Abstract
Identification of cellular proteins, in addition to already known transcription factors such as NF-κB, Sp1, C-EBPβ, NFAT, ATF/CREB, and LEF-1, which interact with the HIV-1 LTR, is critical in understanding the mechanism of HIV-1 replication in monocytes/macrophages. Our studies demonstrate upregulation of pyruvate kinase isoform M2 (PKM2) expression during HIV-1SF162 infection of monocyte/macrophages and reactivation of HIV-1 in U1 cells, a macrophage model of latency. We observed that HIV-1SF162 infection of monocyte/macrophages and reactivation of HIV-1 in U1 cells by PMA resulted in increased levels of nuclear PKM2 compared to PMA-induced U937 cells. Furthermore, there was a significant increase in the nuclear dimeric form of PKM2 in the PMA-induced U1 cells in comparison to PMA-induced U937 cells. We focused on understanding the potential role of PKM2 in HIV-1 LTR transactivation. Chromatin immunoprecipitation (ChIP) analysis in PMA-activated U1 and TZM-bl cells demonstrated the interaction of PKM2 with the HIV-1 LTR. Our studies show that overexpression of PKM2 results in transactivation of HIV-1 LTR-luciferase reporter in U937, U-87 MG, and TZM-bl cells. Using various truncated constructs of the HIV-1 LTR, we mapped the region spanning −120 bp to −80 bp to be essential for PKM2-mediated transactivation. This region contains the NF-κB binding site and deletion of this site attenuated PKM2-mediated activation of HIV-1 LTR. Immunoprecipitation experiments using U1 cell lysates demonstrated a physical interaction between PKM2 and the p65 subunit of NF-κB. These observations demonstrate for the first time that PKM2 is a transcriptional co-activator of HIV-1 LTR.
The cellular tropism of HIV-1 is controlled predominantly by the viral envelope sequences that regulate viral entry and the LTR sequences (Reed-Inderbitzin and Maury, 2003). HIV-1 replication in T-cell and monocyte/macrophage is controlled by the 5′ LTR of HIV-1 (Rohr et al., 2003; Sadowski and Mitchell, 2005; Kilareski et al., 2009). The HIV-1 LTR functions as a classic promoter that harbors a TATAA box and DNA sequences that facilitate regulation by host cellular factors such as TFIID and Sp1, and viral regulatory proteins such as Tat, Nef, and Vpr (Ahmad and Venkatesan, 1988; Harrich et al., 1989; Southgate et al., 1990; Sakaguchi et al., 1991; Wang et al., 1995). Transcription factors, such as NF-κB, Sp1, C-EBPβ, NFAT, ATF/CREB, and LEF-1, are known to positively regulate HIV-1 LTR (Pereira et al., 2000). In contrast, many cellular factors, such as YY1, ZNF10, and HSP70 binding protein 1 (HspBP1), are known to regulate negatively HIV-1 LTR (Margolis et al., 1994; Nishitsuji et al., 2015; Chaudhary et al., 2016).
Numerous studies have shown that viral infection, such as HIV-1, Herpes simplex virus (HSV-1), hepatitis C virus (HCV), human cytomegalovirus (HCMV), and adenovirus 5 (Ad5), alters host cell metabolism (Mazzon and Mercer, 2014; Goodwin et al., 2015; Sanchez and Lagunoff, 2015). The major metabolic pathways that are altered upon infection by viruses, such as HIV-1, HCMV, HCV, and HSV-1, are glycolysis, pentose phosphate pathway, fatty acid synthesis, and glutaminolysis (Munger et al., 2006, 2008; Chan et al., 2009; Diamond et al., 2010; Hollenbaugh et al., 2011; Vastag et al., 2011; Barrero et al., 2013; Hegedus et al., 2014; Sen et al., 2015). This alteration in cellular metabolism is advantageous as it increases free nucleotide pool, amino acid, and fatty acid production that are required for rapid viral replication and virion assembly. We have shown earlier that HIV-1 Vpr modulates macrophage glycolytic, and TCA cycle enzymes (Barrero et al., 2013), and HIV-1 infection in macrophages modulates hexokinase-1 expression and apoptosis (Sen et al., 2015). Therefore, elucidation of virus-mediated alteration in host cell metabolism can lead to novel therapeutic approaches for targeted inhibition of specific cellular metabolic pathways.
There are four mammalian pyruvate kinase (PK) isoforms PKM1, PKM2, PKR, and PKL (Imamura and Tanaka, 1982). PK catalyzes the conversion of phosphoenolpyruvate (PEP) and ADP to pyruvate and ATP (Imamura and Tanaka, 1982). PKM2 is allosterically regulated by both metabolites and intracellular signaling pathways (Israelsen and Vander Heiden, 2015). Studies have shown that PKM2 exists in the cytoplasm predominantly as a monomer while the assembly into a tetramer in the cytoplasm is a prerequisite for glycolysis (Israelsen and Vander Heiden, 2015). However, PKM2 dimer acts as a protein kinase in the nucleus (Spoden et al., 2009; Gao et al., 2012; Wong et al., 2015). Several independent studies demonstrated nuclear translocation of PKM2 in response to different stimuli (Yang et al., 2011, 2012a,b; Lv et al., 2013; Salani et al., 2015). Also, nuclear PKM2 is known to regulate gene expression (Yang et al., 2011, 2012b; Gao et al., 2012; Wang et al., 2014). Our earlier studies using SILAC-based proteomic analysis of macrophages transduced with Vpr showed significant upregulation of PKM2 in macrophages (Barrero et al., 2013). Studies have also shown upregulation of PKM2 expression in HIV-1 infected CD4+ T cells, HIV-1 infected human astrocytes treated with cocaine, HIV-1 infected macrophages, and T-cells expressing HIV-1 Tat (Reynolds et al., 2006; Chan et al., 2007; Rivera-Rivera et al., 2012; Rodríguez-Mora et al., 2015). However, its role in activation of HIV-1 LTR remains unknown.
Therefore, we reasoned that HIV-1 infection of monocyte/macrophages and activation of HIV-1 replication in a macrophage model of HIV-1 latency may induce expression of PKM2 and modulate HIV-1 promoter activity. Herein, we report that HIV-1 induces the expression of PKM2 in monocyte/macrophages, which then translocate to the nucleus. Subsequently, it interacts with the p65 subunit of NF-κB to induce HIV-1 promoter activity and thereby enhancing viral gene transcription and viral biogenesis.
Materials and Methods
Cell lines
The U937 human monocytic cell line was obtained from ATCC (Manassas, VA). The U1 cell line, a subclone of U937, chronically infected with HIV-1 harbors two copies of provirus DNA integrated into the genome (Folks et al., 1987) was obtained from Dr. Thomas Folks through the NIH AIDS Reagent Program, Division of AIDS, NIAID, USA. The TZM-bl reporter cell line, a HeLa derivative containing multiple integrated copies of HIV-1 LTR-Luc (Wei et al., 2002) was obtained from Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc through the NIH AIDS Reagent Program. The U-87 MG cell line was obtained from ATCC.
Cell culture
The U937 and U1 cells were grown in RPMI medium supplemented with 10% FBS and penicillin-streptomycin. TZM-bl and U-87 MG cells were cultured in DMEM supplemented with 10% FBS and penicillin-streptomycin. The cultures were incubated at 37°C in a 5% CO2 in air atmosphere.
HIV-1 virus preparation, infection, and quantitation
Buffy coats were provided by the Comprehensive NeuroAIDS center (CNAC) Basic Core I under the approval of the Temple University Institutional Review Board (IRB). No informed consent was obtained as the study was classified as exempt (category 4) by the IRB. PBMCs were isolated from buffy coats using ficoll gradient centrifugation method and cultured in RPMI medium containing 10% FBS, 10 μg/ml gentamicin and 5 μg/ml phytohaemagglutinin (PHA) from Sigma–Aldrich (St. Louis, MO). HIV-1 SF162 was obtained from Dr. Jay Levy through the NIH AIDS Research and Reagent Program. Infectious virus was prepared from PBMCs induced by PHA-induced (5 μg/ml, 48 h) and titered by TCID50 assay in TZM-bl cells.
