Abstract
The essential element zinc plays critical roles in biology. High zinc homeostasis mechanisms are beginning to be defined in animals, but low zinc homeostasis is poorly characterized. We investigated low zinc homeostasis in Caenorhabditis elegans because the genome encodes 14 evolutionarily conserved Zrt, Irt-like protein (ZIP) zinc transporter family members. Three C. elegans zipt genes were regulated in zinc-deficient conditions; these promoters contained an evolutionarily conserved motif that we named the low zinc activation (LZA) element that was both necessary and sufficient for activation of transcription in response to zinc deficiency. These results demonstrated that the LZA element is a critical part of the low zinc homeostasis pathway. Transcriptional regulation of the LZA element required the transcription factor ELT-2 and mediator complex member MDT-15. We investigated conservation in mammals by analyzing LZA element function in human cultured cells; the LZA element-mediated transcriptional activation in response to zinc deficiency in cells, suggesting a conserved pathway of low zinc homeostasis. We propose that the pathway for low zinc homeostasis, which includes the LZA element and ZIP transporters, acts in parallel to the pathway for high zinc homeostasis, which includes the HZA element, HIZR-1 transcription factor and cation diffusion facilitator transporters.
INTRODUCTION
Zinc is an essential element for all life, and homeostatic mechanisms are critical to ensure a steady supply of zinc. One function of zinc in biology has been well established by biochemical and structural studies - biological systems exploit the unique chemistry of zinc by using it as a cofactor of enzymes from all six major functional classes and as a structural cofactor for many proteins, such as zinc finger transcription factors (1,2). This use of zinc is widespread, as 10% of the human proteome is predicted to bind zinc (3). A second function that is beginning to be defined is the use of zinc as a second messenger. Pulsatile release of zinc has been observed in the extracellular space, described as the zinc spark, and the intracellular space, described as the zinc wave (4,5). The biological requirements for zinc are illustrated by the pathologies caused by zinc deficiency. The human genetic disorder acrodermatitis enteropathica is caused by a mutation in the ZIP4 gene that encodes a ZIP family zinc transporter; reduced activity of this key transporter impairs intestinal absorption of zinc and results in zinc deficiency (6–9). In humans, severe deficiency of zinc leads to defects in the epidermal, gastrointestinal, immune, and nervous systems as well as diminished growth and reproductive ability (1,2,6). Human zinc deficiency is a widespread problem that is primarily caused by inadequate dietary zinc and is estimated to affect 2 billion people (6,7,10). Zinc-deficient cells fail to divide and differentiate, thus impairing the growth of plants and animals. Because zinc is both essential and toxic in excess, mechanisms have evolved to maintain zinc within an appropriate range, described as zinc homeostasis. Characterizing mechanisms of zinc homeostasis is an important objective that has implications for understanding fundamental biological processes and improving human health.
In animals, mechanisms of high zinc homeostasis are beginning to be defined, but low zinc homeostasis remains largely unexplored. Two major families of zinc transporters play important roles in zinc homeostasis in eukaryotes: the cation diffusion facilitator (CDF, ZnT, SLC30) and the Zrt-, Irt-like proteins (ZIP, SLC39) (11,12). CDF family members function to decrease the cytoplasmic concentration by transporting zinc from the cytoplasm to the lumen of vesicles or the extracellular space. By contrast, ZIP family members function to increase the cytoplasmic concentration by importing zinc across the plasma membrane or releasing it from intracellular storage sites. Mammals express 10 CDF and 14 ZIP proteins, each with a unique pattern of tissue expression and subcellular localization (11–13). In mammals, high levels of zinc cause transcriptional activation of metallothionein genes and specific CDF transporter genes. This transcriptional response requires the metal response element (MRE) in gene promoters and the trans-acting Metal regulatory Transcription Factor 1(MTF) (14–16). The nematode Caenorhabditis elegans has been demonstrated to be a useful and relevant model organism to study zinc homeostasis, based on its simple anatomy, powerful genetic techniques and conservation of key genes including metallothioneins, CDF and ZIP (17–24). In C. elegans, high zinc homeostasis also involves transcriptional activation of metallothionein and cdf genes. This response is mediated by the high zinc activation (HZA) element in promoters and the HIZR-1 nuclear receptor transcription factor, which is the sensor for high zinc and the master regulator of high zinc homeostasis (25,26). Mammals also display a transcriptional response to zinc deficiency, since the mRNAs of multiple ZIP family members, such as ZIP2, ZIP4, ZIP8 and ZIP10, accumulate when cells are exposed to zinc-deficient conditions (27–29). However, mechanisms of low zinc homeostasis are not well defined, since the sensor for low zinc, the promoter element that mediates transcriptional activation and the corresponding transcription factor have not been identified. In C. elegans there are no reports describing the mechanism of low zinc homeostasis.
To use C. elegans as a model system to characterize the homeostatic response to zinc deficiency, we searched for zip genes that are transcriptionally regulated by zinc deficiency. We discovered that zipt-2.1, zipt-2.3 and zipt-7.1 are strongly activated by zinc deficiency. By analyzing the promoters of these genes, we discovered a novel DNA enhancer, the low zinc activation (LZA) element, that is necessary and sufficient for transcriptional activation in response to zinc-deficient conditions. By searching the C. elegans genome for LZA elements, we identified candidate zinc regulated genes. Several of these genes were demonstrated to be transcriptionally activated by zinc deficiency, documenting the predictive value of the LZA element. Two trans-acting factors that mediate this response were identified, the ELT-2 transcription factor that is the master regulator of intestinal cell identity and the mediator subunit MDT-15. We demonstrated that the LZA element was sufficient to mediate transcriptional activation in response to zinc deficiency in human cells, suggesting that C. elegans and humans have a conserved mechanism of low zinc homeostasis. These results define a new pathway for low zinc homeostasis that is conserved in animals and functions in parallel to the pathway for high zinc homeostasis.
MATERIALS AND METHODS
Caenorhabditis elegans culture and strains
Caenorhabditis elegans strains were cultured at 20°C on nematode growth medium (NGM) dishes seeded with Escherichia coli OP50, except as noted. For zinc deficiency studies, noble agar minimum media (NAMM) was supplemented with N,N,N',N'-tetrakis (2-pyridylmethyl)ethane-1,2-diamine (TPEN), a zinc-specific chelator (Sigma-Aldrich), and dishes were seeded with 5× concentrated OP50 (19). The wild-type C. elegans and parent of all transgenic strains was the Bristol N2 strain (30). The strain WU1500 contains the mutation hizr-1(am286), which is a nonsense mutation that causes a strong loss-of-function in the nuclear receptor transcription factor (25).
Cell culture
The human-derived HEK293T cell line was cultured in Dulbecco’s modified Eagle’s medium (ThermoFisher) containing 10% fetal bovine serum and 1000 U/ml and 1000 g/ml penicillin and streptomycin, respectively (Invitrogen, Carlsbad, CA, USA). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Identification of the LZA element
To identify DNA motifs shared by genes that were transcriptionally activated by zinc deficiency, we utilized the Multiple Em for Motif Elicitation (MEME) version 4.11.2 computational program with the default settings (31). DNA sequences are numbered by defining the A of the start codon as +1. We used the following C. elegans sequences: zipt-7.1 (−847 to −1), zipt-2.3 (−5076 to −1) and zipt-2.1 (−2199 to −1). The UCSC genome browser was used to identify genes homologous to C. elegans zipt-2.1, zipt-2.3 and zipt-7.1 in Caenorhabditis brenneri, Caenorhabditis briggsae and Caenorhabditis japonica and to obtain the promoter regions of these nine genes, defined as the region upstream of the ATG start codon to the coding sequence of the next closest gene.
Plasmid DNA construction and transgenic strain generation
Using C. elegans wild-type genomic DNA as a template, the polymerase chain reaction (PCR) was used to amplify DNA fragments with Phusion polymerase (New England Biolabs) upstream of the coding sequence of zipt-2.3. Amplified DNA was ligated into pBluescript SK+ along with DNA encoding the green fluorescent protein and DNA containing the unc-54 3′ untranslated region. Plasmids pND3, pND7 and pND8 included the regions −2199 to −1, −217 to −1 and −150 to −1, respectively. Starting with plasmid pND3, the zipt-2.3 sequences were modified by using overlap extension PCR to replace the wild-type sequence with a sequence of the same length and base pair composition but with a scrambled order. Plasmids pND10, pND11, pND12 and pND13 modified LZA2 −175 to −156, −177 to −170, −169 to −162 and −161 to −153, respectively. Plasmid pND9 modified LZA1 −236 to −217. Plasmid pND14 modified LZA1 and LZA2 (−236 to −217 and −175 to −156). The pes-10 promoter plasmids were generated by PCR amplifying the following promoter fragments of zipt-2.3: pND15 contains three copies of a 200-bp sequence containing both LZA1 and LZA2 (−290 to −90); pND16 contains three copies of a 70 bp sequence containing only LZA1 (−270 to −200), and pND17 contains three copies of a 110 bp sequence containing only LZA2 (−200 to −90). These fragments were ligated into plasmid pPD107.94 (a gift from A. Fire). All plasmid sequences were confirmed by standard DNA sequencing.
