Abstract
Sensing of cytosolic nucleotides is a critical initial step in the elaboration of type I interferon. One of several upstream receptor cGAS (cyclic-GMP-AMP synthase) binds to cytosolic DNA and generates di-cyclic nucleotides that act as secondary messengers. These secondary messengers bind directly to Stimulator of Interferon Genes (STING). STING recruits TANK binding kinase 1 (TBK1) which acts as a critical node that allows for efficient activation of interferon regulatory factors (IRFs) to drive the anti-viral transcriptome. NLRC3 is a recently characterized nucleotide-binding domain, leucine rich repeat containing protein (NLR) that negatively regulates the type I interferon pathway by inhibiting subcellular redistribution and effective signaling of STING, thus blunting the transcription of type I interferons. NLRC3 is predominantly expressed in lymphoid and myeloid cells. IQGAP1 was identified as a putative interacting partner of NLRC3 through yeast two hybrid screening. Here we show that IQGAP1 associates with NLRC3 and can disrupt the NLRC3:STING interaction in the cytosol of human epithelial cells. Furthermore, knock down of IQGAP1 in THP1 and HeLa cells causes significantly more interferon-β production in response to cytosolic nucleic acids. This result phenocopies NLRC3 deficient macrophages and fibroblasts and shRNA knock down of NLRC3 in THP1 cells. Our findings suggest IQGAP1 is a novel regulator of type I interferon production possibly via interacting with NLRC3 in human monocytic and epithelial cells.
Introduction
The vertebrate innate immune system is characterized by germ-line encoded pattern recognition receptors (PRRs) that can directly bind to or indirectly sense pathogen associated molecular patterns (PAMPs). The activation of PRRs by their cognate PAMPs initiates several diverse signaling cascades such as the nuclear factor kappaB (NF-κB), mitogen-activated protein kinase (MAPK) and interferon regulatory factors (IRF). These pathways lead to robust cytokine or interferon secretion that is specific for the type of pathogen. Most PRRs have been grouped into 5 super-families based on amino acid sequence homology and domain organization. These families include the Toll-like receptors (TLRs), C-type lectin receptors (CLRs), RIG-I-like receptors (RLRs), AIM2-like receptors (ALRs) and the NOD-like receptors or nucleotide binding domain leucine rich repeat containing proteins (NLRs) (1). Individual PRRs occupy many distinct extra- and intracellular locations. Several PRR and their signaling molecules have been shown to dynamically redistribute within the cell to more efficiently effect their function which ultimately leads to a rapid immune response (2).
The secretion of type I interferon is a critical step in the initiation of the immune response to cytosolic DNA, RNA and nucleotide metabolites. The mechanisms of type I interferon production have been studied extensively (3). Recent evidence has identified key signaling molecules and both positive and negative regulators of this pathway. There is a manifold array of cytosolic sensors of nucleic acids that induce type I interferon in response to viral and bacterial infections. For example, cGAS (cyclic GMP-AMP synthase) binds directly to double-stranded DNA (dsDNA) in the cytosol in a sequence independent manner by interacting with the sugar phosphate backbone of DNA. Therefore cGAS can bind dsDNA from multiple sources such as DNA viruses (HSV), eukaryotic nuclear DNA (calf thymus) and prokaryotic DNA (bacteria) insofar as they are located in the host cytosol. Binding of dsDNA, ATP and GTP to cGAS causes dimerization and dramatic conformational changes to open the active site of cGAS to generate cyclic GAMP (cyclic GMP-AMP) which is a type of cyclic dinucleotide (CDN) that can act as secondary messenger (4–6). CDNs bind directly to the ER-transmembrane protein STING (stimulator of interferon genes, also known as MITA, MYPY, ERIS and TMEM173) causing its dimerization (7–11). The STING dimers can interact with and activate TBK1 (TNFR-associated NF-κB kinase (TANK)-binding kinase) at the mitochondrial associated ER-membrane (MAMs). Active TBK1 can phosphorylate IRF3 and IRF7 to promote dimerization and nuclear translocation. Nuclear IRFs in combination with NF-κB subunits activates interferon stimulated gene (ISG) transcription (12). Intracellular bacterial pathogens such as Listeria monocytogenes and Microbacteria tuberculosis generate unique CDNs (cyclic di-AMP and GMP) as metabolic byproducts bypassing the requirement for cGAS activity (13–16). Both host and pathogen derived CDNs bind directly to STING, inducing different conformational changes that ultimately lead to IRF3/7 activation (7).
Both host and pathogen can negatively regulate the inflammatory response to CDNs. STING dynamically relocalizes from the ER via the Golgi to cytosolic vesicles possibly via an atypical autophagy response. The redistribution of STING is mediated by several autophagy mediated signaling proteins, ULK1/ATG1 and ATG9, to possibly act as a brake on type I interferon production (17, 18). In the absence of Atg9 STING is targeted for proteosomal degradation via endogenous RNF5 (RING-finger protein 5) dependent ubiquitination (19). Additionally, Herpes simplex virus (HSV) has been shown to encode several open reading frames that inhibit STING-dependent IFNb production. For example, vIRF1 can inhibit TBK1 binding to STING and prevent IRF3 phosphorylation; additionally, vIRF1 has been shown to inhibit p300 histone acetylation which blocks IRF3 recruitment of histone modifying enzymes (20). Many other pathogen escape pathways have been described suggesting that targeted inhibition of type I interferon provides a selective advantage for the pathogen (21).
It is important to note that there is growing evidence that NLR proteins are pleiotropic and can act in many pathways. Several NLRs have been shown to bind directly to PAMPs and act as bona fide PRRs (22–24). Others appear to have regulatory roles that indirectly influence pathogen sensing or the regulation of inflammation. However, there is evidence that supports the model that NLR proteins respond cooperatively or redundantly to the same pathogen or that single NLRs can regulate disparate pathways that may be cell or species specific(25). For example, recent evidence has suggested that NLRC3 can also act as a negative regulator of the PI3K-mTOR pathway in epithelial cells, which is involved in colorectal carcinogenesis(26) as well as affecting the type I interferon and proinflammatory pathways. Likewise, NLRX1 has been shown to be involved in negatively regulating multiple tumorigenic pathways(27–29) as well as other inflammatory pathways(23, 28, 30–32). These recent advances in the field have challenged the model that single NLR proteins are pathogen or pathway specific. NLR proteins are characterized by a central Nucleotide Oligomerization Domain (NOD) that is composed of a NACHT domain (NAIP (neuronal apoptosis inhibitor protein), C2TA (MHC class 2 transcription activator), HET-E (incompatibility locus protein from Podospora anserina) and TP1 (telomerase-associated protein)) or NBD (nucleotide binding domain), two Helical Domains (HD1 and HD2) and a Winged Helix Domain (WHD), preceded by an amino-terminus (N) effector domain (either CARD (caspase recruitment domain), PYD (pyrin domain), BIR (baculovirus IAP repeat) or undefined domains) and a series of variable leucine rich repeats (LRRs) at the carboxy terminus (25). Several NLRs have been shown to assemble into large macromolecular complexes: inflammasomes (NLRP1, NLRP3, NLRC4/NAIP and others), enhanceosomes (CIITA, NLRC5 and possibly NLRP3 (33)), NODosomes (NOD1 and NOD2) and a TRAFasome (NLRC3) (34, 35). The formation of these complexes is mediated by the scaffolding ability of individual NLRs to recruit diverse sets of interacting proteins. These protein-protein interactions can be mediated by each of the three domains of NLRs facilitating the recruitment of multiple binding partners, possibly in series or in tandem.
NOD2 (36–41), NLRX1 (42–46) NLRP4 (47) and NLRC3 (48) have been implicated in sensing viral infections in vitro and in vivo. However, the mechanisms are poorly understood and debated within the field (49, 50). Future studies will be needed to address the precise roles of individual NLR proteins in response to viral pathogens. These studies will help further define the cell-type, pathogen and spatiotemporal specific responses. NLRC3 is a poorly characterized NLR that negatively regulates several pathways in many different cell types. NLRC3 contains an amino-terminal CARD-like region, a central NBD and a series of multiple LRRs. The Ting laboratory has shown that NLRC3 can act as a negative regulator of several signaling pathways in T lymphocytes (51), macrophages (52) and epithelial/fibroblast cells (48). NLRC3 was shown to interact with TRAF6 via a TRAF2 consensus binding site in human myeloid and epithelial cells. In mice, this interaction inhibited the NF-κB dependent signaling during experimental colitis induced by cecal ligation puncture (52). In a different system it was shown that recombinant purified NLRC3 binds directly to STING and NLRC3 coimmunoprecipates both STING and TBK1. In HeLa cells this interaction inhibited the redistribution of STING to the perinuclear region and blunted the TBK1-dependent phosphorylation of IRF3. In vivo NLRC3 was necessary for limiting type I interferon in response to lethal HSV infection. Nlrc3−/− mice infected with HSV showed decreased morbidity and enhanced innate immunity (48). This data suggests that NLRC3 is a negative regulator of virus induced interferon responses.
Due to the diverse nature of the inhibitory ability of NLRC3 we sought to identify novel protein-protein interaction pathways associated with NLRC3 by yeast two-hybrid screening. These screens yielded several candidate proteins. Here we show that IQGAP1, a scaffolding protein involved in diverse cellular processes specifically interacts with NLRC3 and can disrupt the NLRC3:STING interaction. IQGAP1 is a highly-conserved protein that has multiple protein-protein interaction domains. The role of IQGAP1 in the immune is not well understood, but it may function to distribute signaling molecules to specific subcellular regions within the cell or facilitate the coordination of positive and negative regulators(53). In human transformed cells IQGAP1 has been shown to negatively regulate cytokine secretion in response to T cell receptor ligation in Jurkat T cells(54) by negatively regulating filamentous actin at the immunological synapse, while it positively regulates degranulation of NK-like cells(55) by controlling the microtubule organizing center. Additionally, IQGAP1 has been shown to be involved in IL-1β secretion in bone marrow-derived macrophages in response to Yersinia pestis(56). IQGAP1 and NLRC3 are expressed in hematopoietic and non-hematopoeitic tissues and partially colocalize in the cytosol/cytoplasm within epithelial cells. IQGAP1 shRNA mediated knockdown in THP1 and HeLa cells phenocopy NLRC3 in response to cytosolic DNA stimulation, implying that the NLRC3:IQGAP1 complex inhibits cGAS-dependent cytosolic DNA sensing. We show that NLRC3 specifically interacts with IQGAP1 and this interaction can inhibit the NLRC3:STING interaction. These data support the hypothesis that the NLRC3:IQGAP1 complex may act as a negative regulator of type 1 IFN, inhibiting the cells ability to mount a hyper interferon response possibly acting as a biological rheostat. These data are the first to illustrate a role for IQGAP1 in the type 1 interferon signaling pathway.
