Abstract
Newly synthesized histones H3 and H4 undergo a cascade of maturation steps to achieve proper folding and to establish post-translational modifications prior to chromatin deposition. Acetylation of H4 on lysines 5 and 12 by the HAT1 acetyltransferase is observed late in the histone maturation cascade. A key question is to understand how to establish and regulate the distinct timing of sequential modifications and their biological significance. Here, we perform proteomic analysis of the newly synthesized histone H4 complex at the earliest time point in the cascade. In addition to known binding partners Hsp90 and Hsp70, we also identify for the first time two subunits of the histone acetyltransferase inhibitor complex (INHAT): PP32 and SET/TAF-Iβ. We show that both proteins function to prevent HAT1-mediated H4 acetylation in vitro. When PP32 and SET/TAF-Iβ protein levels are down-regulated in vivo, we detect hyperacetylation on lysines 5 and 12 and other H4 lysine residues. Notably, aberrantly acetylated H4 is less stable and this reduces the interaction with Hsp90. As a consequence, PP32 and SET/TAF-Iβ depleted cells show an S-phase arrest. Our data demonstrate a novel function of PP32 and SET/TAF-Iβ and provide new insight into the mechanisms regulating acetylation of newly synthesized histone H4.
INTRODUCTION
Histones, the major proteins associated with DNA in eukaryotic cells, are produced in a tightly regulated manner (1,2). The bulk of histone synthesis occurs during S phase of the cell cycle to provide the necessary supply to package the newly replicated DNA into chromatin. After their synthesis, and before their translocation to the nucleus, newly synthesized histones H3 and H4 are processed in a cascade of maturation steps that allows them to acquire their correct folding and to establish distinct post-translational modifications (3,4). This cascade is comprised of at least five biochemically-defined complexes, each one characterized by a different set of histone chaperones and other binding partners (3–5). The earliest detected modification, methylation of H3K9 by the enzyme SetDB1 (SET domain, Bifurcated 1), occurs during translation while H3 is still associated to the ribosome (5). Then, histones H3 and H4 individually associate with the heat shock proteins Hsc70 (Complex Ia) and Hsp90/70 (Complex Ib), respectively. Subsequently, histones H3 and H4 form a heterodimer, which is mediated by the histone chaperone tNASP (testicular nuclear autoantigenic sperm protein) and Hsp90 (Complex II). The acetylation of lysines 14 and 18 of histone H3, a newly synthesized mark described in human cells (6,7), is imposed in this complex by an uncharacterized acetyltransferase. The H3–H4 dimer next associates with the histone chaperones ASF1 (anti-silencing function 1) and sNASP (somatic nuclear autoantigenic sperm protein), and the acetyltransferase HAT1 (histone acetyltransferase 1) (Complex III). It is in this complex that HAT1 acetylates H4 on lysines 5 and 12. Importantly, this reaction requires that lysines 8 and 16 of the histone H4 are unacetylated (8–10), consistent with the observation that these modifications are largely absent on newly synthesized histones (7). This diacetylated pattern (H4K5K12ac) is an evolutionary conserved mark of newly synthesized histones (10,11). Finally, histones H3–H4 interact with Importin4 and ASF1 to translocate to the nucleus (Complex IV) prior to chromatin assembly (3,4).
Newly synthesized histones H3–H4 display distinct modifications (7) that are imposed in a sequential manner while passing through each one of the chaperone complexes (3,4). Previous studies indicated that the H4K5K12ac is the last modification imposed before histones translocate to the nucleus (3,4). Here, we explore the mechanism that regulates this temporal modification. We find that in the first detected cytosolic histone H4 complex, Complex Ib, histone H4 associates with two subunits of the INHAT complex (inhibitor of acetyltransferases): the myeloid leukemia associated oncoprotein SET/TAF-Iβ and a member of a nuclear phosphoprotein family PP32 protein. The INHAT complex inhibits the p300 and PCAF acetyltransferase activity on histone and non-histone substrates by binding and occluding accessibility of the lysine residues for acetylation (12–15). Consequently, HAT-dependent transcription is repressed (14–16). In vitro studies showed that the subunits SET/TAF-Iβ and PP32 bind preferentially to unacetylated histones H3 and H4 (16,17). In line with this, we find that both SET/TAF-Iβ and PP32 block the establishment of HAT1 mediated H4K12ac. Importantly, PP32 and SET/TAF-Iβ not only inhibit the formation of H4K12ac, but also block acetylation of other lysine residues, such as H4K8 and H4K16. We show that hyperacetylated newly synthesized histone H4 is less stable at higher temperatures, likely as the result of an impaired interaction between acetylated histone H4 with Hsp90, supported by the lack of interaction between Hsp90 and an acetylated H4 (aa1–20) peptide. This aberrant acetylation also impacts cell cycle progression. Together, our data support a model where the INHAT subunits PP32 and SET/TAF-Iβ cooperate to ensure a proper spatio-temporal control over acetylation of newly synthesized histone H4.
MATERIALS AND METHODS
Antibodies
HA (Sigma clone 12CA5), HAT1 (Abcam ab12164), Histone H3 (Abcam ab7834), Histone H4 (Abcam ab10158), H4ac (Millipore 06-866), H4K5ac (Millipore 07-327), H4K8ac (Millipore 06-760), H4K12ac (Millipore 07-595), H4K16ac (Millipore 07-329), Hsp70 (Cell Signalling 4876), Hsp90 (Santa Cruz sc-7947), NASP (Dr Almouzni), PP32 (Abcam ab5991), SET/TAF-Iβ (Abcam ab1183).
Purification of Complex Ib
Seven grams of cytosolic HeLa extracts (donated by D. Reinberg) were precipitated with 20% ammonium sulfate and the supernatant precipitated with 40% ammonium sulfate. Complex Ib was purified from the supernatant fraction, by loading it sequentially twice onto a Tosoh Bioscience TSK-DEAE-5PW column equilibrated with a buffer containing 50 mM KCl. The column was washed with the same buffer and eluted with a linear gradient of salt from 50 mM KCl to 500 mM KCl and then 200 mM KCl to 500 mM KCl. Each purification step was analyzed by Western blot with antibodies against H4 and Hsp90. The Complex Ib peak fractions from the last DEAE-5PW column (250–280 mM KCl) were pooled and analyzed by mass spectrometry after TCA precipitation.
Mass spectrometry
Peptides derived from the trypsin digestion of the complexes were extracted, reverse phase HPLC-separated and then analyzed by tandem mass spectrometry (MS/MS) in Orbitrap LTQ spectrometer as previously described (18). Spectra were acquired and analyzed by Scaffold software, as described previously (7,19).
