Abstract
By analyzing multi-tissue gene expression and genome-wide genetic variation data in samples from a vervet monkey pedigree, we generated a transcriptome resource and produced the first catalogue of expression quantitative trait loci (eQTLs) in a non-human primate model. This catalogue contains more genome-wide significant eQTLs, per sample, than comparable human resources, and reveals sex and age-related expression patterns. Findings include a master regulatory locus that likely plays a role in immune function, and a locus regulating hippocampal long non-coding RNAs (lncRNAs), whose expression correlates with hippocampal volume. This resource will facilitate genetic investigation of quantitative traits, including brain and behavioral phenotypes relevant to neuropsychiatric disorders.
Efforts to understand how genetic variation contributes to common diseases and quantitative traits increasingly focus on the regulation of gene expression. Most loci identified through genome-wide association studies (GWAS) lie in non-coding genome regions1, and are enriched for eQTLs; SNPs regulating transcript levels, primarily of nearby genes2. This observation suggests that eQTL catalogs may signpost variants responsible for GWAS signals3.
Normal function of complex organisms depends on tightly regulated gene expression at specific developmental stages in specific cell types. Existing human eQTL datasets likely miss information relevant to understanding disease, as most known human eQTLs have been identified in adults, largely from lymphocytes or lymphoblastoid cell lines4,5. This lack is particularly striking for neuropsychiatric disorders, given the inaccessibility of brain in living individuals and the enormous modifications occurring in it across development6.
The Genotype Tissue Expression (GTEx) project, using samples from post-mortem donors7, has begun to remedy the lack of human data connecting genotypic variation and multi-tissue transcriptome variation. The GTEx eQTL catalog is the most extensive such resource available7. However limitations of GTEx, inherent to human research, motivate the generation and investigation of equivalent resources from model organisms. Advantages of model systems include: (1) feasibility of controlling for inter-individual heterogeneity in environmental exposures and minimizing the interval between death and tissue preservation; (2) practicability of obtaining sizable numbers of multi-tissue samples across development; and (3) opportunity to systematically phenotype individuals carrying particular eQTL variants. The similarities between humans and non-human primates (NHP) in behavior, neuroanatomy, and brain circuitry8,9,10, make NHP eQTLs particularly valuable for illuminating neuropsychiatric disorders.
We report here, in Caribbean vervets (Chlorocebus aethiops sabaeus) from the Vervet Research Colony (VRC) extended pedigree, the first NHP resource combining genotypes from whole genome sequencing (WGS)11, multi-tissue expression data across post-natal development, controlled environmental exposures (Online Methods), and quantitative phenotypes relevant to human brain and behavior. Caribbean vervets are Old World monkeys whose population expanded dramatically from a founding bottleneck occurring when West African vervets were introduced to the Caribbean in the 17th Century10; genetic variation has drastically declined in Caribbean vervet populations, enriching them for numerous deleterious alleles.
Through necropsies performed under uniform conditions, we obtained brain and peripheral tissue samples from captive VRC vervets. Using these resources we have delineated cross-tissue RNA sequencing (RNA-Seq) based expression profiles for seven of these tissues, across multiple developmental stages from birth to adulthood. We identified numerous local and distant eQTLs in each tissue, and validated a locus associated with multiple distant eQTLs, observed previously using pedigree-wide microarrays12. Additionally, we demonstrated the relevance of vervet eQTLs to higher-order traits; hippocampus-specific local eQTLs regulate a set of lncRNAs associated with hippocampal volume, a phenotype related to neuropsychiatric disorders13.
Results
We investigated two datasets. Dataset 1, described previously12, consists of gene expression levels obtained by hybridizing all available VRC, whole blood-derived, RNA samples (N=347) to Illumina HumanRef-8 v2 microarrays, which we used because no vervet arrays are available. After filtering out probe sequences not represented in the vervet genome14 or containing common vervet SNPs11, we estimated expression levels at 6,018 probes, corresponding to 5,586 unique genes (Supplementary Data 1, Supplementary Table 1). Dataset 2 consists of RNA-Seq reads from seven tissues collected under identical conditions from each of 58 VRC monkeys (representing 10 developmental stages, from birth through adulthood, Online Methods). Five of these tissues play prominent roles in cognitive and behavioral phenotypes15–17: Brodmann area 46 [BA46], a cytoarchitectonically defined region encompassing most of dorsolateral prefrontal cortex (DLPFC); hippocampus; caudate nucleus, a component of dorsal striatum; pituitary gland; and adrenal gland. The other two tissues (cultured skin fibroblasts and whole blood) are relatively accessible, and thus widely used in studies aimed at identifying biomarkers. We assessed expression of 33,994 annotated genes, but minimized spurious signals by excluding genes expressed in < 10% of individuals or at lower than one read per tissue(Supplementary Table 2). Principal components analysis (PCA) of Dataset 2 showed that, overall, expression levels clustered more by tissue than by individual (Supplementary Fig. 1). In hierarchical clustering, however, adrenal, pituitary, and fibroblasts cluster separately from brain and blood (data not shown); in GTEx, in contrast, blood clusters separately from other tissues. While most genes were expressed in multiple tissues, 137 genes demonstrated strong expression in only one tissue (Supplementary Table 3).
Sources of Variation in Multi-tissue Expression Data
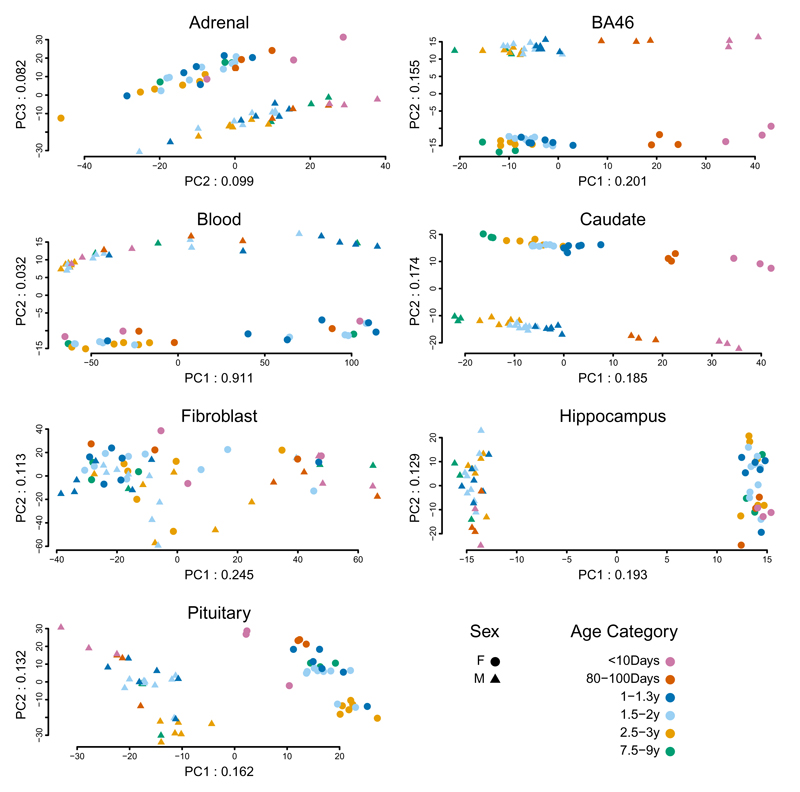
The availability (Dataset 2), of multiple samples from both sexes at each age point enabled us to examine developmental trajectories and sex differences in gene expression. To maximize our ability to observe patterns, we conducted PCA on the expression of the 1,000 most variable genes, separately by tissue (Fig. 1). Comparison of the ranks of expression of the orthologs of these genes in matched tissues in humans and rhesus macaques yielded Spearman correlations of between ~0.5-0.8 and ~0.3-0.4, respectively (Supplementary Note and Supplementary Tables 4-6).
Fig. 1.
PCA of 1,000 genes with the most variable expression levels. Analysis was performed separately by tissue; sample size was 60 animals for adrenal, blood, fibroblasts, and pituitary and 59 for BA46, caudate, and hippocampus. Numbers in the labels for x and y axes indicate the proportion of total variance accounted for by that PC.