Using pND3 as a template, PCR was used to amplify the wild-type promoter of zipt-2.3, which was ligated into the pGL3 promoter vector (Promega) upstream of a SV40 promoter sequence that minimally drives the expression of a modified firefly luciferase to generate the plasmid pND18. Using pND10 as a template, PCR was used to amplify the promoter of zipt-2.3 that contains the mutated LZA2 sequence, which was ligated into pGL3 to generate the plasmid pND19.
Transgenic animals containing extrachromosomal arrays were generated by injecting a plasmid mixed with pCJF104 as a co-injection marker into wild-type animals (32). The pCJF104 plasmid contains the promoter of myo-3 fused to the coding sequence of mCherry (33). Transgenic animals were selected based on mCherry expression in the body wall muscles. For microscopy, animals were paralyzed in a drop of 1mM sodium azide on a 2% agarose pad on a microscope slide; bright field and fluorescence was visualized using a Zeiss Axioplan 2 microscope equipped with a Zeiss AxioCam MRm digital camera. To compare fluorescence between different strains and culture conditions, images were captured using identical settings and exposure times.
Quantitative real-time PCR (qRT-PCR)
We performed quantitative real-time PCR (qRT-PCR) as previously described with minor modifications (34). Mixed-stage populations of C. elegans were collected by washing and then cultured for 16 h on NAMM dishes seeded with concentrated OP50 and supplemented with 0 or 40 μM TPEN. Animals were collected by washing, and RNA was isolated using the TRIzol reagent (Invitrogen) and treated with Dnase I. cDNAs were synthesized using the High Capacity cDNA Reverse Transcription kit according to manufacturer's protocol (Applied Biosystems). PCR was performed using an Applied Biosystems 7900 thermocycler and iTaq Universal SYBR Green Supermix (Bio-Rad). In all cases, the transcript level was normalized to the transcript level of a reference gene (ama-1 for endogenous genes or mCherry for GFP transgenes) in the same sample. Fold change was determined by dividing the normalized transcript level at 40 μM TPEN by the normalized transcript level at 0 μM TPEN. Sequences of oligonucleotide primers used for qRT-PCR are listed in Supplementary Table S2.
Human HEK293T cells were seeded in 24-well plates at a density to achieve ∼80% confluence after 2 days. When cells achieved this confluency, we replaced the standard growth medium with growth medium containing 0 or 40 μM TPEN and 10 μM Caspase inhibitor III (CalBio). After 3 h incubation, the cells were trypsinized and collected. mRNA was isolated and analyzed by qRT-PCR as described above. The reference gene for human cells was Heat Shock Protein 90 Alpha Family Class B Member 1 (HSPCB).
Identification of candidate LZA elements in the C. elegans genome
We used the Regulatory Sequence Analysis Tools (RSAT) website to obtain the predicted promoter sequence of all annotated C. elegans genes. The predicted promoter sequence is defined as the DNA region from the start codon upstream to the closest gene or −3000 bp. This list of DNA sequences was defined as the C. elegans promoterome and was input into the Find Individual Motif Occurrences (FIMO) program as the sequence database (35). The LZA position weight matrix identified by MEME was used to search the promoterome sequence database to find candidate LZA elements. These were ranked by a scoring system that is computed by summing the appropriate entries from each column of the position-dependent scoring matrix that represents the motif (Supplementary Table S1).
RNA interference
RNA interference experiments were performed as previously described (20). The E. coli HT115(DE3) RNAi bacteria or control bacteria containing the empty L4440 vector were grown in Luria Broth containing ampicillin overnight. The culture was diluted into LB containing ampicillin, grown for 6 h, seeded onto NGM dishes containing ampicillin and isopropyl β-D-1-thiogalactopyranoside (1 mM) and the dishes were cultured at room temperature for 2 days. L1 stage animals were transferred to these dishes, cultured until the L4 stage, transferred to dishes containing 0 or 40 μM TPEN and 5× concentrated OP50 bacteria, cultured overnight and analyzed for GFP expression.
Dual-Glo luciferase reporter assay
HEK293T cells were seeded in 96-well plates to achieve a confluency of ∼70% after 2 days, which we considered optimal for transient transfection. Transfection was performed using Lipofectamine 2000 (ThermoFisher) in a mixture containing 95 ng of plasmid pND18 or pND19 and 5 ng of pRL-TK as a control for transfection efficiency. After 48 h, the medium was replaced with medium containing 0 or 40 μM TPEN and 10 μM Caspase inhibitor III (CalBio), and the cells were incubated for 3 h. To measure luciferase activity, we used the Dual-Glo luciferase kit according to manufacturer’s instructions with minimal modifications. Briefly, 50 μl of the Dual-Glo reagent was added to each well, the plate was incubated for 30 min and luminescence was measured by a Biotek Synergy H1 microplate reader. The Stop-N-Glo reagent was added to each well to quench the firefly luciferase reaction and begin the Renilla luciferase reaction. The plate was incubated for 30 min, and luminescence was measured again. Firefly luciferase values were normalized to the values of Renilla luciferase in the same sample. Each experiment included three technical replicates (three wells treated identically), and three independent biological experiments were also performed.
Statistical analysis
All data were analyzed using the two-tailed unpaired Student’s t-test, and differences with P < 0.05 were considered to be significant.
RESULTS
Identification of a conserved element in the promoters of genes regulated by zinc deficiency
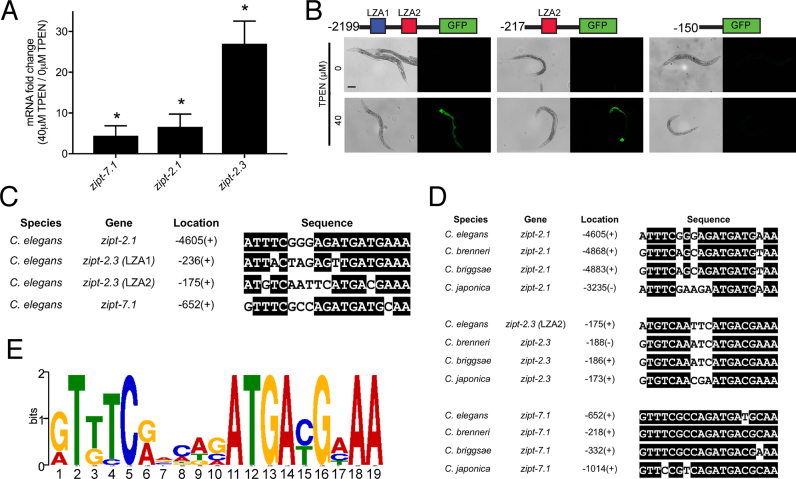
The C. elegans genome encodes 14 predicted members of the ZIP family, and these genes were recently designated zipt (Supplementary Figure S1A). Based on the model that some ZIP proteins function as zinc transporters that increase the level of cytoplasmic zinc, we hypothesized that some zipt genes are regulated by low levels of zinc, thereby promoting zinc homeostasis. To investigate this hypothesis, we used the zinc chelator N,N,N',N'-tetrakis (2-pyridylmethyl)ethane-1,2-diamine (TPEN) to create zinc-deficient culture conditions. A mixed-stage population of wild-type animals was cultured on standard medium, transferred to NAMM dishes containing 0 or 40 μM TPEN for 16 h and harvested for RNA preparation. The mRNA levels of 14 zipt genes were determined by qRT-PCR (Supplementary Figure S1B). Eleven genes did not display significant regulation, whereas zipt-7.1, zipt-2.1 and zipt-2.3 displayed between 4- and 30-fold increased mRNA levels when cultured with TPEN compared to control (Figure 1A). Increased mRNA levels might result from increased transcription or decreased mRNA degradation. To distinguish these models, we analyzed the promoter of zipt-2.3. We generated transgenic animals containing a plasmid with 2199 bp upstream of the start codon of zipt-2.3 fused to the GFP coding sequence. Transgenic animals cultured with TPEN displayed strong induction of GFP, suggesting regulation occurs at the level of transcriptional activation. To begin to identify the regulatory element, we generated transgenic animals with plasmids containing 217 or 150 bp of the zipt-2.3 promoter. The 217 bp promoter fragment was sufficient to mediate activation in response to TPEN treatment, whereas the 150 bp promoter fragment was not sufficient (Figure 1B). These results indicate that a regulatory element between −217 and −150 bp of the promoter of zipt-2.3 was necessary for transcriptional activation in response to zinc-deficient conditions.
Figure 1.
Identification of the LZA element. (A) A population of mixed-stage, wild-type animals were cultured with 0 or 40 μM TPEN for 16 h. RNA was analyzed by qRT-PCR. Values are the ratio of mRNA levels at 40 μM TPEN/0 μM TPEN, an indication of transcriptional activation and are the average of three biological replicates; error bars display standard deviation. Each gene displayed significantly increased mRNA levels at 40 μM TPEN (*P < 0.05). (B) Diagrams of the zipt-2.3 promoter region (not to scale) extending 2199, 217 or 150 bp upstream of the translation start site fused to the GFP coding region. Boxes show LZA1 element (blue) and LZA2 element (red). Transgenic animals containing these constructs were cultured with 0 or 40 μM TPEN for 16 h and imaged with bright field to display worms (left) or fluorescence to display GFP expression (right). Scale bar indicates 100 μm. (C) An alignment of LZA elements from the promoters of Caenorhabditis elegans zipt-2.1, zipt-2.3 and zipt-7.1. Identical residues are highlighted in black. Location is the position of the first base pair of the LZA element relative to the ATG start codon, and +/− refers to the orientation relative to the DNA strand that is transcribed. (D) Sequence alignments of LZA elements in the promoter regions of zipt-2.1, zipt-2.3 and zipt-7.1 in the Caenorhabditis species elegans, brenneri, briggsae and japonica identified by MEME. For zipt-2.3, the LZA2 element in C. elegans is conserved in three other species, and the LZA1 element is not conserved. (E) A position weight matrix of the LZA element based on 12 motifs shown in D. The height of each nucleotide represents the frequency scaled in bits.