Freshly isolated PBMCs were induced with PHA for 48 h (5 μg/ml) then incubated overnight with 10,000 TCID50/106 cells (MOI ~0.01) of HIV-1 SF162. Infection levels were quantified in the supernatants collected at various time points by p24Gag capture ELISA (ABL, Inc., Rockville, MD).
Induction of viral replication in U1 cells
U1 and U937 cells were induced to differentiate into macrophages using phorbol myristate acetate (PMA) (Sigma–Aldrich) as described earlier (Sen et al., 2015). PMA induction of U1 cells also induces HIV-1 replication and viral biogenesis (Sen et al., 2015). Viral levels in culture supernatants were measured by p24Gag capture ELISA (ABL, Inc.).
Preparation of cell lysates and estimation of protein concentration
Cells were lysed in lysis buffer supplemented with protease inhibitor cocktail (Pierce Biotechnology, Rockland, IL) as described earlier (Barrero et al., 2013; Sen et al., 2015). Protein concentration in cell lysates and nuclear and cytoplasmic fractions were determined using BCA protein assay reagent (Pierce Biotechnology).
Preparation of nuclear and cytoplasmic extracts
The nuclear and cytoplasmic extracts were prepared from human monocyte/macrophages (mock and HIV-1 infected), U937, and U1 cells using NE-PER Nuclear and Cytoplasmic extraction kit (Pierce Biotechnology) as per the manufacturer’s protocol.
Western blot analysis
Western blot analyses were performed as previously described (Barrero et al., 2013; Sen et al., 2015). In brief, 50 μg of protein were resolved by SDS–PAGE, transferred to PVDF membranes by electroblotting. The membranes were blocked with 5% powdered milk in Tris-buffer saline solution (pH 7.6) containing 0.05% Tween-20 (TBS/T). Membranes were incubated with primary antibodies (PKM2, α-tubulin, Grb2, 1:1,000 dilution) overnight at 4°C, washed, and then incubated with appropriate secondary antibodies conjugated to Licor IR dyes for 1 h. Images were captured using Odyssey CLx (LI-COR Biosciences, Lincoln, NE) and analyzed using Image Studio Software (LI-COR Biosciences).
Native SDS–PAGE
Native SDS–PAGE was done using 4–20% pre-cast acrylamide gradient gel (Bio-Rad, Carlsbad, CA) and gel running buffer (0.25 M Tris-base and 1.92 M glycine, 0.05% SDS). Gels were run at constant current (20 mA) at 4°C for 120 min, after which proteins were transferred to PVDF by electroblotting and probed for PKM2 as described in the earlier section.
HIV-1 LTR reporter constructs and cDNA expression vectors
The HIV-1 LTR-LUC plasmids used were described earlier (Chipitsyna et al., 2006). The expression plasmid pCMV-p65-V5-tag was described previously and kindly provided by Dr. Carl Sasaki, National Institute on Aging, National Institutes of Health, Baltimore, MD (Sasaki et al., 2005). The expression plasmid for PKM2 was obtained from Open Biosystems (GE Healthcare Bio-Sciences, Pittsburgh, PA). Endotoxin-free plasmid DNAs were prepared using PureYield Plasmid Maxiprep System (Promega, Madison, WI).
Transfection and luciferase assay
U937 cells in suspension were transfected with 0.3 μg of reporter plasmid HIV LTR-LUC full-length construct and cotransfected with 0.6 μg of PKM2 cDNA using Fugene HD (Promega). U87MG cells were transfected with 0.25 μg of reporter plasmid HIV LTR-LUC/full-length or deletion constructs and cotransfected with 0.25 μg of PKM2 cDNA or p65 cDNA using Fugene 6 (Promega). The amount of DNA used for each transfection was normalized with pcDNA3 vector plasmid. For assessing effects of PMA, U87MG cells were transfected with 0.25 μg of reporter plasmid HIV LTR-LUC/−120/+66 and cotransfected with 0.25 μg of PKM2 cDNA or p65 cDNA using Fugene 6 (Promega). The amount of DNA used for each transfection was normalized with pcDNA3 vector plasmid. Twenty-four hours post-transfection, media was changed and supplemented with complete media or media containing PMA (100 μg/ml). The TZM-bl cells were transfected 0.25 μg of pcDNA3 empty vector or 0.25 μg of PKM2 cDNA expression vector using Fugene HD (Promega). Cell extracts were prepared 48 h after transfection, and luciferase assay (Promega) was performed as described earlier (Maranto et al., 2011).
Chromatin immunoprecipitation (ChIP) assay
TZM-bl (5 × 106) were treated with 10 nM PMA for 4 h at 37°C. U1 cells (5 × 106) were treated with 10 nM PMA for 24 h at 37°C. Cells were then cross-linked with formaldehyde, harvested, and ChIP assay was done as recommended by the manufacturer (Upstate Biotechnology, Inc., Lake Placid, New York). The sheared chromatin was then immunoprecipitated with PKM2 and p65 antibody (positive control) or normal IgG as isotype control. Antibody-bound chromatin complexes were reverse cross-linked with 0.3 M NaCl at 65°C, and treated with RNAse A, and proteinase K. DNA was extracted using phenol: chloroform extraction and subject to PCR amplification to check for the occupancy of PKM2 and p65 on the HIV-1 LTR promoter. The following sets of primers were used for PCR amplification, forward 5′-TAGAGTGGA GGTTTGACAGCCG-3′ (−201 to −180) and reverse 5′-GTACAGGCAAAAAGCAGCTGCT-3′ (−24 to −3) (for TZM-bl chromatin); and forward: 5′-TTAGCAGAACTACA CACCAGGGCC-3′ (−374 to −351) and reverse 5′-CCGAGAG CTCCCAGGCTCAGATCT-3′ (+19 to +44) (for U1 chromatin).
Co-immunoprecipitation assay
U1 cells (5 ×107) were differentiated with 10 nM PMA overnight, washed with 1×PBS and replenished with fresh media. After 72 h, cells were lysed in ice-cold RIPA buffer containing 50 mM Tris-HCl pH7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS, in the presence of HALT-Phosphatase-protease inhibitor (Pierce, Rockland, IL).
The cell lysates were centrifuged at 12,000 rpm at 4°C for 15 min. The protein concentration in the supernatant was estimated using BCA protein assay (Pierce). One milligram of protein aliquot of the lysate was pre-cleared by adding 1 μg of rabbit IgG, together with 20 μl of resuspended volume of Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Santa Cruz, CA) by incubating at 4°C for 30 min, followed by centrifugation at 12,000 rpm for 10 min and the supernatant was collected. Antibody against NF-κB p65 (1:100) was added to the supernatant and the mixture was rotated at 4°C overnight. As a negative control, the supernatants were incubated with IgG. On the following day, Protein A/G PLUS-Agarose beads (20 μl) were added to each sample and incubated for 2 h at 4°C. The beads were then spun down, washed three times with PBS, boiled in 2 × SDS sample buffer, and separated by SDS–PAGE. The separated proteins were transferred to PVDF membrane by electroblotting. Immunoblot analysis was done using p65 and PKM2 antibodies as described in the section on Western blot.
Statistical analysis
The statistical analysis was performed using Student’s t-test. The results are plotted as mean ±SD of two or three experiments. A P-value <0.05 was considered statistically significant.
