Abstract
Human telomeres and promoter regions of genes fulfill a significant role in cellular aging and cancer. These regions comprise of guanine and cytosine-rich repeats, which under certain conditions can fold into G-quadruplex (G4) and i-motif structures, respectively. Herein, we use UV, circular dichroism and NMR spectroscopy to study several human telomeric sequences and demonstrate that G4/i-motif-duplex interconversion kinetics are slowed down dramatically by 2′-β-fluorination and the presence of G4/i-motif-duplex junctions. NMR-monitored kinetic experiments on 1:1 mixtures of native and modified C- and G-rich human telomeric sequences reveal that strand hybridization kinetics are controlled by G4 or i-motif unfolding. Furthermore, we provide NMR evidence for the formation of a hybrid complex containing G4 and i-motif structures proximal to a duplex DNA segment at neutral pH. While the presence of i-motif and G4 folds may be mutually exclusive in promoter genome sequences, our results suggest that they may co-exist transiently as intermediates in telomeric sequences.
INTRODUCTION
Human telomeric DNA is composed of tandem guanine-rich repeats (TTAGGG)n along with complementary cytosine-rich repeats (CCCTAA)n (1). The G-rich sequence forms G4 structures consisting of square-planar stacked guanine tetrads stabilized by Hoogsteen hydrogen bonds and mono- or divalent cations (Figure 1) (2–4). In the past two decades, G4s have gained considerable attention due to their significant stability under physiological conditions. In addition, their formation in cells has been implicated in several key biological processes (5,6). It has been suggested that G4 structures formed at telomeric ends of chromosomal DNA and stabilized by specific ligands might inhibit the activity of telomerase or lead to telomere dysfunction (7–11). Telomerase, a ribonucleoprotein, is the enzyme responsible for telomere maintenance by elongating the G-rich 3′-overhang (12). Recent studies have shown that telomerase can recognize and extend the unwound 3′-end of intermolecular parallel G4 structures and not the intramolecular antiparallel conformation (13). The occurrence of DNA and RNA G4 structures in human cells has been visualized utilizing structure-specific antibodies (14,15) and smart fluorescent probes (16). Interestingly, recent studies have shown that conserved Zika virus RNA sequences adopt stable, parallel-stranded G4 folds (17). Therefore, G4-binding molecules may be utilized to inhibit viral replication.
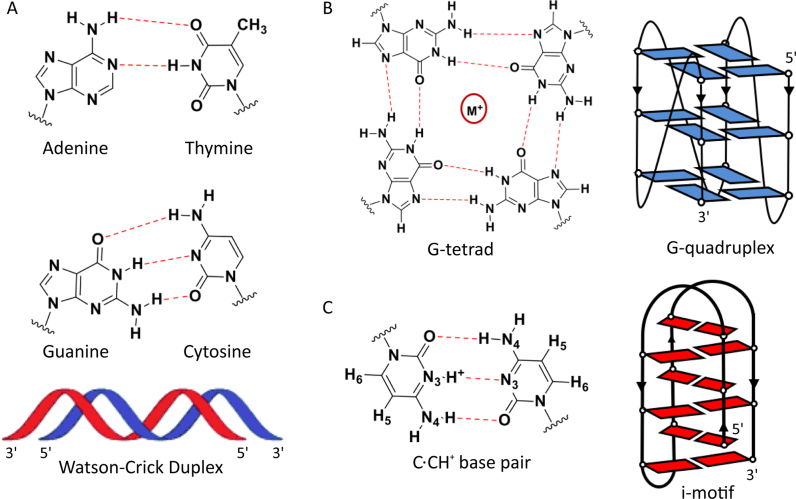
Figure 1.
(A) A:T and C:G Watson-Crick base pairs (top) and schematic representation of an antiparallel duplex (bottom). The G-rich strand is represented in blue and the C-rich strand in red. (B) G-quartet (left) and schematic representation of an intramolecular G-quadruplex (right). (C) C:CH+ base pair (left) and schematic representation of an intramolecular i-motif (right).
Under acidic conditions, the complementary C-rich strand folds into i-motif structures consisting of two parallel duplexes intercalated in an antiparallel orientation and held together by hemiprotonated cytosine-cytosine (C:CH+) base pairs (Figure 1) (18–20). Several factors have been found to stabilize these structures at physiological conditions such as molecular crowding, single-walled carbon nanotubes (21), silver cations (22), and negative supercoiling (23). We have shown that sugar modifications, in particular 2′F-araC substitutions (Figure 2), significantly stabilize telomeric and centromeric i-motif structures at neutral pH (24). To date, several i-motif structures had been reported in numerous oncogene promoter regions, such as c-MYC (25), BCL2 (26,27), VEGF (28) and RET (29), in addition to telomeric (30) and centromeric regions (31,32). Furthermore, two recent reports show that a number of genomic sequences fold into i-motif structures at neutral pHs (33,34). Even more recently, it has been demonstrated that i-motif structures in a DNA template can cause Klenow fragment DNA polymerase to stall immediately before the i-motif-forming region, suggesting that i-motifs could impede DNA replication or repair (35).
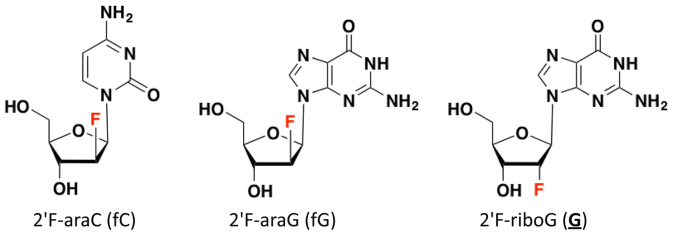
Figure 2.
Structures of 2′-deoxy-2′-fluoro-arabinocytidine (2′F-araC, left), 2′-deoxy-2′-fluoro-arabinoguanosine (2′F-araG, middle) and 2′-deoxy-2′-fluoro-guanosine (2′F-riboG, right).
The telomeric G-rich 3′-overhang is a substrate of telomerase; however, no extension products are observed for the C-rich telomeric repeat due to the lack of complementarity between the C-rich strand and the RNA template of telomerase (36). In blunt-ended telomeric duplexes where the two strands are fully hybridized, the G-rich strand is no longer a substrate of the enzyme (37,38). Therefore, it is important to determine whether the transient formation of i-motif structures in the C-rich strand would free up the G-rich strand for elongation by, or inhibition of telomerase. For this reason, G4/i-motif-duplex interconversion is a process of significant importance (39–43). The kinetics of G4/i-motif unfolding and of duplex formation at pH 5.0 were first studied by Mergny and Phan in 2002 (44). Several other reports regarding G4/i-motif-duplex competition have been conducted, confirming that the competitive equilibria between G4, i-motif, and Watson-Crick (WC) duplexes are primarily driven by the nature of the cation, the pH, and the sequence (45–48).
In this study, we aim to stabilize parallel intramolecular G4 structures and tune the G4/i-motif-duplex interconversion rate at neutral pH by stabilizing i-motif and G4 structures simultaneously. For this purpose, C- and G-rich sequences modified with 2′F-araC, 2′F-araG, and 2′F-riboG residues were synthesized (Table 1 and Figure 2). 2′-Deoxy-2′-F-arabinonucleic acids (2′F-ANA) modifications can modulate G4 stability and topology by favoring the parallel propeller G4 topology over competing conformers, while 2′F-ribose modifications (2′F-RNA) disrupt the conformation of the furanose ring leading to less stable quadruplexes (49–51). We show that 2′F-araG and 2′F-araC modified oligonucleotides provide very stable G4 and i-motif structures in potassium buffer at neutral pH, and that these structures pose a large kinetic barrier for duplex formation.
