Abstract
PUF (Pumilio/fem-3 mRNA binding factor) proteins, a conserved family of RNA-binding proteins, recognize specific single-strand RNA targets in a specific modular way. Although plants have a greater number of PUF protein members than do animal and fungal systems, they have been the subject of fewer structural and functional investigations. The aim of this study was to elucidate the involvement of APUM23, a nucleolar PUF protein in the plant Arabidopsis, in pre-rRNA processing. APUM23 is distinct from classical PUF family proteins, which are located in the cytoplasm and bind to 3′UTRs of mRNA to modulate mRNA expression and localization. We found that the complete RNA target sequence of APUM23 comprises 11 nt in 18S rRNA at positions 1141–1151. The complex structure shows that APUM23 has 10 PUF repeats; it assembles into a C-shape, with an insertion located within the inner concave surface. We found several different RNA recognition features. A notable structural feature of APUM23 is an insertion in the third PUF repeat that participates in nucleotide recognition and maintains the correct conformation of the target RNA. Our findings elucidate the mechanism for APUM23’s-specific recognition of 18S rRNA.
INTRODUCTION
In eukaryotes, PUF (Pumilio/fem-3 mRNA binding factor) proteins are a conserved family of RNA-binding proteins with modular structured repeat domains. Their reported molecular functions are translational repression or activation, mRNA localization (1) and pre-ribosomal RNA (pre-rRNA) processing (2,3). Classical PUF proteins are located in the cytoplasm and regulate the stability and translation of their target mRNAs by binding to specific sequences in the 3′ untranslated regions (UTRs) (4). The PUF binding motif normally contains a conserved UGUR sequence (where R is a purine) at the 5′ end (5). The typical RNA-binding domain in PUF proteins is known as a PUF repeat or Pumilio homology domain (PUM-HD) (6). Structural studies have shown that canonical PUF proteins typically have eight tandem PUF repeats with one pseudo-repeat at each terminus and that they fold into crescent shapes (6–15). In most cases, they recognize their 8-nt specific RNA targets in a modular and anti-parallel way, with each PUF repeat binding specifically to a single RNA base such that PUF repeats 1–8 sequentially bind to RNA bases 8–1 (6,10–12). However, there are exceptions. Some PUF proteins bind RNA targets longer than eight bases, with the extra RNA bases flipping away from the PUF–RNA interface (9,13,16,17); and some comprise more than eight repeats and are therefore able to bind RNA targets longer than eight bases, or even structured DNA or RNA (3,17).
PUF repeats normally contain approximately 40 residues and fold into an arrangement comprising three α-helices (6). Their RNA base recognition specificity is determined by three key residues within the five-residue motif in the second helix of each PUF repeat, known as the tripartite recognition motif (TRM) (18–22). The first and fifth residues bind the edge of the RNA base via hydrogen bonding or van der Waals contacts, and the second residue makes a stacking interaction with two adjacent bases (23). The third and fourth residues can be any hydrophobic residues. In Pumilio1 and FBF-2 (6,16), the motifs (notated as the residues in TRM positions 1, 5 and 2) bind as follows: S1E5-N2 or S1E5-H2 motif with guanine, N1Q5-H2 or N1Q5-Y2 motif with uracil and C1Q5-R2 or S1Q5-R2 motif with adenine. Cytosine-recognition PUF repeats are rarely seen in nature, although early studies have reported PUF repeats engineered for specific C-recognition with (G, A, S, T or C)1R5 motifs (18,19). Further RNA-recognition codes of PUF proteins have also been identified (21,22,24).
The number of PUF proteins varies between species, with only a small number in vertebrates (4) and a greater number in plants (25). For example, the plant Arabidopsis alone encodes more than 25 PUF proteins, named Arabidopsis Pumilios (APUMs) (25–27). Despite this, PUF proteins in plants have been the subject of fewer structural and functional investigations than those in animal and fungal systems.
APUM23 is a recently characterized nucleolar PUF protein with critical roles in leaf development, plant organ polarity and pre-rRNA processing (2,28), but how it regulates these processes remains largely unknown. According to phylogenetic analysis, orthologs of APUM23 include yeast nucleolar protein 9 (Nop9) and human NOP9. It has been predicted that APUM23 contains ten PUF repeats rather than the typical eight (25). A recent study found that the target of APUM23 is a unique 10-mer sequence (5′-GAAUUGACGG-3′) in the 18S rRNA (29), unlike the cognate ‘UGUR’ RNAs typical of cytoplasmic PUF proteins. However, how APUM23 specifically recognizes its RNA target remains unknown.
Our understanding of the molecular mechanisms that underlie the functions of APUM23 is limited by the lack of high-resolution structure of APUM23 PUF repeats. In this study, therefore, we focused on the structure of APUM23 and its RNA target. We solved several crystal structures for APUM23–RNA complexes (wild-type (WT) and mutant) and found that APUM23 has 10 PUF repeats and folds into a C-shaped structure. There is an insertion within its third PUF repeat, located at the inner concave surface. APUM23 also has an insertion within the eighth PUF repeat, located at the side face. We also found that the complete RNA target sequence of APUM23 comprises 11 nt (5′-GGAAUUGACGG-3′) at positions 1141–1151 in 18S rRNA, rather than the 10 nt that had previously been suggested (29). The insertion of the third PUF repeat may function in both RNA base recognition and RNA conformation stabilization. Thus, this study elucidated the structural basis for the specific recognition of 18S rRNA by APUM23.
