of eIF5, 5MP and potentially other factors such as eIF2A. Our study using the cancer genomic database suggests that eIF5 or 5MP1 levels probably govern non-AUG initiaion programs through regulation by oncogenes including cMYC and ATF4 (Figure 7 and Supplementary Figures S9 and S10).
In contrast to the findings suggesting that non-AUG initiation is elevated in mouse ES cells (14) or SOX2-induced premalignant tumor cells (15), our data suggest that the efficiency of non-AUG translation is mostly constant in various cell types and organisms. However, the latter does not exclude the possibility that non-AUG initiation, which may be implicated in tumorigenesis, is transient and induced by increase in eIF5 copy number or specific signaling events (e.g. cMyc activation). Given the important role of cMyc in cell reprogramming and tumorigenesis (50,51), it would be important to investigate the role of the longer, CUG-initiated form of cMyc, shown here to be up-regulated by eIF5 expression (Figure 6). For example, the longer cMyc isoform might serve as an activator of 5MP1 transcription, thereby shutting down the transient, non-AUG upregulation, and simlutanously promoting ATF4 translation (Figure 7 and Supplementary Figure S9). ATF4, in turn, confers stress-resistance to cancer cells (52). Again, it would be pertinent to study the effect of the proposed enrichment of AUG-initiated (vs. CUG-initiated) cMyc isoform on oncogenic transcriptional programs in this context (Figure 7C). We believe that the ribosome profiling data presented in this study provides the wealth of information in order to effectively identify such key proteins encoded by non-AUG regulated genes.
ACCESSION NUMBER
GEO ID: GSE102786.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Masa Sokabe, Chris Fraser (UC Davis), Jon Lorsch (NIGMS, NIH), Nahum Sonenberg and Yuri Svitkin (McGill University) for discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Innovative Award from Terry Johnson Cancer Center; KSU; KU-COBRE Protein Structure and Function Pilot Grant [P30GM110761]; NSF Research Grant [1412250 to K.A.]; US National Institutes of Health [R01AG042400, R01GM1222814 to S.-B.Q.]; Chelsea Moore, Sarah Gillaspie and Eric Aube were K-INBRE scholars [P20GM103418]; S.-B. Q. is a HHMI Faculty Scholar [55108556]; I.T. and M.W. are recipients of Chercheur Boursier-Junior 2 salary support from FRQ-S. KA’s visit to CiRA was funded by JSPS Fellowship for Foreign Scientist Invitation and Heiwa Nakajima Foundation (to H. Saito). Funding for open access charge: KSU open access fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hinnebusch A.G., Ivanov I.P., Sonenberg N.. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016; 352:1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Obayashi E., Luna R.E., Nagata T., Martin-Marcos P., Hiraishi H., Singh C.R., Erzberger J.P., Zhang F., Arthanari H., Morris J. et al. Molecular landscape of the ribosome pre-initiation complex during mRNA scanning: structural role for eIF3c and its control by eIF5. Cell Rep. 2017; 18:2651–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pestova T.V., Lorsch J.R., Hellen C.U.T.. Mathews MB, Sonenberg N, Hershey JWB. Translational Control in Biology and Medicine. 2007; NY: Cold Spring Harbor Lab Press; 87–128. [Google Scholar]
- 4. Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014; 83:779–812. [DOI] [PubMed] [Google Scholar]
- 5. Asano K. Why is start codon selection so precise in eukaryotes. Translation. 2014; 2:e28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Asano K., Clayton J., Shalev A., Hinnebusch A.G.. A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev. 2000; 14:2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Asano K., Shalev A., Phan L., Nielsen K., Clayton J., Valasek L., Donahue T.F., Hinnebusch A.G.. Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001; 20:2326–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamamoto Y., Singh C.R., Marintchev A., Hall N.S., Hannig E.M., Wagner G., Asano K.. The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3 and eIF4G. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:16164–16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sinvani H., Haimov O., Svitkin Y., Sonenberg N., Tamarkin-Ben-Harush A., Viollet B., Dikstein R.. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab. 2015; 21:479–492. [DOI] [PubMed] [Google Scholar]
- 10. Tamarkin-Ben-Harush A., Vasseur J.-J., Debart F., Ulitsky I., Dikstein R.. Cap-proximal nucleotides via differential eIF4E binding and alternative promoter usage mediate translational response to energy stress. eLife. 2017; 6:e21907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991; 266:19867–19870. [PubMed] [Google Scholar]
