Abstract
In cyanobacteria, nitrogen homeostasis is maintained by an intricate regulatory network around transcription factor NtcA. Although mechanisms controlling NtcA activity appear to be well understood, its regulon remains poorly defined. To determine the NtcA regulon during the early stages of nitrogen starvation for the model cyanobacterium Synechocystis sp. PCC 6803, we performed chromatin immunoprecipitation, followed by sequencing (ChIP-seq), in parallel with transcriptome analysis (RNA-seq). Through combining these methods, we determined 51 genes activated and 28 repressed directly by NtcA. In addition to genes associated with nitrogen and carbon metabolism, a considerable number of genes without current functional annotation were among direct targets providing a rich reservoir for further studies. The NtcA regulon also included eight non-coding RNAs, of which Ncr1071, Syr6 and NsiR7 were experimentally validated, and their putative targets were computationally predicted. Surprisingly, we found substantial NtcA binding associated with delayed expression changes indicating that NtcA can reside in a poised state controlled by other factors. Indeed, a role of PipX as modulating factor in nitrogen regulation was confirmed for selected NtcA-targets. We suggest that the indicated poised state of NtcA enables a more differentiated response to nitrogen limitation and can be advantageous in native habitats of Synechocystis.
INTRODUCTION
Cyanobacteria perform oxygenic photosynthesis and play key roles in the global carbon and nitrogen cycles (1,2). They are continuously exposed to environmental fluctuations, such as changes in nutrient availability, light conditions or temperature, and have developed sophisticated mechanisms to sense and respond to these fluctuations to maintain their metabolic homeostasis. This is also the case for nitrogen, an essential element necessary for the synthesis of molecular building blocks, such as amino acids and nucleotides. Its deficiency results in a gradual decrease in transcripts encoding for components of photosynthesis, e.g. the tricarboxylic acid cycle (TCA), the Calvin–Benson cycle, and protein synthesis (3–5). To counteract nitrogen limitation, systems for high-affinity nitrogen uptake and sugar catabolic genes are induced, including those involved in the pentose phosphate pathway (OPP) or glycogen metabolism (3,4). Cyanobacteria prefer ammonium as a nitrogen source, although they can also use nitrate, nitrite, urea and some amino acids. In addition, many cyanobacteria are able to fix N2 (2). Nitrogen compounds acquired by cyanobacteria are converted to ammonium, which is then incorporated into the carbon skeleton of 2-oxoglutarate (2-OG) through the glutamine synthetase (GS)-glutamine oxoglutarate aminotransferase (GOGAT) cycle. Low intracellular ammonium levels during nitrogen starvation limit the turnover of the GS-GOGAT cycle resulting in increased levels of 2-OG, which serves as an indicator of an unbalanced C/N ratio within the cells (6).
In cyanobacteria, the global regulator for nitrogen assimilation and metabolism is NtcA, a transcription factor belonging to the CRP (cAMP receptor protein) family (7,8). NtcA is highly conserved in cyanobacteria and controls the cellular response to nitrogen availability (including nitrogen fixation in diazotrophic cyanobacteria) by binding as dimer to a consensus sequence GTAN8TAC within the promoter of its target’s genes (8). In the absence of ammonium, NtcA activates the expression of genes for nitrogen assimilation pathways, including urtA, nirA, ntcB and glnA (8). NtcA also acts as a transcriptional repressor of some genes, such as gifA and gifB, that encode for the GS inactivating factors IF7 and IF17, respectively (8,9). Under nitrogen depletion, the accumulation of the metabolite 2-OG stimulates DNA-binding of NtcA as well as the transcriptional modulation of targets genes by NtcA. Maximal activation of NtcA requires the subsequent binding of the coactivator, PII interacting protein (PipX), a small monomeric protein conserved among several cyanobacteria (10). This interaction is modulated by both 2-OG levels and the signal transduction protein PII, an integrator of the nitrogen and carbon balance in bacteria and plants (11). When nitrogen is abundant, PII binds to PipX to counteract NtcA activity at low 2-OG levels. However, under conditions of low nitrogen abundance (high 2-OG levels), PII binds 2-OG in a cooperative manner with adenosine triphosphate (ATP) and is phosphorylated (1). This causes the release of the PipX, and its interaction with NtcA, stabilizing the active 2-OG-bound conformation of NtcA (12).
Previous studies based on transcriptomic and bioinformatic predictions have attempted to identify putative binding sites of NtcA in different cyanobacteria (13–15). In Synechocystis sp. PCC 6803 (hereafter Synechocystis), which serves as a model cyanobacterium and promising microbial cell factory, 48 putative binding sites were computationally predicted in an early study; but no experimental validation of these sites was carried out (13). For Anabaena sp. PCC 7120, two genome-wide studies, one based on chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) analysis, and another based on the identification of transcriptional start sites (TSS) by RNA sequencing (RNA-seq) under nitrogen-depleted conditions, showed a great discrepancy in the number of possible NtcA-regulated genes. The RNA-seq data obtained by Mitschke et al. (15) suggested 158 TSS as potential NtcA targets, while the ChIP-seq analysis of NtcA performed by Picossi et al. (14) returned 2424 putative NtcA binding DNA regions, 865 of them ascribed to promoter regions. This striking discrepancy in the number of NtcA targets is not surprising, as separate application of RNA-Seq and ChIP-Seq provides only incomplete evidence for the regulatory activity of transcription factors. Although RNA-seq alone can faithfully detect changes in expression, it remains unclear whether these changes are related to the transcription factor of interest or reflect secondary effects caused by downstream events. In contrast, ChIP-seq alone can capture (differential) binding of transcription factors, but does not provide an indication of whether this causes activation or repression of nearby genes. A combination of these complementary high-throughput techniques, however, overcomes the limitations of individual approaches and can determine the NtcA regulon with unprecedented resolution. Through integration of differential expression and chromosomal binding location, we could identify not only in vivo functional NtcA binding sites, but also whether NtcA binding repressed or induced gene transcription during the early phase of nitrogen starvation. We identified 51 genomic regions bound by NtcA in ammonium-replete conditions, and 141 regions after 4 h of nitrogen starvation. Parallel transcriptome profiling revealed 669 genes as differentially expressed between these two conditions. Integration of NtcA binding and RNA-seq data classified 51 genes as being directly activated by NtcA and 28 as being directly repressed, including eight non-coding RNAs (ncRNAs). Direct target genes encoded mainly for proteins known to be involved in nitrogen and carbon metabolism, photosynthesis, respiration and transport, as well as various proteins without functional annotation. Interestingly, we observed significant differential expression for some genes, despite unchanged NtcA binding in their promoter regions as well as NtcA binding with only delayed expression changes of associated genes. Both observations suggest involvement of additional regulatory elements and potential different states of NtcA. A modifying role of PipX was specifically examined in selected NtcA target genes, confirming that PipX assists NtcA with nitrogen control in Synechocystis.
MATERIALS AND METHODS
Cyanobacterial strains and growth conditions
A glucose-tolerant strain of Synechocystis sp. PCC 6803 was grown in flask culture at 30°C under constant illumination (45 μmol photons m−2 s−1) on a rotatory shaker in liquid BG110C medium (16), supplemented with 10 mM NH4Cl and 20 mM TES (BG110C−NH4). The pipX(ssl0105)-disruptant mutant (ΔpipX) was grown under the same conditions, except that 50 μg ml−1 of kanamycin was added. To induce nitrogen starvation, flask cultures of Synechocystis cells, growing in BG110C−NH4, at linear growth phase (3–6 μg Chl/ml; Supplementary Figure S1) were collected, washed twice with BG110C and resuspended in BG110C medium under the same growth conditions for 4 h. Samples in the control treatment were also washed twice in BG110C supplemented with NH4 and re-suspended in BG110C-NH4 medium. For all experiments, nitrogen starvation and ammonium-cultured cells were resuspended in the same medium volume at similar cell density.
Construction of ΔpipX mutant Synechocystis strain
A 1639-bp DNA fragment, lacking a 211-bp internal fragment of pipX, was constructed by two-step polymerase chain reaction (PCR), using oligonucleotides pairs pipX-5’-HindIII and pipX-3’-XbaI, as well as pipX-deletion-for and pipX-deletion-rev (Supplementary Table S1). The resulting 1617-pb HindIII-XbaI restriction fragment was cloned into pBS II KS(+). An antibiotic resistance C.K1 cassette, which confers kanamycin resistance (Kmr), was inserted into a BamHI site in the positive transcription orientation to generate pPipX. Transformation of Synechocystis cells with pPipX was carried out. Correct integration of the C.K1 cassette and total segregation of the mutant chromosomes into the Synechocystis ΔpipX mutant strain was confirmed by PCR (Supplementary Figure S2).
Chromatin immunoprecipitation and sequencing procedure
Aliquots of 250 ml of ammonium and nitrogen-starved cultures were used for chromatin immunoprecipitation. To achieve protein–DNA crosslinking, formaldehyde was added to these cultures, yielding a final concentration of 1% and incubated for 15 min at room temperature, with occasional gentle shaking. The crosslinking reaction was terminated by adding 125 mM of glycine followed by a 5-min incubation at room temperature, with occasional gentle shaking. Next, cells were filtered, washed with cold TBS (20 mM Tris–HCl, pH 7.4, 140 mM NaCl) and collected in tubes (50 ml of culture per tube). Cell lysis was carried out, as previously described (14). The lysate was sonicated (15 cycles of 10 s at 10% amplitude, with 40 s on ice between cycles) to fragment chromosomal DNA into sequences of sizes between 100 and 400 bp. Cell debris was removed by centrifugation (15 min at 10 000 × g, 4°C). Chromatin was collected before immunoprecipitation to serve as a control input sample. Immunoprecipitation of NtcA-bound chromatin by an anti-NtcA antibody (generated in our laboratory using purified NtcA protein from Synechocystis, injected in rabbits according to standard immunization procedures) was carried out, as described by Picossi et al. (14). This protocol was repeated four times, using cells from independent inductions. The resulting DNA was pooled on the same DNA purification column (miniElute; QIAGEN, Hilden, Germany) to obtain 40 and 20 ng samples from ammonium and nitrogen-depleted conditions, respectively. Prior to construction of sequencing libraries, quantitative Real Time PCR (qRT-PCR) was performed to assess the enrichment of the promoter region of glnA and glnB in the immunoprecipitated samples, compared with the control input sample. Enrichment of a promoter region in the ChIP sample was determined using the percent input method: 100*2∧[Ct(IP-sample)−Ct(Adjusted-Input)], with the control input sample adjusted to give 100%, given that 5% of starting chromatin was used. A CFX Connect RT-PCR machine (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and ssoFast EvaGreen Supermix (Bio-Rad Laboratories) were used for qRT-PCR. The sequences of the primers used for the qRT-PCR to validate ChIP are listed in Supplementary Table S2.
Illumina libraries (Illumina, San Diego, CA, USA) were prepared from 32.5 and 12.5 ng of immunoprecipitated DNA from ammonium and nitrogen-depleted samples, respectively, as well as from the two controls DNA (input samples), using the Illumina TruSeq ChIP-seq DNA sample preparation kit. DNA sequencing was performed on the Illumina HiSeq 2500 platform, using single-end 50 bp sequencing. A total of 153 894 213 reads were obtained for four samples (Supplementary Table S3). The Finnish Microarray and Sequencing Centre (FMSC, Turku, Finland) conducted all DNA-sequencing and library preparation.
ChIP-seq peak calling analysis
Raw reads were mapped against the Synechocystis genome (NCBI Reference sequences: NC_000911.1 (chromosome), NC_005229.1 (plasmid pSYSM), NC_005230.1 (plasmid pSYSA), NC_005231.1 (plasmid pSYSG) and NC_005232.1 (plasmid pSYSX)) using Bowtie2 (17). The resulting BAM files were processed using SAMTools and BEDTools (18,19). BAM files were normalized for visual inspection in the Integrative Genomics Viewer (IGV) (20), using BamCoverage from DeepTools2 (21). Peaks for NtcA were identified by two peak calling algorithms: MACS (v1.4.1) (22) and BayesPeaK (v1.22.0) (23). In both algorithms, the background noise of unspecific binding was modeled using the input data (control DNA). Surrounding genes were retrieved using the Bioconductor package ChIPseeker (v1.6.7) (24). Peaks were also visually inspected for artefacts, and false positives were removed.
RNA preparation and differential expression analysis
RNA from Synechocystis cultures was extracted using the PGTX 95 RNA extraction protocol, described in Pinto et al. (25). Samples for RNA-seq were taken simultaneously from the same cultures used for the chromatin immunoprecipitation experiments. The quantity and quality of total RNA were evaluated using RNA electropherograms (Agilent 2100 Bioanalyzer; Agilent Technologies, Santa Clara, CA, USA). The Ribo-Zero Magnetic Kit for Bacteria (Epicentre; Illumina) was applied to remove ribosomal RNA from each sample. Two biological replicates for each condition of RNA samples were analyzed using the Ion PGM Template Hi-Q OT2 kit and Ion Touch 2 Instrument (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and subsequently sequenced on the Personal Genome Machine (Ion PGM; Thermo Fisher Scientific), with reagents from Ion Hi-Q Sequencing kit (STAB VIDA, Lda, Lisbon, Portugal), following the manufacturer’s instructions (Supplementary Table S4). Reads were aligned against the NCBI genome sequence for Synechocystis (NC_000911.1, NC_005229.1, NC_005230.1, NC_005231.1 and NC_005232.1), applying the Torrent Mapping Alignment Program (TMAP; available from https://github.com/iontorrent/TMAP). Raw read counts were calculated using the HTSeq Python script from HTSeq-count (26). The Bioconductor DESeq package from R software (27) was used to detect differentially expressed genes under ammonium repletion and nitrogen starvation. An adjusted P-value of <0.1 was considered to be significant. For Gene Ontology (GO) enrichment analysis, the GSEA tool (28) and Synergy (29) (available at http://synergy.plantgenie.org), were applied. Network construction and visualisation were carried out using Cytoscape 3.2.0 (30).