Results
HIV-1 induces PKM2 expression in monocytes/macrophages
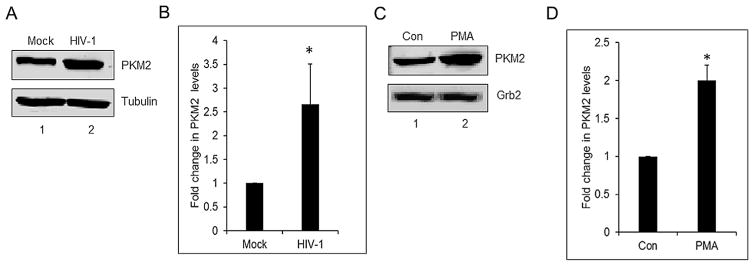
Our earlier studies demonstrated that HIV-1 Vpr induces PKM2 expression in macrophages (Barrero et al., 2013). Therefore, we explored whether HIV-1 infection of monocytes/macrophages and reactivation of HIV-1 expression in chronically infected U1 cells modulates expression of PKM2. We assessed the expression levels of PKM2 by Western blot of cell lysates prepared from mock-infected and HIV-1SF162 infected monocyte/macrophages after infection for 6 days wherein the p24 levels in the media were 71 ± 10.3 ng/ml. A significant increase (~2.6-fold) in the expression of PKM2 was seen in HIV-1 infected cells in comparison to the mock-infected cells (Fig. 1A and B). Since estimation of total levels of PKM2 in HIV-1 infected monocytes and macrophages limit our ability to discriminate whether induction occurred exclusively in productively infected cells, in uninfected bystander cells or both populations, we used a cell line model to mimic human HIV-1 infected macrophages. U1 cells are U937 cells chronically infected with HIV-1 (Folks et al., 1987) and used widely to study the effects of HIV-1 in macrophages (Fernandez Larrosa et al., 2008; Olivares et al., 2009). U1 cells were treated with PMA for differentiation into macrophages and induction of HIV-1 replication. Cell supernatants were assessed for p24 levels by ELISA 72 h after induction. The p24 level in PMA-induced U1 cells was 8929 ± 652 ng/ml. Western blot of cell lysates showed a ~2.0-fold higher level of PKM2 in comparison to the control cells (Fig. 1C and D).
Fig. 1.
HIV-1 induces PKM2 expression in monocytes/macrophages and in PMA-induced U1 cells. (A) Western blot of PBMC lysates. Lane 1: mock infected (mock); lane 2: infected with HIV-1 SF162 (HIV-1) showing PKM2 and α-tubulin protein expression from whole cell lysates after infection for 6 days. (B) Densitometric analysis of PKM2 expression levels normalized to α-tubulin levels plotted as fold change in PKM2 levels. *P <0.05. (C) Western blot of undifferentiated U1 cell lysates. Lane 1: (Con) and lane 2: PMA-treated U1 cells (PMA) showing PKM2 and Grb2 protein expression in cell lysates after 72 h of differentiation. (D) Densitometric analysis of PKM2 expression levels normalized to Grb2 levels, plotted as fold change in PKM2 levels. *P <0.05.
HIV-1 replication modulates nuclear translocation of PKM2
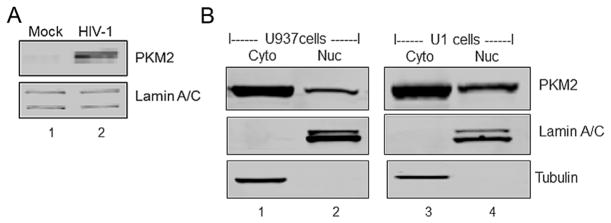
Since earlier studies demonstrated nuclear translocation of PKM2 in response to different agents (Yang et al., 2011, 2012; Lv et al., 2013; Salani et al., 2015), we assessed whether induction of HIV-1 replication by PMA modulates the levels of PKM2 in the nucleus. Western analysis of nuclear fractions prepared from monocytes/macrophages infected with HIV-1SF162 showed a threefold increase (P < 0.05) in the level of nuclear PKM2 in comparison to nuclear extracts from mock-infected cells (Fig. 2A). A similar analysis of nuclear extracts from U1 cells induced with PMA showed a twofold increase (P <0.05) in nuclear PKM2 (Fig. 2B) in comparison to U937 cell nuclear extract wherein a 1.5-fold increase was observed (Fig. 2B).
Fig. 2.
HIV-1 induces nuclear translocation of PKM2 in monocytes/macrophages and PMA induced U937 and U1 cells. (A) Western blot of PBMC lysates. Lane 1: mock infected (mock). Lane 2: infected with HIV-1 SF162 (HIV-1) showing levels of PKM2 and lamin A/C protein in nuclear fractions obtained from cells after infection for 6 days. (B) Western blot of cytoplasmic fraction and nuclear fractions. Lane 1: cytoplasmic fraction (Cyto) and lane 2: nuclear fraction (Nuc) of U937 cells differentiated with PMA. Lane 3: cytoplasmic fraction (Cyto) and lane 4: nuclear fraction (Nuc) of U1 cells differentiated with PMA. Western blots showing levels of PKM2, tubulin in cytoplasmic fraction and lamin A/C protein in nuclear fractions.
HIV-1 modulates PKM2 oligomeric state
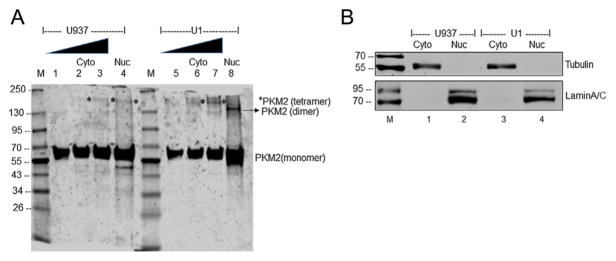
Having observed approximately twofold increase in nuclear PKM2 in U1 cells, we questioned whether there is an alteration in the oligomeric status of PKM2. As mentioned earlier, the enzymatically active tetrameric form of PKM2 located in the cytoplasm converts PEP to pyruvate (Israelsen and Vander Heiden, 2015). The dimeric form of PKM2 in the nucleus has been suggested to act as a protein kinase and regulate gene transcription (Spoden et al., 2009; Wong et al., 2015). Native gel electrophoresis of cytoplasmic fractions (Fig. 3A, lanes 1–3 and 5–7) and nuclear fraction (Fig. 3A, lanes 4 and 8) isolated from PMA induced U937 and U1 cells demonstrated an increase in the nuclear dimeric form in U1 cells (Fig. 3A, lane 8) in comparison to the nuclear extract from U937 cells (Fig. 3A, lane 4). However, in the cytoplasmic fractions, the tetrameric form (*) was predominant (Fig. 3A, lanes 2, 3 and 6, 7). The purity of the nuclear and cytoplasmic fractions used in these studies is shown using Lamin A/C and α-tubulin as markers, respectively (Fig. 3B).
Fig. 3.
Oligomerization status of PKM2 in cytoplasmic and nuclear fractions of PMA differentiated U937 and U1 cells. (A) Lane 1: 1 μg of cytoplasmic extract from U937 cells; lane 2: 2.5 μg of cytoplasmic extract from U937 cells; lane 3: 5 μg of cytoplasmic extract from U937 cells; lane 4: 40 μg of nuclear extract from U937 cells; Lane 5: 1 μg of cytoplasmic extract from U1 cells; lane 6: 2.5 μg of cytoplasmic extract from U1 cells; lane 7: 5 μg of cytoplasmic extract from U1 cells; lane 8: 40 μg of nuclear extract from U1 cells. *Tetrameric form of PKM2. Lane M: Molecular weight marker. (B) Western blot analysis of the cytoplasmic (lane 1) and nuclear fractions (lane 2) isolated from PMA induced U937, and cytoplasmic (lane 3) and nuclear fractions (lane 4) isolated from PMA induced U1 cells used in panel A to demonstrate the purity of fractionation. Tubulin was used as cytoplasmic marker and lamin A/C as a nuclear marker.