MATERIALS AND METHODS
Plasmid constructions
The cDNA of APUM23 was amplified from cDNA of Arabidopsis thaliana that was generously provided by Lab of Professor Yong Ding (University of Science and Technology of China, Hefei, China). The PUF repeats of APUM23 (residues 85-655) was cloned into a modified pET28a (Novagen) vector without a protease cleavage site after the 6 × His tag (p28a) vector. All mutants were generated using the MutanBEST kit (Takara) and verified by DNA sequencing.
Protein expression and purification
All proteins were expressed in Escherichia coli Bl21-Gold (DE3) cells (Novagen). For native proteins, cells were cultured in LB medium at 310 K to an OD600 of 0.8–1.0, subsequently shifted to 289 K and induced with 0.4 mM isopropyl β-D-thiogalactopyranoside (IPTG) for 24 h. For Se-Met labeled proteins, cells were cultured in LeMaster and Richards minimal medium (LR medium) at 310 K to an OD600 of 0.8–1.0, then transferred to LR medium that contained Val, Ile, Leu, Phe, Trp, Thr, Lys at the concentration of 50 mg/l and Se-Met at the concentration of 60 mg/l. After 30 min at 310 K, cells were shifted to 289 K and induced with 0.4 mM IPTG for 24 h. All proteins were purified by Ni-NTA affinity chromatography (GE Healthcare) after lysing cells by sonication in 20 mM Tris (pH 8.0) and 500 mM NaCl. After clarification at 13 000 rpm in an R22A2 Hi-tachi rotor (Hitachi, himac CR22G, Japan) for 30 min at 277 K, the supernatants were eluted in buffer containing 30–500 mM imidazole (pH 7.5) at 293 K. The eluted His-tag proteins were further purified using a Superdex 200 column (GE Healthcare) in buffer containing 20 mM Tris (pH 7.5) and 200 mM NaCl with no reducing agent at 293 K.
RNA preparation
RNA oligomers were purchased from Takara Bio, Inc., and dissolved in diethyl pyrocarbonate (DEPC)-treated water to a final concentration of 2 mM.
Crystallization and data collection
The native or Se-Met labeled APUM2385-655 with an N-terminal 6 × His tag was concentrated to ∼20 mg/ml in buffer of 15 mM Tris (pH 7.5), 200 mM NaCl and 2 mM Dithiothreitol (DTT). The native or Se-Met labeled APUM2385-655–RNA complex was prepared by mixing 10 mg/ml (final concentration) proteins with RNA in a mole ratio of 1:1.5. All crystals were formed at 293 K via sitting drop method with 1 μl each of protein–RNA complexes and reservoir solutions. The crystals of native APUM2385-655–GAAUUGACGG complex were formed in mother liquor containing 0.15 M DL-Malic acid (pH 7.0) and 20% w/v Polyethylene glycol 3350 via sitting drop method. The crystals of Se-Met labeled APUM2385-655–GAAUUGACGG complex were formed in mother liquor containing 0.1 M Calcium acetate hydrate, 0.1 M Sodium cacodylate (pH 5.5) and 12% w/v Polyethylene glycol 8000. The crystal of the APUM2385-655–GGAAUUGACGG and APUM2385-655–GGAGUUGACGG were also formed in mother liquor containing 0.15 M DL-Malic acid (pH 7.0) and 20% w/v Polyethylene glycol 3350. The crystal of the APUM2385-655–GGAUUUGACGG was formed in mother liquor containing 0.2 M Sodium malonate (pH 7.0) and 20% w/v Polyethylene glycol 3350. The crystal of APUM23Delete-GGAAUUGACGG was formed in mother liquor containing 0.2 M Lithium sulfate monohydrate, 0.1 M Bis-Tris (pH 6.5) and 25% w/v Polyethylene glycol 3350. The cryoprotectant for crystals was 80% reservoir solution supplemented with 20% glycerol. The crystals were preserved in liquid nitrogen stream at 100 K. X-ray diffraction data of the crystals were collected at 100 K. X-ray diffraction data for the crystals were collected on beamline 17U1, 18U1 and 19U1 of the Shanghai Synchrotron Radiation Facility (SSRF). The data collection statistics are listed in Supplementary Tables S1 and 2.
Structure determination and refinement
Data were processed and scaled using HKL2000 and CCP4 program stackage (30). Phase was solved by the single-wavelength anomalous dispersion method using Shelx C/D/E (31) and phenix.autosol wizard (32) with the Se-Met labeled APUM2385-655–GAAUUGACGG complex data. The initial model was built automatically by program Buccaneer (33). The structure of native APUM2385-655–GAAUUGACGG complex was determined by program Phaser MR (34) using the model of Se-Met labeled APUM2385-655–GAAUUGACGG complex as the search model. The other structures were using the refined native APUM2385-655–GAAUUGACGG structure as the search model. The structures of native APUM2385-655–RNA complexes were further modified and refined through a combination of the Phenix suite (35) and Coot (36). The refinement statistics are listed in Supplementary Tables S1 and 2. All structure figures were prepared with PyMOL (37).
Isothermal titration calorimetry (ITC)
Isothermal titration calorimetry (ITC) assays were carried out on a MicroCal iTC200 calorimeter (GE Healthcare) at 298 K. The buffer for proteins and RNA oligomers was 50 mM Tris (pH = 7.5), 200 mM NaCl. The titration protocol consisted of 20 injections of RNA (0.1–0.15 mM) into protein (0.01 mM). We performed three trials for each ITC titration. Curve fitting to a one-binding-site model was performed by ITC data analysis module of Origin7.5 (OriginLab) provided by the manufacturer. The parameters and fitted curves are shown in Table 1 and Supplementary Figure S7.