Hierarchical complete-linkage clustering was performed with Cluster 3.0 (31). Clusters were visualized using Java TreeView software (32). Differential expression was displayed as heat maps, using a color range from yellow to blue.
Potential target genes of NsiR7 were identified with the IntaRNA algorithm v.2.0.2 (33), using default parameters and a window of 275 nt around the start codon (200 upstream and 75 downstream). A threshold P-value of <0.005 was used.
Northern blot analysis
Total RNA was isolated from 30 ml samples of Synechocystis cultures in the mid-exponential growth phase (3 to 4 μg chlorophyll ml−1). Extractions were performed by vortexing cells in presence of phenol–chloroform and acid-washed baked glass beads (0.25–0.3 mm diameter), as described by Garcia-Dominguez and Florencio (34). An aliquot of 5 μg of total RNA was loaded per lane, and electrophoresed in 1.2% agarose denaturing formaldehyde gels (35). For the ncRNA analysis, RNA samples (5–10 μg) were separated on 6% urea-polyacrylamide gels for 3 h at 25 mA. In both cases, migrated gels were later transferred to nylon membranes (Hybond N-Plus; GE Healthcare, Little Chalfont, UK). Prehybridization, hybridization and washes were carried out in accordance with GE Healthcare instruction manuals. Probes for northern blot hybridization were prepared by PCR, using primers shown in Supplementary Table S2. DNA probes were 32P-labeled, with a random-primer kit (Amersham Biosciences, GE Healthcare), using [α-32P] dCTP (3000 Ci/mmol). Hybridization signals were quantified with a Cyclone Plus storage phosphor scanner (PerkinElmer, Inc., Waltham, MA, USA). Each experiment was replicated at least twice.
Western blot analysis
To prepare western blot analyses, 5 μg of total proteins from soluble extracts were fractionated on sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS/PAGE) and immunoblotted (35) with antibodies against: thioredoxin A (1:3000) (36); IF7 and IF17 (1:2000) (37); PII (1:4000) and GS III (1:20000) (34). ECL Prime (GE Healthcare) was used to detect the different antigens, with anti-rabbit secondary antibodies conjugated to horseradish peroxidase (1:25 000).
Glutamine synthetase assay
GS activity was determined in situ by a Mn2+-dependent γ-glutamyl-transferase assay in cells permeabilized with mixed alkyltrimethylammonium bromide (MTA) (38). The same assay, but without MTA, was performed in cells from the same samples in parallel. One unit of GS activity corresponds to the amount of enzyme that catalyses the synthesis of 1 μmol min−1 of γ-glutamylhydroxamate.
Gel retardation assays
DNA fragments used in the binding assays were obtained by PCR, with the corresponding oligonucleotides pairs (Supplementary Table S4). DNA probes, including NtcA-binding consensus sequences, were cut with NotI restriction enzyme, generating fragments of ∼200 bp. These fragments were end-labeled with [α-32P]dCTP, using Sequenase version 2.0 enzyme. The GST-NtcA fusion protein was expressed and purified, as previously described (39). DNA radiolabeled fragments (0.5 nM) were incubated with purified NtcA (0.1–0.4 μM), and with 2-OG (0.6 mM), when indicated. The binding reaction with the corresponding DNA fragment was carried out in a final volume of 15 μl within binding buffer (12 mM HEPES-NaOH pH 8.0, 8 mM Tris–HCl pH 8.0, 10% (w/v) glycerol, 0.5 mM ethylenediaminetetraacetic acid pH 8.0, 100 mM KCl, 2 mM MgCl, 0.05 μg/μl poly (dI–dC), 0.01 μg/μl bovine serum albumin and 1 mM dithiothreitol (DTT)). These mixtures were incubated at 25°C for 20 min, and the DNA–protein complexes were separated on non-denaturing 6% (w/v) polyacrylamide gel. Gels were dried and imaged using a Cyclone Plus storage phosphor scanner (PerkinElmer).
Quantitative real-time polymerase chain reaction analysis
The qRT-PCR was performed in an iQTM5 multicolor RT-PCR detection system (Bio-Rad), in a 10 μl reaction volume using the ssoFast EvaGreen Supermix (Bio-Rad). The sequences of primers used for the qRT-PCR for RNA-seq validation are listed in Supplementary Table S2. The efficiency of the PCR was calculated using the program LinRegPCR (40). Normalized data were calculated by dividing the average of at least three replicates of each sample from the candidate and the reference gene, rnpB.
RESULTS
Nitrogen starvation response of Synechocystis
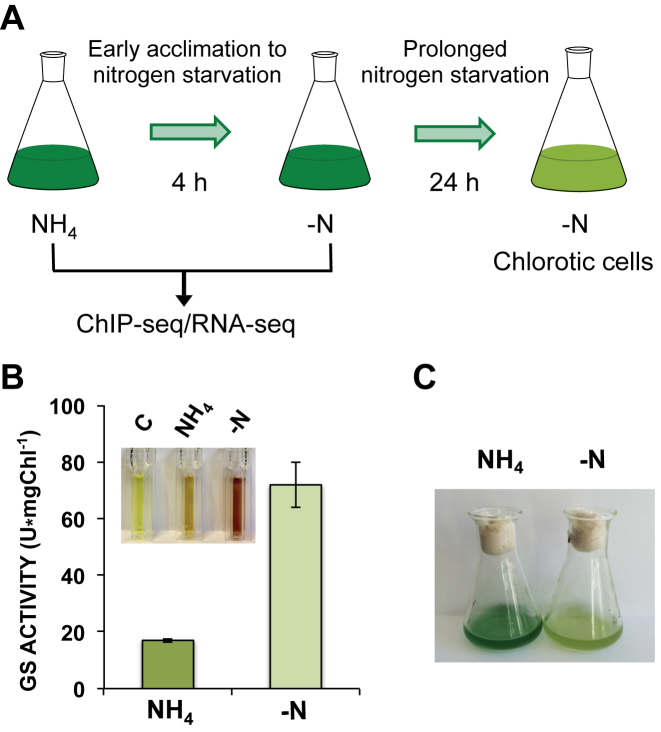
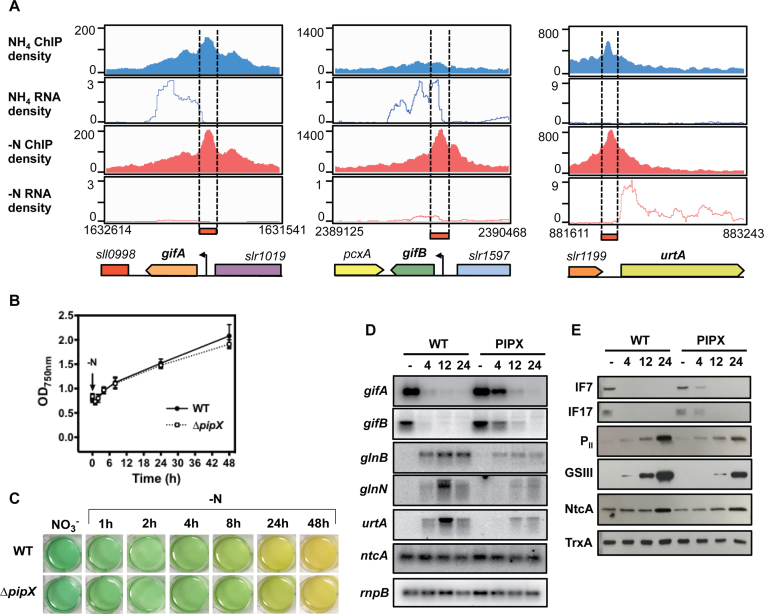
To characterize the NtcA regulon in response to nitrogen deprivation in Synechocystis sp. PCC 6803, we combined the two powerful genome-wide profiling techniques, RNA-seq and ChIP-seq. Samples were taken from wild-type (WT) cells grown in a medium with ammonium (NH4) as nitrogen source and after their incubation in a medium depleted of combined nitrogen (−N) for 4 h (Figure 1A). GS activity was measured to verify that nitrogen starvation was induced under the experimental conditions. Results obtained for GS activity were consistent with a transition from a nitrogen-rich medium to a nitrogen-depleted medium, showing higher activity after 4 h (71.92 ± 7.93 U/mg*chl−1) in the depleted medium (Figure 1B). After 24 h in a nitrogen-depleted medium, cell growth arrested and a yellow appearance (bleaching) typical of the chlorotic cultures was observed (Figure 1C), consistent with degradation of the phycobilisomes (PBS) related to prolonged nitrogen starvation.
Figure 1.
Nitrogen starvation conditions for Synechocystis sp. PCC 6803. (A) Schematic representation of the experimental procedure to capture the early acclimation to the nitrogen starvation examined by RNA-seq and ChIP-seq in this study, before prolonged starvation leads to chlorosis. (B) In situ analysis of GS activity from Synechocystis cells growing in BG11C+NH4 or after incubation for 4 h in nitrogen-free BG11C medium. A sample of 1 ml of Synechocystis culture was used for each assay. Transferase activity was measured after 5 min of incubation. Activity measures represent arithmetic means of three independent experiments, and their standard deviation values. C, control without adding cells. (C) Image of Synechocystis cultures used for RNA-seq and ChIP-seq experiments, after growing in either BG11C+NH4 or nitrogen-free BG11C medium for 24 h.
Transcriptional profiling by RNA-seq of nitrogen starvation response
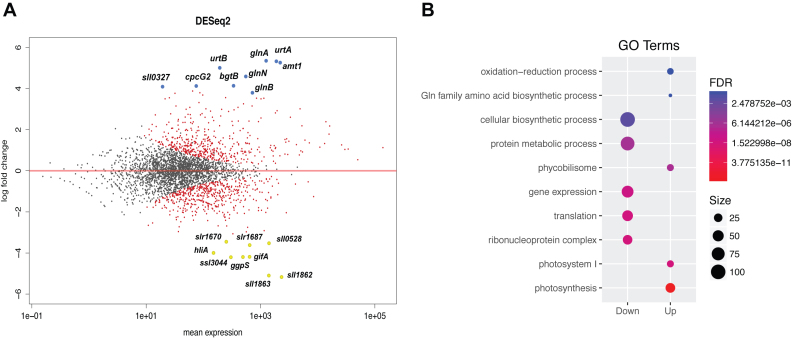
To study the transcriptional response after the transition from an NH4-replete to a N-depleted medium, total RNA obtained under both conditions was sequenced. Samples taken after 4 h of nitrogen starvation were compared with samples taken at the zero time point (with NH4; Figure 2A). There were 1080 genes significantly regulated (adj.P < 0.1; Supplementary Table S6), of which 669 genes showed more than a 2-fold change in expression. Of these latter ones, 332 and 337 genes were up- and downregulated, respectively (Supplementary Tables S7 and 8). RNA-seq analysis also enabled detection of small open reading frames (ORFs) and ncRNAs, which were not captured by previous DNA microarray analyses (3,4). In total, 21 ncRNAs were detected as differentially expressed, including NsiR4 (Supplementary Tables S7 and 8), which has been recently identified as playing a role in nitrogen control in Synechocystis (41). To identify processes and pathways involved in the transcriptional response to nitrogen starvation, we performed GO analysis of the differentially expressed genes. Our results reveal that downregulated genes tended to be associated with biosynthetic processes, gene expression and translation, indicating a reduction of overall protein synthesis (Figure 2B). Upregulated genes were significantly enriched in genes encoding proteins involved in nitrogen assimilation. Remarkably, genes related to the photosynthetic machinery, especially those coding for photosystem I (PSI) and PBS, were also overrepresented among the upregulated genes.
Figure 2.
Global response to nitrogen starvation in Synechocystis sp. PCC 6803. (A) MA plot: a scatterplot of log2-fold-change (−N/NH4) versus average expression in log2 scale for each gene, produced using the DEseq2 package. Dots shown in red indicate differentially expressed genes with adj. P-value of < 0.1. The nine genes with the highest induction and repression are shown in blue and yellow, respectively. (B) GO enrichment analysis of differentially expressed genes under nitrogen starvation conditions. Only GO of biological processes and cellular components having a false-discovery rate of <0.1 are shown.
Nitrogen assimilation
Adjustment of nitrogen assimilation pathways is crucial for survival under nitrogen-limited conditions. This is reflected in the expression pattern, with 24 genes involved in nitrogen uptake and assimilation being differentially expressed after nitrogen depletion (Table 1). Seven of them (glnA, urtA, amt1, urtB, glnN, bgtB and amt2) are among the 10 most-induced genes (Figure 2A). They include genes encoding the high-affinity nitrate/nitrite transporters (nrtBACD), the glutamine permease (bgtA and bgtB), the urea transport system (urtADBC) and the ammonium permeases (amt1 and amt2). Thus, after 4 h of nitrogen starvation, all scavenging nitrogen systems were activated to compensate for the loss of combined nitrogen from the media.
