Abstract
RNase R is a conserved exoribonuclease in the RNase II family that primarily participates in RNA decay in all kingdoms of life. RNase R degrades duplex RNA with a 3′ overhang, suggesting that it has RNA unwinding activity in addition to its 3′-to-5′ exoribonuclease activity. However, how RNase R coordinates RNA binding with unwinding to degrade RNA remains elusive. Here, we report the crystal structure of a truncated form of Escherichia coli RNase R (residues 87–725) at a resolution of 1.85 Å. Structural comparisons with other RNase II family proteins reveal two open RNA-binding channels in RNase R and suggest a tri-helix ‘wedge’ region in the RNB domain that may induce RNA unwinding. We constructed two tri-helix wedge mutants and they indeed lost their RNA unwinding but not RNA binding or degrading activities. Our results suggest that the duplex RNA with an overhang is bound in the two RNA-binding channels in RNase R. The 3′ overhang is threaded into the active site and the duplex RNA is unwound upon reaching the wedge region during RNA degradation. Thus, RNase R is a proficient enzyme, capable of concurrently binding, unwinding and degrading structured RNA in a highly processive manner during RNA decay.
INTRODUCTION
RNase R is a typical member of the RNase II family of ribonucleases present in all kingdoms of life (1). The RNase II family of ribonucleases bears a conserved RNB domain with 3′-to-5′ exoribonuclease activity and cleaves RNA during messenger RNA turnover (2–4) and ribosomal RNA maturation (5,6). Two RNase II family proteins—RNase R and RNase II—are present in Escherichia coli and both participate in RNA decay (2,7). In yeast, the RNase II family protein Rrp44 is associated with the multi-protein exosome complex in cytoplasm, whereas DSS1 is associated with the helicase Suv3 in mitochondria, and both proteins play a primary role in RNA decay (8,9). In humans, the RNase R homologs Dis3L and Dis3L1 are also exosome components participating in RNA processing and degradation, whereas Dis3L2 is an exosome-independent protein that degrades microRNA and non-coding RNA (10–13). Malfunctions of Dis3L and Dis3L2 are linked to human diseases, including multiple myeloma (14) and Perlman syndrome (15), indicative of the crucial roles the RNase II family of exoribonucleases play in RNA metabolism (16).
The roles of RNase R and RNase II in RNA decay have been extensively characterized in E. coli (4,17). Messenger RNAs are first processed by pyrophosphohydrolase RppH (18) to remove the 5′-end pyrophosphate and then they are cleaved by endoribonucleases such as RNase E (19) and RNase III (20). The fragmented RNAs are further degraded by 3′-to-5′ exoribonucleases, such as RNase R (4), RNase II (7) and PNPase (17). RNase II only degrades linear RNA, whereas RNase R and PNPase are responsible for degrading structured RNAs such as those with repetitive sequences (4). PNPase requires a DEAD-box helicase RhlB to unwind RNA and promote its exoribonuclease activity in degrading structured RNA (21). In contrast, RNase R can work independently to degrade duplex RNA with a 3′-overhang, suggesting that RNase R is a unique enzyme that possesses both RNA degrading and unwinding activities (22).
Genetic studies further support that RNase R acts as a helicase. Single deletion of either PNPase, RNase II or RNase R elicits only a mild effect on E. coli survival. However, double deletion of RNase R and PNPase is lethal, evidencing their overlapping function in degrading structured RNA (17). Moreover, overexpression of RNase R can rescue the temperature-sensitive phenotype resulting from deletion of CsdA (an RNA helicase that participates in the cold-shock response), evidencing that RNase R can compensate for the function of a helicase (22). RNase R also interacts with the SmpB–tmRNA complex and participates in the trans-translational process, representing a pathway to rescue stalled ribosomes under cold-shock conditions (23). These findings demonstrate that both the RNA unwinding and degrading roles of RNase R are crucial for cell growth and survival in E. coli.
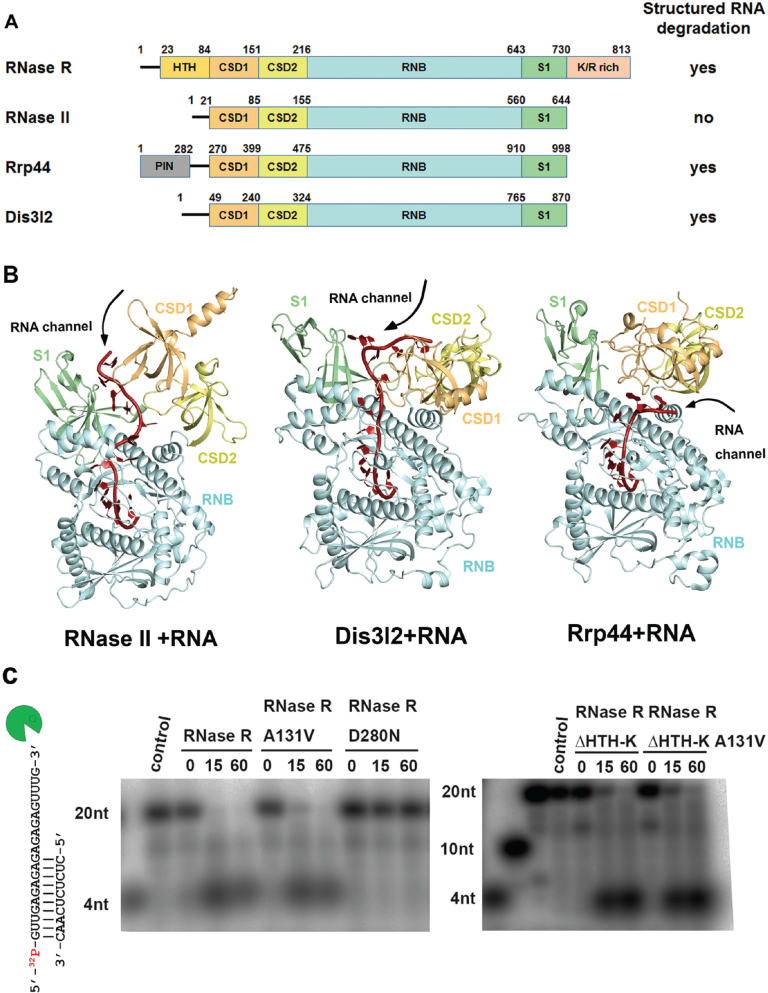
Besides its RNB exoribonuclease domain, RNase R also consists of a helix-turn-helix (HTH) domain, two cold shock domains (CSD1 and CSD2), an S1 domain and a K/R-rich domain (Figure 1A). Other RNase II family proteins (such as RNase II, Rrp44 and Dis3l2) share with RNase R the RNB domain, as well as the auxiliary domains at N- and C-terminal regions, including the CSD1, CSD2 and S1 domains (Figure 1A and Supplementary Figure S1). The RNB domain in RNase R is suggested to play a primary role in RNA unwinding and degradation, whereas the remaining auxiliary domains are responsible for RNA binding (24,25). Duplex RNA is suggested to be unwound upon reaching a ‘wedge’ region in the RNB domain, and the unwound 3′-end RNA strand is further pulled into the RNB domain and degraded in the active site (26). Single-molecule fluorescence analysis has revealed that the RNA unwinding energy for Rrp44 is provided by RNA hydrolysis, which differs from classical helicases that acquire energy from ATP hydrolysis (27).