PKM2 interacts with HIV-1 LTR
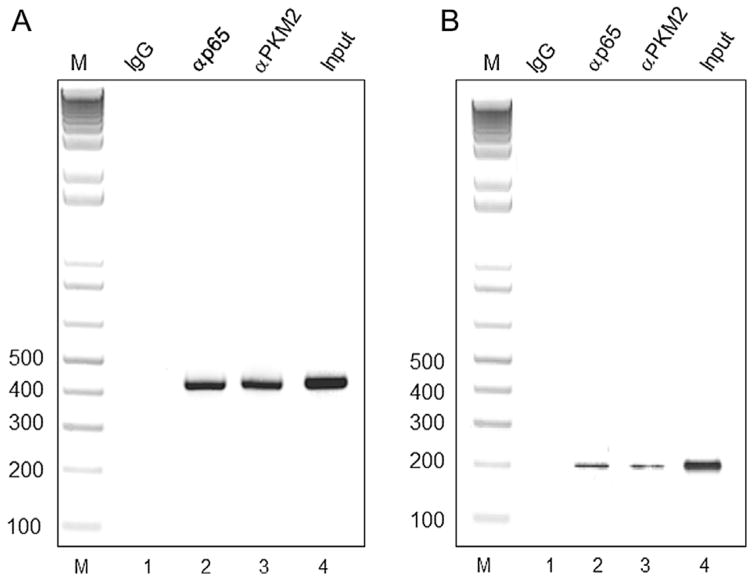
To demonstrate in vivo occupancy of PKM2 at the chromatin level, we used TZM-bl and U1 cells that harbor integrated copies of HIV-1 LTR in their genome (Folks et al., 1987; Wei et al., 2002). Cells were induced with PMA to ensure increased nuclear PKM2 and NF-κB p65 expression. Cross-linked chromatin from these cells was immunoprecipitated with PKM2 and p65 (positive control) antibodies. PCR amplification of immunoprecipitated chromatin show significant enrichment of amplified products using PKM2 and p65 antibody over IgG with U1 and TZM-bl chromatin, respectively (Fig. 4A and B). The results thus demonstrate that PKM2 interacts with the HIV-1 LTR.
Fig. 4.
(A) The interaction of PKM2 and p65 in vivo with HIV-1 LTR in U1 cells induced with PMA was analyzed using ChIP analysis. DNA isolated from chromatin immunoprecipitated with Lane 1: normal rabbit IgG (negative control, IgG); Lane 2: p65 antibodies (p65), and lane 3: PKM2 antibodies (PKM2) was amplified by PCR using primers described in the materials and methods. Lane 4: Total chromatin used as the input (input) and amplified by PCR. Lane: M: DNA size ladder (B). The interaction of PKM2 and p65 in vivo with HIV-1 LTR in TZM-bl cells induced with PMA was analyzed using ChIP analysis. DNA isolated from chromatin immunoprecipitated with Lane 1: normal rabbit IgG (negative control, IgG); Lane 2: p65 antibodies (p65) and lane 3: PKM2 antibodies (PKM2) was amplified by PCR using primers described in the materials and methods. Lane 4: Total chromatin used as the input (lane: input) and amplified by PCR. Lane: M: DNA size ladder.
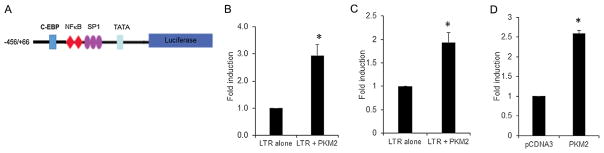
Ectopic expression of PKM2 induces the HIV-1 LTR promoter activity
The observation that HIV-1 infection of monocyte/macrophages and activation of HIV-1 replication in U1 cells up regulates PKM2 expression and also induces PKM2 nuclear translocation prompted us to assess its effect on HIV-1 LTR transactivation. HIV-1 LTR/LUC reporter gene construct harboring a full-length promoter (−456/+66 bp) (Fig. 5A) and PKM2 expression vector were cotransfected into U937 and U-87 MG cells (Fig. 5B and C). TZM-bl cells were transfected with pcDNA3 empty vector or with PKM2 expression vector since this cell line has integrated copies of HIV-1 LTR (Fig. 5D). Analysis of luciferase activity in cell lysates demonstrates that PKM2 activates HIV-1 LTR in all cell models. The results show that in U937, U-87 MG, and TZM-bl cells overexpression of PKM2 induces HIV-LTR reporter gene activity by ~2.9-fold, ~2-fold, and ~2.6-fold respectively (Fig. 5B–D). These observations demonstrate that PKM2 can transactivate HIV-1 LTR.
Fig. 5.
(A) Schematic representation of the HIV-1 LTR/LUC construct. The −456/+66 construct harbors the C/EBP site, two NF-κB, three SP1 sites, and the TATAA box. (B) Fold induction of HIV-1 LTR (−456/+66) promoter activity in U937 cells transiently cotransfected with 0.3 μg of HIV-1 LTR/Luc reporter vector and 0.6 μg of an empty pcDNA3 expression vector (LTR alone), and 0.3 μg of HIV-1 LTR/Luc reporter vector and 0.6 μg expression vector for PKM2 (LTR + PKM2). RLU/s was normalized to protein concentration and expressed as fold induction wherein the activity in HIV-1 LTR/Luc alone transfected cells is set at 1. Statistical significance (*P <0.007 between LTR alone and LTR + PKM2) was analyzed by Student’s t-test. (B) Fold induction of HIV-1 LTR (−456/+66) promoter activity in U-87 MG cells transiently cotransfected with 0.25 μg of HIV-1 LTR/Luc reporter vector and 0.25 μg of an empty expression vector (LTR alone), and 0.25 μg of HIV-1 LTR/LUC and 0.25 μg expression vector for PKM2 (LTR + PKM2). RLU/s was normalized to protein concentration and expressed as fold induction wherein the activity in HIV-1 LTR/Luc alone transfected cells is set at 1. Statistical significance (*P <0.001 between LTR alone and LTR + PKM2) was analyzed by Student’s t-test. (C) Transient transfection of TZM-bl cells harboring integrated copies of HIV-1 LTR/LUC. pcDNA represents cells transfected with 0.25 μg of pcDNA3 empty vector and PKM2 represents cells transfected with 0.25 μg of PKM2 expression vector. RLU/s was normalized to protein concentration and expressed as fold induction wherein the activity in pcDNA3 alone-transfected cells is set at 1. Statistical significance (*P <0.002 between pcDNA3 and PKM2) was analyzed by Student’s t-test.
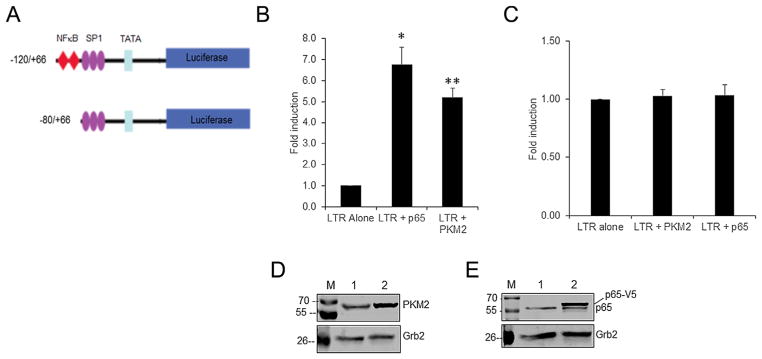
The NF-κB binding site is essential for PKM2-mediated activation of HIV-1 LTR
To map the region that is involved in the induction of HIV-1 LTR by PKM2, we used U-87 MG cells as a model. U-87 MG cells were co-transfected with −120/+66 and −80/+66 luciferase reporter constructs and PKM2 expression plasmid. The −120/+66 construct is a truncated LTR that contains both NF-κB site and SP1 sites while the −80/+66 construct lacks the NF-κB site but retains the SP1 sites (Fig. 6A). The promoter activities in cell lysates were compared with the basal level of activity of each promoter construct co-transfected with empty pcDNA3 vector. The results show that PKM2 overexpression induces the −120/+66 promoter activity by ~6.8-fold (Fig. 6B) but does not induce the −80/+66 promoter activity (Fig. 5C). As a positive control, we also co-transfected the −120/+66 and −80/+66 luciferase reporter constructs with a p65 expression plasmid. The result shows that overexpression of p65 induces the −120/+66 construct (Fig. 6B) but fails to induce the −80/+66 construct (Fig. 6C) that is devoid of NF-κB binding sites. The results of these experiments suggest that the region spanning −120/+66 contains the putative binding site for PKM2-mediated transactivation. The overexpression levels of PKM2 and p65 are shown by Western blots in Figure 6D and E, respectively. These observations suggest that the NF-κB binding site is essential for PKM2-mediated activation of the HIV-1 LTR.