Table 1. Selected genes involved in nitrogen assimilation which expression is altered after 4 h of nitrogen starvation.
| Gene | Symbol | Function | Log2 (ratio) | P-value |
|---|---|---|---|---|
| Regulators of nitrogen metabolism | ||||
| ssl0707 | glnB | nitrogen regulatory protein PII | 3.83 | 2.6E-101 |
| ssl1911 | gifA | GS inactivating factor IF7 | −4.20 | 4.3E-96 |
| sll1515 | gifB | GS inactivating factor IF17 | −2.32 | 4.3E-23 |
| Nitrate/Nitrite assimilation | ||||
| slr0898 | nirA | ferredoxin–nitrite reductase | 1.86 | 1.6E-06 |
| sll1450 | nrtA | nitrate transport | 3.45 | 2.7E-25 |
| sll1451 | nrtB | nitrate transport | 2.51 | 1.5E-05 |
| sll1452 | nrtC | nitrate transport | 2.39 | 8.7E-19 |
| sll1453 | nrtD | nitrate transport | 2.16 | 2.0E-06 |
| Glutamine/glutamate assimilation | ||||
| slr1756 | glnA | glutamate–ammonia ligase GSI | 5.38 | 9.2E-191 |
| slr0288 | glnN | glutamate–ammonia ligase GSIII | 4.64 | 4.5E-117 |
| sll1502 | gltB | glutamate synthase large subunit | 0.96 | 9.4E-09 |
| sll1027 | gltD | glutamate synthase small subunit | 0.50 | 1.4E-02 |
| slr0710 | gdhA | glutamate dehydrogenase (NADP+) | 0.93 | 2.1E-03 |
| slr1735 | bgtA | Component of ABC-type Bgt permease | 1.27 | 1.8E-05 |
| sll1270 | bgtB | Component of ABC-type Bgt permease | 4.19 | 7.0E-81 |
| Ammonium asimilation | ||||
| sll0108 | amt1 | ammonium/methylammonium permease | 5.32 | 2.8E-216 |
| sll1017 | amt2 | ammonium/methylammonium permease | 4.12 | 2.7E-15 |
| Urea assimilation | ||||
| slr0447 | urtA | Component of ABC-type urea transport system | 5.36 | 7.7E-212 |
| sll0764 | urtD | Component of ABC-type urea transport system | 1.48 | 6.5E-09 |
| slr1200 | urtB | Urea transport system permease protein | 5.19 | 8.1E-71 |
| slr1201 | urtC | Urea transport system permease protein | 3.33 | 4.7E-17 |
Nitrogen in the form of ammonium is incorporated into amino acids by sequential action of GS and glutamate synthase (GOGAT) (Figure 3). Significant upregulation was observed for glnA and glnN encoding GS type I and III, respectively (Figure 3). The upregulation of GS coincided with a strong downregulation of genes encoding the GS inactivating factors IF7 (gifA) and IF17 (gifB) (Figure 3). Both NADH-GOGAT and Ferredoxin-GOGAT convert glutamine to glutamate, with 2-OG provided by the isocitrate dehydrogenase (icd). This reaction constitutes a crossroad between nitrogen and carbon metabolism (Figure 3). Transcripts of gltB and gltD encoding the large and the small subunits of the NADH-GOGAT were slightly induced (Figure 3 and Table 1). Remarkably, icd together with the genes encoding the phosphoenol pyruvate carboxylase (ppc) and the pyruvate kinase (pyk1) genes were also upregulated, facilitating enhanced synthesis of 2-OG in response to nitrogen starvation (Figure 3; Supplementary Tables S6 and 7). Surprisingly, expression of ntcA and pipX was not significantly affected, despite the observed differential expression of known NtcA targets (i.e. gifB, gifA glnA glnB, amt1 and icd). In contrast, transcripts encoding the regulatory protein PII (glnB) were accumulated upon nitrogen depletion. PII is involved in the regulation of central metabolism processes, integrating signals of cellular carbon, nitrogen and energy balances by binding 2-OG and ATP (42).
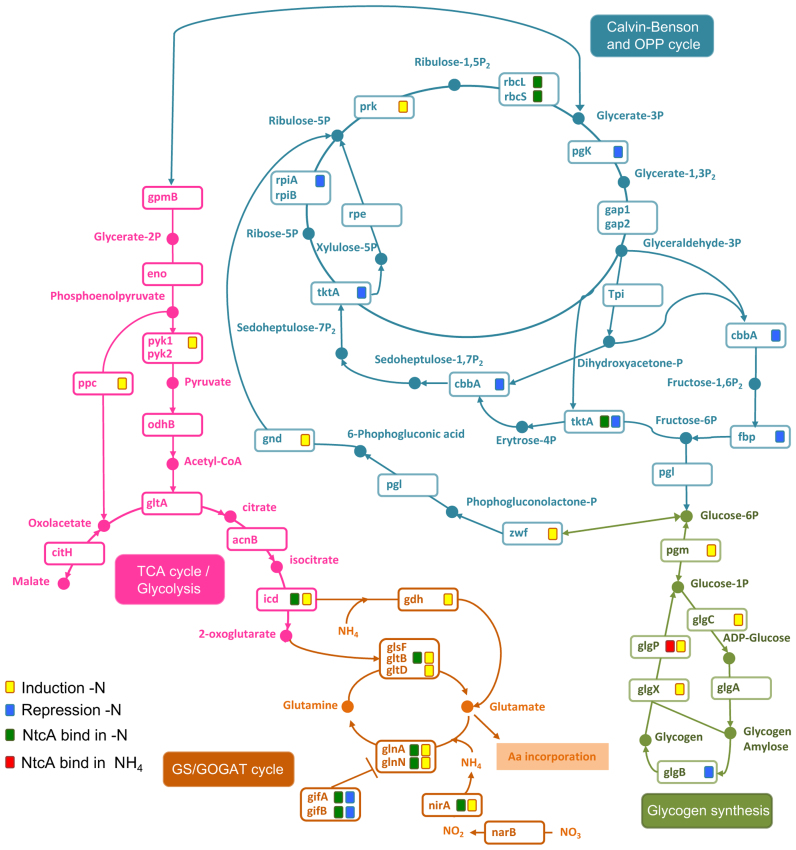
Figure 3.
Changes in expression of genes involved in nitrogen assimilation and carbon metabolism. This diagram shows metabolic pathways and metabolites of the TCA and GS/glutamine oxoglutarate amidotransferase (GOGAT) cycle, glycolysis, gluconeogenesis, pentose phosphate pathway (OPP) and glycogen metabolism. These pathways were predicted from the KEGG database (www.genome.jp/kegg/pathway.html). NtcA target genes obtained from ChIP-seq analysis are shown for −N (green square) or NH4 (red square) treatments. Genes encoding metabolic enzymes are shown below; narB, nitrate reductase; nirA, nitrite reductase, gifA, GS-inactivating factor; gifB, GS-inactivating factor; glnA, GS type I; glnN, GS type III; gltD, glutamate synthase small subunit; gltB, GOGAT; glsF, ferredoxin-dependent glutamate synthase, gdhA, glutamate dehydrogenase; icd, isocitrate dehydrogenase; acnB, aconitate hydratase; citH, malate dehydrogenase; gltA, citrate synthase; odhB, pyruvate dehydrogenase; pyk1–2, pyruvate kinase; eno, enolase; ppc, phosphoenolpyruvate (PEP) carboxylase; gpmB, phosphoglycerate mutase; pgk phosphoglycerate kinase; gap1, glyceraldehyde-3-phosphate dehydrogenase (catabolic reaction); gap2, glyceraldehyde-3-phosphate dehydrogenase (anabolic reaction); tpi, glycogen isoamylase; cbbA, fructose-bisphosphate aldolase; fbp, fructose-1,6-bisphosphatase; pgl, 6-phosphogluconolactonase; pgm, phosphoglucomutase; glgC glucose-1-phosphate adenylyltransferase; glgP, glycogen phosphorylase; glgX, glycogen isoamylase; glgA, glycogen synthase; glgB, 1,4-alpha-glucan branching enzyme; zwf, glucose-6-phosphate dehydrogenase; gnd, 6-phosphogluconate dehydrogenase; tktA, transketolase; rpiA or B, ribose-5-phosphate isomerase; rpe, pentose-5-phosphate-3-epimerase; prk, phosphoribulokinase; rbcL, ribulose bisphosphate carboxylase large subunit; rbcS, ribulose bisphosphate carboxylase small subunit.
Photosynthesis, carbon assimilation and central carbon metabolism
We observed a major increase in the expression of genes encoding for almost all subunits of PSI and PBS. In addition, several subunits of the ATP synthase (atpI, atpC, atpH) and photosystem II (PSII), namely psbO, psbU, psbK, psbT and psbB, were upregulated (Supplementary Table S7). In contrast, CO2 fixation appeared to be diminished after nitrogen starvation, as genes encoding the bicarbonate transporter (Na+/HCO3− symport), (bicA), as well as the carbon-concentrating mechanism proteins, ccmN and ccmM, were repressed (Supplementary Table S8). Additionally, ndhF3-ndhD3-cupA encoding subunits of the NDH–1MS complex associated with high-affinity CO2 uptake were downregulated (43,44) (Supplementary Table S6). Although the expression of the genes encoding the two subunits of the ribulose-1,5,-bisphosphate carboxylase/oxygenase (RuBisCo) remained unaltered, expression of other genes encoding enzymes of the Calvin-Benson cycle was downregulated, especially for prk, pgk, tktA and rpiA (Figure 3 and Supplementary Table S8).
Genes involved in sugar and glycogen metabolism displayed divergent patterns. Genes encoding sugar catabolic enzymes were induced under nitrogen starvation, while genes for sugar anabolism were downregulated. In this case, increased expression of the oxidative pentose phosphate pathway (OPP) genes zwf, talB and gnd was observed, whereas expression of fbp (slr0952) encoding for the fructose-1,6-bisphosphatase, which acts exclusively in the direction of the gluconeogenesis, was diminished (Figure 3; Supplementary Tables S7 and 8). Similar divergence was recorded for glycogen metabolism. Both glgX (slr1857) and glgP (slr1356) involved in the catabolism of glycogen were upregulated; while glgB and glgC, which participate in the synthesis of this molecule, were down- and upregulated, respectively (Figure 3; Supplementary Tables S7 and 8).
To confirm and validate differential expression detected by RNA-seq, qRT-PCR was carried out for genes related to nitrogen and carbon metabolism (glnA, glnB, glnN, amt1, gifB, icd and rre37). For these genes, RNA-seq and qRT-PCR data provided very consistent results (Supplementary Figure S3).
ChIP-seq analysis of NtcA binding
ChIP with anti-NtcA antibodies enabled in vivo genome-wide detection of NtcA binding sites. For quality control, enrichment of the promoter regions of two bona fide NtcA targets, glnA and gifB, within the immunoprecipitated fraction was confirmed by qRT-PCR. Primers against the promoter region of the ORF, slr1875, which is not regulated by NtcA, served as a negative control. Our qRT-PCR results confirmed the specificity of the immunoprecipitation, as strong enrichment of the two known binding sites for NtcA was measured, especially after nitrogen depletion, in contrast to the negative control (Supplementary Figure S4).
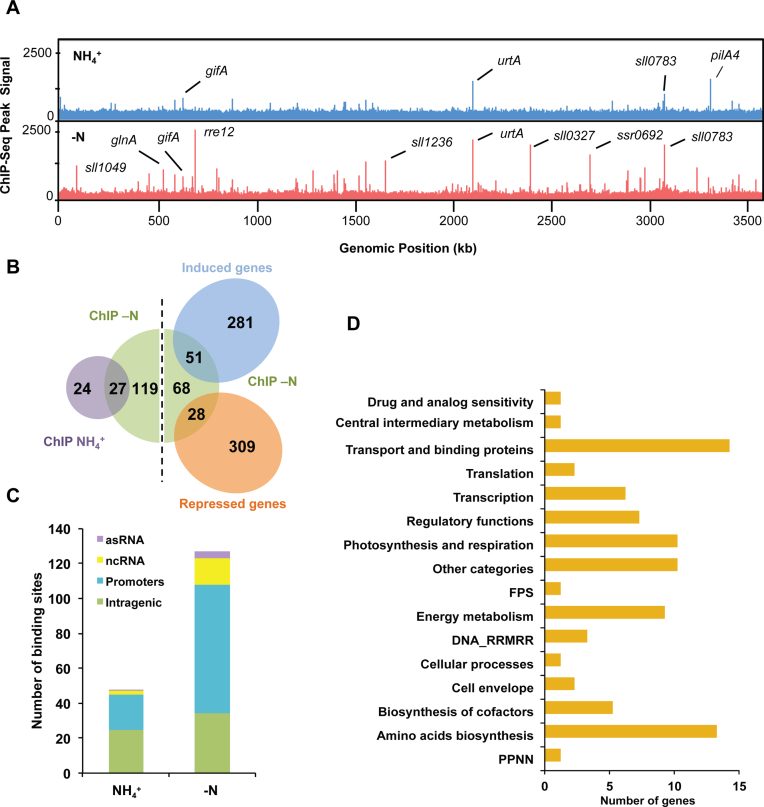
For comprehensive detection of binding sites, two different peak-calling methods were applied, MACS (22) and BayesPeak (23). NtcA binding was identified, if peaks in the sequence coverage were detected by at least one of the two peak-calling algorithms and passed visual inspection. We identified 51 NtcA binding peaks in NH4 and 141 peaks after 4 h of nitrogen depletion. Notably, 27 peaks were detected under both conditions (Figure 4A and B; Supplementary Tables S9 and 10). Three-quarters of the binding regions (151/192) were shorter than 450 bp, consistent with DNA fragment sizes obtained after DNA sonication. Thirty-two regions were slightly larger in size (500–850 bp), while only one region extended over more than 1 kbp (1200 bp). Visualization of this latter peak, revealed two peaks that overlapped each other within the bidirectional-promoter region for both sll0783 and slr0821 genes. Interestingly, sll0783 gene is reported to be involved in polyhydroxybutyrate accumulation under nitrogen starvation conditions in Synechocystis (45).
Figure 4.