Labeling and isolation of newly synthesized histone H4
Seventy percent confluent HeLa cells were transfected with a Flag-H4 expressing plasmid. Twenty four hours later, cells were incubated for 1 h with fresh methionine and cysteine free DMEM. Cells were then pulsed for 10 min with 25 μM of AHA (l-azidohomoalanine). Cytosolic extraction was performed as described (20). To conjugate AHA-labeled proteins to Biotin, cytosolic extracts were conjugated to biotin-alkyne by using the ‘Click-iT™ Protein Reaction Buffer Kit’ following the manufacturer's instructions (Invitrogen™, USA). To perform the Flag-immunoprecipitation and Streptavidin pulldown, 600 μg of cytosolic extracts conjugated to biotin-alkyne were incubated with 30 μl of anti-Flag agarose beads 1 h at 4°C, in the presence of 0.05% NP40. Then, beads were washed three times with Wash Buffer (10 mM Tris pH 7.9, 50 mM KCl and 0.05% NP40). One third of the total beads were stored for western blot analyses and the remaining two-thirds were eluted by incubating 1 h, at room temperature and rotation, with 0.4 μg/μl of Flag peptide in Wash Buffer. Then, the eluted material was mixed with 20 μl of Streptavidin-agarose beads, incubated 1 h at 4°C and washed 3 times with Wash Buffer. Samples were analyzed by Western blot against HSP90, PP32, SET, H4 and streptavidin–HRP to visualize AHA-labeled proteins conjugated to biotin.
Protein purification
Bacterially expressed His-tagged PP32, His-tagged Hsp90 and His-tagged SET/TAF-Iβ were purified by standard nickel affinity purification. Bacterially expressed untagged histone H4 was purified as described (21).
siRNA treatment
Either 10 nM of human I2PP2A (SET/TAF-Iβ) siRNA duplex (Santa Cruz, sc-43856), human ANP32A (PP32) siRNA duplex (Santa Cruz, sc-40696), or negative control siRNA (Silencer Negative Control #1 siRNA, Ambion) were transfected with Lipofectamine 2000 (Invitrogen) for 72 h, according to the manufacturer's instructions. For cell cycle distribution analysis, 72 h post-transfection, cells were fixed with 70% ethanol ON at –20°C, and then incubated ON at 4°C with 40 μg/ml RNase A. The DNA content was analyzed by flow cytometry, staining the cells with 40 μg/ml propidium iodide (Life Technologies). Histograms obtained were analyzed by FlowJo software (Vx 0.7, Tree Star).
Histone acetyltransferase assay
The assay was performed using 0.11 μM of purified HAT1 and 3.2 μM of recombinant histone H4 in the presence of the acetylation buffer containing 50 mM Tris–HCl, pH 8.0, 50 mM KCl, 0.1 mM acetylCoA, 10% glycerol, 0.1 mM EDTA, 10 mM sodium butyrate, 1 mM DTT, 0.2 mM PMSF. When using PP32 or SET/TAF-Iβ proteins on the assay, proteins were pre-incubated with histone H4 15 min at 4°C before adding HAT1. The reaction was incubated at 30°C for 30 min and stopped by adding 10% SDS. The reaction products were loaded onto nitrocellulose membranes, dried and analyzed by Dot blot against H4K12ac.
Thermal stability assay
20 μg of cytosolic extract derived from either untreated or siPP32 treated HeLa cells were incubated at increasing temperatures, as indicated, for 20 min. After this, samples were centrifuged at 10 000 rpm for 10 min at 4°C. Supernatant and pellet were separated and the pellet resuspended in 1× SDS-PAGE sample buffer.
Fluorescence spectroscopy
Fluorescence spectra were acquired at room temperature on a Fluorolog-3 FL3C22 spectrofluorimeter (Horiba Jovin Yvon) (excitation: 280 nm, emission: 300–450 nm) in 25 mM Na-phosphate, pH 7.2, 300 mM NaCl and 1 mM DTT, using quartz cuvettes with 20 mm light path (Hellma Analytics). To evaluate interaction between Hsp90 and full length H4, 11.1 μM of HSP90 and a range between 0.5 to 8.34 μM of H4 were used. To evaluate interaction between Hsp90 and either unmodified (Millipore 12-347) or acetylated (Millipore 12-353) peptides containing amino acids 1–20 of histone H4, 55.5 μM of Hsp90 and a range between 1.0 and 26.5 μM of H4 peptides were used. In either case, interactions were evaluated by quenching of intrinsic fluorescence emission of Hsp90. To obtain the maximum fluorescence intensity peak and the integrated fluorescence intensity of each acquired spectra, each one were subjected to a non-linear least square fitting to an Asymmetric double sigmoidal function in the OriginLab 8.0 software. Curve fitting for Kd calculations was accomplished using the GraphPad Prism 6.0 software assuming one site-binding by the equation (ΔF = F0 – FQ = ΔFmax × [Q]/(Kd + [Q]) + NS[Q]), where ΔF is the difference between initial fluorescence (F0) and the fluorescence after ligand addition (FQ) observed fluorescence intensity, ΔFmax, the fluorescence intensity at the start of the titration (F0) and at saturating concentrations of the ligand (Q), NS, the nonspecific fluorescence signal and Kd the dissociation constant of the ligand with Hsp90.
RESULTS
Purification of endogenous Complex Ib
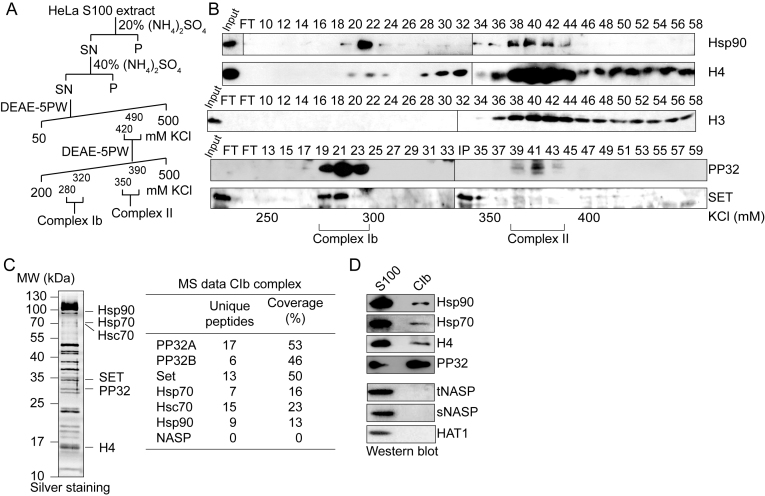
To gain insight into the role of Complex Ib in the processing of newly synthesized histone H4, we sought to identify novel components of this complex. For this, we purified Complex Ib from HeLa S100 cytosolic extracts. We previously showed that H3–H4 histones derived from S100 extracts mainly correspond to newly synthesized histones (7). We purified Complex Ib using four chromatographic steps including two ammonium sulfate cuts and two anion exchange columns following the elution of histone H4 (Figure 1A). We performed Western blot analyses of fractions derived from the last purification step, the anion exchange DEAE-5PW column, for the known components of Complex Ib, Hsp90 and H4 (Figure 1B). The elution profile shows two H4 peaks, a main peak eluting in fractions 40–42 corresponding to Complex II, and a minor peak eluting in fraction 22 corresponding to Complex Ib. Consistent with previous results, Hsp90 co-elutes with both histone H4 peaks (3,4). Importantly, histone H3 co-elutes with Complex II (fractions 40–42), and not with Complex Ib (fractions 21–22).
