Abstract
RNase III enzymes cleave double stranded (ds)RNA. This is an essential step for regulating the processing of mRNA, rRNA, snoRNA and other small RNAs, including siRNA and miRNA. Arabidopsis thaliana encodes nine RNase III: four DICER-LIKE (DCL) and five RNASE THREE LIKE (RTL). To better understand the molecular functions of RNase III in plants we developed a biochemical assay using RTL1 as a model. We show that RTL1 does not degrade dsRNA randomly, but recognizes specific duplex sequences to direct accurate cleavage. Furthermore, we demonstrate that RNase III and dsRNA binding domains (dsRBD) are both required for dsRNA cleavage. Interestingly, the four DCL and the three RTL that carry dsRBD share a conserved cysteine (C230 in Arabidopsis RTL1) in their dsRBD. C230 is essential for RTL1 and DCL1 activities and is subjected to post-transcriptional modification. Indeed, under oxidizing conditions, glutathionylation of C230 inhibits RTL1 cleavage activity in a reversible manner involving glutaredoxins. We conclude that the redox state of the dsRBD ensures a fine-tune regulation of dsRNA processing by plant RNase III.
INTRODUCTION
RNasesIII are endonucleases involved in double-stranded (ds)RNA cleavage. They play an essential role in processing of ribosomal RNA, in sno- and snRNA biogenesis and RNA decay. Furthermore, RNase III Drosha and Dicer proteins cleave likewise precursors of small RNA involved in RNA interference (RNAi) pathway. RNase III proteins are structurally diverse. Depending on the isoform, they have a single or two RNase III domain and up to six other protein domains, including one or two dsRBD (double stranded RNA Binding Domain), PAZ and helicase (1,2).
In Arabidopsis thaliana, two families of RNase III are found (3). The first is the Dicer-like (DCL) family composed of four members (4). DCL1 is involved in the production of ancient ∼21-nt miRNA derived from non-perfectly folded hairpin precursor transcribed from MIR genes. DCL2 produces 22-nt endo-siRNA, while DCL4 produces 21-nt recently evolved miRNA and ta-siRNA (5,6). They also participate in the plant defense by processing viral/transgene siRNA (7). In contrast to DCL1, 2 and 4, which are implicated in post transcriptional gene silencing (PTGS) pathways, DCL3 plays a central role in transcriptionional gene silencing (TGS). DCL3 produces 24-nt siRNA, essential for guiding DNA methylation and directing chromatin reorganization in the RNA directed DNA Methylation (RdDM) pathway (8). The second family is the RNase Three Like (RTL) which consist of five members, RTL1 through 3, RTL4/NFD2 and RTL5/RNC1 (3,9). In contrast to DCL proteins, the role of the RTL proteins remains poorly understood.
RTL1 and 2 localize in both the nucleus and the cytoplasm (3). RTL2 has one RNase III and two dsRBD, forms homodimers through disulfides bonds and is required for processing of 45S pre-rRNA (3). Furthermore RTL2 modulates siRNA expression and DNA methylation of hundreds of specific coding and non-coding genes (10). RTL1 is structurally simpler, containing single RNase III and dsRBD domains. In contrast to RTL2, which is ubiquitously expressed, the RTL1 gene is only expressed weakly in roots (3). Remarkably, RTL1 is induced under viral stress conditions (11). Plants over-expressing RTL1 lack siRNAs produced by DCL2, DCL3 and DCL4 from 6000 endogenous loci, but show unchanged levels of miRNAs produced by DCL1 (11). Together, these results indicate that perfectly paired dsRNA precursor molecules are cleaved by RTL1 before they are processed by DCL2, DCL3 or DCL4. Oddly, a handful of siRNAs over-accumulates in plants overexpressing RTL1 or are found at different size variants compared to wild-type (WT) controls, suggesting that, occasionally, dsRNA precursor molecules can still be processed by DCL(s) protein(s) after an initial processing by RTL1 (11).
Although the molecular bases of RNA recognition and cleavage by RNase III from bacteria and in yeast are relatively well known (1,2) much less is known about RNase III from higher eukaryotic organism. In mammalian cells, Drosha cleaves dsRNA and generates overhang that in turn are recognized by Dicer to produces 24-nt RNA duplexes by a counting ruler mechanism (12,13). In Arabidopsis, bulged stem structures are required for DCL1 activity, whereas DCL2 and DCL4 function through a ruler mechanism (14–16). On the other hand, DCL3 has better affinity for short dsRNA (14,17,18), likely explaining why Pol IV-RDR2-derived precursors, which are generally 30-60-nt long, are predominantly processed by DCL3 (17,18). Nevertheless, the production of some Pol IV-RDR2-DCL3-dependent siRNAs depends on RTL2 (10), while other depends on DCL4 (19), suggesting that the longest Pol IV-RDR2-derived precursors need to be processed by RTL2 or DCL4 to reach the size allowing cleavage by DCL3. Regarding RTL1-3, the essential residues in the RNase III domain are well conserved indicating a similar mechanism of dsRNA catalysis (3,10,11). The molecular features controlling and/or regulating the RNase III activity remain largely unknown not only for RTL but also for Dicer proteins.
Here, we describe molecular RNA cleavage site determinants of RTL1 and demonstrate that a conserved cysteine in the dsRBD of all RNase III family members from Arabidopsis and other plants species, is required for cleavage activity. This cysteine can be glutathionylated which inhibits the activity of RTL1. We also show that RTL1 can be deglutathionylated by glutaredoxins (GRXs), restoring the cleavage activity of RTL1. Our finding indicates that the redox state of the RNase III protein domains might play a major role in controlling the fate of coding and non-coding RNAs in plants.
MATERIALS AND METHODS
Production of RTL1 constructs
Cloning and production of His-RTL1, His-RTL1mR3 and His-RTL2 protein for in vitro assay was previously described (3,11). The C230S and C260S mutant versions of His-RTL1 were obtained using the primers C230fw/C230rev and C260fw/C260rev respectively. The ΔdsRBD version of His-RTL1 was obtained by introducing a stop codon at position 627 using the primers dsRBDfw and dsRBDrev. The His-R1D2 and His-R2D1 swap versions were generated from His-RTL1 and His-RTL2. Recombinant proteins were purified using Talon Metal Affinity Resin following Clontech's instructions and dialyzed against Sample Buffer (20 mM Tris–HCl, pH 7.5, 100 mM NaCl, 20% glycerol, 1 mM ethylenediaminetetraacetic acid, 1 mM Dithiothréitol (DTT)).
The 35S:RTL1 and 35S:RTL1-Myc constructs derived from RTL1 genomic sequences have been described previously (11). To obtain the 35S:RTL1ΔdsRBD-Myc construct the dsRBD was amplified using primers attB1RTL1-F and attB2RTL1-R3-R and cloned into the pDONR207 (Gateway® Technology-Invitrogen). The 35S:RTL1 construct was made using the His-RTL1 construct as a template. Similarly, the 35S:RTL1C230S and 35S:RTL1C260S were generated using the His-RTL1C230S and His-RTL1C260S constructs as templates, respectively.
The 35S:DCL1-RDD and 35S:DCL1-RD constructs were made using genomic sequences. DCL1 fragments starting at the beginning of the second RNase III domain and ending either at the end of the first dsRBD (35S:DCL1-RD) or at the stop codon (35S:DCL1-RDD) were amplified using primers attB1DCL1RDD-F and attB2DCL1RD-R or attB2DCL1RDD-R. Recombination into pGWB2 through the gateway LR recombinase reaction (Invitrogen) created 35S:DCL1-RDD and 35S:DCL1-RD. Point mutations in the conserved cysteine of the first DCL1 dsRBD (C1742, corresponding to RTL1 C230) was perfomed using the DCL1-RDm-F and DCL1-RDm-R primers.
Cloning of the 3′UTR At3g18145 mRNA sequence
Cloning of a cDNA sequence encoding 3′UTR At3g18145 was performed by reverse transcription polymerase chain reaction (RT-PCR) using total RNA and UTRrt primers, followed by a PCR amplification (Phusion, Finnzymes) with UTRfw and p1 primers and ligation into pGEMT-Easy vector (Promega). The mutated version of the RNA substrate 1 (V1-V4) were obtained using the primers M1csFV1, M1csRV1, M2csFV1, M2csRV1, M1csFV2, M1csRV2, M2csFV2, M2csRV2, M1csFV3, M1csRV3, M2csFV3, M2csRV3, M2csFV4, M2csRV4, M2csFV5, M2csRV5.
RNA-related methods
In vitro transcribed RNA for primer extension and reverse transcription in circle (cRT) was obtained with Riboprobe in vitro Transcription Systems (Promega) using linear pGemT-3′UTR_At3g18145. The [32P]CTP-labeled RNA substrates were transcribed from linear pBSIIk+ plasmids containing rcr1 sequences (GeneCust). Primer extension (20) analysis was performed using in vitro transcribed RNA treated or untreated with His-RTL1 and specific 5′ end labeled primers p1, p2 and p7. For Reverse Transcription in circle (cRT) reactions, His-RTL1 treated RNA was incubated with T4 RNA ligase (Promega) for 1 h at 37°C. The reaction was then used to perform RT with UTRrt primers, followed by 42 cycles PCR with p1/p8 primers. PCR products were purified on 2% agarose gel, cloned and sequenced. Total RNA from Wt and RTL1-Flag plants was prepared using TriZol reagent (GE Healthcare, Littler Chalfont, Buckinghamshire, UK). All RNA samples were then treated with Turbo DNase (Ambion) to eliminate contaminant DNA (21).
His-RTL1 cleavage assays
In vitro assay were performed as previously described (11). To determine cleavage activity of specific RNA targets, treated and untreated RNA (∼50 ng) was used to carry out RT-PCR (Kit Promega) with p3/p4 and p5/p6 primers to detect At3g18145 RNA sequences or with tas2fw/tas2rev, mir159afw/mir159arev, I4fw/I4rev, mir828afw/mir828arev, U3fw/U3rev primers to detects TAS, MIR159, Intron4 At2g33255, MIR828 and U3 snoRNA sequences. Alternatively, 4 μl of diluted (1:10) radiolabeled rcr1 and rcr2 RNA probes were mixed with recombinant His-RTL1 (∼200 ng) proteins and incubated 30 min at 37°C. The reaction was stopped by heating samples at 70°C for 5 min and the products of reaction analyzed on a 6% denaturing polyacrylamide gel.
Cleavage in crude protein extracts
All lines were derived from A. thaliana Columbia (Col 0) ecotype. Plants 35S-RTL1 and dcl3 were described previously (11). Seeds were sown on 1× Murashige and Skoog medium (MS containing 1% sucrose) and left for 2 days at 4°C to synchronize. Plants were then grown in controlled growth chambers under a 16 h light/ 8 h dark cycle at 21°C for 2 weeks. For treatment with GSSG (Sigma-Aldrich), 2 week-old plant seedlings were transferred to petri dishes containing 5 ml of liquid MS/2 medium complemented with 10 mM GSSG for 24 h. Before harvesting, plants were extensively washed to remove the excess of GSSG. Glutathione contents were determined on these samples by using the recycling enzymatic assay described in (22). Each measurement included four biological repetitions.