Among the seven vervet tissues, the patterns in BA46 and caudate display the clearest association with development; PC1 (20.1% of BA46 variability and 18.5% of caudate variability) distinguishes the vervets, nearly linearally, by age. All tissues except fibroblast show sharply demarcated expression patterns between males and females; on PC1 (hippocampus and pituitary, 19.3% and 16.2% of variability, respectively), on PC2 (BA46, caudate and blood, 15.5%, 17.4%, and 3.2% of variability, respectively), and on PC3 (adrenal, 8.2% of variability).
As an initial, descriptive exploration of the biology underlying these patterns, we identified, in brain and endocrine tissues, the genes in the top and bottom 10% of the distribution of PC loadings on PCs 1, 2, and 3 (200 genes per tissue, per PC). We evaluated the known functions of these genes, which contribute most to the variance explained by the PCs in relation to sex (BA46, caudate, hippocampus, pituitary, and adrenal, Supplementary Table 7, Supplementary Note) or age (BA46 and caudate, Supplementary Table 8).
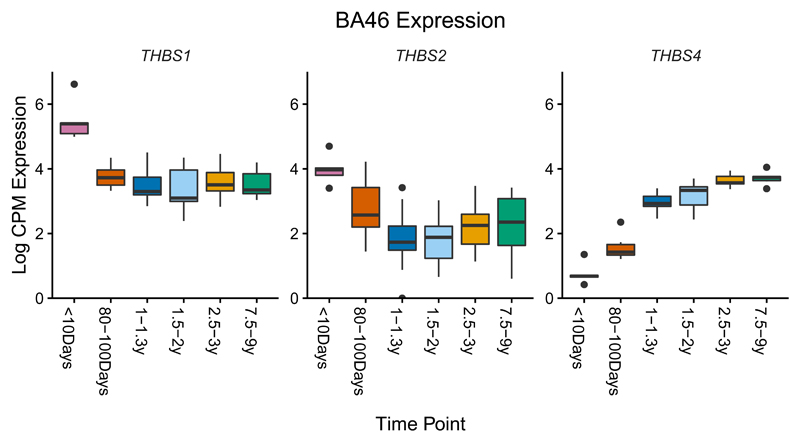
Age-related expression patterns in BA46 and caudate highlight numerous genes essential for nervous system development or implicated in human diseases. For example, three thrombospondin genes controlling synaptogenesis show a clear developmental pattern in BA46; THBS1 and THBS2 are upregulated in neonates, while THBS4, a gene upregulated during human brain evolution18, shows increasing expression across development (Fig. 2). Striking age-related expression patterns in BA46 and caudate are observed for other notable genes (Supplementary Fig. 2, Supplementary Note); orthologs of these genes in human and rhesus macaque brain tissues that are most equivalent to vervet BA46 and caudate (Online Methods) show patterns are similar to, but less pronounced than those in vervet (Supplementary Fig. 3, 4). Given the PCA results showing an age-related component to gene expression variation that differs by tissue, we conducted a differential expression analysis, using age as both a continuous and a categorical predictor in two different linear models. Nearly 8,000 genes across all seven tissues show significant differential expression by age for either analysis, mostly with very small effects (Supplementary Table 9)
Fig. 2.
Boxplot of log counts per million (CPM) expression in samples of BA46 from 58 animals vs. timepoint, for three genes with a strong relationship between expression pattern and age. The inter-quartile range defines the height of the box, and whiskers extend to 1.5x the inter-quartile range. Outliers are indicated as individual points. In each box, the median is represented by the horizontal black bar.
To evaluate whether cell-type heterogeneity influences interpretation of our expression and eQTL results for blood and brain tissues, we conducted a transcriptional deconvolution analysis of these tissues, using published data19,20 (Supplementary Fig.5). We estimated the diversity of cell types per sample in each tissue by calculating entropy, observing that blood has substantially higher diversity of cell types than do the three brain tissues (Supplementary Fig. 5).
We also examined the relationship between the proportion of specific cell types and developmental stage. For BA46 and hippocampus, the proportion of Oligodendrocyte Precursor cells decreases as age increases, as observed previously in human21; in caudate, the proportion of this cell type increases with age. Similarly, the proportion of neurons increases with age in BA46 and hippocampus, and decreases with age in caudate. (Supplementary Fig. 6-9). We found no correlation between estimated cell proportions and major PC axes in any tissue. These estimated proportions may not fully reflect in vivo cellular composition, but any bias would remain relatively systematic across animals and so unlikely to confound other analyses.
We evaluated the effect of RNA-Seq sample batch on transcriptomic profiles and PC patterns (Supplementary Note). As batch showed association with expression profiles in pituitary and adrenal (PC2) and caudate and pituitary (PC3), we included it as a covariate in eQTL analyses.
Identification of eQTLs
Whole genome sequencing (WGS) of 721 VRC monkeys provided the first NHP genome-wide, high-resolution genetic variant set11: 497,163 WGS-based SNPs that tag common variation genome-wide. Using these SNPs we conducted separate GWAS of Datasets 1 and 2 to identify local (probes/genes < 1 Mb from an associated SNP) and distant (all other probe/gene-SNP associations) eQTLs in each dataset. Covariates in all eQTL analyses included age, sex, and batch.
Using SOLAR22, we identified significant estimated heritability for 3,417 probes in Dataset 1 (out of the 6,018 filtered probes that we evaluated, corresponding to 5,586 unique genes) at a false discovery rate (FDR) threshold < 0.01 (Supplementary Data 1, 2). A GWAS of each heritable probe identified one or more significant eQTLs at 461 local and 215 distant probes (Bonferroni-corrected thresholds of 4.8 x 10-8 for local and 1.5 x 10-11 for distant eQTLs, Table 1, Supplementary Data 3). Approximately 35% of probes with a significant eQTL (173/498) displayed at least one local and one distant significant association.
Table 1. Gene expression data sets. The number of probes/genes with at least one significant local and distant eQTL (at Bonferroni corrected thresholds) are presented. We have 80% power to detect distant eQTLs accounting for 15% of the variability in expression in Dataset 1 and 66% of the variability in Dataset 2.
| Tissue | Probes/genes analyzeda | Local eQTLb | Distant eQTLc | %Distant eQTL on same chr |
|---|---|---|---|---|
|
Dataset 1: Microarray | ||||
| Blood | 3,417 | 461 | 215 | 80.8% |
|
Dataset 2: RNA-seq | ||||
| Adrenal | 25,187 | 555 | 80 | 54.5% |
| BA46 | 27,530 | 307 | 30 | 81.8% |
| Blood | 33,776 | 60 | 4 | 100.0% |
| Caudate | 28,249 | 441 | 47 | 69.0% |
| Fibroblast | 22,328 | 239 | 43 | 33.2% |
| Hippocampus | 26,957 | 361 | 45 | 70.6% |
| Pituitary Gland | 27,236 | 596 | 80 | 77.5% |
microarray dataset (Dataset 1) with an initial set of 22,184 probes on Illumina HumanRef-8 v2 (6,018 probes passed filters described in Supplementary Table 1; 3,417 were heritable); RNA-Seq (Dataset 2) with an initial set of 33,994 genes annotated in vervet
Local eQTL are eQTL that are within 1 Mb of the gene. Bonferroni threshold for Dataset 1: 4.8 x 10-8; Bonferroni threshold for Dataset 2: 6.5 x 10-10
Distant eQTL are more than 1 Mb away from the gene, and may be on the same or a different chromosome. Bonferroni threshold for Dataset 1: 1.5 x 10-11; Bonferroni threshold for Dataset 2: 5.3 x 10-13
In Dataset 2 we observed, for each of the five solid tissues, between 361-596 genes with local eQTLs and 30-80 genes with distant eQTLs. For blood and fibroblasts, 60 and 239 genes showed local eQTLs and 4 and 43 genes showed distant eQTLs, respectively, all at Bonferroni corrected thresholds (6.5 x 10-10 [local] and 5.3 x 10-13 [distant], Table 1, Supplementary Data 4). The paucity of eQTLs in blood likely reflects heterogeneity in the proportions of different cell types in this tissue, as identified in deconvolution analyses (Supplementary Fig. 1, 5). The paucity of eQTLs in fibroblasts has no obvious explanation, although we analyzed fewer genes, overall in fibroblasts than in tissues with cellular heterogeneity. At Bonferroni thresholds, we had 80% power to detect a significant local eQTL accounting for 11% of variability in expression in Dataset 1, and accounting for 55% of variability in expression in Dataset 2. For about 70% of Bonferroni-significant eQTLs (local and distant and in all tissues), the SNPs demonstrating association had minor allele frequency > 30% (Supplementary Table 10).