Figure 1.
PP32 and SET/TAF-Iβ are components of the Complex Ib. (A) Scheme illustrating the purification procedure for obtaining Complex Ib from HeLa S100 extracts. (B) Western blots of fractions derived from the last DEAE-5PW column, as indicated. The corresponding salt concentration of the fractions and the elution of Complex Ib and Complex II are specified at the bottom. Complex Ib elutes in fractions 21–22 and Complex II in fractions 40–42. An additional uncharacterized histone H3 and H4 peak is observed from fraction 48. (C) Left, silver staining of the enriched Complex Ib, corresponding to the pool of fractions 19–23 derived from the last DEAE-5PW column. Right, mass spectrometry (MS) data of the enriched Complex Ib, indicating the number of peptides identified for each protein and the coverage. (D) Western blot of the enriched Complex Ib, corresponding to the pool of fractions 19–23 derived from the last DEAE-5PW column.
The INHAT subunits PP32 and SET/TAF-Iβ are novel components of Complex Ib
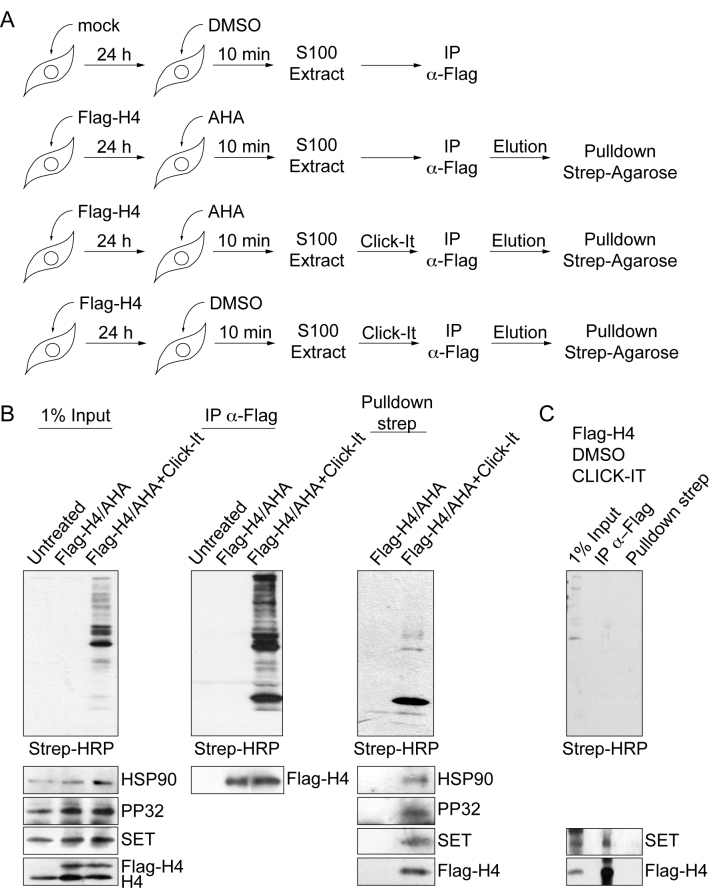
To identify novel components, we analyzed the peak of Complex Ib by mass spectrometry (Figure 1C, left). We find several peptides corresponding to Hsp90, Hsp70 and Hsc70 (Figure 1C, right). We do not find peptides from other H4 interacting proteins, such as NASP or chromatin-binding factors, validating the purification procedure. Interestingly, we further detect several peptides corresponding to the PP32 and SET/TAF-Iβ proteins, components of the INHAT complex (15). We confirmed our mass spectrometry results by Western blot analysis with antibodies specific for the proteins Hsp90, Hsp70, H4 and PP32 in Complex Ib (Figure 1D). Consistent with the mass spectrometry results, we do not detect histone H4 interacting proteins of other cytosolic complexes, including tNASP, sNASP and HAT1 (Figure 1D). We confirmed by Western blot the co-elution of INHAT and Complex Ib in the last purification step (Figure 1B, fractions 19–23). In addition, as previously shown (16,17), pull-down assays using recombinant His-tagged PP32 and recombinant histone H4 demonstrate a direct interaction between PP32 and histone H4 (Supplementary Figure S1A–F). We then investigated whether the interaction of PP32 and SET/TAF-Iβ proteins with newly synthesized histone H4 could occur in the cell. For this, we transiently expressed Flag-tagged histone H4 and pulse-labelled all newly synthesized proteins utilizing the methionine analogue AHA 24 h after transfection. After a 10 min pulse we performed the CLICK-IT procedure to crosslink AHA to alkyne-biotin (Figure 2A). We then immunopurified H4 from S100 extracts using Flag-beads. From this pool of soluble H4 we isolated newly synthesized proteins with Streptavidin–agarose beads. Western blot analyses of the input material show that we started from the same amount of material (Figure 2B, left). When using Flag-beads, we could specifically purify the tagged H4 from transfected cells (Figure 2B, middle). After a short pulse labeling using AHA and alkyne–biotin, we could purify the newly synthesized H4 from the soluble H4 pool and found that we could co-purify HSP90, PP32, SET and Flag-H4 (Figure 2B, right). As a control for non-specific protein absorption to the beads, we also performed the Flag-IP and Streptavidin–agarose pull-down with extracts in which we performed the CLICK-IT procedure with DMSO instead of AHA (Figure 2C). Here, we specifically purified the tagged H4 and co-purified SET from transfected cells, but we could not pull-down the proteins with Streptavidin–agarose beads, confirming the specificity of the pull-down and CLICK-IT reaction. Taken together, we conclude that PP32 and SET/TAF-Iβ proteins indeed associate with newly synthesized H4 in the cytosolic Complex Ib.
Figure 2.
PP32 and SET/TAF-Iβ associates with newly synthesized histone H4. (A) Scheme illustrating the procedure utilized to label and isolate newly synthesized proteins. (B) Western blot analysis of 1% of the input material (left), purified Flag-immunoprecipitated proteins (middle), and Streptavidin–agarose pulled-down proteins derived from the Flag-immunoprecipitated material (right), as indicated. Top panels were developed with Streptavidin-HRP to visualize AHA-labeled proteins conjugated to biotin AHA–biotin. Bottom panels were developed as indicated. (C) Western blot analysis of 1% of the input material, purified Flag-immunoprecipitated proteins and Streptavidin–agarose pulled-down proteins derived from the Flag-immunoprecipitated material, with extracts in which the CLICK-IT procedure was performed with DMSO instead of AHA. Top panel was developed with Streptavidin–HRP to visualize AHA-labeled proteins conjugated to biotin AHA–biotin. Bottom panels were developed as indicated.