Fig. 6.
(A) Schematic representation of the HIV-1 LTR/LUC deletion constructs. The −120/+66 construct harbors two NF-κB, three SP1 sites, and TATAA box. The −80/+66 construct harbors only three SP1 sites and TATAA box. (B) Fold induction of HIV-1 LTR (P−120/+66) promoter activity in U-87 MG cells transiently cotransfected with 0.25 μg HIV-1 LTR/Luc reporter vector and 0.25 μg of an empty expression vector (LTR alone), or an expression vector for p65 (LTR + p65) or PKM2 (LTR + PKM2). RLU/s was normalized to protein concentration and expressed as fold induction wherein the activity in HIV-1 LTR/Luc alone transfected cells is set at 1. Statistical significance (*P <0.002 between LTR alone and LTR + p65 transfected cells; **P <0.003 between LTR alone and LTR + PKM2 transfected cells) was analyzed by Student’s t-test. (C) Fold induction of HIV-1 LTR (−88/+66) promoter activity in U-87 MG cells transiently cotransfected with 0.25 μg HIV-1 LTR/Luc reporter vector and 0.25 μg of an empty expression vector (LTR alone) or 0.25 μg of an expression vector for PKM2 (LTR + PKM2) or p65 (LTR + p65). Cells were harvested 48 h post-transfection to measure luciferase activity. RLU/s was normalized to protein concentration and expressed as fold induction wherein the activity in HIV-1 LTR/Luc alone transfected cells is set at 1. (D) Representative Western blot analysis of expression levels of PKM2 in U-87 MG cells. Lane 1: pcDNA 3 empty vector; lane 2: PKM2 cDNA vector. (E) Representative Western blot analysis of expression levels of p65 in U-87 MG cells. Lane 1: pcDNA empty vector; lane 2: p65 cDNA vector. p65 denotes the endogenous p65 protein and p65-V5 represents the V5-tagged p65 protein.
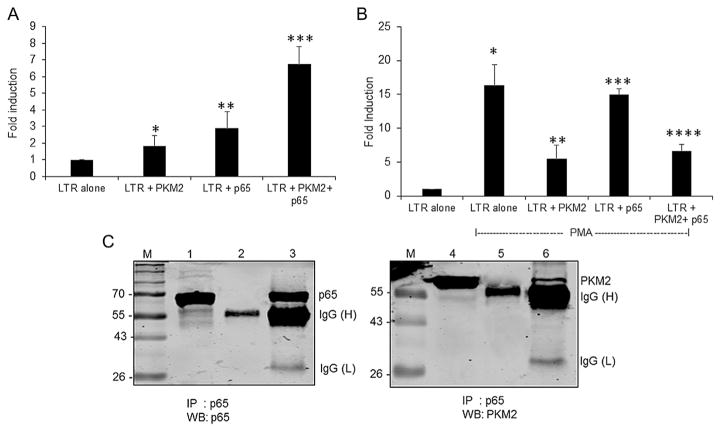
PKM2 and NF-κB activates HIV-1 LTR synergistically
To elucidate the mechanistic details of the stimulatory effect of PKM2 on LTR-mediated reporter expression, we used bioinformatic analysis to predict any DNA binding motifs in PKM2 using iDNA-Prot (Lin et al., 2011) and DNAbinder (Kumar et al., 2007). The analysis revealed that PKM2 has no DNA-binding motifs and hence not a DNA-binding protein. Therefore, we hypothesized that PKM2 might have an indirect influence on the HIV-1 LTR promoter activation. PKM2 is known to interact with other proteins, like β-catenin, and some transcriptions factors, including Oct-4, HIF-1α, and NF-κB p65 subunit, to exert its function (Luo et al., 2011; Yang et al., 2011; Morfouace et al., 2014; Xu et al., 2015). We considered the possibility that PKM2 might interact with the p65 subunit of NF-κB since this transcription factor plays a critical role in HIV-1 LTR activation. Thus, we assessed the effects of PKM2 and p65 overexpression on HIV-1 LTR expression in U-87 MG cells. The results showed that both p65 and PKM2 independently activated HIV-1 LTR by 2.9- and 1.9-fold, respectively. However, co-expression of p65 and PKM2 induced the HIV-1 LTR by 6.8-fold (Fig. 7A). Since PMA is known to stimulate NF-κB p65 subunit and PKM2 dimerization (Amini et al., 2002; Kaminski et al., 2014; Chaman et al., 2015), we assessed whether PMA could modulate the synergistic effects observed with PKM2 and p65 overexpression. The results showed that PMA alone induced HIV-1 LTR activity by 16-fold (Fig. 7B), while PKM2 and p65 overexpression individually in combination with PMA stimulated LTR activity by 5.5-fold and 15-fold, respectively. Surprisingly, overexpression of p65 and PKM2 together in conjunction of PMA stimulated HIV-1 LTR activity by 6.6-fold, similar to that observed in the absence of PMA (Fig. 7A and B). These observations suggest that PKM2 synergistically activates p65-mediated HIV-1 LTR activation.
Fig. 7.
(A) Synergistic activation of HIV-1 LTR by p65 and PKM2. Fold induction of HIV-1 LTR (−120/+66) promoter activity in U-87 MG cells transiently cotransfected with 0.25 μg of HIV-1 LTR/Luc reporter vector and 0.5 μg of an empty expression vector (LTR alone), or 0.25 μg of an expression vector for p65 (LTR + p65), or 0.25 μg of PKM2 (LTR + p65), and 0.25 μg of p65 and PKM2 expression vectors (LTR + p65 + PKM2). Cells were harvested 48 h post transfection to measure luciferase activity. RLU/s was normalized to protein concentration and expressed as fold induction wherein the activity in HIV-1 LTR/Luc alone transfected cells is set at 1. Statistical significance (*P <0.034 between LTR alone and LTR + PKM2 transfected cells; **P <0.016 between LTR alone and LTR+ p65; ***P <0.0003 between LTR alone and PKM2 + p65 transfected cells) was analyzed by Student’s t-test. (B) Fold induction of HIV-1 LTR (−120/+66) promoter activity in U-87 MG cells transiently cotransfected with 0.25 μg of HIV-1 LTR/Luc reporter vector and 0.5 μg of an empty expression vector (LTR alone), or 0.25 μg of an expression vector for p65 (LTR + p65), or 0.25 μg of PKM2 (LTR + p65), and 0.25 μg of p65 and PKM2 expression vectors (LTR + p65 + PKM2) and then treated with PMA for 24 h post transfection. Cells were harvested 48 h post-transfection to measure luciferase activity. RLU/s was normalized to protein concentration and expressed as fold induction wherein the activity in HIV-1 LTR/Luc alone transfected cells is set at 1. Statistical significance (*P <0.0004 between LTR alone and LTR alone transfected cells treated with PMA; **P <0.008 between LTR alone and LTR + PKM2 transfected cells treated with PMA; ***P <0.00005 between LTR alone and LTR + p65 transfected cells treated with PMA; ****P <0.003 between LTR alone and LTR+ p65 and PKM2 transfected cells treated with PMA) was analyzed by Student’s t-test. (C) PKM2 interacts with NF-κB p65 in U1 cells. Left panel: IP-Western analysis of whole cell lysates from PMA induced U1 cells immunoprecipitated with p65 antibodies or isotype specific antibodies and immunoblotted with p65 antibodies. Lane 1: whole cell lysate, lane 2: cell lysates IP with isotype specific antibody, and lane 3: cell lysates IP with p65 antibodies. Right panel: IP-Western analysis of whole cell lysates from PMA induced U1 cells immunoprecipitated with p65 antibodies or isotype specific antibodies and immunoblotted with PKM2 antibodies. Lane 1: whole cell lysate, lane 2: cell lysates IP with isotype specific antibody, and lane 3: cell lysates IP with p65 antibodies. IgG(H): heavy chain, IgG(L): light chain.