Figure 1.
Domain organization and structure comparison of the RNase II family of exoribonucleases. (A) Overall domain organization of the RNase II family proteins: RNase R, RNase II, Rrp44 and Dis3l2. RNase R, Rrp44 and Dis3l2 can degrade RNA with secondary structures, but RNase II cannot, as indicated at right. (B) Crystal structures of the RNA-bound form of RNase II (PDB: 2IX1), Dis3l2 (PDBID: 4PMW) and Rrp44 (PDB: 2VNU). The open top and side channels are marked on the structures. (C) Wild-type RNase R and A131V mutant (100 nM) unwound and degraded the 5′-end-32P-labeled dsRNA (2.5 nM), whereas the D280N mutant could not degrade dsRNA with a 3′ overhang (left panel). The truncated mutant RNase R ΔHTH-K and its A131V mutant (crystallized protein in this study) retained their activity in dsRNA unwinding and degradation (right panel).
Similar to RNase R, Rrp44 and Dis3l2 can degrade structured RNA with a 3′ overhang, so this subgroup of RNase II-family proteins possesses both RNA unwinding and degrading activities (Figure 1A) (11). Crystal structures of Dis3l2 (11) and RNase II (28) bound to single-stranded RNA reveal that the CSD1 and S1 domains form an RNA-binding channel (referred to hereafter as the ‘top channel’), through which RNAs are threaded into the active site of the RNB domain (Figure 1B). In contrast, the crystal structure of RNA-bound Rrp44 (8) shows a different binding mode, with single-stranded RNAs being guided into the active site on being bound between the CSD1 and RNB domains (referred to hereafter as the ‘side channel’) (Figure 1B). Crystal structures of Rrp44 in complex with exosomes also reveal an open side channel bound with single-stranded RNA (29,30). Therefore, it still remains unclear how duplex RNA with a 3′ overhang is bound and unwound by the RNase II family of exoribonucleases.
To understand how RNase R degrades RNA, here, we report the crystal structure of RNase R at a resolution of 1.85 Å. In contrast to the apo-form of RNase II containing only one open top channel, the crystal structure of RNase R reveals two open top and side channels. We also reveal a tri-helix ‘wedge’ region within the RNB domain in RNase R by truncation and mutation studies that is crucial for RNA unwinding but not degrading activity. Based on our structural and biochemical data, we provide a working model for how RNase R binds, unwinds and degrades structured RNA to promote RNA decay. These studies illuminate how the RNase II family protein, RNase R, has evolved into an elegant enzyme possessing both RNA unwinding and degrading activities.
MATERIALS AND METHODS
Expression vector construction
The full-length RNase R gene was amplified by PCR from E. coli BL21 (DE3) using the forward primer: 5′-GGGATATCCATATGTCACAAGCTTTCCAG-3′ and the reverse primer: 5′-CCGGAATTCTCACTCTGCCACTTTTTTCT-3′. The truncated RNase R ΔHTH-K (residues 87 to 725) was amplified from the full-length gene using the forward primer: 5′-GGGAATTCCATATGGGTACCGTTATTGGCCACC-3′ and the reverse primer: 5′-CCGCAATTCTCAGATCAGGCTAAAGTCGATTTT-3′. The PCR products were inserted into EcoRI/NdeI sites of the pET-28a expression vector (Invitrogen) to produce proteins with N-terminal 6xHistidine tags. The wedge mutants RNase R Δ3H and 1H were constructed by overlapping extension polymerase chain reaction (OE-PCR) (31). For Δ3H, fragments with residues 487–545 were deleted using the primers 5′-TTCCGTATTCACGACGGCAGCCAGGCGATTTACGATCCACAAAACC-3′ and 5′-CTGCCGTCGTGAATACGGAACAGTGCCGGTTCTTTCGCTTTCTCAACG-3′. For the 1H mutant, OE-PCR was carried out in two steps; first, residues 524–535 in RNase R were replaced by residues 458–465 of RNase II and then residues 536–545 in RNase R were replaced by residues 466–473 of RNase II. The primers used for the first step were 5′-GCGGAGCTGCTGGAGGCGCAACCAACTGGTTTCCTCGACCAGGCGATTTACGATCC-3′ and 5′-GGATCGTAAATCGCCTGGTCGAGGAAACCAGTTGGTTGCGCCTCCAGCAGCTCCGC-3′, whereas the primers used for the second step were 5′-ACTGGTTTCCTCGACAGCCGCATTCGTCGCTTCCAGTCACAGGCGATTTACGATCCAG-3′ and 5′-CGGCTGTCGAGGAAACCAGTTGGTTGCGCCTCCAGCAGCTCCGC-3′.
Protein expression and purification
All the constructs were transformed into E. coli (BL21) cells (Stratagene) incubated at 37°C. When OD600 reached 0.6, protein expression was induced using 1.0 mM IPTG and overnight incubation at 18°C. After centrifugation, cells were collected and resuspened in buffer A (25 mM Tris–HCl, pH7.5, 500 mM NaCl, 5–10 mM imidazole, 5 mM beta-ME, 1 mM PMSF), then ruptured by a microfluidizer (Microfluidics M-110P). The supernatant was applied to the HisTrap HP column (GE HealthCare) prior to being equilibrated by buffer A. Bound proteins were eluted using an imidazole gradient between buffer A and buffer B (20 mM Tris–HCl, pH 8.0, 500 mM NaCl, 300 mM imidazole). Eluted protein fractions were combined and (NH4)2SO4 was added up to 1.5 M. Protein samples were loaded into a reverse-phase phenyl column (GE HealthCare) equilibrated by the phenyl buffer (25 mM Tris–HCl pH 8.0, 1.5 M (NH4)2SO4). Proteins were then eluted with a gradient of 1.5 to 0 M (NH4)2SO4. The purified RNase R was concentrated and applied to a gel filtration column (GE HealthCare, Superdex 200 Increase 10/300 GL) in a running buffer of 20 mM Tris–HCl, pH 7.5 and 400 mM NaCl. Collected protein samples were concentrated to ∼15 mg/ml and stored at –20°C.