Genome-wide NtcA DNA binding analysis. (A) NtcA binding across the genome was compared for NH4 (blue track) or −N (red track) conditions. The x-axis indicates the genomic position of the ChIP-seq peaks, while the y-axis indicates the read count after each dataset was normalized using BamCoverage (Ramirez, F. 2016). Names of genes assigned to peaks with high read count are also shown. B. Venn diagram showing overlap of genes with significant binding by NtcA in +NH4 versus −N conditions. Overlap of genes differentially expressed (P-value < 0.1 and fold change > 2) after nitrogen depletion versus genes with significant binding by NtcA are also shown. (C) Distribution of NtcA binding peaks for NH4 and −N conditions. NtcA peaks were classified into four categories: intragenic region (green), gene promoter (blue), ncRNA promoter (yellow) and antisense promoter (purple). (D) Genes with NtcA binding peaks assigned to NH4 or −N conditions, were grouped into functional categories according to the CyanoBase classification. FPS: Fatty acid, phospholipid and sterol metabolism; PPNN: Purines, pyrimidines, nucleosides and nucleotides; DNA_RRMRR: DNA replication, restriction, modification, recombination and repair.
The majority of chromosomal NtcA-IP peaks (73%) in the −N medium were located immediately upstream of gene coding sequences, ncRNAs and asRNAs, consistent with preferential NtcA binding to promoter regions under nitrogen-depleted conditions (Figure 4C). In contrast, only 46% of chromosomal peaks correspond to putative promoter regions in the NH4 condition, and of those 92% were also present in the −N condition (Figure 4C). Interestingly, we detected 24 peaks that were present exclusively under NH4 conditions. The vast majority of these peaks occurred within intragenic locations, with only four in promoter regions.
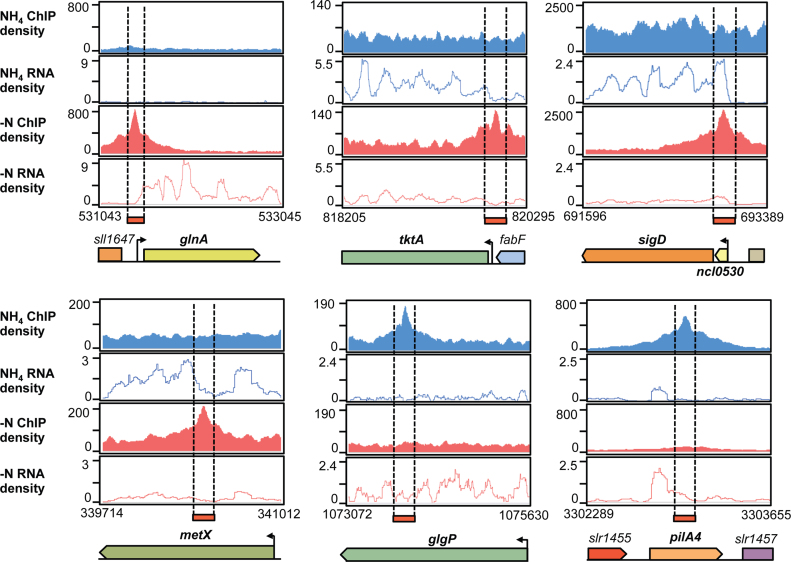
NtcA binding in promoter regions was assigned to transcribed genes, based on experimentally defined TSS in Synechocystis (5,46) and on gene expression data from our RNA-seq analysis. An unambiguous assignment to a single TSS was not possible for six of the NtcA binding peaks, because of the presence of nearby flanking genes. These peaks were assigned to both genes. In nine cases, we observed NtcA binding close to small RNAs, which were co-expressed with down-stream genes of the same transcriptional unit; a genetic feature that has recently been termed ‘actuaton’ (47). For these transcriptional structures, both coding genes and small RNAs were considered as NtcA-targets. Collectively, 18 ncRNAs and 5 asRNAs had an NtcA binding region within their promoter regions, with the exception of ncRNA ncl0530 and the downstream gene sigD, with its peak located within the ncl0530 sequence (Figure 5).
Figure 5.
Read coverage of potential NtcA targets detected by parallel differential profiling. The ChIP-seq and RNA-seq density profiles are shown in blue for NH4 and in red for −N treatments. Examples of genes included in the NtcA regulon are displayed: NtcA activated (glnA) or repressed (tktA) genes, long transcripts containing ncRNA and downstream protein-coding genes that are transcribed by read-through over the ncRNA terminator (ncl0530-sigD); and genes with internal peaks (metX, glgB and pilA4). Values on the x-axis are the genomic coordinates. Arrows indicate transcriptional start sites (TSS) obtained from Mitschke et al. (46).
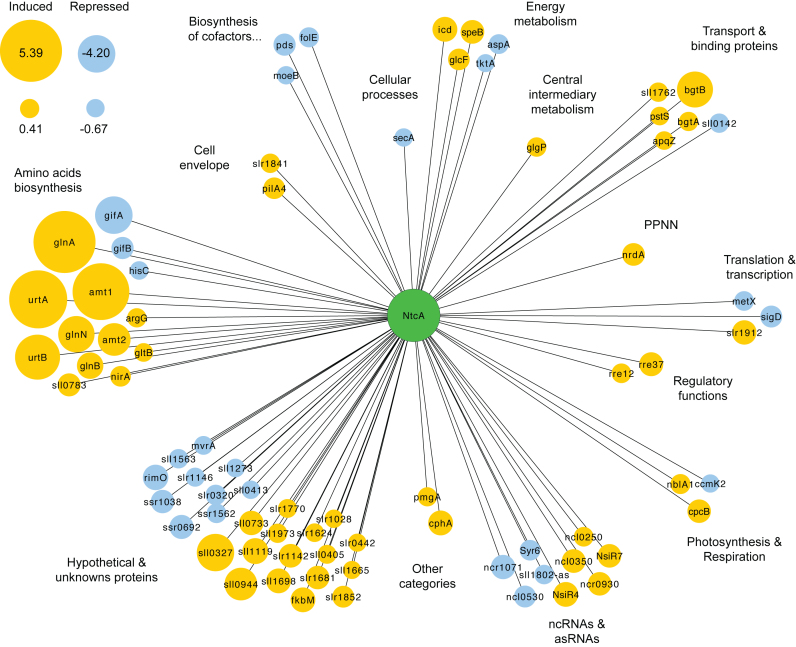
Genes identified as NtcA targets were classified into 16 functional categories (Figure 4D). About a third of the assigned genes (48) encode unknown or hypothetical proteins. The most represented functional categories were ‘amino acids biosynthesis’ and ‘transport and binding’. Both functional categories include most of the well-known NtcA-regulated genes, such as glnA, glnB, gifA, as well as several genes coding for the nitrogen uptake system (amt1, amt2, urtABCD, bgtA, bgtB, nrtABCD). Interestingly, NtcA binding sites were also found upstream of ten genes coding regulatory proteins. These included PipX (49), response regulator Rre37 (50), PII (34) and NtcA, but also newly identified ones, i.e. the response regulators Rre8, Rre12 and SphR, and the predicted transcription factor Sll0782.
Electrophoretic mobility shift assays (EMSA) were performed to validate the capability of NtcA to bind in vitro to DNA fragments of binding regions determined by ChIP-seq (Supplementary Figure S5). As a positive control, the promoter region of glnA, which contains a verified NtcA binding site, was used (51). In contrast, a DNA fragment containing the promoter region of nrsR, which has not been described as an NtcA target, was used as the negative control. We selected seven binding regions, including those with high (sll0327, urtA, pilA4, ncl0350) and low (gltB, metX, glgP) fold enrichment, as identified by ChIP-seq analysis (for both NH4 or −N conditions; Supplementary Tables S9 and 10). A strong positive correlation between ChIP-seq fold enrichment and NtcA EMSA affinities was found, indicating that our ChIP-seq analysis produced reliable results (Supplementary Figure S5).
Parallel differential profiling of transcriptome and NtcA binding sites
Occupancy of transcription factor binding sites alone gives only a weak indication of their regulatory potential. Therefore, we used parallel profiling of changes in both NtcA binding and transcription in response to nitrogen depletion. To define the NtcA regulon and to determine its dynamics, we intersected potential direct targets of NtcA, with their observed differential expression. This approach identified genes for which changes in NtcA binding were indicative of changes in transcriptional activity. We detected 79 genes (including ncRNAs and asRNAs) that fulfilled this criterion; they define the direct NtcA regulon in early phase of nitrogen starvation. Fifty-one of these genes were upregulated and 28 were downregulated, after 4 h of nitrogen depletion (Figure 4B and Table 2). Read coverage from RNA-Seq and ChIP-seq is shown for representative genes in Figure 5. The direct NtcA regulon (Figure 6) included eight enzymes involved in amino acid biosynthesis (glnA, glnN, nirA, argG, gltB, hisC, icd, aspA), six nitrogen compound transporter-related genes (urtA, urtB, amt1, amt2, bgtA, bgtB) and four regulators of nitrogen metabolism (gifA, gifB, glnB, rr37). Genes coding for proteins involved in the synthesis of the two main nitrogen storage pools in cyanobacteria, cphA and cpcB for cyanophycin and phycocyanin, respectively, were also targeted and regulated by NtcA, as previously described in Anabaena sp. PCC 7120 (14,52). Additionally, several genes that encode proteins related to carbon metabolism displayed NtcA-dependent regulation: transketolase (tktA; Figures 3 and 5), glycolate oxidase subunit (glcF), carbon dioxide concentrating mechanism protein (ccmk2), Sll0783 protein required for accumulation of polyhydroxybutyrate in Synechocystis (45), and glycogen phosphorylase (glgP; Figures 3 and 6). In particular, glgP contains an intragenic NtcA-binding peak under NH4-replete condition suggesting a repressive role of NtcA, triggered by binding to an intragenic region and blocking transcription in nitrogen-rich media. This correlates well with the observed diminished expression of glgP in NH4, compared with the −N conditions (Figure 5). Similar potential NtcA-regulation by intragenic binding was observed for genes, such as pilA4 in NH4, or metX (Figure 5) and secA under −N conditions. We were particularly interested in identifying new NtcA-regulated genes. Our study reveals that NtcA coordinates a wide range of functions to cope with the early acclimation of nitrogen starvation, given that NtcA targets included genes coding for porins (apqZ and slr1841), the periplasmic protein involved in phosphate uptake (pstS), and the photomixotrophic growth protein A (pmgA). Furthermore, the NtcA regulon comprises 26 genes encoding hypothetical or unknown proteins, as well as asRNA sll1802-as and eight ncRNAs (Figure 6).
Table 2. The NtcA regulon.