PKM2 interacts with NF-κB p65 subunit
To further examine whether PKM2 directly interacts with p65, we performed co-immunoprecipitation (Co-IP) using cell extracts prepared from PMA induced U1 cells. Cell lysates were immunoprecipitated with anti-p65 antibodies or isotype-matched rabbit IgG followed by Western blot for p65 and PKM2 (Fig. 7C). The results show the presence of co-immunoprecipitating PKM2 (Fig. 7C, lane 6) in cell lysate immunoprecipitated with p65 antibodies but not in cell lysate immunoprecipitated with isotype-matched rabbit IgG (Fig. 7C, lane 5).
Collectively, these results suggest that PKM2 and p65 interaction is essential for PKM2-p65 complex binding to HIV-1 LTR and upregulation of HIV-1 LTR-mediated reporter gene expression.
Discussion
The expression of HIV-1 gene products essential for the biogenesis of HIV-1 is regulated by the interaction of numerous DNA binding proteins with cis-acting DNA sequences present in the LTR (Pereira et al., 2000). Numerous studies have shown that HIV-1 manipulates the host cellular machinery for viral biogenesis (Brass et al., 2008; König et al., 2008; Zhou et al., 2008; Yeung et al., 2009). In addition, HIV-1 also induces significant alterations in the metabolism of infected cells (Hollenbaugh et al., 2011; Hegedus et al., 2014). PKM2 levels are upregulated by HIV-1 infection in different cell types such as T-cells, macrophage, and astrocytes (Reynolds et al., 2006; Chan et al., 2007; Rivera-Rivera et al., 2012; Rodríguez-Mora et al., 2015). However, the functional significance of PKM2 upregulation in HIV-1 pathogenesis is unknown.
In this study, we report that the expression of PKM2 protein is increased in monocyte/macrophages infected with HIV-1 and in latently infected U1 cells induced with PMA. The mechanism of induction of PKM2 by HIV-1 infection is unknown. However, we have demonstrated earlier that HIV-1 Vpr induces the expression of PKM2 in an HIF-1α-dependent manner in macrophages (Barrero et al., 2013). We also observed that viral replication enhances nuclear translocation of PKM2 and increases the amount of dimeric form of PKM2 in the nuclear fraction in PMA stimulated U1 cells. The mechanisms involved in the nuclear translocation of PKM2 are numerous and include phosphorylation of PKM2 at residue S37 by ERK2 (Yang et al., 2012), hydroxylation of residues P403 and P408 by PHD3 (Luo et al., 2011), and acetylation of residue K433 by p300 acetyltransferase (Lv et al., 2013). However, the mechanism(s) involved in HIV-1-mediated nuclear translocation and dimerization of PKM2 needs further investigation.
Elucidation of the non-metabolic function of PKM2 in the nucleus has revealed that it regulates gene expression by acting as a protein kinase (Gao et al., 2012; Wang et al., 2014). PKM2 is known to phosphorylate Stat3 at Y705 and histone H3 at T11 (Gao et al., 2012; Yang et al., 2012b). Furthermore, PKM2-mediated phosphorylation of H3-T11 induces Myc gene transcription following dissociation of HDAC3 (Yang et al., 2012b). In the context of HIV-1 latency, it is known that activation of NF-κB (p65-p50) is negatively regulated by HDAC3 through deacetylation. Studies have shown that HDAC2 and HDAC3 inhibition leads to the activation of HIV-1 in HeLa P4/R5 cell line model of latency (Keedy et al., 2009). It remains to be investigated in future studies whether nuclear translocation of PKM2 following stimulation of U1 cells with PMA results in phosphorylation of histone H3 and this, in turn, results in HIV-1 LTR induction following dissociation of HDAC3.
In this study, the functional significance of PKM2 in HIV-1 replication is shown by the interaction of PKM2 with HIV-1 LTR at the chromatin level and by upregulation of HIV-1 LTR by PKM2 overexpression. The early phase of HIV-1 transcription that is Tat-independent is initiated by the interaction of host transcription factors, such as NF-κB, SP1, and RNAP II, with cis-regulatory DNA sequences in the HIV-1 LTR (Kilareski et al., 2009). Among them, NF-κB proteins, such as p65 and p50, play a fundamental role in the transactivation of HIV-1 LTR (Nabel and Baltimore, 1987; Roulston et al., 1995). Studies have shown that NF-κB-mediated regulation of HIV-1 LTR is also modulated by interaction of NF-κB subunits with other cellular proteins. In this context, we have demonstrated that Rad51 hijacks the NF-κB pathway to enter the nucleus and in cooperation with p65 stimulates transcription of HIV-1 promoter by interacting with the NF-κB sequence (Kaminski et al., 2014). Likewise, the Werner syndrome helicase protein has been shown to interact physically with p50 and RelA to induce HIV-1 LTR in an NF-κB-dependent manner (Mizutani et al., 2015), and interferon-regulatory factor 1 (IRF-1) is known to form a complex with NF-κB and bind to κB sites on the LTR (Sgarbanti et al., 2008). The observation that PKM2 elicits it action through the NFκB enhancer region located in the regions −81 to −91 and −95 to −104 relative to the transcriptional start site prompted us to further investigate the mechanism of PKM2-mediated transactivation of HIV-1 LTR. In this regard, we have demonstrated that PKM2 had a synergistic effect with p65 on HIV-1 LTR induction and PKM2 physically interacts with the p65 subunit of NF-κB. A recent study has shown that PKM2 binds to the rel homology domain (amino acids 1–304) of p65, to enhance p65 interaction in EGR1 promoter to stimulate EGR1 expression (Xu et al., 2015).
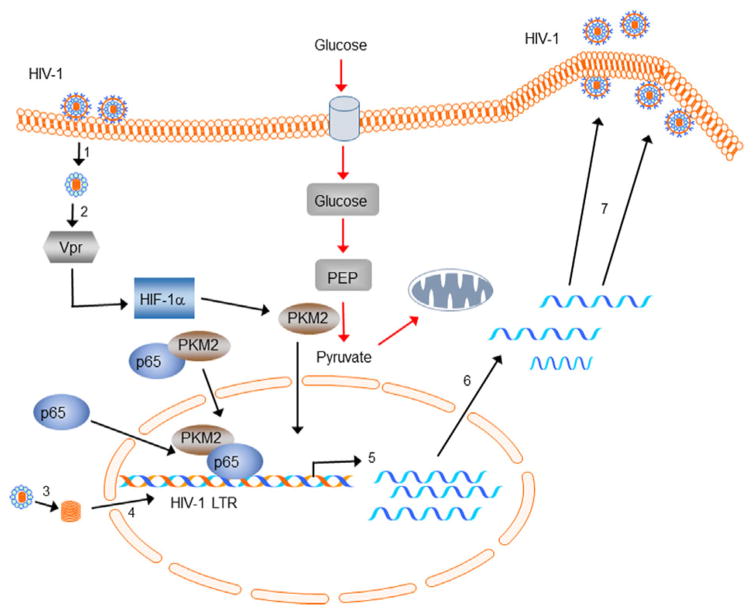
Based on these observations, we postulate that following HIV-1 infection of monocyte/macrophages and virus entry and core release (step 1), most likely virion-associated Vpr (step 2) in an HIF-1α-dependent manner induces PKM2 (Barrero et al., 2013). Following reverse transcription, proviral DNA (step 3) is imported into the nucleus and integrated into the host genome (step 4). Viral infection or reactivation of integrated virus (latent infection) results in the induction and nuclear translocation of PKM2 and p65 independently from the cytoplasm or by forming a functional complex (p65/PKM2) in the cytoplasm and subsequent nuclear translocation. p65/PKM2 interacts with the kB sites on HIV-1 LTR and synergistically activates transcription (step 5). Fully spliced mRNAs and partially spliced and unspliced RNAs are then transported to the cytoplasm and translated (step 6). Unspliced RNA is transported to the cell membrane, where Env, Gag, and Gag-Pol polyproteins assemble and incorporates into new virus particles (step 7). The new virus particle is then released after maturation (Fig. 8).