Crystallization and structure determination
RNase R was crystallized by the hanging-drop vapor diffusion method at room temperature by mixing 1 μl of protein sample (15 mg/ml RNase R ΔHTH-K, 20 mM Tris–HCl, pH7.5 and 400 mM NaCl) and 1 μl reservoir solution (modified from Hampton PEG/Ion 2 Screen, number 14: 8% Tacsimate, pH6.0 and 15% Polyethylene glycol 3350). X-ray diffraction data were collected at BL-13C1 at the National Synchrotron Radiation Research Center (NSRRC), Hsinchu, Taiwan, and the collected data were processed and scaled by HKL2000 (32). The structure was solved by the molecular replacement method using yeast Rrp44 (PDB id: 2VNU) as the search model by the AutoMR function in the program Phenix (33). The structure model was built using Coot and refined by Phenix, containing two RNase R molecules in the asymmetric unit. Disordered regions not built in the model included residues 102–105, 132–136 and 428–431 in chain A molecule, and residues 101–105, 127–137 and 429–431 in chain B molecule.
RNase activity assays
For the RNA degradation experiments shown in Figure 1C, RNase R proteins (100 nM), including the full-length RNase R, RNase R A131V, RNase R D280N, ΔHTH-K and ΔHTH-K A131V, were incubated with the dsRNA (2.5 nM) at 37°C for 0–60 min in a reaction buffer containing 20 mM Tris–HCl (pH 7.5), 100 mM NaCl, 1 mM DTT and 0.25 mM MgCl2. The dsRNA was prepared by annealing a 5′-end 32P-labeled 20-nucleotide RNA with a sequence of 5′-GUUGAGAGAGAGAGAGAGUUUG-3′ with a 10-nucleotide RNA with a sequence of 5′-CUCUCUCAAC-3′ to generate a 10-basepair dsRNA with a 10-nucleotide 3′ overhang. The RNA degradation reactions were stopped by adding 2× urea loading dye (Thermo) at different time points (0, 15 and 60 min) and the samples were loaded on a 20% TBE–urea denaturing gel for gel-eletctrophresis shown in Figure 1C.
For the RNA degradation experiments shown in Figure 3, the 5′-end 32P-labeled single-stranded RNA (ssRNA_40, 2.5 nM, 5′-GCUGUCUAGAGACUAUCGAUUUUUUUUUUUUUUUUUUUUU-3′) and the stem–loop RNA with a 20-nt 3′ overhang (slRNA_20, 2.5 nM, 5′-GGCCCGCGAAAGCGCGGGCCAAAAAAAAAAAAAAAAAAAA-3′) were incubated respectively with RNase R proteins (100 nM) in 10 μl of a reaction buffer containing 20 mM Tris–HCl (pH 7.5), 100 mM NaCl, 0.25 mM MgCl2 and 1 mM DTT at 37°C. The RNA degradation reactions were stopped by adding Novex® TBE-Urea Sample Buffer at the indicated times, 0–60 min. RNA degradation patterns were analyzed by 7.5 M urea/20% (w/v) polyacrylamide denaturing PAGE which was exposed on a FujiFilm Image plate and detected by Typhoon FLA 9000 (GE HealthCare).
Figure 3.
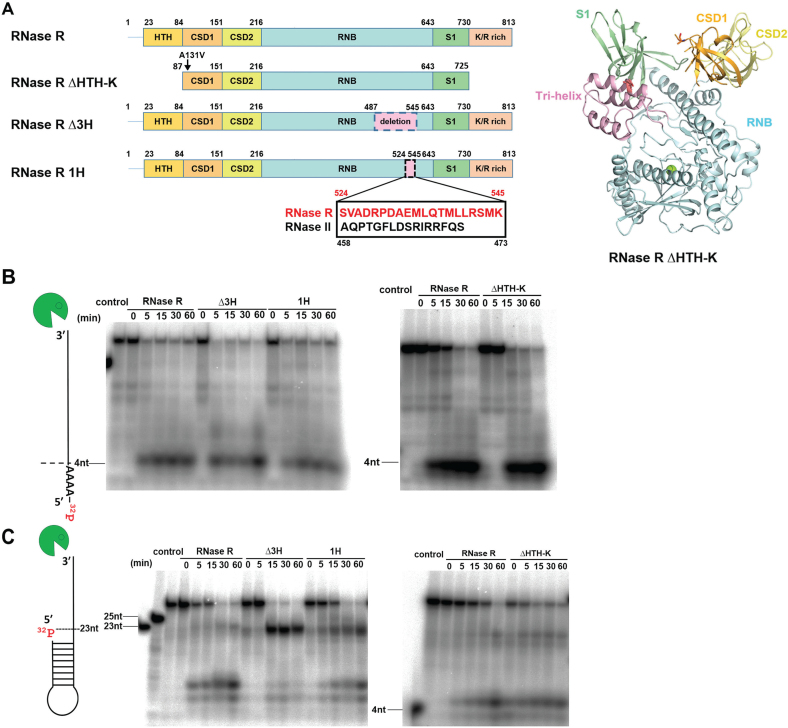
A tri-helix ‘wedge’ region in the RNB domain of RNase R is crucial for its RNA unwinding activity. (A) Two mutants were constructed; a tri-helix region (residues 487–545) was deleted from the tri-helix mutant RNase R Δ3H, whereas a 21-residue helix (residues 524–545) was replaced by the corresponding helix from RNase II (residues 458–473) in the RNase R 1H mutant. The tri-helix region is colored in pink in the crystal structure of RNase R ΔHTH-K displayed in the right panel. (B) RNA degradation assays show that RNase R and its mutants Δ3H, 1H and ΔHTH-K (100 nM) fully degraded the 40-nt single-stranded RNA (2.5 nM) to a final product of ∼4 nt. (C) Full-length RNase R and truncation mutant ΔHTH-K degraded the stem–loop RNA with a 3′ overhang to a final product of ∼4 nt, but the wedge mutants Δ3H and 1H could not fully degrade the stem–loop RNA and generated major digested products of ∼23 nt.
