Abstract
Of the four human Argonaute (AGO) paralogs, only AGO2 has been shown to have slicer activity. The others (AGO1, AGO3 and AGO4) have been thought to assemble with microRNAs to form slicer-independent effector complexes that bind target mRNAs and silence gene expression through translational repression and deadenylation but not cleavage. Here, we report that recombinant AGO3 loaded with miR-20a cleaves complementary target RNAs, whereas AGO3 loaded with let-7a, miR-19b or miR-16 does not, indicating that AGO3 has slicer activity but that this activity depends on the guide RNA. Our cleavage assays using chimeric guides revealed the significance of seed sequence for AGO3 activity, which depends specifically on the sequence of the post-seed. Unlike AGO2, target cleavage by AGO3 requires both 5′- and 3′-flanking regions. Our 3.28 Å crystal structure shows that AGO3 forms a complete active site mirroring that of AGO2, but not a well-defined nucleic acid-binding channel. These results demonstrating that AGO3 also has slicer activity but with more intricate substrate requirements, explain the observation that AGO3 has retained the necessary catalytic residues throughout its evolution. In addition, our structure inspires the idea that the substrate-binding channel of AGO3 and consequently its cellular function, may be modulated by accessory proteins.
INTRODUCTION
Argonaute proteins are key players in microRNA (miRNA)-mediated gene silencing (1,2). They assemble with small RNAs to form ribonucleoproteins referred to as the RNA-induced silencing complexes (RISCs), which in turn degrade target mRNAs (3,4). Humans have four Argonaute paralogs, AGO1, AGO2, AGO3 and AGO4, sharing about 80% identity in their amino acid sequences (5). AGO2 was discovered as the only paralog retaining slicer activity (6,7). The others serve as slicer-independent paralogs capable of RNA silencing through translational repression and deadenylation but not cleavage (8). Thus, AGO3 has been thought to be slicer-deficient, despite the fact that its PIWI domain possesses the catalytic DEDH tetrad (D, E and H refer to aspartic acid, glutamic acid and histidine, respectively) (9). The observation that AGO3 has retained the catalytic tetrad throughout its molecular evolution lends intrigue as to its defective slicer activity and has motivated several studies.
Two groups independently reported that AGO2 chimeras whose PIWI domain was swapped with that of AGO3 cleaved target RNAs, demonstrating that the AGO3 PIWI domain indeed has slicer activity if coupled to motifs I and II in the N-terminus of AGO2 (10,11). In a reciprocal experiment, AGO3 showed an AGO2-like slicer activity by replacing its motifs I and II with the corresponding ones of AGO2. Therefore, it was postulated that the AGO3-characteristic N-terminal motifs inactivate its slicer activity and that unidentified AGO3-accessory proteins might trigger structural changes that enable correct alignment of the target RNA and the catalytic center (11).
In this report, we demonstrate that recombinant AGO3 purified from insect cells has slicer activity but only on select RNAs bearing 5′- and 3′-flanking sequences to the region of guide-target complementarity. Our structure of AGO3-RISC identified the complete catalytic site and the presence of an imperfect nucleic acid-binding channel. Our striking discovery of the slicer activity of AGO3 itself solves a long-standing enigma.
MATERIALS AND METHODS
Cloning, expression and purification of AGO3
The gene encoding human AGO3 was cloned into a pFastBac™HTB vector (Invitrogen) and overexpressed using the Bac-to-Bac Baculovirus Expression System with Trichoplusia ni cells (Expression Systems). The cell extract was prepared by homogenization in Buffer A (10 mM phosphate buffer pH 7.3, 500 mM NaCl, 40 mM imidazole, 10 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor cocktail (Sigma)). The supernatant was loaded onto a nickel-Sepharose high-performance column (GE Healthcare), washed with Buffer A, and eluted with a linear gradient to 50% Buffer B (10 mM phosphate buffer pH 7.3, 500 mM NaCl, 1.5 M imidazole, 10 mM β-mercaptoethanol). The sample was dialyzed overnight with TEV protease against Buffer C (10 mM phosphate buffer pH 7.3, 500 mM NaCl, 10 mM β-mercaptoethanol), and the cleaved His6-tag was removed by loading the sample onto a nickel-Sepharose high-performance column (GE Healthcare).
The flow-through sample was dialyzed against Buffer D (10 mM Tris–HCl pH 8.0, 80 mM KCl, 10 mM β-mercaptoethanol) and then loaded onto Mono Q 5/50 GL (GE Healthcare) equilibrated with buffer E (10 mM Tris–HCl pH 8.0, 50 mM KCl, 10 mM β-mercaptoethanol). The flow-through sample was collected and dialyzed against buffer F (20 mM CHES buffer pH 9.0, 300 mM NaCl, 10 mM β-mercaptoethanol), followed by ultrafiltration. The concentrated sample was loaded onto a HiLoad 16/600 Superdex 200 column (GE Healthcare) in Buffer G (20 mM CHES buffer pH 9.0, 300 mM NaCl, 5 mM dithiothreitol (DTT)). After concentration, the purified AGO3 was stored at −80°C.
Crystallization, structure determination and refinement
AGO3 crystals were obtained by hanging-drop vapor-diffusion at 20°C in 100 mM succinate-phosphate-glycine buffer pH 4.4, 23% PEG2000 and 4% PEG400. Crystals were flash-frozen in liquid nitrogen after being soaked in cryoprotectant buffer (100 mM succinate-phosphate-glycine buffer pH 4.4, 25.3% (w/v) PEG2000, 4.4% (w/v) PEG400, 20% (v/v) glycerol). Diffraction datasets were collected at the NE-CAT beamlines (Advanced Photon Source, Chicago) and indexed, integrated and scaled with NE-CAT RAPD automated protocol (https://rapd.nec.aps.anl.gov/rapd). The phase was obtained by molecular replacement with PHASER using the AGO2 structure (PDB ID: 4OLA) as a search model. The entire model was improved with iterative cycles of refinement with Phenix (12). All figures of structures were generated using PyMol (http://www.pymol.org/). The coordinates for the current structure (PDB ID: 5VM9) have been deposited in the PDB.
Cell culture and cell lysate preparation
HEK293T cells were cultured in Dulbecco’s Modified Eagle Medium (Gibco) with 10% (v/v) fetal bovine serum, 1% (w/v) penicillin and streptomycin at 37°C with 5% CO2. The cells were harvested 48 hours after transfection with 10 μg of pCAGEN plasmid encoding FLAG-tagged AGO protein using the TransIT-X2® system (Mirus).