We considered whether genotypic variation within the vervet pedigree could confound the effects of age in generating the strong loadings on genes in age-associated PCs in BA46 and caudate. Among the 200 genes with such loadings, in caudate, 37 genes showed evidence of eQTLs, using the more liberal FDR controlling procedure. For these 37 genes, we modeled expression as a function of both age and genotype, using the most significant eQTLs, and found that genotype could not account for the age-association (data not shown). Similarly, in BA46, 26 genes showed evidence of an eQTL, for only one of which (LOC103219658) could genotype partially account for the age-association. Using genes without age effects as reference (Supplementary Table 9), we observed that genes with age-related patterns are depleted for eQTLs (Supplementary Table 11); this finding agrees with predictions that purifying selection causes such depletion in genes that are important at specific developmental timepoints23.
Comparison to Human eQTLs
While the eQTLs summarized in Table 1 are genome-wide significant at Bonferroni thresholds, we also applied FDR-controlling procedures, to expand the list of local eQTLs for more exploratory investigations, and to make our results comparable to those of GTEx (Table 2). We controlled FDR for eGenes at 0.05 (Online Methods), accounting for multiple testing using a hierarchical error controlling procedure developed for multi-tissue eQTL analysis24. We applied this same procedure to GTEx eQTLs to facilitate comparisons between the datasets.
Table 2. Comparison of specific genes with local eQTL in Vervet Dataset 2 to GTEx. For each tissue we present the number of genes with at least one significant local eQTL in Vervet (at FDR thresholds).
| Tissue | Vervet number of individuals | # Local eQTL Vervet Genesa | GTEx number of individuals | GTEx number of eGenesa | # Vervet Genes with Human Ortholog | # Orthologous Genes Tested in GTExb | % Tested Genes p<0.05 | % Tested Genes p <.05/# tested Genesc | % Tested Genes significant genome-wide in GTExd |
|---|---|---|---|---|---|---|---|---|---|
| Adrenal | 58 | 2932 | 126 | 2915 | 1828 | 1674 | 100% | 28.7% | 18.2% |
| Blood | 58 | 574 | 338 | 5438 | 264 | 229 | 100% | 70.7% | 38.9% |
| Caudate | 57 | 3140 | 100 | 2396 | 1737 | 1548 | 100% | 24.6% | 14.1% |
| Hippocampus | 58 | 2437 | 81 | 1405 | 1436 | 1296 | 100% | 18.4% | 9.2% |
| Pituitary | 58 | 3395 | 87 | 2222 | 1863 | 1743 | 100% | 20.7% | 13.0% |
The number of eGenes found in the multi-tissue hierarchical FDR procedure applied to vervet Dataset 2 and to GTEx.
Vervet genes with a human ortholog that were not tested in GTEx were filtered by their QC procedures
The threshold for significance corrected for the number of genes compared between Vervet and GTEx (column 7).
Genes were declared significant by GTEx at an FDR of 0.05.
Despite having a smaller sample size than GTEx V6, we identify more local eQTLs (at FDR thresholds applied to both datasets, Online Methods) for the five solid tissues evaluated in both resources (Table 2). The larger number of local eQTLs in the vervets likely reflects the more homogenous environment of colonied NHPs compared to humans, and the more uniform tissue collection process in this study. Specific vervet and GTEx eQTLs overlap, substantially. All vervet genes with a genome-wide significant eQTL (FDR <0.05) also display a human eQTL in the same tissue (p< 0.05), given that the gene has a known human ortholog and was tested in GTEx. Using instead GTEx’s defined significance threshold for orthologous genes (FDR < 0.05), an average of 19% of vervet eQTLs display a human eQTL (Table 2). Restricting the comparison to Bonferroni-significant local eQTLs, an average of 23% of vervet eQTLs also have an eQTL in the same tissue in GTEx (Supplementary Table 12).
We additionally compared our local eQTL results for brain tissues to the Open Access version of human eQTLs from DFPLC, available from CommonMind Consortium (CMC)25. Almost 90% of vervet brain local eQTL genes with human orthologs in the CMC dataset have a local eQTL at FDR<0.05 in that dataset (Supplementary Note and Supplementary Table 13).
eGene Sharing Among Tissues
In all tissues except blood, tissue-specific locally regulated eGenes (genes with a significant local eQTL, see Online Methods) are more common than local eGenes shared among tissues (Supplementary Fig. 10). Adrenal and pituitary, organs inter-regulated in the same neuroendocrine pathway, display the largest number of shared local eGenes (300). The three brain regions share 239 such eGenes, while 229 eGenes are shared across all tissues but blood, and 82 eGenes are shared across all seven tissues.
Genomic Distribution of eQTLs
Regulatory variants occur most frequently in functional genomic regions26. Vervet local eQTLs are clearly enriched in regions encompassing exons, introns and adjacent flanks and depleted in intergenic regions (Supplementary Fig. 11, Supplementary Table 14). As in other primates27, vervet eQTLs are enriched around gene boundaries (transcription start site [TSS] and transcription end site [TES]) (Supplementary Fig. 12).
We used previously published chromatin immunoprecipitation with DNA sequencing (ChIP-Seq) data28,29 to evaluate eQTL distribution in H3K4me3 enriched regions (promoters) and H3K27ac enriched regions (which include acetylated promoters and enhancers). As H3K4me3 marks are typically conserved across tissues we analyzed them using vervet liver data29. As enhancer marks are more tissue specific29–31 we analyzed H3K27ac marks in both vervet liver and available brain data (caudate and prefrontal cortex) from rhesus macaque28,29. The promoter regions show stronger enrichment for vervet local eQTLs than either genic or H3K27ac-enriched regions (Supplementary Fig. 11, Supplementary Table 14).
Validation of Distant eQTLs
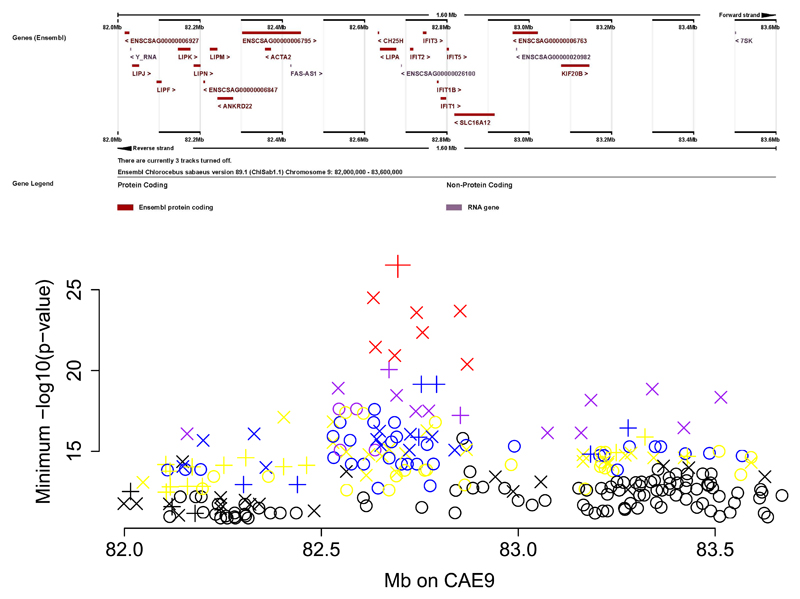
Dataset 1 is well-powered for discovery of distant eQTLs. Among 215 genes for which we observed genome-wide significant associations to one or more distant eQTLs, a locus on CAE9 in which 76 SNPs across a ~500 Kb region displayed genome-wide significant local eQTL signals, stood out for showing association to multiple unlinked genes. For each of these 76 SNPs we identified genome-wide significant distant eQTLs at between five and 14 genes, on different vervet chromosomes (2,127 total distant SNP-gene associations, Fig. 3, Supplementary Table 15).
Fig. 3.
Master regulatory locus on vervet chromosome CAE 9. Upper panel: Ensembl view of the CAE 9 region. Lower panel: The minimum –log10(p-value) for each SNP in association analyses vs. expression in 347 animals of microarray probes on different chromosomes. The symbols are color-coded to represent the number of probes significantly associated to each SNP: 1-2 probes (black), 3-4 probes (yellow), 5-6 probes (blue), 7-10 probes (purple), 11-14 probes (red). Symbols indicate the p-value from analysis of expression in Dataset 2 (RNA-Seq). Cross: p<2.35e-05; X: p<0.001; circle: p>0.001. The large red X at the top of the plot is CAE9_82694171.