RNA binding by fluorescence polarization assays
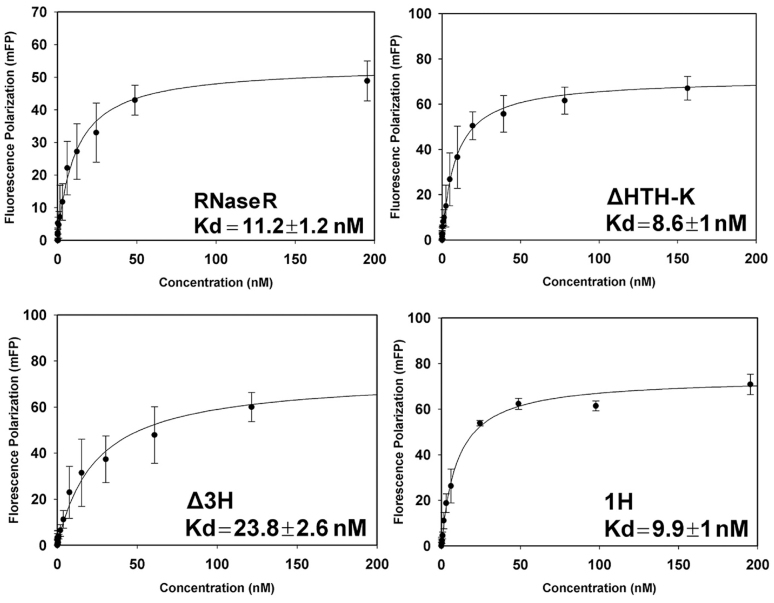
RNA binding by RNase R was determined by measuring the changes in fluorescence polarization using a Paradigm plate reader (Molecular Devices). The RNA with a sequence of 5′-Cy3-AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA-3′, labeled at the 5′-hydroxyl group with Cyanine-3 (MD-Bio) at a 10 nM final concentration was titrated with the indicated concentrations of RNase R in binding buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl and 25 mM EDTA). Reactions were prepared at 37°C and incubated for 15 min, then read at 595 nm at an excitation of 535 nm. The RNA-binding affinities of different RNase R mutants were calculated by fitting the plot shown in Figure 4 to a one-site saturation-binding model using SigmaPlot (34).
Figure 4.
RNA-binding affinities of RNase R mutants measured by fluorescence polarization assays. A 5′-end Cyanine-3-labeled single-stranded RNA (30 nucleotides, polyA) was used as the substrate to measure the binding affinity with RNase R proteins in the presence of EDTA. The ssRNA binds RNase R with a Kd of 11.2 ± 1.2 nM for the full-length RNase R, 8.6 ± 1.0 nM for RNase R ΔHTH-K, 23.8 ± 2.6 nM for RNase R Δ3H, and 9.9 ± 1.0 nM for RNase R 1H. The average of three independent experiments is shown with error bars representing one standard deviation. Kd values with standard errors were obtained by fitting the curve to a one-site saturation-binding model using SigmaPlot (34).
RESULTS
RNase R degrades duplex RNA with a 3′ overhang
To understand how RNase R unwinds and degrades RNA, we expressed and purified various forms of RNase R, including full-length RNase R (residues 1–813) and the truncated RNase R ΔHTH-K (residues 87–725) (see Figure 2A). However, after DNA sequencing, we found that the expression vector for RNase R ΔHTH-K carried a spontaneous mutation (A131V). We further mutated V131 back to A131 to generate the RNase R ΔHTH-K with the wild-type sequence. We also mutated the catalytic residue D280 to N in the two constructs to prepare inactive RNase R for co-crystallization experiments with RNA as it has been shown that D280A mutation in RNase R could generate an inactive exoribonuclease (35,36). The recombinant N-terminal His-tagged proteins were purified by chromatographic methods and they were eluted as monomers in the last step of size exclusion chromatography with a molecular weight of ∼92 kDa for full-length RNase R (calculated MW: 92 105 Da) and ∼75 kDa for RNase R ΔHTH-K (calculated MW: 72 381 Da) (Supplementary Figure S2).
Figure 2.
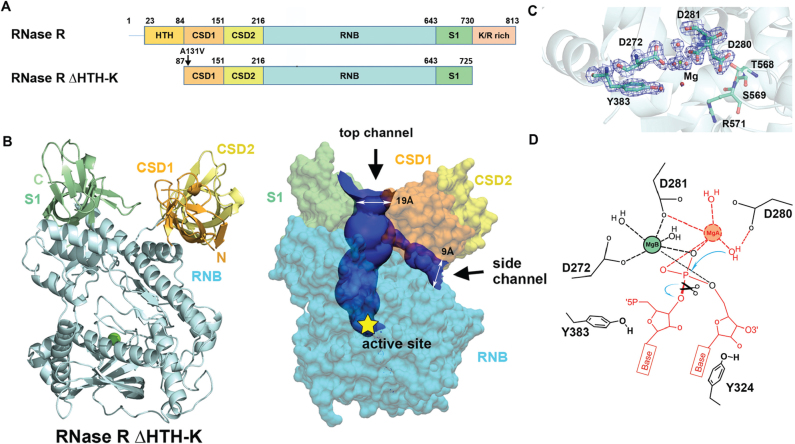
Crystal structure of RNase R ΔHTH-K reveals two open channels and a Mg2+-bound active site. (A) Domain structures of the full-length RNase R and the truncated RNase R ΔHTH-K. (B) Crystal structure of RNase R ΔHTH-K reveals that the RNB domain (sky blue) is capped with S1 (light green), CSD1 (orange yellow) and CSD2 (yellow) domains (PDB entry: 5XGU). The Mg2+ located in the active site is shown as a green sphere. The top and side RNA-binding channels displayed in the right panel were calculated by HOLE2 (39). (C) The 2Fo - Fc Fourier map of the active site in the RNB domain contoured at 1.9σ (blue). The Mg (B) ion is displayed as a green sphere, whereas water molecules are displayed as red spheres. (D) The proposed two-metal-ion-dependent hydrolysis mechanism of RNase R. Only one Mg2+ (B site) that bound to D272 and D281 is observed in the crystal structure. The second Mg2+ (A site) is only bound upon RNA binding. D280 functions as the general base to deprotonate the Mg(A)-coordinated water for nucleophilic attack on a scissile phosphate. The RNA and Mg2+ atoms displayed in red are not observed in the crystal structure of the apo-form of RNase R.
To test the RNA degrading activity of the recombinant proteins, we incubated the full-length RNase R, A131V or D280N mutant, as well as the RNase R ΔHTH-K with or without A131V mutation, with 32P-labeled double-stranded RNA with a 10-nucleotide 3′ overhang. Full-length RNase R and A131V mutant efficiently unwound and degraded the double-stranded RNA with a 10-nucleotide 3′ overhang, but the D280N mutant did not degrade the dsRNA, suggesting the indispensable role of the D280 residue in RNA degradation (Figure 1C). Moreover, RNase R ΔHTH-K with or without A131V mutation degraded the dsRNA, with a slightly lower activity as compared to the full-length RNase R, suggesting that the truncation and A131V mutation did not significantly affect the enzyme activity (Figure 1C).