- 12. Martin-Marcos P., Cheung Y.-N., Hinnebusch A.G.. Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol. Cell. Biol. 2011; 31:4814–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ivanov I.P., Firth A.E., Michel A.M., Atkins J.F., Baranov P.V.. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 2011; 39:4220–4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ingolia N.T., Lareau L.F., Weissman J.S.. Ribosome profiling of mouse embryonic stem cells reveals the complexity of mammalian proteomes. Cell. 2011; 147:789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sendoel A., Dunn J.G., Rodriguez E.H., Naik S., Gomez N.C., Hurwitz B., Levorse J., Dill B.D., Schramek D., Molina H. et al. Translation from unconventional 5′ start sites drives tumour initiation. Nature. 2017; 541:494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Starck S.B., Jiang V., Pavon-Eternod M., Prasad S., McCarthy B., Pan T., Shastri N.. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC Class I. Science. 2012; 336:1719–1723. [DOI] [PubMed] [Google Scholar]
- 17. Huang H., Yoon H., Hannig E.M., Donahue T.F.. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997; 11:2396–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loughran G., Sachs M.S., Atkins J.F., Ivanov I.P.. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 2011; 40:2998–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marintchev A., Wagner G.. eIF4G and CBP80 share a common origin and similar domain organization: Implications for the structure and function of eIF4G. Biochemistry. 2005; 44:12265–12272. [DOI] [PubMed] [Google Scholar]
- 20. Rothenburg S., Seo E.J., Gibbs J.S., Dever T.E., Dittmar K.. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat. Struct. Mol. Biol. 2009; 16:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh C.R., Watanabe R., Zhou D., Jennings M.D., Fukao A., Lee B.-J., Ikeda Y., Chiorini J.A., Fujiwara T., Pavitt G.D. et al. Mechanisms of translational regulation by a human eIF5-mimic protein. Nucleic Acids Res. 2011; 39:8314–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marash L., Liberman N., Henis-Korenblit S., Sivan G., Reem E., Elroy-Stein O., Kimchi A.. DAP5 promotes Cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol. Cell. 2008; 30:447–459. [DOI] [PubMed] [Google Scholar]
- 23. Yoffe Y., David M., Kalaora R., Povodovski L., Friedlander G., Feldmesser E., Ainbinder E., Saada A., Bialik S., Kimchi A.. Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes Dev. 2016; 30:1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sugiyama H., Takahashi K., Yamamoto T., Iwasaki M., Narita M., Nakamura M., Rand T.A., Nakagawa M., Watanabe A., Yamanaka S.. Nat1 promotes translation of specific proteins that induce differentiation of mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hiraishi H., Oatman J., Haller S., Blunk L., McGivern B., Morris J., Papadopoulos E., Guttierrez W., Gordon M., Bokhari W. et al. Essential role of eIF5-mimic protein in animal development is linked to control of ATF4 expression. Nucleic Acids Res. 2014; 42:10321–10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li S., Chai Z., Li Y., Liu D., Bai Z., Li Y., Li Y., Situ Z.. BZW1, a novel proliferation regulator that promotes growth of salivary mucoepidermoid carcinoma. Cancer Lett. 2009; 284:86–94. [DOI] [PubMed] [Google Scholar]
- 27. Kozel C., Thompson B., Hustak S., Moore C., Nakashima A., Singh C.R., Reid M., Cox C., Papadopoulos E., Luna R.E. et al. Overexpression of eIF5 or its protein mimic 5MP perturbs eIF2 function and induces ATF4 translation through delayed re-initiation. Nucleic Acids Res. 2016; 44:8704–8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Young S.K., Wek R.C.. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J. Biol. Chem. 2016; 291:16927–16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Acker M.G., Kolitz S.E., Mitchell S.F., Nanda J.S., Lorsch J.R.. Reconstitution of yeast translation initiation. Methods Enzymol. 2007; 430:111–145. [DOI] [PubMed] [Google Scholar]
- 30. Nanda J., Saini A.K., Munoz A.M., Hinnebusch A.G., Lorsch J.R.. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal pre-initiation complex. J. Biol. Chem. 2013; 288:5316–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee B., Udagawa T., Singh C.S., Asano K.. Yeast phenotypic assays on translational control. Methods Enzymol. 2007; 429:139–161. [DOI] [PubMed] [Google Scholar]
- 32. Acker M.G., Kolitz S.E., Mitchell S.F., Nanda J.S., Lorsch J.R.. Reconstitution of yeast translation initiation. Methods Enzymol. 2007; 430:111–145. [DOI] [PubMed] [Google Scholar]