Because we obtained Dataset 2 using a different platform from Dataset 1, and from a mostly non-overlapping sample (only 6 vervets were in both datasets), we evaluated it for replication of the CAE 9 distant eQTLs, recognizing the limited power of this smaller dataset. Considering the percent of variance accounted for by the distant eQTLs in Dataset 1 (Supplementary Table 15), we have 82% power to identify eQTLs in Dataset 2, with 58 animals, when the SNP accounts for ≥35% of expression variance, using a significance threshold (p<2.35 x 10-5) accounting for 2,127 tests. Two genes, ST7 (31 SNPs) and YPEL4 (22 SNPs) replicate association at this threshold, with estimated regression coefficients for these 53 SNP-gene associations being similar in magnitude and direction in the two datasets (Supplementary Table 16). We confirmed eight distant associations (RANBP10, LCMT1, ST7, TMEM57, YPEL4, NARF, STXBP1, DEDD2) across the two datasets, with at least one SNP demonstrating association at p<0.05 (Supplementary Table 15).
These results indicate that the CAE 9 eQTL is a master regulatory locus (MRL). This genomic segment contains a cluster of acid lipase genes and interferon-inducible genes, including IFIT1B (Interferon-Induced Protein With Tetratricopeptide Repeats 1B), a gene implicated in viral resistance in vervets, but not humans32. The same SNPs contributing to the MRL are also genome-wide significant local eQTLs for IFIT1B; GTEx reports no significant local eQTLs for IFIT1B in human blood.
Expression of IFIT1B correlates strongly with expression of the distant genes regulated by this eQTL (Supplementary Note, Supplementary Table 17). We conducted mediation analyses in Dataset 1 for a SNP (CAE9_82694171) that, at Bonferroni corrected significance thresholds, is both a distant eQTL for all 14 genes and a local eQTL for IFIT1B (Supplementary Table 18). This SNP accounts for 19-37% of the variance in expression level of the 14 genes not on CAE 9. When we conditioned these analyses on IFIT1B expression, the magnitude of distant associations diminished substantially, the variance accounted for by this SNP dropping to ≤10% for all 14 genes. These results indicate that IFIT1B, under direct control of a local eQTL on CAE 9, influences expression of 14 other genes spread across the genome. Such mediation by local eQTLs of distant eQTLs provides a further validation of the latter loci33.
Hippocampus eQTLs in a Region Linked to Hippocampal Volume
As an initial investigation of the impact of vervet eQTLs on higher order traits we focused on MRI-based hippocampal volume, a highly heritable trait in the VRC (h2 =0.95)34, for which the strongest QTL signal genome-wide (peak LOD score 3.42) lies in an ~8.3 Mb segment of CAE 18. Power simulations (SOLAR) indicate that, in the VRC pedigree, quantitative trait data for 347 vervets (the number with hippocampal volume data) provide 80% power to detect a locus with LOD=2 when locus-specific heritability is > 45%.
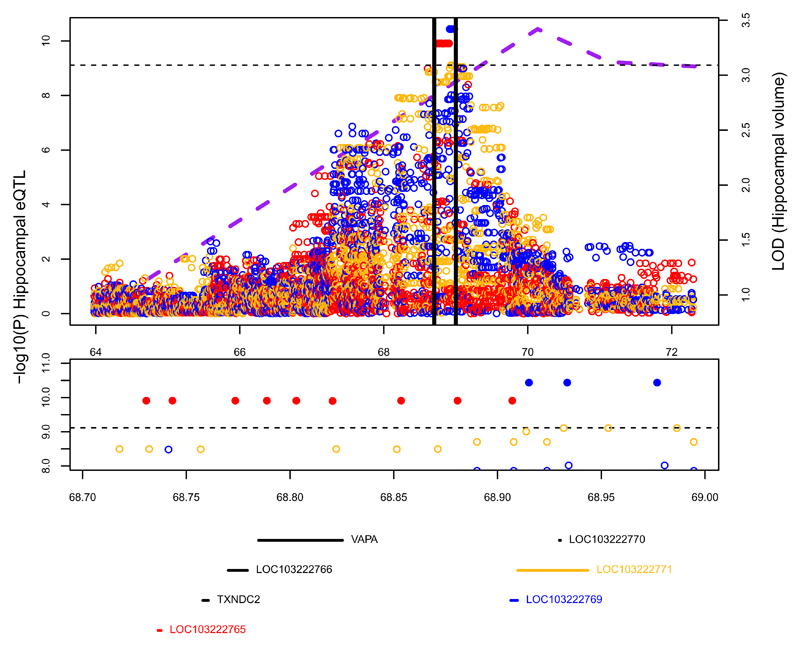
In the center of the broad region around this linkage peak, two hippocampus-specific local eQTLs were genome-wide significant (Bonferroni threshold, Fig. 4). These SNPs reside in, and regulate expression of, two lncRNAs located 168 Kb apart: LOC103222765 (nine associated SNPs) and LOC103222769 (three associated SNPs). An additional lncRNA, LOC103222771, situated two bp from LOC103222769, shows hippocampal specific association to six SNPs at a significance level (p < 10-9) just above the genome-wide threshold. While all three genes display hippocampus-specific eQTLs, the genes themselves are expressed across all seven tissues that we analyzed, and show no significant sex or age specific differences in expression patterns (data not shown). The incomplete database annotation of lncRNAs35 limits comparative analyses of such genes among primates; a BLAST search found a homolog for LOC103222765 in the white-tufted-ear marmoset and one for LOC103222771, in the crab-eating macaque. While LOC103222765 overlaps a coding gene (RAB31), LOC103222769 and LOC103222771 do not overlap exons of any coding genes36.
Fig. 4.
Hippocampal volume QTL and local hippocampal eQTLs in RNA-Seq analysis. Top panel: purple dotted line is the multipoint LOD score for hippocampal volume (measured in 347 animals). Circles represent evidence for association of SNPs to hippocampal expression in 58 animals of three genes: LOC103222765 (red), LOC103222769 (blue) and LOC103222771 (gold). Solid circles indicate genome-wide significant associations. The region between the black vertical lines is blown up in the middle and bottom panels. The horizontal dotted line represents the genome-wide significant threshold for local eQTLs. Middle panel: SNPs with –log10(p-value)>8 for association to expression in hippocampus, color codes are as in the top panel. Bottom panel: Genes sited between 68.7 and 69 Mb (the eQTL region). Color codes are as in the top panel. The Pearson correlations for expression between these three genes are: LOC103222765-LOC103222769 r=-0.16; LOC103222765-LOC103222771 r=0.32; LOC103222769-LOC103222771 r=0.60.
Given the physical proximity of these lncRNAs, we used multivariate conditional analyses to evaluate whether the regulation of these genes depends on a single or multiple independent eQTLs. For each lncRNA we designated a “lead SNP” (the SNP most significantly associated to its expression, Supplementary Table 19). For both LOC103222769 and LOC103222771, modeling expression as a function of both lead SNPs diminished the significance levels for both SNPs (Supplementary Table 19), suggesting that one eQTL regulates both genes. Modeling LOC103222765 expression as a function of its lead SNP and the lead SNP of the other two genes, the lead SNP for LOC103222765 remains significant, while the other two lead SNPs are non-significant, confirming the “distinctness” of this signal (Supplementary Table 19). This analysis suggests two eQTLs in this region; one associated with LOC103222765, and the second associated with LOC103222769 and LOC103222771.