RNase R is a fully functional exoribonuclease that degrades duplex RNA with a 3′ overhang in the absence of ATP (27). However, it has been suggested that the helicase activity of RNase R is promoted by ATP, as an RNase R inactive mutant D272N unwound duplex RNA with a 3′ overhang in the presence of ATP (37,38). To clarify this issue, we further tested the RNA unwinding activity of the RNase R inactive mutant D280N in the presence of ATP (5 mM). In comparison to the wild-type RNase R which unwound and degraded duplex RNA with or without the presence of ATP, RNase R D280N could not unwind or degrade the duplex RNA with an 10-nucleotide 3′ overhang in the presence of ATP (see Supplementary Figure S3). Taken together, our results suggest that RNase R is an ATP-independent exoribonuclease in degrading duplex RNA with a long 3′ overhang.
Crystal structure of RNase R
Both the full-length RNase R and the truncated mutants with or without the D280N or A131V mutation were screened for crystallization conditions. Only RNase R ΔHTH-K with A131V mutation yielded crystals that appeared within two weeks using the hanging drop vapor diffusion method. The mutation of A131V in RNase R ΔHTH-K promoted crystallization but it had no effect on the degrading activity in comparison to the one with the wild-type sequence (Figure 1C). The crystal structure of RNase R ΔHTH-K with A131V mutation was determined at a resolution of 1.85 Å by molecular replacement using the structure of Rrp44 (PDB entry: 2VNU) as the search model. The final model contained two RNase R molecules in one asymmetric unit in the monoclinic P21 unit cell, with each molecule containing one Mg2+ ion bound in the active site. The mutated site V131 was located in a disordered loop, and so it remains unclear why the mutation promoted protein crystallization. The final model has an R-factor of 22.22% for 124 300 reflections and an R-free of 24.02% for 2005 reflections. All the refinement statistics are listed in Table 1.
Table 1. X-ray data collection and refinement statistics for RNase R ΔHTH-K.
| Data collection statistics | |
| Wavelength (Å) | 1.0 |
| Space group | P21 |
| Cell dimensions (a, b, c) (Å) | 73.69, 120.86, 83.63 |
| (β) (°) | 91.61 |
| Resolution (Å) | 30–1.85 (1.92–1.85)* |
| Observed/unique reflections | 124 328/48 331 |
| Data redundancy | 4.5 (4.5)* |
| Completeness (%) | 99.14 (99.9)* |
| R sym (%) | 8.9 (44.5)* |
| I/σ(I) | 11.8 (3.7)* |
| Refinement statistics | |
| Resolution range | 29.67–1.85 |
| Reflections (work/test) | 124 300/2005 |
| R-work/R-free (%) | 22.22/ 24.02 |
| Number of atoms (protein/water) | 9,278/736 |
| Average B-factor (protein/solvent) (Å2) | 22.11/29.96 |
| RMSD in bond length (Å)/bond angle (o) | 0.08/1.165 |
| Ramachandran plot& (%) | |
| Favorable | 99.8 |
| Allowed | 0.2 |
| Disallowed | 0.0 |
*The statistics for the highest resolution shell (1.92-1.85 Å) are shown in parentheses.
&Ramachandran plot statistics were calculated by PROCHECK.
The domain arrangement of RNase R is similar to those of RNase II and Rrp44, with the CSD1, CSD2 and S1 domains capping the top of the RNB domain (see Figure 2B). For clarity, only one RNase R model (chain A in PDB entry 5XGU) was used for structural comparison, as the two molecules in the asymmetric unit matched well (average RMSD of 0.5 Å for 618 Cα atoms). Superimposition of the Cα atoms in the RNB domain of RNase R with those of RNase II or Rrp44 gave, respectively, an average RMSD of 2.3 Å (residues 156–560) and 2.6 Å (residues 134–555), suggesting that their RNB domains share a similar overall structure. However, when only the RNB domains were superimposed, the auxiliary domains of RNase R—CSD1, CSD2 and S1—were positioned and oriented differently than those in RNase II and Rrp44, with average RMSD of 23.1 and 12.1 Å for CSD1, 18.1 and 6.9 Å for CSD2, and 12.3 and 6.9 Å for the S1 domain, respectively. These differences suggest that the RNA-binding domains might be flexible or that these exoribonucleases might have different RNA binding modes. Moreover, the overall domain arrangement of RNase R more closely resembles that of Rrp44 than that of RNase II.
Two RNA-binding channels in RNase R
The crystal structure of RNase R reveals a unique feature that differs from the other reported crystal structures of the RNase II family exoribonucleases in that RNase R has not one but two open channels, i.e. a top channel between the S1 and CSD1 domains and a side channel between the RNB and CSD1 domains (see the channels calculated by HOLE2 (39) in Figure 2B). The top channel has a width of ∼19 Å that is sufficient for accommodating a duplex RNA, whereas the side channel is narrower with a width of ∼9 Å. However, RNase II has only one open top channel and the side channel is closed in both apo- and ssRNA-bound forms (see Figure 1B). On the contrary, Rrp44 has only one open side channel and the top channel is closed due to blocking by the interactions between CSD1 and S1 domains. Previous mutation and truncation studies on RNase R support that the S1, CSD1 and CSD2 domains are involved in binding to RNA (24). Therefore, we suggest that RNase R has two open channels, i.e. top and side, that can serve as RNA-binding sites.
Hydrolysis mechanism of RNase R
A close look at the active site in RNase R reveals a Mg2+ ion bound by residues D272 and D281 that matches the same position of the Mg2+ ion in the crystal structures of RNase II (bound by D201 and D210, PDB entry: 2IX1) and Rrp44 (bound by D549 and D552, PDB entry: 2VNU). RNase II family proteins use a two-metal ion catalytic mechanism for RNA hydrolysis (28) but, in the crystal structure of the apo-form of RNase R, only one Mg2+ (B site) ion was observed. The second Mg2+ ion (A site) was not present in the structure as it is only bound upon RNA binding. We propose a hydrolysis mechanism for RNase R that is based on previous studies on RNase II (40), whereby D280 functions as the general base to deprotonate the Mg(A)-coordinated water for nucleophilic attack on a scissile phosphate (Figure 2D). This explains why mutation of D280 to N produces an inactive mutant in terms of RNA degradation (Figure 1C). Moreover, Y324 and Y383 are matched at the same positions as Y253 and Y492 in RNase II that stacks or interacts with the 3′-end nucleotides (28). These two tyrosine residues thus likely share a similar role as the ones in RNase II responsible for clamping the 3′-end nucleobases and setting the end-product (36). The bound RNA substrate is thereby hydrolyzed to produce the cleaved products of a nucleoside monophosphate and an RNA fragment with a 5′-end phosphate and a 3′-end OH group.