Protein crude extracts were prepared as previously described (14). Briefly, plant material (500 mg), was homogenized and extracted in protein extraction buffer (20 mM Tris–HCl pH 7.5, 4 mM MgCl2, 5 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml leupeptin and 1 μg/ml pepstatin A). Soluble fractions were cleared by centrifugation at 20 000 g for 20 min at 4°C (twice).
Single stranded RNA (ssRNA) were purchase to GenScript (NY, USA) and end-labelled by T4-PNK and [γ-32P]-ATP. The sense and antisense sequences are shown in Supplementary Figure S1. For dsRNA annealing, equal amount of end-labelled ssRNA (∼ 2 pmoles) were incubated in 10 mM Tris–HCl pH 7.5 and 100 mM NaCl at 90°C for 5 min. After turning off the heat, the samples where maintained for 10 min and then transferred to 22°C for 10 extra min. [γ-32P]-ATP dsRNA was purified by phenol/chloroform and ethanol precipitated.
32P-dsRNA (∼0.2 pmoles) was incubated with 15 μl of crude protein extract, 5 μl of 4× cleavage buffer (40 mM Tris–HCl pH 7.5, 200 mM NaCl 16 mM MgCl2), 6 mM ATP, 2 mM GTP and 20U RNase OUT (Invitrogen) for 45 min at 37°C. After incubation, the RNA were purified by phenol/chloroform, ethanol precipitated and analyzed on 10% denaturing polyacrylamide gel electrophoresis (PAGE) with 8% urea.
Nicotiana benthamiana agroinfiltrations
The 10 ml cultures of Agrobacterium tumefaciens C58C1 (pMP90) carrying the plasmids of interest were grown overnight at 28°C and then centrifuged at 6000 rpm. The bacterial pellets were suspended at 10 mM MgCl2, 10 mM MES pH5.2, 150 μM acetosyringone solution to a final OD600 of 1.0. The bacterial solution was incubated at 20°C for 3 h and then used to infiltrate leaves. Leaves were harvested 3 days after infiltration.
Protein analysis
For crosslinking studies, His-RTL1 recombinant protein (∼100–500 ng) was incubated in 0.2 M TEA, pH 8 with or without 2 mM DMP (1 h at 4°C). The crosslink reaction was stopped by adding Tris–HCl pH 8 to a final concentration of 50 mM. The products of reaction were analyzed by western blot using α-RTL1 antibodies. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting were performed as described previously (20). The membrane was hybridized with 1:1000 dilution of α-RTL1 Rat polyclonal antibodies (Eurogentec). Alternatively, protein samples were suspended in loading buffer (30 mM Tris pH 6.8, 12.5% Glycerol, 0.005% Bromophenol Blue) without SDS and DTT and analyzed at 4°C on a 12% non-denaturing gel. For glutathionylation studies, the samples were incubated at room temperature for 30 min with or without GSSG or GSH+H2O2. For deglutathionylation test, the samples were further incubated at room temperature for 20 min with or without the glutaredoxin system (5 mM NADPH, 2 μM GRXC1 (or GRXC2), 0.45 units yeast Glutathione Reductase (GR, Sigma-Aldrich).
Gel filtration chromatography
His-RTL1 recombinant proteins (∼400 μg) were fractionated using Superdex 75 10/300 GL (GE Healthcare) column equilibrated in Protein Buffer (50 mM Tris pH 7,5, 5 mM MgCl2) containing 150 mM NaCl, 500 mM NaCl or 500 mM NaCl/100 mM DTT. The protein standards were conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa) and ribonuclease A (13.7 kDa) (GE Healthcare).
Bioinformatics analyses
The amino acid sequences of Dicer-like proteins were obtained from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), and RTL1 and RTL2 sequences from (3). The dsRBD were identified using Motif Scan software (http://myhits.isb-sib.ch/cgi-bin/motif_scan) and aligned using by ClustalW2 software (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Secondary structure of RNA were obtained using Mfold (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form). RTL1 modeling has been performed using Mouse dicer sequence structure (3c4b.1.A) (23), Swiss Model (http://swissmodel.expasy.org/) and the 3D representation was featured using the Genious software.
LC-MS/MS analysis
Purified His-RTL1 recombinant protein was digested in-solution using trypsin enzyme, digestion peptides were purified prior to nanoLC-MS/MS analysis on a nanoHPLC-Q Exactive Plus system (Thermo Fisher Scientific, Bremen, Germany) and MS/MS spectra were interpreted as described in detail in Supplementary methods. Spectra and fragmentation tables identifying glutathionylated peptides are provided in Supplementary Figure S6.
Circular dichroism
Synthetic gene coding RTLI protein was cloned into a pET28–6xHis-SUMO vector. The C230S mutation was introduced in the plasmid using site–directed PCR mutagenesis according to standard protocols. RTLI and RTLI C230S proteins were expressed in Escherichia coli BL21(DE3) Magic cells. Cells were induced with 0.4 M IPTG at OD600 = 0.8–0.9, grown overnight at 18°C, harvested by centrifugation and lysed by sonication in buffer containing 20 mM NaH2PO4 (pH 7.5), 250 mM NaCl, 20 mM imidazole, 5% glycerol and 5 mM β-mercaptoethanol (buffer A). The lysate was clarified by centrifugation at 40 000 rpm and loaded onto a 5 ml Ni-NTA (HiTrap, GE Healthcare) column equilibrated in buffer A. After washing with buffer A containing 40 mM imidazole, protein was eluted with buffer A containing 300 mM imidazole. The His-SUMO tag was removed by overnight incubation with 6xHis-SENP protease. Following ammonium sulfate precipitation, protein was dissolved in buffer A and re-applied onto Ni-NTA column. The flow-through that contained the protein of interest was concentrated on an Amicon Centrifugal Filter Device (Millipore). The protein was further purified on a Superdex 200 size exclusion column (GE Healthcare) in a buffer that contained 20 mM NaH2PO4 and 100 mM NaCl. Peak fractions were concentrated using 10 kDa cut-off centricon (Milipore) to 4.7 mg/ml. For glutathionylation, protein samples at 0.14 mM concentration were incubated at room temperature for 30 min with 20 mM GSSG in the presence of 1mM DTT. The unbound GSSH was removed by exchanging buffer to 20 mM NaH2PO4, 100mM NaCl using centricon (Milipore).
The circular dichroism (CD) spectra of each protein were recorded intriplicates in a Jasco J-750 spectropolarimeter using cell of path length 0.02 cm. The protein concentration was 1.15 mg/ml for both WT and MUT samples untreated with GSSG and 0.65 mg/ml for samples treated with GSSG. Spectra were corrected for the solvent CD signal. The secondary structure content was determined from the data in the far UV range (190–240 nm using) the CONTIN software package (24) available from DichroWeb server (25) with SP175 reference dataset (26).
Primers
All oligonucleotide sequences are provided in Supplementary Table S1.
RESULTS
RTL1 cleaves long dsRNA precursor sequences
We previously showed that ectopic expression of RTL1 prevents the production of thousands of siRNAs and of certain recently evolved miRNAs, but not of old conserved miRNAs, suggesting that RTL1 cleaves near-perfectly paired dsRNA (11). We also showed that a functional RNase III domain is required to observe this effect. However, RTL1 cleavage sites remained uncharacterized. To determine if RTL1 degrades dsRNA randomly or recognizes specific sequences, we took advantage of the At3g18145 3′UTR, which adopts a near-perfect dsRNA structure (Figure 1A). In WT plants, DCL1 processes this dsRNA into a 21-nt species referred to as miR3440b, whereas in 35S:RTL1 plants, constitutive expression of RTL1 results in the production of a unique 24-nt siRNA molecule in replacement of the 21-nt miR3440b (11). This is likely due to RTL1-mediated cleavage of the At3g18145 3′UTR into a shorter dsRNA for which DCL3 has a better affinity (14,17,18).
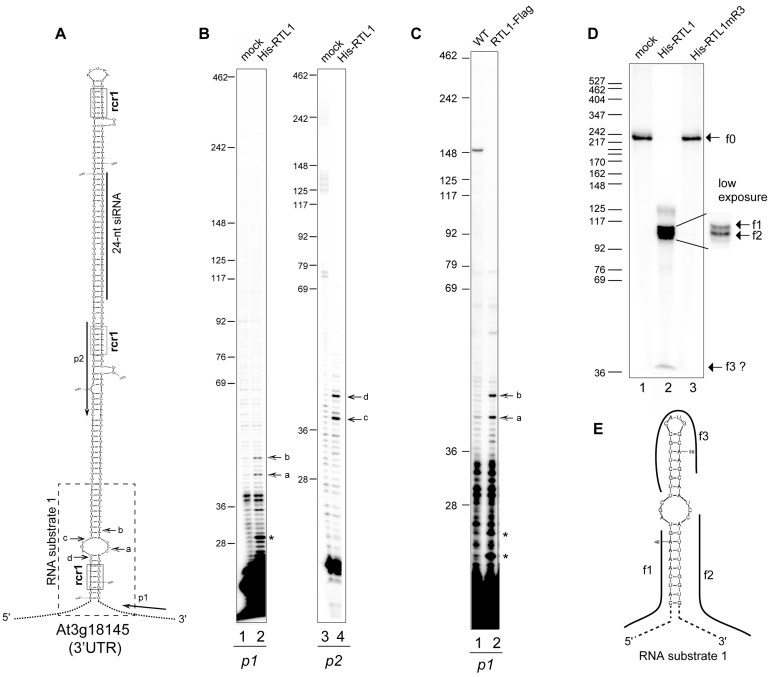
Figure 1.
RTL1 site-specifically cleaves a near-perfect duplex structure in vitro and in planta. (A) Left, the hairpin structure predicted from 3′UTR At3g18145 mRNA sequence is shown. The positions of p1 and p2 primers used in primer extension experiments are indicated. The dashed rectangle indicates the sequence corresponding to the RNA substrate-1 shown in (E). The sequence corresponding to a 24-nt small RNA is shown by a bar. Arrows show cleavage sites a–d mapped by primer extension. The RNA duplex consensus region rcr1 are boxed (B) Primer extension analysis using in vitro transcribed 3′UTR -At3g18145 and p1 or p2 primers. Arrows (a–d) show mapped cleavage sites in both strands. (C) Primer extension analysis on total RNA extracted from Col0 and overexpressing RTL1-Flag #1 plants with p1 primer. Arrows show the rcr1 cleavage positions. (D) Cleavage assay of 32P-CTP RNA substrate-1 using His-RTL1 or His-RTL1m3 proteins. Arrows indicate non cleaved RNA substrate-1 (f0) and cleavage fragment products (f1–f3). A low exposure insert is shown to better visualize the f1 and f2 fragments. DNA size markers are indicated on the left. (E) The predicted 32P-CTP RNA substrate-1 structure and f1–f3 products are represented.