Table 1. Thermodynamic parameters of the WT and mutants of the APUM23 to different RNAs by ITC experiments.
| Protein APUM23 | RNA 5′ to 3′ | ΔH kcal/mol | ΔS cal/mol/K | K D nM | K D (mean) nM | K rel # |
|---|---|---|---|---|---|---|
| 36 | −85 | 21 | ||||
| WT | GGAAUUGACGG | 38 | −91 | 20 | 21 ± 2 | 1 |
| 36 | −84 | 23 | ||||
| 35 | −82 | 27 | ||||
| WT | GAAUUGACGG | 33 | −76 | 29 | 32 ± 8 | 1.5 |
| 38 | −95 | 41 | ||||
| 39 | −96 | 12 | ||||
| WT | GGAUUUGACGG | 39 | −94 | 11 | 13 ± 3 | 0.6 |
| 39 | −96 | 17 | ||||
| 24 | −47 | 35 | ||||
| WT | GGAAAUGACGG | 27 | −58 | 44 | 43 ± 7 | 2.0 |
| 25 | −50 | 49 | ||||
| 26 | −61 | 1236 | ||||
| F398A | GGAAUUGACGG | 29 | −71 | 2725 | (1.9 ± 0.7) × 103 | 90 |
| 28 | −68 | 1783 | ||||
| 28 | −67 | 1422 | ||||
| Q401E | GGAAUUGACGG | 22 | −48 | 1143 | (1.1 ± 0.3) × 103 | 52 |
| 21 | −41 | 840 | ||||
| 27 | −62 | 205 | ||||
| Delete | GGAAUUGACGG | 29 | −66 | 116 | (0.160 ± 0.04) × 103 | 7.6 |
| 28 | −64 | 160 | ||||
| 34 | −86 | 943 | ||||
| Delete | GAAUUGACGG | 30 | −73 | 1248 | (1.1 ± 0.2) × 103 | 52 |
| 32 | −80 | 980 |
# K rel reports the affinity of WT or mutant APUM23 for the RNA sequence, relative to the affinity of WT APUM23 for the 11-mer WT RNA 5′-GGAAAUUGACGG-3′.
The ITC experiments were carried out at 293K and all proteins were in buffer conditions: 50 mM Tris (pH 7.5), 200 mM NaCl. We performed three trials for each ITC titration. WT, F398A, Q401E and Delete represent APUM2385-655, APUM2385-655 F398A mutant, APUM2385-655 Q401E mutant and APUM2385-655 without the insertion in PUF repeat R3, respectively. The average dissociation constant (KD mean) values (±standard deviation) were derived from three independent experiments. The fitted curves are shown in Supplementary Figure S7.
RESULTS
Overall structure of the APUM2385-655–GGAAUUGACGG complex
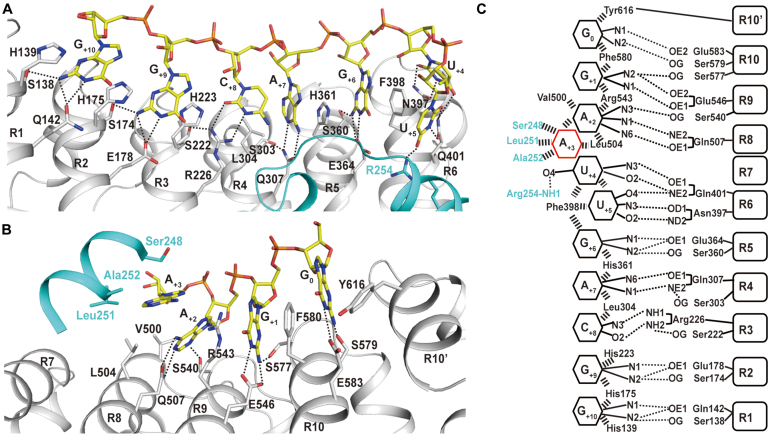
As reported previously (29), Arabidopsis thaliana APUM23 selectively interacts with a 10-mer RNA target (5′-GAAUUGACGG-3′) in 18S rRNA. To gain a structural insight into how APUM23 recognizes 18S rRNA, we performed extensive crystallization trials to determine the structure of the APUM23–RNA complex. We finally determined a 2.55 Å-resolution crystal structure of the APUM2385-655–GAAUUGACGG complex (Supplementary Figure S1 and Table S1) and found that the complete target of APUM23 was the 11-mer RNA 5′-GGAAUUGACGG-3′ at positions 1141–1151 in 18S rRNA (referred to hereafter as ‘11-mer WT RNA’). This is conserved in 18S rRNA across species (Supplementary Figure S2). Thus, we determined the crystal structure of the APUM2385-655–GGAAUUGACGG complex at a resolution of 2.5 Å (Figure 1A and Supplementary Table S1). APUM23 comprises ten PUF repeats and is folded into a twisted C shape, with one pseudo-repeat at each terminus. Eight of the 10 PUF repeats comprise approximately 40 amino acid residues; they share a similar structure, in the form of a typical arrangement of three α-helices. The structures of the other two PUF repeats (R3 and R8) differ from those of typical PUF repeats (Supplementary Figure S3). R8 contains 61 amino acid residues (residues 466–526), with an insertion between the first and second helix. This insertion folds into an α-helix at the side face of the C-shaped structure (Figure 1A). A remarkable feature of the structure of APUM2385-655 is the unique characteristics of R3. This PUF repeat contains 74 amino acid residues (residues 213–286) and includes an insertion (residues 235–270; the electron density of residues 258–270 was missing) located between the second and third helix. The insertion comprises a disordered region and two short α-helices (αi1 and αi2), located at the inner concave surface, that participate in the recognition of RNA bases (Figure 1A and Supplementary Figure S3).