Substrate preparation
The siRNA duplex of miR-20a was generated by mixing the 5′ phosphorylated synthetic guide with an equal amount of the 5′ hydroxyl passenger RNAs (Dharmacon) in dsRNA annealing buffer (30 mM HEPES-KOH pH 7.5, 100 mM KOAc, 1 mM EDTA). The mixture was incubated at 90°C for 5 min and slowly cooled to 23°C overnight. The annealed siRNA duplexes were separated from ssRNAs using 20% (w/v) polyacrylamide native gel and eluted from gel slices in 300 mM NaCl overnight. The extracted siRNA duplexes were ethanol-precipitated, recovered and stored in dsRNA annealing buffer at 4°C until they were used in the cleavage assays. The siRNA duplex of miR-20a was quantified by measuring the absorbance at the wavelength of 260 nm with the molar extinction coefficient (M−1cm−1) of 442 500. Genes encoding the 60-, 58-, 46- and 40-nt target RNAs were cloned into a pUC19 vector, in vitro transcribed by T7 RNA polymerase and cap-labeled with [α-32P]GTP (3000 Ci mmol−1) using ScriptCap m7G Capping System (CellScript). The synthetic 21- and 23-nt targets were 5′ end-labeled using Optikinase (Affymetrix) with [γ-32P]ATP (3000 Ci mmol−1) and gel purified. All the RNAs used in this study are shown in Supplementary Table S1.
In vitro AGO cleavage assays
48 hours after transfection with pCAGEN plasmid encoding FLAG-AGO, the HEK293T cells were harvested and weighed. For the cleavage assay of AGO3 and its E638A mutant, the cells from three 10 cm plates were resuspended with a 5-fold excess volume (v/w) of lysis buffer (30 mM HEPES-KOH pH 7.4, 100 mM KOAc, 2 mM Mg(OAc)2, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, EDTA-free protease inhibitor cocktail) followed by lysis with ultrasonication. For the cleavage assay of AGO2 and its E637A mutant, we used one 10 cm plate as the source of enzyme. The lysates were centrifuged at 17 000 × g for 20 min. The supernatant was incubated with 20 nM siRNA duplexes at 37°C for 2 hours for RISC assembly and subsequently mixed with 50 μl of anti-FLAG M2 beads (Sigma-Aldrich) for 2 hours at 25°C. The beads were washed eight times with (30 mM HEPES-KOH pH 7.4, 300 mM NaCl, 100 mM KOAc, 2 mM Mg(OAc)2, 1% Triton K-100). Cleavage reaction at 37°C was initiated by addition of 10 nM target RNAs. The reaction was directly quenched with 40 μl of quenching dye (7 M urea, 1 mM EDTA, 0.05% (w/v) xylene cyanol, 0.05% (w/v) bromophenol blue, 10% (v/v) phenol), and resolved by 16% (w/v) polyacrylamide/7 M urea polyacrylamide gel electrophoresis (PAGE). The gels placed on filter paper were dried and analyzed by a phosphor imager.
Cleavage assays using recombinant AGO proteins were performed in 1 × reaction buffer (25 mM HEPES-KOH pH 7.5, 5 mM MgCl2, 50 mM KCl, 5 mM DTT, 0.2 mM EDTA, 0.05 mg ml−1 bovine serum albumin (BSA) (Ambion), 5 U/μl RiboLock RNase Inhibitor (Thermo Scientific)). A final 50 nM 5′ phosphorylated synthetic single-stranded guide RNAs were incubated with recombinant AGO proteins for 1 hour at 37°C for RISC assembly and followed by adding 5′-32P labeled short target RNAs or 5′-cap-labeled target RNAs for the cleavage reaction. The reaction was directly quenched with 20 μl urea quench dye after 1 hour incubation at 37°C. The cleavage products were resolved on the 16% (w/v) polyacrylamide/7 M urea PAGE.
Western blot analyses
The samples were separated by 4–12% gradient Bis-Tris gels (Life Technologies) and blotted onto Amersham Hybond ECL nitrocellulose membranes (GE Healthcare). Anti-FLAG antibody (F1804, Sigma, 1:1000 dilution) was used as the primary antibody. Then, anti-mouse antibody (IRDye 800CW Goat anti-Mouse IgG, LI-COR, 1:16 000 dilution) was used as the secondary antibody to detect FLAG-tagged AGO proteins in the total extract as well as in the immunoprecipitates. The fluorescence from the secondary antibody was measured by the LI-COR Odyssey CLX imaging system. Anti-α tubulin antibody (Santa Cruz, 1:1000 dilution) was used for detection of α-tubulin in the total extract. The western blot analyses were performed in triplicate.
Filter binding assay
The recombinant AGO proteins were incubated with 50 nM of 5′-32P labeled synthetic single-stranded guide RNA in 1 × reaction buffer (25 mM HEPES-KOH pH 7.5, 5 mM MgCl2, 50 mM KCl, 5 mM DTT, 0.2 mM EDTA, 0.05 mg/ml BSA (Ambion) and 5 U/μl RiboLock RNase Inhibitor (Thermo Scientific)) for 1 h at 37°C for RISC assembly. The samples were applied to Amersham Hybond ECL nitrocellulose membranes (GE Healthcare). The membranes were washed 10 times with 100 μl of 1 × binding buffer (25 mM HEPES-KOH pH 7.5, 10 mM MgCl2, 3 mM DTT, and 125 mM NaCl) and then the dried membranes were analyzed by phosphor imager.