Primer extension with primers p1 and p2 revealed that the At3g18145 3′UTR is cleaved at least at two positions by His-tagged RTL1 protein in vitro (Figure 1 and Supplementary Figure S1). Extension of p1 (Figure 1A) resulted in two major products of ∼43-nt (a) and ∼47-nt (b) showing cleavage at two positions (∼4-nt apart) in the base of the hairpin structure (Figure 1B). When using primer p2, extension resulted also in two major products of ∼38-nt (c) and ∼42-nt (d), suggesting cleavage in the complementary RNA strand. Note that cleavage sites c and d are 2-nt distant of cleavage sites b and a respectively (Figure 1B), indicating that RTL1 likely generates overhangs ends (Figure 1A). Analysis performed using 35S:RTL1-Flag plants with primer p1 confirmed cleavage at positions a and b in planta (Figure 1C). These signals were not detected in WT plants because RTL1 is not expressed in WT seedlings (3,11). Extension of p1 detected also products of ∼29-nt and 24-nt and/or ∼22-nt (shown by asterisk) in vitro and/or in planta. However, none of these signals were detected with primer p2 and therefore their significance remains undefined.
Primer extension results were confirmed using an in vitro transcribed RNA substrate-1, which contains the base of the hairpin structure of the At3g18145 3′UTR sequence (Figure 1A and E). When RNA substrate-1 is incubated with the His-RTL1 protein, two major fragments of ∼110-nt (f1) and ∼105-nt (f2) are generated, revealing endonucleolytic cleavage (Figure 1D). Fragments f1 and f2 correspond respectively to the 5′ and 3′ single stranded RNA fragments generated after cleavage at a/b and c/d positions (Figure 1A). The ∼240-nt fragment (f0) corresponds to the non-digested RNA substrate-1 while the ∼20-nt fragment (f3) might correspond to the top of the cleaved RNA substrate-1 (Figure 1D and Supplementary Figure S1a and c). The His-RTL1mR3 protein, which is mutated in two residues of the conserved RNase III motif (E89A/D96A), did not cleave RNA substrate-1 neither the At3g18145 3′UTR sequence after in vitro cleavage and RT-PCR analysis (Supplementary Figure S1a and (11)). To conclude, the results show that RTL1 behaves like an endoribonuclease and suggest that it cleaves dsRNA targets on both strands near a loop structure and generates overhang ends.
RTL1 cleaves rcr1 near to a conserved 5-nt RNA duplex sequence
In yeast RNase III recognizes substrates by interacting with the dsRNA structure of RNA hairpins that exhibit an AGNN tetraloop (27,28), while in E. coli it recognizes at least two specific sequences termed proximal and distal boxes in the dsRNA structure (1). The terminal stem loop or bulge structures in the At3g18145 3′UTR do not contain any obvious AGNN tetraloop sequence. However, the cleavage site signals (a–d) detected by primer extension are positioned near to a CAAAA::GUUUU RNA duplex that is strikingly similar to the proximal box of 4 nucleotides recognized by E. coli RNase III (1). This motif is found at two additional positions in the At3g18145 3′UTR hairpin structure (Figure 1A) and in 4431 out of the 6102 dsRNA precursors targeted by RTL1 identified previously (11) (Supplementary Figure S2 and Table S2). We named this motif rcr1 (for RTL1 consensus region1).
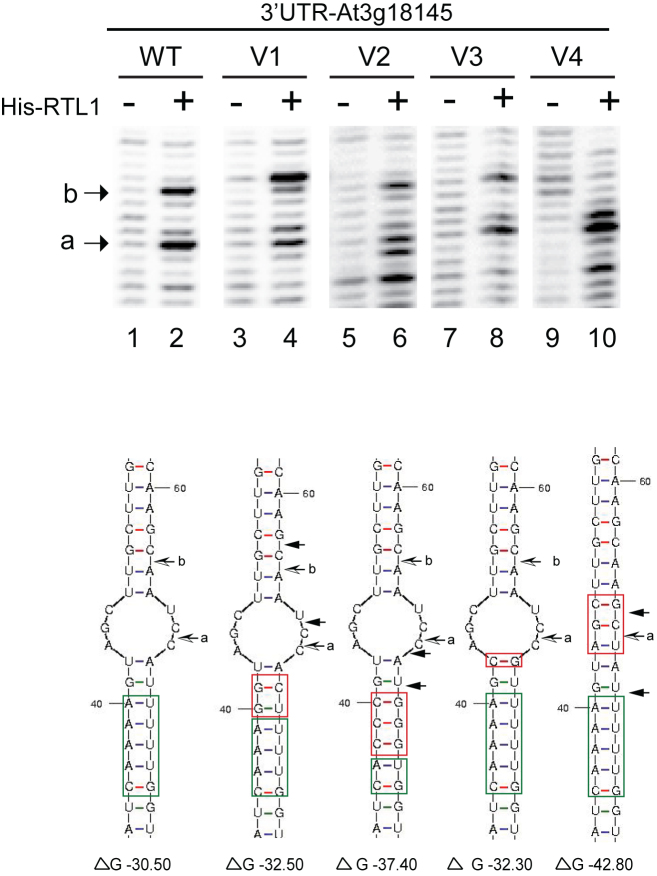
To attest the significance of the CAAAA::GUUUU RNA duplex and the bulge structure, we mutated the RNA substrate-1 in the stem and in the loop structure (Figure 2). The RNA substrate-1 V1, V2 and V3 are conservative structural mutations while the RNA substrate-1 V4 is a structural disruptive mutation. In vitro cleavage assay and primer extension analysis reveal that none of these mutations inhibits His-RTL1 activity in vitro. However, the position of cleavage is affected in RNA substrate-1 V1, V2 and V4. The cleavages occurs 1, 2 and/or 3 nt up or down stream from WT cleavage sites. Note that cleavage activity at site a is less affected compared with site b. The major effect is observed with the RNA substrate-1 containing one or three mutated A:U pairs in the CAAAA::GUUUU RNA duplex (V1 and V2) or with the RNA substrate-1 with deleted bulge structure (V4). Inversely, a mutation that does not affect RNA duplex or bulge structures (V3) does not affect the position of the cleavage site. Thus, the CAAAA::GUUUU RNA duplex and the bulge structure are required to direct site-specific cleavage of RNA by RTL1.
Figure 2.
RNA sequence and structure requirements for RTL1 cleavage. Primer extension analysis using in vitro transcribed 3′UTR RNA (WT and mutated V1-V4) incubated with His-RTL1 protein (+) or buffer alone (−). In the version V1 the A::U and the G::U base pairs within and next to the RNA duplex motif were mutated to G::U and G::C respectively; in V2, three of the A::U base within the RNA duplex motif were mutated to C::G; in V3 the U:: A base pair located in the loop structure was mutated to C::G and in the -V4, the UCG nucleotides in loop structure were mutated to GCU. The Wt and mutated sequences are green and red boxed respectively. Arrow shows cleavage site mapped with p1 primer. Note the two novel cleavage sites (black arrows) adjacent to the a and b sites (black and white arrows) in V1 and the novel cleavage sites adjacent to the a site in V2 and V4. The ΔG (kcal/mol) determined by RNA Mfold for each RNA substrate is indicated.
RTL1 dsRBD is essential for cleavage activity and substrate specificity
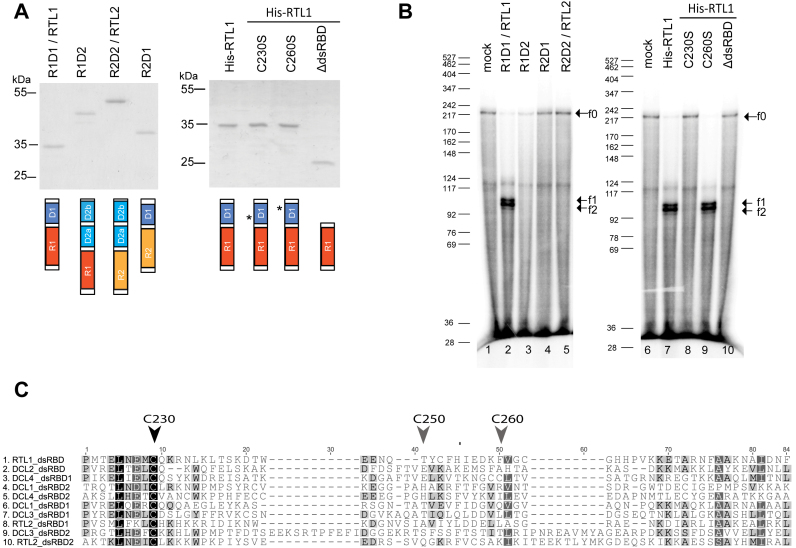
Both RNase III and dsRBD are required for substrate recognition and cleavage activity of E. coli and yeast RNase III enzymes (1,2,29,30). We have shown that mutations in the RNase III domain abolish RTL1 activity (Supplementary Figure S1 and (11)). However, if the RTL1-dsRBD is also required for cleavage activity has not been tested yet. To address this question, His-RTL1 protein deleted of the dsRBD (His-RTL1ΔdsRBD) was stably produced in E. coli (Figure 3A). In vitro cleavage assay show that His-RTL1ΔdsRBD protein does not cleave RNA substrate-1; only the f0 fragment is observed, the f1 and f2 fragments indicate of cleavage are observed only with His-RTL1 (Figure 3B).
Figure 3.
The native dsRBD and conserved cysteine C230 are required for RTL1 cleavage of RNA in vitro. (A) Coomassie blue staining and schematic representation of Wt His-RTL1 (R1D1), His-RTL2 (R2D2), swapped His-R1D2 and His-R2D1 or His-RTL1 with mutated cysteines C230S and C260S or truncated HisRTL1ΔdsRBD proteins. Red and yellow boxes correspond to RNase III domains while blue and light blue boxes correspond to dsRBD from RTL1 (R1 and D1) and RTL2 (R2 and D2a and b) proteins respectively. Asterisks show position of mutated Cys 230 and Cys260. (B) 32P-CTP RNA substrate-1 was incubated with 100 ng of His-RTL1(R1D1), His-R1D2, His-R2D1, His-RTL2 (R2D2), His-RTL1C230S, His-RTL1C260S or His-RTL1ΔdsRBD (lane 10) recombinant proteins or with buffer alone. Arrowhead indicates full-length RNA substrate (f0) and major cleavage fragment products (f1 and f2). DNA size markers are indicated on the left. (C) Amino acid sequence alignment of dsRBD from RTL1 (F4JK37), RTL2 (Q9LTQ0) and DCL1 (NP_171612), DCL2 (NP_566199), DCL3 (NP_189978) and DCL4 (NP_197532). Vertical arrows heads show highly conserved Cys230 and non-conserved Cys250 and Cys260 in the RTL1 sequence.