Figure 1.
Overall structure of the APUM2385-655–GGAAUUGACGG complex. (A) The overall structure of the complex is represented as cartoon cylindrical helices, with the pseudo-repeats at each terminus labeled R1’ and R10’, and the PUF repeats labeled R1 to R10. The insertion is colored in cyan and the two short α-helices are labeled αi1 and αi2. (B) The 2Fo-Fc electron density map around the 11-mer WT RNA 5′-GGAAUUGACGG-3′, contoured at 1.0 sigma. Nucleotides G1140 to G1150 are labeled as G0 to G+10. (C) The overall structure of the Nop9Delete-GGAAUUGACGG complex is represented as cartoon cylindrical helices (PDB code: 5WTY). The insertion in the R3 PUF repeat has been removed.
During the preparation of this manuscript, reports were published on the apo and RNA-bound structures of yeast Nop9 (17,38). The apo structure of yeast Nop9 was first reported in 2016 as having 11 PUF repeats with one N-terminal pseudo-repeat (38). However, the more recent study reported that yeast Nop9 contained 10 PUF repeats with two terminal pseudo-repeats in its RNA-bound structure (17). In the present study, we adopted this latter definition and compared APUM23 and yeast Nop9 in their recognition of the same RNA sequence (Figure 1C). The sequence position of RNA 5′-GGAAUUGACGG-3′ differs between Arabidopsis (G1141–G1151) and Saccharomyces cerevisiae (G1140–G1150), so we labeled the 11 nt as G0 to G+10 to facilitate the description of the differences in RNA recognition (Figure 1B). A search of the Dali server showed that the structure of APUM23 resembles that of yeast Nop9 (Z-score 25.4, sequence identity 18%).The Cα atom root mean square deviation between the RNA-bound structure of APUM23 and yeast Nop9 is 6.673 Å (389–389 Cα atoms, calculated by PyMOL). The most remarkable structural difference between yeast Nop9 and APUM23 is found in the PUF repeat R3. In both APUM23 and yeast Nop9, repeat R3 contains an insertion between the second and the third helix; however, this insertion was invisible in the apo structure of yeast Nop9 (38) and was removed (Nop9Delete) from the Nop9–RNA structure (17).
All the RNA bases of 11-mer WT RNA are positioned below APUM23’s inner concave surface and the insertion in PUF repeat R3 (Supplementary Figure S4). C-shaped APUM23 interacts in a modular way with the RNA bases of G0 to A+2 and U+5 to G+10 through PUF repeats R10 to R8 and R6 to R1, respectively, (Figure 2). R7 is not responsible for RNA interactions, and A+3 flips away from the inner concave surface; however, APUM23 stabilizes A+3 via a hydrophobic pocket that is formed mostly by the insertion (discussed in detail below), and interacts with U+4 via both the insertion (Arg-254) and R6 (Phe-398 and Gln-401), which also recognizes U+5. In addition, A+3 makes stacking interactions with A+2, and U+4 with U+5.
Figure 2.
Recognition of target RNA by APUM23. (A and B) Interactions between APUM23 and the target RNA. The dotted lines represent hydrogen bonds. (C) Schematic representation of APUM23–RNA interactions. Hydrogen bonds and stacking interactions are shown as dotted lines and stacked bars, respectively.
Yeast Nop9 and APUM23 recognize the same target RNA in different ways (Supplementary Table S3). In the Nop9Delete–RNA structure, A+3 does not stack with A+2, and U+4 and U+5 flip away from the inner concave surface of Nop9, so their uracil rings do not interact with Nop9Delete. APUM23 has only one PUF repeat (R7) not involved in specific RNA recognition of RNA; Nop9Delete has two, R6 and R7. Furthermore, APUM23 and Nop9 use different motifs to recognize some of the same RNA bases (Supplementary Table S3).
Taken together, the ten PUF repeats and the insertion in PUF repeat R3 enable APUM23 to specifically recognize the 11-mer 18S rRNA sequence 5′-GGAAUUGACGG-3′.
Modular recognition of G0, G+6, A+7, C+8, G+9 and G+10
G0, G+6 and G+9 are recognized by the previously reported typical S1E5 motif in PUF repeats R10, R5 and R2, respectively (6). The side chain of S1 (the residue in TRM position 1) interacts with the N2 group of guanine via hydrogen bonding, and the side chain of E5 interacts with the N1 group, or the N1 and N2 groups, of guanine (Supplementary Figure S5a, c and d). G+10 is recognized by an S1Q5 motif in PUF repeat R1, which normally specifies adenine (6). Nevertheless, the interaction network is identical to that for the recognition of G by the S1E5 motif (Supplementary Figure S5e). The S1Q5 motif for guanine recognition has previously been identified (8,39).
PUF repeat R4 has an S1Q5-L2 motif, which recognizes A+7 through the side chain of Gln-307 (position 5); the side chain of Ser-303 stabilizes the conformation of Gln-307 via a hydrogen bond interaction (Supplementary Figure S5h).