Materials and Methods
Cell Culture and Transfections
Human Embryonic Kidney (HEK293T), SW480 and HeLa cells were maintained in DMEM (Sigma) supplemented with 10% Fetal Bovine Serum (Invitrogen), 20 units penicillin, 20 mg of streptomycin, 1X sodium pyruvate (Gibco), and 146 mg of L-glutamine cocktail (Gibco). THP1 and THP1-Dual™ Cells (Invivogen) were grown in RPMI (Gibco) supplemented as described above. All cells were grown at 37 °C and 5 % CO2.
HEK293T, SW480 and HeLa cells were seeded into 6-well plates at a density between 1.0–5.0×105 cells per well and transfected with epitope tagged constructs using Lipofectamine 2000 (Invitrogen) at a ratio of 3:1 according to the manufacturer’s protocol. Cells were processed 18–24 hours after transfection.
Stable Knockdown in human cell lines
HeLa, THP1 and THP1-Dual™ cells were transduced with shRNA lentiviral particles produced from plasmid DNA for human shRNA for IQGAP1 (Mission™ TRCN0000298928, TRCN0000298930, TRCN0000298931, Sigma Aldrich) according to the manufacturer’s protocol. The IQGAP1 shRNA-transduced clones were selected with 0.6μg/ml puromycin. To determine efficiency of knockdown, RNA was isolated from cells using Trizol Reagent (Life Technologies) following manufacturer protocol and mRNA levels were assessed by RT-PCR (data not shown). IQGAP1 protein expression levels were evaluated by immunobloting with rabbit anti-IQGAP1 (1:5000; Abcam) antibody.
Co-immunoprecipitations, Subcellular Fractionation and Western Blotting
Cells were lysed in 1X PBS and 1% Triton-X 100 and 0.05 % Sodium Deoxycholate supplemented with protease inhibitor cocktail for 30 minutes at 4°C. The lysates were then cleared by centrifugation (14000 × g, 15 min at 4°C) and incubated with mouse anti-Flag M2 antibody (Sigma, 1:1000) for 4 hours at 4°C and immunopreciptated using Protein A/G Ultralink Resin (Pierce) for 24 hours shaking at 4°C. Immunoprecipitates were washed three times in phosphate buffered saline and 1% Triton X-100 and 0.05% deoxycholate. Samples were analyzed using SDS-PAGE (using 4–15% Mini-PROTEAN® TGX™ gels). Gels were then transferred to nitrocellulose membrane (Bio-rad). Immunoblots were probed with anti-Flag M2 conjugated to horseradish peroxidase (HRP) (Sigma, 1:5000), anti-HA conjugated to HRP (Sigma,1:5000), rabbit anti-GFP (Sigma, 1:5000), goat anti-rabbit IgG HRP (Santa Cruz,1:5000). Transfected SW480 cells were fractionated using subcellular fractionation kit for cultured cells (Pierce) per manufacturer’s suggested protocol. Rabbit mAbs to PKM2, Vimentin, CoxIV and H3 (Cell Signaling, 1:1000) were used to probe immunoblots of subcellular fractions.
Immunofluorescence
SW480 and HeLa cells were seeded in a 6-well dish containing glass coverslips at a density of 5.0×105 cells per well and transfected as previously described. After 24 hours the cells were fixed using 4% formaldehyde (Sigma) in 1XPBS for 10 minutes, then washed three times with 1X PBS and blocked with PBTN (1X PBS, 1% normal goat serum, and 0.1% Triton-X 100) for 15 minutes. Cells were then stained with mouse anti-Flag M2 (Sigma, 1:1000) or rat anti-Flag (Novus, 1:1000), rat anti-HA (Roche, 1:1000), chicken anti-HA (Abcam, 1:1000) mouse anti-HA (Sigma, 1:1000) or rabbit anti-HA (Cell Signaling, 1:1000), rabbit anti-V5 (Cell Signaling, 1:1000) or mouse anti-V5 (Life Technologies, 1:1000). We observed no differences in staining between different species and/or isotypes of antibodies used in our studies (data not shown). Primary antibodies to HSP60, NUP98, PKM2 were purchased from Cell Signaling Technologies (1:300). Phalloidin conjugated to either Alexa Fluor 488, 568 or 647 was used (Life Technologies, 1:1000). Rabbit anti-Tomm20 (Abcam, 1:1000) was used to identify mitochondria. Species and isotype matched goat secondary antibodies conjugated to various Alexa Fluor dyes were used (Life Technologies, 1:5000) with DAPI nuclear counterstain. Cells were imaged using a Nikon E400 epifluorescent microscope equipment with a camera or a Leica SP8 confocal microscopes equipped with 5 laser lines or with a white light laser at Franklin and Marshall College or the Imaging Core at Pennsylvania State College of Medicine, respectively.
SEAP/Luciferase Reporter Analysis and ELISA
THP1-Dual™ Cells were seeded into 12 well plates at the density of 5×105 cells per well and transfected with 5 μg/mL of 2′2′ or 3′3′ cGAMP, 1μg/mL of ISD or HSV DNA (Invivogen) with Lipofectamine 3000 per the manufacturer’s suggested protocol. Secreted Embryonic Alkaline Phosphatase (SEAP) and Lucia™ activity were measured 24 hours after induction using QUANTI-Blue™ (Invivogen) and QUANTI-Luc™ (Invivogen) according to the manufacturer’s protocol. Tissue culture supernatants from stimulated cells were assayed for TNF-α, IL-1β (BD Biosciences) and IFN-β (Antigenix) by sandwich ELISA using the manufacturors’ suggested protocol.
Vector Construction
The full length IQGAP1 open reading frame was amplified from a plasmid template using a mixture Platinum Pfx and Taq polymerases(Life Technologies) and cloned into the pCR8/TOPO/TA (Life Technologies) by TOPO TA cloning. The truncations of IQGAP1 and NLRC3 were amplified from full length template using the primers listed in Table 1 (Integrated DNA Technologies) and cloned into the pCR8/TOPO/TA (Life Technologies) base vector. Recombination reactions were performed into modified FLAG or Flourescent protein Gateway destination vectors (described in (57)) using LR Clonase II (Life Technologies). Generation of the NLRC3 plasmid has been previously described (51). ColorfulCell was a gift from Pierre Neveu (Addgene plasmid # 62449), pEGFP-IQGAP1 was a gift from David Sacks (Addgene plasmid # 30112), pCAG-mGFP was a gift from Connie Cepko (Addgene plasmid # 14757), epitope tagged MAVS, human STING and human TBK1 were provided by J. Ting University of Chapel Hill. STING was subcloned into pCDNA3.1D to provide a C-terminal V5 tag. All vector sequences were verified by bidirectional sequencing.
Table 1.
Primer sequences of constructs used
| Gene | Forward primer (5′-->3′) | Reverse primer (5′-->3′) |
|---|---|---|
| IQGAP1 full length | ATGTCCGCCGCAGACGAGGTTGACGGGC | TTACTTCCCGTAGAACTTTTTGTTGAGAAGG |
| IQGAP1 RGCT | ATGAAAAAGTCAAAATCTGTAAAGG | TTACTTCCCGTAGAACTTTTTGTTGAGAAGG |
| NLRC3 full length | ATGAGGAAGCAAGAGGTGCGGACGGGC | TCACATTTCAACAGTGCACGTGGGAGC |
| NLRC3 N term | ATGAGGAAGCAAGAGGTGCGGACGGGC | GTCATTGCTGCAGGGCCCCAGC |
| NLRC3 NBD | TCAAGGATACAGAGGCACCGCAAGGCCC | GTCCAGCCTGAGCTTCCGGCAGTAGAGC |
| NLRC3 NNBD | ATGAGGAAGCAAGAGGTGCGGACGGGC | GTCCAGCCTGAGCTTCCGGCAGTAGAGC |
| QPCR NLRC3 F | CGGAGAACCAGATCAGTAACAA | CCTTGTGGTCCAATGGAGTTA |
| QPCR HPRT | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT |
Yeast Two-Hybrid Screening
A yeast two-hybrid screen was completed by Hybrigenics (Paris, France) using a truncation of human NLRC3 consisting of amino acids 1–600 as the bait and a human leukocyte and activated mononuclear cell cDNA library as the prey. Additionally, two small-scale library screens were performed in our laboratory using MAV203 cells (Life Technologies). These screens used full length human NLRC3 as the bait and a normalized human universal library (Dual Systems) as the prey. In brief, 10μg of both bait and prey DNA were added to 250μl of MAV203 cells in dulpicate. To each tube a polyethylene glycol/lithium acetate solution was added and tubes were incubated for 30 minutes in a 30°C water bath. After the addition of sterile DMSO the cells were heat shocked at 42°C for 20 minutes, pelleted, and resuspended in sterile PBS. 400μl of cells were plated onto –Leu/-Trp/-His/-Ura plates and incubated at 30°C. The resulting colonies were picked, placed into 100μl of LiOAc+1% SDS and heated at 70°C for 5 minutes. Following the addition of 300μl of 70% ethanol the cells were pelleted at 15000 × g for 3 minutes and washed with 70% ethanol. The pellets were resuspended into 100μl sterile water and spun at 15000 × g for 15 seconds to pellet debris. PCR was then performed using Dream TAQ DNA polymerase (Thermo Scientific) and 1μl of the supernatant of the resuspended pellet. Bidirectional sequencing was performed using 5μl of the PCR product to identify the interactors of human NLRC3.