Proteomics
Proteins were diluted in 100 mM ammonium bicarbonate to a final concentration of 30 μg in 100 μL volume. A total of 50 μl dithiothreitol (5 mg/ml in 100 mM ammonium bicarbonate) was added and samples were incubated at 37°C for 30 min. Free cysteines were subsequently carboxymethylated by the addition of 50 μl iodoacetamide (5 mg/ml in 100 mM ammonium bicarbonate) and incubation at 37°C for 30 min. Proteins were digested overnight at 37°C with 1 μg trypsin (Trypsin Gold, mass spectrometry grade, Promega). Trifluoroacetic acid (TFA) was added to a final concentration of 0.1% and peptides were desalted by solid phase extraction (SPEC Pt C18, Agilent). A nanoACQUITY UPLC (Waters) coupled to an Orbitrap Elite mass spectrometer (Thermo Fisher) was used for liquid chromatography-tandem mass spectrometry (LC-MS/MS). A 200 ng peptide sample in 1 μl of 0.1% TFA was injected, trapped for 15 min and separated on a Peptide CSH C18 capillary column (130 Å, 1.7 μm, 75 μm × 150 mm, Waters) at a flow rate of 0.3 μl per min. Water with 0.1% formic acid was used as solvent A and acetonitrile with 0.1% formic acid was used as solvent B. The following program was used for peptide elution: 5 to 35% B in 40 min, 35 to 40% B in 5 min, 40 to 85% B in 5 min, hold at 85% B for 5 min, re-cycle to 5% B in 1 min. MS was performed in the Orbitrap analyzer and MS/MS of the ten most abundant peaks at a given elution time was performed in the ion trap analyzer. Data were processed by Proteome Discoverer V1.4 (Thermo Fisher) and searched against the combined UniProt databases for Homo sapiens, T. ni and Drosophila melanogaster using a precursor mass tolerance of 10 ppm, and a fragment mass tolerance of 0.8 Da. Proteins were identified based on the following criteria: peptide confidence high, and ≥2 peptide spectral matches.
Native mass spectrometry
AGO2 was buffer-exchanged into 200 mM ammonium acetate twice using a Micro Bio-Spin P-6 column (Bio-Rad; 6 kD cut-off) and AGO3 was buffer-exchanged into 200 mM ammonium acetate by ultrafiltration (Pierce 30K MWCO, 0.5 ml, Thermo Fisher) because the protein was found to adhere to the beads in the Micro Bio-Spin P-6 column. Samples were measured on a Q Exactive Plus EMR (Thermo Fisher) using nano-electrospray ionization. The instrument settings were as follows: maximum injection time = 100 ms, HCD = 20 V, In-source CID = 20 V, source DC offset = 45 V, injection flatapole DC = 10 V, inter flatapole lens = 8 V, bent flatapole CD = 6 V, transfer multipole DC = 0 V, C-trap entrance lens = 0V, EMR mode on, trapping gas pressure = 6.
RESULTS
Only limited guide RNAs activate AGO3 for RNA cleavage
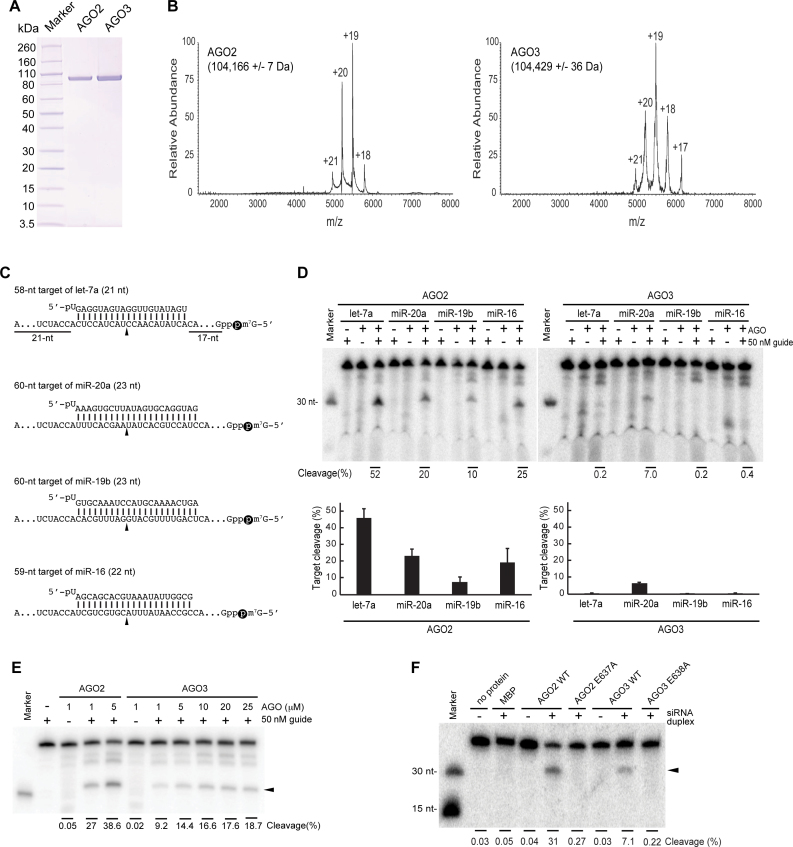
Recombinant AGO2 and AGO3 were purified from insect cells and analyzed by native mass spectrometry (Figure 1A and B) and by bottom-up proteomics. The proteins and corresponding peptides identified in the AGO2 and AGO3 samples are listed in Supplementary Information Excel Spreadsheets. Each of purified proteins was incubated with a synthetic single-stranded let-7a, miR-20a, miR-19b or miR-16, followed by addition of a 5′-cap-bearing target RNA, in which there are 5′ (17 nucleotides (nt)) and 3′ (21 nt) extensions flanking the complementary sequence to each guide (Figure 1C). AGO2 showed slicer activity regardless of the type of guide RNA, albeit with different cleavage efficiencies (Figure 1D, left). In contrast, the slicer activity of AGO3 was observed only when loaded with miR-20a (Figure 1D, right). AGO3 loaded with miR-19 or miR-16 did not cleave RNAs, which is consistent with the previous study using those guide RNAs (7). These results suggest the possibility that AGO3 cleaves RNAs but the intrinsic slicer activity depends on the guide RNA. Although the slicing activity of AGO3 was weaker than AGO2, the cleavage of the 5′-cap-bearing 60-nt target RNA occurred in a concentration-dependent manner (Figure 1E). However, AGO2, but not AGO3, cleaved a short target of miR-20a lacking both flanking sequences (Supplementary Figure S1A and B), consistent with findings reported previously (11). We performed a filter-binding assay to examine whether the observed difference in target cleavage between AGO2 and AGO3 could be attributed to a significant difference in their miRNA loading efficiencies. Recombinant AGO2 or AGO3 was incubated with a 5′-[32P]-labeled single-stranded miR-20a and then spotted on a nitrocellulose filter, to separate bound and free RNA. AGO2 and AGO3 bound comparable amounts of miR-20a (Supplementary Figure S1C), indicating that purified recombinant AGO2 and AGO3 retain similar loading efficiencies.
Figure 1.