Table 1. Apparent Tm values (°C) of modified C-and G-rich sequences.
| Code | Sequence (5′-3′) | T m | ΔTm |
|---|---|---|---|
| 22G0 | A GGG TTA GGG TTA GGG TTA GGG | 65.6 ± 0.1 | — |
| 22G1 | A (fG)GG TTA (fG)GG TTA (fG)(fG)G TTA (fG)GG | 74.2 ± 0.9 | +8.6 |
| 22G2 | A (fG)GG TTA (fG)(fG)G TTA (fG)GG TTA (fG)GG | 73.8 ± 0.2 | +8.2 |
| 22G3 | A (fG)GG TTA (fG)(fG)G TTA (fG)(fG)G TTA (fG)GG | 80.2 ± 0.5 | +14.6 |
| 22G4 | A GGG TTA GGG TTA GGG TTA GGG | 53.4 ± 0.2 | –12.2 |
| 22G6 | A GGG TTA GGG TTA GGG TTA GGG | 57.6 ± 0.2 | –8.0 |
| 22G7 | A GGG TTA GGG TTA GGG TTA GGG | 59.3 ± 0.2 | –6.3 |
| 22C0 | CCC TAA CCC TAA CCC TAA CCC T | n.d. | — |
| 22C1 | C(fC)(fC) TAA C(fC)(fC) TAA C(fC)(fC) TAA C(fC)(fC) T | 25.7 ± 0.1 | — |
| 22C2 | ((fC)(fC)(fC) TAA)3 (fC)(fC)(fC) T | 30.0 ± 0.3 | — |
| 35G0 | C ACA GAT GCG TTT A GGG TTA GGG TTA GGG TTA GGG | 56.0± 0.2 | — |
| 35G3 | C ACA GAT GCG TTT A (fG)GG TTA (fG)2G TTA (fG)2 G TTA (fG)GG | 75.0± 0.1 | +19.0 |
| 35C0 | CCC TAA CCC TAA CCC TAA CCC T TTT CGC ATC TGT G | n.d. | — |
| 35C2 | (fC)3 TAA (fC)3 TAA (fC)3 TAA (fC)3 T TTT CGC ATC TGT G | 31.3± 0.2 | — |
All melting curves were recorded in 20 mM KPi and 70 mM KCl, pH 7.0. 22G-rich sequences were recorded at 295 nm, at a ramp of 0.2 °C/min, and 10 μM strand concentration. 35G-rich sequences were recorded at 295 nm, 0.5 °C/min, and 4 μM strand concentration. 22C- and 35C-rich sequences were recorded at 265 nm, at a ramp of 0.5 °C/min, and 4 μM strand concentration. (fG): 2′F-araG, G: 2′F-riboG, (fC): 2′F-araC, and n.d. not detected.
MATERIALS AND METHODS
Oligonucleotide synthesis and purification
Oligonucleotide synthesis was performed on an ABI 3400 DNA synthesizer (Applied Biosystems) at 1 μmol scale on Unylinker (Chemgenes) CPG solid support. Thymidine (dT), deoxycytidine (N-acetyl) (dC), and deoxyadenosine (N-Bz) (dA) phosphoramidites were used at 0.1 M concentration in acetonitrile and coupled for 110 s and deoxyguanosine (N-ibu) (dG) was coupled for 300 s. 0.15 M concentration was used for 2′F-araC which was coupled for 600 s. 2′F-araG and 2′F-riboG were used at 0.15 M concentration and coupled for 900 s. After completion of synthesis, the CPG was transferred to a 1.5 ml screw-cap eppendorf. Cleavage of the oligonucleotide from the CPG and removal of the nucleobase protecting groups were achieved by adding 1000 μl aqueous ammonium hydroxide for 16 h at 55 °C. The deprotection solution was centrifuged and decanted from the CPG. Samples were vented for 30 min, chilled in dry ice and evaporated to dryness. Crude oligonucleotides were purified by anion exchange HPLC on an Agilent 1200 Series instrument using a Protein-Pak DEAE 5PW column (7.5 × 75 mm) at a flow rate of 1 ml/min using a gradient of 0–24% lithium perchlorate buffer (1.0 M) over 30 min at 60 °C. Under these conditions, the desired peaks eluted between 22 and 24 min. Samples were desalted on NAP-25 desalting columns according to manufacturer protocol. The extinction coefficients of 2′F-araC, 2′F-araG and 2′F-riboG were assumed to match those of the unmodified deoxynucleosides. Masses were verified by high resolution LC–MS.
Thermal melting experiments
UV thermal denaturation data were obtained on a Varian CARY 100 UV-visible spectrophotometer equipped with a Peltier temperature controller. The concentration of oligonucleotides used was 10 μM for the G-rich single strands, 4 μM for the C-rich sequences. The total concentration for the duplexes was 2 μM. Samples were dissolved in appropriate buffer as indicated in the text. Concentrations were determined after quantitating the samples by UV absorbance at λ = 260 nm. Samples were heated to 90 °C for 10 minutes, then cooled slowly to room temperature, and stored at 5 °C for at least 18 h before the measurements were performed. Denaturation curves were acquired at 295 nm and 0.2 °C/min for G4s, 265 nm for i-motifs and at 260 nm for the duplexes at a rate of 0.5 °C/min. In order to avoid water condensation on the cuvettes at low temperatures, samples were kept under a stream of dry nitrogen at temperatures below 12 °C. The dissociation temperatures were calculated as the midpoint of the transition (Tm) values using the first derivatives of the experimental data.
Circular dichroism experiments
Circular dichroism (CD) studies were performed on a JASCO J-810 spectropolarimeter using a 1 mm path length cuvette. Temperature was maintained using the Peltier unit within the instrument. Spectra were recorded from 350 to 230 nm at a scan rate of 100 nm/min and a response time of 2.0 s with three acquisitions recorded for each spectrum. The spectra were normalized by subtraction of the background scan with buffer. Data were smoothed using the means-movement function within the JASCO graphing software. Oligonucleotide solutions for CD measurements were prepared with 20 mM potassium phosphate and 70 mM KCl buffer (pH 5.8, 6.2, 6.6, 7.0 and 7.4) in a similar manner to that used for UV melting. The concentration of oligonucleotides used was 10 μM for the G-rich sequences at 25 °C, 20 μM for the C-rich sequences at 5 °C, and 15 μM for the duplexes at 5 °C. CD melting temperature experiments were performed on an Applied Photophysics CD spectrophotometer using a 1 mm path length cuvette. Spectra were recorded from 350 to 230 nm at a heating rate of 0.5 °C/min. Oligonucleotide solutions for CD melting experiments were either 40 or 100 μM in 20 mM KPi and 70 mM KCl buffer pH 7.0.
NMR experiments
Samples for NMR experiments were dissolved in 9:1 H2O/D2O in 20 mM potassium phosphate and 70 mM KCl buffer, pH 7.0. The sample concentration for 1D NMR kinetic experiments was 0.1 mM. The samples were annealed before running the NMR experiments as indicated in the main text. Spectra were acquired on Bruker Avance spectrometers operating at 600 MHz equipped with a cryoprobe. Two-dimensional spectra were recorded at 0.5 mM strand concentration in the same buffer. NOESY spectra were acquired with mixing times of 50 and 100 ms. TOCSY spectra were recorded with the standard MLEV-17 spin-lock sequence and a mixing time of 80 ms. Water suppression was achieved by including a WATERGATE module in the pulse sequence prior to acquisition.