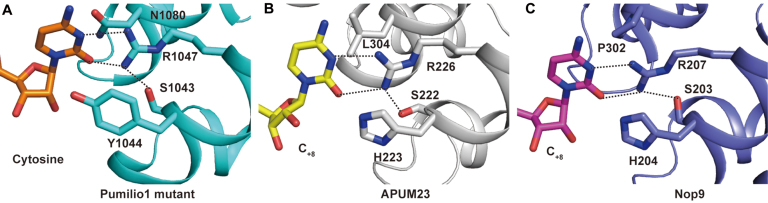
Cytosine-recognition PUF repeats are rarely seen in nature. The PUF repeat R3 has an S1R5-H2 motif in both APUM23 and Nop9. The NH1 and NH2 groups of R5 form hydrogen bond interactions with the N3 and O2 groups of cytosine, and the side chain of S1 stabilizes the conformation of R5 via a hydrogen bond interaction (Figure 3).
Figure 3.
APUM23 and Nop9 each possess a natural cytosine-recognition PUF repeat. (A–C) Recognition of cytosine by R6 PUF repeat of a mutant of Pumilio1, R3 PUF repeat of APUM23 and R3 PUF repeat of Nop9, respectively. The dotted lines represent hydrogen bonds.
Recognition of G+1, and A+2
In general, PUF proteins recognize target RNAs in a modular fashion, with each repeat binding to a single base, and a single RNA base forms hydrogen bond interactions with the TRM residues of only a single PUF repeat. However, in our complex, G+1 forms hydrogen bonds with residues in the two PUF repeats R9 and R10 (Supplementary Figure S5b), and A+2 forms hydrogen bonds with residues in both R8 and R9 (Supplementary Figure S5g).
PUF repeat R9 contains the A1E5-R2 motif. However, we were unable to observe any interaction between G+1 and Ala-542 in R9; instead, the N2 group of G+1 forms a hydrogen bond with the OG group of Ser-577 in R10. At the same time, the N1 and N2 groups of G+1 form hydrogen bonds with the OE1 and OE2 groups of Glu-546, respectively. Thus, the G+1 recognition code is the S1’E5-R2 motif, in which the S1’ structurally mimics the function of the residue at TRM position 1.
PUF repeat R8 contains a C1Q5-L2 motif in α-helix 2. However, the N3 group of A+2 does not interact with Cys-503 in R8 but forms a hydrogen bond with the OG group of Ser-540 in R9. At the same time, the N1 and N6 groups of A+2 interact with the OE1 and NE2 group of Gln-507 in R8, respectively. Thus, the A+2 recognition code is S1’Q5-L2. A+2 also makes stacking interactions with A+3 and Val-500 in R8.
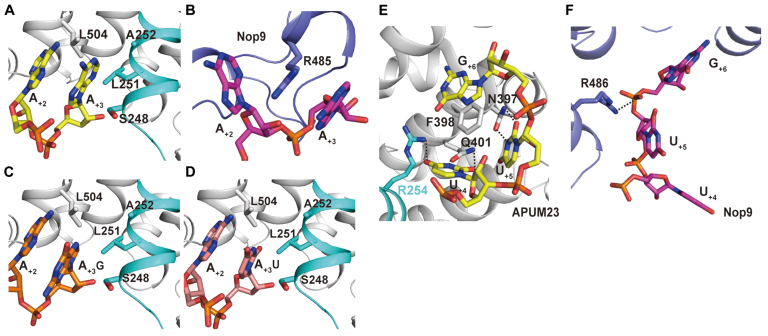
The insertion in PUF repeat R3’s involvement in the recognition of A+3
A+3 is not recognized by the APUM23 five-amino acids SGVVA motif in the second α-helix of PUF repeat R7, as would be expected. Instead, the A+3 adenine is stabilized by a hydrophobic pocket that comprises Ser-248, Leu-251 and Ala-252 in the αi2 helix of the insertion, Leu-504 in R8, and a stacking interaction with A+2 (Figure 4A). Yeast Nop9 makes a stacking interaction only with A+3 via the side chain of Arg-485 in PUF repeat R8 (Figure 4B). To investigate whether APUM23 shows a preference for A+3, we further solved the crystal structures of the APUM2385-655–GGAUUGACGG (11-mer A+3U) and APUM2385-655–GGAGUGACGG (11-mer G+3U) complexes at resolutions of 2.10 Å and 2.75 Å, respectively (Supplementary Table S2). The U+3 and G+3 positions showed no significant difference from that of A+3 (Figure 4C and D). ITC results showed that the binding affinity of APUM2385-655 for the 11-mer A+3U RNA (5′-GGAUUUGACGG-3′) was ∼1.6 times higher than that for 11-mer WT RNA (Table 1 and Supplementary Figure S7a and c). These results indicate that APUM23 has no selectivity for A+3.
Figure 4.
Recognition of A+3, U+4 and U+5 by APUM23. (A and B) Recognition of A+3 by APUM23 and Nop9. The insertion in the R3 PUF repeat of APUM23 is colored in cyan. (C and D) Positions of A+3G and A+3U in the crystal structures of APUM2385-655 in complex with 11-mer A+3G 5′-GGAGUUGACCG-3′ and 11-mer A+3U 5′-GGAUUUGACCG-3′, respectively. (E) APUM23 recognizes U+4 and U+5 through the R6 PUF repeat and the insertion in this repeat. Phe-398 has interactions with U+4, U+5 and G+6. (F) There are no interactions between Nop9 and U+4 and U+5.
Recognition of U+4 and U+5 by PUF repeat R6 and the insertion in R3
U+5 is typically recognized by the N1Q5-F2 motif in R6 (Figure 4E). The side chain of Asn-397 (position 1) forms hydrogen bonds with the O2 and N3 groups of U+5, the side chain of Gln-401 (position 5) forms hydrogen bonds with the O4 group of Gln-401 (position 5), and the benzene ring of Phe-398 (position 2) stacks in parallel between U+5 and G+6.