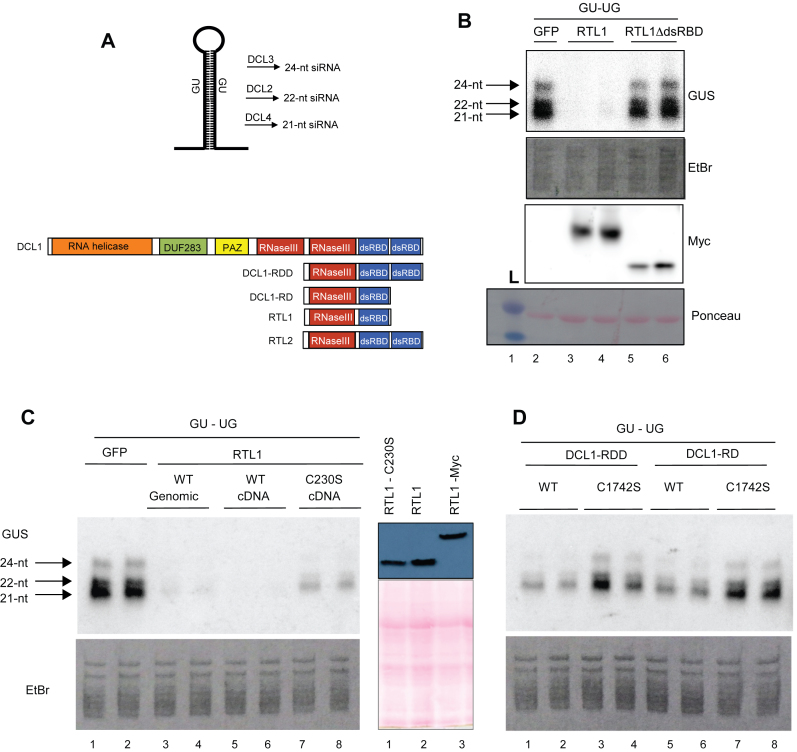
To verify that the absence of dsRBD is detrimental for RTL1 activity in planta, a 35S:GU-UG transgene expressing an inverted repeat hairpin RNA (Figure 4A), was introduced transiently into Nicotiana benthamiana leaves together with either a 35S:RTL1-Myc or 35S:RTL1ΔdsRBD-Myc construct or 35S:GFP as a control (Figure 4B). As previously reported (11), 35S:GU-UG + 35S:GFP infiltrated leaves accumulate high levels of 21- and 24-nt GU siRNAs, whereas 35S:GU-UG + 35S:RTL1 infiltrated leaves do not accumulate detectable levels of GU siRNAs (Figure 4B and C), indicating that RTL1 prevents the accumulation of transgene siRNAs deriving from an inverted repeat. In 35S:GU-UG + 35S:RTL1ΔdsRBD-Myc infiltrated leaves, the level of 21- and 24-nt GU siRNA is similar to that observed in 35S:GU-UG + 35S:GFP infiltrated leaves (Figure 4B). Note that western blot analysis reveals a slightly lower accumulation of the RTL1ΔdsRBD-Myc protein compared with RTL1-Myc (Figure 4B), but this is unlikely to explain the absence of effect of the 35S:RTL1ΔdsRBD-Myc construct on GU siRNA production. Rather, these results indicate that the dsRBD is essential for RTL1 activity in planta.
Figure 4.
The native dsRBD and its conserved cysteine C230 are required for RTL1 cleavage of RNA in planta. (A) Top, schematic representation of the GU-UG RNA and of the siRNAs produced from the processing of the dsRNA stem by DCL2, DCL3 and DCL4. The GU-UG siRNA construct contains promoter (p35) and terminator (t35S) sequences derived from 35S Cauliflower mosaic virus (CaMV) Bottom, schematic representation of native DCL1, RTL1 and RTL2 and of the truncated proteins DCL1-RD and DCL1-RDD mimicking RTL1 and RTL2, respectively. (B) Nicotiana benthamiana leaves co-infiltrated with a 35S:GU-UG construct (GU-UG) and either a control 35S-driven construct (GFP), a WT-tagged 35S:RTL1-Myc construct (RTL1) or a mutant 35S:RTL1ΔdsRBD-Myc construct lacking the dsRBD (RTL1ΔdsRBD). Low molecular weight RNAs were extracted and hybridized with a GU probe. Ethidium bromide-stained RNAs are shown as loading control. Proteins were extracted and hybridized with a Myc antibody. Ponceau staining (pink dye) is shown as protein loading control (lanes 2–6). L, indicates protein ladder. (C). Left, N. benthamiana leaves co-infiltrated with a 35S:GU-UG construct and either a control 35S-driven construct (GFP), a genomic-based or cDNA-based WT 35S:RTL1 construct (WT genomic and WT cDNA), 35S:RTL1 constructs mutated at cysteine 230 (C230S). Right, western blot analysis show comparable expression of 35S:RTL1 C230), 35S:RTL1 and 35S:RTL1-Myc constructs in N. benthamiana leaves. Proteins were extracted and hybridized with α RTL1 antibody. Ponceau staining (pink dye) is shown as protein loading control (lanes 1–3). (D) Nicotiana benthamiana leaves co-infiltrated with a 35S:GU-UG construct and either a control 35S-driven construct (GFP), a genomic-based or cDNA-based WT 35S:RTL1 construct (WT genomic and WT cDNA), 35S:RTL1 constructs mutated at cysteine 230 (C230S) and truncated 35S:DCL1 constructs (DCL1-RDD and DCL1-RD) carrying WT DCL1 sequence (WT) or a mutation at cysteine 1742 (C1742S). Note that RNA samples shown in (C) and d were run on a unique gel, so that the control 35S:GU-UG + 35S:GFP for (D) is visible on (C).
To further address the specificity of the RTL1 RNase III and dsRBD domains, swap experiments were performed between the RNase III and dsRBD domains of Arabidopsis RTL1 and RTL2 to generate the R1D2 construct carrying the RTL1-RNase III (R1) and RTL2-dsRBD (D2) domains, and the R2D1 construct, carrying the RTL2-RNase III (R2) and RTL1-dsRBD (D1) domains (Figure 3A and Supplementary Figure S3). We tested the activity of R1D2 and R2D1 proteins in vitro using RNA substrate-1 (Figure 3B). Whereas His-RTL1 fusion protein is able to cleave RNA substrate-1, visualized by the presence of the f1 and f2 fragments, neither His-R1D2 nor His-R2D1 proteins, which are stably produced in E. coli (Figure 3A), were able to cleave RNA substrate-1 (Figure 3B). Together, these results indicate that RTL1 cleavage activity and substrate specificity requires both RTL1 RNase III and intrinsic dsRBD domain organization.
A conserved Cysteine of RTL and DCL dsRBD is essential for cleavage activity
A cysteine of the RTL1-dsRBD (Cys230) is conserved among the dsRBD of RTL and DCL proteins (Figure 3C), but not in HYL1/dsRBD proteins, which carry a dsRBD but no RNase III domain (Supplementary Figure S4a), suggesting that Cys230 could be required for the RNase III activity of RTL and DCL enzymes. Cys230 is also conserved among all plant RTL1 proteins (Supplementary Figure S4b). The RTL1dsRBD contains two additional cysteines (Cys250 and Cys260), but they are not conserved among the dsRBD of RTL and DCL proteins (Figure 3C) and not among the other plant RTL1 proteins (Supplementary Figure S4b). To determine if the highly conserved Cys230 is required for RTL1 activity, the His-RTL1 was mutated to change cysteine 230 into a Serine. CD analysis revealed that this mutation does not alter the secondary structure of RTL1 (Table 1 and Supplementary Figure S10). As a control, the non-conserved Cysteine 260 was changed into Serine. The resulting His-RTL1 C230S and His-RTL1 C260S were stably produced in E. coli (Figure 3A). Whereas His-RTL1 C260S cleaved RNA substrate-1 as efficiently as His-RTL1, His-RTL1 C230S failed to cleave RNA substrate-1 (Figure 3B), indicating that the Cys230 of RTL1-dsRBD is required for RTL1 cleavage activity in vitro.
Table 1. CD spectra secondary structure analysis, of RTL1 and RTL1 C230S treated or not with 20 mM glutathione (GSSG).
| α-helices (%) | β-helices (%) | Turns (%) | NRMSD | |
|---|---|---|---|---|
| RTL1 | 44.4 | 3.5 | 14.4 | 0.20 |
| RTL1 C230S | 48.2 | 1.2 | 10.7 | 0.21 |
| RTL1 (20 mM GSSG) | 31.9 | 12.6 | 15.7 | 0.09 |
| RTL1 C230S (20 mM GSSG) | 48.6 | 2.2 | 14.6 | 0.19 |
NRMSD is normalized root mean square deviation.
To determine if Cys230 is also important for RTL1 activity in planta, a 35S:RTL1 construct derived from the RTL1 cDNA was mutagenized to change the Cysteine 230 into Serine. At first, co-transfection of 35S:GU-UG + 35S:RTL1 revealed that the 35S:RTL1 construct derived from the RTL1 cDNA is as efficient as the 35S:RTL1 construct derived from the RTL1 genomic DNA, which was previously used (Figure 4B). In contrast, the 35S:RTL1 C230S construct failed to prevent the accumulation of 21- and 24-nt GU siRNAs (Figure 4C). The level of 21- and 24-nt GU siRNAs in 35S:GU-UG + 35S:RTL1 C230S infiltrated leaves was lower than that observed in 35S:GU-UG + 35S:GFP infiltrated leaves, which could reflect either an instability of the RTL1 C230S protein or a residual activity of the RTL1 C230S protein. However, this is unlikely based on the CD experiments (Table 1 and Supplementary Figure S10). In addition, western blot analysis revealed that the RTL1 C230S protein accumulates at almost the level of the native RTL1 protein (Figure 4c). Therefore, it is more likely that the RTL1 C230S protein has reduced activity, indicating that Cys230 is required for efficient RTL1 activity in planta.
Since RTL1-Cys230 is conserved among the dsRBD of RTL and DCL proteins, we examined whether this cysteine also is essential for the cleavage activity of DCL proteins. At first, we examined whether truncated versions of DCL1 that structurally mimic RTL1 or RTL2 exhibit RTL1-like or RTL2-like activity (Figure 4D). For this purpose, we transformed N. benthamiana leaves with 35S:GU-UG + 35S:DCL1-RD, a construct carrying the second RNase III domain and the first dsRBD of DCL1, or 35S:DCL1-RDD, a construct carrying the second RNase III domain and the two dsRBD of DCL1 (Figure 4A). Both 35S:DCL1-RD and 35S:DCL1-RDD constructs reduced the accumulation of GU siRNAs, indicating that they exhibit RTL1-like activity (Figure 4D). However, the effect of 35S:DCL1-RD and 35S:DCL1-RDD was weaker than that observed with the 35S:RTL1 construct, suggesting that RTL1 has a higher affinity for dsRNA substrates than DCL proteins or a better RNase III activity than the truncated DCL1 proteins. Remarkably, changing DCL1-Cys1742 (the equivalent of RTL1-Cys230, Figure 3C) into Serine in 35S:DCL1-RD and 35S:DCL1-RDD constructs allowed 21- and 24-nt GU siRNAs to accumulate at the level observed in 35S:GU-UG + 35S:GFP infiltrated leaves (Figure 4C), indicating that DCL1-Cys1742 is as important as RTL1-Cys230 for cleavage activity.