Reverse Transcriptase-polymerase chain reaction and Q-PCR, Barcoding and microarray database searches
Human multiple tissue cDNA panels I and II (Clontech), were used as templates to determine expression patterns of NLRC3 and IQGAP1. Briefly, 1–2μls of cDNA was amplified using Phusion 2X master mix. The cycling conditions were as follows: 94°C for 10 seconds, 55°C for 15 seconds, and 72°C for 30 seconds for 25 cycles (GAPDH), 30 cycles (IQGAP1) or 33 cycles (NLRC3). Total RNA was harvested from Jurkat, THP1, HeLa and HEK293T cells using Trizol reagent (Life Technologies) and treated with DNAse I (Sigma Aldrich) and reversed transcribed using RevertAid reverse transcriptase (Life Technologies). QPCR was performed with primers for NLRC3 and HPRT using AmpliTaq (Applied Biosystems) on a Bio-Rad CFX96 machine. Values were calculated by relative expression to HPRT using ΔCt method. Nextbio (https://www.nextbio.com/) database was searched for IQGAP and NLRC3. Raw data was downloaded and specific cell types were graphed. Molecular barcodes were obtained from The Gene Expression 3.0 Program (barcode.luhs.org) utilizing the “Probe Set Pages” option. The HGU133plus2(Human)v3 platform was used and the tissue graph was selected to show only Immune Cells and Tissues from Affymetrix arrays. The affinity probe sets examined were IQGAP1 (200791_s_at) and NLRC3 (236295_s_at). The resulting graphs showing Z-scores ± median absolute deviation (MAD) were used.
Statistical analysis
Two-way ANOVA with Tukey’s post hoc statistical analyses were done using Prism 5.0 for Macintosh. Data are represented as mean ± standard deviation of triplicates. Asterisks represent p values < .05
Results
Yeast two- hybrid screen
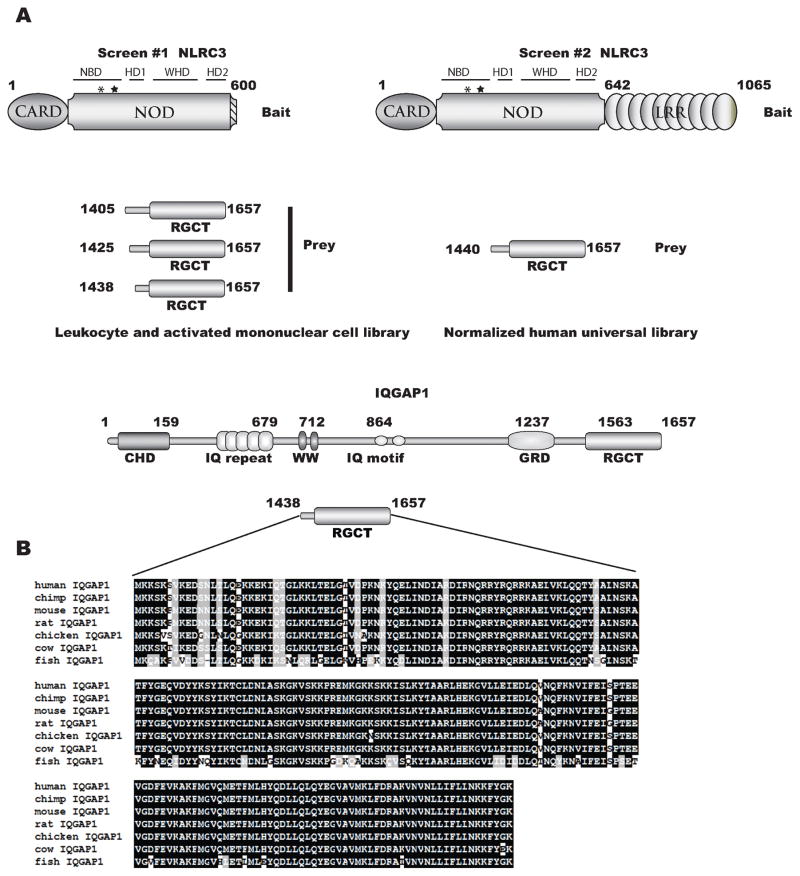
To investigate putative protein interactions with NLRC3 we performed yeast two-hybrid screens. A carboxy-terminal truncation (amino acids 1–600, containing the CARD and a majority of the NBD) and full length NLRC3 were used to screen a human activated peripheral mononuclear cell cDNA library and a universal human cDNA library, respectively. We initially performed a yeast two-hybrid screen with a bait construct lacking the LRR because the LRR is a potential auto-inhibiting structure that might mask potential interacting partners. Recent crystal structures and functional studies of mouse Nlrc4 and rabbit NOD2 support the hypothesis that the LRRs maintain NLRs in a closed or inhibited confirmation possibly to prevent premature activation (58, 59). Based on molecular modeling of NLRC3 on rabbit NOD2, (data not shown), the truncation construct used for our screens should contain the known structural domains (see Figure 1A) that correspond to a functional NOD. However, the crystal structure for NLRC3 has not been published and the exact three-dimensional conformation of the NOD is unknown. In our studies, two separate libraries were used; first, to enrich for immune system specific interactions we screened a human leukocyte and activated mononuclear cell library. Additionally, we screened a human universal cDNA library to potentially uncover nonhematopoietic interacting proteins. Over 107 clones were screened. Seven clones from the independent screens identified the carboxy terminus of IQGAP1 as a putative interacting region strongly suggesting that this interaction may occur in multiple cell types. Figure 1A depicts the bait and prey constructs from our screens. IQGAP1 is a well-characterized scaffold protein involved in many disparate cellular responses. Sequence analysis of yeast colonies revealed that the RasGAP carboxy terminal domain (RGCT) of IQGAP1 interacts with the NNBD region of NLRC3. The RGCT domain is highly conserved (>91% identity and 95% similarity) in vertebrate evolution (Figure 1B). Likewise the NBD region of NLRC3 is well-conserved (77% identity and 84% similarity) in between humans and mice (51).
Figure 1.
NLRC3 yeast two-hybrid constructs. Schematic representation of both bait (NLRC3) and prey (IQGAP1) constructs used to screen with a human leukocyte and activated mononuclear cell or human universal cDNA libraries. Abreviations used for NLRC3 are CARD= caspase recruitment and activation domain, NOD= nucleotide oligomerization domain, NBD= nucleotide binding domain, HD1= helical domain 1, WHD= winged helix domain, HD2= helical domain 2, LRR= leucine rich repeats; the asterisk and star represent the approximate locations of the Walker A and Walker B motifs. Abreviations used for IQGAP1 are CHD= calponin homology domain, GRD= rasGAP related domain, RGCT= rasGAP carboxy terminus. Amino acid alignment represents the minimal binding region of IQGAP1, amino acids 1438–1657.
Expression profiling of IQGAP1 and NLRC3
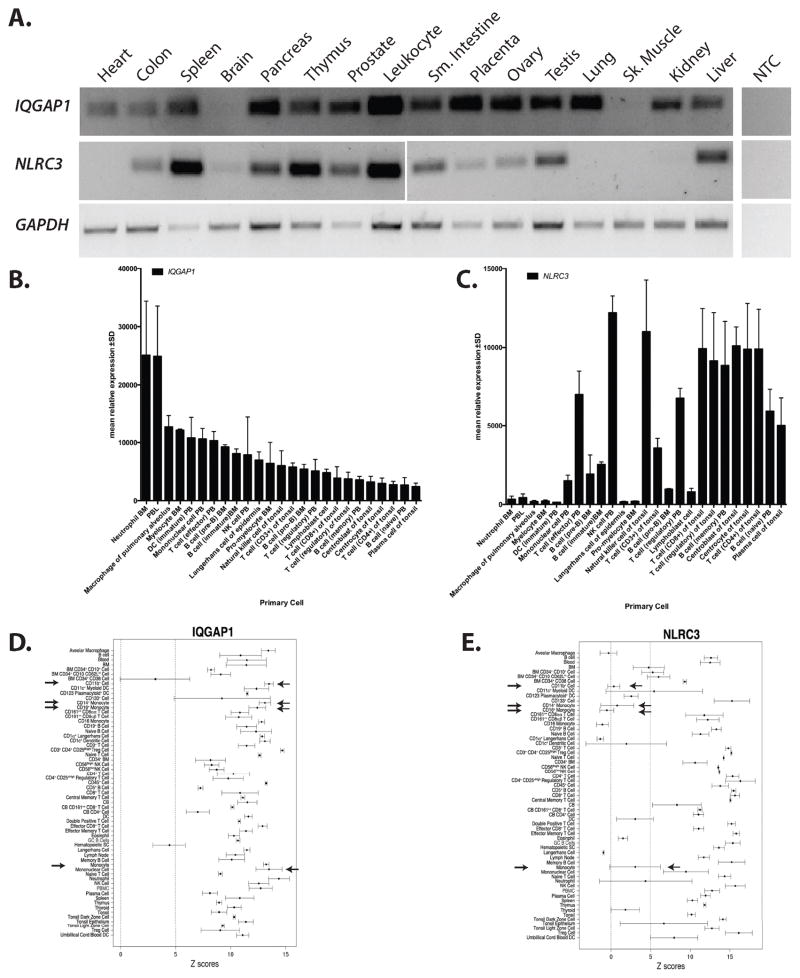
Previous data has shown that NLRC3 is expressed in human innate and adaptive immune cells. NLRC3 was most abundantly expressed in Jurkat T cells, with modest expression in Raji B cells, HL-60 and THP1 monocytic cells. Bioinformatic mining of Human Gene Atlas supported these findings(51). The initial characterization of IQGAP1 demonstrated that it was expressed in many different tissues, with the exception of brain(60). Additionally, it has been shown that IQGAP1 is expressed in many tumors and transformed cells lines including Jurkat T cells(54) and HL-60 cells(61). We have also verified that IQGAP1 is expressed in THP1 and HeLa cells (Supplemental Figure 1). To confirm and extend the initial expression profiling of both NLRC3 and IQGAP1 we used endpoint RT-PCR on commercially available human cDNA panels. We chose to focus on human cells, cell lines and tissues. The results are shown in Figure 2A. IQGAP1 has a broad tissue expression profile, but was not expressed in the brain and skeletal muscle. NLRC3 has a more restricted expression profile; it is highly expressed in immunological tissues such as spleen, thymus, liver and leukocytes. We did detect NLRC3 in non-hematopoietic tissues such as the prostate, small intestine and ovary, although this result might reflect tissue resident immune cells. These results are consistent with expression profiles generated by the Human Protein Atlas(62).
Figure 2.