Unexpectedly, U+4 is also recognized by R6 (Figure 4E). The side chain of Gln-401 forms hydrogen bonds with the O2 and N3 groups of U+4 and the benzene ring of Phe-398 interacts vertically with U+4 (Figure 4E). The side chain of Arg-254 in the αi2 helix of the insertion in PUF repeat R3 forms a hydrogen bond with the O4 group of U+4. Thus, the recognition motif for U+4 is R1’Q5-F2. However, there are no interactions between Nop9 and the bases of U+4 and U+5 (Figure 4F).
The ITC results showed that the binding affinity of APUM2385-655 for the 11-mer U+4A RNA (5′-GGAAAUGACGG-3′) was approximately half that of the WT 11-mer WT RNA (Table 1 and Supplementary Figure S7a and d). This may have been because of the loss of the hydrogen bond interaction with Gln-401. Phe-398 and Gln-401 display more interactions with the RNA bases than the residues in other TRMs (Figure 4E). Phe-398 has interactions with U+4, U+5, and G+6, and Gln-401 has interactions with U+4 and U+5. These interactions indicate that Phe-398 and Gln-401 play important roles in the recognition of the 11-mer RNA target. ITC results suggested that the binding affinity of the 11-mer WT RNA for APUM23Q401E and APUM23F398A were ∼52-fold and 90-fold weaker, respectively, than that for APUM2385-655 (Table 1 and Supplementary Figure S7a, e and f).
Taken together, these findings indicate that APUM23 PUF repeat R6 is involved in the recognition of both U+4 and U+5.
The insertion in PUF repeat R3 blocks G0 from stacking with A+3
To investigate the importance of this insertion further, we expressed an APUM23 mutant without the insertion (APUM23Delete). Our ITC results showed that the binding affinity of the 11-mer WT RNA for APUM23Delete was ∼7.6-fold weaker than that for APUM2385-655 (Table 1 and Supplementary Figure S7a and g). We also obtained a 2.8-Å resolution crystal structure of APUM23Delete–GGAAUUGACGG (Figure 5A and Supplementary Table S2). Apart from the insertion, the structure of APUM23Delete is identical to that of APUM2385-655. G+1 to G+10 are located in the same locations in the APUM23Delete–RNA structure as in the APUM2385-655–RNA structure (Figure 5A). Surprisingly, G0 in the APUM23Delete–RNA structure is not recognized by APUM23 R10; instead, it bends round to stack with A+3 (Figure 5B). By superpositioning the APUM2385-655–GGAAUUGACGG and APUM23Delete–GGAAUUGACGG structures, we observed a steric clash between the insertion and the bent-around G0 (Figure 5C). This indicates that the presence of the insertion blocks G0 from bending.
Figure 5.
The insertion blocks G0 from stacking with A+3. (A) Superposition of the crystal structure of APUM2385-655-GGAAUUGACGG (gray, with the insertion in cyan) and APUM23Delete-GGAAUUGACGG (in magenta). (B) The 2Fo-Fc electron density map around G0 to G+10 in the crystal structure of APUM23Delete-GGAAUUGACGG, contoured at 1.0 sigma. (C) G0 bends to stack against A+3. The steric clash is highlighted by the red oval.
The ITC results suggested that the binding affinity of APUM23Delete for 11-mer WT RNA was ∼6.8-fold higher than that for the 10-mer RNA 5′-GAAUUGACGG-3′ (Table 1 and Supplementary Figure S7g and h). This may be due to A+3 instability caused by the loss of G0 and the insertion.
These results indicated that the insertion in APUM23 PUF repeat R3 has functions that include both RNA recognition and RNA conformation maintenance.
DISCUSSION
Three main PUF protein structural features have previously been reported (Figure 6). The classic crescent-shaped PUF proteins contain eight PUF repeats and choose their specific single-stranded RNA target via modular interactions with the RNA bases (6). The L-shaped Puf-A/Puf6 contains 11 PUF repeats and binds single- or double-stranded RNA via interactions with the phosphate backbone (3). The C-shaped yeast Nop9 contains ten PUF repeats and interacts with single-stranded RNA and the duplex region of the internal transcribed spacer 1 (ITS1) pre-rRNA or the 11-mer single-stranded 18S rRNA (17,38). In the present study, we solved the crystal structure of APUM23 in complex with 11-mer 5′-GGAAUUGACGG-3′ single-stranded RNA. This crystal structure showed that the third PUF repeat of APUM23 contains an insertion located at the inner concave surface. However, the crystallization of the Nop9–RNA complex did not include this insertion. Furthermore, the conformation of the same single-stranded target RNA (5′-GGAAUUGACGG-3′) differs in the structures of APUM23–RNA and Nop9–RNA, especially from A+3 to U+5. We speculate that this difference in RNA conformation is not caused by the removal of the insertion, for three reasons. First, the sequential difference between APUM23 and yeast Nop9 results in the difference in their target RNA conformations. The RNA recognition residues in the insertion in APUM23 PUF repeat R3 are not conserved in yeast Nop9 (Supplementary Figure S6), suggesting that the insertion of Nop9 may not interact with A+3 and U+4. In APUM23, the R6 PUF repeat stabilizes the conformation of both U+4 and U+5 through direct interactions. However, R6 in Nop9 has no interaction with U+4 or U+5, leaving them with greater flexibility. Second, removal of the insertion in APUM23 only affects the conformation of G0, which bends round to stack with A+3. In the APUM23Delete–RNA structure, PUF repeat R6 still recognizes U+4 and U+5, and G0 replaces the insertion to stabilize the conformation of A+3. Third, APUM2385-655 showed similar binding affinity toward both the 10-mer RNA (5′-GAAUUGACGG-3′) and the 11-mer RNA (5′-GGAAUUGACGG-3′). In contrast, the binding affinity of APUM23Delete for the 10-mer RNA is much weaker than that for the 11-mer RNA, possibly caused by the instability of A+3 resulting from the loss of both the insertion and G0. These findings indicate that the main function of the insertion in APUM23 is to stabilize the conformation of A+3 and to block G0 from bending. Taken together, the insertion in APUM23 plays roles in both stabilizing the conformation of A+3 and maintaining the right conformation of G0. However, the insertion in Nop9 may not participate in the recognition of 5′-GGAAUUGACGG-3′. Further investigation is needed to elucidate the physiological function of this insertion in APUM23 and yeast Nop9.