Gel electrophoresis experiments
Native gel electrophoresis was performed utilizing 24% polyacrylamide and TAE (Tris, Acetic acid, EDTA) buffer, pH 7.0. Running buffer was 1× TAE. The gels were run at 280 V and 10 °C for 4 h and 30 min. Annealed samples were heated to 90 °C for 10 min and then slowly cooled down to room temperature. For the pre-folded-mixed samples, 22G- and 22C-rich single strands were annealed separately and then mixed in equimolar amounts without further annealing. Gels were stained using SYBR Gold and visualized using Bio-Rad Gel Doc XR and controlled with the Image Lab software package.
RESULTS AND DISCUSSION
Structural properties of 22mer cytosine and Guanine-Rich telomeric single strands
G4, i-motif, and WC duplexes can be distinguished via nuclear magnetic resonance (NMR) since they possess distinct imino proton chemical shifts that originate from different base pairing configurations (52). Hence, the imino protons of base pairs stabilizing i-motifs (C:CH+), duplex (A:T and C:G), and G-tetrads appear at 15–16 ppm, 12–14 ppm, and 10–12 ppm, respectively. The relative stability of these structures is readily assessed via UV melting temperature experiments. Melting of duplex, i-motif, and G4 structures were monitored at 260, 265, and 295 nm, respectively (53,54). Like NMR, circular dichroism (CD) provides a convenient means to distinguish between these structures since their spectra exhibit characteristic bands (55–57). Therefore, NMR, UV melting, and CD experiments served as complementary techniques to assess the stability and determine the kinetics of G4/i-motif-duplex interconversion (Figure 3).
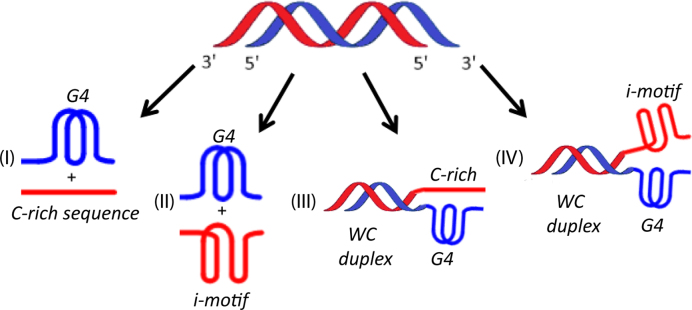
Figure 3.
Models representing G4/i-motif-duplex interconversion of (I) 22mer human telomeric repeat forming G4, (II) 22mer human telomeric repeat forming G4 and i-motif secondary structures due to 2′F-ANA modifications, (III) 35mer human telomeric repeat forming a duplex and G4 and (IV) 35mer human telomeric repeat forming a duplex, a G4, and an i-motif within the same complex.
1H-NMR and CD experiments performed on the single stranded G-rich sequences confirm the formation of G4 structures. In all cases, buffers contained 20 mM KPi and 70 mM KCl at pH 7.0 (unless explicitly noted otherwise). 1H-NMR spectra of 2′F-araG modified strands show signals in the 10–12 ppm range, which are characteristic of G4 formation (Supplementary Figure S1). These imino signals are still observed at 45 °C indicating the formation of thermally stable G4 conformations. The CD spectrum of 22G0 shows two maxima suggesting the presence of two conformers: an antiparallel conformation with a positive band at 295 nm and a parallel conformer with a positive band at 260 nm (Supplementary Figure S2A). The incorporation of 2′F-araG and 2′F-riboG modifications triggers a shift in the spectra leading to a major positive signal at 260 nm and a negative signal at 240 nm, characteristic of a single parallel G-quadruplex (Supplementary Figure S2). Among the 2′F-araG modified sequences, the CD spectrum of 22G2 reveals a small shoulder at 295 nm suggesting the presence of a minor species folded in an antiparallel fashion. All 2′F-araG modified strands are more stable compared to the control (22G0), as indicated by the increase in melting temperature, with the highest Tm observed for 22G3 (ΔTm = +14.6 °C) (Table 1 and Supplementary Figure S3). 2′F-riboG modified G-rich strands are less stable compared to the unmodified sequence (Table 1).
UV-Melting experiments performed at different oligonucleotide concentrations (2–100 μM) had little effect on the Tm, thus confirming the formation of monomeric G4 structures in the case of 22G1 and 22G3 (Supplementary Table S1). The presence of multiple conformations in the case of 22G2 and the decreased stability of the 2′F-riboG modified G-rich strands led us to exclude these sequences from further interconversion studies.
At neutral conditions, the unmodified single-stranded C-rich sequence does not form stable i-motif structures even at high concentrations (100 μM, Supplementary Table S1). However, the presence of 2′F-araC modifications provokes significant i-motif stabilization (Supplementary Figure S4), in agreement with our recent report (24). The most stable construction, 22C2 (Tm = 30.0 °C), was utilized in the G4/i-motif-duplex interconversion studies. 22C2 adopts an intramolecular folding topology since the Tm obtained from CD melting temperature experiments at 2, 40 and 100 μM was ∼30.0 °C (Supplementary Table S1).
Structural properties of cytosine and guanine-rich telomeric duplexes (annealing versus mixing)
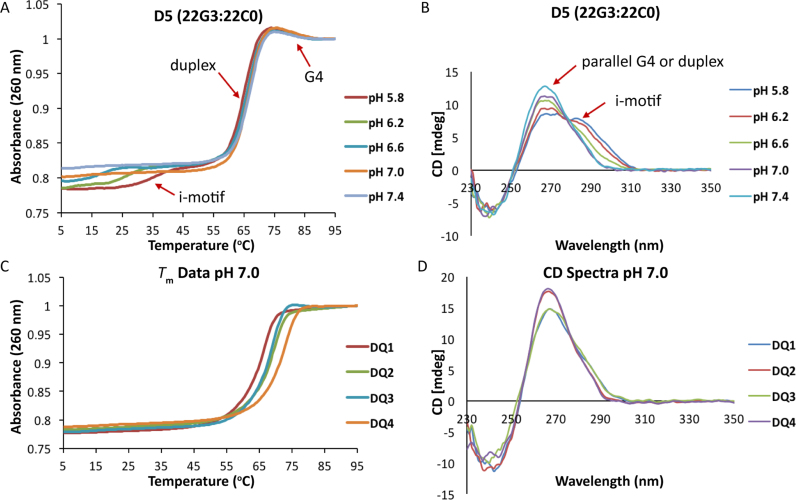
UV-melting temperature and circular dichroism experiments were performed at pH 5.8, 6.2, 6.6, 7.0 and 7.4 (and monitored at 260 nm) after annealing equimolar quantities of the 22C- and 22G-rich strands (i.e. a 1:1 mixture of the complementary strands was heated to 90 °C for 10 min and then slowly cooled down to room temperature, mix-heat-cool). Most of the melting curves display more than one transition (Table 2 and Supplementary Table S2). Looking closely at the melting curves (Figure 4A and Supplementary Figures S5 and S6), two hyperchromic and one hypochromic transitions are observed: a major, pH-independent melting transition corresponding to duplex unfolding and two minor transitions associated with the unfolding of i-motif (lowest pH-dependent Tm values) and G4 structures (highest pH-independent Tm values). To confirm that the minor transitions correspond to the formation/melting of i-motif and G4 structures, UV-melting experiments for the constituent isolated 22G- and 22C-rich strands were performed at different pH values (Supplementary Table S3). The Tm values observed for 22C0 and 22C2 decrease as the pH of the sample increases from 5.8 to 7.4 and were very similar to the values obtained for the first transition in the duplex melting experiments (Supplementary Tables S2 and S3). Similarly, the Tm values observed for the G-rich sequences are comparable to those obtained for the hypochromic transition, confirming that this transition corresponds to the melting of G4 structures. CD experiments as a function of pH were performed for all the samples containing 2′F-ANA modified C- and G-rich sequences (Figure 4B and Supplementary Figure S7). The samples containing 22C0 hybridized to 22G0 or to 2′F-ANA modified G4s, all exhibit a clear positive band at 285 nm at pH 5.8 and 6.2. This band decreases in magnitude as the pH increases to 7.0 and 7.4, reflecting the progressive unfolding of the i-motif structure with increasing pH. However, in the case of 22C2, this band was found in the CD spectra up to pH 7.0. Moreover, in all of the studied samples, a positive band appears between 270–260 nm with a negative band ∼240 nm. To unequivocally determine which species were forming under these conditions, 1D NMR spectra were recorded at neutral pH. The spectra obtained after mixing and annealing equimolar quantities of the 22G-rich and 22C-rich strands exhibit imino signals in the 12–14 ppm region, indicating that the formation of WC duplex structures is favored under slow annealing conditions and at higher concentrations, which is expected since duplex formation is concentration-dependent (Supplementary Figure S8). The well-known ability of 2′F-ANA modifications to stabilize duplex structures can be noticed in the presence of clear imino signals for the modified duplexes at higher temperatures (58). This is also consistent with the higher Tm values of the main melting transition in the modified duplexes (Table 2 and Supplementary Figure S8).