- 33. Algire M.A., Maag D., Lorsch J.R.. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005; 20:251–262. [DOI] [PubMed] [Google Scholar]
- 34. Uno S., Masai H.. Efficient expression and purification of human replication fork-stabilizing factor, Claspin, from mammalian cells: DNA-binding activity and novel protein interactions. Genes Cells. 2011; 16:842–856. [DOI] [PubMed] [Google Scholar]
- 35. Lee S., Liu B., Lee S., Huang S.-X., Shen B., Qian S.-B.. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nanda J.S., Cheung Y.-N., Takacs J.E., Martin-Marcos P., Saini A.K., Hinnebusch A.G., Lorsch J.R.. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 2009; 394:268–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cui Y., Dinman J.D., Kinzy T.G., Peltz S.W.. The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol. Cell. Biol. 1998; 18:1506–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martin-Marcos P., Nanda J., Luna L.E., Wagner G., Lorsch J.R., Hinnebusch A.G.. β-hairpin loop of eIF1 mediates 40S ribosome binding to regulate initiator tRNAMet recruitment and accuracy of AUG selection in vivo. J. Biol. Chem. 2013; 288:27546–27562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Asano K., Sachs M.S.. Translation factor control of ribosome conformation during start codon selection. Genes Dev. 2007; 21:1280–1287. [DOI] [PubMed] [Google Scholar]
- 40. Luna R.E., Arthanari H., Hiraishi H., Nanda J., Martin-Marcos P., Markus M., Arabayov B., Milbradt A., Luna L.E., Seo H.-C. et al. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β. Cell Rep. 2012; 1:689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Imataka H., Olsen S., Sonenberg N.. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 1997; 16:817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weser S., Riemann J., Seifart K.H., Meissner W.. Assembly and isolation of intermediate steps of transcription complexes formed on the human 5S rRNA gene. Nucleic Acids Res. 2003; 31:2408–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu W.-F., Zhang A., He G.-J., Yan Y.-B.. R3H domain stabilizes poly(A)-specific ribonuclease by stabilizing the RRM domain. Biochem. Biophys. Res. Commun. 2007; 360:846–851. [DOI] [PubMed] [Google Scholar]
- 44. Takayama S., Reed J.C.. Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 2001; 3:237–241. [DOI] [PubMed] [Google Scholar]
- 45. Greenhough J., Papadakis E.S., Cutress R.I., Townsend P.A., Oreffo R.O.C., Tare R.S.. Regulation of osteoblast development by Bcl-2-associated athanogene-1 (BAG-1). Sci. Rep. 2016; 6:33504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wethmar K., Smink J.J., Leutz A.. Upstream open reading frames: molecular switches in (patho)physiology. Bioessays. 2010; 32:885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Uhlmann-Schiffler H., Rössler O.G., Stahl H.. The mRNA of DEAD box protein p72 is alternatively translated into an 82-kDa RNA helicase. J. Biol. Chem. 2002; 277:1066–1075. [DOI] [PubMed] [Google Scholar]
- 48. Luna R.E., Arthanari H., Hiraishi H., Akabayov B., Tang L., Cox C., Markus M.A., Luna L.E., Ikeda Y., Watanabe R. et al. The interaction between eukaryotic initiation factor 1A and eIF5 retains eIF1 within scanning preinitiation complexes. Biochemistry. 2013; 52:9510–9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Singh C.R., Lee B., Udagawa T., Mohammad-Qureshi S.S., Yamamoto Y., Pavitt G.D., Asano K.. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 2006; 25:4537–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takahashi K., Yamanaka S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126:663–676. [DOI] [PubMed] [Google Scholar]
- 51. Iwafuchi-Doi M., Zaret K.S.. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014; 28:2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ye J., Kumanova M., Hart L.S., Sloane K., Zhang H., De Panis D., Bobrovnikova-Marjon E., Diehl J.A., Ron D., Koumenis C.. The GCN2-ATF4 pathway is critical for tumor cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010; 29:2082–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ivanov I.P., Loughran G., Sachs M.S., Atkins J.F.. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc. Natl. Acad. Sci. U.S.A. 2010; 107:18056–18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stewart J.D., Cowan J.L., Perry L.S., Coldwell M.J., Proud C.G.. ABC50 mutants modifly translation start codon selection. Biochem J. 2015; 467:217–229. [DOI] [PubMed] [Google Scholar]
- 55. Tzani I., Ivanov I.P., Andreev D.E., Dmitriev R.I., Dean K.A., Baranov P.V., Atkins J.F., Loughran G.. Systematic analysis of the PTEN 5′ leader identifies a major AUU initiated proteoform. Open Biol. 2016; 6:150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.