IQGAP1 and NLRC3 are preferentially expressed in the immune system. RT-PCR was used to determine the tissue expression of human IQGAP1 and NLRC3. GAPDH was used as a positive control. BioGPS was queried for human IQGAP1 and NLRC3 expression in specific immune cells. The top overlapping cells and tissues for IQGAP1 and NLRC3 were graphed. B) Microarray results show relative IQGAP1 expression is highest in myeloid cells with moderate levels in lymphocytes. C) Relative NLRC3 expression is highest in lymphocytes with moderate expression in myeloid cells. Arrows have been used to illustrate the difference expression levels in primary monocytic cells. Abbreviations in panel A, B and C: NTC= no template control, BM=bone marrow, PBL=peripheral blood leukocyte, PB=peripheral blood, NK= natural killer cell. D and E) Across tissue expression for IQGAP1 (probe 200791_s_at) and NLRC3 (probe 236295_s_at), using immune cells and tissues, are shown. Z-scores ± median absolute deviation is plotted. A dashed line represents a z-score of 5 which signifies that a given transcript is expressed in a tissue. Arrows depict cells of monocyte/macrophage lineage. Abbreviations used in panel C and D: BM=bone marrow, DC=dendritic cell, Treg= regulatory T cell, CB=cord blood, DC=dendritic cell, GC=germinal center, NK=natural killer, PBMC=peripheral blood mononuclear cell. F) QPCR analysis of NLRC3 expression in human transformed cells lines. Values are normalized to HPRT levels using ΔCt method. Asterisk= p<.05 when compared to NTC
To further assess the possible expression profile of human NLRC3 and IQGAP1 in purified immune cells we screened available microarray databases for expression profiles of IQGAP1 and NLRC3. We found that the relative expression patterns of IQGAP1 and NLRC3 have partial overlap in the hematopoietic compartment with expression in primary lymphoid and myeloid cells (Figure 2B and C). Highest relative expression of IQGAP1 was seen in bone marrow neutrophils and peripheral blood leukocytes and lowest relative expression in lymphocytes, in contrast NLRC3 has high relative expression in lymphocytes and lower relative expression in myeloid cells.
To investigate absolute expression profiles of NLRC3 and IQGAPQ1 in human cells we screened an additional database: The Barcode 3.0(63). We explored the absolute expression levels of each of these transcripts in primary immune cells and tissues to determine if transcripts were silenced or expressed (63). In Figure 2D and E, gene expression barcoding for immune tissues and cells is illustrated as a function of Z-score. A Z-score above 5 suggests a high likelihood that a gene is expressed in that cell or tissue (63). This barcoding demonstrates that IQGAP1 and NLRC3 are expressed in the spleen and thymus (Figure 2D and E) consistent with our expression data in Figure 2A. However, purified cell populations are less clear. For example, IQGAP1 has robust and significant expression in myeloid/monocytic cells (see arrows Figure 2D). NLRC3 has consistent expression in lymphoid cells, but the expression in myeloid cells is unclear, possibly due to its lower level of expression (see arrows in Figure 2E). These data suggest a reciprocal and complex relationship between IQGAP1 and NLRC3 expression levels in single cell types, which might be dependent upon the biological roles of these proteins. QPCR analysis of NLRC3 expression in cell lines used in this study is consistent with previous reports (51): Jurkat cells express highest levels of NLRC3, while THP1 cells express modest amounts. HeLa cells express low but detectable levels of NLRC3 under our experimental conditions (Figure 2F). Despite the high relative expression of NLRC3 in T lymphocytes there have been no published in vivo phenotypes using mouse models. Phenotypic results have only been described in myeloid and epithelial/fibroblast cells (26, 48, 52). Therefore, we have focused our initial investigation using myeloid (THP1) and epithelial cells (HeLa), which express moderate to low levels of NLRC3, but relatively high levels of IQGAP1.
IQGAP1 RGCT interacts with NLRC3 via the NBD
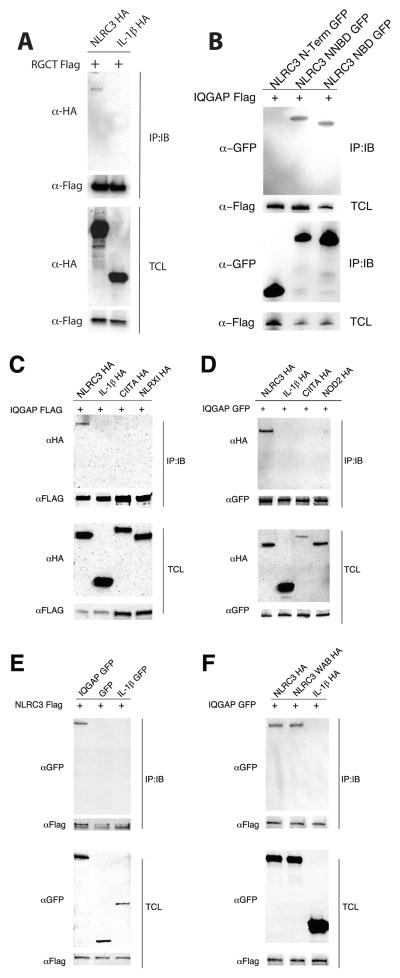
We investigated the potential interaction of IQGAP1 and NLRC3 in human cells by coimmunoprecipitation. We generated IQGAP1 RGCT constructs and NLRC3 truncation constructs, CARD (residues 1–60) only referred to as N, the CARD+NBD (residues 1–646) referred to as NNBD and the NBD only (residues 61–646) to confirm the yeast two-hybrid interaction data. As shown in Figure 3A epitope tagged (FLAG) IQGAP1 RGCT interacts with full-length epitope-tagged (HA) NLRC3 but not HA-tagged IL-1β in HEK293T cells. Likewise, full length FLAG-tagged IQGAP1 interacts with both the NBD and NNBD of NLRC3 but not the N-terminal domain when expressed as GFP-fusion constructs. This data demonstrates that the region of NLRC3 sufficient for mediating the interaction with IQGAP1 spans amino acid 60–600, which encodes most of the NBD. This region of NLRC3 has also been shown to directly interact with STING (48). To further explore the interaction between IQGAP1 and NLRC3, we co-transfected full length constructs into HEK293T cells. FLAG- and GFP-tagged IQGAP1 specifically interacted with HA-tagged NLRC3 but not other NLR proteins (Figure 3C and D). Two related NLR proteins NOD2 and CIITA, which contain a CARD and CARD-like regions, and NLRX1, which is involved in antiviral responses do not interact with IQGAP1 under these conditions. Likewise, FLAG-tagged NLRC3 coimmunoprecipitates a GFP-IQGAP1 fusion construct but not GFP (Figure 3E). These data, using multiple epitope tags and reciprocal immunoprecipitations, suggest that IQGAP1 specifically interacts with NLRC3 but not other NLR proteins (NLRX1, CIITA or NOD2) or irrelevant proteins (GFP and IL-1β), in human epithelial cells. Although the yeast-two hybrid data suggests the NLRC3 directly interacts with IQGAP1, we cannot rule out the possibility of additional proteins may influence directly or indirectly the binding of NLRC3 to IQGAP1 in human cells.
Figure 3.
IQGAP1 specifically interacts with NLRC3 in human cells. HEK293T cells were transiently transfected with different epitope tagged full length and truncation versions of IQGAP1 and NLRC3. Lysates were immunoprecipitated for IQGAP1 and immunoblots were probed to detect NLRC3 or vice versa. A) The RGCT domain (amino acids 1438–1657) of IQGAP1 interacts with full length NLRC3. B) The NBD (amino acids 60–646) of NLRC3 interacts with full length IQGAP1. C and D) Full length IQGAP1 (either amino terminal FLAG or GFP tagged) interacts with HA-tagged NLRC3 but not with HA tagged IL-1β, CIITA, NLRX1 nor NOD2. E) Reciprocal co-immunoprecipitations confirm the interaction between IQGAP1 and NLRC3 but not an irrelevant protein such as IL-1β. F) Mutations in the Walker A and Walker B motifs (WAB) do not alter the ability of NLRC3 to interact with IQGAP1. Images are representative of triplicate experiments.
One yeast two-hybrid screen (screen #1 in Figure 1) and our co-immunoprecipitation (Figure 3B) experiments in HEK293T cells suggested that the NBD of NLRC3 was sufficient to mediate an interaction with IQGAP1 RGCT domain. The NBD of NLRs contains the highly-conserved Walker A (GKS/T) and Walker B (DGLD) motifs necessary for binding and hydrolyzing nucleotide triphosphates such as ATP and GTP (64). Several NLR family members have been shown to act as ATPases or GTPases (22, 65). The nucleotide binding cycle of NLR proteins is hypothesized to regulate NLR function by promoting homotypic oligomerization or heterologous protein interaction. To test if the nucleotide binding cycle is involved in facilitating the binding of IQGAP1 to NLRC3, we generated mutations in the NLRC3 cDNA. A double mutant NLRC3 consisting of Walker A (corresponding to G150K151→A150A151) and Walker B mutations (D219G220L221D222 →A219G220L221A222) was constructed and cotransfected with FLAG-tagged IQGAP1 into HEK293T cells. This putative loss-of-function construct retains its ability to interact with IQGAP1 to similar levels as wild type NLRC3 which suggests the predicted nucleotide binding cycle of NLRC3 is not necessary for binding IQGAP1 (Figure 3F).