Table 2. Apparent Tm values (°C) of telomeric duplexes.
| Code | Duplex | T m pH 7.0 | Code | Duplex | T m pH 7.0 |
|---|---|---|---|---|---|
| D1 | 22G0:22C0 | 10.8± 0.2 | D7 | 22G4:22C0 | 66.0± 0.1 |
| 64.7± 0.2 | |||||
| D2 | 22G0:22C2 | 26.5± 1.5 | D8 | 22G4:22C2 | 70.8± 0.2 |
| 69.2± 0.3 | |||||
| D3 | 22G1:22C0 | 11.1 | DQ1 | 35G0:35C0 | 66.2 ± 0.2 |
| 67.0± 0.3 | |||||
| 78.1± 0.4 | |||||
| D4 | 22G1:22C2 | 29.2 | DQ2 | 35G0:35C2 | 69.5± 0.1 |
| 72.1± 0.4 | |||||
| 83.5 | |||||
| D5 | 22G3:22C0 | 11.3± 0.6 | DQ3 | 35G3:35C0 | 69.1± 0.3 |
| 66.3± 0.1 | |||||
| 80.8± 0.1 | |||||
| D6 | 22G3:22C2 | 26.4± 1.3 | DQ4 | 35G3:35C2 | 72.9± 0.2 |
| 71.9± 0.2 | |||||
| 83.8± 0.3 |
Duplex concentration is 2 μM. Tm were recorded at 260 nm and 0.5 °C / min in 20 mM KPi and 70 mM KCl, pH 7.0.
Figure 4.
(A) UV-melting curves of 22mer sample D5 (2 μM) recorded at 260 nm and 0.5 °C / min. (B) CD spectra of D5 (15 μM) recorded at 5 °C. (C) UV-melting curves of DQ (2 μM) recorded at 260 nm and 0.5 °C / min, pH 7.0. (D) CD spectra of DQ (15 μM) recorded at 5 °C, pH 7.0. Buffer conditions: 20 mM KPi and 70 mM KCl.
NMR kinetic experiments of 22mer duplexes
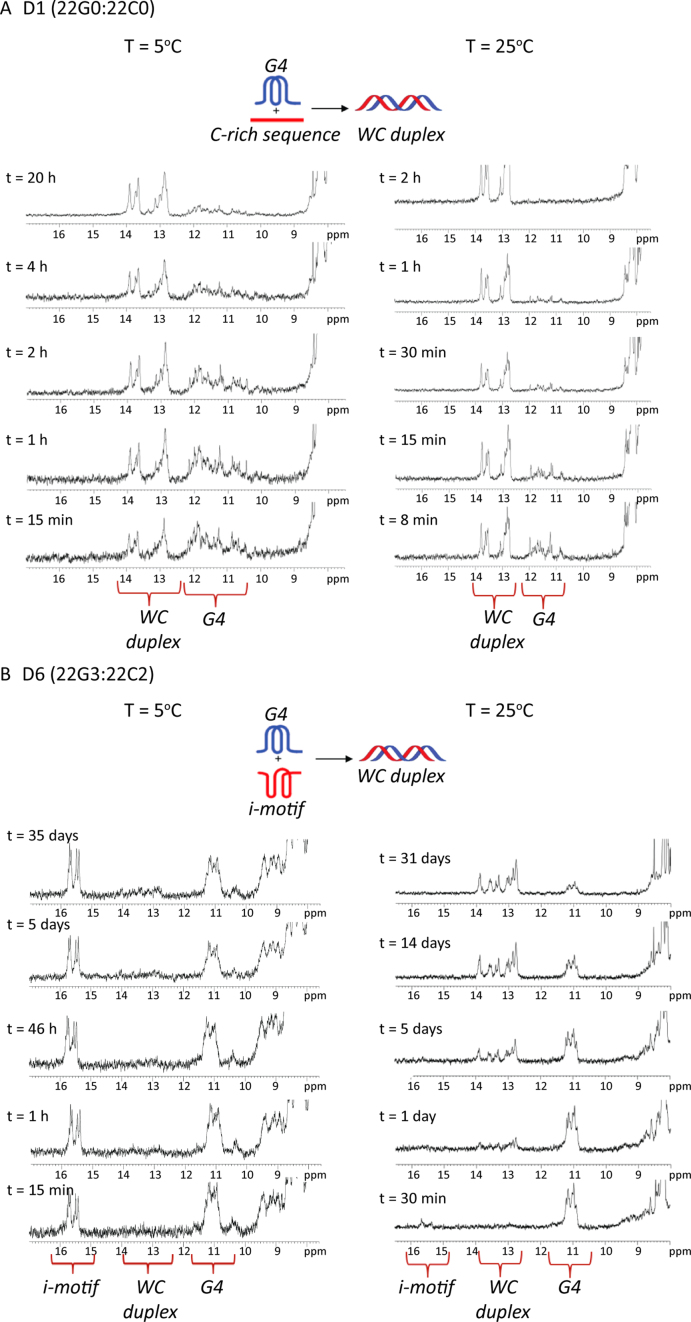
The kinetics of duplex formation were monitored by following the signal changes in the imino region of the NMR spectra (Figure 5, Supplementary Figures S9 and S10). The 22G- and 22C-rich single strands were annealed separately and then mixed in equimolar amounts without further annealing (heat-cool-mix). Changes in the relative populations of duplex, G4, and i-motif species over time were followed by monitoring the change in their relative signal intensities. In the control 22mer sample (D1), the intensity of G4 imino signals decreases within hours at 5 °C and within minutes at 25 °C. After 20 h (5 °C) and 2 h (25 °C), the G4 imino signals vanish completely, indicating that the two strands are fully hybridized, forming a duplex structure (Figure 5). Incorporation of 2′F-ANA modifications in any of the strands slows down the hybridization process dramatically. Indeed, at 5 °C, when only the C-strand is modified (D2, Supplementary Figure S9B), i-motif signals are present after 5 days. Likewise, when only the G-strand is modified (D5, Supplementary Figure S9C), G4 imino signals are visible after 35 days at 5 °C. Fluorination in the G-strand gives rise to G4 structures that are more stable than the duplexes formed when the G-strand hybridizes with the unmodified C-strand. This is consistent with the Tm data (Table 2) where the G4 transition always occurred at higher temperatures compared to the duplex. This effect is even more pronounced when the two strands are modified (D6, Figure 5 and Supplementary Figure S9D); G4 and i-motif imino signals are observed after 35 days with only very minor WC imino signals detectable.