Fig. 8.
Schematic representation of the non-metabolic role of PKM2 in HIV-1-infected macrophages. HIV-1 infection of monocyte/macrophages and virus entry and core release (step 1). Virion-associated Vpr (step 2) in an HIF-1α-dependent manner induces PKM2 (Barrero et al., 2013). Following reverse transcription, proviral DNA (step 3) is imported into the nucleus and integrated into the host genome (step 4). Viral infection or reactivation of integrated virus (latent infection) results in the induction and nuclear translocation of PKM2 and p65 independently from the cytoplasm or by forming a functional complex (p65/PKM2) in the cytoplasm and subsequent nuclear translocation. p65/PKM2 interacts with the kB sites on HIV-1 LTR and synergistically activates transcription (step 5). Fully spliced mRNAs and partially spliced and unspliced RNAs are then transported to the cytoplasm and translated (step 6). Unspliced RNA is transported to the cell membrane, where Env, Gag, and Gag-Pol polyproteins assemble and incorporates into new virus particles (step 7). The new virus particle is then released after maturation.
Our observations suggest an important role of nuclear translocation of PKM2 and its interaction with HIV-LTR at the chromatin level to activate HIV-1 LTR activity and subsequently enhance viral biogenesis. Since the dimeric form of nuclear PKM2 is known to moonlight as a transcriptional regulator, targeting PKM2 dimerization could have potential translational relevance in inhibiting HIV-1 replication. In this regard, small-molecule PKM2 activators, such as TEPP-46, have therapeutic potential since it enhances association of PKM2 subunits into stable tetramers (Anastasiou et al., 2012) to promote glycolysis and not PKM2 dimer formation. Since we have shown that there is an increase in the dimeric form of PKM2 in the nucleus of HIV-1 infected cells, TEPP-46 or other small molecule activators of PKM2 can, therefore, attenuate HIV-1 biogenesis in macrophages.
In conclusion, we demonstrate for the first time that PKM2 is a non-metabolic regulator of HIV-1 transcription.
Acknowledgments
Contract grant sponsor: NIH/NIDA;
Contract grant numbers: 1P01DA037830, 5R01DA033213-05, P30MH092177.
This work was supported by an NIH/NIDA grant 1P01DA037830 and in part by an NIH/NIDA grant (5R01DA033213-05) to PKD. This study utilized services offered by core facilities of the Comprehensive NeuroAIDS Center (CNAC, NIMH grant number P30MH092177) Lewis Katz School of Medicine at Temple University. We thank Dr. Kamel Khalili for his support and constructive criticisms/discussions. We are grateful to Martyn K. White for critically reading the manuscript and editorial help.
Literature Cited
- Ahmad N, Venkatesan S. Nef protein of HIV-1 is a transcriptional repressor of HIV-1 LTR. Science. 1988;241:1481–1485. doi: 10.1126/science.3262235. [DOI] [PubMed] [Google Scholar]
- Amini S, Clavo A, Nadraga Y, Giordano A, Khalili K, Sawaya BE. Interplay between cdk9 and NF-kappaB factors determines the level of HIV-1 gene transcription in astrocytic cells. Oncogene. 2002;21:5797–5803. doi: 10.1038/sj.onc.1205754. [DOI] [PubMed] [Google Scholar]
- Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, Yang H, Mattaini KR, Metallo CM, Fiske BP, Courtney KD, Malstrom S, Khan TM, Kung C, Skoumbourdis AP, Veith H, Southall N, Walsh MJ, Brimacombe KR, Leister W, Lunt SY, Johnson ZR, Yen KE, Kunii K, Davidson SM, Christofk HR, Austin CP, Inglese J, Harris MH, Asara JM, Stephanopoulos G, Salituro FG, Jin S, Dang L, Auld DS, Park HW, Cantley LC, Thomas CJ, Vander Heiden MG. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero CA, Datta PK, Sen S, Deshmane S, Amini S, Khalili K, Merali S. HIV-1 Vpr modulates macrophage metabolic pathways: A SILAC-based quantitative analysis. PLoS ONE. 2013;8:e68376. doi: 10.1371/journal.pone.0068376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Chaman N, Iqbal MA, Siddiqui FA, Gopinath P, Bamezai RN. ERK2-pyruvate kinase axis permits phorbol 12-myristate 13-acetate-induced megakaryocyte differentiation in K562Cells. J Biol Chem. 2015;290:23803–23815. doi: 10.1074/jbc.M115.657411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Qian WJ, Diamond DL, Liu T, Gritsenko MA, Monroe ME, Camp DG, 2nd, Smith RD, Katze MG. Quantitative analysis of human immunodeficiency virus type 1-infected CD4+ cell proteome: Dysregulated cell cycle progression and nuclear transport coincide with robust virus production. J Virol. 2007;81:7571–7583. doi: 10.1128/JVI.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Sutton JN, Jacobs JM, Bondarenko A, Smith RD, Katze MG. Dynamic host energetics and cytoskeletal proteomes in human immunodeficiency virus type 1-infected human primary CD4 cells: Analysis by multiplexed label-free mass spectrometry. J Virol. 2009;83:9283–9295. doi: 10.1128/JVI.00814-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary P, Khan SZ, Rawat P, Augustine T, Raynes DA, Guerriero V, Mitra D. HSP70 binding protein 1 (HspBP1) suppresses HIV-1 replication by inhibiting NF-κB mediated activation of viral gene expression. Nucleic Acids Res. 2016;44:1613–1629. doi: 10.1093/nar/gkv1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipitsyna G, Sawaya BE, Khalili K, Amini S. Cooperativity between Rad51 and C/EBP family transcription factors modulates basal and Tat-induced activation of the HIV-1 LTR in astrocytes. J Cell Physiol. 2006;207:605–613. doi: 10.1002/jcp.20612. [DOI] [PubMed] [Google Scholar]
- Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, Metz TO, Camp DG, 2nd, Waters KM, Smith RD, Rice CM, Katze MG. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez Larrosa PN, Croci DO, Riva DA, Bibini M, Luzzi R, Saracco M, Mersich SE, Rabinovich GA, Martinez Peralta L. Apoptosis resistance in HIV-1 persistently-infected cells is independent of active viral replication and involves modulation of the apoptotic mitochondrial pathway. Retrovirology. 2008;5:19. doi: 10.1186/1742-4690-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folks TM, Justement J, Kinter A, Dinarello CA, Fauci AS. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238:800–802. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin CM, Xu S, Munger J. Stealing the keys to the kitchen: Viral manipulation of the host cell metabolic network. Trends Microbiol. 2015;23:789–798. doi: 10.1016/j.tim.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrich D, Garcia J, Wu F, Mitsuyasu R, Gonazalez J, Gaynor R. Role of SP1-binding domains in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1989;63:2585–2591. doi: 10.1128/jvi.63.6.2585-2591.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus A, Kavanagh Williamson M, Huthoff H. HIV-1 pathogenicity and virion production are dependent on the metabolic phenotype of activated CD4+ T cells. Retrovirology. 2014;11:98. doi: 10.1186/s12977-014-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbaugh JA, Munger J, Kim B. Metabolite profiles of human immunodeficiency virus infected CD4+ T cells and macrophages using LC-MS/MS analysis. Virology. 2011;415:153–159. doi: 10.1016/j.virol.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Tanaka T. Pyruvate kinase isozymes from rat. Methods Enzymol. 1982;90:150–165. doi: 10.1016/s0076-6879(82)90121-5. [DOI] [PubMed] [Google Scholar]