In vitro cleavage assays of AGO2 and AGO3. (A) SDS-PAGE analysis of the recombinant AGO2 and AGO3 purified from insect cells and used in the functional studies depicted in panels D and E. The gel was stained with Coomassie blue. (B) Native mass spectra of recombinant AGO2 and AGO3. Molecular weights with standard deviations for the main species as well as the charge states are indicated. The molecular weights correspond to those expected for the AGO proteins with bound RNA. (C) Guide and target RNAs used in D–F. The cap-labels are depicted as black spheres. (D) RNA Cleavage by AGO2 and AGO3 with different guide RNAs. Top: representative gel images of in vitro cleavage assay with the recombinant AGO2 and AGO3. The cap-labeled targets were incubated with either of 1 μM recombinant AGO2 or AGO3 for 1 h and resolved on 16% (w/v) polyacrylamide/7 M urea PAGE. Bottom: average of three experiments are shown in bar graphs with error bars calculated as standard deviation. (E) A representative gel image of in vitro cleavage assay with the recombinant AGO2 (1–5 μM) and AGO3 (1–25 μM). The cap-labeled 60-nt target of miR-20a was incubated with either of recombinant AGO2 or AGO3 for 1 h and resolved on 16% (w/v) polyacrylamide/7M urea PAGE. (F) A representative gel image of in vitro cleavage assay with the catalytic mutant of immunopurified FLAG-tagged AGO2 and AGO3 as well as their wild-type counterpart. Lysates of HEK293T cells expressing either of FLAG-tagged proteins were pre-incubated with a siRNA-like duplex of miR-20a for 2 h, followed by immunoprecipitation with anti-FLAG beads. The purified RISCs were incubated with the cap-labeled 60-nt target of miR-20a for 60 min. The reactions were resolved on a 16% (w/v) polyacrylamide/7 M urea PAGE.
To test whether loading of siRNA duplexes enables AGO3 to cleave RNAs, the lysate of HEK293T cells expressing FLAG-tagged AGO2 or FLAG-tagged AGO3 was pre-incubated with the siRNA-like duplex of miR-20a whose passenger strand lacked a 5′-monophosphate to preferentially load the guide strand into FLAG-AGO. Subsequently, FLAG-AGO2 and -AGO3 in the crude lysate were immunopurified with anti-FLAG beads and incubated with the 5′-cap-labeled 60-nt target of miR-20a (Figure 1C and Supplementary Figure S1D). Mass spectrometry analysis confirmed that there was no cross-contamination of any of the immunopurified samples used for cleavage assays (Supplementary Information Excel Spreadsheets). Both FLAG-AGO2 and FLAG-AGO3 cleaved the target RNAs (Figure 1F). Next, we verified that the observed target RNA cleavage was due to the slicer activity of AGO3. The catalytic glutamate mutants of FLAG-AGO2 and FLAG-AGO3 were expressed in HEK293T cells, validated to confirm no cross-contamination, and tested for their slicer activity (Supplementary Information Excel Spreadsheets, and Supplementary Figure S1D). No cleavage product was detected when the target of miR-20a was incubated with FLAG-AGO2 E637A or FLAG-AGO3 E638A (Figure 1F), proving that the cleavage product generated by immunopurified FLAG-AGO3 was due to the presence of its catalytic tetrad. Altogether, these biochemical assays demonstrate that AGO3 cleaves approximately 60-nt target RNAs only when loaded with miR-20a.
Conventional guide-target complementary rules are not applicable to RNA cleavage by AGO3
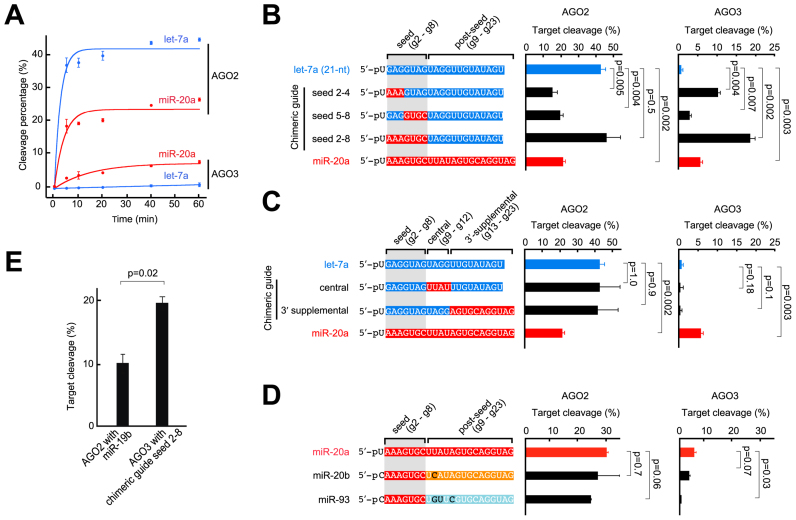
Our in vitro cleavage assay shows that AGO3 is catalytically activated by miR-20a but not by let-7a (Figure 1D). We performed a time course cleavage assay and confirmed that let-7a and miR-20a have different impacts on the slicer activities of AGO2 and AGO3 (Figure 2A). This observation is not consistent with the accepted model that the slicer activity of Argonaute proteins exclusively depends on the base complementarity between the loaded guide and target RNAs, regardless of the guide sequence because Argonaute proteins do not recognize base moieties of the guide RNA except for the 5′ end nucleotide.
Figure 2.
Influence of the seed and post-seed regions on the slicer activity of AGO3. (A) In vitro slicer activities of recombinant AGO2 and AGO3 with miR-20a and let-7a. Aliquots of the reaction were sampled at 0, 10, 20, 40 or 60 min. Their slicer activities with let-7a and miR-20a are drawn as blue and red curves, respectively. (B) RNA cleavage of AGO2 (left) and AGO3 (right) loaded with chimeric guides. Either of the seed 2–4, 5–8 or 2–8 of miR-20a was incorporated into let-7a. Nucleotides of let-7a and miR-20a are colored in blue and red, respectively. (C) RNA cleavage of AGO2 (left) and AGO3 (right) loaded with chimeric guides. Either of the central or 3′-supplementary site of miR-20a was incorporated into let-7a. The color codes are the same as B. (D) RNA cleavage of AGO2 (left) and AGO3 (right) loaded with miR-17 families. The post-seed regions are shown in different colors. The different nucleotides are colored in black. (E) RNA cleavage by AGO2 with miR-19b and by AGO3 with chimeric guide seed 2–8. The mean ± standard deviation for three biological replicates is shown. Student's t-test values are shown in B–E.