RTL1 dimerizes essentially through its RNase III domain
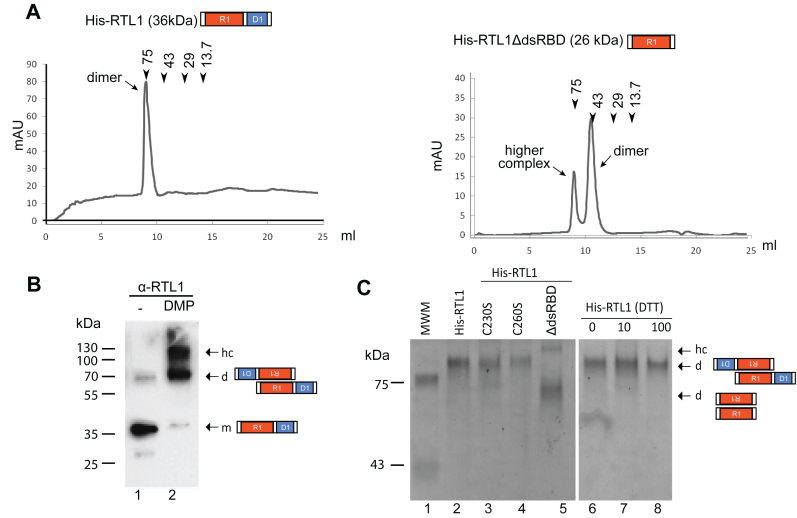
RNase III proteins act as homodimers to forms the RNase III catalytic center (31–33). To verify RTL1 homodimerization, His-RTL1 protein was fractionated on gel filtration (Figure 5). A single peak of estimated 75 kDa was detected, suggesting association of the two 36 kDa His-RTL1 monomers (Figure 5A). In agreement, His-RTL1 protein treated with the cross-linking agent dimethyl pimelimidate (DMP), used to stabilize protein-protein interactions, migrated on SDS-PAGE near the 70 kDa protein marker.
Figure 5.
RTL1 dimerizes through the N-terminal domain. (A) Superdex 75 Gel filtration chromatography of His-RTL1 and His-RTL1ΔdsRBD proteins in 150 mM NaCl buffer conditions. The peak positions of conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa) and ribonuclease A (13.7 kDa) are indicated by arrows (B) His-RTL1 protein untreated (lane 1) or treated (lane 2) with DMP was analyzed by western blot using α-RTL1 antibodies (C) Coomassie Blue staining of His-RTL1, His-RTL1 C230S or His-RTL1 C260S and His-RTL1ΔdsRBD migrated on native gel in absence of DTT or in the presence of increased amount of DTT. In B and C, arrows indicate positions of monomer (m), dimers (d) and higher order structures (hc) according to standard molecular weight markers.
The higher molecular weight protein complex (hc) detected in the DMP-treated protein fraction (Figure 5B) is likely due to artefactual aggregation induced by the DMP treatment because hc are not detected on gel filtration. Note also that while most of non DMP-treated His-RTL1 migrates near to the 35 kDa markers (the monomer), a fraction of the His-RTL1 migrates near to the 70 kDa marker (the homodimer) (Figure 5B).
Because cysteine residues can be easily oxidized to form dimers through disulfide bonds, we investigated the role of RTL1 cysteines in RTL1 dimerization. The His-RTL1 C230S and His-RTL1 C260S proteins were treated with DTT and then migrated in a non-denaturing-PAGE (Figure 5C). His-RTL1C230S and His-RTL1C260S migrated slower than the 75 kDa protein marker indicating that both mutated proteins are able to form homodimers at similar efficiency than is His-RTL1 (Figure 5C). RTL1 sequence contains three cysteines in the dsRBD and one in the RNase III (Cys195) domain (Supplementary Figure S5), which might form intra- or inter-molecular disulfide bonds. To assure that none of the cysteines in RTL1 are required for dimerization, we treated His-RTL1 with DTT to inhibit any disulfide bonds (Figure 5C). Analysis on non-denaturing-PAGE revealed that His-RTL1 homodimerizes even at 100 mM DTT, confirming that RTL1 does not require disulfide bonds to form homodimers. Furthermore, we demonstrates that RTL1 form also homodimers in planta, as observed by western blot of immunoprecipitated RTL1-Flag protein and gel filtration of 35S::RTL1-Flag plant extracts. This analysis revealed also that RTL1 might associates with other proteins to form high molecular weight protein complex (Supplementary Figure S6).
Bacterial RNase III dimerizes through its RNase III domain (34,35). To verify if this is also true for RTL1, the His-RTL1 lacking the dsRBD (His-RTL1ΔdsRBD), which is 26 kDa (Figure 3A), was fractionated on gel filtration. A major protein peaks between the 43 and 75 kDa markers was detected (Figure 5A), indicating that the RNase III domain of RTL1 is sufficient for homodimerization. Note a second protein peak eluting earlier than the 75 kDa marker, suggesting the occurrence of higher molecular weight organization (Figure 5A). Similarly, analysis of His-RTL1ΔdsRBD protein in non-denaturing PAGE (Figure 5C), revealed bands larger than 43 and 75 kDa, indicating also formation of His-RTL1ΔdsRBD dimers and other higher molecular complexes. Note that hc-RTL1 are detected only in presence of DMP or with truncated RTL1ΔdsRBD protein (Figure 5A and B), indicating that RTL1 exist mainly as a stable homodimer. Together, these results demonstrate that RTL1 dimerization requires the N-terminal sequence, containing the RNase III domain, but not the dsRBD neither disulfide bonds.
RTL1 activity is regulated by glutathionylation
Based on current models of RNase III::RNA complexes (34,35), it is unlikely that the conserved Cys230 is involved in the formation of a homodimer by an intermolecular disulfide bond (Figure 5). Accordingly, increasing concentrations of a strong oxidant H2O2 does not induce dimerization of the His-RTL1 protein (Supplementary Figure S7b). However, cysteine residues are prone to other post-transcriptional modifications like sulfenylation, glutathionylation, nitrosylation which can affect the structure and/or the activity of the protein (36,37). Glutathionylation occurs by formation of a mixed disulfide bond with the small molecular weight thiol containing glutathione.
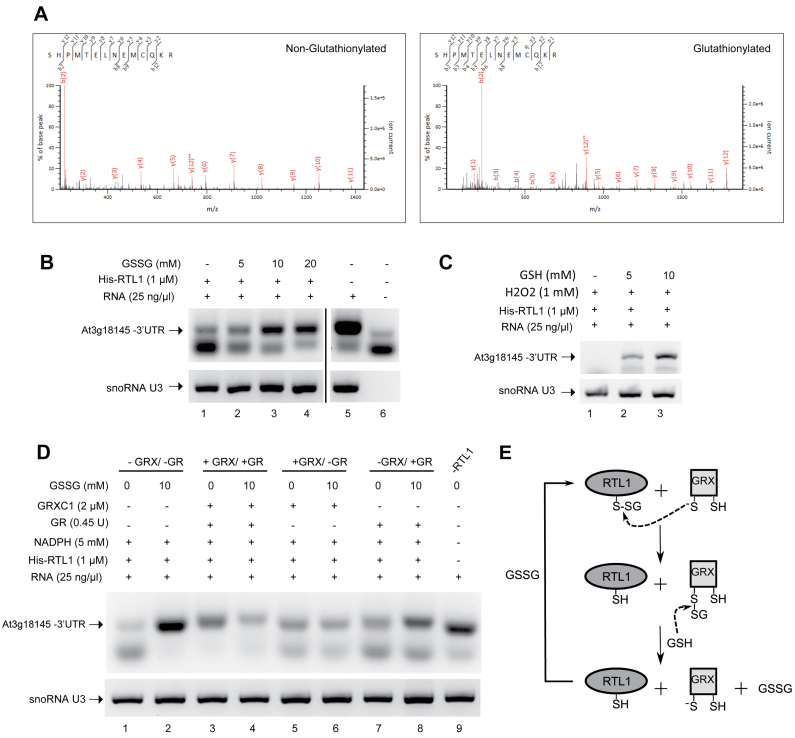
We investigated if RTL1 can be glutathionylated by treating His-RTL1 protein with oxidized glutathione (GSSG) and using mass spectrometry analyses. After trypsin digestion, peptides were analyzed by LC-MS/MS (Figure 6A). This reveals a glutathione adduct on two different Cys230-containing peptides (with and without a missed trypsin cleavage) detected, based on a 305-Da mass increase of the modified peptides when compared to the parent peptide. These experiments also attest that Cys230 can be glutathionylated in vitro. Two additional glutathione adducts were also identified at positions Cys195 and Cys250 in the RNase III and dsRBD domains respectively, which are non-conserved amino acids (Supplementary Figure S8).
Figure 6.
Glutathionylation of RTL1 affects RNase III cleavage activity. (A) His-RTL1 treated with 5 mM GSSG was trypsin digested and analyzed by nanoLC-MSMS. The panels show fragmentation spectra matching peptides with either unmodified (left) or with glutathionylated C230 (right). (B) Effect of reduced glutathione (GSSG) on cleavage of the 3′UTR sequence by His-RTL1. The protein samples were incubated at room temperature for 30 min with 0–20 mM GSSG before cleavage assay. (C) Effect of oxidized glutathione (GSH) on cleavage of the 3′UTR sequence by His-RTL1 in the presence of H2O2. The samples were incubated at room temperature for 30 min with 1 mM H2O2 and 0–10 mM GSH. (D) Reactivation of cleavage activity His-RTL1 by deglutathionylation with GRXC1. The samples were incubated at room temperature for 30 min with 10 mM GSSG or buffer only. The samples were further incubated at room temperature for 20 min with or without the GRX system (5 mM NADPH, 2 μM GRXC1, 0.45 units GR). Full system is absent in lanes 1–2 while present in lanes 3–4. GR was omitted in lanes 5–6 and no GRXC1 was added in lanes 7–8. Lane 9 is without RTL1 protein. (E) Schematic representation of the likely process of deglutathionylation of His-RTL1 by GRX based on a classical deglutathionylation model. The thiolate form of the GRX catalytic Cys attacks the disulfide of the glutathionylated Cys residue, releasing the reduced peptide, and becoming glutathionylated. A molecule of GSH reduces the glutathionylated thiol of GRX, releasing the thiolate catalytic Cys and generating a GSSG molecule. RT-PCR reactions were performed using total RNA from 14 days-old Arabidopsis thaliana plants and primers p1/p9 and U3fw/U3rev.