Figure 5.
1H-NMR monitored kinetic experiments for (A) D1 and (B) D6 at 5 °C and 25 °C in a 1:1 mixture at pH 7.1 in 20 mM KPi and 70 mM KCl.
A comparison of the different kinetics of duplex formation at 25 °C indicates that full duplex formation is achieved after approximately 11 days when the C-strand alone is modified (D2, Supplementary Figure S10B). When both strands are modified, the speed of duplex formation is inversely proportional to the number of 2′F-araG residues incorporated (Supplementary Figure S10Cversus S10D). The G4 signals in D2 at 25 °C are sharp and co-exist with the WC imino signals for 11 days. On the other hand, the G4 signals of D2 at 5 °C are very broad and disappear after ∼20 h. This might be due to aggregation of G4 at lower temperatures leading to broad imino signals of the unmodified G-rich strand. These broad signals tend to disappear at a faster rate compared to the G4 signals at 25 °C.
When the G-strand is unmodified and the C-strand is modified (D2; 22G0:22C2), the rate-determining step for duplex formation at 5 °C is the unfolding of 2′F-ANA modified i-motif. When the G-strand is modified and the C-strand is unmodified (D5; 22G3:22C0), the unfolding of 22G3 is rate-determining. When both strands are modified (D6; 22G3:22C2), the i-motif and G4 structures persist with no duplex formation observed even after 35 days at 5 °C. Therefore, at 5 °C, G4 and/or i-motif unfolding are the rate determining steps for duplex formation and the slowdown effect is exacerbated when modifications are incorporated in the G-strand rather than in the C-strand. When the temperature was raised to 25 °C, 22C2 is partially unfolded (Tm = 30 °C) and the G4/i-motif to duplex interconversion rate is dominated by G4 unfolding. Other studies have shown that i-motif unfolding represents a large kinetic barrier to duplex formation (59); however, this is the first time that this effect has been demonstrated at neutral pH.
Properties of 35mer cytosine and guanine-rich single strands and duplexes
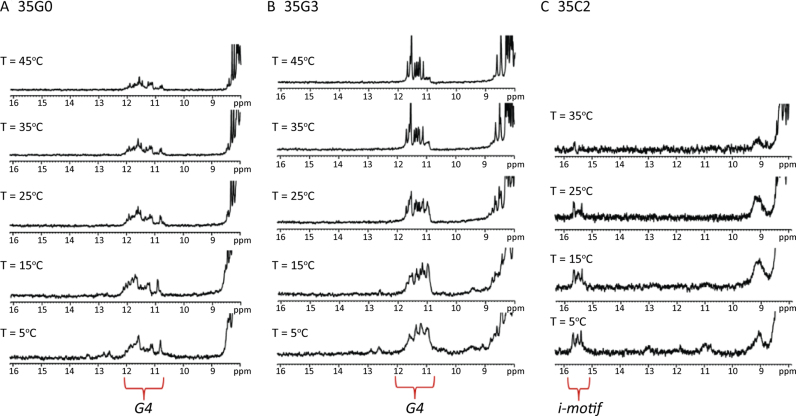
In order to study a scenario where i-motif and G4 structures are in close proximity, longer constructs were prepared. These constructs consist of the same 22-nt sequences studied above, but linked to a 13-nt stretch of mixed base composition (Table 1, Figure 3III and IV). A TTT spacer between the G4/i-motif domain and the duplex domain was inserted to allow for some structural flexibility and to prevent duplex destabilization caused by the proximal G4 and i-motif structures (60). Similar to the data obtained for the 22-nt single-stranded sequences, the CD spectra of the control G-rich strand (35G0) exhibits the characteristic signature of an antiparallel G4 structure, while the 2′F-araG modified strand (35G3) possesses a positive band at 260 nm and a negative band at 240 nm, indicating the formation of a parallel G4 (Supplementary Figure S11). As expected, the stability of the 35C-rich sequences is pH-dependent (Table 1). The CD spectra of the 2′F-araC modified structures at pH 7.0 indicate the formation of i-motifs. To confirm the formation of G4 and i-motif secondary structures in those constructs, 1H-NMR melting experiments were performed (Figure 6) and reveal broad signals for the protons of 35G0, indicating a dynamic equilibrium between multiple conformers. On the other hand, 35G3 exhibits sharp and well-dispersed imino signals, in line with the formation of a single, parallel conformer as indicated by the CD spectra (Supplementary Figure S11). In the case of 35C2, i-motif imino signals are clearly observed up to 25–35 °C and at neutral pH. Therefore, the presence of single-stranded, 13-nt long overhangs does not prevent the formation of stable i-motif and G4 structures.
Figure 6.
1H-NMR melting experiments at pH 7.1 in 20 mM KPi and 70 mM KCl. G4 imino signals show at 10–12 ppm and i-motif imino signals at 15–16 ppm.
When 35G- and 35C-rich strands were annealed together, they hybridized to form WC duplexes. The UV melting curves show only one transition corresponding to duplex dissociation (Figure 4C). The Tm values obtained are comparable with those obtained for the 22-nt duplexes (Table 2). This is likely due to the presence of three T:T mismatches within the duplexes which can be detected in the 1H-NMR spectra as imino signals in the 10–11 ppm range (Supplementary Figure S12).