The tri-helix region in the RNB domain is the ‘wedge’ for RNA unwinding
It has been reported that RNase R can degrade structured RNA by only the RNB domain, suggesting that this domain is not only responsible for RNA degradation but also for RNA unwinding (24). To further investigate which region in the RNB domain is responsible for RNA unwinding, we compared the crystal structures of RNA-bound RNase II, Dis3l2 and Rrp44 and found that a tri-helix region in the RNB domain interacts with RNA in Dis3l2 and Rrp44. However, this region in RNase II (displayed in pink in Figure 3A) does not interact with RNA, indicating that it might be involved in RNA unwinding in Dis3l2 and Rrp44. To test the hypothesis that this tri-helix region contributes to the RNA unwinding activity of RNase R, we constructed a tri-helix deletion mutant (RNase R Δ3H) and a single-helix replacement mutant (RNase R 1H). The tri-helix region (residues 487–545) in the Δ3H mutant was deleted, whereas a 21-residue helix (residues 524–545) in the 1H mutant was replaced by the corresponding helix (residues 458–473) of RNase II (Figure 3A). These two tri-helix mutants, Δ3H and 1H, shared similar circular dichroism spectra to that of wild-type RNase R (Supplementary Figure S4A) and they all had comparable thermal melting points of about 65.7–70.0°C (Supplementary Figure S4B), suggesting that the truncation and mutations did not disturb the overall protein folding.
To test the RNA degrading activity of these wedge mutants, we incubated full-length and mutated RNase R with 32P-labeled single-stranded 40-nt RNA and a stem–loop RNA with a 3′ overhang in the presence of magnesium ions. Full-length RNase R and the truncation mutant ΔHTH-K degraded the single-stranded RNA to a final product of ∼4 nucleotides. The wedge mutants Δ3H and 1H also degraded the single-stranded RNA to a final product of ∼4 nucleotides with a similar activity compared to the full-length RNase R (Figure 3B). On testing stem–loop RNA with a 3′ overhang, full-length RNase R and ΔHTH-K degraded RNA into the stem region and generated the final product of ∼4 nt, whereas the wedge mutant Δ3H could not fully degrade the stem–loop RNA and produced major digested products of ∼23 nt and minor products of ∼4 nt (Figure 3C). The wedge 1H mutant degraded the stem–loop RNA slightly better than Δ3H mutant, generating the major digested products of ∼23 nt and ∼4 nt. Taken together these results show that the RNase R wedge mutants Δ3H and 1H could fully degrade single-stranded RNA without secondary structure, but they lost partially their RNA unwinding activity so they could not completely unwind and degrade the stem–loop RNA.
To further test the RNA-binding activity of RNase R wedge mutants, we measured the binding affinity between RNase R and a 5′-end Cyanine-3-labeled single-stranded RNA in the absence of Mg2+ (by adding 25 mM of EDTA) by fluorescence polarization assays. Full-length RNase R and ΔHTH-K bound ssRNA tightly, with dissociation constants (Kd) of 11.2 ± 1.2 nM and 8.6 ± 1.0 nM, respectively. The RNase R wedge mutants Δ3H and 1H also bound ssRNA with a high affinity; Kd values of 23.8 ± 2.6 nM and 9.9 ± 1.0 nM, respectively (Figure 4). These results show that deletion of the tri-helix region or replacement of one helix in the RNB domain does not significantly disrupt the RNA-binding ability of RNase R. Our activity assays thus support that the tri-helix region in RNase R is critical for RNA unwinding but not for RNA binding or degradation. Therefore, this tri-helix region may serve as the ‘wedge’ to unwind duplex RNA.
DISCUSSION
How are structured RNAs unwound and degraded by RNase R? Based on the crystal structure of the RNase R truncation mutant ΔHTH-K, we identified two open RNA-binding channels. Both of these channels, top and side, are used as RNA-binding channels by RNase II family exoribonucleases; RNase II and Dis 312 bind RNA in the top channel, whereas Rrp44 binds it in the side channel (Figure 1C). Our result suggests that both of the open channels in RNase R contain positively charged residues that could be RNA-binding sites. The top channel has a width of ∼19 Å that is wide enough for accommodating a duplex RNA, and on the other hand, the side channel has a width of ∼9 Å that is sufficient for binding of a single-stranded RNA. However the auxiliary RNA-binding domains in RNase R, including HTH, CSD1, CSD2, S1 and K/R rich domains, could be flexible, and the widths of the channels may be changed upon RNA binding. RNase R degrades RNA in a highly processive manner, with one RNase R molecule being able to unwind over 500 bp of a structured substrate, as measured by single-molecule optical-trapping assay (41). The two open RNA-binding channels support the notion that RNA is tightly enclosed by RNase R and, therefore, it is difficult to dissociate RNA from the enzyme during degradation. Hence, compared to PNPase that also encloses RNA with a ring-like structure but has much more limited processivity, RNase R uses its two RNA-binding channels to ensure that RNA remains firmly associated with the enzyme during RNA degradation.
We also identified a tri-helix wedge region in the RNB domain that is involved in RNA unwinding but not in RNA binding or degradation (see Figures 4 and 5). The wedge mutants, Δ3H and 1H, degraded the stem–loop RNA we tested close to the duplex region to produce a final product of a duplex RNA with a ∼3-nucleotide overhang (Figure 4). This result suggests that if the RNA unwinding activity of RNase R is inhibited, it can only degrade the 3′ overhang of a structured RNA molecule up to a limit of three nucleotides. A similar result was observed for Rrp44 digestion of a cross-linked RNA duplex that could not be unwound, which produced a duplex with a 3-nucleotide overhang (27). Moreover, the RNase R 1H mutant lost partially its RNA unwinding activity, even though only one helix (residues 524–545) was replaced by the corresponding one from RNase II (residues 458–473). This result suggests that this helix in RNase R plays a crucial role in determining the RNA unwinding activity of RNase R.
Figure 5.