In order to determine the impact of glutathionylation on the RTL1 cleavage activity, we measured the His-RTL1 activity in presence of increasing concentrations of GSSG (Figure 6B) or in the presence of a H2O2+GSH mix which can trigger glutathionylation (Figure 6C). His-RTL1 activity was determined using total RNA and RT-PCR to detect cleavage of At3g18145 3′UTR. Both treatments progressively inhibited the RTL1 cleavage activity, observed by increased RT-PCR signal corresponding to non-cleaved At3g18145 3′UTR, indicating that glutathionylation negatively impacts RTL1 activity (Figure 6B and C; Supplementary Figure S9a). Glutathionylation is a reversible post-transcriptional modification which can be removed by non- enzymatic or enzymatic mechanisms. GRX are known to be major enzymes involved in deglutathionylation (38). In order to know if GRX is able to recover the enzymatic activity of glutathionylated RTL1, we incubated GSSG-inactivated RTL1 with recombinant cytosolic GRXC1 (Figure 6D) or GRXC2 (Supplementary Figure S10b) proteins. These GRX have been previously described to exhibit deglutathionylation activities using GR and glutathione (GSH) as electron donors (39). GRXC1 and GRXC2 are both able to recover the cleavage activity of the glutathionylated RTL1, indicating that both of them are active to deglutathionylate RTL1 (Figure 6D and E; Supplementary Figure S9b). Indeed, RT-PCR signals are much weaker in the GSSG samples reactions treated with GRX compared with samples non-treated with GRX. Note also that GR alone does not restore RTL1 cleavage activity as efficiently as GRX or GRX+GR. Altogether, these data show that RTL1 cleavage activity is redox regulated by glutathionylation/deglutathionylation of a specific Cys230 residue in the dsRBD.
Finally, to verify the impact of glutathionylation on the RTL1 structure, we performed circular dichroism analysis on His-RTL1 and His-RTL1 C230 proteins treated or not with GSSG (Table 1). This analysis show that the mutation C230S does not induce a major secondary structure changes of His-RTL1. Indeed, the percent of total α, β and Turn structures remains similar in His-RTL1 C230S (48.2, 1.2 and 10.7) compared to His-RTL1 (44.4, 3.5 and 14.4). In contrast, the percent of total α and β helix is strongly affected in His-RTL1 (31.9 and 12.6), but not in His-RTL1 C230S (48.6 and 2.2), treated with GSSG compared to untreated proteins (Figure 7A and Supplementary Figure S10). Altogether, this analysis revealed that glutathionylation of C230 induces a major secondary structure conformational change of RTL1.
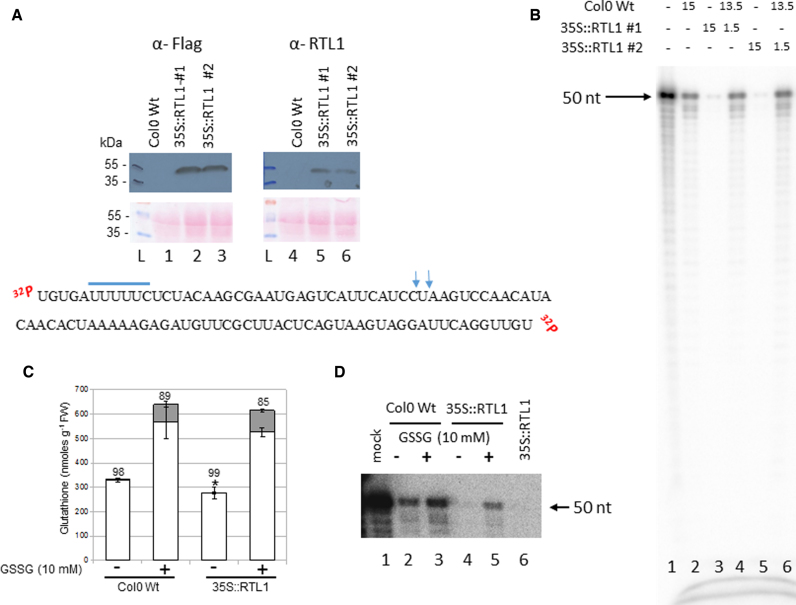
Figure 7.
Plant GSSG treatment affects RTL1 cleavage activity. (A) Top, proteins from Col0 WT and 35S::RTL1 (#1 and #2) plants were extracted and hybridized with α-Flag and α-RTL1 antibodies. Ponceau staining (pink dye) is shown as protein loading control (lanes 1–3 and 4–6). L, indicates protein ladder. Bottom, sequence of dsRNA substrate-2. The putative rcr motif is underlined and the vertical arrows show DCL3/DCL4 cleavage site. (B) [γ-32P]-ATP RNA substrate-2 was incubated with either WT (lanes 2), 35S:RTL1-Flag #1 (lane 3) or 35S:RTL1-Flag #2 (lane 5) protein extracts or with WT + 35S:RTL1-Flag #1 (lane 4) or WT + 35S:RTL1-Flag #2 (lane 6) protein extracts. Reactions were performed either with 15 μl, 1.5 μl or 15 μl + 1.5 μl. Lane 1 shows [γ-32P]-ATP RNA substrate-2 only. (C) Glutathione levels in 2-weeks old WT and 35S:RTL1-Flag #1 plantlets treated or not with 10 mM GSSG. Asterisks indicate a significant difference (P ≤ 0.01) between total glutathione levels of WT and 35S:RTL1 plants. Reduced and oxidized glutathione levels are indicated by white and gray bars, respectively. The percentage reduction state of glutathione is indicated above bars. Error bars represent SD (n = 4). (D) [γ-32P]-ATP RNA substrate-2 was incubated with WT and 35S:RTL1-Flag #1 protein extracts prepared from treated (lanes 3 and 5) or not (lanes 2 and 4) with 10 mM GSSG respectively. Lane 1, shows [γ-32P]-ATP RNA substrate-2 only and Lane 6, cleavage reaction using 35S:RTL1-Flag #1 protein extract not submitted to GSSG buffer and is used as a positive control.
Glutathionylation controls RTL1 activity in plants
RTL1 targets more than 6000 endogenous siRNA-producing loci. RTL1-mediated cleavage generally prevents the production of siRNAs, probably by providing RNA substrates for degradation by exonucleases in planta (11). Here we addressed the significance of the RTL1 in the degradation of siRNA-producing RNA precursors and the effect of glutathionylation in the RTL1 activity in planta.
For this study we designed a new RNA substrate based in the TAS1A sequence, which has been used to detect DLC3 and DCL4 activities in crude protein extracts from A. thaliana seedlings (14,40). In contrast to At3g18145 3′UTR-derived small RNA, TAS1A siRNAs do not accumulate in the 35S:RTL1 plants, suggesting that TAS1A dsRNA substrates are degraded following RTL1-mediated cleavage. We carried out RTL1 cleavage assay using 5′end labeled 50 nt TAS1A dsRNA (RNA substrate-2) and crude protein extracts prepared from WT and 35S:RTL1-Flag (#1 and #2) Arabidopsis seedlings (Figure 7). The TAS1A dsRNA is 50 nt long and contains rcr motif and a DCL3/4 cleavage site and the 35S:RTL1-Flag #1 and #2 plants accumulate the RTL1-Flag protein (Figure 7A and (11)).
First we determined the processing of RNA substrate-2 in WT and 35S:RTL1-Flag protein extracts (Figure 7B). When the RNA substrate-2 was incubated with 35S:RTL1-Flag extracts (15 μl), only trace amount of the 5′ end labelled 50 nt RNA substrate-2 were detected (lanes 3 and 5) compared with WT extracts (lane 2). In contrast, when RNA substrate-2 was incubated with 1/10 of 35S:RTL1-Flag extract (1.5 μl, lane 4 and 6), the amount of RNA substrate-2 was similar to that detected in WT extracts alone, indicating that cleavage of RNA substrate-2 by RTL1 is dose dependent. It is worth mentioning that we did not detect accumulation of small size RNA in any of these samples, indicating that this cleavage product is probably degraded or quickly processed in undetectable (unlabeled) small RNAs.
Next, to show that glutathionylation is detrimental for RTL1 activity in planta, we treated Col-0 WT and 35S:RTL1-Flag plants with 10 mM GSSG. As expected, the treatment led to a marked accumulation of both reduced and oxidized glutathione in plants and switched the glutathione redox state to a more oxidized status (Figure 7C). Then, we examined cleavage of RNA substrate-2 on total protein extracts (Figure 7D). In the 35S:RTL1-Flag plants, treatment with GSSG inhibited RTL1 activity, observed by an increased amount of RNA substrate-2 (lane 5), compared to RTL1 activity in untreated plants (lane 4). Interestingly, treatment with GSSG also affects accumulation of RNA substrate-2 in WT (lane 3) compared to untreated plants (lane 2). This indicates that glutathionylation inhibits extra RNase activities that cleaves RNA substrate-2, likely DCL3 and/or DCL4.
Altogether, these results show that RTL1 cleaves dsRNA and generate substrates for degradation and that glutathionylation control RTL1 activity in planta.
DISCUSSION
We previously demonstrated that RTL1 acts as an RNase III enzyme with siRNA suppressor activity. As a result, RTL1 overexpression strongly reduces levels of endogenous siRNA produced by DCL2-4 (11). Therefore, fine-tune control of RTL1 cleavage activity is a crucial step to regulate RNA accumulation through the small RNA and/or RNA degradation pathways.
Our results show that RTL1 cleaves dsRNA substrates at specific positions and generates fragments with 2-nt 3′ or 5′ overhangs (Figures 1 and 2; Supplementary Figures S1 and 2). Subsequently, the RTL1 products with 3′overhangs can be processed by Dicer to generate siRNAs. Accordingly, recent reports demonstrated that DLC3 selects shorter rather than longer dsRNA for cleavage (17,18). Dicer via its PAZ domain can recognize 3′ overhang ends generated by RTL1 cleavage and cuts every 21–24 nt in a ruler-like mechanism (33,41). However, overexpression of RTL1 reduces the amount of thousands of siRNAs (11), indicating that RTL1 products are degraded and subsequently not available for Dicer cleavage. Indeed, we demonstrated that RTL1 induces degradation of TAS1A RNA precursors in 35S:RTL1-Flag protein extracts (Figure 7).