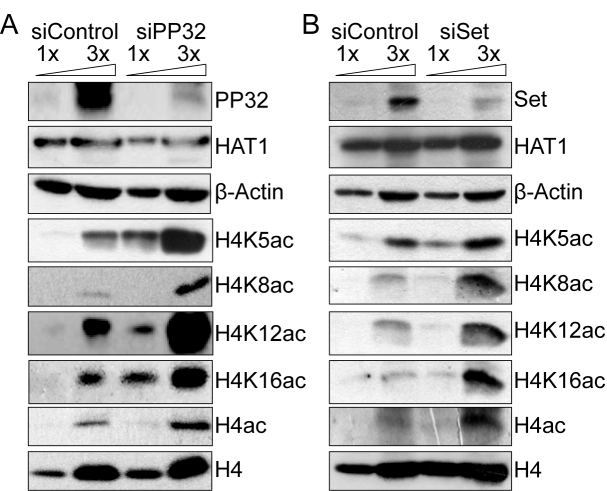
PP32 and SET/TAF-Iβ regulates the levels of acetylation on newly synthesized histone H4 in vivo
We next examined whether PP32 and SET/TAF-Iβ proteins could block acetylation of histone H4 in vivo. Using siRNA (siPP32 and siSET) we reduced the cellular levels of PP32 and SET/TAF-Iβ proteins. Western blot analyses confirm the depletion of siPP32 and siSET relative to siControl (Figure 3A and B, respectively). Importantly, while an antibody that recognizes any acetylated lysine on the H4 tail suggests hyperacetylated histone H4 in each case (Figure 3A and B, H4ac), further examination of H4K5ac and H4K12ac shows a specific increase in the siPP32/siSET extracts (Figure 3A and B). We confirmed that this effect is not merely due to changes in the HAT-1 protein level (Figure 3A and B). Intriguingly, we also observe hyperacetylation of H4K8 and H4K16 (Figure 3A and B) in the siPP32/siSET extracts, marks that are associated with nucleosomal histone H4 rather than newly synthesized histones (7). However, we do not observe changes in histone H3 acetylation (Supplementary Figure S2). These findings suggest that, by binding to histone H4, PP32 and SET/TAF-Iβ proteins have a broad effect occluding acetylation of all H4 tail lysine residues. These data suggest that an aberrant acetylation pattern will arise if PP32 and SET/TAF-Iβ proteins are perturbed. Taken together, our results indicate that PP32 and SET/TAF-Iβ are critical players to regulate the acetylation state of newly synthesized histone H4 in vivo.
Figure 3.
PP32 and SET/TAF-Iβ proteins regulate newly synthesized H4 acetylation levels in vivo. Western blots of 10 and 30 μg of S100 extracts derived from either siControl and siPP32 (A) or siSET/TAF-Iβ (B) treated HeLa cells.
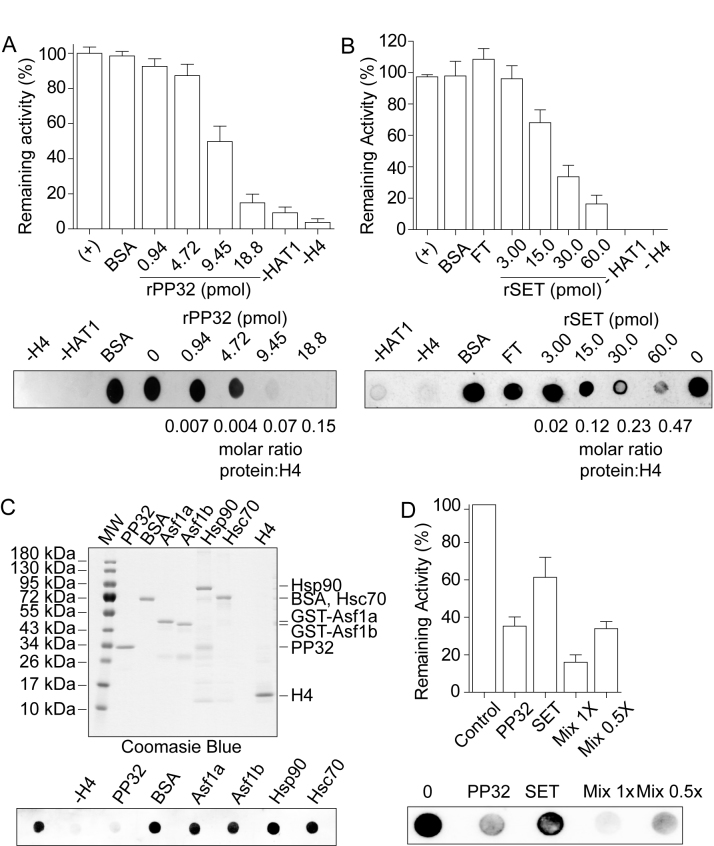
PP32 and SET/TAF-Iβ proteins block HAT1-mediated H4 acetylation in vitro
We next explore whether PP32 and SET/TAF-Iβ proteins block HAT1-mediated H4 acetylation, given that HAT1 is the enzyme implicated in establishing this mark (22). We first performed in vitro acetylation assays by incubating recombinant histone H4 with purified native HAT1 (Supplementary Figure S1D and E) and revealed histone H4 lysine 12 acetylation (H4K12ac). In the absence of HAT1, H4 or acetyl-CoA, we observe minimal background levels, validating the assay (Supplementary Figure S1G). Accordingly, H4K12 acetylation increases in a HAT1 dose-dependent manner (Supplementary Figure S1G), consistent with previous reports (10). We then titrated PP32 or SET/TAF-Iβ proteins (Supplementary Figure S1A, B and E) and incubated them with a constant H4 concentration for 15 min before initiating the reaction by adding HAT1. We observe that both proteins block HAT1 activity in a dose-dependent manner (Figure 4A and B). Although PP32 appears to be more active than SET/TAF-Iβ, we cannot rule out the possibility that the recombinant SET/TAF-Iβ is less active due to other issues. This blocking activity is PP32 and SET/TAF-Iβ specific, as similar amount of BSA do not affect the acetylation (Figure 4A and B). To corroborate their specificity, we further tested the following known histone binding proteins: ASF1a, ASF1b, Hsp90 and Hsc70, on their ability to inhibit acetylation and find that only PP32 and SET/TAF-Iβ impact H4 acetylation (Figure 4C). Finally, PP32 and SET/TAF-Iβ have an additive effect on inhibiting HAT1 mediated H4K12 acetylation (Figure 4D). Taken together, our results indicate that PP32 and SET/TAF-Iβ proteins inhibit HAT1 mediated H4K12ac mark in vitro.
Figure 4.
PP32 and SET/TAF-Iβ proteins block HAT1 mediated H4 acetylation in vitro. Acetylation assay performed using 0.128 nmol of recombinant histone H4 and purified HAT1 in the presence or absence of increasing amounts of recombinant PP32 (A) and recombinant SET/TAF-Iβ (B), followed by H4K12ac Dot-blot detection. Top: graph of the remaining HAT1 activity. 100% activity correspond to the HAT1 mediated H4 acetylation in the absence of PP32 and SET/TAF-Iβ. Bottom: a representative H4K12ac dot blot HAT1 assay. FT corresponds to the flow through material of the recombinant SET/TAF-Iβ Ni+2-beads purification. (C) Top: Coomassie blue stained gel of the different recombinant proteins used in the acetylation assay. Bottom: acetylation assay performed as in (A), pre-incubating H4 as indicated. (D) Acetylation assay performed using recombinant histone H4 and purified HAT1 in the presence of 8 pmol of recombinant PP32, 12 pmol of recombinant SET/TAF-Iβ, and a mix of 8 pmol of recombinant PP32 and 12 pmol of recombinant SET/TAF-Iβ (Mix 1×), and 4 pmol of recombinant PP32 and 6 pmol of recombinant SET/TAF-Iβ (Mix 0.5×).