The current and previous structures revealed that in the RISC conformation, the base moieties of guide positions 2–4 (g2-g4) are exposed to solvent in order to scan target RNAs (9,13–16). The recent structure of AGO2-RISC in complex with a short target showed that the nucleic acid-binding channel is widened with the incoming target that pairs to the g2-g8 (17). Thus, the g2-g4 and g5-g8 nucleotides serve as the primary and secondary seeds, respectively, in terms of target recognition and subsequent cleavage. To test the influence of the seed sequence on target cleavage, we independently replaced these two seed segments on let-7a with the corresponding parts of miR-20a. The resultant chimeric guides were loaded into recombinant AGO2 and AGO3, and mixed with their cognate cap-labeled target RNAs (Supplementary Figure S2A). Intriguingly, the 2-nt replacement at g2 and g4, which provided the corresponding seed 2–4 of miR-20a, lowered the slicer activity of AGO2 (Figure 2B, left and Supplementary Figure S2B). Another 2-nt replacement at g7 and g8, forming a seed 5–8 of miR-20a, also reduced the slicer activity. However, a concomitant replacement of both segments, forming a seed 2–8 of miR-20a, resulted in a comparable slicer activity to that observed with let-7a (Figure 2B, left and Supplementary Figure S2B). This indicates that the slicer activity of even AGO2 is affected to some degree by the sequence of guide RNA. In contrast, the incorporation of the seed 2–4 of miR-20a into let-7a dramatically enhanced the slicer activity of AGO3 (Figure 2B, right and Supplementary Figure S2C). The miR-20a-like seed 5–8 also increased the activity to some extent. In addition, the incorporation of the entire seed sequence (g2-g8) of miR-20a boosted the slicer activity of AGO3 even more (Figure 2B, right and Supplementary Figure S2C). These results indicate that slicer activities of both AGO2 and AGO3 are influenced by the sequence of the seed region, although the influence is more significant in AGO3.
To examine the impact of the sequence in the post-seed region (g9-g23) on slicer activity, the central (g9-g12) or 3′-supplementary site (g13-g23) of miR-20a was incorporated into let-7a. The resultant chimeric guides were loaded into recombinant AGO2 and AGO3, followed by addition of the cognate cap-labeled target RNAs (Supplementary Figure S2A). Neither of the replacements affected the slicer activities of AGO2 (Figure 2C, left and Supplementary Figure S2B), indicating that if the guide has the seed of let-7a, the impact on the slicer activity of AGO2 is hardly affected by the sequence of the post-seed. A similar result was seen with AGO3, although the weak background slicer activity observed with the let-7a guide precludes confident analysis of any small effects on activity with these chimeric guides (Figure 2C, right and Supplementary Figure S2C). Nonetheless, the guide including the seed of let-7a does not catalytically activate AGO3 regardless of the sequence of the post-seed. Since miRNAs are categorized by the sequence of the seed region (g2-g8), the possibility that AGO3 could exhibit slicer activity with other related guide RNAs was tested (18). miR-20a shares its seed sequence with other miR-17 families such as miR-20b and miR-93, which contain different nucleotide(s) in the post-seed region (Figure 2D). We tested whether these two guide RNAs can cause catalytic activation of slicer activity. AGO2 loaded with either miR-20b or miR-93 showed a slightly lower slicer activity compared to miR-20a (Figure 2D, left and Supplementary Figure S2D). On the other hand, AGO3 cleaved RNAs with miR-20b but not with miR-93 (Figure 2D, right and Supplementary Figure S2E). These results suggest that even if the guide has the seed of miR-20a, the impact on the slicer activities of AGO3 is manipulated by the sequence of the post-seed region. Supporting this, the chimeric guide whose seed of miR-20a was fused with the post-seed of let-7a boosted the slicer activities of AGO2 and AGO3, relative to that of the original miR-20a guide (Figure 2B).
In our cleavage assay, miR-19b was the least efficient guide RNA in terms of catalytic activation of AGO2 (Figure 1D). On the other hand, the chimeric guide whose seed 2–8 was derived from miR-20a while the rest was from let-7a (referred to as ‘seed 2–8’) provided the highest slicer activity for AGO3 (Figure 2B, right). Compared side-by-side, the slicer activity of AGO3 with this chimeric guide showed 2-fold the target cleavage by AGO2 with miR-19b (Figure 2E and Supplementary Figure S2F). This indicates that AGO3 is capable of cleaving RNAs more efficiently than AGO2 depending on the type of their loaded guide RNAs.
AGO3-RISC has an incomplete nucleic acid-binding channel
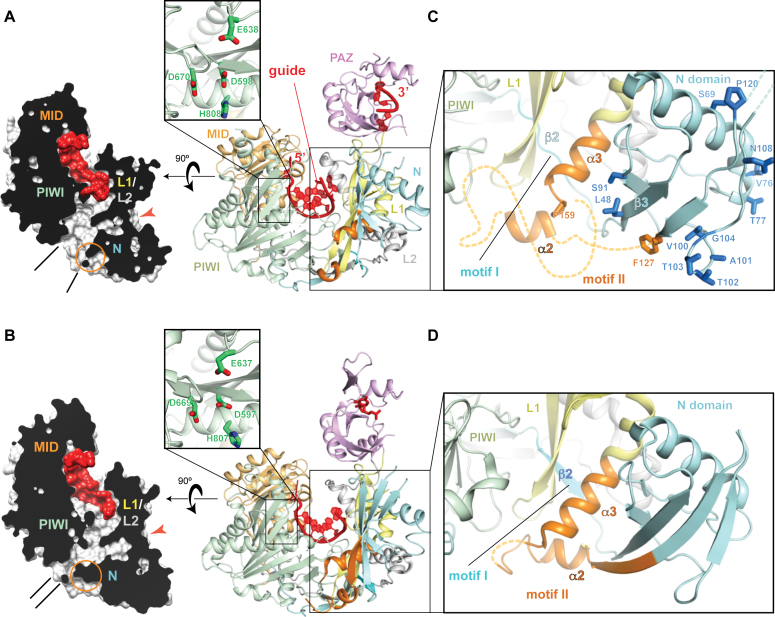
To understand the molecular mechanism of target cleavage, we determined the crystal structure of AGO3 in complex with guide RNA (Figure 3A, Supplementary Figure S3 and Table 1). The asymmetric unit contained two non-crystallographic symmetry (NCS) molecules. The current structure of guide-bound AGO3 revealed that both NCS molecules completed the catalytic DEDH tetrad, a hallmark of the active conformation seen in AGO2 (insets of Figure 3A and B). This indicates that loading of guide RNA converts AGO3 into the catalytically active conformation. The overall AGO3-RISC structure was quite similar to that of AGO2, but a cross section along the nucleic acid-binding channel showed that AGO3 has a much wider channel exit between the N and PIWI domains, compared to AGO2 (Figure 3A and B, left). This difference is attributed to part of the AGO3 N domain, where an AGO3-unique insertion between α2 and α3 was completely disordered (Figure 3C). Note that α2 had a weak electron density in one of the NCS molecules (Supplementary Figure S4). Due to such instability, the nucleic acid-binding channel of AGO3-RISC remains imperfect at one edge. In contrast, the corresponding part of the AGO2 N domain is well-ordered (Figure 3D). Because in most cases AGO2 is a superior slicer compared to AGO3 (Figure 1D), an important inference is that a well-defined nucleic acid-binding channel is essential for efficient target cleavage by Argonaute proteins. These structural data could explain the previous observation that swapping of only the N domain from AGO2 conferred detectable slicer activity on AGO3 (10,11).