| Treatment | Regulated gene | Symbol | Peak start | Peak end | Region | log2 ratio (-N/NH4) | Gene function | TSS located inside peak |
|---|---|---|---|---|---|---|---|---|
| −N | slr1756 | glnA | 531291 | 531451 | promoter | 5.380 | glutamate–ammonium ligase | 531399 |
| −N, NH4 | slr0447 | urtA | 2098581 | 2098991 | promoter | 5.363 | periplasmic protein, ABC-type urea transport system substrate-binding protein | 2098760 |
| −N | sll0108 | amt1 | 2971052 | 2971551 | promoter | 5.318 | ammonium/methylammonium permease | 2971398 |
| −N, NH4 | slr1200 | urtB | 881702 | 882201 | promoter | 5.185 | urea transport system permease protein | 881923 |
| −N, NH4 | sll0327 | 2389552 | 2390401 | promoter | 5.141 | unknown protein | 2390100c | |
| −N | slr0288 | glnN | 2128602 | 2128851 | promoter | 4.643 | glutamate–ammonium ligase | 2128736 |
| −N | sll1270 | bgtB | 1115933 | 1116332 | promoter | 4.195 | periplasmic substrate-binding and integral membrane protein of the ABC-type Bgt permease | 1116162c |
| −N | sll1017 | amt2 | 401952 | 402201 | promoter | 4.123 | ammonium/methylammonium permease | 402151 |
| −N | sll0944 | 2267152 | 2267301 | promoter | 4.094 | hypothetical protein | 2267292 | |
| −N | ssl0707 | glnB | 2152602 | 2152751 | promoter | 3.826 | nitrogen regulatory protein PII | 2152741 |
| −N | sll1119 | 874502 | 874651 | promoter | 3.823 | hypothetical protein | 874644 | |
| −N, NH4 | slr2002 | cphA | 1447927 | 1448046 | promoter | 3.121 | cyanophycin synthetase | 1448016 |
| −N | slr1289 | icd | 282874 | 283225 | promoter | 3.043 | isocitrate dehydrogenase (NADP+) | 283033 |
| −N | slr0909 | 2785363 | 2785603 | intragenic | 2.874 | unknown protein | ||
| −N | slr1912 | 615686 | 616010 | promoter | 2.874 | anti-sigma F factor antagonist | 615878 | |
| −N | ncl0540/sll1698 | NsiR4/ | 1289152 | 1289451 | promoter | 2.861 | hypothetical protein | 1255935 |
| −N | sll0733 | 3419302 | 3419651 | promoter | 2.783 | unknown protein | 3419527 | |
| −N, NH4 | sll1330 | Rre37 | 3295152 | 3295451 | promoter | 2.704 | two-component system response regulator OmpR subfamily | 3295334 |
| −N | slr1142 | 801102 | 801851 | promoter | 2.657 | hypothetical protein | 801482 | |
| −N | ncl0350/sll1077 | /speB | 801102 | 801851 | promoter | 2.657 | Agmatinase | 801420 |
| −N | ncr0210 | NsiR7 | 450202 | 450501 | promoter | 2.633 | ncRNA | 450322 |
| −N, NH4 | sll1577 | cpcB | 727802 | 727951 | promoter | 2.614 | phycocyanin beta subunit | 727919 |
| −N, NH4 | sll0783 | 3071502 | 3072751 | promoter | 2.509 | unknown protein | 3072049 | |
| −N | slr1164 | nrdA | 1927974 | 1928364 | promoter | 2.438 | ribonucleotide reductase subunit alpha | 1928191 |
| NH4 | slr1852 | 1190528 | 1191867 | intragenic | 2.225 | unknown protein | ||
| −N, NH4 | slr1456 | pilA4 | 3302952 | 3303101 | intragenic | 2.151 | type 4 pilin-like protein, or general secretion pathway protein G | |
| −N | sll1831 | glcF | 625122 | 625461 | Intragenic TSS | 2.146 | glycolate oxidase subunit (Fe-S) protein | 625336 |
| −N | slr0898 | nirA | 2768902 | 2769151 | promoter | 1.861 | ferredoxin–nitrite reductase | 2769102 |
| −N, NH4 | slr1841 | 958152 | 958451 | promoter | 1.364 | probable porin | 958082 | |
| NH4 | sll1356 | glgP | 1073751 | 1074000 | intragenic | 1.347 | glycogen phosphorylase | |
| −N | sll1762 | 1233657 | 1233801 | promoter | 1.331 | putative polar amino acid transport system substrate-binding protein | 1233755c | |
| −N | sll1973 | 1576602 | 1576801 | promoter | 1.304 | hypothetical protein | 1576662 | |
| −N | slr0585 | argG | 3533652 | 3534001 | promoter | 1.276 | argininosuccinate synthetase | 3533815 |
| −N | slr1735 | bgtA | 1317495 | 1317738 | promoter | 1.275 | ATP-binding subunit of the ABC-type Bgt permease | 1317670 |
| −N | sll1968 | pmgA | 909452 | 909601 | promoter | 1.214 | photomixotrophic growth related protein | 909579c |
| −N | slr1770 | 549805 | 550138 | intragenic | 1.006 | hypothetical protein | ||
| −N | sll1502 | gltB | 482302 | 482501 | promoter | 0.955 | NADH-dependent glutamate synthase large subunit | 482370c |
| −N | slr1624 | 2034892 | 2035528 | promoter | 0.909 | hypothetical protein | 2035343c | |
| −N | sll0680 | pstS | 2641802 | 2641951 | promoter | 0.877 | phosphate-binding periplasmic protein precursor (PBP) | 2641853 |
| −N | slr2057 | apqZ | 1412702 | 1412951 | promoter | 0.850 | water channel protein | 1412833 |
| −N | slr1028 | 647502 | 647651 | intragenic | 0.705 | unknown protein | ||
| −N | sll0254 | crtL | 1507002 | 1507101 | intragenic | 0.705 | probable phytoene dehydrogenase Rieske iron-sulfur component | |
| NH4 | sll1665 | 268551 | 268700 | promotor | 0.472 | unknown protein | 3304862 | |
| −N, NH4 | slr0442 | 2081302 | 2081501 | intragenic | 0.471 | unknown protein | ||
| −N | sll1070 | tktA | 819816 | 820281 | promoter | −0.399 | transketolase | 820067 |
| −N | sll1049 | 90302 | 90801 | promoter | −0.399 | hypothetical protein | 90594c | |
| −N | sll0616 | secA | 2648652 | 2648801 | intragenic | −0.671 | preprotein translocase SecA subunit | |
| −N | sll1536 | moeB | 2041452 | 2041601 | promoter | −0.707 | molybdopterin biosynthesis MoeB protein | 2041560 |
| NH4 | sll0413 | 2544264 | 2545469 | intragenic | −0.802 | hypothetical protein | 2545087 | |
| −N | sll1958 | hisC | 1411852 | 1412351 | promoter | −0.831 | histidinol phosphate aminotransferase | 1412183 |
| −N | sll1273 | 1109887 | 1110178 | intragenic | −0.891 | unknown protein | ||
| −N, NH4 | slr0616 | mvrA | 2948302 | 2948751 | promoter | −1.035 | methyl viologen stress protein | 2948503 |
| −N | ncr1071 | 2215852 | 2216151 | promoter | −1.127 | ncRNA | 2215968 | |
| −N | slr1146 | 811052 | 811551 | promoter | −1.175 | hypothetical protein | 811318 | |
| NH4 | sll0142 | 2197197 | 2198566 | intragenic | −1.338 | probable cation efflux system protein | ||
| −N | sll1563 | 1964452 | 1964601 | intragenic | −1.358 | unknown protein | ||
| −N | sll0927 | metX | 340352 | 340501 | intragenic | −1.474 | S-adenosylmethionine synthetase | |
| −N | ssr1562 | 3235202 | 3235651 | promoter | −1.477 | hypothetical protein | 3235428 | |
| −N | slr1705 | aspA | 732302 | 732451 | promoter | −1.538 | aspartoacylase | 732376 |
| −N | slr0426 | folE | 2717952 | 2718051 | promoter | −1.632 | GTP cyclohydrolase I | 2718008 |
| NH4 | slr0320 | 2267622 | 2269004 | intragenic | −1.777 | hypothetical protein | ||
| −N | ncl0880 | Syr6 | 1816295 | 1816865 | promoter | −1.859 | ncRNA | 1816624c |
| −N | ncl0530/sll2012 | /sigD | 1255752 | 1255940 | intragenic | −2.248 | group2 RNA polymerase sigma factor SigD | 1204108 |
| −N, NH4 | slr1254 | pds | 1397352 | 1397951 | promoter | −2.311 | phytoene dehydrogenase | 1397780 |
| −N | sll1515 | gifB | 458552 | 459201 | promoter | −2.324 | GS inactivating factor IF17 | 458961 |
| −N, NH4 | ssr1038 | 2949752 | 2950201 | promoter | −2.597 | unknown protein | 2949923 | |
| −N | ssr0692 | 2695446 | 2695626 | promoter | −2.738 | hypothetical protein | 2695575 | |
| −N, NH4 | slr0082 | rimO | 2885852 | 2886301 | promoter | −2.975 | hypothetical protein | 2886040 |
| −N, NH4 | ssl1911 | gifA | 631902 | 632351 | promoter | −4.204 | GS inactivating factor IF7 | 632192 |
| −N, NH4 | ncl0250/sll1291 | /rre12 | 692659 | 693007 | promoter | 1.462 | two-component response regulator PatA subfamily | 692816 |
| −N | Ncl0930/slr1681 | 1969237 | 1969498 | promoter | 2.618 | unknown protein | 1969319 | |
| −N | sll0405 | 2552752 | 2552901 | intragenic | 1.100 | unknown protein | ||
| −N | sll1802-as | 840302 | 840791 | promoter | −1.405 | Antisense RNA | 840574 | |
| −N | sll1028 | ccmK2 | 219070 | 219290 | intragenic | −0.626 | carbon dioxide concentrating mechanism protein |
List of genes differentially expressed after nitrogen starvation that contain an NtcA binding site. The condition in which NtcA peaks were higher is highlighted in bold.
Figure 6.
NtcA regulatory network and its transcriptional dynamics for the early stages of nitrogen depletion. Target genes are represented by nodes that are colored based on gene expression after 4 h of nitrogen depletion: yellow for upregulated genes, and blue for downregulated genes. Node sizes correspond to the magnitudes of expression changes as a log2 expression ratio (−N/NH4). Genes are grouped according to the functional categories obtained from CyanoBase. PPNN: purines, pyrimidines, nucleosides and nucleotides.
Sequence analysis of NtcA-bound promoters
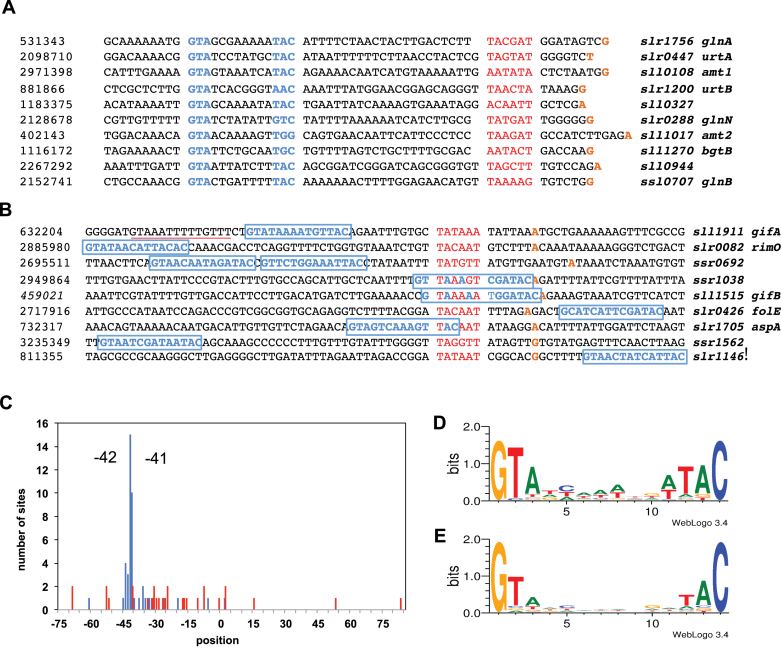
The NtcA consensus binding sequence GTAN8TAC was first described in the NtcA-regulated promoters of Synechococcus (53). Promoters activated by NtcA tend to contain a consensus sequence, centered close to position −41 with respect to the TSS, although actuating NtcA binding in positions further upstream has been reported in some cases (53). Conversely, NtcA-mediated repression is thought to be caused by NtcA binding at positions incompatible with correct assembly and positioning of the RNA polymerase. This model is supported by a promoter analysis of the NtcA regulon for Synechocystis. Promoters of the 10 most-induced genes of the NtcA regulon showed a highly conserved GTAN8TAC motif, centered close to position −41.5. Notably, GTA at positions 1−3 was strictly conserved, and only little variation was observed for TAC at positions 12−14 (Figure 7A). In the case of the top ten repressed genes, the potential NtcA binding sites displayed greater derivation from the consensus sequence, and were widely distributed along the promoter (Figure 7B). These patterns were also found, when we examined NtcA binding positions for the entire regulon. While 71% of binding sites of induced genes were concentrated at positions −41 and −44, binding for repressed genes ranged from −69 to +83, with respect to the TSS, with slight accumulation around −30 (Figure 7C). This finding confirms previous observations that the distance between the NtcA binding site and the TSS determines the regulatory mode of NtcA (53).
Figure 7.
Sequence analysis of NtcA binding sites. (A and B) Promoter regions having NtcA binding sites for ten most strongly induced and repressed genes, respectively. Nucleotides confirming the putative NtcA consensus-binding sequence are indicated in blue, potential −10 promotor elements in red and transcriptional start sites (TSS) in orange. An alternative NtcA binding site for the gifA promoter at position −45 is underlined in red. (C) Distribution of positions of NtcA binding sites identified for genes of the NtcA regulon. The location of NtcA binding was determined by matching the NtcA consensus sequence. The relative position was defined as the distance of the seventh nucleotide of a putative NtcA binding site to the TSS. Blue and red bars indicate the frequency of these positions for induced and repressed genes, respectively. (D) NtcA binding motif defined by target genes of the NtcA regulon. (E) Binding motif found using all the putative NtcA binding sites identified by ChIP-seq analysis. (Representation by WebLogo 3.0) (54).
In addition to examining the positioning of the established consensus binding sequence, studying the NtcA regulon provides the opportunity to refine the NtcA binding motif for Synechocystis. Using the WebLogo application (54), we obtained an approximately palindromic motif of length 14 nt, with a high prevalence of G, T and C at positions 1, 2 and 14; and medium prevalence of A, T and A at positions 3, 12 and 13 (Figure 7D). In most of the remaining positions (4–11), a weak tendency towards A or T was noticed. Subsequently, we extended this analysis to include all NtcA binding regions, determined in our ChIP-seq analysis. For this enlarged set, the information content of the motif deteriorated, although it still resembled the consensus sequence GTAN8TAC (Figure 7E). Interestingly, the prevalence of the flanking nucleotides (G at 1, C at 14) increased, demonstrating their importance for NtcA dimer binding, even when other nucleotides were less conserved.
Noncoding RNAs regulated by NtcA
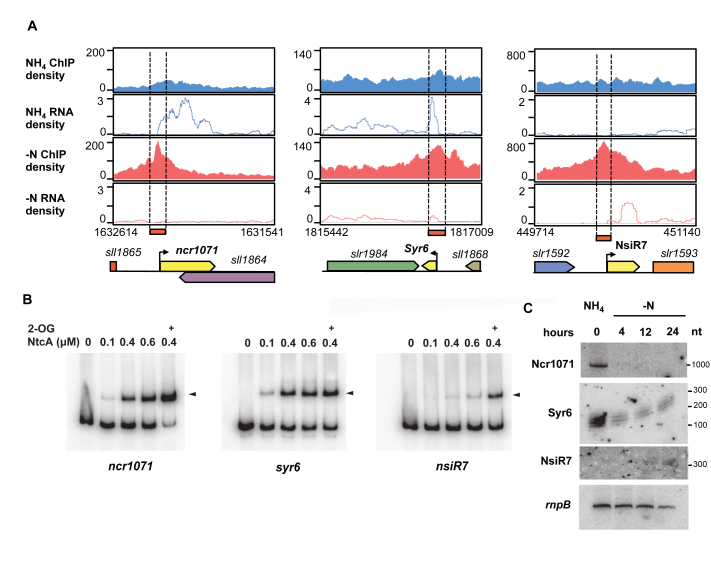
Several NtcA binding peaks were assigned to promoter regions of ncRNAs and asRNAs. In total, 18 ncRNAs and 5 asRNAs with assigned NtcA binding were identified in our ChIP-seq analysis (Supplementary Tables S9 and 10). Eight of these ncRNAs were also differentially regulated; thus, we added these to the defined NtcA regulon. These included NsiR4, which has been previously reported to be involved in nitrogen assimilation control by targeting IF7 (41). Notably, three of these NtcA targeted ncRNAs were transcribed from their own exclusive transcriptional units (ncr1071, ncl0880, ncr0210; Figure 8A). Specifically, ncr1071 codes for a predicted 1057-nt long ncRNA, while ncl0880 (also called Syr6) and ncr0210 (also called NsiR7) have been reported to belong to the 33 most abundant ncRNAs in Synechocystis (5,47). Since these ncRNAs were transcribed from their own transcriptional units, they are unlikely to be by-products of the expression of other genes; thus, we decided to analyze them further.