Figure 6.
APUM23 varies in its structural features. (A–D) Cartoon representations of the crystal structures of Pumilio1 (PDB code: 1M8W), Puf-A (PDB code: 4WZR), Nop9 (PDB code: 5SVD) and APUM23. Insertions in each protein are colored in cyan.
Our complex structure included some other atypical structural features. For instance, PUF proteins usually recognize target RNAs in a modular way, with a single RNA base interacting with only one PUF repeat; however, in our complex, A+2 interacts with both R8 and R9, and G+1 with both R9 and R10. In addition, R6 is involved in the recognition of both U+4 and U+5. One advantage of PUF proteins is that they can be engineered as RNA-binding tools to monitor and control the RNA metabolism in living cells (40). Our structural data may offer an alternative template for developing such engineered RNA-binding tools.
Pre-rRNA processing is a pivotal process during eukaryotic ribosome biogenesis. In reaching their final forms, pre-rRNAs undergo multiple cleavages, modifications, and refolding, performed by endo- and exonucleases, small nucleolar RNAs, and other regulatory proteins, such as yeast Nop9 and APUM23 (41–44). Although the sequence identity between them is only 18%, APUM23 and Nop9 share some functional similarity; APUM23 expression in yeast can partially rescue Nop9 deletion (28). APUM23 and Nop9 are both involved in regulating the cleavage of 35S pre-rRNA and the mature of 18S rRNA (2,28,45). However, how they regulate pre-rRNA processing through direct interaction with 18S rRNA remains unknown. We are eager to identify other partners (protein or RNA) of APUM23 to elucidate its roles in the regulation of pre-rRNA processing. It has been proposed that Nop9 protects 20S pre-rRNA in the nucleolus from premature cleavage by maintaining the endonuclease Nob1 binding site in ITS1 (38). Whether APUM23 has other rRNA binding sites remains to be investigated.
DATA AVAILABILITY
The atomic coordinates and structure factors (codes: 5WZG, 5WZH, 5WZI, 5WZJ and 5WZK) have been deposited in the Protein Data Bank (http://wwpdb.org/). Primary data are available upon request· from the corresponding author.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of Beamline BL17U, BL18U and BL19U at SSRF for the assistance in data collection. We also thank Dr Fudong Li for help with the model building of Se-Met labeled APUM23.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [2016YFA0500700]; Grant of the Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010101]; Chinese National Natural Science Foundation [31330018]; China Postdoctoral Science Foundation [2015M582009, 2016T90579]. Funding for open access charge: Ministry of Science and Technology of China [2016YFA0500700]; Grant of the Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010101]; Chinese National Natural Science Foundation [31330018]; China Postdoctoral Science Foundation [2015M582009, 2016T90579].
Conflict of interest statement. None declared.
REFERENCES
- 1. Quenault T., Lithgow T., Traven A.. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011; 21:104–112. [DOI] [PubMed] [Google Scholar]
- 2. Abbasi N., Kim H.B., Park N.i., Kim H.S., Kim Y.K., Park Y.I., Choi S.B.. APUM23, a nucleolar Puf domain protein, is involved in pre‐ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010; 64:960–976. [DOI] [PubMed] [Google Scholar]
- 3. Qiu C., McCann K.L., Wine R.N., Baserga S.J., Hall T.M.T.. A divergent Pumilio repeat protein family for pre-rRNA processing and mRNA localization. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:18554–18559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wickens M., Bernstein D.S., Kimble J., Parker R.. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002; 18:150–157. [DOI] [PubMed] [Google Scholar]
- 5. Lu G., Dolgner S.J., Hall T.M.. Understanding and engineering RNA sequence specificity of PUF proteins. Curr. Opin. Struct. Biol. 2009; 19:110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang X., McLachlan J., Zamore P.D., Hall T.M.T.. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002; 110:501–512. [DOI] [PubMed] [Google Scholar]
- 7. Wang X., Zamore P.D., Hall T.M.T.. Crystal structure of a Pumilio homology domain. Mol. Cell. 2001; 7:855–865. [DOI] [PubMed] [Google Scholar]
- 8. Gupta Y.K., Nair D.T., Wharton R.P., Aggarwal A.K.. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure. 2008; 16:549–557. [DOI] [PubMed] [Google Scholar]
- 9. Miller M.T., Higgin J.J., Hall T.M.. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 2008; 15:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu D., Stumpf C.R., Krahn J.M., Wickens M., Hall T.M.T.. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:20192–20197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu G., Hall T.M.. Alternate modes of cognate RNA recognition by human PUMILIO proteins. Structure. 2011; 19:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qiu C., Kershner A., Wang Y., Holley C.P., Wilinski D., Keles S., Kimble J., Wickens M., Hall T.M.T.. Divergence of Pumilio/fem-3 mRNA binding factor (PUF) protein specificity through variations in an RNA-binding pocket. J. Biol. Chem. 2012; 287:6949–6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Valley C.T., Porter D.F., Qiu C., Campbell Z.T., Hall T.M.T., Wickens M.. Patterns and plasticity in RNA-protein interactions enable recruitment of multiple proteins through a single site. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6054–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilinski D., Qiu C., Lapointe C.P., Nevil M., Campbell Z.T., Tanaka Hall T.M., Wickens M.. RNA regulatory networks diversified through curvature of the PUF protein scaffold. Nat. Commun. 2015; 6:8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jenkins H.T., Baker-Wilding R., Edwards T.A.. Structure and RNA binding of the mouse Pumilio-2 Puf domain. J. Struct. Biol. 2009; 167:271–276. [DOI] [PubMed] [Google Scholar]