Figure 3.
AGO3 has an immature nucleic acid binding channel. (A and B) Right: overall structure of AGO3-RISC (A, current structure: PDB ID: 5VM9) and AGO2-RISC (B, PDB ID: 4OLA). The bound guide RNA is depicted as a ribbon model (red). The catalytic residues are drawn as stick models (green). Left: cross-section (black) of the surface model along the nucleic acid-binding channel (white). The bound guide RNAs are colored in red. Structural differences due to the disordered motifs and the widths of their channel exit are highlighted with orange circles and black lines, respectively. The loose (AGO3) and tight (AGO2) connections between the N and L1/L2 domains are highlighted with orange arrowheads. (C and D) Structural differences around the N domain between AGO3 (C) and AGO2 (D). Disordered regions in the motif II are shown as dotted lines (orange). Residues unique to AGO3 N domain are shown as sticks (dark blue).
Table 1. Data collection and refinement statistics.
| AGO3-RISC (PDB ID: 5VM9) | |
|---|---|
| Data collection | |
| Space group | P43 |
| Cell dimensions | |
| a, b, c (Å) | 68.4, 68.4, 408.8 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 136.3–3.28 (3.46–3.28) |
| R pim | 0.086 (0.408) |
| I/σI | 7.5 (2.1) |
| Completeness (%) | 100 (99.8) |
| Redundancy | 13.6 (12.4) |
| Refinement | |
| Resolution (Å) | 67.48–3.28 |
| No. reflections | 28557 |
| R work/Rfree | 18.71/23.91 |
| No. atoms | |
| Protein | 12 557 |
| RNA | 564 |
| Water | 0 |
| B-factors | |
| Protein | 96.8 |
| RNA | 118.9 |
| Water | |
| R.M.S. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.846 |
| Ramanchandran plot | |
| Favored (%) | 93.50 |
| Allowed (%) | 5.77 |
| Disallowed (%) | 0.72 |
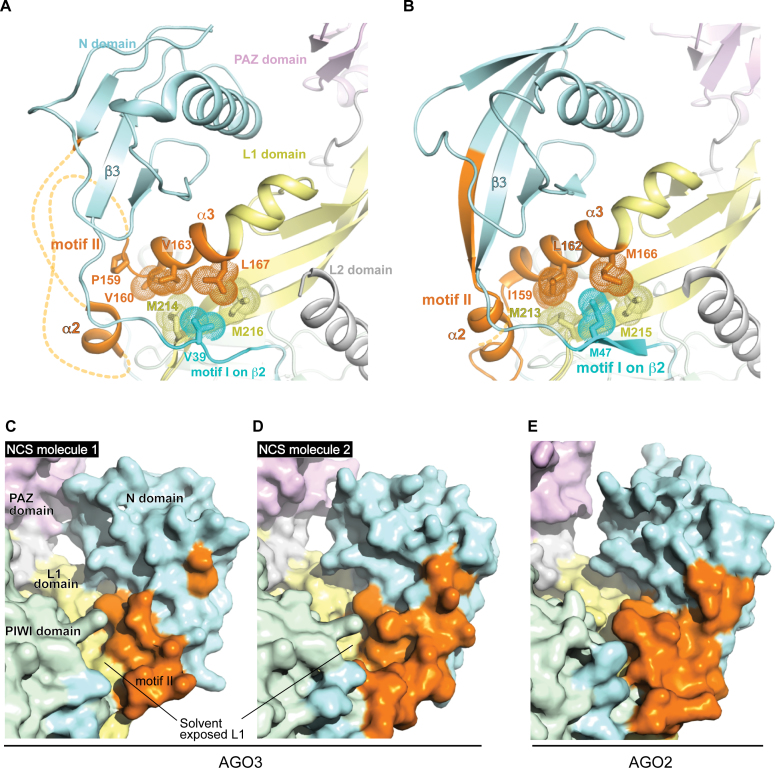
Although earlier studies reported that the intrinsic activity of AGO3 PIWI domain is masked by its characteristic motifs I and II (10,11), the molecular mechanism of this effect remained unknown. Our current structure and the comparison with AGO2 address this gap, especially by analysis of their local structures where the two motifs generate a hydrophobic core to hinge the hook-shaped N domain on the L1 domain (Figure 4A and B). M47 in AGO2 motif I, which is critical to target cleavage and RISC assembly (19), inserts its side chain into the hydrophobic pocket composed of nonpolar, long aliphatic amino acid residues from motif II (Figure 4B). As a result, the tight packing reinforces the connection between the N and L1/L2 domains (Figure 3B, left and Figure 4E). In contrast, the corresponding residues in the AGO3-characteristic motifs are short, nonpolar amino acid residues (Figure 4A), thus engendering a looser packing in the region connecting the N and L1/L2 domains (Figure 3A, left and Figure 4C and D). Most of these non-polar, short residues in AGO3 are also conserved in AGO1 (Supplementary Figure S5). Swapping these characteristic motifs in AGO3 with those of AGO2 results in the increase of the structural rigidity between the N and L1/L2 domains, and presumably accounts for the activity observed in these protein chimeras (10,11).
Figure 4.
Difference in the structural rigidity between AGO3 and AGO2. (A and B) Packing of nonpolar residues between the motifs I and II in AGO3 (A) and AGO2 (B). Van der Waals surface areas are depicted as dots. For clarity, the bound guide RNA and part of the L2 domain are not shown. (C–E) Surface models of the AGO3 NCS molecules 1 (C) and 2 (D), and AGO2-RISC (PDB ID: 4OLA) (E). The color codes are the same as Figure 3. For clarity, bound RNAs are not shown.