We reasoned that these three genes might contain similar sequence element(s) in the promoter region that regulate transcriptional activation in zinc-deficient conditions. To identify such an element, we utilized the computational tool MEME that identifies related DNA motifs in multiple sequences (31). Figure 1C displays the highest scoring motif found in the three promoters; the motif spans 19 bp and has nine highly conserved positions. We named this motif the LZA element. The LZA element was located 4065 bases upstream of the translation start site of zipt-2.1 and 652 bases upstream of the translation start site of zipt-7.1 in C. elegans. Two motifs were identified in the promoter of zipt-2.3, positioned 236 and 175 bp upstream of the translation start site, and LZA2 is in the region necessary for transcriptional activation. If the LZA element mediates transcriptional activation in zinc-deficient conditions, then we predict it will be conserved during evolution. To investigate this possibility, we identified the homologs of zipt-7.1, zipt-2.1 and zipt-2.3 in three additional Caenorhabditis species (C. brenneri, C. briggsae and C. japonica) and analyzed the predicted promoter regions upstream of the translation start sites. MEME detected one LZA element in the promoter of all three genes in all three Caenorhabditis species; for zipt-2.3 the LZA2 element appears to be conserved whereas the LZA1 element is not conserved (Figure 1D). The conservation of the LZA elements appears to be a specific feature of these promoters, because the flanking sequences 20 bases upstream and downstream of the predicted LZA elements did not display substantial conservation (Supplementary Figure S2). Figure 1E shows a weight matrix generated based on twelve conserved motifs from these three genes and four species. This evolutionary conservation in a non-coding region suggests that the LZA element has an important functional role.
The LZA element was necessary for transcriptional activation in response to zinc deficiency
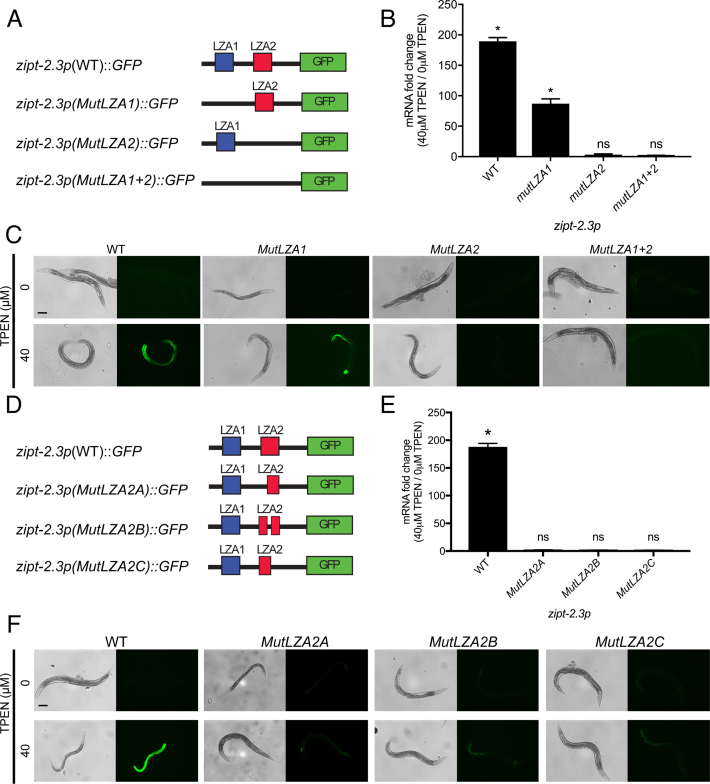
To rigorously analyze the function of the LZA element, we performed a mutational analysis of the zipt-2.3 promoter because it displayed the highest activation in response to zinc-deficient conditions. A plasmid containing 2199 bp of the wild-type zipt-2.3 promoter driving expression of the GFP coding region was modified by randomizing 24 bp of sequence including LZA1, LZA2 or both LZA1 + LZA2 (Figure 2A). Randomizing the sequence of the LZA element maintains the base pair composition and the spacing of elements in the promoter, but is predicted to abrogate interactions with sequence-specific LZA element binding proteins. Four strains of transgenic animals were generated, each containing a multicopy array with one of the plasmids. These strains were cultured in medium with 40 μM TPEN for 16 h to induce zinc deficiency or standard zinc-replete medium as a control. GFP mRNA levels were analyzed by qRT-PCR, and GFP protein expression was analyzed by fluorescence microscopy. A strain with the wild-type promoter displayed a dramatic increase in mRNA levels in zinc-deficient conditions, about 200-fold, and a readily detectable increase in GFP fluorescence in intestinal cells (Figure 2B and C). The fold activation of this reporter construct exceeded the fold activation of the endogenous zipt-2.3 gene, probably because there are multiple copies of the reporter construct. A strain with the LZA1 mutation displayed strong transcriptional activation, about 100-fold and readily detectable increased GFP fluorescence. The extent of mRNA activation was less than wild-type but not statistically different. Thus, LZA1, which is not evolutionarily conserved and is somewhat divergent from the LZA element consensus sequence, was not necessary for transcriptional activation. By contrast, strains containing the LZA2 mutation and the LZA1 + LZA2 mutation did not display GFP fluorescence in response to zinc deficiency, and mRNA analysis revealed significantly reduced activation (Figure 2B and C). These data suggest that the LZA2 element, which is evolutionarily conserved and closely matches the LZA element consensus sequence, is necessary for transcriptional activation of zipt-2.3 in zinc-deficient conditions.
Figure 2.
The LZA element was necessary for transcriptional activation of zipt-2.3 in response to zinc deficiency. (A and D) Diagrams (not to scale) of the zipt-2.3 promoter beginning at base pair −2199 and extending to the ATG translation start codon (black line). Boxes show LZA1 element (blue), LZA2 element (red) and the coding region of green fluorescent protein (green). Mutations created by randomizing the sequence of LZA1 or LZA2 elements are indicated by black lines. MutLZA2A, B and C affect base pairs 1–6, 7–14 and 15–19, respectively, of the 19 bp LZA weight matrix shown in Figure 1E. (B and E) A mixed stage population of transgenic animals containing multicopy arrays of the reporter constructs were cultured on medium with 40 μM TPEN to induce zinc deficiency or control, zinc replete medium (0 μM TPEN). GFP mRNA levels were analyzed by qRT-PCR. Bars depict the change in mRNA levels as the ratio between 40 and 0 μM TPEN, with positive values indicating transcriptional activation in zinc-deficient conditions. Values represent the average and error bars are the standard deviation of three independent biological replicates (*P < 0.01). (C and F) Images show transgenic animals at the young adult stage cultured with 0 or 40 μM TPEN; bright field images (left) show worm morphology, and fluorescent images (right) show GFP fluorescence. GFP signals were captured with identical settings and exposure times. Scale bar indicates 100 μm.
To further dissect the function of different portions of the LZA2 element, we used site–directed mutagenesis to randomize the sequence of base pairs 1–6, 7–14 or 15–19 (Figure 2D). Transgenic strains containing plasmids with these mutant promoters displayed almost no transcriptional activation in zinc-deficient conditions and very low levels of GFP fluorescence, a significant difference compared to the wild-type promoter (Figure 2E and F). Thus, mutations of base pairs 1–6, 7–14 and 15–19 disrupted the function of the LZA element similar to mutation of base pairs 1–19, indicating that functionally critical base pairs occur throughout the LZA sequence.