NMR kinetic experiments of 35mer duplexes
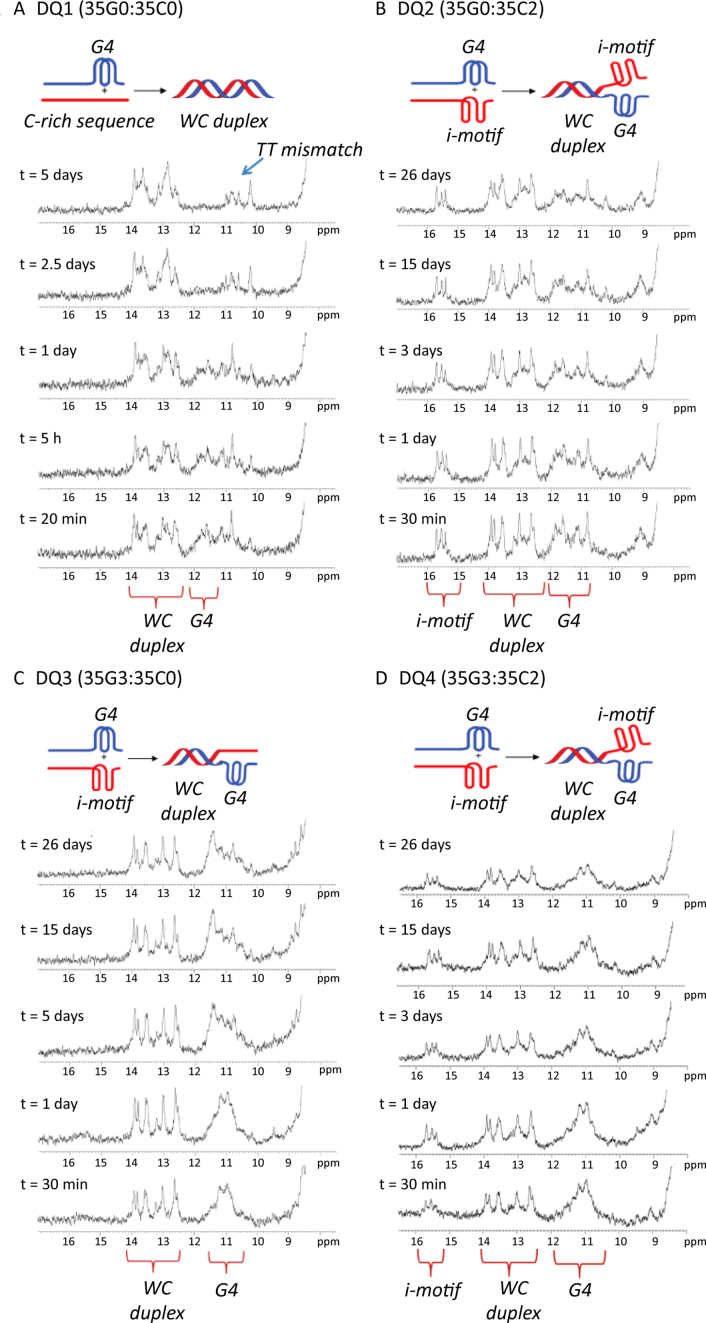
The kinetics of duplex formation upon mixing the pre-folded 35mer G4 and i-motif structures were monitored by 1H-NMR spectroscopy (Figure 7). Some imino signals from WC base pairs are observed immediately after mixing, corresponding to hybridization of the mixed-base complementary ends. However, like the 22mer sequences, NMR experiments show that full hybridization of the 35 nucleotides takes much longer when 2′F-ANA nucleotides are included in any of the strands. For example, in the native 35mer duplex (35G0:35C0), full duplex formation is achieved after two days at T = 5 °C (Figure 7A). In DQ3, where the C-rich strand does not form stable i-motif structures and the G4 structure is very stable (Tm of 35G3 = 75.0 °C, Table 1), G4 and WC imino signals can still be observed after 26 days (Figure 7C). This is in agreement with the results obtained previously by Ren et al. on the same 22G-rich sequence attached to a Dickerson-Drew dodecamer tail, reporting “little or no cross talk" between the G4 structure and the proximal 12-nt duplex (61). Therefore, the presence of G4 structures does not prevent duplex formation in the adjacent overhang. In cases where the C-rich strands form stable i-motif structures (Figure 7B and D), G4 and i-motif imino signals can be detected along with WC imino signals arising from overhang hybridization. Very clear i-motif, G4, and WC imino signals can still be observed one month after mixing the pre-folded 35C-rich and 35G-rich strands. Therefore, given the long-term kinetic stability observed for the modified G4 and i-motif structures, 2′F-ANA modifications can be utilized as a convenient means to “trap" these structures at neutral conditions.
Figure 7.
1H-NMR spectra acquired at different times and at 5 °C for a 1:1 mixture of (A) DQ1 (35G0:35C0), (B) DQ2 (35G0:35C2), (C) DQ3 (35G3:35C0) and (D) DQ4 (35G3:35C2). Buffer conditions: 20 mM KPi and 70 mM KCl, pH 7.1.
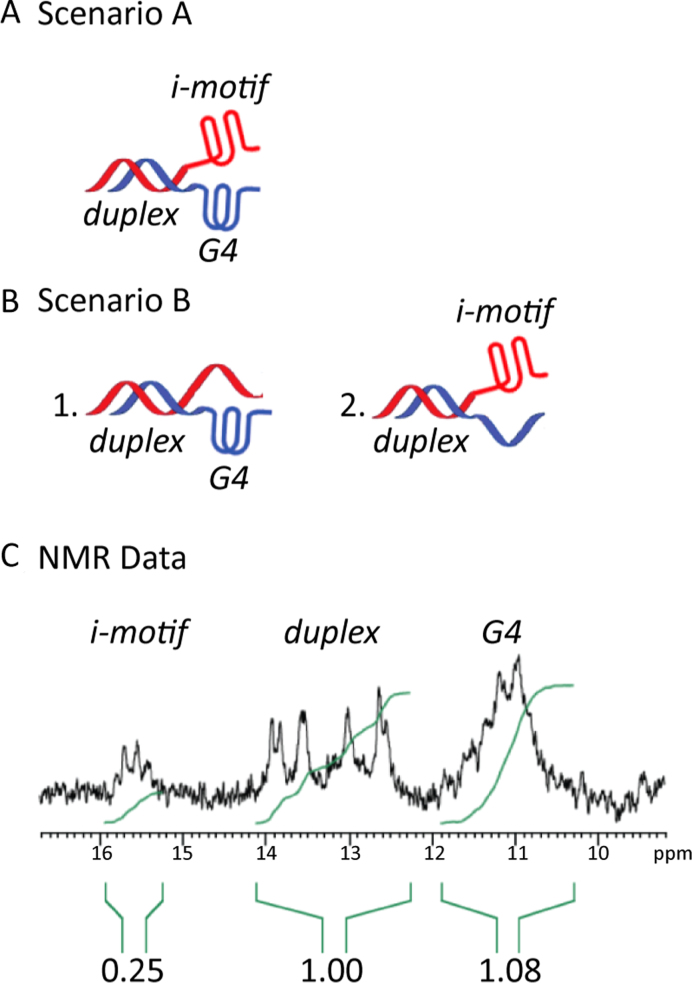
Although no definite conclusions can be reached regarding the structure of 35G3:35C2, the data obtained is consistent with the existence of a hybrid complex wherein a DNA duplex, a G4, and an i-motif co-exist in the same complex at neutral pH (Figure 8A). Gel electrophoretic experiments showed that all strands form dimeric complexes, without substantial presence of monomers or higher order structures (Supplementary Figure S13). The pre-folded-mixed samples (lanes 10–13) show one band with very similar mobility to the annealed duplexes. We do not expect full hybridization since under the gel conditions (pH 7.0, T = 10 °C), 35G0 and 35G3 exhibit high melting temperatures (56 °C and 75 °C, respectively) and therefore should be in the folded state. Although, the presence of complexes in which either the G4 or the i-motif is unfolded cannot be completely disregarded (Figure 8B), some of the NMR data obtained indicate that the complex in which the three secondary structures occur simultaneously is significantly populated. As shown in Figure 8C, the G-rich strand is mainly folded, since the ratio of signal intensities between Watson-Crick and G4 imino signals is very close to the one expected for scenario A (1:1.2, corresponding to 10 WC base pairs and 12 G4 iminos). The situation for the C-strand, however, is less clear since the overall intensity of i-motif imino signal is lower than the expected ratio (0.6:1, corresponding to 6 C:CH+ base pairs). Although these imino protons may be affected by exchange with water to a greater extent than WC and G4 imino protons, some population of partially unfolded species may still be present (Figure 8B.1). It can be observed from the 2D NMR of 35C2 that most cytosine H5 protons exhibit cross-peaks with amino protons in the 8.5–9.5 ppm region and with imino protons ∼15.5 ppm, indicating that they are involved in C:CH+ base pairs (Supplementary Figure S14A). Moreover, the number and intensities of cytosine H5-H6 cross-peaks did not change upon complex formation; such a change would be expected if a significant population of unfolded C-rich strand was present (Supplementary Figure S14B). Finally, the poor signal dispersion of the 2D spectra of the complex 35G3:35C2 makes the sequential assignment impossible; however, comparison of the cytosine signals involved in C:CH+ base-pairs of 35C2 isolated and in the complex with 35G3 strand, suggests that the overall population of i-motif does not decrease significantly upon complex formation (Supplementary Figures S14A and S15) as would be expected for the putative hybrid structure.
Figure 8.