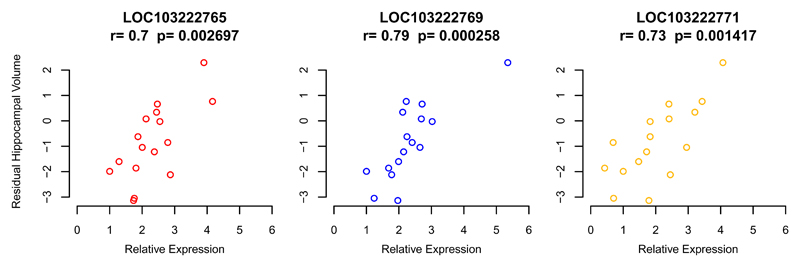
We observed, in six vervets with both MRI and RNA-Seq data, a positive correlation between hippocampal expression of LOC103222765, LOC103222769 and LOC103222771, and hippocampal volume. To extend this observation, we assessed, using an independent platform, quantitative real-time PCR, LOC103222765, LOC103222769 and LOC103222771 hippocampal expression in these six vervets and 10 additional vervets with both hippocampal RNA and MRI. In this expanded sample, we identified significant positive correlations (Fig. 5) between LOC103222765, LOC103222769 and LOC103222771 expression and hippocampal volume. While the above data suggest that genetic variation in this region regulates these lncRNAs and also has a strong impact on the MRI phenotype, colocalization analysis37 does not support the hypothesis that a single variant accounts for both the genome-wide linkage (MRI) and GWAS (eQTL) findings (8.2% posterior probability).
Fig. 5.
Correlation in 16 animals of hippocampal volume (MRI) with hippocampal expression of LOC103222765 (left), LOC103222769 (middle) and LOC103222771 (right). The expression data are from qRT-PCR. Quantification was performed using the relative standard curve method, with the reference gene HPRT1 used as an endogenous control for normalization of the interpolated lncRNA quantities. Hippocampal volume measurements are residuals from a regression on covariates of age and sex. “r” is the Pearson correlation coefficient, and the p-value tests the null hypothesis that r=0. The Pearson correlation between expression of these three genes are: LOC103222765-LOC103222769 r=0.56; LOC103222765-LOC103222771 r=0.64; LOC103222769-LOC103222771 r=0.63.
Discussion
We describe here the first NHP resource for investigating the genetic contribution to inter-individual variation in multi-tissue gene expression across development. This resource complements GTEx38,39, but is differentiated from it by study designs that are infeasible in human research. Notably, the age-based sampling enabled delineation of tissue-specific expression profiles in relation to developmental trajectories. These profiles illuminate biological processes associated with expression patterns of particular genes. For example, several genes critical in synapse formation and postnatal myelination of the central nervous system40–43 contribute to the near linear age-related pattern observed in BA46 and caudate, and suggest that the observed expression pattern reflects this process. Conversely, the lack of such a developmentally specific pattern in the hippocampus may relate to the lifelong generation of functional neurons in this tissue that underpins its functions in learning and memory44,45.
Three factors increased the signal-to-noise ratio of vervet eQTL analyses, relative to human studies: (i) homogeneity of environmental exposures; (ii) greater control over necropsy conditions; and (iii) restricted genetic background of the population. These factors enabled us to identify 385 genes with genome-wide significant distant eQTLs, including the MRL at IFIT1B.
The function of IFIT1B, one of a cluster of five IFIT genes, is poorly understood. It is a paralog of IFIT1, which is involved in innate antiviral immunity in mammals, broadly46, and in regulation of gut microbiota in mouse47. In some mammals IFIT1B contributes to discrimination between “self versus non-self” transcripts based on the lack of 2’ O-methylation on mRNA 5’ caps in viruses, a so-called cap0 structure32. Vervet IFIT1B recognizes and inhibits replication of viruses with cap0-mRNAs, while human IFIT1B lacks this function32. This functional divergence of IFIT1B antiviral activity may reflect the divergence of the human lineage from that of other primates, in exposures and adaptations to particular pathogens, including arboviruses responsible for diseases such as encephalitis, dengue, and yellow fever.
Investigations of genes regulated by IFIT1B in vervet might reveal mechanisms for its role in defense against viral pathogens. While these genes do not act together in any annotated pathway, the products of several of them have immune functions. For example, RANBP10, a transcriptional coactivator, promotes viral gene expression and replication in HSV-1 infected cells48. SUGT1, a cell cycle regulator, is the homolog of SGT1, an essential component of innate immunity in plants and mammals49,50, while TMEM57 shows genome-wide significant association in human to blood markers of inflammation51.
Just as GTEx data help refine signals from human GWAS of complex traits5, we used vervet hippocampal eQTLs to identify a set of lncRNAs as candidate genes for hippocampal volume. The genetic and environmental homogeneity of the relatively small vervet study sample likely facilitated these findings, and supports multi-tissue vervet eQTL studies as a strategy for identifying loci with a large impact on higher-order phenotypes, generally. The tissues examined to date are a fraction of those available from the same vervets; the investigations reported here can be extended to an additional 60 brain regions and 20 peripheral tissues.
Expanding tissue resources in NHPs, generally, will create additional opportunities to identify biomedically relevant eQTLs9,52. The abundance of natural Caribbean vervet populations, and their genetic near-identity to the samples we analyzed, make them uniquely valuable for maximizing the value of our eQTL resource 10,12. Each lead SNP for the eQTLs associated with hippocampal volume in the VRC is common in Caribbean populations (Supplementary Note). We anticipate that our eQTL database will enhance interpretation of well-powered GWAS that can be conducted in these populations for a wide range of complex traits.
Online Methods
Study Sample
The monkeys in this study were from the Vervet Research Colony (VRC), established by UCLA during the 1970’s-1980’s from 57 founder animals wild-captured in St. Kitts and Nevis10. MRI phenotypes were obtained before the VRC moved to Wake Forest School of Medicine in 2008 (Supplementary Note). All vervets in this study were captive-born, mother-reared and socially-housed in large, indoor-outdoor enclosures, in matrilineal groups that approximated the social structure of wild vervet populations. They had a uniform exposure to light and darkness and were fed a standardized diet.
Gene Expression
Two gene expression datasets were collected. Dataset 1 consisted of microarray (Illumina HumanRef-8 v2) assays of whole-blood RNA in 347 vervets. Dataset 2 consisted of RNA-Seq data from seven tissues assayed in 60 animals. Six vervets were in both Datasets. No randomization was applied in allocating animals to Datasets and investigators were not blinded to the allocation of animals to Datasets.
Dataset 1: Microarrays From Whole Blood
The microarray dataset has been described previously12. For details on RNA extraction, cDNA synthesis, and initial data processing, see Supplementary Note. To obtain a set of probes usable in vervet from the Illumina HumanRef-8 v2 microarray, we used the vervet reference sequence to select probes containing no vervet indels and demonstrating ≤ five mismatches, with a maximum of one mismatch in the 16 nt central portion of the probe. To prevent bias in expression measurement due to SNP interference with hybridization, we excluded probes targeting sequences with common SNPs identified in the VRC. A total of 11,001 probes passed these filters (Supplementary Table 1). Illumina provides a “detection p-value” for detection of a given probe in a specific individual (with p<0.05 considered significant). We analyzed 6,018 probes with detection p-values of p<0.05 in at least 5% of vervets, and tested 3,417 significantly heritable probes for eQTL association. Expression data were inverse-normal transformed prior to analysis.
Dataset 2: RNA-Seq Data from Seven Tissues
Tissues harvested during experimental necropsies (Wake Forest School of Medicine IACUC protocol A09-512) were obtained from 60 vervets representing 10 developmental stages, ranging from neonates (7 days), through infants (90 days and one year), young juveniles (1.25, 1.5, 1.75, 2 years old), subadults (2.5, 3 years old) to adults (4+ years old), with six vervets (3 male and 3 female) from each developmental time point. Two vervets (a 1.75 year old female and a 7 day old male) for which we did not have WGS data were excluded from this study. Altogether, we included 11 vervets below one year old, 23 vervets between one to two years old, and 24 vervets between two and four years old, 29 males and 29 females. For details regarding tissue collection and RNA preparation procedures, see Supplementary Note.
For all vervets we conducted RNA-Seq in seven tissues: three brain tissues (BA46, caudate and hippocampus), two neuroendocrine tissues (adrenal and pituitary) and two peripheral tissues (blood and fibroblasts). From purified RNA, we created two types of cDNA libraries (Supplementary Note); poly-A RNA (fibroblasts, adrenal and pituitary) and total RNA (blood, caudate, hippocampus, BA46) libraries (Supplementary Table 20, Supplementary Note). For one vervet in which the RNA-Seq data indicated a mix-up between the caudate and BA46 samples, we excluded data from these two tissues in all analyses.