Figure 8.
Regulation of non-coding RNAs by NtcA. (A) Visualization of NtcA-binding peaks assigned to ncr1071, Syr6 and NsiR7. Their ChIP-seq and RNA-seq density profiles are shown in blue for the NH4 condition and in red for the −N conditions. (B) Electrophoretic mobility shift assay verification of NtcA binding sites. Promoter sequences of ncr1071, syr6 and nsiR7 were PCR amplified (primers listed in Supplementary Table S5) before being mixed with 0.1, 0.4, 0.6 μM of NtcA. An aliquot of 0.6 mM of 2-oxoglutarate (2-OG) was added, when indicated. (C) Northern blot analysis of expression of Ncr1071, Syr6 and NsiR7. Total RNA was isolated from WT cells transferred from BG11C supplemented with ammonium to nitrogen-free BG11C medium for 24 h. The filters were hybridized with Ncr1071, Syr6 and NsiR7 probes and subsequently stripped and rehybridized with rnpB probe as a control.
The RNA-seq data showed the accumulation of NsiR7 transcripts after 4 h of nitrogen depletion, having an NtcA binding site upstream at position −43 from the TSS, consistent with activation by NtcA (Figure 8A and Supplementary Table S10). In contrast, both ncr1071 and syr6 were downregulated and their NtcA binding sites were found at positions +3 and +16, respectively (Figure 8A and Supplementary Table S9). To experimentally validate the NtcA binding peaks detected by ChIP-Seq, we performed EMSA experiments, which clearly showed NtcA binding to promoter sequences of these selected ncRNAs (Figure 8B). We also carried out northern blot experiments to examine the expression levels of these ncRNAs over a prolonged period of nitrogen starvation. For this purpose, Synechocystis cultures grown in NH4 were shifted to nitrogen-depleted media, and samples were collected after 4, 12 and 24 h. Both ncr1071 and syr6 were strongly repressed after 4 h of nitrogen starvation; although in the case of syr6, repression was not complete and residual transcription was still observed (Figure 8C). nsiR7 showed a weak induction that was detected by northern blot, after only 12 h.
To predict potential targets of these three ncRNAs, we used two RNA target predictions programs: CopraRNA (55) and IntaRNA (33). CopraRNA has better accuracy than IntaRNA since it uses phylogenetic conservation in its scoring system (56), but this implies that presence of homologous sRNA sequence from distinct organisms is necessary. For both ncr1071 and syr6 homologous where found in other cyanobacteria but not for nsiR7 that is only present in Synechocystis. Thus CopraRNA was applied for Ncr1071 and Syr6, while IntaRNA was only used for NsiR7. Assuming repression as the dominant mode of regulation by ncRNAs, we expected that genes under the control of repressed ncRNAs after nitrogen depletion (Ncr1071 and Syr6) would show increased expression in our RNA-seq data; while those under the control of induced NsiR7 would show decreased expression. In the case of Ncr1071, four predicted targets were significantly induced: sll1451 encoding the nitrate transport protein NrtB, slr2136 encoding the GcpE protein homolog involved in the terpenoid biosynthesis and slr0151 and sll1219 encoding the hypothetical proteins Slr0151 and Sll1219, respectively (Supplementary Table S11). Alternatively, it may act as an asRNA to sll1864 coding for a chloride channel protein. In the case of Syr6, also four of the predicted targets displayed increased expression: ssl2598 (psbH) encoding the photosystem II reaction center protein H, slr0079 encoding the general secretion pathway protein E (GspE), sll1533 encoding the twitching motility protein PilT and sll0376 encoding the hypothetical protein Sll0376 (Supplementary Table S11). In the case of NsiR7, the only predicted target that was repressed after nitrogen step-down was the transcription factor nusA (slr0743).
Role of PipX in nitrogen regulation in Synechocystis
For a number of NtcA binding sites, the normalized peak height was preserved during nitrogen starvation, suggesting a similar NtcA binding affinity under both conditions. Surprisingly, the clearest example was observed in the promoter region of IF7 coding gene gifA. Despite significant expression change in gifA (−4.2 log FC; Supplementary Tables S8 and 12), following transition to a nitrogen-free medium, its NtcA peak height remained almost identical, with less 1% differential fold enrichment (DFE) between both conditions (Figure 9A). This contrasts with gifB encoding IF17 (Figure 9A), for which its downregulation (−2.3 log fold change, FC; Supplementary Table S8) was correlated with an NtcA binding peak present only under nitrogen starvation. Given that promoters of both GS inactivation factors contain NtcA binding sites at positions considered repressive for transcription (9), the persistence of NtcA binding under nitrogen-replete conditions, when gifA is highly induced, is remarkable. Intriguingly, we found several other NtcA targets (some of them well established) with unexpectedly small changes in their NtcA binding, considering their large expression changes. These included rre37 (23% DFE, 2.7 log FC), urtA (24% DFE, 5.3 log FC; Figure 9A) and urtB (<38% DFE, 5.1 log FC). Collectively, eight genes having more than a two log FC in expression showed less than 40% DFE (Supplementary Table S12).
Figure 9.
The role of PipX in the NtcA-regulated genes. (A) Read coverage of NtcA-binding peaks assigned to gifA, gifB and urtA. Their ChIP-seq and RNA-seq density profiles are shown in blue for NH4 and in red for −N conditions. (B) Growth of Synechocystis WT and ΔpipX strains after nitrogen step-down. Cells of WT and ΔpipX strains cultivated to their midlog growth phase in BG11C medium supplemented with NH4 were transferred to nitrogen-free BG11C medium.Their growth was measured at OD750 for 48 h. (C) Images of cyanobacterial cell suspensions of Synechocystis WT and ΔpipX obtained during growth curve analysis (panel B) at indicated times. (D) Northern blot analysis of expression of gifA, gifB, glnB, glnN, urtA and ntcA in response to nitrogen starvation. Total RNA was isolated from WT cells transferred from BG11C supplemented with ammonium to nitrogen-free BG11C medium for 24 h. The filters were hybridized with gifA, gifB, glnB, glnN, urtA and ntcA probes and subsequently stripped and rehybridized with rnpB probe as a control. (E) Western blot analysis of IF7, IF17, PII, GSIII and NtcA in response to nitrogen starvation. Cells were grown in BG11C supplemented with ammonium to midlog growth phase and then transferred to nitrogen-free BG11C medium and cultivated for 24 h. Samples of 10 μg of total proteins from soluble extracts were separated by 15% SDS/PAGE and analyzed by western blots to detect IF7, IF17, PII, GSIII, NtcA and TrxA.
It has previously been demonstrated that high cytoplasmic concentrations of NtcA alone are not sufficient to promote activation of selected target genes (57). In fact, activation of several NtcA-dependent genes, including the amt1 or nir operon, has been directly stimulated by 2-OG in Synechococcus elongatus PCC 7942 (58). Later studies revealed that 2-OG enhances complex formation between NtcA and PipX, which is crucial for activation of NtcA target genes in S. elongatus PCC 7942 and Anabaena sp. PCC 7120 under nitrogen-depleted conditions (49,59). Whether this is the case for Synechocystis remains unclear, as the role of PipX in nitrogen control in this cyanobacterial model has not been fully established.
To investigate the role of PipX in the regulation of the NtcA-controlled genes in Synechocystis, especially those having promoters with similar affinity to NtcA under both conditions (NH4 and −N), a pipX-deficient mutant (ΔpipX) was generated (see ‘Materials and Methods’ section and Supplementary Figure S2). Cells of Synechocystis WT and ΔpipX cultivated in BG11C medium supplemented with ammonium were shifted to a nitrogen-free BG11C medium and cultivated for an additional 48 h. Although the growth kinetics for both strains did not show significant differences during this nitrogen-starvation treatment, chlorosis of the cells in the ΔpipX strain was slightly delayed, compared with WT (Figure 9B and C). A similar observation was made for a pipX-deficient mutant of S. elongatus PCC 7942 (10). Subsequently, we analyzed the transcript and protein accumulation of several genes involved in nitrogen assimilation, including IF7 and IF17, GSIII, PII and the urea transport protein UrtA. Consistent with a role of PipX as an NtcA enhancer, both transcriptional activation and repression of the selected NtcA targets were delayed in the ΔpipX strain. This delay was also reflected in their corresponding protein levels (Figure 9D and E). Furthermore, transcriptional induction of glnN, glnB and urtA was weaker in the ΔpipX strain, suggesting that PipX enables faster and more efficient adaptation to nitrogen starvation. Such a function might be especially important, given that transcription of ntcA remained unaltered in both strains during our treatment with only a small increase in protein levels after 24 h, in agreement with previous report (3). Remarkably, we did not find any differences in the transcriptional expression of gifA between both strains under nitrogen-replete condition, indicating that PipX is not acting as a coactivator of NtcA for gifA transcription. A possible explanation for the persistent NtcA binding peak within the promoter of gifA in NH4-replete media might be the presence of a degenerate NtcA consensus sequence GTAAATTTTTGTTT at position −45. At this position, NtcA could bind and enhance transcription of gifA (Figure 7). Stimulated by 2-OG or other factors, NtcA could slide to the binding site at position −33, acting as a repressor, when nitrogen is limited.
DISCUSSION
To understand how cyanobacteria respond to varying conditions in the environment, it is important to have comprehensive models of their underlying transcriptional regulation. Here, we present the first genome-wide analysis of a transcription factor regulon for Synechocystis, based on both ChIP-seq and RNA-seq techniques. Combining these two high-throughput approaches permitted us not only to identify the regulon of NtcA, but also to capture the activity of this key transcriptional regulator of nitrogen. We were able to identify 48 and 121 NtcA-bound regions in the Synechocystis genome under nitrogen-eplete conditions and after 4 h of nitrogen step-down, respectively. In parallel, we obtained transcriptional profiles of Synechocystis under the same experimental conditions. This enabled us to classify target genes into three categories: activated, repressed or unaffected by NtcA. Activated and repressed genes were used to define the NtcA regulon during the early phase of nitrogen starvation. They included various genes involved in nitrogen and carbon metabolism or photosynthesis, as well as several ncRNAs. Our ChIP-seq detected 25 previously known or predicted NtcA targets in Synechocystis (Supplementary Table S13), demonstrating the sensitivity of our approach. Importantly, we identified 67 new NtcA targets within the defined NtcA regulon, expanding the scope of the NtcA regulon during the early phase of nitrogen acclimation in Synechocystis. The higher proportion of intragenic NtcA binding sites under nitrogen-replete conditions indicates a distinct role for NtcA under this condition. Finally, expression analyses of functionally relevant NtcA targets in a pipX mutant strain revealed the importance of PipX for efficient transcriptional regulation by NtcA in Synechocystis during nitrogen starvation.
Transcriptional adaptation to early stages of nitrogen starvation
The RNA-seq analysis revealed that transcription of genes involved in transcription and translation, biosynthetic process and protein synthesis were downregulated, indicating a reduction of overall protein synthesis, as previously reported for nitrogen deprivation (3–4,60). Major changes in transcript levels were observed for genes involved in nitrogen uptake and metabolism (Table 1 and Figure 2), including many NtcA targets: glnA, glnB, amt1, urtA, gifA and gifB. However, genes coding for both NtcA and the nitrogen coactivator PipX were not affected (Figure 9). Absence of induction of NtcA under nitrogen starvation condition has been previously observed (3); suggesting that post-transcriptional regulation of NtcA plays a more important role than the amount of NtcA per se. Surprisingly, expression of the PSI- and PBS-related genes was induced. This finding is consistent with a previously reported transcriptome profiling after 6 h of nitrogen starvation by Krasikov et al. (3), in which PSI- and PBS-related genes were transiently activated, while cells proliferated at a normal growth rate. In contrast, Osanai et al. observed in their microarray study of nitrogen starvation that photosynthetic genes were repressed (4). The discrepancies between these studies might be related to differences in culture conditions prior or during nitrogen starvation or in a strain-specific genetic background. For example, illumination intensity used by Osanai (70 μmol photons−2 s-1) was 55 and 40% greater than used in our experiment (45 μmol photons−2 s−1) or by Krasikov et al. (50 μmol photons−2 s−1), respectively. In any case, the observed transient upregulation of PBS- and PSI-related genes may indicate increased cyclic electron transport around PSI as early stress response. This hypothesis is reinforced by the induction of ndhD1 (slr0331), which codes for a component of the NADH dehydrogenase complex involved in PSI cyclic electron flow, and some subunits of ATPase synthase (atpI, atpC and atpH; Supplementary Table S6). Increased cyclic electron flow could enhance ATP synthesis to provide the necessary energy required for nitrogen assimilation pathways. However, this phase is only transitional to a more severe response to nitrogen starvation, in which growth decreases and degradation of the PBS leads to a yellow appearance of cyanobacterial cultures. This acclimation process, known as chlorosis or bleaching, requires the expression of the cotranscribed nblA genes (nblA1 and nblA2) (5). In fact, expression of nblA2 (ssl0453) after 4 h of nitrogen starvation was observed, suggesting that a transcriptional response towards PBS degradation and long-term acclimation had already started.