Subcellular distribution of IQGAP1 and NLRC3
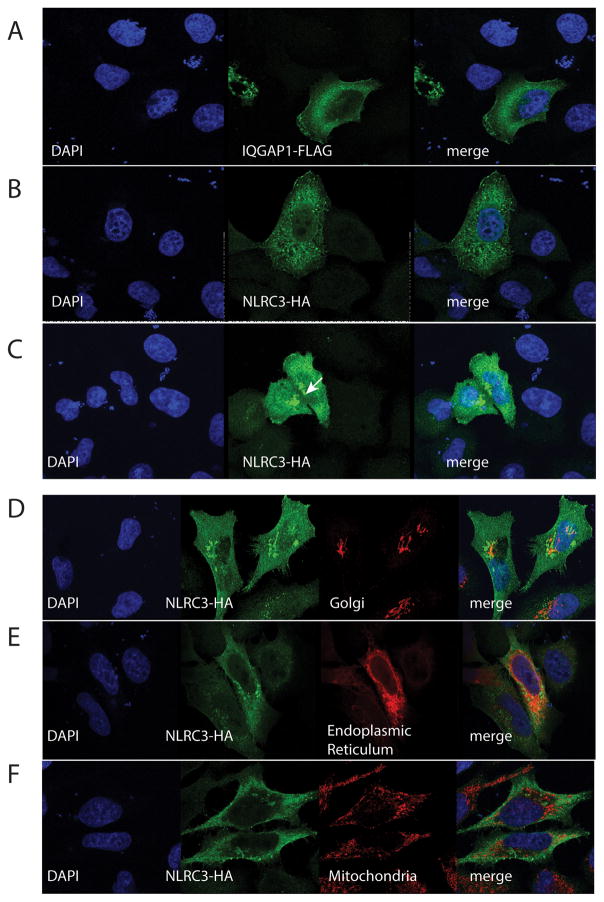
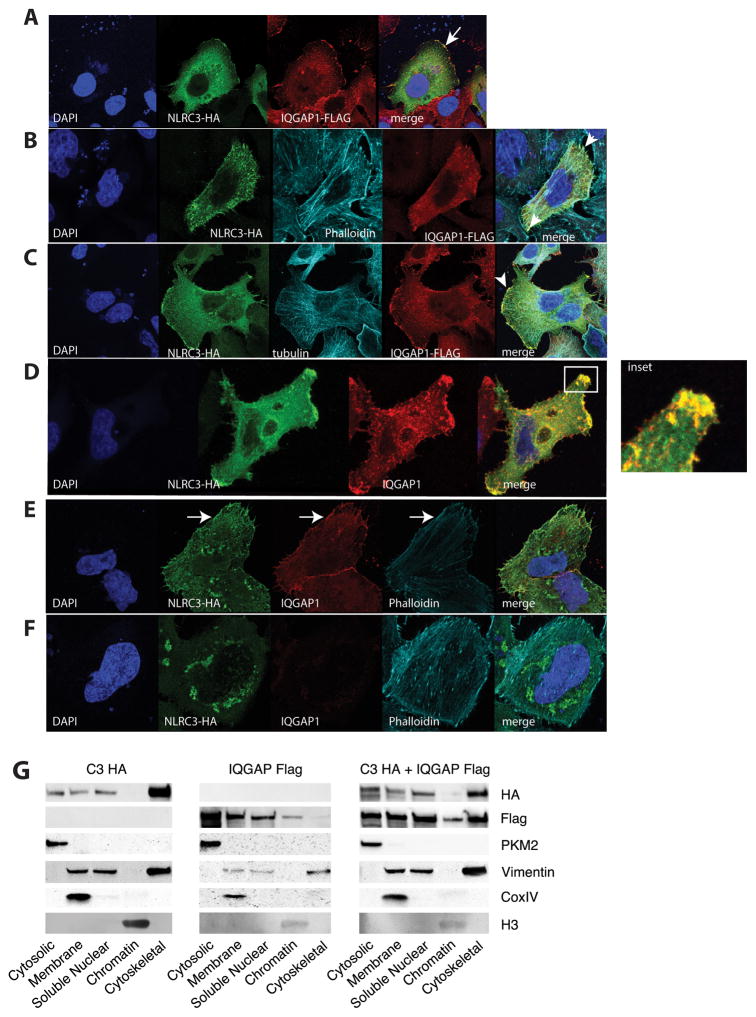
Based on the biochemical interaction between IQGAP1 and NLRC3, we were interested in exploring if NLRC3 and IQGAP1 colocalize in human cells. NLRC3 is a predicted cytosolic protein that has been shown to interact with TBK1 and STING in HEK293T cells in vitro and as purified recombinant protein in solution (48). This interaction inhibits TBK1-dependent activation of IRFs. NLRC3 has little to no effect on MAVS (Mitochondria Antiviral Signaling also known as VISA, IPS1 and CARDIF)-dependent activation of ISRE- and NFκ-B-dependent luciferase activity (48). IQGAP1, due to its scaffolding nature, has been implicated in many diverse processes in the cell. As such, it localizes to the cytoskeleton, the cytoplasm and the nucleus (66). To explore the subcellular localization of these proteins, HeLa cells were transiently transfected with recombinant FLAG-tagged IQGAP1 or HA-tagged NLRC3 and visualized with confocal microscopy (Figure 4). Consistent with previous reports epitope tagged IQGAP1 showed enrichment at the cell cortex but also showed diffuse cytosolic staining (Figure 4A). Epitope tagged NLRC3-HA showed variable subcellular distribution in individual cells. In Figure 4B NLRC3-HA shows diffuse cytosolic staining with regions that are consistent with the cell cortex and the endoplasmic reticulum. However, over multiple transfections and different epitope tags overexpressed NLRC3 forms discreet and large puncta or aggregates (Figure 4C see arrow). We were interested in determining if these puncta colocalized to the endoplasmic reticulum, Golgi apparatus or the mitochondria as these organelles have been described as signaling platforms. HeLa cells transfected with NLRC3-HA were stained with an anti-golgin-97 antibody to detect the Golgi apparatus (Figure 4D), were contransfected with an endoplasmic reticulum targeted mClover fluorescent protein (Figure 4E) or were stained with an anti-Tomm20 antibody to stain the mitochondria (Figure 4F). These puncta do not appear to colocalize with the mitochondria and there is only partial overlap with the endoplasmic reticulum and minimal colocalization with the Golgi apparatus. These results were confirmed with SW480 cells transfected with the ColorfulCell vector (see supplemental Figure 2). To determine if NLRC3 and IQGAP1 colocalize in epithelial cells we cotransfected HeLa cells. As seen in Figure 5A, both IQGAP1 and NLRC3 show enrichment at the cell cortex (see arrow) as well as diffuse cytosolic/cytoplasmic staining. IQGAP1 has been shown to interact with both actin filaments and microtubules. We next investigated if the presumed cortical staining colocalized with actin filaments. In Figure 5B, we counterstained with phalloidin in our cotransfected HeLa cells. In these cells NLRC3 and IQGAP1 partially colocalizes to the actin cortex, but not the microtubules (Figure 5C). To explore this colocalization in more physiological conditions, we transfected NLRC3-HA and stained for endogenous IQGAP1. Figure 5D shows distinct and robust colocalization of NLRC3-HA with endogenous IQGAP1 (see inset). We have not been able to find a suitable NLRC3 antibody that works well in immunofluorescence, so we have been unable to detect endogenous NLRC3. HeLa cells that have been transduced with shRNA scramble sequence (Figure 5E) and a shRNA that targets IQGAP1 (Figure 5F) were used to show that our staining of endogenous IQGAP1 is specific (see also supplemental Figure 1). When these HeLa cells are transfected with NLRC3-HA we see a reduced amount of localization to the cell cortex (see arrow in Figure 5E) in the absence (or reduction) of endogenous IQGAP1. These data suggest that NLRC3 can be localized to the cell cortex, possibly via interactions with IQGAP1. These data are consistent with previous studies on IQGAP1, but a novel finding for NLRC3.
Figure 4.
Confocal microscopy of HeLa cells transfected with epitope tagged IQGAP1-FLAG or NLRC3-HA. Cells were fixed, permeabilized and stained with antibodies specific to the epitope tag followed by species-specific Alexa Fluor 488 and Alexa 568 secondary antibodies. Cells are representative of more than three independent transfections. An endoplasmic reticulum specific fusion protein (Addgene Plasmid #56310) was used to highlight the endoplasmic reticulum. The Golgi Apparatus was stained with an anti-Golgin-97 antibody; whereas the mitochondria was identified with an anti-Tomm20 antibody. The white arrow represents NLRC3-HA aggregates seen in a percentage of cells
Figure 5.
Confocal microscopy of HeLa cells co-transfected with FLAG-tagged IQGAP1 and HA-tagged NLRC3 (A) and stained for endogenous proteins: B) phalloidin conjugated to Alexa 647 C) tubulin D) Endogenous IQGAP1 boxed inset shows robust cortical colocalization with NLRC3. E) shRNA scramble F) shIQGAP1 HeLa cells were transfected with NLRC3-HA and stained for endogenous IQGAP1 and epitope tagged NLRC3 and phalloidin. White arrows represent cortical actin staining of NLRC3 and IQGAP1 in panel E. G) SW480 cells were transfected with epitope tagged NLRC3, epitope tagged IQGAP1 and cotransfected. Cells were fractionated using subcellular fractionation kit. Immunoblots were probed with antibodies to specific proteins in each fraction. Images are representative of at least three individual replicates.
To confirm our confocal microscopy data, we performed biochemical subcellular fractionation by differential lysis on SW480 cells transfected with IQGAP1 or NLRC3 or both constructs to determine if localization is altered by either protein. In Figure 6G we show that NLRC3 and IQGAP1 have diverse subcellular distributions in single transfections. These proteins localize to the cytosol, membrane, nucleus and cytoskeleton. In a small percentage of cells we have observed NLRC3 localized to the nucleus (data not shown). There are several reported instances of unexpected nuclear localization of presumably cytosolic NLRs such as NOD2 and NLRP3 (33, 67), this result might reflect a generalized ability of NLR proteins to translocate into the nucleus. Consistent with our microscopy data, there is a fraction of the pool of NLRC3 proteins that is distributed to the cytoskeleton fraction. The relative amount of cytoskeletal NLRC3 varied between independent experiments, but was always present. We detect a small relative amount of IQGAP1 in the cytoskeletal fraction as predicted. However, the amount of cytoskeletal IQGAP1 increases in the presence of co-transfected NLRC3.
Figure 6.
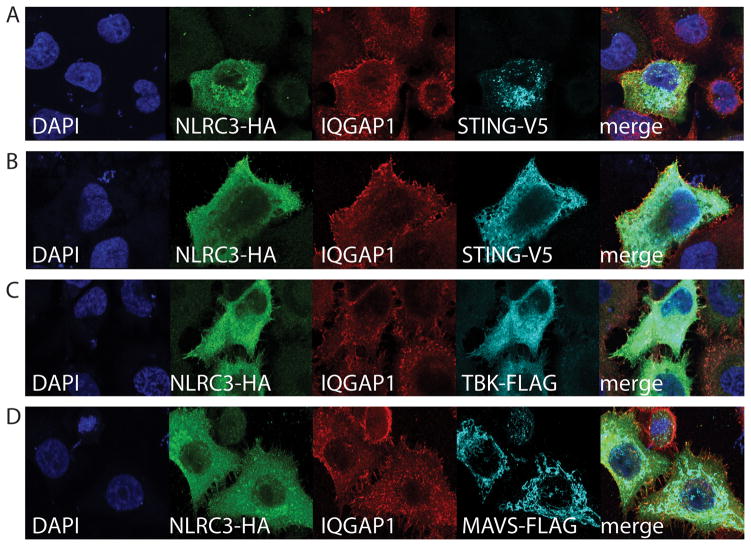
IQGAP1 and NLRC3 show partial colocalization with recombinant STING, TBK1 and not MAVS. HeLa cells were transiently transfected different expression constructs. Panels A–D) Confocal microscopy of HeLa cells co-transfected with NLRC3 and associated expression constructs. Panels A and B show two distinct colocalization patterns of STING: (A) puncta and (B) endoplasmic reticulum.