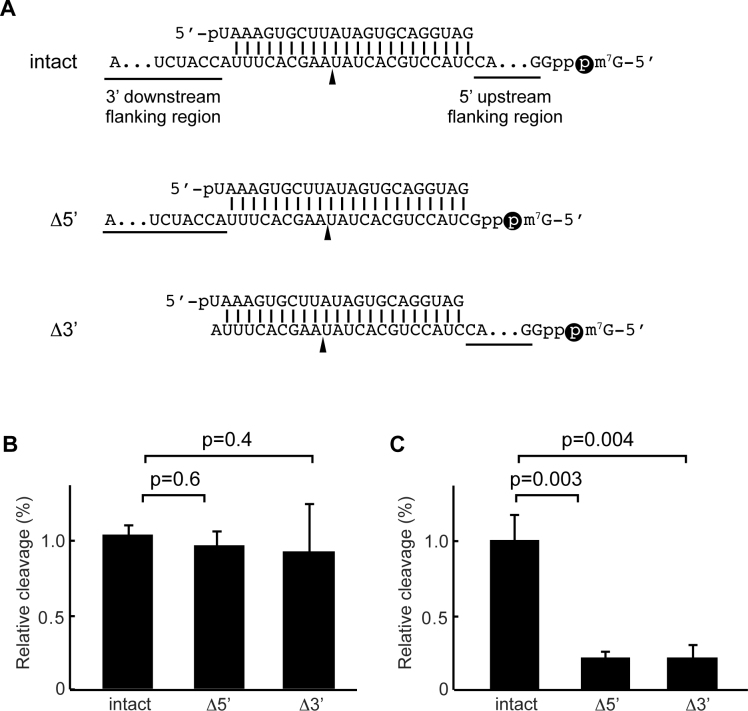
AGO3 requires both flanking regions to cleave the target site
Our in vitro assay showed that AGO3 cleaved the 60-nt but not the 23-nt target of miR-20a (Figure 1E and Supplementary Figure S1B), likely reflecting the importance of flanking regions for the target cleavage. To test this idea, recombinant AGO2 and AGO3 were programmed with a single-stranded miR-20a guide, followed by incubation with the 60-nt target RNA or a shorter target lacking either the 5′ upstream or 3′ downstream flanking sequence (Figure 5A). AGO2 sufficiently cleaved the target RNA regardless of the presence of the flanking sequences (Figure 5B and Supplementary Figure S6). However, deletion of either the 5′ or 3′ flanking region drastically decreased the slicer activity of AGO3 (Figure 5C and Supplementary Figure S6), indicating that both flanking regions of the target site are necessary for cleavage by AGO3. Given that the nucleic acid-binding channel of RISC accommodates only the guide and its complementary target region, both the 5′-upstream and 3′-downstream flanking regions of the bound target mRNAs are left outside the channel. Those flanking regions may interact with a surface in the context of the bound AGO3-RISC and thus could reinforce the formation of the correct structural orientation required to enhance target cleavage.
Figure 5.
AGO3 requires flanking regions to cleave the target sufficiently. (A) Cap-labeled (black sphere) targets of miR-20a used in B and C. Two targets, Δ5′ and Δ3′, were designed by deletion of the 5′ upstream and 3′ downstream flanking sequences from the 60-nt target of miR-20a (intact). (B and C) Influence of flanking regions on the target cleavage by AGO2 (B) and AGO3 (C). The mean ± standard deviation for three biological replicates is shown. Student's t-test values are shown in B and C.
DISCUSSION
In this study, we discovered that AGO3 cleaves RNAs (Figure 1D–F), which answered a longstanding question of why slicer-deficient AGO3 retains the catalytic tetrad. However, why has this activity not been identified since the first reports in 2004 that AGO2 was the only slicer? (6,7) One of the reasons is that the study of AGO3 activity was always accompanied by a significant concern that any observed slicer activity might be attributed to a contamination of endogenous AGO2. To rule out this possibility, we performed mass spectrometry analysis for all of the AGO3 samples used for our in vitro cleavage assays, demonstrating no detectable cross-contamination in our purified fractions (Figure 1B and Supplementary Information Excel Spreadsheets). In addition, we confirmed that the slicer activity of AGO3 observed in this work disappeared when the AGO3 catalytic glutamate was altered to alanine (Figure 1F). These results demonstrate that the observed cleavage was caused by AGO3 itself. To our knowledge, this is the first report that AGO3 has slicer activity.
We also revealed that the activity of AGO3 depends on the type of guide RNA (Figure 1D). This property seems to be another factor that made the study of AGO3’s slicer activity difficult. Indeed, in our in vitro study, we detected little slicer activity from AGO3 loaded with let-7a, whereas similarly purified AGO2 readily cleaved the cognate target (Figure 2A, blue curves). However, loading of miR-20a activated AGO3 for RNA cleavage (Figures 1D–F and 2A, red curves). Our cleavage assay using chimeric guides demonstrated the significance of the seed of miR-20a for the slicer activity of AGO3 (Figure 2B right). In contrast, miR-19b, miR16, and a guide complementary to luciferase gene, which were used in the previous studies (6,7), share none or only one nucleotide within the seed region with miR-20a. These findings could explain why previous studies using only a few specific guide RNAs did not detect any cleavage activity of AGO3 and thus concluded that AGO3 was catalytically inactive (6,7).
Our biochemical experiments demonstrate that AGO3 is not always catalytically activated even though the guide and target are fully complementary, indicating that the molecular mechanism of RNA cleavage is different from the conventional rules applicable to AGO2. Consistent with previous genetic analysis, AGO3 alone does not seem to be a suitable slicer to cleave mRNAs in place of AGO2. Given the versatility of Argonaute proteins (20–22), the AGO3 slicer activity discovered here may be utilized for different biological processes beyond RNA silencing. We note that humans have more than 4000 miRNAs (miRBase, release 21) (23), and thus there are many untested guide RNAs that may confer slicer activity on AGO3, similar to miR-20a. Further studies are clearly required to understand the physiological relevance of the slicer activity of AGO3.