The LZA element was sufficient to promote transcriptional activation in zinc-deficient conditions
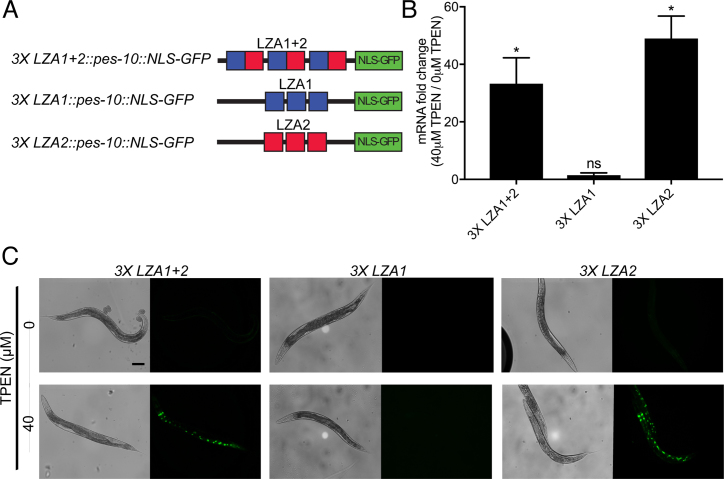
To determine if the LZA element is sufficient to mediate transcriptional activation in zinc-deficient conditions, we introduced the motif into a minimal worm promoter that expresses nuclear-localized GFP in the intestinal cells. We chose the pes-10 promoter because it is not zinc regulated and it has been used to successfully study multiple cis-regulatory elements (26,36). We generated plasmids with three copies of the following zipt-2.3 promoter fragments positioned upstream of the pes-10 promoter: (i) 200 bp containing both LZA1 and LZA2 (−290 to −90); (ii) 70 bp containing only LZA1 (−270 to −200); (iii) 110 bp containing only LZA2 (−200 to −90) (Figure 3A). Transgenic strains containing multicopy arrays of these plasmids were cultured on control, zinc replete medium or medium containing 40 μM TPEN for 16 h to induce zinc deficiency. The strain containing LZA1+2 elements displayed no GFP fluorescence when cultured in control medium, but displayed bright GFP expression in the nuclei of intestinal cells when cultured in zinc-deficient conditions. Furthermore, GFP mRNA levels increased approximately 30-fold in zinc-deficient conditions (Figure 3B and C). Thus, the combination of LZA1 and LZA2 elements was sufficient to mediate transcriptional activation in zinc-deficient conditions. A pes-10 promoter containing only the LZA2 element also mediated robust activation in zinc-deficient conditions, with the mRNA levels increasing about 40-fold and the appearance of bright expression of GFP protein. By contrast, the pes-10 promoter containing only the LZA1 element did not mediate increased mRNA levels. This strain displayed a low level of nuclear GFP fluorescence in several posterior intestinal cells of animals in zinc-replete and zinc-deficient conditions (Figure 3B and C). These results indicate that the LZA2 element, but not the LZA1 element, was sufficient to mediate transcriptional activation in zinc-deficient conditions, just as the LZA2 element, but not the LZA1 element, was necessary for transcriptional activation in zinc-deficient conditions.
Figure 3.
The LZA element was sufficient to mediate transcriptional activation in response to zinc deficiency in a heterologous promoter. (A) Diagrams (not to scale) of the pes-10 promoter (black line). Boxes show the LZA1 element (blue), the LZA2 element (red) and the coding region of green fluorescent protein with a nuclear localization sequence (green). (B) A mixed stage population of transgenic animals containing multicopy arrays of the reporter constructs were cultured on medium with 40 μM TPEN to induce zinc deficiency or control, zinc replete medium (0 μM TPEN). GFP mRNA levels were analyzed by qRT-PCR. Bars depict the change in mRNA levels as the ratio between 40 and 0μM TPEN, with positive values indicating transcriptional activation in zinc-deficient conditions. Values represent the average and standard deviation of 3 independent biological replicates (*P < 0.01). (C) Images show transgenic animals at the young adult stage cultured with 0 or 40 μM TPEN; bright field images (left) show worm morphology, and fluorescent images (right) show GFP fluorescence. GFP signals were captured with identical settings and exposure times. Green puncta are the nuclei of intestinal cells. Scale bar indicates 100 μm.
The LZA element can be used to predict genes that are transcriptionally activated by zinc-deficient conditions
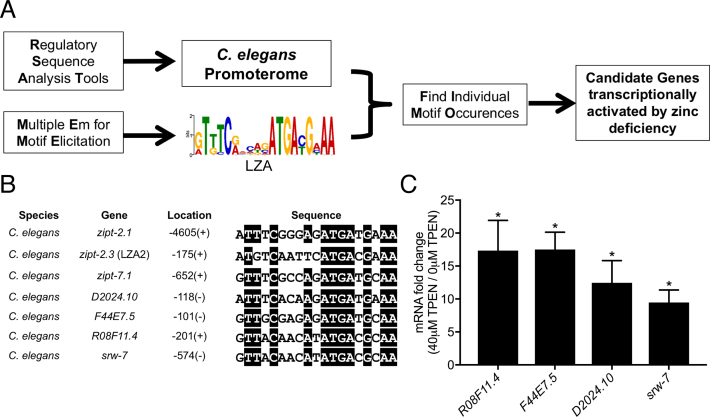
The identification of multiple zipt genes with LZA elements in the promoter, and the demonstration that the LZA element is necessary and sufficient for transcriptional activation in zinc-deficient conditions, led us to hypothesize that additional genes regulated by low zinc conditions will contain an LZA element in the promoter. To identify such genes, we used the RSAT website to obtain the promoterome of C. elegans (37). This database was assembled by collecting the upstream sequence from each annotated gene in the C. elegans genome, beginning at the ATG start codon of the identified gene and ending at the stop codon of the closest upstream gene or −3000 bp. We defined the LZA element as the weight matrix shown in Figure 1E. The bioinformatic tool FIMO was used to search through the C. elegans promoterome for occurrences of the LZA element (35). This approach generated a list of genes whose promoters contained a predicted LZA element, and the list is ranked based on the similarity of the identified sequence to the LZA weight matrix (Figure 4A). Supplementary Table S1 shows the top 50 candidates from this ranked list. To begin to determine if this list contains genes that are regulated by zinc deficiency, we isolated mRNA from a mixed-stage population of wild-type animals cultured on NAMM plates containing 0 or 40 μM TPEN for 16 h and analyzed the mRNA levels of 15 of the top 30 predicted genes. Four of these genes displayed significant transcriptional activation in zinc-deficient conditions compared to zinc-replete control conditions (Figure 4C). R08F11.4 displayed an 18-fold increase, F44E7.5 displayed a 19-fold increase, D2024.10 displayed a 13-fold increase, and srw-7 displayed a 10-fold increase. The LZA elements in these four promoters contain all nine of the invariant residues in the LZA weight matrix, reinforcing the importance of these residues (Figure 4B). We generated a LZA weight matrix using the seven C. elegans genes shown in Figure 4B that were experimentally demonstrated to be induced by zinc-deficient conditions (Supplementary Figure S3); the weight matrix was very similar but not identical to the weight matrix shown in Figure 1E. These results demonstrate that the presence of an LZA element can be used to successfully predict transcriptional activation of genes in response to zinc-deficient conditions.
Figure 4.
The LZA element can be used to predict genes regulated by zinc deficiency in Caenorhabditis elegans. (A) An overview of the strategy used to identify candidate genes regulated by zinc deficiency. The Regulatory Sequence Analysis Tools database was used to obtain the predicted promoters of all C. elegans genes (C. elegans promoterome). MEME was used to generate the LZA weight matrix. The Find Individual Motif Occurrences program was used to search the promoterome for matches to the LZA weight matrix, generating a list of candidate genes. (B) Sequence alignments of the LZA elements in the promoter regions of C. elegans genes zipt-7.1, zipt-2.1 and zipt-2.3, which were used to generate the LZA weight matrix, and D2024.10, F44E7.5, R08F11.4 and srw-7, which were identified in the genome-wide LZA element search. Identical residues are highlighted in black. (C) A population of mixed-stage, wild-type animals were cultured with 0 or 40 μM TPEN for 16 h. RNA was analyzed by qRT-PCR. Values are the ratio of mRNA levels at 40 μM TPEN/ 0 μM TPEN, an indication of transcriptional activation, and are the average of three biological replicates; error bars display standard deviation. Each gene displayed significantly increased mRNA levels at 40 μM TPEN (*P < 0.01).
The LZA element-mediated transcriptional activation in human cells in response to zinc deficiency
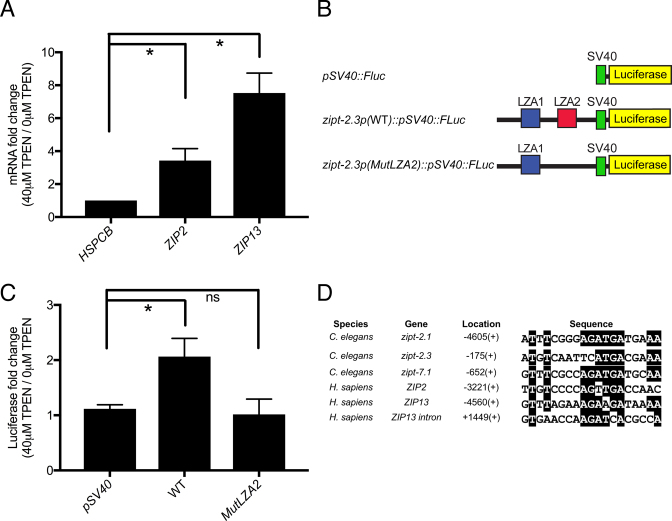
To explore the possibility that the function of the LZA has been conserved during animal evolution, we analyzed human embryonic kidney (HEK293T) cells. To identify human ZIP genes that are activated by zinc deficiency, we cultured HEK293T cells in the presence and absence of 40 μM TPEN for 3 h, purified RNA and measured the levels of all 14 identified human ZIP transcripts by qRT-PCR. Transcripts of two human ZIP genes, ZIP2 and ZIP13, displayed significantly higher levels in zinc-deficient conditions compared to control, zinc-replete conditions (Figure 5A). These results confirm previous reports of ZIP family members whose mRNA is induced by zinc deficiency (27–29) and validate that our experimental conditions are suitable for performing functional tests of gene activation in conditions of zinc deficiency. To identify LZA elements in these genes, we used the program MEME to search the promoters and first introns of human ZIP2 and ZIP13 for matches to the LZA weight matrix. We identified one candidate LZA element in the promoter of ZIP2 and two in ZIP13, one in the promoter and one in the first intron (Figure 5D). These results indicate that there may be functional conservation of the LZA element in mediating transcriptional activation in response to zinc deficiency in C. elegans and human cells.