NLRC3 has been shown to interact with TBK1 and STING and possibly disrupt the redistribution of STING from the MAM to the perinuclear region (18, 68). We next investigated if known NLRC3 interacting proteins colocalized to specific regions in HeLa cells. FLAG-tagged TBK1, V5-tagged STING and FLAG-tagged MAVS were transfected with NLRC3-HA (see Figure 6) and counterstained with antibodies to endogenous IQGAP1. Transfected STING constructs show two distinct staining patterns that may reflect its activation status. Figure 6A shows putatively active STING and panel B shows STING with endoplasmic reticulum staining typical of non-activated STING. The localization of all of these proteins occurred in the absence of stimulation; however, overexpressed proteins might be aberrantly activate signaling pathways. These data possibly do not differentiate between homeostatic versus activation induced interaction or localizations. NLRC3 and IQGAP1 showed minimal to no colocalization with MAVS in HeLa cells (Figure 6D). Either form of STING (Figure 6A or B) did not colocalize to the cell cortex along with a fraction of NLRC3 and IQGAP1. TBK1 showed diffuse cytosolic or cytoplasmic staining that did not clearly colocalize at the cortex with IQGAP1 and NLRC3 (Figure 6A–C). However, there is a portion of the cellular pool of both IQGAP1 and NLRC3 that has diffuse cytosolic/cytoplasmic staining which might colocalize with TBK1 and/or STING. These data suggest that there might be individual pools of NLRC3 that interact with different signaling or scaffolding proteins.
IQGAP1 is involved in cytosolic nucleotide sensing and type I interferon production
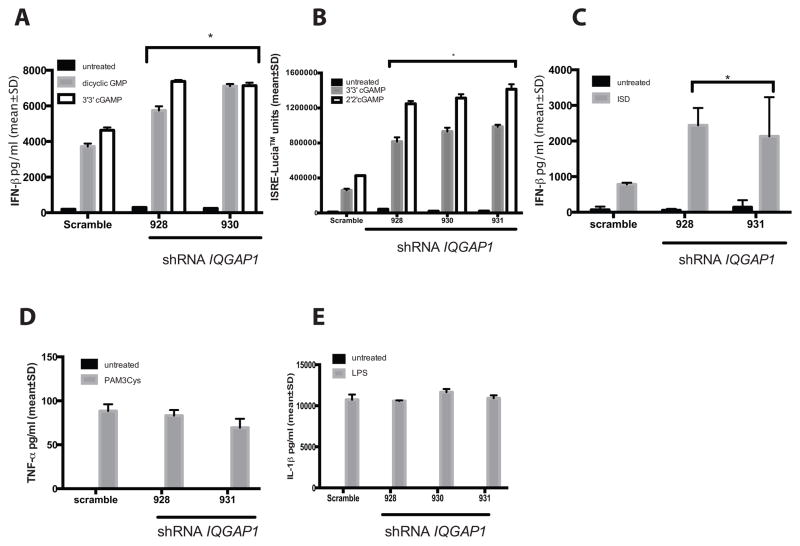
We chose to use THP1 and HeLa cells to investigate the functional significance of the NLRC3 and IQGAP1 interaction in vitro. THP1 cells have been shown to express both NLRC3 and IQGAP1 (see Figure 2F and supplemental Figure 1). Additionally, the function of NLRC3 has been investigated using THP1 cells as well as mouse bone marrow-derived macrophages(52) and mouse embryonic fibroblasts(48). We generated IQGAP1 shRNA knockdown in THP1, THP1-Dual™ and HeLa cells (Figure 7). THP1 Dual™ cells are engineered with a secreted luciferase (Lucia™) gene fused to the ISG54 minimal promoter with five tandem ISREs and a secreted embryonic alkaline phosphatase gene (SEAP) driven by the minimal IFN-β promoter fused to five NF-κB responsive sites and three c-Rel binding sites to allow for rapid detection of both IRF and NF-κB dependent transcription. We investigated the role of IQGAP1 in all three cell lines. First, we investigated if IQGAP1 is necessary for regulated responses to intracellular nucleic acids that are potent activators of type I interferon. To this end, we stimulated THP1, THP1-dual™ and HeLa shRNA cells with transfected dicyclic nucleotides (2′2′ and 3′3′ cGAMP) or immunostimulatory DNA (ISD), and measured IFN-β secretion (A and C) or Lucia™ activity (B). In the absence (or with significant reduction) of IQGAP1, all cell lines secreted significantly higher levels of IFN-β (or ISG54 driven-Lucia™). NLRC3 has also been shown to affect proinflammatory cytokine secretion during cecal ligation puncture model of experimental colitis (52). We next stimulated THP1 (Figure 7D) and THP1-Dual™ (Figure 7E) cells with bacterial PAMPs to induce a NF-κB-dependent proinflammatory cytokine response and measured IL-6 or IL-1β production. IQGAP1 appears to be dispensable for TLR responses, as there was no significant difference between IQGAP1 knockdown and scramble cells. These data suggest that IQGAP1 functions specifically to inhibit the production of type 1 interferon and not proinflammatory cytokines.
Figure 7.
shRNA mediated knockdown of IQGAP1 in different cell lines induces hyperactivation of the IFN-β pathway. In panels A–C) Stable integrants (panel A and D represent THP-1 cells, panel B and E represent THP-1 dual cells and panel C represents HeLa cells) were selected and stimulated as indicated. Interferon or cytokine levels were measured by ELISA and secreted Lucia™. Values represent means ± standard deviation of triplicate samples. Data are representative of at least three biological replicates. * represents a p<.05 when compared to Scramble shRNA cells. Panels D and E show no significant differences relative to the scramble shRNA.
IQGAP1 disrupts the NLRC3:STING interaction
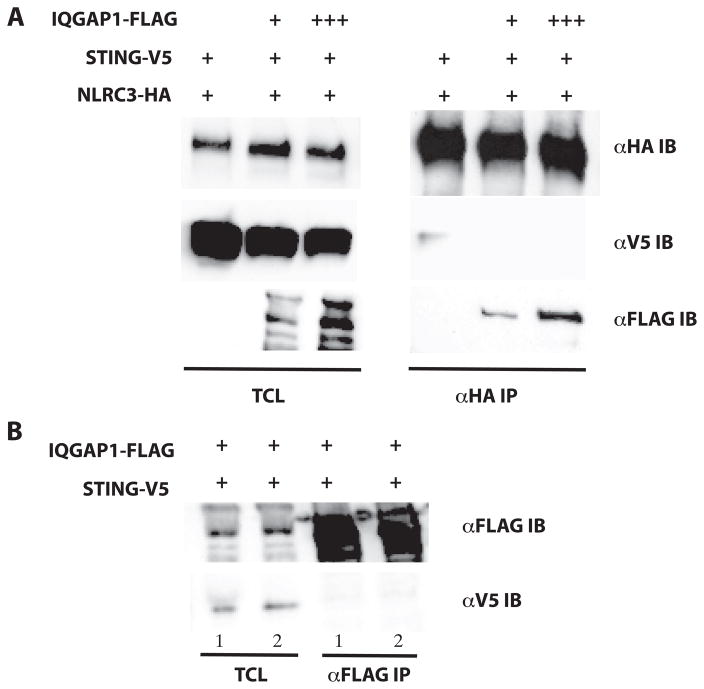
Our colocalization studies (see Figure 5A–F) clearly demonstrate at least a portion of the cellular pool of NLRC3 is localized to the cell cortex, but this localization appears to be independent of STING and TBK1(Figure 6A–C). This observation coupled with the overlapping interaction location of both STING and IQGAP1 to the NBD of NLRC3 led us to speculate that IQGAP1 could regulate the NLRC3:STING complex. To explore this possible mechanism, we examined if increasing amounts of over-expressed IQGAP1 could modulate the interaction of NLRC3 with STING. As shown in Figure 8A, in the presence of over-expressed IQGAP1, NLRC3 no longer interacts with over-expressed STING. We next investigated if IQGAP1 could interact with STING in the absence of over-expressed NLRC3. Under two different stringencies, we were unable to detect STING interaction with IQGAP1 (see Figure 8B). These data suggest that IQGAP1 has the ability to interact with NLRC3, and this interaction can affect NLRC3:STING interaction. How these dynamic interactions negatively regulate type 1 interferon production is not well understood.
Figure 8.
IQGAP1 sequesters NLRC3 to the actin cytoskeleton. A) HEK293T cells were transiently transfected with epitope tagged IQGAP1, NLRC3 and STING. Lysates were immunoprecipitated for NLRC3-HA and blotted for interacting proteins. Data are representative of three replicates. B) IQGAP1 and STING were cotransfected in the absence of NLRC3-HA and immunoprecipitated for IQGAP1 and blotted for STING. Cells were lysed in stringent (PBS with 1% Triton X-100 + 0.5% Sodium Deoxycholate shown in lane 1) and less stringent (PBS with 1% Triton X-100 shown in lane 2) conditions. Data are representative of three replicates.