- Israelsen WJ, Vander Heiden MG. Pyruvate kinase: Function, regulation and role in cancer. Semin Cell Dev Biol. 2015;43:43–51. doi: 10.1016/j.semcdb.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski R, Wollebo HS, Datta PK, White MK, Amini S, Khalili K. Interplay of Rad51 with NF-κB pathway stimulates expression of HIV-1. PLoS ONE. 2014;9:e98304. doi: 10.1371/journal.pone.0098304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J Virol. 2009;83:4749–4756. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilareski EM, Shah S, Nonnemacher MR, Wigdahl B. Regulation of HIV-1 transcription in cells of the monocyte-macrophage lineage. Retrovirology. 2009;6:118. doi: 10.1186/1742-4690-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Gromiha MM, Raghava GPS. Identification of DNA-binding proteins using support vector machines and evolutionary profiles. BMC Bioinformatics. 2007;8:463. doi: 10.1186/1471-2105-8-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WZ, Fang JA, Xiao X, Chou KC. IDNA-Prot: Identification of DNA binding proteins using random forest with grey model. PLoS ONE. 2011;6:e24756. doi: 10.1371/journal.pone.0024756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N, Jiang Y, Zhou X, Li TT, Guan KL, Lei QY, Xiong Y. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell. 2013;52:340–352. doi: 10.1016/j.molcel.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranto J, Rappaport J, Datta PK. Role of C/EBP-β, p38 MAPK, and MKK6 in IL-1β-mediated C3 gene regulation in astrocytes. J Cell Biochem. 2011;112:1168–1175. doi: 10.1002/jcb.23032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DM, Somasundaran M, Green MR. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol. 1994;68:905–910. doi: 10.1128/jvi.68.2.905-910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzon M, Mercer J. Lipid interactions during virus entry and infection. Cell Microbiol. 2014;16:1493–1502. doi: 10.1111/cmi.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T, Ishizaka A, Furuichi Y. The Werner protein acts as a coactivator of nuclear factor κB (NF-κB) on HIV-1 and interleukin-8 (IL-8) promoters. J Biol Chem. 2015;290:18391–18399. doi: 10.1074/jbc.M115.657155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfouace M, Lalier L, Oliver L, Cheray M, Pecqueur C, Cartron PF, Vallette FM. Control of glioma cell death and differentiation by PKM2-Oct4 interaction. Cell Death Dis. 2014;5:e1036. doi: 10.1038/cddis.2013.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- Nishitsuji H, Sawada L, Sugiyama R, Takaku H. ZNF10 inhibits HIV-1 LTR activity through interaction with NF-κB and Sp1 binding motifs. FEBS Lett. 2015;589:2019–2025. doi: 10.1016/j.febslet.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Olivares I, Ballester A, Lombardia L, Dominguez O, Lopez-Galindez C. Human immunodeficiency virus type 1 chronic infection is associated with different gene expression in MT-4, H9 and U937 cell lines. Virus Res. 2009;139:22–31. doi: 10.1016/j.virusres.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira LA, Bentley K, Peeters A, Churchill MJ, Deacon NJ. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JL, Mahajan SD, Bindukumar B, Sykes D, Schwartz SA, Nair MP. Proteomic analysis of the effects of cocaine on the enhancement of HIV-1 replication in normal human astrocytes (NHA) Brain Res. 2006;1123:226–236. doi: 10.1016/j.brainres.2006.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Rivera L, Perez-Laspiur J, Colón K, Meléndez LM. Inhibition of interferon response by cystatin B: Implication in HIV replication of macrophage reservoirs. J Neurovirol. 2012;18:20–29. doi: 10.1007/s13365-011-0061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Mora S, Mateos E, Moran M, Martín MÁ, López JA, Calvo E, Terrón MC, Luque D, Muriaux D, Alcamí J, Coiras M, López-Huertas MR. Intracellular expression of Tat alters mitochondrial functions in T cells: A potential mechanism to understand mitochondrial damage during HIV-1 replication. Retrovirology. 2015;12:78. doi: 10.1186/s12977-015-0203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr O, Marban C, Aunis D, Schaeffer E. Regulation of HIV-1 gene transcription: From lymphocytes to microglial cells. J Leukoc Biol. 2003;74:736–749. doi: 10.1189/jlb.0403180. [DOI] [PubMed] [Google Scholar]
- Roulston A, Lin R, Beauparlant P, Wainberg MA, Hiscott J. Regulation of human immunodeficiency virus type 1 and cytokine gene expression in myeloid cells by NF-kappa B/Rel transcription factors. Microbiol Rev. 1995;59:481–505. doi: 10.1128/mr.59.3.481-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Mitchell DA. TFII-I and USF (RBF-2) regulate ras/MAPK-responsive HIV-1 transcription in t cells. Eur J Cancer. 2005;41:2528–2536. doi: 10.1016/j.ejca.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M, Zenzie-Gregory B, Groopman JE, Smale ST, Kim SY. Alternative pathway for induction of human immunodeficiency virus gene expression: Involvement of the general transcription machinery. J Virol. 1991;65:5448–5456. doi: 10.1128/jvi.65.10.5448-5456.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salani B, Ravera S, Amaro A, Salis A, Passalacqua M, Millo E, Damonte G, Marini C, Pfeffer U, Sambuceti G, Cordera R, Maggi D. IGF1 regulates PKM2 function through Akt phosphorylation. Cell Cycle. 2015;14:1559–1567. doi: 10.1080/15384101.2015.1026490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez EL, Lagunoff M. Viral activation of cellular metabolism. Virology. 2015;479–480:609–618. doi: 10.1016/j.virol.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki CY, Barberi TJ, Ghosh P, Longo DL. Phosphorylation of RelA/p65 on serine 536 defines an I{kappa}B{alpha}-independent NF-{kappa}B pathway. J Biol Chem. 2005;280:34538–34547. doi: 10.1074/jbc.M504943200. [DOI] [PubMed] [Google Scholar]
- Sen S, Kaminiski R, Deshmane S, Langford D, Khalili K, Amini S, Datta PK. Role of hexokinase-1 in the survival of HIV-1-infected macrophages. Cell Cycle. 2015;14:980–989. doi: 10.1080/15384101.2015.1006971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgarbanti M, Remoli AL, Marsili G, Ridolfi B, Borsetti A, Perrotti E, Orsatti R, Ilari R, Sernicola L, Stellacci E, Ensoli B, Battistini A. IRF-1 is required for full NF-kappaB transcriptional activity at the human immunodeficiency virus type 1 long terminal repeat enhancer. J Virol. 2008;82:3632–3641. doi: 10.1128/JVI.00599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southgate C, Zapp ML, Green MR. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature. 1990;345:640–642. doi: 10.1038/345640a0. [DOI] [PubMed] [Google Scholar]
- Spoden GA, Rostek U, Lechner S, Mitterberger M, Mazurek S, Zwerschke W. Pyruvate kinase isoenzyme M2 is a glycolytic sensor differentially regulating cell proliferation, cell size and apoptotic cell death dependent on glucose supply. Exp Cell Res. 2009;315:2765–2774. doi: 10.1016/j.yexcr.2009.06.024. [DOI] [PubMed] [Google Scholar]
- Vastag L, Koyuncu E, Grady SL, Shenk TE, Rabinowitz JD. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7:e1002124. doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HJ, Hsieh YJ, Cheng WC, Lin CP, Lin YS, Yang SF, Chen CC, Izumiya Y, Yu JS, Kung HJ, Wang WC. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α-mediated glucose metabolism. Proc Natl Acad Sci USA. 2014;111:279–284. doi: 10.1073/pnas.1311249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Mukherjee S, Jia F, Narayan O, Zhao LJ. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem. 1995;270:25564–25569. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015;356:184–191. doi: 10.1016/j.canlet.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Xu Q, Liu LZ, Yin Y, He J, Li Q, Qian X, You Y, Lu Z, Peiper SC, Shu Y, Jiang BH. Regulatory circuit of PKM2/NF-κB/miR-148a/152-modulated tumor angiogenesis and cancer progression. Oncogene. 2015;356:184–191. doi: 10.1038/onc.2015.6. [DOI] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012a;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012b;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of Jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]