Figure 5.
The LZA element functioned in human cells. (A) HEK293T cells were cultured in growth medium containing 0 or 40 μM TPEN for 3 h, and RNA was analyzed by qRT-PCR. Values are the ratio of mRNA levels at 40 μM TPEN/ 0 μM TPEN, an indication of transcriptional activation and are the average of three biological replicates and the standard deviation. In response to zinc deficiency, transcripts of the control gene Heat Shock Protein 90 Alpha Family Class B Member 1 (HSPCB) did not accumulate, whereas transcripts of ZIP2 and ZIP13 displayed significant accumulation (*P < 0.01). (B) Diagrams (not to scale) of the SV40 promoter (green) driving expression of the coding region of Firefly luciferase protein (yellow). The zipt-2.3 promoter region extended from −2199 to the ATG start codon (black line) and was cloned upstream of the SV40 promoter. Boxes show the LZA1 element (blue) and the LZA2 element (red); the LZA2 element was mutated by randomizing the order of 24 base pairs (MutLZA2). (C) Plasmids were co-transfected with a plasmid expressing renilla luciferase as a control for transfection efficiency into HEK293T cells. Forty-eight hours post-transfection, cells were incubated with 0 or 40 μM TPEN for 3 h and extracts were analyzed for firefly and renilla luciferase enzyme activity. Values are the ratio of normalized firefly luciferase enzyme activity at 40 μM TPEN/0 μM TPEN, an indication of transcriptional activation and are the average of three biological replicates and the standard deviation (*P < 0.01). (D) Sequence alignments of the LZA elements in the promoter regions of Caenorhabditis elegans genes zipt-7.1, zipt-2.1 and zipt-2.3, which were used to generate the LZA weight matrix, the promoter regions of human ZIP2 and ZIP13, and the intron of human ZIP13, which were identified in gene-specific searches for LZA elements. Identical residues are highlighted in black. Location is the position of the first base pair of the LZA element relative to the ATG start codon, and +/− refers to the orientation relative to the DNA strand that is transcribed.
To directly test the function of the LZA element in human cells, we cloned the promoter region of C. elegans zipt-2.3 into a plasmid containing the SV40 promoter driving expression of firefly luciferase. The plasmid was transiently transfected into HEK293T cells along with a plasmid driving expression of Renilla luciferase as a control for transfection efficiency (Figure 5B). These cells were then cultured in TPEN-containing medium or control medium lacking TPEN for 3 h, and the level of firefly luciferase enzyme activity was measured and normalized to the level of Renilla luciferase enzyme activity from the same samples; the level of firefly luciferase enzyme activity is an indirect measure of mRNA levels and transcriptional activity. The unmodified plasmid displayed a similar level of firefly luciferase activity in zinc deficient and control medium, indicating that the SV40 promoter is not regulated by zinc deficiency (Figure 5C). By contrast, the promoter that contained the zipt-2.3 sequence displayed a statistically significant, 2-fold increase in firefly luciferase activity in zinc deficient medium compared to control medium. These results indicate that the zipt-2.3 promoter region is sufficient to confer transcriptional activation in response to zinc deficiency in human cells. To test the role of the LZA element in this response, we used site directed mutagenesis to scramble the sequence of LZA2. The mutated promoter displayed a similar level of luciferase activity in zinc deficient and control medium, a result that was not statistically different from the unmodified plasmid (Figure 5C). Thus, the LZA element was necessary for the transcriptional activation response to zinc deficiency. These findings indicate that the mechanism used by C. elegans to transcriptionally respond to zinc-deficient conditions appears to be conserved in human cells.
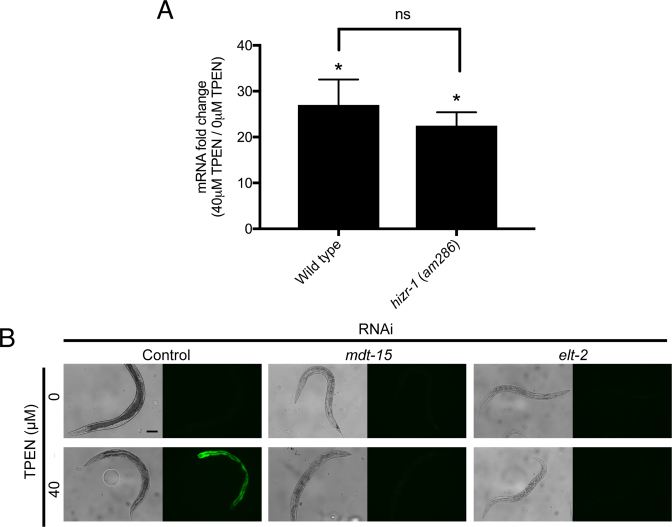
The mediator subunit gene mdt-15 and the GATA transcription factor gene elt-2 were necessary for transcriptional activation in response to zinc deficiency
To identify proteins that promote transcriptional regulation of LZA element-containing promoters, we used a candidate approach. In C. elegans, transcriptional activation in response to high dietary zinc is mediated by the HZA element and requires the HIZR-1 nuclear receptor transcription factor that binds the HZA element, the ELT-2 GATA transcription factor that binds GATA elements in genes expressed in intestinal cells and the MDT-15 subunit of the mediator complex that is necessary for a variety of transcriptional responses to environmental cues (25,26,38). Searches for genes that contain a HZA element identified zipt-2.3, raising the possibility that hizr-1 may play a regulatory role (25,26). To test this possibility, we cultured a mixed stage population of hizr-1(lf) mutant animals on medium containing 0 or 40 μM TPEN for 16 h and used qRT-PCR to measure the level of zipt-2.3 mRNA. hizr-1(lf) and wild-type animals both displayed robust accumulation of zipt-2.3 mRNA in zinc-deficient conditions, suggesting that hizr-1 is not necessary for this transcriptional regulation (Figure 6A). To determine if elt-2 or mdt-15 play a regulatory role, we analyzed transgenic animals containing zipt-2.3p(WT)::gfp as a reporter. To reduce gene activity, we cultured animals with bacteria expressing double stranded RNA from the elt-2 or mdt-15 genes or control RNAi bacteria. These animals were then transferred to medium containing 40 μM TPEN for 16 h and GFP expression was analyzed by fluorescence microscopy. Compared to control RNAi bacteria, animals exposed to both mdt-15 and elt-2 RNAi bacteria demonstrated a dramatic reduction of GFP expression (Figure 6B). These results suggest that mdt-15 and elt-2 are necessary for the transcriptional activation of zipt-2.3 mRNA in zinc-deficient conditions. We propose that ELT-2 protein binds to GATA elements in the zipt-2.3 promoter, and MDT-15 is part of the mediator complex that is assembled at this promoter in response to zinc-deficient conditions (Figure 7B).
Figure 6.
mdt-15 and elt-2 were necessary for transcriptional activation of zipt-2.3 in zinc-deficient conditions. (A) Populations of mixed-stage, wild-type or hizr-1(am286) animals were cultured with 0 or 40 μM TPEN for 16 h. RNA was analyzed by qRT-PCR. Values are the ratio of mRNA levels at 40 μM TPEN/0 μM TPEN, an indication of transcriptional activation, and are the average of three biological replicates and the standard deviation. zipt-2.3 mRNA displayed significant accumulation to similar levels in both genotypes. (B) Transgenic animals containing zipt-2.3p(WT)::GFP were cultured on the indicated RNAi bacteria from the L1 to the young adult stage and then transferred to medium containing 0 or 40 μM TPEN for 16 h. Bright field images (left) show worm morphology, and fluorescent images (right) show GFP fluorescence. GFP signals were captured with identical settings and exposure times. Scale bar indicates 100 μm.
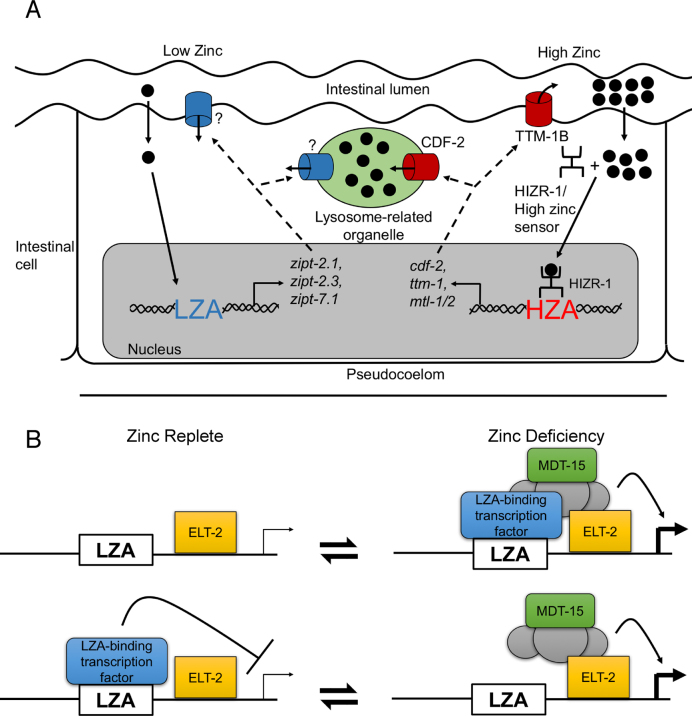
Figure 7.