Discussion
Macrophages and epithelial cells detect cytosolic nucleotides and mount a robust type I interferon (IFNα/β) response to limit pathogen spread. Cells respond by activating the IRF family of transcription factors to induce the antiviral transcriptome. TBK1 is the keystone kinase that integrates numerous upstream cytosolic nucleic acid sensors. TBK1 phosphorylates IRF3 and/or IRF7, which allows it to dimerize and translocate into the nucleus. One upstream sensor in particular, cGAS, directly binds cytosolic DNA to generate dicyclic nucleotides. These intermediates can directly bind to STING and allowing it to activate TBK1. However, sustained or aberrant type I interferon production is associated with several autoimmune and autoinflammatory diseases: broadly termed interferonopathies (69). Distinct host pathways, such as miR-146a (70), SOCS-1 (71) and DUBA (72) among others (73), can downregulate type I interferon responses, possibly to limit immunopathology associated with overzealous responses. STING is targeted for degradation via multiple pathways. UNC-51-like kinase (ULK1) has been shown to phosphorylate STING at S366 to facilitate its lysosomal degradation in a negative-feedback loop to inhibit prolonged responses (17). Additionally, RNF5 facilitates K48-linked ubiquitination of STING on the K150 residue after viral infection. This modification allows for ubiquitin-dependent proteosomal degradation (19). Previously, NLRC3 has been shown to inhibit type I interferon production by disrupting the dynamic redistribution of STING between the ER and MAM and inhibiting its interaction with TBK1 (48). However, the molecular mechanisms that control NLRC3 inhibition of STING remain poorly characterized. In this study we have shown that NLRC3 interacts with IQGAP1 using a yeast two-hybrid screen and coimmunoprecipitation experiments in human cell lines. We identified a carboxy-terminal fragment of IQGAP1 that specifically interacts with the NBD, albeit independently of the nucleotide binding cycle, of NLRC3 (Figure 3). The RGCT domain of IQGAP1 has been shown to interact with multiple protein partners, for example, E-cadherin, β-catenin, Clip170, Clasp2 (cytoplasmic linker associated protein 2), and APC (adenomatous polyposis coli) (74). The RGCT domain is responsible for anchoring IQGAP1 to the membrane at the cell periphery and has been shown to dynamically bind small GTPases, such as Cdc42, in a phosphorylation dependent manner at the leading edge of the plasma membrane (75).
IQGAP1 regulates many diverse cellular processes in different cell types and tissues, notably in cell motility and invasion via cytoskeletal rearrangement, but also cotranscriptional regulation with β-catenin. NLRC3 and IQGAP1 have similar, but not necessarily overlapping expression profiles in unstimulated cells. Both have increased relative expression in lymphocyte and myeloid cells, further suggesting a possible endogenous role in vivo (Figure 2). Additionally, both NLRC3 and IQGAP1 are expressed, albeit at low levels for NLRC3, in epithelial cells. We have demonstrated that NLRC3 and IQGAP1 colocalize in human epithelial cells (Figure 4–6). In the adaptive immune system IQGAP1 negatively regulates T-cell receptor signaling and filamentous actin dynamics. In the absence of IQGAP1, Jurkat T cells and primary mouse T lymphocytes secrete significantly more IL-2 and IFN-γ (54). It is interesting to note that knockdown of NLRC3 in Jurkat T cells also show increased IL-2 production and CD25 expression levels (51). This effect might be mediated via IQGAP1, but this hypothesis remains to be tested. In macrophages mouse IQGAP1 has been shown to be important in phagocytosis and phagocytic cup formation in macrophages during incubation with avidin-coated beads (76) and LPS-induced Rac1 activation during phagocytosis (77). This effect requires the interaction of IQGAP1 with the actin nucleating protein, diaphanous-related formin (Dia1). The rearrangement of the host cell cytoskeleton is a crucial mechanism by which invasive bacteria enter cells. The pathogenic bacterium, Salmonella enterica co-opts IQGAP1 to alter Rac1 and MAPK signaling to facilitate chronic infection in epithelial cells (78), suggesting that IQGAP1 is necessary for invasion.
The movement of signaling proteins in sensing cytosolic nucleic acids is dynamic and highly coordinated. The activated STING/TBK1 complex is trafficked from the ER and Golgi apparatus, via ATG9, to the endosomes/lysosomes containing IRF and NF-κB transcription factors in a VPS34-dependent manner (17). Movement of STING is thought to be mediated via posttranslational palmitoylation at two key cysteine residues (C88 and C91) at the Golgi apparatus (79). This trafficking allows for efficient transcription factor activation and subsequent antiviral transcriptional program. TBK1 molecules are recruited to larger macromolecular signaling complexes upon activation via interactions with scaffolding and adaptor proteins such as TANK, SINTBAD, NAP1, TAPE and IFIT3 (80). The latter two of which are involved in TBK1-dependent type I interferon production in response to viral infection. Specifically, TBK1 has been shown to relocate from the cytosol to mitochondria in response to DNA virus infection or transfection of DNA into the cytosol of HeLa and HepG2 cells but not RAW264.7, L929 or T-23 cells(81). These data suggest that TBK1 subcellular localization is cell type- or stimulus-dependent. This clustering of TBK1 to specific subcellular locations facilitates autophosphorylation and subsequent substrate targeting. Nontheless, the mechanisms by which TBK1 and STING dynamically redistribute within the subcellular space is largely uknown. Our data suggest that IQGAP1 with its known scaffolding ability, its diverse subcellular distribution and its novel interaction with NLRC3 might function to regulate TBK1 or STING activity or location and downstream activation of the IRF3 and/or IRF7.
The subcellular localization of NLR proteins is as diverse as the cellular pathways with which they are associated. NOD2 has been shown to localize to the plasma membrane via interactions with ERBIN (67, 82) and FRMBP2(83). Additionally, NOD2 has been shown to localize to endosomes (possibly via SLC15A3 or SLC15A4) in dendritic cells (84) and autophagosomes (via ATG16L1) in HeLa cells and MEFs (85) in response to invasive bacterial infection or stimulation with the bacterial PAMP, muramyl dipeptide the known ligand for NOD2. Also, NOD2 has been shown to localize to the mitochondria where it interacts with MAVS in response to ssRNA virus (36). The molecular mechanisms by which NOD2 is distributed to different subcellular locations is not well understood and how the subcellular distribution relates to the pathophysiology related to the NOD2-dependend pathologies (Crohn’s disease and early onset sarcoidosis) is under investigation. Surprisingly, we show that a portion of NLRC3 is localized to the cell cortex in HeLa cells, possibly via its interaction with IQGAP1, a known actin and tubulin binding protein. How this specific subcellular localization affects NLRC3 function is unknown but might relate to compartmentalization or sequestration of signaling molecules to inhibit or dampen chronic or aberrant activation of the type I interferon pathway.
We show that IQGAP1 is necessary for appropriate responses to cytosolic nucleotides. Type I interferon production was significantly increased in THP1 and HeLa cells that stably express shRNA to IQGAP1. However, this effect was specific for type I interferon, THP1 cells expressing shRNA to IQGAP1 did not secrete significantly different amounts of proinflammatory cytokines, IL-1β or TNF-α. A limitation of our studies is the reliance on ectopic and overexpression of IQGAP1, NLRC3 and TBK1 in particular and siRNA. In an attempt to overcome these limitations we used human HAP1 cells that were engineered via Cas9/CRISPR to be deficient in IQGAP1. In our experiments the parental HAP1 cell line was refractory to stimulation with immunostimulatory nucleic acids and was not amenable to transfection (data not shown). Nonetheless, our data is the first demonstration that IQGAP1 negatively regulates type I interferon production, possibly via its interaction with NLRC3. The role of IQGAP1 as a scaffolding protein is well documented and the ability of NLRC3 to interact with the RGCT domain adds to the growing number of IQGAP1 interacting proteins. These data help better define the molecular mechanisms of NLR-mediated interferon inhibition that is necessary to maintain homeostasis. Investigating the role of additional regulatory proteins in the NLR/IFN axis will be critical to our understanding of host-pathogen interactions.
Supplementary Material
Acknowledgments
We thank Jenny PY Ting, University of North Carolina, for the plasmids encoding MAVS, STING, and TBK1. We are grateful to Wade Edris for technical assistance with confocal microscopy at the Imaging Core at PennState College of Medicine and Heather Hoffmann for technical assistance.
This work was supported by National Institutes of Health Grant R15 DK098754 (B.K.D.), startup funds from Franklin & Marshall College (to B.K.D.), and an Undergraduate Science Education Award from the Howard Hughes Medical Institute to Franklin & Marshall College (52007538). This material is based upon work supported by the National Science Foundation under Grant No. 1626073. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health nor the National Science Foundation.