- 16. Wang Y., Opperman L., Wickens M., Hall T.M.T.. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:20186–20191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang B., Ye K.. Nop9 binds the central pseudoknot region of 18S rRNA. Nucleic Acids Res. 2017; 45:3559–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dong S., Wang Y., Cassidy-Amstutz C., Lu G., Bigler R., Jezyk M.R., Li C., Hall T.M., Wang Z.. Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains. J. Biol. Chem. 2011; 286:26732–26742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Filipovska A., Razif M.F., Nygard K.K., Rackham O.. A universal code for RNA recognition by PUF proteins. Nat. Chem. Biol. 2011; 7:425–427. [DOI] [PubMed] [Google Scholar]
- 20. Adamala K.P., Martin-Alarcon D.A., Boyden E.S.. Programmable RNA-binding protein composed of repeats of a single modular unit. PNAS. 2016; 113:E2579–E2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hall T.M. De-coding and re-coding RNA recognition by PUF and PPR repeat proteins. Curr. Opin. Struct. Biol. 2016; 36:116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Campbell Z.T., Valley C.T., Wickens M.. A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nat. Struct. Mol. Biol. 2014; 21:732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koh Y.Y., Wang Y., Qiu C., Opperman L., Gross L., Tanaka Hall T.M., Wickens M.. Stacking interactions in PUF-RNA complexes. RNA. 2011; 17:718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hall T.M. Expanding the RNA-recognition code of PUF proteins. Nat. Struct. Mol. Biol. 2014; 21:653–655. [DOI] [PubMed] [Google Scholar]
- 25. Tam P.P., Barrette-Ng I.H., Simon D.M., Tam M.W., Ang A.L., Muench D.G.. The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010; 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Francischini C.W., Quaggio R.B.. Molecular characterization of Arabidopsis thaliana PUF proteins–binding specificity and target candidates. FEBS J. 2009; 276:5456–5470. [DOI] [PubMed] [Google Scholar]
- 27. Abbasi N., Park Y.-I., Choi S.-B.. Pumilio Puf domain RNA-binding proteins in Arabidopsis. Plant Signal. Behav. 2014; 6:364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang T., Kerstetter R.A., Irish V.F.. APUM23, a PUF family protein, functions in leaf development and organ polarity in Arabidopsis. J. Exp. Bot. 2014; 65:1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang C., Muench D.G.. A Nucleolar PUF RNA-binding Protein with Specificity for a Unique RNA Sequence. J. Biol. Chem. 2015; 290:30108–30118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A.. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sheldrick G.M. A short history of SHELX. Acta Crystallogr. A. 2008; 64:112–122. [DOI] [PubMed] [Google Scholar]
- 32. Zwart P.H., Afonine P.V., Grosse-Kunstleve R.W., Hung L.-W., Ioerger T.R., McCoy A.J., McKee E., Moriarty N.W., Read R.J., Sacchettini J.C.. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 2008; 426:419–435. [DOI] [PubMed] [Google Scholar]
- 33. Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006; 62:1002–1011. [DOI] [PubMed] [Google Scholar]
- 34. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DeLano W.L. The PyMOL molecular graphics system. 2002; http://www.pymol.org.
- 38. Zhang J., McCann K.L., Qiu C., Gonzalez L.E., Baserga S.J., Hall T.M.. Nop9 is a PUF-like protein that prevents premature cleavage to correctly process pre-18S rRNA. Nat. Commun. 2016; 7:13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheong C.G., Hall T.M.. Engineering RNA sequence specificity of Pumilio repeats. PNAS. 2006; 103:13635–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y., Wang Z., Tanaka Hall T.M.. Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism. FEBS J. 2013; 280:3755–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Woolford J.L. Jr, Baserga S.J.. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013; 195:643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weis B.L., Kovacevic J., Missbach S., Schleiff E.. Plant-Specific Features of Ribosome Biogenesis. Trends Plant Sci. 2015; 20:729–740. [DOI] [PubMed] [Google Scholar]
- 43. Henras A.K., Plisson-Chastang C., O’Donohue M.F., Chakraborty A., Gleizes P.E.. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA. 2015; 6:225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fernandez-Pevida A., Kressler D., de la Cruz J.. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdiscip. Rev. RNA. 2015; 6:191–209. [DOI] [PubMed] [Google Scholar]
- 45. Thomson E., Rappsilber J., Tollervey D.. Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA. 2007; 13:2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors (codes: 5WZG, 5WZH, 5WZI, 5WZJ and 5WZK) have been deposited in the Protein Data Bank (http://wwpdb.org/). Primary data are available upon request· from the corresponding author.