PP32 knock down affects the interaction between histone H4 and Hsp90
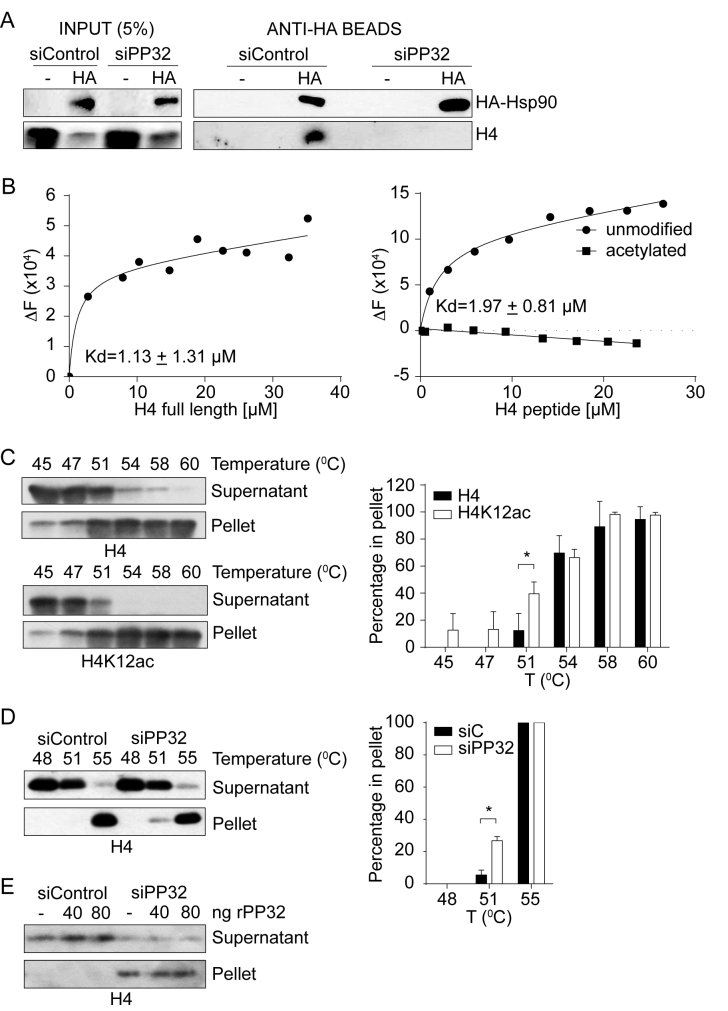
Newly synthesized histones are always escorted by histone chaperones and other proteins to prevent spurious interactions (23,24). Therefore, we speculated that premature acetylation of histone H4 blocks the association with key interacting proteins, leading to changes in the composition of H4 protein-complexes. We focus on the Hsp90 protein because it interacts with histone H4 in Complex Ib, prior to H4 acetylation (3). We ectopically expressed a hemagglutinin (HA)-tagged version of Hsp90 and performed pull-down experiments from cytosolic extracts derived from siControl and siPP32 treated HeLa cells. HA-Hsp90 is pulled-down efficiently from both conditions (Figure 5A). Interestingly, histone H4 co-immunoprecipitates only from siControl, but not from siPP32 derived extracts (Figure 5A). This suggests that acetylation of histone H4 possibly interferes with Hsp90 interaction. However, Hsp90 function itself is also regulated by acetylation in a way that acetylation weakens its affinity for most of its client proteins (25). Therefore, we investigated the impact that histone H4 acetylation has on its interaction with Hsp90. We first monitored intrinsic protein fluorescence of recombinant Hsp90 expressed in bacteria to avoid acetylation (Figure 5B and Supplementary Figure S3). Titration of Hsp90 with histone H4 (tryptophan free protein) reveals a binding affinity typical for chaperone-substrate interactions (KD = 1.13 ± 1.31 μM; Figure 5B, left). We then evaluate the interaction between Hsp90 and either an unmodified or acetylated peptide containing amino acids 1–20 of histone H4. Titration of Hsp90 with the unmodified histone H4 peptide reveals a similar binding affinity as the full-length protein (KD = 1.97 ± 0.81 μM; Figure 5B, right). In contrast, we do not detect an interaction between Hsp90 and the acetylated H4 peptide (Figure 5B, right). Taken together, our results suggest that the interaction between histone H4 and Hsp90 may be negatively affected due to aberrant histone H4 acetylation when the levels of PP32 are reduced.
Figure 5.
PP32 knock-down affects the maturation of newly synthesized histone H4. (A) Western blot of HA-Hsp90 pull-down assay from cytosolic extracts derived from siControl and siHsp90 treated HeLa cells. (B) Graph of the tryptophan fluorescence spectroscopy of Hsp90 upon titrating full length histone H4 (left), or unmodified or acetylated amino acids 1–20 of histone H4 (right). ΔF is the difference between initial fluorescence (F0) and the fluorescence after ligand addition (FQ). KD is the dissociation constant of the ligand with Hsp90. (C) Left, Western blot analysis of thermal stability assay using HeLa S100 extracts. 20 μg of S100 extracts were heated for 20 min at the indicated temperatures and centrifuged at 10 000 × g for 5 min. Supernatant and pellet were analyzed by western blot. Right, graph of the percentage of either total or acetylated histone H4 found on the pellet, taken as 100% of the sum of the supernatant and pellet at each temperature. Standard deviations were taken from three independent experiments. *P < 0.05, Student′s t-test. (D) Thermal stability of S100 extracts derived from siControl and siPP32 HeLa cells, as described in (C). (E) Thermal stability of S100 extracts derived from siControl and siPP32 HeLa cells, as described in (C), in the absence or presence of recombinant PP32.
PP32 affects the stability of newly synthesized histone H4
We then investigated the consequences of reducing the H4–Hsp90 interaction in cells that have lowered PP32 levels. Given that Hsp90 is a heat shock chaperone, we focused on histone H4 stability. It is well documented that histone acetylation increases the solubility of chromatin, but its effect on soluble histones remains an open question (26–28). Our results suggest that in the absence of PP32, newly synthesized histone H4 is not only acetylated at an inappropriate time, but it is aberrantly acetylated on K8 and K16, which contribute to preventing the interaction with Hsp90. Therefore, we hypothesized that newly synthesized histone H4 presenting aberrant acetylation could influence its stability due to a defective Hsp90 interaction. To investigate this, we set up a thermal stability assay, in which cytosolic extracts are incubated for 20 min at increasing temperatures, after which soluble and insoluble proteins are separated by centrifugation. We find that, under basal conditions, histone H4 remains soluble at 45°C, 12% of the protein precipitates at 51°C, and almost all the histone H4 precipitates at 58°C (Figure 5C). However, when we analyzed H4K12ac under the same conditions, 40% of the acetylated protein precipitates at 51°C (Figure 5C), indicating that acetylated newly synthesized histone H4 is less stable than hypoacetylated H4. We then compared the thermal stability of cytosolic histone H4 derived from siControl and siPP32 treated cells. We find that cytosolic histone H4 from siPP32 treated cells is less stable at 51°C compared to siControl cells, as 6% compared to 27% of histone H4 precipitated from siControl and siPP32 cells, respectively (Figure 5D). To confirm that this effect was not due to a direct role that PP32 itself could confer thermal stability, we performed the assay at 51°C while adding back recombinant PP32 (Figure 5E). We observe that histones derived from siPP32 are less stable regardless of the presence of recombinant PP32. Therefore, we conclude that cytosolic histone H4 stability decreases when PP32 protein levels are reduced, likely due to histone H4 hyperacetylation and a decreased interaction with Hsp90.