Models of transcriptional regulation during zinc deficiency. (A) Diagram of an intestinal cell flanked by the intestinal lumen (above) and the pseudocoelom (below). Two dietary zinc-conditions are illustrated: deficiency (left) and excess (right). Zinc (black circles) enters the intestinal cell, and an undefined sensing mechanism causes activation of LZA element containing promoters. ZIPT proteins (blue) might localize to the plasma membrane or lysosome related organelles (LRO, green), but the subcellular localization has not been established. During zinc excess, the HIZR-1 nuclear receptor causes activation of HZA element containing promoters and the CDF-2 protein (red) localizes to the LRO where it functions to store zinc, and the TTM-1B protein (red) localizes to the plasma membrane where it functions to excrete zinc. (B) Two models of LZA element mediated transcriptional regulation. In the upper model, a proposed LZA element binding protein (blue box) is a transcriptional activator that interacts with the LZA element in response to zinc deficiency; this triggers assembly of a transcriptional activation complex including MDT-15 (green box). ELT-2 (yellow box) constitutively binds GATA elements and is permissive for intestinal cell expression. Stimulation is shown by an arrow and thick transcription start site. In the lower model, the LZA-element binding protein is a transcriptional repressor that interacts with the LZA element during zinc-replete conditions. Repression is shown by a bar and thin transcription start site; zinc deficiency abrogates the binding interaction, allowing assembly of a transcriptional activation complex. These two models are not mutually exclusive, since the LZA element binding protein might be converted from a repressor in zinc-replete conditions to an activator in zinc-deficient conditions.
DISCUSSION
Discovery and characterization of the LZA element, an enhancer that mediated transcriptional activation in response to low zinc conditions in C. elegans
Because zinc is essential for multiple functions in biological systems, homeostatic mechanisms have evolved that promote zinc availability when animals are confronted with low dietary zinc. We used C. elegans to investigate these mechanisms by identifying three zipt genes that are transcriptionally activated in response to zinc-deficient conditions. Because ZIP proteins function as zinc transporters and play critical roles in zinc biology, the regulation of these genes is likely to increase zinc concentrations in the cytoplasm and have important physiological consequences. A bioinformatic analysis of these three promoters revealed that each contained a similar, 19 bp motif that we named the LZA element. The LZA element was evolutionarily conserved in the promoters of the zipt-2.1, zipt-2.3 and zipt-7.1 genes in three other Caenorhabditis species, C. brenneri, C. briggsae and C. japonica, suggesting that it has an important function. To directly test whether the LZA element mediates transcriptional activation, we characterized the promoter of C. elegans zipt-2.3, because this gene demonstrated the highest level of mRNA accumulation in response to zinc-deficient conditions. The LZA element was both necessary for transcriptional activation of the zipt-2.3 promoter in zinc-deficient conditions and sufficient to confer low zinc activation on a minimal pes-10 promoter. Because the LZA element can function at variable distances from the transcription start site and in both orientations, we conclude that the LZA element is a DNA enhancer. The 19 bp LZA element contains nine highly conserved positions, including a central ATGA sequence. Critical residues occur throughout the motif, since mutations of base pairs 1–6, 7–14, or 15–19 abrogated activity. This motif has not been previously implicated in zinc homeostasis.
DNA elements that mediate the transcriptional response to low zinc have been described in yeast. In Saccharomyces cerevisiae, the Zap1 transcription factor binds to the Zinc Responsive Element (ZRE) in multiple promoters in response to low zinc conditions, thereby promoting transcriptional activation (39). In Schizosaccharomyces pombe, the Loz1 transcriptional repressor binds to a GN(A/C)GATC element in multiple promoters in zinc-replete conditions; Loz1 binding is lost in low zinc conditions, thereby causing transcriptional activation (40,41). Enhancer elements that mediate transcriptional activation in response to low zinc have not been identified in animals, and the sequence of the LZA element is not similar to the ZRE in S. cerevisiae or Loz1 binding site in S. pombe. Enhancer elements that mediate the response to high zinc have been defined in animals. In mammals, the MTF-1 transcription factor binds to the MRE, promoting transcriptional activation in high zinc conditions (14–16,42). The zinc transcriptional regulatory element (ZTRE) mediates transcriptional repression in high zinc conditions. The ZTRE is bound by the zinc finger protein ZNF658 which is similar to yeast Zap1 (43,44). The sequence of the ZTRE is distinct from the sequence of the LZA element. In C. elegans, the HIZR-1 transcription factor binds the HZA enhancer element to promote transcriptional activation in response to high dietary zinc (25). The sequence of the LZA element is not similar to the sequence of the MRE or HZA element, indicating that animals use separate enhancer elements to mediate transcriptional activation in response to high and low zinc conditions. Furthermore, the HIZR-1 transcription factor that is the master regulator of high zinc homeostasis was not necessary for the transcriptional response to zinc deficiency. These results indicate that the pathway for low zinc homeostasis, which includes the LZA element and ZIPT zinc transporters, functions in parallel to the pathway for high zinc homeostasis, which includes HIZR-1, the HZA element and CDF zinc transporters (Figure 7A).
The GATA transcription factor ELT-2 and the mediator complex subunit MDT-15 were necessary for transcriptional activation in response to zinc-deficient conditions
Enhancer elements typically function by directly interacting with sequence-specific DNA-binding transcription factors. For example, the HIZR-1 transcription factor directly binds the HZA element, thereby activating transcription in response to high dietary zinc (25). Therefore, we hypothesize the existence of an LZA element binding factor that has yet to be identified. The LZA element binding factor might be a transcriptional activator that binds the LZA element during zinc-deficient conditions, analogous to the mechanism of yeast Zap1. Alternatively, the LZA element binding factor might be a transcriptional repressor that binds the LZA element during zinc-replete conditions and dissociates during zinc-deficient conditions, analogous to the mechanism of yeast Loz1 (Figure 7B). (39,40). A variety of strategies can now be used to search for the LZA element binding factor, including biochemical approaches to identify an interacting protein and genetic approaches to identify genes that are necessary for the LZA element to function in transcriptional activation.
The zipt-2.3 promoter was activated specifically in intestinal cells. To explore the basis for this tissue-specific regulation, we analyzed the ELT-2 transcription factor. ELT-2 protein binds GATA elements that are present in the promoters of all genes that are expressed in the intestinal cells, and ELT-2 is a critical regulator of intestinal cell fates (45,46). Reducing the activity of elt-2 after the developmental stage when it is required to form the intestine abrogated the activation of the zipt-2.3 promoter in response to zinc deficiency. The zipt-2.3 promoter contains at least five predicted GATA motifs that might mediate this effect. This result is complementary with previous studies indicating that the ELT-2 transcription factor plays a role in gene expression in response to high levels of zinc, cadmium, iron and heme (26,45–47). Thus, ELT-2 may function in the homeostasis of many metals and metal-related factors. However, ELT-2 expression is not responsive to zinc levels, suggesting that another factor senses and responds to low zinc, while ELT-2 is likely to play a permissive role by mediating intestine specific expression. These findings indicate that multiple enhancers, including GATA and LZA elements, are responsible for tissue specific and environmentally responsive gene expression during low zinc homeostasis.
We propose that the DNA-binding transcription factors assemble a complex of coactivators to promote gene expression. The mediator complex protein MDT-15 is involved in regulating the expression of detoxifying enzymes in response to ingested materials, including metals and MDT-15 is necessary for transcriptional activation of the HZA element in response to high zinc (26, 38). MDT-15 is the homolog of the Med15 protein in other organisms that has been proposed to interact with a variety of DNA binding protein factors (48–50). Thus, we hypothesized that mdt-15 might play a role in the response to low zinc. Consistent with this model, MDT-15 was necessary for transcriptional activation in response to zinc deficiency, suggesting that the mediator complex is assembled at the zipt-2.3 promoter to activate transcription in response to low zinc. One possible model is that ELT-2 binding to a GATA element and an undefined LZA element binding protein recruit the mediator complex to activate transcription in response to zinc-deficient conditions. Alternatively, the undefined LZA element binding protein represses transcription in zinc-replete conditions and dissociates in zinc-deficient conditions, allowing the mediator complex to assemble and activate transcription (Figure 7B).
Identification of genes regulated by zinc deficiency in C. elegans
We used two approaches to identify genes that are regulated by zinc-deficient conditions. First, a candidate approach was used to focus on the 14 zipt genes, because the ZIP family of metal transporters are established to play critical roles in zinc biology. Three zipt genes displayed significant mRNA accumulation in animals exposed to zinc-deficient conditions, whereas 11 zipt genes did not display substantial regulation. This is a novel demonstration in C. elegans that zipt genes are regulated in zinc-deficient conditions. The degree of regulation was substantial, with mRNA levels of zipt-2.3, zipt-2.1 and zipt-7.1 increasing 27-, 6- and 4-fold, respectively. In the case of the zipt-2.3 gene, our results indicate that mRNA accumulation is due to increased transcription, since the promoter region is sufficient to confer this property on a heterologous GFP mRNA. The second approach took advantage of the newly discovered LZA element; we hypothesized that this motif is present in the promoters of additional genes that are transcriptionally activated in response to zinc-deficient conditions. The genome of C. elegans is extensively characterized, and we used bioinformatic techniques to search for LZA elements in predicted promoter regions of each gene and rank discovered motifs based on their degree of similarity to the LZA weight matrix. Among the genes with the highest scoring LZA elements were zipt-7.1 and zipt-2.3, as expected. We analyzed a subset of high ranking genes, and four displayed significant mRNA accumulation in zinc-deficient conditions. These results support the conclusion that the LZA element mediates transcriptional regulation in zinc-deficient conditions and establishes that the LZA weight matrix has enough information to successfully predict novel genes induced by zinc deficiency in C. elegans. Thus, we have identified seven genes that are substantially induced by zinc-deficient conditions. The ZIP transporters likely function to increase the cytoplasmic levels of zinc, thus promoting low zinc homeostasis. The gene srw-7 encodes a predicted G-protein coupled receptor that is homologous to human genes GPR139 and GPR142. The genes R08F11.4, F44E7.5 and D2024.10 are not clearly homologous to human genes. The function of these four newly identified genes in C. elegans has not been reported, so their functional role during zinc deficiency remains to be analyzed. Based on the observed regulatory control, we hypothesize that these four novel genes might act by a variety of mechanisms to promote physiological processes that are adaptive in the face of zinc deficiency. These results establish the foundation for future experiments to determine the complete set of genes that are regulated by zinc-deficient conditions and the functional significance of these genes for zinc homeostasis.