RNA-Seq reads were aligned to the vervet genomic assembly Chlorocebus_sabeus 1.1 by the ultrafast STAR aligner53 using our standardized pipeline. STAR was run using default parameters, which allow up to ten mismatches. Gene expression was measured as total read counts per gene. For paired-end experiments we considered total fragments. Fragment counts aligning to known exonic regions (based on NCBI Chlorocebus sabaeus Annotation Release 100) were quantified using the HTSeq package54. The counts for all 33,994 genes were then combined; lowly expressed genes (mean in raw counts of < 1 across all samples) and genes detected in < 10% of individuals were filtered out. The calcNormFactors function in the edgeR package55 was applied to normalize counts. Finally, an inverse-normal transform was applied to counts per million, prior to analysis.
Deconvolution analysis was performed in vervet brain and blood tissue using available references for these tissues. For brain tissues, gene signatures were obtained from Zhang et al.20; for blood, cell type specific markers were taken from datasets built into the CellMix package19. Cell type composition for each tissue was evaluated using the CellMix R package.
Datasets for comparative expression analysis between species
We performed comparative analysis of gene expression between vervet brain samples, GTEx, and age-matched samples from Allen Brain Atlas (ABA) datasets; BrainSpan (human RNA-seq data, see URLs) and the NIH Blueprint NHP Atlas (rhesus macaque microarray data, see URLs)6,52, (Supplementary Tables 21, 22) Matching the three vervet brain tissues to the most closely corresponding available tissues in the other species (Supplementary Table 23), we compared overall expression profiles between these species, and inspected developmental expression patterns of selected genes.
Overall mean levels of expression were compared between species using a rank correlation. GTEx and BrainSpan were compared to vervet, independently. For the GTEx comparison, vervet tissues were matched to the five available corresponding tissues: adrenal, blood, caudate, hippocampus and pituitary. Analyses involving the two ABA datasets were limited to the three brain regions most closely related to the brain tissues analyzed in vervets (Supplementary Table 23). As the rhesus macaque dataset included only males, we limited comparisons to male vervets.
For each of the three dataset comparisons, vervet raw counts were first converted to RPKM values using the edgeR R package55. GTEx and human ABA counts were already normalized to RPKM values; rhesus macaque counts had been normalized using an RMA approach52. Mean expression was then calculated by tissue for each dataset. For ABA datasests, mean expression was calculated by tissue type and time point, according to matched age groups (Supplementary Tables 21, 22). Vervet gene names were converted to their corresponding human orthologs to ensure gene names matched between vervet and comparison datasets; Genes with no human ortholog were excluded. Additionally, genes not present in both vervet and the comparison species dataset were also removed. Variances were then calculated for each gene across the five or three different vervet tissues, for GTEx and ABA comparisons, respectively. The top 1,000 genes with the highest variances were then selected for rank-rank correlation testing. The base R function cor.test was used to perform correlation testing.
Real-time quantitative PCR (qPCR)
Real-time quantitative PCR was performed in two steps. First, reverse transcription (RT) was performed using the SuperScript® III First-Strand Synthesis System (Life Technologies) following the manufacturer’s protocol for priming with random hexamers. Custom primers and hydrolysis probes were designed for each lncRNA and three candidate reference genes (Hypoxanthine phosphoribosyltransferase 1, HPRT1; Glyceraldehyde 3-phosphate dehydrogenase, GAPDH; and Beta-2-Microglobulin, B2M) using the Custom TaqMan® Assays Design Tool (Applied Biosystems, Supplementary Table 24). Expression analyses were conducted on the LightCycler™ 480 platform (Roche) using the iTaq® Universal Probes Supermix (Bio-Rad). All qPCR reactions were carried out in triplicate; reactions containing water instead of cDNA were included as negative controls. cDNA samples were diluted 1:5 with water, and a five-point standard curve of four-fold dilutions was prepared for each gene using pooled cDNA as the template. Stability of each candidate reference gene was evaluated using the NormFinder software (v5) in R56. Quantification was performed using the relative standard curve method, with the geometric mean of the most stably expressed reference genes (GAPDH and HPRT1) used as an endogenous control for normalization of the interpolated lncRNA quantities. Finally, relative expression levels were generated by dividing the normalized lncRNA quantities by the corresponding quantity in one experimental sample which served as a calibrator. For additional experimental details and complete primer and probe sequence information see Supplementary Note.
Hippocampal Volume
Estimates of hippocampal volume were measured in 347 vervets >2 years of age using MRI. Details of the image acquisition and processing protocol were described previously34 and are outlined in Supplementary Note. Prior to genetic analysis, hippocampal volume was log transformed, regressed on sex and age (SOLAR22), and residuals used as the final phenotype.
Genotype Data
Genotypes were generated through WGS, as described previously11. Genotypes from 721 VRC vervets that passed QC procedures can be queried via the EVA at EBI. Two genotype data sets were used11: (1) The Association Mapping Set consists of 497,163 SNPs on the 29 vervet autosomes. This set has, on average, 198 SNPs per Mb of vervet sequence, with a maximal gap of 5 Kb between adjacent SNPs. (2) The Linkage Mapping Set consists of 147,967 SNPs on the 29 vervet autosomes. This set has, on average, 58.2 SNPs per Mb of vervet sequence, with an average gap of 17.5 Kb between adjacent SNPs.
The software package Loki57, which implements Markov Chain Monte Carlo methods, was used to estimate multipoint identical by decent (MIBD) allele-sharing among all vervet family members from the genotype data. As long stretches of IBD were evident among these closely related animals, density 9,752 SNPs of the 148K set were sufficient to evaluate MIBD at 1cM intervals. The correspondence between physical and genetic positions of vervet SNPs was established by interpolation using 360 markers from the vervet STR linkage map58, for which physical and genetic position was known.
Statistical Analysis
Principal Components Analysis (PCA)
The top 1,000 most variable genes were selected for each tissue (Dataset 2), and PCA applied to log2-transformed counts per million, using the singular value decomposition and the prcomp function in. Expression was mean-centered prior to analysis. We examined genes in the top and bottom 10% of the distribution of PC loadings on PCs 1, 2, or 3 (200 genes per tissue, per PC) where these loadings are taken from the eigen-decomposition of the expression matrix. The gene loadings represent the amount that gene contributes to the PC value for that sample on the axis in question.
Differential Expression Analysis of Age
We conducted a differential expression analysis, using age as both a continuous and a categorical predictor in two different linear models, and inverse-normal transformed gene expression as the outcome. Both analyses were performed separately by tissue; sample size was 60 animals for adrenal, blood, fibroblasts, and pituitary and was 59 for BA46, caudate, and hippocampus.
Mapping of Gene Expression and Hippocampal Volume Phenotypes
For the higher-order phenotype, hippocampal volume, we anticipated having power only to detect loci with a strong effect, and therefore evaluated it using linkage analysis. For gene expression traits we expected power to identify relatively small effects and therefore applied genome-wide association analyses.
Heritability and Multipoint Linkage Analysis
We estimated familial aggregation (heritability) of traits using SOLAR, which implements a variance components method to estimate the proportion of phenotypic variance due to additive genetic factors. This model partitions total variability into polygenic and environmental components. The environmental component is unique to individuals while the polygenic component is shared between individuals as a function of their pedigree kinship. If the variance in phenotype Y due to the polygenic component is designated as σg2 and the environmental component as σe2, then in this model Var(Y) = σg2 + σe2, and the covariance between phenotype values of individuals i and j is Cov(Yi,Yj)=2 φij σg2, where φij is the kinship between individuals i and j.
Genome-wide multipoint linkage analysis of hippocampal volume was also implemented in SOLAR, which further partitions the genetic covariance between relatives for each trait into locus-specific heritability and residual genetic heritability. Linkage analysis was performed at 1cM intervals using the likelihood ratio statistic.
Association Analysis
Association between specific SNPs and gene expression phenotypes was evaluated using EMMAX59. EMMAX employs a linear mixed model approach, where SNP genotype is a fixed effect, and correlation of phenotype values among individuals is accounted for using an identity-by-state approximation to kinship. Association analyses used 497,163 SNP markers, and for both Dataset 1 and Dataset 2 included age (in Dataset 2, age corresponds to developmental stage), sex, and sample batch as covariates. It is common to try to account for unmeasured factors influencing global gene expression by including probabilistic estimation of expression residuals (PEER) factors as covariates60. We considered the controlled nature of the study environment and experimental design to preclude the need for this adjustment.