Schematic representation of the proposed scenarios: Scenario (A) where a duplex moiety co-exists in the same complex with G4 and i-motif. Scenario (B) where a duplex moiety co-exists with a C-rich ssDNA and a G4 (B1), a G-rich ssDNA and an i-motif (B2). (C) Imino region of the 1H-NMR spectrum of 35G3:35C2 recorded at T = 5 °C after 30 min from mixing the two strands and showing the values resulting from the integration of i-motif, duplex, and G4 imino signals.
Although studies on promoter regions suggest that G4s and i-motifs are mutually exclusive when the G4/i-motif are on opposite strands at the same location of the duplex (40–43), the scenario here mirrors the telomeric region where the two structures are located at the duplex termini and where steric constraints are more relaxed (60,61). Further characterization studies of these and other structures (e.g. G4/i-motif flanked by two duplex overhangs) are underway.
CONCLUSION
In conclusion, 2′F-ANA modifications slow down G4 and i-motif unfolding, thus providing an excellent tool to study G4/i-motif-duplex interconversion processes at neutral pH. When 2′F-ANA modified i-motif and G4 structures are pre-folded, their unfolding presents a significant barrier to duplex formation. This effect is more pronounced in the presence of G4/i-motif-duplex junctions, leading to very long-lasting intermediates in which i-motifs, G4s, and duplex structures may co-exist. While the presence of i-motif and G4 folds may be mutually exclusive under certain conditions, our results suggest that they may co-exist transiently as intermediates in telomeric sequences. Hybrid structures such as the ones studied here represent attractive tools for the discovery of selective ligands that can specifically stabilize i-motif structures, or target the G4/i-motif-duplex interface, which can in turn interfere with telomerase activity (61,62). In this context, 2′F-ANA modification may become a useful method for building stable supramolecular DNA nanostructures based on the co-existence of i-motif and G4 complexes (63), and for modulating the response of i-motif based nanodevices (64,65).
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MINECO [BFU2014-52864-R]; Natural Sciences and Engineering Research Council of Canada. Funding for open access charge: NSERC Discovery (to M.J.D.); MINECO [BFU2014-52864-R to C.G.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Blackburn E.H., Epel E.S., Lin J.. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science. 2015; 350:1193–1198. [DOI] [PubMed] [Google Scholar]
- 2. Sen D., Gilbert W.. Formation of parallel 4-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. [DOI] [PubMed] [Google Scholar]
- 3. Williamson J.R., Raghuraman M.K., Cech T.R.. Mono-valent cation induced structure of telomeric DNA—the G-Quartet model. Cell. 1989; 59:871–880. [DOI] [PubMed] [Google Scholar]
- 4. Mergny J.L., De Cian A., Ghelab A., Sacca B., Lacroix L.. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005; 33:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lipps H.J., Rhodes D.. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009; 19:414–422. [DOI] [PubMed] [Google Scholar]
- 6. Hansel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279–284. [DOI] [PubMed] [Google Scholar]
- 7. Mergny J.L., Helene C.. G-quadruplex DNA: a target for drug design. Nat. Med. 1998; 4:1366–1367. [DOI] [PubMed] [Google Scholar]
- 8. Neidle S., Read M.A.. G-quadruplexes as therapeutic targets. Biopolymers. 2001; 56:195–208. [DOI] [PubMed] [Google Scholar]
- 9. De Cian A., Lacroix L., Douarre C., Temime-Smaali N., Trentesaux C., Riou J.F., Mergny J.L.. Targeting telomeres and telomerase. Biochimie. 2008; 90:131–155. [DOI] [PubMed] [Google Scholar]
- 10. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Cian A., Cristofari G., Reichenbach P., De Lemos E., Monchaud D., Teulade-Fichou M.P., Shin-Ya K., Lacroix L., Lingner J., Mergny J.L.. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:17347–17352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao Y., Sfeir A.J., Zou Y., Buseman C.M., Chow T.T., Shay J.W., Wright W.E.. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009; 138:463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moye A.L., Porter K.C., Cohen S.B., Phan T., Zyner K.G., Sasaki N., Lovrecz G.O., Beck J.L., Bryan T.M.. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat. Commun. 2015; 6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Biffi G., Di Antonio M., Tannahill D., Balasubramanian S.. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014; 6:75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laguerre A., Wong J.M.Y., Monchaud D.. Direct visualization of both DNA and RNA quadruplexes in human cells via an uncommon spectroscopic method. Sci. Rep. 2016; 6:32141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fleming A.M., Ding Y., Alenko A., Burrows C.J.. Zika virus genomic RNA possesses conserved G-quadruplexes characteristic of the flaviviridae family. ACS Infect. Dis. 2016; 2:674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gehring K., Leroy J.L., Gueron M.. A tetrameric DNA-structure with protonated cytosine. Cytosine base-pairs. Nature. 1993; 363:561–565. [DOI] [PubMed] [Google Scholar]
- 19. Brazier J.A., Shah A., Brown G.D.. I-motif formation in gene promoters: unusually stable formation in sequences complementary to known G-quadruplexes. Chem. Commun. 2012; 48:10739–10741. [DOI] [PubMed] [Google Scholar]
- 20. Berger I., Egli M., Rich A.. Inter-strand C-H center dot center dot center dot O hydrogen bonds stabilizing four-stranded intercalated molecules: Stereoelectronic effects of 04′ in cytosine-rich DNA. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:12116–12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li X., Peng Y., Ren J., Qu X.. Carboxyl-modified single-walled carbon nanotubes selectively induce human telomeric i-motif formation. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:19658–19663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Day H.A., Huguin C., Waller Z.A.E.. Silver cations fold i-motif at neutral pH. Chem. Commun. 2013; 49:7696–7698. [DOI] [PubMed] [Google Scholar]
- 23. Sun D., Hurley L.H.. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009; 52:2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abou Assi H., Harkness R.W.V, Martin-Pintado N., Wilds C.J., Campos-Olivas R., Mittermaier A.K., Gonzalez C., Damha M.J.. Stabilization of i-motif structures by 2′-beta-fluorination of DNA. Nucleic Acids Res. 2016; 44:4998–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Simonsson T., Pribylova M., Vorlickova M.. A nuclease hypersensitive element in the human c-myc promoter adopts several distinct i-tetraplex structures. Biochem. Biophys. Res. Commun. 2000; 278:158–166. [DOI] [PubMed] [Google Scholar]
- 26. Kendrick S., Akiyama Y., Hecht S.M., Hurley L.H.. The i-motif in the bcl-2 P1 promoter forms an unexpectedly stable structure with a unique 8:5:7 loop folding pattern. J. Am. Chem. Soc. 2009; 131:17667–17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kang H.J., Kendrick S., Hecht S.M., Hurley L.H.. The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014; 136:4172–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo K., Gokhale V., Hurley L.H., Sun D.. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008; 36:4598–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guo K., Pourpak A., Beetz-Rogers K., Gokhale V., Sun D., Hurley L.H.. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007; 129:10220–10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Phan A.T., Gueron M., Leroy J.L.. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000; 299:123–144. [DOI] [PubMed] [Google Scholar]