The homeostatic response to low zinc in eukaryotes has been characterized most extensively in Saccharomyces sp. and in Arabidopsis thaliana (40,41,51,52), whereas relatively little is known in animals. In S. cerevisiae, over 80 direct targets of the Zap1 transcription factor have been identified; some target genes directly affect zinc levels, such as ZIP transporters, whereas others mediate adjustments that promote survival in a low zinc environment, such as expression of enzymes that do not require a zinc cofactor (53). In mammals, genes encoding ZIP family members, including ZIP2 and ZIP10, are transcriptionally activated during zinc deficiency (27,28). The mRNA of ZIP4 is stabilized during zinc deficiency to increase expression of the transporter, and the levels of ZIP7 protein are increased in zinc deficiency (29,54). The results described here indicate that the regulation of ZIP transporters is a highly conserved mechanism of low zinc homeostasis and establish a new animal model that can be used to develop a comprehensive understanding of low zinc homeostasis.
Evolutionary conservation of LZA element function in human cells
To test the function of the LZA element in mammals, we first established appropriate zinc-deficiency conditions using the HEK293T cell line derived from human embryonic kidney cells. As with C. elegans we focused on the ZIP genes as candidates for regulatory control. Following 3 h of treatment with the zinc chelator TPEN, the mRNAs of human ZIP2 and ZIP13 displayed significant accumulation, whereas the other twelve ZIP genes did not display substantial regulation. Regulation of ZIP2 in response to zinc deficiency has been described in keratinocytes, human peripheral blood mononuclear and THP-1 cells, and our observation in kidney cells supports the conclusion that this gene plays a widespread role in homeostatic regulation (27,55). Caenorhabditis elegans zipt-2.1 and zipt-2.3, which are also regulated by zinc deficiency, are homologous to the human ZIP2 gene (27,56). Based on these observations, we hypothesized that there is a conserved mechanism of regulation of ZIP2 genes and a conserved function of ZIP2 proteins in the homeostatic response to low zinc-conditions in humans and C. elegans. To begin to test this hypothesis, we engineered the promoter of C. elegans zipt-2.3 into a plasmid with a minimal SV40 promoter and monitored expression in human cells. The C. elegans promoter was sufficient to mediate transcriptional activation in response to zinc-deficient conditions in human cells. Furthermore, site-directed mutagenesis demonstrated that the LZA element was necessary for this regulatory control. These results indicate the existence of an evolutionarily conserved mechanism in worms and humans that is capable of recognizing the LZA element and promoting transcriptional activation in response to zinc deficiency. The promoter regions of human ZIP2 and ZIP13 contain candidate LZA elements, suggesting these may contribute to regulation during zinc deficiency, but the function of these candidate LZA elements has not been analyzed experimentally.
While multiple ZIP genes have been reported to be regulated by zinc-deficient conditions in mammals, there is limited information about the mechanism of this transcriptional response. For ZIP10, it is proposed that the MTF-1 transcription factor represses transcription in zinc-replete conditions and loss of repression causes transcriptional activation in zinc-deficient conditions (57). For ZIP4, it is proposed that KLF-4, a kruppel-like zinc finger transcription factor (58), binds to the promoter of ZIP4 and activates transcription in zinc-deficient conditions (59–61). The identification of the LZA element, a novel enhancer that mediates the response to zinc-deficient conditions in C. elegans and human cells, is an important contribution to the understanding of low zinc homeostasis in animals and establishes the experimental foundation for the identification of the low zinc sensor and DNA binding transcription factor that mediates this conserved response.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Caenorhabditis Genetics Center for providing strains, Krupa Deshmukh for assistance with phylogenetic analysis and Kurt Warnhoff for discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health [R01GM068598–09 to K.K.]. Funding for open access charge: National Institute of Health [R01GM068598–09 to K.K.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gaither L.A., Eide D.J.. Eukaryotic zinc transporters and their regulation. Biometals. 2001; 14:251–270. [DOI] [PubMed] [Google Scholar]
- 2. Vallee B.L., Falchuk K.H.. The biochemical basis of zinc physiology. Physiol. Rev. 1993; 73:79–118. [DOI] [PubMed] [Google Scholar]
- 3. Andreini C., Banci L., Ivano B., Rosato A.. Counting the zinc-proteins encoded in the human genome. J. Proteome Res. 2005; 5:196–201. [DOI] [PubMed] [Google Scholar]
- 4. Yamasaki S., Sakata-Sogawa K., Hasegawa A., Suzuki T., Kabu K., Sato E., Kurosaki T., Yamashita S., Tokunaga M., Nishida K. et al. . Zinc is a novel intracellular second messenger. J. Cell Biol. 2007; 177:637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim A.M., Bernhardt M.L., Kong B.Y., Ahn R.W., Vogt S., Woodruff T.K., O’Halloran T. V.. Zinc sparks are triggered by fertilization and facilitate cell cycle resumption in mammalian eggs. ACS Chem. Biol. 2011; 6:716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hambridge K. Zinc and chromium in human nutrition. J. Hum. Nutr. 1978; 32:99–110. [PubMed] [Google Scholar]
- 7. Prasad A.S. Discovery of human zinc deficiency: 50 years later. J. Trace Elem. Med. Biol. 2012; 26:66–69. [DOI] [PubMed] [Google Scholar]
- 8. Wang K., Zhou B., Kuo Y.-M., Zemansky J., Gitschier J.. A novel member of a zinc transporter family is defective in acrodermatitis enteropathica. Am. J. Hum. Genet. 2002; 71:66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Küry S., Dréno B., Bézieau S., Giraudet S., Kharfi M., Kamoun R., Moisan J.-P.. Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat. Genet. 2002; 31:239–240. [DOI] [PubMed] [Google Scholar]
- 10. Hambidge M. Human zinc deficiency. J. Nutr. 2000; 130:1344S–1349S. [DOI] [PubMed] [Google Scholar]
- 11. Kambe T., Suzuki T., Nagao M., Yamaguchi-Iwai Y.. Sequence similarity and functional relationship among eukaryotic ZIP and CDF transporters. Genomics Proteomics Bioinformatics. 2006; 4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kambe T., Tsuji T., Hashimoto A., Itsumura N.. The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol. Rev. 2015; 95:749–784. [DOI] [PubMed] [Google Scholar]
- 13. Lichten L.A., Cousins R.J.. Mammalian zinc transporters: nutritional and physiologic regulation. Annu. Rev. Nutr. 2009; 29:153–176. [DOI] [PubMed] [Google Scholar]
- 14. Giedroc D.P., Chen X., Apuy J.L.. Metal response element (MRE)-binding transcription factor-1 (MTF-1): structure, function, and regulation. Antioxid. Redox Signal. 2001; 3:577–596. [DOI] [PubMed] [Google Scholar]
- 15. Brugnera E., Georgiev O., Radtke F., Heuchel R., Baker E., Sutherland G.R., Schaffner W.. Cloning, chromosomal mapping and characterization of the human metal-regulatory transcription factor MTF-1. Nucleic Acids Res. 1994; 22:3167–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heuchel R., Radtke F., Georgiev O., Stark G., Aguet M., Schaffner W.. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J. 1994; 13:2870–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jakubowski J., Kornfeld K.. A local, high-density, single-nucleotide polymorphism map used to clone Caenorhabditis elegans cdf-1. Genetics. 1999; 153:743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bruinsma J.J., Jirakulaporn T., Muslin A.J., Kornfeld K.. Zinc ions and cation diffusion facilitator proteins regulate ras-mediated signaling. Dev. Cell. 2002; 2:567–578. [DOI] [PubMed] [Google Scholar]
- 19. Bruinsma J.J., Schneider D.L., Davis D.E., Kornfeld K.. Identification of mutations in Caenorhabditis elegans that cause resistance to high levels of dietary zinc and analysis using a genomewide map of single nucleotide polymorphisms scored by pyrosequencing. Genetics. 2008; 179:811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roh H.C.C., Collier S., Guthrie J., Robertson J.D.D., Kornfeld K.. Lysosome-related organelles in intestinal cells are a zinc storage site in C. elegans. Cell Metab. 2012; 15:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roh H.C., Collier S., Deshmukh K., Guthrie J., Robertson J.D., Kornfeld K.. ttm-1 encodes CDF transporters that excrete zinc from intestinal cells of C. elegans and act in a parallel negative feedback circuit that promotes homeostasis. PLoS Genet. 2013; 9:e1003522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murphy J.T., Bruinsma J.J., Schneider D.L., Collier S., Guthrie J., Chinwalla A., Robertson J.D., Mardis E.R., Kornfeld K.. Histidine protects against zinc and nickel toxicity in Caenorhabditis elegans. PLoS Genet. 2011; 7:e1002013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Warnhoff K., Murphy J.T., Kumar S., Schneider D.L., Peterson M., Hsu S., Guthrie J., Robertson J.D., Kornfeld K.. The DAF-16 FOXO transcription factor regulates natc-1 to modulate stress resistance in Caenorhabditis elegans, linking insulin/IGF-1 signaling to protein N-terminal acetylation. PLoS Genet. 2014; 10:e1004703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dietrich N., Tan C.-H., Cubillas C., Earley B.J., Kornfeld K.. Insights into zinc and cadmium biology in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2016; 611:120–133. [DOI] [PubMed] [Google Scholar]