PP32 knock down affects cell cycle progression
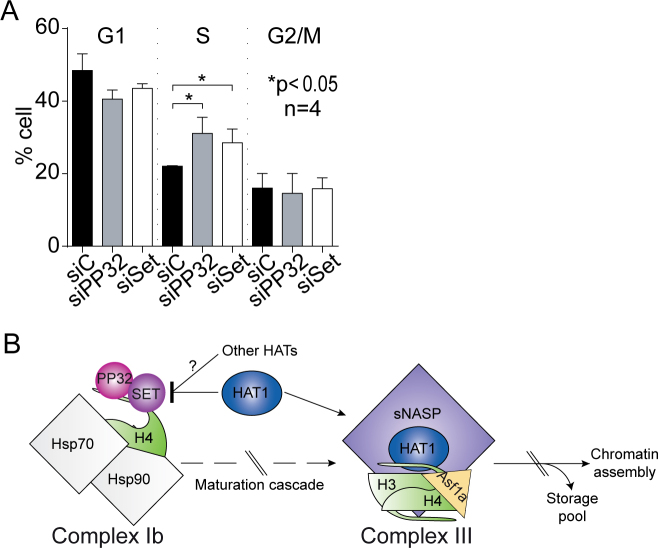
Our results suggest that PP32 is involved in the proper maturation of newly synthesized histone H4 by preventing premature and aberrant acetylation. Therefore, we hypothesized that histone H3–H4 deposition may be altered upon siPP32. We monitored newly synthesized histone deposition in vivo utilizing HeLa cell lines stably expressing H3.1- and H3.3-SNAP tagged proteins and quench-chase-pulse experiments (29). The SNAP tag enables covalent labeling with a cell-permeable small molecule in vivo. To distinguish parental from newly synthesized histones, parental histone H3 are quenched with a non-fluorescent blocking molecule. A chase time permits the synthesis of new SNAP-H3 variant proteins, which are subsequently labeled with a fluorescent probe (TMR) (29). We tested the incorporation of the newly synthesized histone H3 variants H3.1 and H3.3 (Supplementary Figure S4). As a control, we confirmed that depleting the H3.3 chaperone HIRA (histone regulation A) (30) using siRNA leads to a decrease in H3.3, but not H3.1, deposition (Supplementary Figure S4) (29). We then investigated the effect of siPP32 (Supplementary Figure S4) and find a small, but statistically significant, defect on the incorporation of both H3.1 and H3.3 variants when compared to siControl (Supplementary Figure S4). These data suggest that perturbation of PP32 may impact histone H3–H4 deposition. Further work will address whether this impact is more pronounced at defined windows of the cell cycle and if it plays a particular role during critical phases in an organism's life cycle, such as during development and in particular pathologies. In line with this, we examined the consequences of knocking down PP32 and SET/TAF-Iβ on cell cycle distribution. Flow cytometry analysis of DNA content shows that cells accumulated in S-phase 72 h after siPP32 (31% of cells) and siSET/TAF-Iβ (29% of cells), as compared to control cells (22% of cells) (Figure 6A). This resembles the S-phase arrest observed upon perturbation of CAF1 and ASF1 (31–33). Together, our data support a model that PP32 and SET/TAF-Iβ, by regulating newly synthesized histone H4 metabolism, may contribute to regulate histone deposition and cell cycle progression.
Figure 6.
PP32 knock-down affects cell cycle progression. (A) Cell cycle profiles of HeLa cells treated with siControl, siPP32, and siSET/TAF-Iβ. (B) Proposed model for the regulation of newly synthesized histone H4 acetylation. After synthesis, unacetylated histone H4 interacts with the heat shock proteins Hsp90 and Hsp70 and two members of the INHAT complex: PP32 and SET/TAF-Iβ. The binding of PP32 and SET/TAF-Iβ protects histone H4 from premature acetylation by HAT1, as well as for non-specific acetylation mediated by other HATs. When PP32 protein levels are diminished, histone H4 is acetylated at early steps of the maturation cascade, and it no longer can interact with Hsp90 affecting its stability and maturation.
DISCUSSION
Acetylation of lysine is a cellular-wide dynamic post-translational modification that mediates interactions with other proteins and can impact genome stability and function (34). In one example, acetylation of tubulin is implicated in regulating microtubule assembly (35), while acetylation of Hsp90 weakens its affinity for most of its client proteins (25). Histone acetylation plays critical roles both in the context of a nucleosome but also on soluble histones. Following biosynthesis, histones H3 and H4 proceed through a maturation cascade of complexes that ensure their proper folding and post-translational modification state prior to chromatin assembly or storage. Notably, the H4K5K12 acetylation mark, established by the HAT1 acetyltransferase, is observed late in the histone maturation cascade, suggesting that there are regulatory mechanisms that prevent premature acetylation. In addition, acetylation of other lysine residues is not observed on newly synthesized histone H4. Here, we reveal that the inhibitor of HAT (INHAT) complex subunits PP32 and SET/TAF-Iβ play key roles in regulating the timing and residue specificity of acetylating newly synthesized histones (Figure 6B). We observe an interaction between newly synthesized H4 and the INHAT subunits PP32 and SET/TAF-Iβ early in the maturation cascade prior to H4 acetylation. Through depletion experiments, we find that the INHAT subunits impede acetylation of H4, not only of the residues K5 and K12, known to be acetylated at later stages of its maturation, but also residues that are not commonly acetylated on newly synthesized histone H4, including K8 and K16. Moreover, we observe that aberrant H4 acetylation impacts stability, its interaction with Hsp90, and cell cycle progress. Whether this effect is due to premature H4K5K12 acetylation, aberrant H4K8K16 acetylation, or both warrants further investigation.