Nitrogen depletion in cyanobacteria leads to downregulation of genes related to carbon fixation and induction of sugar catabolic genes (3–4,60,48). Here, we observed the induction of genes involved in the oxidative pentose phosphate (zwf, talB and gnd), as well as catabolism (glgX and glgP) and anabolism (glgC) of glycogen (Figure 3). Expression of genes for glycogen degradation, which are likely not to be active during nitrogen starvation, is in agreement with an anticipatory state of chlorotic Synechocystis cells described recently by Klotz et al. (61). Cells anticipate future recovery from nitrogen starvation by preparing proteins necessary for fast resuscitation without having to synthesize the corresponding enzymes de novo.
Strikingly, 33 genes involved in regulatory functions responded to nitrogen step-down, suggesting that acclimation to nitrogen depletion is a process that requires extensive transcriptional reprogramming. This is corroborated by our observation that several genes coding for two-component systems were affected by nitrogen deprivation. Two-component systems enable cells to respond to both environmental and intracellular changes. Six histidine kinases and six response regulators were upregulated, including rre37, which is involved in the activation of sugar catabolism under nitrogen starvation (62,63). The complexity of the nitrogen starvation response is highlighted by the large number of genes involved in heavy metal and oxidative stress, which altered their expression in response to nitrogen depletion. These included genes encoding the copper sensing system CopRS (sll0789, sll0790 and their respective copies in the plasmid pSYSX: slr6040 and slr6041), the ferric uptake regulator Fur (sll0567), the peroxide stress response transcriptional regulator PerR (slr1738), and the LexA repressor (sll1636). Furthermore, many genes coding for metal importer systems were downregulated, such as the ATPases for copper (ctaA and pacS), cobalt (coaT) and zinc (ziaA), as well as iron import proteins FutA1 (futA1) and FutA2 (futA2). Such widespread adjustment of metal uptake could be the consequence of diminished protein synthesis following nitrogen deprivation. Given that up to a third of the total microbe proteome contains metal cofactors (64), reduction in metal uptake could be an important response to avoid build-up of excess free metals in the cytosol that would lead to detrimental reactions.
The asymmetric distribution of the NtcA binding peaks in NH4 and −N media points to condition-dependent modus operandi of NtcA
Comparative ChIP-seq analysis enabled us to characterize changes in the binding behavior of NtcA. It revealed that NtcA binds to 141 DNA regions (121 in the chromosome and 20 in the plasmids) after 4 h of nitrogen step-down, but also to a large number of regions (51 with 48 in the chromosome and 3 in the plasmids) in nitrogen-replete medium. Its unexpected binding in vivo under nitrogen-replete conditions supports in vitro measurements showing that 2-OG is not absolutely required for NtcA binding to DNA (12). The 169 binding sites identified and assigned to 157 genes, included the vast majority of currently known NtcA target genes for Synechocystis, indicating the high sensitivity of our ChIP-Seq experiment. Further support of its reliability was provided by validation of NtcA binding to target regions with different features (intragenic or promoter binding region, different peak enrichment, etc.; Supplementary Figure S5).
Under nitrogen starvation conditions, most of the binding sites were located upstream of gene coding regions (73% of peaks; Supplementary Tables S9 and 10), consistent with preferential binding of NtcA to promoter regions. Unexpectedly, this preference changed in ammonium-rich medium, with 54% of binding peaks located at intragenic positions. This highly significant change in the distribution of binding loci (P = 0.00116, Fisher’s exact test) could indicate a condition-dependent functional mode for NtcA in Synechocystis. Under nitrogen depletion, many genes involved in nitrogen metabolism and other cellular functions were induced or repressed by NtcA, supporting its established role as the master regulator of nitrogen control in cyanobacteria. In contrast, the role of NtcA in the presence of ammonium has remained elusive. The lower number of peaks and the high proportion of intragenic binding sites for NH4 compared to −N treatment could simply indicate that NtcA rests in an inactive state under nitrogen-replete conditions, as previously suggested (57). However, other potential scenarios could exist, given that extensive binding of transcription factors to intragenic regions has been reported in recent years, based on genome-wide profiling. For example, substantial intragenic binding has been detected for OmpR in Salmonella enterica, GlxR in Corynebacterium glutamicum and RutR in Escherichia coli (65–67). Remarkably, a high proportion of intragenic binding sites have also been observed in a previous ChIP-seq analysis for NtcA in Anabaena sp. PCC 7120 (14). Hence, an examination of the functional relevance of intergenic binding is clearly vindicated. Closer inspection of intragenic binding sites detected by our ChIP-seq experiment revealed the existence of the canonical NtcA consensus sequence, located upstream of internal TSS in some cases (i.e. ssll0377, sll0735, slr1065, slr1864, glcF; Supplementary Tables S9 and 10). In other cases, our transcriptomic data suggest that NtcA could act as a repressor, blocking transcription elongation (Figure 5) in a similar manner to that reported for CodY in Bacillus subtilis (68). Candidates for this type of transcriptional repression by NtcA are: slr1852, glgP and pilA4 under nitrogen-replete conditions; or slr1028, slr0442, secA, sll1273, mrvA and metX under nitrogen starvation (Figure 5 and Table 2). Such a regulatory mode, however, cannot be generalized, as we found other cases, such as sll0142 in NH4 or slr0909 in −N treatments, which showed increased expression and stronger intragenic NtcA binding—an observation that eludes an obvious explanation. In summary, examination of intragenic binding sites supports a model in which NtcA is not simply inactive under nitrogen-replete conditions, but in which it remains functionally important, albeit with a different modus operandi. Such a model is consistent with previous ChIP–chip and ChIP-seq studies of E. coli and Anabaena suggesting that the CRP transcriptional regulator family could work not only as a canonical transcriptional regulator, but also as a chromosome-shaping protein by binding to multiple low affinity sites (14,69).
NtcA targets in nitrogen control and cellular metabolic processes
Parallel profiling with ChIP-seq and RNA-seq techniques led to the definition of a direct target regulon for NtcA, having unprecedented resolution. In total, 79 genes were identified to have NtcA binding peaks with significantly altered expression after 4 h of nitrogen step-down (Figure 5). Analysis of their respective gene functions revealed that NtcA plays a role in the coordination of cellular processes beyond nitrogen metabolism. Indeed, various other metabolic processes (biosynthesis of cofactors, cellular processes, carbon metabolism, energy metabolism, central intermediary metabolism, photosynthesis and respiration) were dominant among these NtcA targets (Figure 6). Additionally, a small proportion of the NtcA regulon was associated with other biological functions, such as cell envelope, translation and transcription, or regulatory functions (Figure 6).
A considerable number of NtcA targets lack current annotation. In particular, 27 genes correspond to hypothetical and unknown proteins. They are prime candidates for further experimental investigation to enhance our understanding of nitrogen control and metabolism, and to extend the functionality of the NtcA regulon. For instance, genes sll0327 and ssr0692 showed levels of induction and repression, similar to those involved in nitrogen metabolism, suggesting related roles (Figure 6). Likewise, sll0327 codes for a small protein containing 139 amino acids, and is a closed homolog to the amino acid transport system permease component LivM of Microcystis aeruginosa (58.1% identity; 72.1% similarity). Meanwhile, ssr0692 was reported to be highly induced under conditions of low CO2 i.e. conditions which resemble a relative high abundance of nitrogen (70). It may be involved in the accumulation of the NDH-1 complex under low carbon/high nitrogen, given that Ssr0692 interacts with NdhH, according to yeast-two-hybrid data (71). Furthermore, the expression of both sll0327 and ssr0692 is strongly correlated with known NtcA target genes across a large set of conditions compiled in CyanoEXpress—a platform for a gene expression meta-analysis in Synechocystis (72). This is a remarkable finding, since CyanoEXpress does not include expression data for nitrogen starvation. Thus, this correlation is solely based on differential expression of NtcA target genes under other conditions. Strikingly, we found that sll0327 together with bgtB, glnA and urtB (all members of the NtcA regulon) are highly upregulated under iron limitation and downregulated under carbon limitation or oxidative stress (Supplementary Figure S6A). Conversely, ssr0692 together with gifA and gifB are downregulated under iron limitation and upregulated under carbon limitation or oxidative stress (Supplementary Figure S6B). This suggests that sll0327 and ssr0692 have a common regulation, along with other bona fide members of the NtcA regulon. In addition, the strong correlation of NtcA targets observed in gene expression meta-analysis points to potential regulatory activity of NtcA under a wider range of environmental perturbations.
Finally, asRNAs and ncRNAs were the third most abundant class in the NtcA expression network. Although differential expression of asRNAs and ncRNAs during acclimation to different nitrogen stresses in cyanobacteria has been documented in several recent studies (15,41,61), only one ncRNA, NsiR4, has been experimentally validated to be under the control of NtcA to date (41). Our results greatly expand the repertoire of NtcA regulated ncRNAs. Eight ncRNAs were included in the NtcA regulon (Figure 6) and we have experimentally validated three of them: Ncr1071, NsiR7, Syr6 (Figure 8). Interestingly, NsiR7 is also induced under conditions of carbon limitation and was recently suggested as an NdhR-regulated sRNA (73,74). Thus, NsiR7 might play an important role in the integration of signals integration related to the intracellular carbon and nitrogen status.
NtcA binding defines poised states of transcriptional regulation in Synechocystis, which are modulated by PipX and additional factors
Surprisingly, almost half of the genes having assigned NtcA binding were not transcriptionally affected after 4 h of nitrogen step-down. However, comparison with a longer time series study of nitrogen starvation (3) revealed that many of the initially unaffected genes were activated or repressed at later time points (Supplementary Figure S7). Examples are the induction of genes coding for the urease gamma subunit (ureA), the serine/threonine kinase (spkD) or repression of the PSII oxygen evolving complex protein (psbP2) after 12 h of nitrogen depletion. Even longer delays in transcriptional response were detected for the repression of pipX (24 h) and the induction of the ferrichrome-iron receptor coding gene fhuA (96 h). Importantly, late transcriptional responses are not simply the consequence of delayed NtcA accumulation, given that both NtcA transcript and protein levels remained unaltered after nitrogen step-down. Rather, these findings indicate that NtcA binding in vivo may not be sufficient in certain cases to evoke immediate full transcriptional control, but defines a poised regulatory state, which correlates with later changes in expression. Such poised regulation suggests the need for additional factors, as previously proposed (14,57). Further support for this model was gained through comparison of the ChIP-seq data obtained under NH4 and −N treatments. We identified 14 NtcA targets having assigned NtcA binding peaks that did not show significant changes, but were significantly differentially expressed after nitrogen depletion. Our examination of a pipX mutant supports the role of PipX as an NtcA modulating factor in Synechocystis. Transcriptional regulation of NtcA-controlled genes and their corresponding proteins levels were delayed in the pipX mutant strain compared to the WT strain, consistent with a role of PipX as an NtcA enhancer, as reported in other cyanobacteria (10,49,59). This mechanism could explain why several genes showed strong transcriptional change after 4 h of nitrogen starvation, without an equivalent variation in the effective affinity of NtcA to their promoters (Supplementary Table S12). This indicates that interaction of NtcA with other factors is essential to activate/repress target genes.
It is tempting to speculate on the physiological function of a poised regulatory state for NtcA. One advantage could be that it prepares the transcriptional machinery for a more extensive response, but without a full commitment. It would provide a temporal buffer, which is especially advantageous under conditions of fluctuating nitrogen availability, where premature commitment to unmitigated chlorosis might prove to be adverse. In this way, some NtcA targets, for which no differential expression was recorded after 4 h of nitrogen depletion, might constitute a ‘latent NtcA regulon’; in contrast to the NtcA regulon defined herein, which is activated during early acclimation.
In conclusion, this study provides the first genome-wide captures of the dynamics of in vivo NtcA binding events and the regulatory network that they define. Application of parallel differential profiling enabled us to distinguish direct from indirect regulation, extending the scope of the NtcA regulon. Finally, indications of a poised state for NtcA and of regulatory activity in a wide range of conditions beyond nitrogen limitation offer exciting avenues for further investigations.
DATA AVAILABILITY
RNA-seq and ChIP-seq datasets are available at GEO accession (GSE97291).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Trudi Semeniuk and Maria Isabel Ortiz Marchena for critical reading of the manuscript, and Alicia Muro Pastor and Elvira Olmedo Verd for support in ncRNAs experiments.
Author contributions: J.G-L., M.H-P. and M.E.F. designed the experiments, while J.G-L., R.R-R. and M.I.M-P. conducted the experimental work. J.G-L. analyzed the data, carried out the literature research and wrote the manuscript. J.G-L., M.H-P., M.I.M-P., F.J.F. and M.E.F. contributed to interpretation and discussion of the results, and the review/editing before submission. J.G-L and M.E.F. supervised the study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Portuguese Funding through FCT (Fundaçao para a Ciência e a Tecnologia) projects [PTDC/BIA-MIC/4418/2012, IF/00881/2013, UID/BIM/04773/2013–CBMR, UID/Multi/04326/2013–CCMAR]; Ministerio de Economia y Competitividad (MINECO) Grants [BFU2013–41712, BIO2016–75634 to F.J.F.]; Junta de Andalucía-European Regional Funds (FEDER) [Group BIO-284, P12-BIO-1119]. Funding for open access charge: School of Biomedical & Healthcare Sciences, Plymouth University Peninsula Schools of Medicine and Dentistry.
Conflict of interest statement. None declared.