- 25. Warnhoff K., Roh H.C., Kocsisova Z., Tan C.-H., Morrison A., Croswell D., Schneider D.L., Kornfeld K.. The nuclear receptor HIZR-1 uses zinc as a ligand to mediate homeostasis in response to high zinc. PLoS Biol. 2017; 15:e2000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roh H.C., Dimitrov I., Deshmukh K., Zhao G., Warnhoff K., Cabrera D., Tsai W., Kornfeld K.. A modular system of DNA enhancer elements mediates tissue-specific activation of transcription by high dietary zinc in C. elegans. Nucleic Acids Res. 2015; 43:803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Inoue Y., Hasegawa S., Ban S., Yamada T., Date Y., Mizutani H., Nakata S., Tanaka M., Hirashima N.. ZIP2 protein, a zinc transporter, is associated with keratinocyte differentiation. J. Biol. Chem. 2014; 289:21451–21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ryu M.-S., Lichten L.A., Liuzzi J.P., Cousins R.J.. Zinc transporters ZnT1 (Slc30a1), Zip8 (Slc39a8), and Zip10 (Slc39a10) in mouse red blood cells are differentially regulated during erythroid development and by dietary zinc deficiency. J. Nutr. 2008; 138:2076–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andrews G.K. Regulation and function of Zip4, the acrodermatitis enteropathica gene. Biochem. Soc. Trans. 2008; 36:1242–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974; 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bailey T.L., Elkan C.. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings. Int. Conf. Intell. Syst. Mol. Biol. 1994; 2:28–36. [PubMed] [Google Scholar]
- 32. Mello C.C., Kramer J.M., Stinchcomb D., Ambros V.. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991; 10:3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frøkjær-Jensen C., Wayne Davis M., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.-P., Grunnet M., Jorgensen E.M.. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008; 40:1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davis D.E., Roh H.C., Deshmukh K., Bruinsma J.J., Schneider D.L., Guthrie J., Robertson J.D., Kornfeld K.. The cation diffusion facilitator gene cdf-2 mediates zinc metabolism in Caenorhabditis elegans. Genetics. 2009; 182:1015–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harfe B.D., Fire A.. Muscle and nerve-specific regulation of a novel NK-2 class homeodomain factor in Caenorhabditis elegans. Development. 1998; 125:421–429. [DOI] [PubMed] [Google Scholar]
- 37. Medina-Rivera A., Defrance M., Sand O., Herrmann C., Castro-Mondragon J.A., Delerce J., Jaeger S., Blanchet C., Vincens P., Caron C. et al. . RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 2015; 43:W50–W56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taubert S., Hansen M., Van Gilst M.R., Cooper S.B., Yamamoto K.R.. The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet. 2008; 4:e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao H., Butler E., Rodgers J., Spizzo T., Duesterhoeft S., Eide D.. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem. 1998; 273:28713–28720. [DOI] [PubMed] [Google Scholar]
- 40. Corkins M.E., May M., Ehrensberger K.M., Hu Y.-M., Liu Y.-H., Bloor S.D., Jenkins B., Runge K.W., Bird A.J.. Zinc finger protein Loz1 is required for zinc-responsive regulation of gene expression in fission yeast. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:15371–15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ehrensberger K.M., Corkins M.E., Choi S., Bird A.J.. The double zinc finger domain and adjacent accessory domain from the transcription factor loss of zinc sensing 1 (loz1) are necessary for DNA binding and zinc sensing. J. Biol. Chem. 2014; 289:18087–18096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Radtke F., Heuchel R., Georgiev O., Hergersberg M., Gariglio M., Dembic Z., Schaffner W.. Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J. 1993; 12:1355–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Coneyworth L.J., Jackson K.A., Tyson J., Bosomworth H.J., van der Hagen E., Hann G.M., Ogo O.A., Swann D.C., Mathers J.C., Valentine R.A. et al. . Identification of the Human Zinc Transcriptional Regulatory Element (ZTRE): a palindromic protein-binding DNA sequence responsible for zinc-induced transcriptional repression. J. Biol. Chem. 2012; 287:36567–36581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ogo O.A., Tyson J., Cockell S.J., Howard A., Valentine R.A., Ford D.. The zinc finger protein ZNF658 regulates the transcription of genes involved in zinc homeostasis and affects ribosome biogenesis through the zinc transcriptional regulatory element. Mol. Cell. Biol. 2015; 35:977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. McGhee J.D., Sleumer M.C., Bilenky M., Wong K., McKay S.J., Goszczynski B., Tian H., Krich N.D., Khattra J., Holt R.A. et al. . The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 2007; 302:627–645. [DOI] [PubMed] [Google Scholar]
- 46. Hawkins M.G., McGhee J.D.. elt-2, a second GATA factor from the nematode Caenorhabditis elegans. J. Biol. Chem. 1995; 270:14666–14671. [DOI] [PubMed] [Google Scholar]
- 47. Sinclair J., Hamza I.. A novel heme-responsive element mediates transcriptional regulation in Caenorhabditis elegans. J. Biol. Chem. 2010; 285:39536–39543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guglielmi B., van Berkum N.L., Klapholz B., Bijma T., Boube M., Boschiero C., Bourbon H.-M., Holstege F.C.P., Werner M.. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004; 32:5379–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lariviere L., Seizl M., Cramer P.. A structural perspective on Mediator function. Curr. Opin. Cell Biol. 2012; 24:305–313. [DOI] [PubMed] [Google Scholar]
- 50. Plaschka C., Lariviere L., Wenzeck L., Seizl M., Hemann M., Tegunov D., Petrotchenko E. V, Borchers C.H., Baumeister W., Herzog F. et al. . Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 2015; 518:376–380. [DOI] [PubMed] [Google Scholar]
- 51. Assunção A.G.L., Persson D.P., Husted S., Schjørring J.K., Alexander R.D., Aarts M.G.M.. Model of how plants sense zinc deficiency. Metallomics. 2013; 5:1110–1116. [DOI] [PubMed] [Google Scholar]
- 52. Inaba S., Kurata R., Kobayashi M., Yamagishi Y., Mori I., Ogata Y., Fukao Y.. Identification of putative target genes of bZIP19, a transcription factor essential for Arabidopsis adaptation to Zn deficiency in roots. Plant J. 2015; 84:323–334. [DOI] [PubMed] [Google Scholar]
- 53. Eide D.J. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 2009; 284:18565–18569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Grubman A., Lidgerwood G.E., Duncan C., Bica L., Tan J.-L., Parker S.J., Caragounis A., Meyerowitz J., Volitakis I., Moujalled D. et al. . Deregulation of subcellular biometal homeostasis through loss of the metal transporter, Zip7, in a childhood neurodegenerative disorder. Acta Neuropathol. Commun. 2014; 2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cao J., Bobo J.A., Liuzzi J.P., Cousins R.J.. Effects of intracellular zinc depletion on metallothionein and ZIP2 transporter expression and apoptosis. J. Leukoc. Biol. 2001; 70:559–566. [PubMed] [Google Scholar]
- 56. Dufner-Beattie J., Langmade S.J., Wang F., Eide D., Andrews G.K.. Structure, function, and regulation of a subfamily of mouse zinc transporter genes. J. Biol. Chem. 2003; 278:50142–50150. [DOI] [PubMed] [Google Scholar]
- 57. Lichten L.A., Ryu M.-S., Guo L., Embury J., Cousins R.J.. MTF-1-mediated repression of the zinc transporter Zip10 is alleviated by zinc restriction. PLoS One. 2011; 6:e21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shields J.M., Christy R.J., Yang V.W.. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J. Biol. Chem. 1996; 271:20009–20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liuzzi J.P., Guo L., Chang S.-M., Cousins R.J.. Krüppel-like factor 4 regulates adaptive expression of the zinc transporter Zip4 in mouse small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2009; 296:517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kindermann B., Döring F., Pfaffl M., Daniel H.. Identification of genes responsive to intracellular zinc depletion in the human colon adenocarcinoma cell line HT-29. J. Nutr. 2004; 134:57–62. [DOI] [PubMed] [Google Scholar]
- 61. Kindermann B., Döring F., Budczies J., Daniel H.. Zinc-sensitive genes as potential new target genes of the metal transcription factor-1 (MTF-1). Biochem. Cell Biol. 2005; 83:221–229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.