Colocalization of eQTL and Hippocampal Volume QTL
We evaluated the posterior probability that the hippocampal volume QTL and the hippocampus local eQTLs on CAE 18 share a single, common causal variant using COLOC37. The same variants were tested in both analyses; six vervets overlapped between the two data sets.
Multiple Testing Considerations in eQTL
As our primary error-controlling strategy for eQTL discovery we used a Bonferroni correction to account for multiple testing across genes, SNPs, and tissues. Thresholds for Dataset 2 were more stringent, as it included analysis of multiple tissues and tested more genes than in Dataset 1 (~25K vs. ~3K). In Dataset 1 we analyzed association to 3,417 heritable probes. The local eQTL significance threshold (4.8 x 10-8) was corrected for testing of SNPs within 1 Mb of 3,417 probes. The distant eQTL significance threshold (1.5 x 10-11) accounted for genome-wide. Dataset 2 significance thresholds were constructed in a similar fashion, but also accounted for testing of 191,263 gene-tissue combinations (see Table 1). The RNA-Seq local eQTL threshold was 6.5 x 10-10, and the distant eQTL threshold was 5.3 x 10-13.
To identify multi-tissue eGenes, the tissues in which they are active, and the associated SNPs in each of these tissues, we used TreeBH, a hierarchical testing approach24 which extends the error-controlling procedure characterized in Peterson et al.61 to multi-tissue eQTLs. To apply this method, the hypotheses are grouped into a tree with three levels: genes in level 1, tissues in level 2, and SNPs in level 3. Testing proceeds sequentially starting from the top of the tree in a manner that accounts for each previous selection step. This method controls the FDR of local eGenes (genes whose expression is regulated in at least one tissue by some genetic variants located within 1 Mb of the gene) and of the expected average false discovery proportion of the tissues in which we claim this regulation is present across the discovered eGenes. P-values are defined by building up from the bottom of the tree. Specifically, to obtain a p-value for the null hypothesis of no local regulation for a given gene in a given tissue (corresponding to a hypothesis in level 2 of the tree), we applied Simes’ combination rule62 to the p-values obtained via EMMAX for the hypotheses of no association between the expression of the gene in the tissue and each of the SNPs in the local neighborhood (corresponding to the hypotheses in level 3 of the tree). To obtain a p-value for the null hypothesis of no local regulation for a given gene in any of the tissues under study (corresponding to a hypothesis in level 1 of the tree), we applied Simes’ combination rule to the gene x tissues p-values just described. We then tested the global null hypotheses of no local regulation in any tissue for all the genes in our study, applying the Benjamini Hochberg procedure63 to control the FDR at the 0.05 level. For those genes for which we were able to reject the null hypotheses of no local regulation, we examined the tissue-specific p-values, applying the Benjamini Bogomolov procedure that allows identification of significant findings, controlling for the initial selection64. Finally, the individual SNPs responsible for regulation of the gene in each tissue were identified, again using a selection-adjusted threshold as previously described.24 An R package implementing this procedure is available.65
We compared the number of eGenes identified in each tissue using the above procedure with the results of GTEx (Analysis Release V6; dbGaP Accession phs000424.v6.p1). We downloaded all eQTL association results for tissues in common with our study, and applied this same hierarchical procedure to the GTEx results to identify eGenes.
Association between local eQTLs and genomic features
We estimated possible enrichment of eQTLs in exons, introns, flanking regions, intergenic regions, and regulatory regions using logistic regression in a generalized linear mixed model (GLMM), using the GMMAT software66. We categorized each SNP in two binary dimensions (local eQTL and located in or near a given region). A SNP was considered a local eQTL if it was associated (at Bonferroni thresholds) to gene expression of a gene within 1 Mb, in any tissue, in either Dataset. Local eQTL status was the outcome variable, and a separate GLMM logistic regression performed for each region. A matrix of r2 values among all SNPs was included as a random effect to account for lack of independence among SNPs. GLMMs are computationally demanding and the full set of 497,163 SNPs could not be analyzed in one model. We LD-pruned the SNP data, agnostic to eQTL status and region, and used 18,464 genome-wide SNPs based on LD-pruning 497,163 SNPs at r2<0.6 in 14 unrelated individuals. This SNP set included 1,202 local eQTLs.
Enrichment of local eQTLs in near TSS/TES
Our examination of potential enrichment of local eQTLs near TSS/TES gene regions was descriptive, involving no hypothesis testing. We restricted our summary to the 27,196 genes that were <0.5 Mb in size, and the 426,403 SNPs within 200kb of the TSS/TES of these genes (or in between the TSS/TES). In this set of SNPs, 17,595 were local eQTLs to ≥ 1 of the 27,196 genes (at Bonferroni thresholds), in > 1 tissues in either Dataset, and were within 200 Kb of the TSS/TES of the gene(s) to which they were associated. For each gene, we created 10 Kb distance bins on either side of the TSS/TES, and tallied the proportion of SNPs in the bin that were local eQTLs for the gene. As the distance between TSS and TES varied by gene, we binned distances in this area by deciles of the total distance.
Supplementary Material
Acknowledgements
We thank S. Groman for assistance with tissue resources. T. Chavanne, K. Finnie, M. Long, J. Gardin and D. Swaim for technical assistance with necropsies and tissue collections, and M. Vermunt for sharing rhesus macaque epigenetic data. This work was supported by the following grants, all from the U.S. National Institutes of Health: U54HG00307907 (to RKW); P40RR019963/OD010965 (to JRK); R01RR016300/OD010980 (to NBF); R37MH060233 (to Daniel Geschwind); UL1DE019580 (to Robert Bilder); PL1NS062410 (to Christopher Evans); RL1MH083270 (to JDJ); P30NS062691 (to NBF and GC); R01MH101782 (to CS and EE). RN, BLA, PF acknowledge support from Wellcome Trust PF, BLA, RN acknowledge support from the Wellcome Trust (grant number WT108749/Z/15/Z) and the European Molecular Biology Laboratory. ESW was supported by an EMBO Advanced Fellowship (aALTF1672-2014). AEF was supported by NIH award T32MH073526. Data were generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881 and R37MH057881S1, HHSN271201300031C, AG02219, AG05138 and MH06692. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories and the NIMH Human Brain Collection Core. CMC Leadership: P. Sklar, J. Buxbaum (Icahn School of Medicine at Mount Sinai), B. Devlin, D. Lewis (University of Pittsburgh), R. Gur, C.-Gyu Hahn (University of Pennsylvania), K. Hirai, H. Toyoshiba (Takeda Pharmaceuticals Company Limited), E. Domenici, L. Essioux (F. Hoffman-La Roche Ltd), L. Mangravite, M. Peters (Sage Bionetworks), T. Lehner, B. Lipska (NIMH).
Footnotes
URLs
Vervet genomic assembly: http://www.ncbi.nlm.nih.gov/assembly/GCF_000409795.2
R version 3.2.3 https://www.R-project.org
Treeqtl software http://www.bioinformatics.org/treeqtl/
Rhesus macaque microarray data: © 2015 Allen Institute for Brain Science. Blueprint Non-Human Primate (NHP) Atlas. Available from: http://www.blueprintnhpatlas.org/
Human RNA-Seq brain data: © 2015 Allen Institute for Brain Science. BrainSpan Atlas of the Developing Human Brain. Available from: http://www.brainspan.org/
Author contributions: AJJ, JRK, GMW, KD, RKW, JDJ, WW, RPW, and NBF designed the study. AJJ, IZ, OWC, JD, LAF, SF, AEF, YSH, VR, CAS, JDJ, GC, and RPW produced the data. AJJ, IZ, SKS, CP, RMC, EE, LAF, SF, YSH, VR, CAS, HS, DV, BLA, PF, RN, ESW, JB, TDD, MB, YB, CS, and GC analyzed the data. OWC, JD, and MJJ managed data and samples. AJJ, SKS, and NBF wrote the paper. All authors reviewed the final draft.
Data Availability
The RNA-Seq datasets generated in this study are available in the NCBI Gene Expression Ominibus repository, https://www.ncbi.nlm.nih.gov/gds/?term=PRJNA219198. The genotype and microarray datasets are available in the NCBI Gene Expression Ominibus repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15301 (microarray data) and the EMBL-EBI, https://www.ebi.ac.uk/ena/data/view/ERP008917 (genotype data).