The guide type-dependent slicer activity is attributed to the AGO3-characteristic N-terminal two motifs I and II (10,11). Our current structure revealed that the two motifs shape the incomplete nucleic acid-binding channel at one end between the N and PIWI domains (Figure 3A, left). Therefore, it is reasonable to expect that loading of only specific guide RNAs might ‘activate’ AGO3 for RNA cleavage, but the activity might yet lend itself to modulation through alterations to the rigidity of its channel. A previous study proposed a possibility that binding of unidentified proteins triggers a conformational change to activate AGO3 (11). The current crystal structure revealed that 9 out of 13 AGO3 N domain-specific residues are located in the vicinity of the AGO3-unique insertion (Figure 3C and Supplementary Figure S5). We hypothesize further that such accessory proteins recognize the AGO3-characteristic local structures to manipulate the channel's rigidity. A spatiotemporal repertoire of different accessory proteins may inhibit and enhance the intrinsic slicer activity of AGO3 to enable gene regulation critical for development and differentiation.
A recent study using a single-molecule approach suggested that Argonaute proteins can reshape the binding properties of the loaded guide (24). Meanwhile, previous structures of AGO1 and AGO2 showed that human AGO paralogs have unique local structures, thereby forming slightly different nucleic acid-binding channels (13–16). Here, we have shown the incomplete channel of AGO3, further expanding the variety of substrate-binding channel conformations observed in existing structures (Figure 3). Our biochemical assays showed that if the guide contains the seed of miR-20a, it has a potential for catalytic activation of AGO3. The impact was manipulated by the sequence of the post-seed region (Figure 2D). Given that the post-seed region pairs to the target between the N and PIWI domains, the AGO3-unique N domain or insertion may control the slicer activity in combination with the remainder of the composite channel (Figure 3A). We demonstrated that AGO3 requires flanking regions of the target site for its slicer activity, whereas AGO2 does not (Figure 5B and C). Comparison of the electrostatic surface potentials among human AGO1, AGO2 and AGO3 revealed that they possess positively charged areas on their exterior, although the precise locations of these positively charged patches are slightly different (Supplementary Figure S7). It is possible that the surface potential differences may contribute to recruitment of different AGO-binding partners that could further modulate the activities we have observed here. Alternatively, the association of AGO3-RISC with target RNAs could be stabilized further by other interactions between AGO3 and the flanking regions of the bound target RNA, thereby facilitating target cleavage. These findings prompted us to propose a model that AGO3 employs multiple factors, rather than relying simply on the guide sequence, to recognize target RNAs and cleave them. This idea could expand the definition of target recognition beyond conventional guide-target complementarity alone and may explain how a guide strand directs different AGO-derived RISCs to distinct target RNAs (20). Further structural studies of eukaryotic RISCs will shed light on their varied nucleic acid-binding channels and specialized functions.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 5VM9.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to D.P. Bartel (MIT), V. Gopalan (OSU) and J.E. Jackman (OSU) for useful advice and insights. We thank T-H. Chen and V. Gopalan (OSU) for technical assistance, G. Singh (OSU) for providing HEK293T cells, and the NE-CAT beamlines staff, especially K. Rajashankar and J. Schuermann for advice on data processing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation Fellowship (to F.B.); NE-CAT beamlines of the Advanced Photon Source with support by Department of Energy [GUP-41799, GUP-51294 to K.N.]; PRESTO [JPMJPR13L7 to K.N.]; The Ohio State University Center, an RNA Biology Seed Grant (to K.N.); The Ohio State University Start-up Fund (to K.N.); National Institutes of Health [R01GM113658 to V.H.W., R01GM124320 to K.N.]. Funding for open access charge: The Ohio State University Start-up fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013; 14:447–459. [DOI] [PubMed] [Google Scholar]
- 2. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kobayashi H., Tomari Y.. RISC assembly: coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta. 2015; 1859:71–81. [DOI] [PubMed] [Google Scholar]
- 4. Nakanishi K. Anatomy of RISC: how do small RNAs and chaperones activate Argonaute proteins. Wiley Interdiscip. Rev. RNA. 2016; 7:637–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sasaki T., Shiohama A., Minoshima S., Shimizu N.. Identification of eight members of the Argonaute family in the human genome. Genomics. 2003; 82:323–330. [DOI] [PubMed] [Google Scholar]
- 6. Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J.. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004; 305:1437–1441. [DOI] [PubMed] [Google Scholar]
- 7. Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T.. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004; 15:185–197. [DOI] [PubMed] [Google Scholar]
- 8. Jonas S., Izaurralde E.. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015; 16:421–433. [DOI] [PubMed] [Google Scholar]
- 9. Nakanishi K., Weinberg D.E., Bartel D.P., Patel D.J.. Structure of yeast Argonaute with guide RNA. Nature. 2012; 486:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schurmann N., Trabuco L.G., Bender C., Russell R.B., Grimm D.. Molecular dissection of human Argonaute proteins by DNA shuffling. Nat. Struct. Mol. Biol. 2013; 20:818–826. [DOI] [PubMed] [Google Scholar]
- 11. Hauptmann J., Dueck A., Harlander S., Pfaff J., Merkl R., Meister G.. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat. Struct. Mol. Biol. 2013; 20:814–817. [DOI] [PubMed] [Google Scholar]
- 12. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schirle N.T., MacRae I.J.. The crystal structure of human Argonaute2. Science. 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faehnle C.R., Elkayam E., Haase A.D., Hannon G.J., Joshua-Tor L.. The making of a slicer: activation of human Argonaute-1. Cell Rep. 2013; 3:1901–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elkayam E., Kuhn C.D., Tocilj A., Haase A.D., Greene E.M., Hannon G.J., Joshua-Tor L.. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012; 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakanishi K., Ascano M., Gogakos T., Ishibe-Murakami S., Serganov A.A., Briskin D., Morozov P., Tuschl T., Patel D.J.. Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep. 2013; 3:1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schirle N.T., Sheu-Gruttadauria J., MacRae I.J.. Structural basis for microRNA targeting. Science. 2014; 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kozomara A., Griffiths-Jones S.. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011; 39:D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kwak P.B., Tomari Y.. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012; 19:145–151. [DOI] [PubMed] [Google Scholar]
- 20. Miyazaki Y., Du X., Muramatsu S., Gomez C.M.. An miRNA-mediated therapy for SCA6 blocks IRES-driven translation of the CACNA1A second cistron. Sci. Transl. Med. 2016; 8:347ra394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R., Morozova N., Fenouil R., Descostes N., Andrau J.C. et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012; 19:998–1004. [DOI] [PubMed] [Google Scholar]
- 22. Huang V., Qin Y., Wang J., Wang X., Place R.F., Lin G., Lue T.F., Li L.C.. RNAa is conserved in mammalian cells. PLoS One. 2010; 5:e8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salomon W.E., Jolly S.M., Moore M.J., Zamore P.D., Serebrov V.. Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015; 162:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data bank under accession number 5VM9.