RTL1 likely recognizes a consensus motif within hairpins. This is in contrast to Drosha and Rnt1p proteins, which use a terminal loop as an anchor to cut respectively 14–16 nt or 22 nt away in the stem of RNA substrates (13,27,28). It also contrasts to yeast Dcr1, which starts cleavage at the interior of dsRNA hairpin at arbitrary position to generate 23-nt siRNA products by positioning dimers to adjacent sites (42). Remarkably, the consensus motif we identified is identical to the proximal box required for RNA target recognition and cleavage efficiency by E. coli RNase III (43). Thus, the proximity of a bulge or a loop near to the consensus motif could be an additional determinant for substrate recognition by RTL1 as already observed for other RNase III (13,33,35,44). The fact that the rcr1 motif is not found in all siRNA precursors targeted in 35S:RTL1 plants suggests that RTL1 recognizes other RNA sequences-structures that remain to be identified.
The dsRBD of RTL1 is also essential for RNA cleavage activity (Figures 3 and 4). This is more similar to yeast Rnt1p than E. coli RNase III where the dsRBD seems not to be required for dsRNA cleavage activity (31,45). Remarkably, the single dsRBD of RTL1 is not commutable with the two dsRBD of RTL2, indicating that intrinsic RTL1 dsRBD is required for proper RTL1 activity. It is also notable that the dsRBD of RTL1 contains conserved residues (T154, Q157, E158 and Q161), which form a hydrogen bond with an O2′ hydroxyl of RNA and then required for RNase III cleavage in Aquifex aeolicus (35). These four residues are not conserved in the first dsRBD of RTL2 but are found in its second dsRBD (Supplementary Figure S4 and 5). The conformational repositioning of the dsRBD is required to convert the RNase III–RNA complex from a catalytically inactive to an active form (1,2). We can expect that lack of RNase III cleavage activity in R1D2 is likely due to changes in the organization of two dsRBD. This might disturb the position of essential residues and/or constraining the organization of dsRBD to form an active RTL1-RNA complex. Note that neither the R1D2 swap nor the reciprocal swap exhibit RTL2 activity, indicating that RTL2 also requires its own RNase III and dsRBD domains.
The Cys230 of the dsRBD of RTL1 and the equivalent Cys1742 of the first dsRBD of the DCL1 protein are required for cleavage activity (Figures 3 and 4). This cysteine is not conserved in the HYL1 and/or DRB proteins, which carry a dsRBD but lack an RNase III domain, suggesting that RTL1-Cys230 and DCL1-Cys1742 are not involved in RNA binding interaction but rather in the RNase III activity. Interestingly, this cysteine is conserved in two additional dsRBD proteins, DRB7.1 and DRB7.2 (46,47), which carry an RNase III domain exhibiting a non-canonical RNase III signature (Supplementary Figure S4c). Whether DRB7.1 and DRB7.2 exhibit RNase III activity and whether this activity requires the conserved cysteine remains to be determined.
RTL1 forms homodimers in vitro and in plant extracts (Figure 5 and Supplementary Figure S6). The predicted structural model (Figure 8) reveals that Cys230 from two RTL1 subunits are too far apart to participate in the homodimerization by forming a disulfide bridge. In fact, RTL1 forms dimers throughout hydrophobic interactions among RNase III domains (Supplementary Figure S7a), as others RNases III (31,32,48–50). In contrast, Cys230 is prone to glutathionylation and this modification inhibits RNase III cleavage activity (Figure 6; Supplementary Figures S8 and 9). This modification can be reversed by GRX. Therefore, rather than having a structural role, Cys230 has a regulatory function, switching on or off the catalytic activity of RTL1 in function of its redox state (Figure 8). Interestingly, the Cys230 is surrounded by the four residues that are required for RNase III cleavage in A. aeolicus (35). The glutathione molecule is a tripeptide (Gly-Cys-Glu) that could interfere with the RNA recognition process by creating a sterical bulk or by changing the electronic environment of the protein. CD analysis revealed that glutathionylation strongly affects RTL1 protein secondary structure and likely RNA binding and/or RNase cleavage activities of RTL1 (Table 1 and Supplementary Figure S10). This is in agreement with previous reports showing that glutathionylation might also induce major conformational changes of histone H3 (51), cyclophilin A (52), protein disulfide isomerase (53) and Hsp33 (54) proteins.
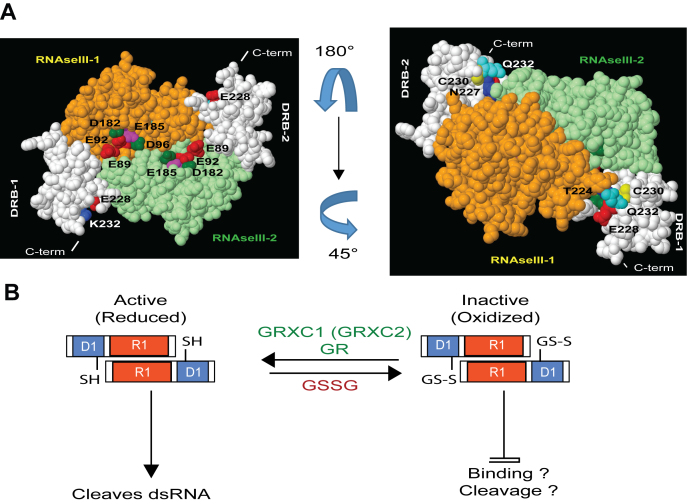
Figure 8.
In silico and functional analysis of RTL1 suggest a novel regulatory mechanism for RNase III activity in plants. (A) Modeled RTL1 (residues 50–284) homodimer based on mouse Dicer (3c4b.1.A) (23). The RNase III domain of two RTL1 molecules are shown in orange and green while both dsRBD are shown in white. In the left panel, the residues E89, E92, D96 and E185 (E37, E40, D44 and E110 in Aquifex aeolicus RNase III) located in the RNase III domain and required for RNA cleavage are shown. In the right panel, the RTL1 homodimer was rotated 180° and 45° to show conserved cysteines C230 in each dsRBD. The residues T224, N227, E228 and Q231 (T154, Q15, E158 and Q161 in A. aeolicus RNase III) located near to the C230 are indicated. These residues are required for RNA binding of Aa-RNase III. (B) In the proposed model, the cysteine C230, which is essential for cleavage activity, is kept in its reduced state (-SH). Glutathionylation of C230 (S-SG) in RTL1 sequence does not affect RTL1 dimerization but it might inhibit dsRNA binding and/or cleavage activity. RTL1 activity inhibition is reversible upon treatment with GRXC1 or GRXC2.
The regulatory role of a cysteine in RNase activity is not unique to RTL1 or RNase III enzymes. Indeed, cysteine residues participate in the formation of disulfide bonds for structural stability and/or activity of other RNases (55–58). Our data suggest that Cys230 glutathionylation/deglutathionylation plays a regulatory role, adjusting RTL1 activity depending on the redox environment. Glutathionylation-dependent redox regulation has been documented for several enzymes, including in plants (37,38). But to our knowledge, this is the first description showing that glutathionylation might control the activity of proteins implicated in RNA metabolism in vitro and in planta (Figures 6 and 7). RTL1 is up-regulated in response to virus infection (11) and it has been demonstrated that virus infection induces the accumulation of reactive oxygen speciess (ROS) (59). Generation of ROS and modification of the glutathione redox state are early and ubiquitous events occurring in response to biotic stress. Thus, oxidative conditions might favor simultaneously accumulation of RTL1 transcripts and repress activity of RTL1 protein. In this context, accumulation of RTL1 can be a response to compensate the lower RTL1 activity. Alternatively, glutathionylation of RTL1 might protect Cys230 from an irreversible oxidation by ROS. Importantly, Cys230 is conserved in dsRBD from all Arabidopsis RNase III enzymes, suggesting a conserved role in the structural bases for double stranded RNA cleavage by RTL and Dicer proteins in plants.
To conclude, our results demonstrate that the RNase III activity of RTL1 depends on specific RNA sequences structures, and explicit organization of RNase III and dsRBD domains and post translational modifications. All these features govern RTL1 cleavage activity of long hairpin structured RNA. Then, generated RNA substrates can be processed either into small RNA by Dicer or degraded by exonucleases/exosome. RTL1 can play a key role in the regulation of coding and non-coding RNA during growth and development and/or in response to stress conditions. Finally, because RTL1 overexpression drastically impairs also the siRNA population in plants, RTL1 protein expression and modifications must be tightly regulated. RTL1 gene expression is relatively low and essentially detected in roots (3,11). Therefore, it would be interesting to determine the developmental and environmental circumstances requiring de-repression of silencing by RTL1. These and other questions should then be addressed to better understand physiological and molecular impact of RNase III in RNA processing of RNA precursors of coding and non-coding RNAs in plants.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Boudvillain for useful discussion concerning RTL1 protein structure organization and M. Delseny for critical reading of the manuscript. We also thank, Mariusz Czarnocki—Cieciura and Krzysztof Skowronek for technical assistance with CD experiments.
Footnotes
Present addresses:
Cyril Charbonnel, UMR CNRS 6293, Clermont Université, INSERM U1103, Clermont-Ferrand, France.
Adnan K. Niazi, CABB, University of Agriculture Faisalabad (UAF), Faisalabad, Pakistan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CNRS; Agence Nationale de la Recherche [ANR-11-BSV6-007 to H.V., M.Z., J.S-V., ANR-12-BSV6-0011 to A.K.N., J.P.R.]; Proteomic French Infrastructure (ProFI) [ANR-10-INSB-08-03]. Funding for open access charge: CNRS; ANR [ANR-11-BSV6-007].
Conflict of interest statement. None declared.