Role of newly synthesized histone H4K5K12 acetylation
Although H4K5K12ac is a well characterized mark of newly synthesized histones, a debate still exists regarding the role of this mark on histone function (10,36,37). Given that all newly synthesized H4 histones carry this pattern, and that this acetylation pattern is removed shortly after deposition onto chromatin (38–40), it has been suggested that H4K5K12ac mediates chromatin assembly. However, genetic studies in yeast suggest that this modification is not essential (41–44). This may be species specific, or perhaps there is functional redundancy of H4K5K12 acetylation with other acetylation sites. In support of the latter idea, combining the mutation H4K5RK12R (resembling unacetylated lysines) with H4K91Q (an acetylation mimic) leads to defects in chromatin reassembly following double-strand DNA breaks (45). Alternatively, H4 acetylation patterns may play a role in the cytoplasmic/nuclear transport of histones given that K5 and K12 are located within the nuclear localization signal of H4. Indeed, in the mold Physarum model the H4K5RK12R mutation blocks H3–H4 nuclear translocation, while H4K5QK12Q favors import (46). Similarly, in HeLa cells, the mutant H4K5QK12Q favors the nuclear transport in an in vitro translocation assay by favoring the importin/histone interaction (3). Interestingly, our data suggest a new role for H4 acetylation in which it regulates a protein-protein interaction by blocking the association with heat shock proteins, thus helping to regulate the timing of the establishment of this mark.
Mechanisms that regulate post-translational modifications
The establishment and maintenance of histone acetylation involves several mechanisms, including the opposing activities of acetyltransferases and deacetylases and histone turnover. Additionally, protein-protein interactions may contribute to the maintenance of modifications. In one example, bromodomains, which recognize and bind acetylated lysines, may limit access to the lysine, preventing enzymatic deacetylation. Indeed, BRD4 (bromodomain containing protein 4) binds acetylated histones at transcriptionally active genes throughout the cell cycle (47) to maintain the transcriptionally active mark (38). In response to DNA damage, BRD4 dissociates, which induces deacetylation of H4K5ac and H4K8ac (48,49) by HDACs. Another example is the regulation of acetylation on non-histone proteins. SET/TAF-Iβ interacts in the nucleus with the Ku70/80 heterodimer, required for the NHEJ (non-homologous end-joining) DNA DSB (double strand break) repair pathway, and inhibits Ku70 acetylation. Upon DNA damage, SET/TAF-Iβ dissociates and releases the Ku complex, which is recruited to DNA DSB sites (13). Soluble histones offer a different context for associating with binding proteins, suggesting that other mechanisms may be involved to regulate the timing of establishing modifications. Here, we reveal a novel mechanism where PP32 and SET/TAF-Iβ associate with a histone complex and occlude HAT1, and potentially other acetyltransferases, from binding and acting on its histone substrate. In this way, PP32 and SET/TAF-Iβ proteins ensure that histone H4 is acetylated at the right time and on the proper lysine residues. When early acetylation occurs, as in the case of PP32 knock down cells depletion, histone H4 cannot interact with Hsp90, affecting its stability and maturation. Although we did not investigate the transit of histone H4 beyond Complex Ib, we speculate that following its association with heat shock proteins and INHAT subunits, histone H4 passes through the other complexes to continue its maturation. On Complex III, histone H4 does no longer interacts with the INHAT subunits, instead it interacts with the enzyme HAT1 to acetylate histone H4 on K5 and K12 (Figure 6B). How PP32 and SET/TAF-Iβ dissociation from histone H4 is regulated will be explored in future work. Investigating whether INHAT and histone H4 binding is regulated in a cell cycle dependent manner will be useful to understand how histone maturation is linked to the cellular requirements of histones. Future work should also address whether this kind of mechanism applies more broadly to other modifications and proteins.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Danny Reinberg for providing S100 extracts, Zhiguo Zhang for providing Flag-H4 expressing plasmid.
Footnotes
Present address: Zachary A. Gurard-Levin, SAMDI Tech, Inc. Chicago, IL 60616, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Comisión Nacional de Ciencia y Tecnología [FONDECYT 1160480 and Basal Project PFB16 to A.L., PCHA/Doctorado Nacional/2016-21161044 to C.R., PCHA/Doctorado Nacional/2014–21140346 to F.S., PCHA/Doctorado Nacional/2014-21140956 to S.H.]; Ligue Nationale contre le Cancer (Equipe labellisée Ligue) (to G.A.); French National Research Agency [ANR-11-LABX-0044_DEEP, ANR-10-IDEX-0001-02 PSL, ANR-12-BSV5–0022-02 ‘CHAPINHIB’, ANR-14-CE16-0009 ‘Epicure’, ANR-14-CE10-0013 ‘CELLECTCHIP’, ANR-16-CE15-0018 ‘CHRODYT’, ANR-16-CE12–0024 ‘CHIFT’ to G.A.]; European Commission [N 678563 ‘EPOCH28’, ERC-2015-ADG- 694694 ‘ChromADICT’ to G.A.]; Deutsche Forschungsgemeinschaft [CRC1064/Z03 to A.I.]. Funding for open access charge: Comisión Nacional de Ciencia y Tecnología [FONDECYT 1160480].
Conflict of interest statement. None declared.
REFERENCES
- 1. MacAlpine D.M., Almouzni G.. Chromatin and DNA replication. Cold Spring Harb. Perspect. Biol. 2013; 5:a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marzluff W.F., Duronio R.J.. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 2002; 14:692–699. [DOI] [PubMed] [Google Scholar]
- 3. Alvarez F., Munoz F., Schilcher P., Imhof A., Almouzni G., Loyola A.. Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 2011; 286:17714–17721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Campos E.I., Fillingham J., Li G., Zheng H., Voigt P., Kuo W.H., Seepany H., Gao Z., Day L.A., Greenblatt J.F. et al. The program for processing newly synthesized histones H3.1 and H4. Nat. Struct. Mol. Biol. 2010; 17:1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rivera C., Saavedra F., Alvarez F., Diaz-Celis C., Ugalde V., Li J., Forne I., Gurard-Levin Z.A., Almouzni G., Imhof A. et al. Methylation of histone H3 lysine 9 occurs during translation. Nucleic Acids Res. 2015; 43:9097–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jasencakova Z., Scharf A.N., Ask K., Corpet A., Imhof A., Almouzni G., Groth A.. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell. 2010; 37:736–743. [DOI] [PubMed] [Google Scholar]
- 7. Loyola A., Bonaldi T., Roche D., Imhof A., Almouzni G.. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell. 2006; 24:309–316. [DOI] [PubMed] [Google Scholar]
- 8. Benson L.J., Phillips J.A., Gu Y., Parthun M.R., Hoffman C.S., Annunziato A.T.. Properties of the type B histone acetyltransferase Hat1: H4 tail interaction, site preference, and involvement in DNA repair. J. Biol. Chem. 2007; 282:836–842. [DOI] [PubMed] [Google Scholar]
- 9. Makowski A.M., Dutnall R.N., Annunziato A.T.. Effects of acetylation of histone H4 at lysines 8 and 16 on activity of the Hat1 histone acetyltransferase. J. Biol. Chem. 2001; 276:43499–43502. [DOI] [PubMed] [Google Scholar]