REFERENCES
- 1. Schwarz R., Forchhammer K.. Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology. 2005; 151:2503–2514. [DOI] [PubMed] [Google Scholar]
- 2. Flores E., Herrero A.. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 2010; 8:39–50. [DOI] [PubMed] [Google Scholar]
- 3. Krasikov V., Aguirre von Wobeser E., Dekker H.L., Huisman J., Matthijs H.C.. Time-series resolution of gradual nitrogen starvation and its impact on photosynthesis in the cyanobacterium Synechocystis PCC 6803. Physiol. Plant. 2012; 145:426–439. [DOI] [PubMed] [Google Scholar]
- 4. Osanai T., Imamura S., Asayama M., Shirai M., Suzuki I., Murata N., Tanaka K.. Nitrogen induction of sugar catabolic gene expression in Synechocystis sp. PCC 6803. DNA Res. 2006; 13:185–195. [DOI] [PubMed] [Google Scholar]
- 5. Kopf M., Klahn S., Scholz I., Matthiessen J.K., Hess W.R., Voss B.. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 2014; 21:527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muro-Pastor M.I., Reyes J.C., Florencio F.J.. Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J. Biol. Chem. 2001; 276:38320–38328. [DOI] [PubMed] [Google Scholar]
- 7. Vega-Palas M.A., Flores E., Herrero A.. NtcA, a global nitrogen regulator from the cyanobacterium Synechococcus that belongs to the Crp family of bacterial regulators. Mol. Microbiol. 1992; 6:1853–1859. [DOI] [PubMed] [Google Scholar]
- 8. Herrero A., Muro-Pastor A.M., Flores E.. Nitrogen control in cyanobacteria. J. Bacteriol. 2001; 183:411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia-Dominguez M., Reyes J.C., Florencio F.J.. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol. Microbiol. 2000; 35:1192–1201. [DOI] [PubMed] [Google Scholar]
- 10. Espinosa J., Forchhammer K., Contreras A.. Role of the Synechococcus PCC 7942 nitrogen regulator protein PipX in NtcA-controlled processes. Microbiology. 2007; 153:711–718. [DOI] [PubMed] [Google Scholar]
- 11. Huergo L.F., Chandra G., Merrick M.. P(II) signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol. Rev. 2013; 37:251–283. [DOI] [PubMed] [Google Scholar]
- 12. Forcada-Nadal A., Forchhammer K., Rubio V.. SPR analysis of promoter binding of Synechocystis PCC6803 transcription factors NtcA and CRP suggests cross-talk and sheds light on regulation by effector molecules. FEBS Lett. 2014; 588:2270–2276. [DOI] [PubMed] [Google Scholar]
- 13. Su Z., Olman V., Mao F., Xu Y.. Comparative genomics analysis of NtcA regulons in cyanobacteria: regulation of nitrogen assimilation and its coupling to photosynthesis. Nucleic Acids Res. 2005; 33:5156–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Picossi S., Flores E., Herrero A.. ChIP analysis unravels an exceptionally wide distribution of DNA binding sites for the NtcA transcription factor in a heterocyst-forming cyanobacterium. BMC Genomics. 2014; 15:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mitschke J., Vioque A., Haas F., Hess W.R., Muro-Pastor A.M.. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20130–20135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rippka R., Deruelles J., Waterbury J.B., Herman M., Stanier R.Y.. Generic assigment, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 1979; 111:1–61. [Google Scholar]
- 17. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thorvaldsdóttir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spyrou C., Stark R., Lynch A.G., Tavaré S.. BayesPeak: Bayesian analysis of ChIP-seq data. BMC Bioinformatics. 2009; 10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu G., Wang L.-G., He Q.-Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 25. Pinto F.L., Thapper A., Sontheim W., Lindblad P.. Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol. Biol. 2009; 10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anders S., Pyl P.T., Huber W.. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mähler N., Cheregi O., Funk C., Netotea S., Hvidsten T.R.. Syn ergy: a web resource for exploring gene regulation in Synechocystis sp. PCC6803. PLoS One. 2014; 9:e113496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cline M.S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B.. Integration of biological networks and gene expression data using Cytoscape. Nat. Potoc. 2007; 2:2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Hoon M.J., Imoto S., Nolan J., Miyano S.. Open source clustering software. Bioinformatics. 2004; 20:1453–1454. [DOI] [PubMed] [Google Scholar]
- 32. Saldanha A.J. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004; 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 33. Busch A., Richter A.S., Backofen R.. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008; 24:2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garcia-Dominguez M., Florencio F.J.. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 1997; 35:723–734. [DOI] [PubMed] [Google Scholar]
- 35. Sambrook J., Fritsch E.F., Maniatis T.. Molecular Cloning: a Laboratory Manual. 1989; 2nd edn, NY: Cold Spring Harbor Laboratory press. [Google Scholar]
- 36. Navarro F., Martín-Figueroa E., Florencio F.J.. Electron transport controls transcription of the thioredoxin gene (trxA) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 2000; 43:23–32. [DOI] [PubMed] [Google Scholar]
- 37. Galmozzi C.V., Fernández‐Avila M.J., Reyes J.C., Florencio F.J., Muro‐Pastor M.I.. The ammonium‐inactivated cyanobacterial glutamine synthetase I is reactivated in vivo by a mechanism involving proteolytic removal of its inactivating factors. Mol. Microbiol. 2007; 65:166–179. [DOI] [PubMed] [Google Scholar]
- 38. Mérida A., Candau P., Florencio F.J.. Regulation of glutamine synthetase activity in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 by the nitrogen source: effect of ammonium. J. Bacteriol. 1991; 173:4095–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Muro-Pastor M.I., Reyes J.C., Florencio F.J.. The NADP+-isocitrate dehydrogenase gene (icd) is nitrogen regulated in cyanobacteria. J. Bacteriol. 1996; 178:4070–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramakers C., Ruijter J.M., Deprez R.H.L., Moorman A.F.. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003; 339:62–66. [DOI] [PubMed] [Google Scholar]
- 41. Klahn S., Schaal C., Georg J., Baumgartner D., Knippen G., Hagemann M., Muro-Pastor A.M., Hess W.R.. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E6243–E6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Forchhammer K. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol. Rev. 2004; 28:319–333. [DOI] [PubMed] [Google Scholar]
- 43. Battchikova N., Eisenhut M., Aro E.M.. Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles. Biochim. Biophys. Acta. 2011; 1807:935–944. [DOI] [PubMed] [Google Scholar]
- 44. Ogawa T., Mi H.. Cyanobacterial NADPH dehydrogenase complexes. Photosynth. Res. 2007; 93:69–77. [DOI] [PubMed] [Google Scholar]
- 45. Schlebusch M., Forchhammer K.. Requirement of the nitrogen starvation-induced protein Sll0783 for polyhydroxybutyrate accumulation in Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 2010; 76:6101–6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mitschke J., Georg J., Scholz I., Sharma C.M., Dienst D., Bantscheff J., Voss B., Steglich C., Wilde A., Vogel J. et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:2124–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kopf M., Hess W.R.. Regulatory RNAs in photosynthetic cyanobacteria. FEMS Microbiol. Rev. 2015; 39:301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Choi S.Y., Park B., Choi I.-G., Sim S.J., Lee S.-M., Um Y., Woo H.M.. Transcriptome landscape of Synechococcus elongatus PCC 7942 for nitrogen starvation responses using RNA-seq. Sci. Rep. 2016; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Valladares A., Rodriguez V., Camargo S., Martinez-Noel G.M., Herrero A., Luque I.. Specific role of the cyanobacterial PipX factor in the heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 2011; 193:1172–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Azuma M., Osanai T., Hirai M.Y., Tanaka K.. A response regulator Rre37 and an RNA polymerase sigma factor SigE represent two parallel pathways to activate sugar catabolism in a cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2011; 52:404–412. [DOI] [PubMed] [Google Scholar]
- 51. Reyes J.C., Muro-Pastor M.I., Florencio F.J.. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J. Bacteriol. 1997; 179:2678–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Picossi S., Valladares A., Flores E., Herrero A.. Nitrogen-regulated genes for the metabolism of cyanophycin, a bacterial nitrogen reserve polymer: expression and mutational analysis of two cyanophycin synthetase and cyanophycinase gene clusters in heterocyst-forming cyanobacterium Anabaena sp. PCC 7120. J. Biol. Chem. 2004; 279:11582–11592. [DOI] [PubMed] [Google Scholar]
- 53. Luque I., Forchhammer K.. Nitrogen assimilation and C/N balance sensing. Cyanobacteria. 2008; 335–382. [Google Scholar]
- 54. Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wright P.R., Richter A.S., Papenfort K., Mann M., Vogel J., Hess W.R., Backofen R., Georg J.. Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E3487–E3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pain A., Ott A., Amine H., Rochat T., Bouloc P., Gautheret D.. An assessment of bacterial small RNA target prediction programs. RNA Biol. 2015; 12:509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Luque I., Vazquez-Bermudez M.F., Paz-Yepes J., Flores E., Herrero A.. In vivo activity of the nitrogen control transcription factor NtcA is subjected to metabolic regulation in Synechococcus sp. strain PCC 7942. FEMS Microbiol. Lett. 2004; 236:47–52. [DOI] [PubMed] [Google Scholar]
- 58. Vazquez-Bermudez M.F., Herrero A., Flores E.. Carbon supply and 2-oxoglutarate effects on expression of nitrate reductase and nitrogen-regulated genes in Synechococcus sp. strain PCC 7942. FEMS Microbiol. Lett. 2003; 221:155–159. [DOI] [PubMed] [Google Scholar]
- 59. Espinosa J., Forchhammer K., Burillo S., Contreras A.. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 2006; 61:457–469. [DOI] [PubMed] [Google Scholar]
- 60. Su Z., Mao F., Dam P., Wu H., Olman V., Paulsen I.T., Palenik B., Xu Y.. Computational inference and experimental validation of the nitrogen assimilation regulatory network in cyanobacterium Synechococcus sp. WH 8102. Nucleic Acids Res. 2006; 34:1050–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Klotz A., Georg J., Bučinská L., Watanabe S., Reimann V., Januszewski W., Sobotka R., Jendrossek D., Hess W.R., Forchhammer K.. Awakening of a dormant cyanobacterium from nitrogen chlorosis reveals a genetically determined program. Curr. Biol. 2016; 26:2862–2872. [DOI] [PubMed] [Google Scholar]
- 62. Osanai T., Kanesaki Y., Nakano T., Takahashi H., Asayama M., Shirai M., Kanehisa M., Suzuki I., Murata N., Tanaka K.. Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 sigma factor sigE. J. Biol. Chem. 2005; 280:30653–30659. [DOI] [PubMed] [Google Scholar]
- 63. Joseph A., Aikawa S., Sasaki K., Teramura H., Hasunuma T., Matsuda F., Osanai T., Hirai M.Y., Kondo A.. Rre37 stimulates accumulation of 2-oxoglutarate and glycogen under nitrogen starvation in Synechocystis sp. PCC 6803. FEBS Lett. 2014; 588:466–471. [DOI] [PubMed] [Google Scholar]
- 64. Cvetkovic A., Menon A.L., Thorgersen M.P., Scott J.W., Poole F.L. 2nd, Jenney F.E. Jr, Lancaster W.A., Praissman J.L., Shanmukh S., Vaccaro B.J. et al. Microbial metalloproteomes are largely uncharacterized. Nature. 2010; 466:779–782. [DOI] [PubMed] [Google Scholar]
- 65. Perkins T.T., Davies M.R., Klemm E.J., Rowley G., Wileman T., James K., Keane T., Maskell D., Hinton J.C., Dougan G.. ChIP‐seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Mol. Microbiol. 2013; 87:526–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jungwirth B., Sala C., Kohl T.A., Uplekar S., Baumbach J., Cole S.T., Pühler A., Tauch A.. High-resolution detection of DNA binding sites of the global transcriptional regulator GlxR in Corynebacterium glutamicum. Microbiology. 2013; 159:12–22. [DOI] [PubMed] [Google Scholar]
- 67. Shimada T., Ishihama A., Busby S.J., Grainger D.C.. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 2008; 36:3950–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Belitsky B.R., Sonenshein A.L.. Roadblock repression of transcription by Bacillus subtilis CodY. J. Mol. Biol. 2011; 411:729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Grainger D.C., Hurd D., Harrison M., Holdstock J., Busby S.J.. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17693–17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Battchikova N., Vainonen J.P., Vorontsova N., Keraänen M., Carmel D., Aro E.-M.. Dynamic changes in the proteome of Synechocystis 6803 in response to CO2 limitation revealed by quantitative proteomics. J. Proteome Res. 2010; 9:5896–5912. [DOI] [PubMed] [Google Scholar]
- 71. Sato S., Shimoda Y., Muraki A., Kohara M., Nakamura Y., Tabata S.. A large-scale protein–protein interaction analysis in Synechocystis sp. PCC6803. DNA Res. 2007; 14:207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hernández-Prieto M.A., Semeniuk T.A., Giner-Lamia J., Futschik M.E.. The transcriptional landscape of the photosynthetic model cyanobacterium Synechocystis sp. PCC6803. Sci. Rep. 2016; 6:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Orf I., Klähn S., Schwarz D.D., Frank M., Hess W.R., Hagemann M., Kopka J.. Integrated analysis of engineered carbon limitation in a quadruple CO2/HCO3–uptake mutant of Synechocystis sp. PCC 6803. Plant Physiol. 2015; 169:1787–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Klähn S., Orf I., Schwarz D., Matthiessen J.K., Kopka J., Hess W.R., Hagemann M.. Integrated transcriptomic and metabolomic characterization of the low-carbon response using an ndhR mutant of Synechocystis sp. PCC 6803. Plant Pysiol. 2015; 169:1540–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP-seq datasets are available at GEO accession (GSE97291).