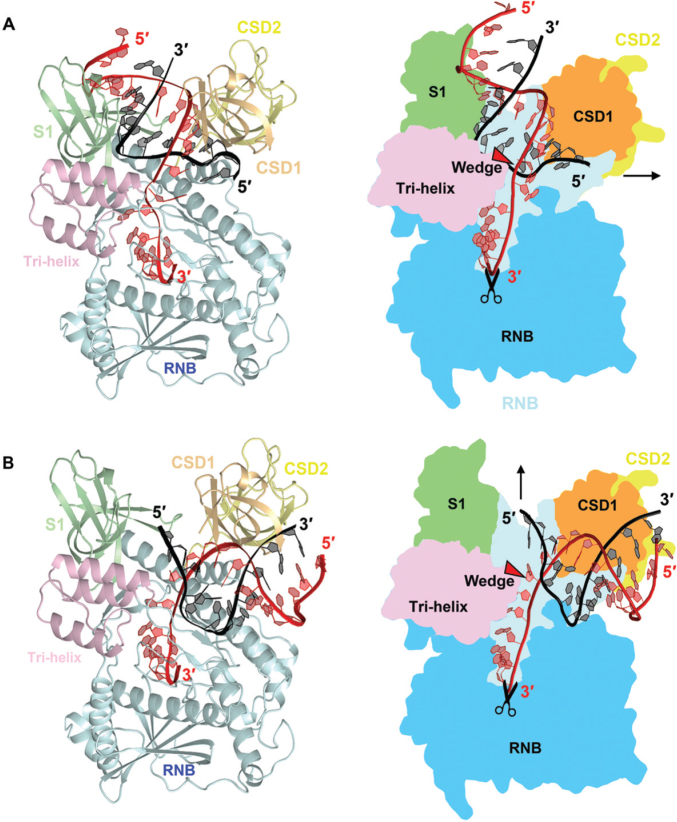
Working models for RNA binding, unwinding and degradation by RNase R. (A) An RNase R-RNA model was built based on the crystal structure of RNase II bound with RNA (PDB: 2IX1). The 3′ overhang of duplex RNA enters all the way into the active site from the top channel. The RNA duplex region is bound by the CSD1 and S1 domains in the top channel and, upon reaching the tri-helix wedge region in the RNB domain, the RNA duplex is unwound. (B) A second RNase R-RNA model was built based on the crystal structure of Rrp44 bound with RNA (PDB: 2VNU). The 3′ overhang of duplex RNA is guided through the side channel into the active site in the RNB domain. The RNA duplex region is bound by the CSD1 and RNB domains in the side channel and, upon reaching the tri-helix wedge region, the RNA is unwound. It should be noted that the side channel in RNase R is not wide enough to accommodate a duplex RNA, however, the CSD1/2 domains may be flexible and change their conformation upon RNA binding.
In combining our structural and biochemical results, we propose two possible RNA unwinding and degradation models for RNase R (Figure 5). The first model is based on the RNase II-RNA complex structure (28), whereby the single-stranded overhang of structured RNA enters from the top channel and is threaded all the way into the active site in the RNB domain (Figure 5A). The duplex region of the RNA is bound by the CSD1 and S1 domains in the top channel and, upon reaching the tri-helix wedge region of the RNB domain, the RNA duplex is unwound. RNA unwinding is driven by RNA hydrolysis, with RNA being continuously degraded in a processive manner that pulls RNA into the catalytic active site of the RNB domain. The unwound 3′ overhang is pulled into the active site for processive RNA degradation, whereas the 5′ non-scissile strand exits from the side channel. This model is supported by a previous study showing that conserved residues in the S1 domain are required for binding of duplex RNA in RNase R (36). Our model is also consistent with the previous results showing that the RNB domain alone in RNase R is capable of degrading duplex RNA with or without a 3′ overhang (24,25), as the tri-helix region is located on the top side of the RNB domain making it possible to be exposed for interacting with RNAs either with or without an overhang when the RNA-binding domains are removed.
The second model is based on the Rrp44-RNA complex structure, whereby the single-stranded overhang of duplex RNA is guided through the side channel into the active site and the RNA duplex region is bound between the CSD1 and RNB domains (see Figure 5B). As the duplex encounters the tri-helix wedge region of the RNB domain, the RNA is unwound and its 3′ overhang is further threaded into the RNB active site for cleavage while the 5′ end of the non-scissile strand exits via the top channel. Similar to the first model, the duplex RNA is bound by the RNA-binding domains, unwound upon reaching the wedge region and cleaved at the active site of the RNB domain. RNA with extensive secondary structures can thus be degraded efficiently by this unique enzyme that concurrently unwinds and degrades RNA molecules. To further verify these two models, co-crystallization of RNase R with structured RNA is underway.
DATA AVAILABILITY
Structural coordinates and diffraction structure factors of RNase R ΔHTH-K have been deposited in the RCSB Protein Data Bank with the PDB ID code 5XGU.