- 31. Garavis M., Escaja N., Gabelica V., Villasante A., Gonzalez C.. Centromeric alpha-satellite DNA adopts dimeric i-motif structures capped by AT Hoogsteen base pairs. Chem. Eur. J. 2015; 21:9816–9824. [DOI] [PubMed] [Google Scholar]
- 32. Garavis M., Mendez-Lago M., Gabelica V., Whitehead S.L., Gonzalez C., Villasante A.. The structure of an endogenous Drosophila centromere reveals the prevalence of tandemly repeated sequences able to form i-motifs. Sci. Rep. 2015; 5:13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wright E.P., Huppert J.L., Waller Z.A.E.. Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 2017; 45:2951–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fleming A.M., Ding Y., Rogers R.A., Zhu J., Zhu J., Burton A.D., Carlisle C.B., Burrows C.J.. 4n-1 Is a “Sweet Spot” in DNA i-motif folding of 2′-deoxycytidine homopolymers. J. Am. Chem. Soc. 2017; 139:4682–4689. [DOI] [PubMed] [Google Scholar]
- 35. Takahashi S., Brazier J.A., Sugimoto N.. Topological impact of noncanonical DNA structures on Klenow fragment of DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:9605–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morin G.B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989; 59:521–529. [DOI] [PubMed] [Google Scholar]
- 37. Lingner J., Cech T.R.. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:10712–10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang H., Blackburn E.H.. De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J. 1997; 16:866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cui Y.X., Kong D.M., Ghimire C., Xu C.X., Mao H.B.. Mutually exclusive formation of G-quadruplex and i-motif is a general phenomenon governed by steric hindrance in duplex DNA. Biochemistry. 2016; 55:2291–2299. [DOI] [PubMed] [Google Scholar]
- 40. Dhakal S., Yu Z.B., Konik R., Cui Y.X., Koirala D., Mao H.B.. G-Quadruplex and i-motif are mutually exclusive in ILPR double-stranded DNA. Biophys. J. 2012; 102:2575–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sutherland C., Cui Y.X., Mao H.B., Hurley L.H.. A mechanosensor mechanism controls the G-quadruplex/i-motif molecular switch in the MYC promoter NHE III1. J. Am. Chem. Soc. 2016; 138:14138–14151. [DOI] [PubMed] [Google Scholar]
- 42. Kaiser C.E., Van Ert N.A., Agrawal P., Chawla R., Yang D., Hurley L.H.. Insight into the complexity of the i-motif and G-quadruplex DNA structures formed in the KRAS promoter and subsequent drug-induced gene repression. J. Am. Chem. Soc. 2017; 139:8522–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brown R.V., Wang T., Chappeta V.R., Wu G., Onel B., Chawla R., Quijada H., Camp S.M., Chiang E.T., Lassiter Q.R. et al. The consequences of overlapping G-quadruplexes and i-motifs in the platelet-derived growth factor receptor beta core promoter nuclease hypersensitive element can explain the unexpected effects of mutations and provide opportunities for selective targeting of both structures by small molecules to downregulate gene expression. J. Am. Chem. Soc. 2017; 139:7456–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Phan A.T., Mergny J.L.. Human telomeric DNA: G-quadruplex, i-motif and watson-crick double helix. Nucleic Acids Res. 2002; 30:4618–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li W., Wu P., Ohmichi T., Sugimoto N.. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002; 526:77–81. [DOI] [PubMed] [Google Scholar]
- 46. Bucek P., Jaumot J., Avino A., Eritja R., Gargallo R.. pH-Modulated Watson-Crick duplex-quadruplex equilibria of guanine-rich and cytosine-rich DNA sequences 140 base pairs upstream of the c-kit transcription initiation site. Chem. Eur. J. 2009; 15:12663–12671. [DOI] [PubMed] [Google Scholar]
- 47. Li W., Miyoshi D., Nakano S., Sugimoto N.. Structural competition involving G-quadruplex DNA and its complement. Biochemistry. 2003; 42:11736–11744. [DOI] [PubMed] [Google Scholar]
- 48. Kumar N., Sahoo B., Varun K.A., Maiti S., Maiti S.. Effect of loop length variation on quadruplex-Watson Crick duplex competition. Nucleic Acids Res. 2008; 36:4433–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peng C.G., Damha M.J.. G-quadruplex induced stabilization by 2′-deoxy-2′-fluoro-D-arabinonucleic acids (2′F-ANA). Nucleic Acids Res. 2007; 35:4977–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lech C.J., Li Z., Heddi B., Phan A.T.. 2′ -F-ANA-guanosine and 2′-F-guanosine as powerful tools for structural manipulation of G-quadruplexes. Chem. Commun. 2012; 48:11425–11427. [DOI] [PubMed] [Google Scholar]
- 51. Martin-Pintado N., Yahyaee-Anzahaee M., Deleavey G.F., Portella G., Orozco M., Damha M.J., Gonzalez C.. Dramatic effect of furanose C2′. substitution on structure and stability: directing the folding of the human telomeric quadruplex with a single fluorine atom. J. Am. Chem. Soc. 2013; 135:5344–5347. [DOI] [PubMed] [Google Scholar]
- 52. Phan A.T., Gueron M., Leroy J.L.. Investigation of unusual DNA motifs. Method Enzymol. 2001; 338:341–371. [DOI] [PubMed] [Google Scholar]
- 53. Mergny J.L., Phan A.T., Lacroix L.. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998; 435:74–78. [DOI] [PubMed] [Google Scholar]
- 54. Leroy J.L., Gueron M., Mergny J.L., Helene C.. Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res. 1994; 22:1600–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kanehara H., Mizuguchi M., Tajima K., Kanaori K., Makino K.. Spectroscopic evidence for the formation of four-stranded solution structure of oligodeoxycytidine phosphorothioate. Biochemistry. 1997; 36:1790–1797. [DOI] [PubMed] [Google Scholar]
- 56. Kypr J., Kejnovska I., Renciuk D., Vorlickova M.. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009; 37:1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Karsisiotis A.I., Hessari N.M., Novellino E., Spada G.P., Randazzo A., da Silva M.W.. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. 2011; 50:10645–10648. [DOI] [PubMed] [Google Scholar]
- 58. Martin-Pintado N., Yahyaee-Anzahaee M., Campos-Olivas R., Noronha A.M., Wilds C.J., Damha M.J., Gonzalez C.. The solution structure of double helical arabino nucleic acids (ANA and 2′F-ANA): effect of arabinoses in duplex-hairpin interconversion. Nucleic Acids Res. 2012; 40:9329–9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mata G., Luedtke N.W.. Fluorescent probe for proton-coupled DNA folding revealing slow exchange of i-motif and duplex structures. J. Am. Chem. Soc. 2015; 137:699–707. [DOI] [PubMed] [Google Scholar]
- 60. Konig S.L., Huppert J.L., Sigel R.K., Evans A.C.. Distance-dependent duplex DNA destabilization proximal to G-quadruplex/i-motif sequences. Nucleic Acids Res. 2013; 41:7453–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ren J., Qu X., Trent J.O., Chaires J.B.. Tiny telomere DNA. Nucleic Acids Res. 2002; 30:2307–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Krauss I.R., Ramaswamy S., Neidle S., Haider S., Parkinson G.N.. Structural insights into the quadruplex-duplex 3′ interface formed from a telomeric repeat: a potential molecular target. J. Am. Chem. Soc. 2016; 138:1226–1233. [DOI] [PubMed] [Google Scholar]
- 63. Cao Y., Gao S., Yan Y., Bruist M.F., Wang B., Guo X.. Assembly of supramolecular DNA complexes containing both G-quadruplexes and i-motifs by enhancing the G-repeat-bearing capacity of i-motifs. Nucleic Acids Res. 2017; 45:26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dong Y.C., Yang Z.Q., Liu D.S.. DNA nanotechnology based on i-motif structures. Acc. Chem. Res. 2014; 47:1853–1860. [DOI] [PubMed] [Google Scholar]
- 65. Yatsunyk L.A., Mendoza O., Mergny J.L.. Nano-oddities:. unusual nucleic acid assemblies for DNA-based nanostructures and nanodevices. Acc. Chem. Res. 2014; 47:1836–1844. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.