REFERENCES
- 1. Nicholson A.W. Ribonuclease III mechanisms of double-stranded RNA cleavage. Wiley Interdiscip. Rev. RNA. 2014; 5:31–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Court D.L., Gan J., Liang Y.H., Shaw G.X., Tropea J.E., Costantino N., Waugh D.S., Ji X.. RNase III: genetics and function; structure and mechanism. Annu. Rev. Genet. 2013; 47:405–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Comella P., Pontvianne F., Lahmy S., Vignols F., Barbezier N., Debures A., Jobet E., Brugidou E., Echeverria M., Saez-Vasquez J.. Characterization of a ribonuclease III-like protein required for cleavage of the pre-rRNA in the 3′ETS in Arabidopsis. Nucleic Acids Res. 2008; 36:1163–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Margis R., Fusaro A.F., Smith N.A., Curtin S.J., Watson J.M., Finnegan E.J., Waterhouse P.M.. The evolution and diversification of Dicers in plants. FEBS Lett. 2006; 580:2442–2450. [DOI] [PubMed] [Google Scholar]
- 5. Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E.. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006; 38:721–725. [DOI] [PubMed] [Google Scholar]
- 6. Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C.. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004; 2:E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bouche N., Lauressergues D., Gasciolli V., Vaucheret H.. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006; 25:3347–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Law J.A., Jacobsen S.E.. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010; 11:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Olmedo G., Guzman P.. Processing precursors with RNase III in plants. Plant Sci. 2008; 175:741–746. [Google Scholar]
- 10. Elvira-Matelot E., Hachet M., Shamandi N., Comella P., Saez-Vasquez J., Zytnicki M., Vaucheret H.. Arabidopsis RNASE THREE LIKE2 Modulates the Expression of Protein-Coding Genes via 24-Nucleotide Small Interfering RNA-Directed DNA Methylation. Plant Cell. 2016; 28:406–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shamandi N., Zytnicki M., Charbonnel C., Elvira-Matelot E., Bochnakian A., Comella P., Mallory A.C., Lepere G., Saez-Vasquez J., Vaucheret H.. Plants encode a general siRNA suppressor that is induced and suppressed by viruses. PLoS Biol. 2015; 13:e1002326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Macrae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A.. Structural basis for double-stranded RNA processing by Dicer. Science. 2006; 311:195–198. [DOI] [PubMed] [Google Scholar]
- 13. Zeng Y., Yi R., Cullen B.R.. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005; 24:138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagano H., Fukudome A., Hiraguri A., Moriyama H., Fukuhara T.. Distinct substrate specificities of Arabidopsis DCL3 and DCL4. Nucleic Acids Res. 2014; 42:1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song L., Axtell M.J., Fedoroff N.V.. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Biol. 2010; 20:37–41. [DOI] [PubMed] [Google Scholar]
- 16. Zhu H., Zhou Y., Castillo-Gonzalez C., Lu A., Ge C., Zhao Y.T., Duan L., Li Z., Axtell M.J., Wang X.J. et al. . Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat. Struct. Mol. Biol. 2013; 20:1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blevins T., Podicheti R., Mishra V., Marasco M., Tang H., Pikaard C.S.. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. Elife. 2015; 4:e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhai J., Bischof S., Wang H., Feng S., Lee T.F., Teng C., Chen X., Park S.Y., Liu L., Gallego-Bartolome J. et al. . A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell. 2015; 163:445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pumplin N., Sarazin A., Jullien P.E., Bologna N.G., Oberlin S., Voinnet O.. DNA methylation influences the expression of DICER-LIKE4 isoforms, which encode proteins of alternative localization and function. Plant Cell. 2016; 28:2786–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saez-Vasquez J., Caparros-Ruiz D., Barneche F., Echeverria M.. A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 2004; 24:7284–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Durut N., Abou-Ellail M., Pontvianne F., Das S., Kojima H., Ukai S., de Bures A., Comella P., Nidelet S., Rialle S. et al. . A duplicated NUCLEOLIN gene with antagonistic activity is required for chromatin organization of silent 45S rDNA in Arabidopsis. Plant Cell. 2014; 26:1330–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. May M.J., Leaver C.J.. Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 1993; 103:621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Du Z., Lee J.K., Tjhen R., Stroud R.M., James T.L.. Structural and biochemical insights into the dicing mechanism of mouse Dicer: a conserved lysine is critical for dsRNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Provencher S.W., Glockner J.. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981; 20:33–37. [DOI] [PubMed] [Google Scholar]
- 25. Whitmore L., Wallace B.A.. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008; 89:392–400. [DOI] [PubMed] [Google Scholar]
- 26. Lees J.G., Miles A.J., Wien F., Wallace B.A.. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006; 22:1955–1962. [DOI] [PubMed] [Google Scholar]
- 27. Chanfreau G., Buckle M., Jacquier A.. Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaudin C., Ghazal G., Yoshizawa S., Elela S.A., Fourmy D.. Structure of an AAGU tetraloop and its contribution to substrate selection by yeast RNase III. J. Mol. Biol. 2006; 363:e322–e331. [DOI] [PubMed] [Google Scholar]
- 29. Henras A.K., Sam M., Hiley S.L., Wu H., Hughes T.R., Feigon J., Chanfreau G.F.. Biochemical and genomic analysis of substrate recognition by the double-stranded RNA binding domain of yeast RNase III. RNA. 2005; 11:1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leulliot N., Quevillon-Cheruel S., Graille M., van Tilbeurgh H., Leeper T.C., Godin K.S., Edwards T.E., Sigurdsson S.T., Rozenkrants N., Nagel R.J. et al. . A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III. EMBO J. 2004; 23:2468–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamontagne B., Tremblay A., Abou Elela S.. The N-terminal domain that distinguishes yeast from bacterial RNase III contains a dimerization signal required for efficient double-stranded RNA cleavage. Mol. Cell. Biol. 2000; 20:1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. March P.E., Gonzalez M.A.. Characterization of the biochemical properties of recombinant ribonuclease III. Nucleic Acids Res. 1990; 18:3293–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W.. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004; 118:57–68. [DOI] [PubMed] [Google Scholar]
- 34. Blaszczyk J., Gan J., Tropea J.E., Court D.L., Waugh D.S., Ji X.. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure. 2004; 12:457–466. [DOI] [PubMed] [Google Scholar]
- 35. Gan J., Tropea J.E., Austin B.P., Court D.L., Waugh D.S., Ji X.. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006; 124:355–366. [DOI] [PubMed] [Google Scholar]
- 36. Guikema B., Lu Q., Jourd’heuil D.. Chemical considerations and biological selectivity of protein nitrosation: implications for NO-mediated signal transduction. Antioxid. Redox Signal. 2005; 7:593–606. [DOI] [PubMed] [Google Scholar]
- 37. Rouhier N., Lemaire S.D., Jacquot J.P.. The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 2008; 59:143–166. [DOI] [PubMed] [Google Scholar]
- 38. Zaffagnini M., Bedhomme M., Marchand C.H., Morisse S., Trost P., Lemaire S.D.. Redox regulation in photosynthetic organisms: focus on glutathionylation. Antioxid. Redox Signal. 2012; 16:567–586. [DOI] [PubMed] [Google Scholar]
- 39. Riondet C., Desouris J.P., Montoya J.G., Chartier Y., Meyer Y., Reichheld J.P.. A dicotyledon-specific glutaredoxin GRXC1 family with dimer-dependent redox regulation is functionally redundant with GRXC2. Plant Cell Environ. 2012; 35:360–373. [DOI] [PubMed] [Google Scholar]
- 40. Seta A., Tabara M., Nishibori Y., Hiraguri A., Ohkama-Ohtsu N., Yokoyama T., Hara S., Yoshida K., Hisabori T., Fukudome A. et al. . Post-translational regulation of the dicing activities of Arabidopsis DICER-LIKE 3 and 4 by inorganic phosphate and the Redox state. Plant Cell Physiol. 2017; 58:485–495. [DOI] [PubMed] [Google Scholar]
- 41. Zhang H., Kolb F.A., Brondani V., Billy E., Filipowicz W.. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002; 21:5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weinberg D.E., Nakanishi K., Patel D.J., Bartel D.P.. The inside-out mechanism of Dicers from budding yeasts. Cell. 2011; 146:262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pertzev A.V., Nicholson A.W.. Characterization of RNA sequence determinants and antideterminants of processing reactivity for a minimal substrate of Escherichia coli ribonuclease III. Nucleic Acids Res. 2006; 34:3708–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liang Y.H., Lavoie M., Comeau M.A., Abou Elela S., Ji X.. Structure of a eukaryotic RNase III postcleavage complex reveals a double-ruler mechanism for substrate selection. Mol. Cell. 2014; 54:431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun W., Jun E., Nicholson A.W.. Intrinsic double-stranded-RNA processing activity of Escherichia coli ribonuclease III lacking the dsRNA-binding domain. Biochemistry. 2001; 40:14976–14984. [DOI] [PubMed] [Google Scholar]
- 46. Clavel M., Pelissier T., Montavon T., Tschopp M.A., Pouch-Pelissier M.N., Descombin J., Jean V., Dunoyer P., Bousquet-Antonelli C., Deragon J.M.. Evolutionary history of double-stranded RNA binding proteins in plants: identification of new cofactors involved in easiRNA biogenesis. Plant Mol. Biol. 2016; 91:131–147. [DOI] [PubMed] [Google Scholar]
- 47. Hiraguri A., Itoh R., Kondo N., Nomura Y., Aizawa D., Murai Y., Koiwa H., Seki M., Shinozaki K., Fukuhara T.. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 2005; 57:173–188. [DOI] [PubMed] [Google Scholar]
- 48. Blaszczyk J., Tropea J.E., Bubunenko M., Routzahn K.M., Waugh D.S., Court D.L., Ji X.. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001; 9:1225–1236. [DOI] [PubMed] [Google Scholar]
- 49. MacRae I.J., Zhou K., Doudna J.A.. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007; 14:934–940. [DOI] [PubMed] [Google Scholar]
- 50. Nagel R., Ares M. Jr. Substrate recognition by a eukaryotic RNase III: the double-stranded RNA-binding domain of Rnt1p selectively binds RNA containing a 5′-AGNN-3′ tetraloop. RNA. 2000; 6:1142–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garcia-Gimenez J.L., Olaso G., Hake S.B., Bonisch C., Wiedemann S.M., Markovic J., Dasi F., Gimeno A., Perez-Quilis C., Palacios O. et al. . Histone h3 glutathionylation in proliferating mammalian cells destabilizes nucleosomal structure. Antioxid. Redox Signal. 2013; 19:1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ghezzi P., Casagrande S., Massignan T., Basso M., Bellacchio E., Mollica L., Biasini E., Tonelli R., Eberini I., Gianazza E. et al. . Redox regulation of cyclophilin A by glutathionylation. Proteomics. 2006; 6:817–825. [DOI] [PubMed] [Google Scholar]
- 53. Townsend D.M., Manevich Y., He L., Xiong Y., Bowers R.R. Jr, Hutchens S., Tew K.D.. Nitrosative stress-induced s-glutathionylation of protein disulfide isomerase leads to activation of the unfolded protein response. Cancer Res. 2009; 69:7626–7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Raman B., Siva Kumar L.V., Ramakrishna T., Mohan Rao C.. Redox-regulated chaperone function and conformational changes of Escherichia coli Hsp33. FEBS Lett. 2001; 489:19–24. [DOI] [PubMed] [Google Scholar]
- 55. Deshpande R.A., Shankar V.. Ribonucleases from T2 family. Crit. Rev. Microbiol. 2002; 28:79–122. [DOI] [PubMed] [Google Scholar]
- 56. Kiritsi M.N., Fragoulis E.G., Sideris D.C.. Essential cysteine residues for human RNase kappa catalytic activity. FEBS J. 2012; 279:1318–1326. [DOI] [PubMed] [Google Scholar]
- 57. Ruoppolo M., Vinci F., Klink T.A., Raines R.T., Marino G.. Contribution of individual disulfide bonds to the oxidative folding of ribonuclease A. Biochemistry. 2000; 39:12033–12042. [DOI] [PubMed] [Google Scholar]
- 58. Thomson J.A., Shirley B.A., Grimsley G.R., Pace C.N.. Conformational stability and mechanism of folding of ribonuclease T1. J. Biol. Chem. 1989; 264:11614–11620. [PubMed] [Google Scholar]
- 59. Hammond-Kosack K.E., Jones J.D.. Resistance gene-dependent plant defense responses. Plant Cell. 1996; 8:1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.