References
- 1.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. doi: 10.1038/nrg3891. [DOI] [PubMed] [Google Scholar]
- 4.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–15. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson G, Powell JE, Marigorta UM. Expression quantitative trait locus analysis for translational medicine. Genome Med. 2015;7:60. doi: 10.1186/s13073-015-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mele M, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–5. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jennings CG, et al. Opportunities and challenges in modeling human brain disorders in transgenic primates. Nat Neurosci. 2016;19:1123–30. doi: 10.1038/nn.4362. [DOI] [PubMed] [Google Scholar]
- 9.Rogers J, Gibbs RA. Comparative primate genomics: emerging patterns of genome content and dynamics. Nat Rev Genet. 2014;15:347–59. doi: 10.1038/nrg3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jasinska AJ, et al. Systems biology of the vervet monkey. ILAR J. 2013;54:122–43. doi: 10.1093/ilar/ilt049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang YS, et al. Sequencing strategies and characterization of 721 vervet monkey genomes for future genetic analyses of medically relevant traits. BMC Biol. 2015;13:41. doi: 10.1186/s12915-015-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jasinska AJ, et al. Identification of brain transcriptional variation reproduced in peripheral blood: an approach for mapping brain expression traits. Hum Mol Genet. 2009;18:4415–27. doi: 10.1093/hmg/ddp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein JL, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–61. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren WC, et al. The genome of the vervet (Chlorocebus aethiops sabaeus) Genome Res. 2015;25:1921–33. doi: 10.1101/gr.192922.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnett MG, Muglia LM, Laryea G, Muglia LJ. Genetic Approaches to Hypothalamic-Pituitary-Adrenal Axis Regulation. Neuropsychopharmacology. 2016;41:245–60. doi: 10.1038/npp.2015.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McEwen BS, Gray JD, Nasca C. 60 YEARS OF NEUROENDOCRINOLOGY: Redefining neuroendocrinology: stress, sex and cognitive and emotional regulation. J Endocrinol. 2015;226:T67–83. doi: 10.1530/JOE-15-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nestler E, Hyman S, Holtzman D, Malenka R. Molecular Neuropharmacology: A Foundation for Clinical Neuroscience. McGraw-Hill Education / Medical; 2015. [Google Scholar]
- 18.Caceres M, Suwyn C, Maddox M, Thomas JW, Preuss TM. Increased cortical expression of two synaptogenic thrombospondins in human brain evolution. Cereb Cortex. 2007;17:2312–21. doi: 10.1093/cercor/bhl140. [DOI] [PubMed] [Google Scholar]
- 19.Gaujoux R, Seoighe C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics. 2013;29:2211–2. doi: 10.1093/bioinformatics/btt351. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–47. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Q, He Z. Comprehensive investigation of temporal and autism-associated cell type composition-dependent and independent gene expression changes in human brains. bioRxiv. 2016 doi: 10.1101/065292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahler N, et al. Gene co-expression network connectivity is an important determinant of selective constraint. PLoS Genet. 2017;13:e1006402. doi: 10.1371/journal.pgen.1006402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogomolov M, Peterson CB, Benjamini Y, Sabatti C. Testing hypotheses on a tree: new error rates and controlling strategies. 2017 doi: 10.1093/biomet/asaa086. arXiv: 1705.07529v1 [stat.ME] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tung J, Zhou X, Alberts SC, Stephens M, Gilad Y. The genetic architecture of gene expression levels in wild baboons. Elife. 2015;4 doi: 10.7554/eLife.04729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vermunt MW, et al. Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat Neurosci. 2016;19:494–503. doi: 10.1038/nn.4229. [DOI] [PubMed] [Google Scholar]
- 29.Villar D, et al. Enhancer evolution across 20 mammalian species. Cell. 2015;160:554–66. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roadmap Epigenomics C et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young RS, et al. The frequent evolutionary birth and death of functional promoters in mouse and human. Genome Res. 2015;25:1546–57. doi: 10.1101/gr.190546.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daugherty MD, Schaller AM, Geballe AP, Malik HS. Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. Elife. 2016;5 doi: 10.7554/eLife.14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce BL, et al. Mediation analysis demonstrates that trans-eQTLs are often explained by cis-mediation: a genome-wide analysis among 1,800 South Asians. PLoS Genet. 2014;10:e1004818. doi: 10.1371/journal.pgen.1004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fears SC, et al. Identifying heritable brain phenotypes in an extended pedigree of vervet monkeys. J Neurosci. 2009;29:2867–75. doi: 10.1523/JNEUROSCI.5153-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 36.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, et al. Imputing Gene Expression in Uncollected Tissues Within and Beyond GTEx. Am J Hum Genet. 2016;98:697–708. doi: 10.1016/j.ajhg.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sargiannidou I, et al. Connexin32 mutations cause loss of function in Schwann cells and oligodendrocytes leading to PNS and CNS myelination defects. J Neurosci. 2009;29:4736–49. doi: 10.1523/JNEUROSCI.0325-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergoffen J, et al. Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science. 1993;262:2039–42. doi: 10.1126/science.8266101. [DOI] [PubMed] [Google Scholar]
- 42.Bond J, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–20. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 43.Tang BS, et al. Small heat-shock protein 22 mutated in autosomal dominant Charcot-Marie-Tooth disease type 2L. Hum Genet. 2005;116:222–4. doi: 10.1007/s00439-004-1218-3. [DOI] [PubMed] [Google Scholar]
- 44.Eriksson PS, et al. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–7. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 45.van Praag H, et al. Functional neurogenesis in the adult hippocampus. Nature. 2002;415:1030–4. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pichlmair A, et al. IFIT1 is an antiviral protein that recognizes 5'-triphosphate RNA. Nat Immunol. 2011;12:624–30. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 47.Brodziak F, Meharg C, Blaut M, Loh G. Differences in mucosal gene expression in the colon of two inbred mouse strains after colonization with commensal gut bacteria. PLoS One. 2013;8:e72317. doi: 10.1371/journal.pone.0072317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato Y, et al. Cellular Transcriptional Coactivator RanBP10 and Herpes Simplex Virus 1 ICP0 Interact and Synergistically Promote Viral Gene Expression and Replication. J Virol. 2016;90:3173–86. doi: 10.1128/JVI.03043-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azevedo C, et al. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–6. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- 50.Mayor A, Martinon F, De Smedt T, Petrilli V, Tschopp J. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat Immunol. 2007;8:497–503. doi: 10.1038/ni1459. [DOI] [PubMed] [Google Scholar]
- 51.Naitza S, et al. A genome-wide association scan on the levels of markers of inflammation in Sardinians reveals associations that underpin its complex regulation. PLoS Genet. 2012;8:e1002480. doi: 10.1371/journal.pgen.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bakken TE, et al. A comprehensive transcriptional map of primate brain development. Nature. 2016;535:367–75. doi: 10.1038/nature18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 57.Heath SC, Snow GL, Thompson EA, Tseng C, Wijsman EM. MCMC segregation and linkage analysis. Genet Epidemiol. 1997;14:1011–6. doi: 10.1002/(SICI)1098-2272(1997)14:6<1011::AID-GEPI75>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 58.Jasinska AJ, et al. A genetic linkage map of the vervet monkey (Chlorocebus aethiops sabaeus) Mamm Genome. 2007;18:347–60. doi: 10.1007/s00335-007-9026-4. [DOI] [PubMed] [Google Scholar]
- 59.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–54. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stegle O, Parts L, Piipari M, Winn J, Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat Protoc. 2012;7:500–7. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peterson CB, Bogomolov M, Benjamini Y, Sabatti C. Many Phenotypes Without Many False Discoveries: Error Controlling Strategies for Multitrait Association Studies. Genet Epidemiol. 2016;40:45–56. doi: 10.1002/gepi.21942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simes RJ. An improved Bonferroni procedure for multiple tests of significance. Biometrika. 1986;73:751–754. [Google Scholar]
- 63.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 64.Benjamini Y, Bogomolov M. Selective inference on multiple families of hypotheses. J R Stat Soc B. 2014;76:297–318. [Google Scholar]
- 65.Peterson CB, Bogomolov M, Benjamini Y, Sabatti C. TreeQTL: hierarchical error control for eQTL findings. Bioinformatics. 2016;32:2556–8. doi: 10.1093/bioinformatics/btw198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen H, et al. Control for Population Structure and Relatedness for Binary Traits in Genetic Association Studies via Logistic Mixed Models. Am J Hum Genet. 2016;98:653–66. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.