- 10. Parthun M.R. Histone acetyltransferase 1: more than just an enzyme. Biochim. Biophys. Acta. 2012; 1819:256–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sobel R.E., Cook R.G., Perry C.A., Annunziato A.T., Allis C.D.. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J.Y., Lee K.S., Seol J.E., Yu K., Chakravarti D., Seo S.B.. Inhibition of p53 acetylation by INHAT subunit SET/TAF-Ibeta represses p53 activity. Nucleic Acids Res. 2012; 40:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim K.B., Kim D.W., Park J.W., Jeon Y.J., Kim D., Rhee S., Chae J.I., Seo S.B.. Inhibition of Ku70 acetylation by INHAT subunit SET/TAF-Ibeta regulates Ku70-mediated DNA damage response. Cell. Mol. Life Sci.: CMLS. 2014; 71:2731–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seo S.B., Macfarlan T., McNamara P., Hong R., Mukai Y., Heo S., Chakravarti D.. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J. Biol. Chem. 2002; 277:14005–14010. [DOI] [PubMed] [Google Scholar]
- 15. Seo S.B., McNamara P., Heo S., Turner A., Lane W.S., Chakravarti D.. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001; 104:119–130. [DOI] [PubMed] [Google Scholar]
- 16. Kutney S.N., Hong R., Macfarlan T., Chakravarti D.. A signaling role of histone-binding proteins and INHAT subunits pp32 and Set/TAF-Ibeta in integrating chromatin hypoacetylation and transcriptional repression. J. Biol. Chem. 2004; 279:30850–30855. [DOI] [PubMed] [Google Scholar]
- 17. Schneider R., Bannister A.J., Weise C., Kouzarides T.. Direct binding of INHAT to H3 tails disrupted by modifications. J. Biol. Chem. 2004; 279:23859–23862. [DOI] [PubMed] [Google Scholar]
- 18. Barth T.K., Schade G.O., Schmidt A., Vetter I., Wirth M., Heun P., Imhof A., Thomae A.W.. Identification of Drosophila centromere associated proteins by quantitative affinity purification-mass spectrometry. Data Brief. 2015; 4:544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bonaldi T., Regula J.T., Imhof A.. The use of mass spectrometry for the analysis of histone modifications. Methods Enzymol. 2004; 377:111–130. [DOI] [PubMed] [Google Scholar]
- 20. Dignam J.D., Lebovitz R.M., Roeder R.G.. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983; 11:1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luger K., Rechsteiner T.J., Flaus A.J., Waye M.M., Richmond T.J.. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997; 272:301–311. [DOI] [PubMed] [Google Scholar]
- 22. Verreault A., Kaufman P.D., Kobayashi R., Stillman B.. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol.: CB. 1998; 8:96–108. [DOI] [PubMed] [Google Scholar]
- 23. Gurard-Levin Z.A., Quivy J.P., Almouzni G.. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 2014; 83:487–517. [DOI] [PubMed] [Google Scholar]
- 24. Loyola A., Almouzni G.. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta. 2004; 1677:3–11. [DOI] [PubMed] [Google Scholar]
- 25. Mollapour M., Neckers L.. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim. Biophys. Acta. 2012; 1823:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wallace R.B., Sargent T.D., Murphy R.F., Bonner J.. Physical properties of chemically acetylated rat liver chromatin. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:3244–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perry M., Chalkley R.. Histone acetylation increases the solubility of chromatin and occurs sequentially over most of the chromatin. A novel model for the biological role of histone acetylation. J. Biol. Chem. 1982; 257:7336–7347. [PubMed] [Google Scholar]
- 28. Wang X., He C., Moore S.C., Ausio J.. Effects of histone acetylation on the solubility and folding of the chromatin fiber. J. Biol. Chem. 2001; 276:12764–12768. [DOI] [PubMed] [Google Scholar]
- 29. Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., Puri A., Schultz D.C., Pchelintsev N.A., Adams P.D., Jansen L.E. et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell. 2011; 44:928–941. [DOI] [PubMed] [Google Scholar]
- 30. Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y.. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004; 116:51–61. [DOI] [PubMed] [Google Scholar]
- 31. Groth A., Ray-Gallet D., Quivy J.P., Lukas J., Bartek J., Almouzni G.. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell. 2005; 17:301–311. [DOI] [PubMed] [Google Scholar]
- 32. Hoek M., Stillman B.. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:12183–12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ye X., Franco A.A., Santos H., Nelson D.M., Kaufman P.D., Adams P.D.. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol. Cell. 2003; 11:341–351. [DOI] [PubMed] [Google Scholar]
- 34. Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M.. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009; 325:834–840. [DOI] [PubMed] [Google Scholar]
- 35. Janke C. The tubulin code: molecular components, readout mechanisms, and functions. J. Cell Biol. 2014; 206:461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keck K.M., Pemberton L.F.. Histone chaperones link histone nuclear import and chromatin assembly. Biochim. Biophys. Acta. 2013; 1819:277–289. [PubMed] [Google Scholar]
- 37. Li Q., Burgess R., Zhang Z.. All roads lead to chromatin: Multiple pathways for histone deposition. Biochim. Biophys. Acta. 2012; 1819:238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Annunziato A.T., Seale R.L.. Histone deacetylation is required for the maturation of newly replicated chromatin. J. Biol. Chem. 1983; 258:12675–12684. [PubMed] [Google Scholar]
- 39. Jackson V., Granner D., Chalkley R.. Deposition of histone onto the replicating chromosome: newly synthesized histone is not found near the replication fork. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:2266–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nagarajan P., Ge Z., Sirbu B., Doughty C., Agudelo Garcia P.A., Schlederer M., Annunziato A.T., Cortez D., Kenner L., Parthun M.R.. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet. 2013; 9:e1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma X.J., Wu J., Altheim B.A., Schultz M.C., Grunstein M.. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:6693–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Megee P.C., Morgan B.A., Mittman B.A., Smith M.M.. Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science. 1990; 247:841–845. [DOI] [PubMed] [Google Scholar]
- 43. Park E.C., Szostak J.W.. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol. Cell. Biol. 1990; 10:4932–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang W., Bone J.R., Edmondson D.G., Turner B.M., Roth S.Y.. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998; 17:3155–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ge Z., Nair D., Guan X., Rastogi N., Freitas M.A., Parthun M.R.. Sites of acetylation on newly synthesized histone H4 are required for chromatin assembly and DNA damage response signaling. Mol. Cell. Biol. 2013; 33:3286–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ejlassi-Lassallette A., Mocquard E., Arnaud M.C., Thiriet C.. H4 replication-dependent diacetylation and Hat1 promote S-phase chromatin assembly in vivo. Mol. Biol. Cell. 2011; 22:245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Devaiah B.N., Singer D.S.. Two faces of brd4: mitotic bookmark and transcriptional lynchpin. Transcription. 2013; 4:13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ai N., Hu X., Ding F., Yu B., Wang H., Lu X., Zhang K., Li Y., Han A., Lin W. et al. Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic Acids Res. 2011; 39:9592–9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu X., Lu X., Liu R., Ai N., Cao Z., Li Y., Liu J., Yu B., Liu K., Wang H. et al. Histone cross-talk connects protein phosphatase 1alpha (PP1alpha) and histone deacetylase (HDAC) pathways to regulate the functional transition of bromodomain-containing 4 (BRD4) for inducible gene expression. J. Biol. Chem. 2014; 289:23154–23167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.