Supplementary Material
ACKNOWLEDGEMENTS
Portions of this research were carried out at the National Synchrotron Radiation Research Center; a national user facility supported by the Ministry of Science and Technology of Taiwan. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Core Facility Program for Biotechnology. We thank Dr Meng-Ru Ho and Dr Meng-Chiao Ho for their assistance in conducting the FP assays at the Biophysics core center at the Institute of Biological Chemistry, Academia Sinica.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
This work was supported by Academia Sinica; and Ministry of Science and Technology, Taiwan, R.O.C. Funding for open access charge: Academia Sinica, Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Reis F.P., Pobre V., Silva I.J., Malecki M., Arraiano C.M.. The RNase II/RNB family of exoribonucleases: putting the ‘Dis’ in disease. Wiley Interdiscip. Rev. RNA. 2013; 4:607–615. [DOI] [PubMed] [Google Scholar]
- 2. Cheng Z., Zuo Y., Li Z., Rudd K., Deutscher M.. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 1998; 273:14077–14080. [DOI] [PubMed] [Google Scholar]
- 3. Lalonde M.S., Zuo Y., Zhang J., Gong X., Wu S., Malhotra A., Li Z.. Exoribonuclease R in Mycoplasma genitalium can carry out both RNA processing and degradative functions and is sensitive to RNA ribose methylation. RNA. 2007; 13:1957–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng Z.F., Deutscher M.P.. An important role for RNase R in mRNA decay. Mol. Cell. 2005; 17:313–318. [DOI] [PubMed] [Google Scholar]
- 5. Bollenbach T.J., Lange H., Gutierrez R., Erhardt M., Stern D.B., Gagliardi D.. RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 2005; 33:2751–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sulthana S., Deutscher M.P.. Multiple exoribonucleases catalyze maturation of the 3′ terminus of 16S ribosomal RNA (rRNA). J. Biol. Chem. 2013; 288:12574–12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arraiano C., Matos R., Barbas A.. RNase II: the finer details of the Modus operandi of a molecular killer. RNA Biol. 2010; 7:276–281. [DOI] [PubMed] [Google Scholar]
- 8. Lorentzen E., Basquin J., Tomecki R., Dziembowski A., Conti E.. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol. Cell. 2008; 29:717–728. [DOI] [PubMed] [Google Scholar]
- 9. Mir S.S., Fiedler D., Cashikar A.G.. Ssd1 is required for thermotolerance and Hsp104-mediated protein disaggregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2009; 29:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang H.M., Triboulet R., Thornton J.E., Gregory R.I.. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013; 497:244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Faehnle C.R., Walleshauser J., Joshua-Tor L.. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature. 2014; 514:252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Staals R.H., Bronkhorst A.W., Schilders G., Slomovic S., Schuster G., Heck A.J., Raijmakers R., Pruijn G.J.. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 2010; 29:2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pirouz M., Du P., Munafo M., Gregory R.I.. Dis3l2-mediated decay is a quality control pathway for noncoding RNAs. Cell Rep. 2016; 16:1861–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robinson S.R., Oliver A.W., Chevassut T.J., Newbury S.F.. The 3′ to 5′ exoribonuclease DIS3: from structure and mechanisms to biological functions and role in human disease. Biomolecules. 2015; 5:1515–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Labno A., Warkocki Z., Kulinski T., Krawczyk P.S., Bijata K., Tomecki R., Dziembowski A.. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res. 2016; 44:10437–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tomecki R., Drazkowska K., Kucinski I., Stodus K., Szczesny R.J., Gruchota J., Owczarek E.P., Kalisiak K., Dziembowski A.. Multiple myeloma-associated hDIS3 mutations cause perturbations in cellular RNA metabolism and suggest hDIS3 PIN domain as a potential drug target. Nucleic Acids Res. 2014; 42:1270–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Donovan W.P., Kushner S.R.. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deana A., Celesnik H., Belasco J.G.. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008; 451:355–358. [DOI] [PubMed] [Google Scholar]
- 19. Mackie G.A. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998; 395:720–723. [DOI] [PubMed] [Google Scholar]
- 20. Court D.L., Gan J., Liang Y.-H., Shaw G.X., Tropea J.E., Costantino N., Waugh D.S., Ji X.. RNase III: genetics and function; structure and mechanism. Ann. Rev. Genet. 2013; 47:405–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coburn GA, Miao X, Briant DJ, Mackie GA. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev. 1999; 13:2594–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Awano N., Rajagopal V., Arbing M., Patel S., Hunt J., Inouye M., Phadtare S.. Escherichia coli RNase R has dual activities, helicase and RNase. J. Bacteriol. 2010; 192:1344–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Venkataraman K., Zafar H., Karzai A.W.. Distinct tmRNA sequence elements facilitate RNase R engagement on rescued ribosomes for selective nonstop mRNA decay. Nucleic Acids Res. 2014; 42:11192–11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vincent H.A., Deutscher M.P.. The roles of individual domains of RNase R in substrate binding and exoribonuclease activity. The nuclease domain is sufficient for digestion of structured RNA. J. Bio. Chem. 2009; 284:486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matos R.G., Barbas A., Goomez-Puertas P., Arraiano C.M.. Swapping the domains of exoribonucleases RNase II and RNase R: Conferring upon RNase II the ability to degrade dsRNA. Proteins Struct. Func. Bioinf. 2011; 79:1853–1867. [DOI] [PubMed] [Google Scholar]
- 26. Vincent H.A., Deutscher M.P.. Insights into how RNase R degrades structured RNA: analysis of the nuclease domain. J. Mol. Biol. 2009; 387:570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee G., Bratkowski M.A., Ding F., Ke A., Ha T.. Elastic coupling between RNA degradation and unwinding by an exoribonuclease. Science. 2012; 336:1726–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frazao C., McVey C.E., Amblar M., Barbas A., Vonrhein C., Arraiano C.M., Carrondo M.A.. Unravelling the dynamics of RNA degradation by Ribonuclease II and its RNA-bound complex. Nature. 2006; 443:110–114. [DOI] [PubMed] [Google Scholar]
- 29. Zinder J.C., Wasmuth E.V., Lima C.D.. Nuclear RNA exosome at 3.1 Å reveals substrate specificities, RNA paths, and allosteric inhibition of Rrp44/Dis3. Mol. Cell. 2016; 64:734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Makino D.L., Baumgartner M., Conti E.. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013; 495:70–75. [DOI] [PubMed] [Google Scholar]
- 31. Bryksin A.V., Matsumura I.. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. BioTechniques. 2010; 48:463–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode, Methods in Enzymology. Macromol. Cryst. A. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 33. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst. D. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 34. Hulme E.C., Trevethick M.A.. Ligand binding assays at equilibrium: validation and interpretation. Brit. J. Pharmacol. 2010; 161:1219–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng Z.F., Deutscher M.P.. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002; 277:21624–21629. [DOI] [PubMed] [Google Scholar]
- 36. Matos R.G., Barbas A., Arraiano C.M.. RNase R mutants elucidate the catalysis of structured RNA: RNA-binding domains select the RNAs targeted for degradation. Biochem. J. 2009; 423:291–301. [DOI] [PubMed] [Google Scholar]
- 37. Hossain S.T., Malhotra A., Deutscher M.P.. How RNase R degrades structured RNA- role of the helicase activity and the S1 domain. J. Biol. Chem. 2016; 291:7877–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hossain S.T., Malhotra A., Deutscher M.P.. The helicase activity of ribonuclease R is essential for efficient nuclease activity. J. Biol. Chem. 2015; 290:15697–15706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smart O.S., Neduvelil J.G., Wang X., Wallace B.A., Sansom M.S.P.. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 1996; 14:354–360. [DOI] [PubMed] [Google Scholar]
- 40. Barbas A., Matos R.G., Amblar M., Lopez-Vinas E., Gomez-Puertas P., Arraiano C.M.. New insights into the mechanism of RNA degradation by Ribonuclease II: identification of the residue responsible for setting the RNase II end product. J. Biol. Chem. 2008; 283:13070–13076. [DOI] [PubMed] [Google Scholar]
- 41. Fazala F.M., Kosloverb D.J., Luisic B.F., Blocka S.M.. Direct observation of processive exoribonuclease motion using optical tweezers. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:15101–15106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structural coordinates and diffraction structure factors of RNase R ΔHTH-K have been deposited in the RCSB Protein Data Bank with the PDB ID code 5XGU.