References
- 1.Cui J, Chen Y, Wang HY, Wang RF. Mechanisms and pathways of innate immune activation and regulation in health and cancer. Hum Vaccin Immunother. 2014;10:3270–85. doi: 10.4161/21645515.2014.979640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–42. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–4. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–7. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–61. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–30. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 8.Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, Li Y, Zhang R, Cheng G, Liu ZJ. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–86. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–45. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, Gu L. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–7. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 11.Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–4. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konno H, Barber GN. The STING controlled cytosolic-DNA activated innate immune pathway and microbial disease. Microbes Infect. 2014;16:998–1001. doi: 10.1016/j.micinf.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen K, Prabakaran T, Laustsen A, Jorgensen SE, Rahbaek SH, Jensen SB, Nielsen R, Leber JH, Decker T, Horan KA, Jakobsen MR, Paludan SR. Listeria monocytogenes induces IFNbeta expression through an IFI16-, cGAS- and STING-dependent pathway. EMBO J. 2014;33:1654–66. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J, Schmid-Burgk JL, Schmidt T, Hornung V, Cole ST, Ablasser A. Mycobacterium tuberculosis Differentially Activates cGAS- and Inflammasome-Dependent Intracellular Immune Responses through ESX-1. Cell Host Microbe. 2015;17:799–810. doi: 10.1016/j.chom.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, Vance RE, Stallings CL, Virgin HW, Cox JS. The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe. 2015;17:811–9. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins AC, Cai H, Li T, Franco LH, Li XD, Nair VR, Scharn CR, Stamm CE, Levine B, Chen ZJ, Shiloh MU. Cyclic GMP-AMP Synthase Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell Host Microbe. 2015;17:820–8. doi: 10.1016/j.chom.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–98. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–92. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong B, Zhang L, Lei C, Li Y, Mao AP, Yang Y, Wang YY, Zhang XL, Shu HB. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 2009;30:397–407. doi: 10.1016/j.immuni.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A. 2015;112:E4306–15. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat Rev Immunol. 2016 doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mo J, Boyle JP, Howard CB, Monie TP, Davis BK, Duncan JA. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J Biol Chem. 2012;287:23057–67. doi: 10.1074/jbc.M112.344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong M, Yoon SI, Wilson IA. Structure and functional characterization of the RNA-binding element of the NLRX1 innate immune modulator. Immunity. 2012;36:337–47. doi: 10.1016/j.immuni.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halff EF, Diebolder CA, Versteeg M, Schouten A, Brondijk TH, Huizinga EG. Formation and structure of a NAIP5-NLRC4 inflammasome induced by direct interactions with conserved N- and C-terminal regions of flagellin. J Biol Chem. 2012;287:38460–72. doi: 10.1074/jbc.M112.393512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–35. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karki R, Man SM, Malireddi RK, Kesavardhana S, Zhu Q, Burton AR, Sharma BR, Qi X, Pelletier S, Vogel P, Rosenstiel P, Kanneganti TD. NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature. 2016 doi: 10.1038/nature20597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koblansky AA, Truax AD, Liu R, Montgomery SA, Ding S, Wilson JE, Brickey WJ, Muhlbauer M, McFadden RM, Hu P, Li Z, Jobin C, Lund PK, Ting JP. The Innate Immune Receptor NLRX1 Functions as a Tumor Suppressor by Reducing Colon Tumorigenesis and Key Tumor-Promoting Signals. Cell Rep. 2016;14:2562–75. doi: 10.1016/j.celrep.2016.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Zhang S, Li F, Qin L. NLRX1 attenuates apoptosis and inflammatory responses in myocardial ischemia by inhibiting MAVS-dependent NLRP3 inflammasome activation. Mol Immunol. 2016;76:90–7. doi: 10.1016/j.molimm.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 29.Coutermarsh-Ott S, Simmons A, Capria V, LeRoith T, Wilson JE, Heid B, Philipson CW, Qin Q, Hontecillas-Magarzo R, Bassaganya-Riera J, Ting JP, Dervisis N, Allen IC. NLRX1 suppresses tumorigenesis and attenuates histiocytic sarcoma through the negative regulation of NF-kappaB signaling. Oncotarget. 2016 doi: 10.18632/oncotarget.8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eitas TK, Chou WC, Wen H, Gris D, Robbins GR, Brickey J, Oyama Y, Ting JP. The nucleotide-binding leucine-rich repeat (NLR) family member NLRX1 mediates protection against experimental autoimmune encephalomyelitis and represses macrophage/microglia-induced inflammation. J Biol Chem. 2014;289:4173–9. doi: 10.1074/jbc.M113.533034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang YG, Fang WL, Wei J, Wang T, Wang N, Ma JL, Shi M. The involvement of NLRX1 and NLRP3 in the development of nonalcoholic steatohepatitis in mice. J Chin Med Assoc. 2013;76:686–92. doi: 10.1016/j.jcma.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, Widman DG, Swanson KV, Wen KW, Damania B, Moore CB, Giguere PM, Siderovski DP, Hiscott J, Razani B, Semenkovich CF, Chen X, Ting JP. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36:933–46. doi: 10.1016/j.immuni.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruchard M, Rebe C, Derangere V, Togbe D, Ryffel B, Boidot R, Humblin E, Hamman A, Chalmin F, Berger H, Chevriaux A, Limagne E, Apetoh L, Vegran F, Ghiringhelli F. The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat Immunol. 2015;16:859–70. doi: 10.1038/ni.3202. [DOI] [PubMed] [Google Scholar]
- 34.Coutermarsh-Ott S, Eden K, Allen IC. Beyond the inflammasome: regulatory NOD-like receptor modulation of the host immune response following virus exposure. J Gen Virol. 2016;97:825–38. doi: 10.1099/jgv.0.000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen IC. Non-Inflammasome Forming NLRs in Inflammation and Tumorigenesis. Front Immunol. 2014;5:169. doi: 10.3389/fimmu.2014.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–80. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou PF, Chang MX, Li Y, Xue NN, Li JH, Chen SN, Nie P. NOD2 in zebrafish functions in antibacterial and also antiviral responses via NF-kappaB, and also MDA5, RIG-I and MAVS. Fish Shellfish Immunol. 2016;55:173–85. doi: 10.1016/j.fsi.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 38.Kim YG, Park JH, Reimer T, Baker DP, Kawai T, Kumar H, Akira S, Wobus C, Nunez G. Viral infection augments Nod1/2 signaling to potentiate lethality associated with secondary bacterial infections. Cell Host Microbe. 2011;9:496–507. doi: 10.1016/j.chom.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jing H, Fang L, Wang D, Ding Z, Luo R, Chen H, Xiao S. Porcine reproductive and respiratory syndrome virus infection activates NOD2-RIP2 signal pathway in MARC-145 cells. Virology. 2014;458–459:162–71. doi: 10.1016/j.virol.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 40.Kapoor A, Forman M, Arav-Boger R. Activation of nucleotide oligomerization domain 2 (NOD2) by human cytomegalovirus initiates innate immune responses and restricts virus replication. PLoS One. 2014;9:e92704. doi: 10.1371/journal.pone.0092704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vissers M, Remijn T, Oosting M, de Jong DJ, Diavatopoulos DA, Hermans PW, Ferwerda G. Respiratory syncytial virus infection augments NOD2 signaling in an IFN-beta-dependent manner in human primary cells. Eur J Immunol. 2012;42:2727–35. doi: 10.1002/eji.201242396. [DOI] [PubMed] [Google Scholar]
- 42.Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, Ranjan P, Monroe KM, Pickles RJ, Sambhara S, Ting JP. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011;34:854–65. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, Lich JD, Heise MT, Chen Z, Ting JP. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–7. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 44.Guo H, Konig R, Deng M, Riess M, Mo J, Zhang L, Petrucelli A, Yoh SM, Barefoot B, Samo M, Sempowski GD, Zhang A, Colberg-Poley AM, Feng H, Lemon SM, Liu Y, Zhang Y, Wen H, Zhang Z, Damania B, Tsao LC, Wang Q, Su L, Duncan JA, Chanda SK, Ting JP. NLRX1 Sequesters STING to Negatively Regulate the Interferon Response, Thereby Facilitating the Replication of HIV-1 and DNA Viruses. Cell Host Microbe. 2016;19:515–28. doi: 10.1016/j.chom.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barouch DH, Ghneim K, Bosche WJ, Li Y, Berkemeier B, Hull M, Bhattacharyya S, Cameron M, Liu J, Smith K, Borducchi E, Cabral C, Peter L, Brinkman A, Shetty M, Li H, Gittens C, Baker C, Wagner W, Lewis MG, Colantonio A, Kang HJ, Li W, Lifson JD, Piatak M, Jr, Sekaly RP. Rapid Inflammasome Activation following Mucosal SIV Infection of Rhesus Monkeys. Cell. 2016;165:656–67. doi: 10.1016/j.cell.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unger BL, Ganesan S, Comstock AT, Faris AN, Hershenson MB, Sajjan US. Nod-like receptor X-1 is required for rhinovirus-induced barrier dysfunction in airway epithelial cells. J Virol. 2014;88:3705–18. doi: 10.1128/JVI.03039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui J, Li Y, Zhu L, Liu D, Songyang Z, Wang HY, Wang RF. NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol. 2012;13:387–95. doi: 10.1038/ni.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, Gregory SM, Zhang Z, Schneider M, Jiang Y, Fitzgerald KA, Ouyang S, Liu ZJ, Damania B, Shu HB, Duncan JA, Ting JP. NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity. 2014;40:329–41. doi: 10.1016/j.immuni.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rebsamen M, Vazquez J, Tardivel A, Guarda G, Curran J, Tschopp J. NLRX1/NOD5 deficiency does not affect MAVS signalling. Cell Death Differ. 2011;18:1387. doi: 10.1038/cdd.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ling A, Soares F, Croitoru DO, Tattoli I, Carneiro LA, Boniotto M, Benko S, Philpott DJ, Girardin SE. Post-transcriptional inhibition of luciferase reporter assays by the Nod-like receptor proteins NLRX1 and NLRC3. J Biol Chem. 2012;287:28705–16. doi: 10.1074/jbc.M111.333146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conti BJ, Davis BK, Zhang J, O’Connor W, Jr, Williams KL, Ting JP. CATERPILLER 16.2 (CLR16.2), a novel NBD/LRR family member that negatively regulates T cell function. J Biol Chem. 2005;280:18375–85. doi: 10.1074/jbc.M413169200. [DOI] [PubMed] [Google Scholar]
- 52.Schneider M, Zimmermann AG, Roberts RA, Zhang L, Swanson KV, Wen H, Davis BK, Allen IC, Holl EK, Ye Z, Rahman AH, Conti BJ, Eitas TK, Koller BH, Ting JP. The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol. 2012;13:823–31. doi: 10.1038/ni.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw AS, Filbert EL. Scaffold proteins and immune-cell signalling. Nat Rev Immunol. 2009;9:47–56. doi: 10.1038/nri2473. [DOI] [PubMed] [Google Scholar]
- 54.Gorman JA, Babich A, Dick CJ, Schoon RA, Koenig A, Gomez TS, Burkhardt JK, Billadeau DD. The cytoskeletal adaptor protein IQGAP1 regulates TCR-mediated signaling and filamentous actin dynamics. J Immunol. 2012;188:6135–44. doi: 10.4049/jimmunol.1103487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanwar N, Wilkins JA. IQGAP1 involvement in MTOC and granule polarization in NK-cell cytotoxicity. Eur J Immunol. 2011;41:2763–73. doi: 10.1002/eji.201040444. [DOI] [PubMed] [Google Scholar]
- 56.Chung LK, Philip NH, Schmidt VA, Koller A, Strowig T, Flavell RA, Brodsky IE, Bliska JB. IQGAP1 is important for activation of caspase-1 in macrophages and is targeted by Yersinia pestis type III effector YopM. MBio. 2014;5:e01402–14. doi: 10.1128/mBio.01402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Croy HE, Fuller CN, Giannotti J, Robinson P, Foley AV, Yamulla RJ, Cosgriff S, Greaves BD, von Kleeck RA, An HH, Powers CM, Tran JK, Tocker AM, Jacob KD, Davis BK, Roberts DM. The Poly(ADP-ribose) Polymerase Enzyme Tankyrase Antagonizes Activity of the beta-Catenin Destruction Complex through ADP-ribosylation of Axin and APC2. J Biol Chem. 2016;291:12747–60. doi: 10.1074/jbc.M115.705442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, Deng H, Wang J, Chang J, Chai J. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–5. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 59.Maekawa S, Ohto U, Shibata T, Miyake K, Shimizu T. Crystal structure of NOD2 and its implications in human disease. Nat Commun. 2016;7:11813. doi: 10.1038/ncomms11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weissbach L, Settleman J, Kalady MF, Snijders AJ, Murthy AE, Yan YX, Bernards A. Identification of a human rasGAP-related protein containing calmodulin-binding motifs. J Biol Chem. 1994;269:20517–21. [PubMed] [Google Scholar]
- 61.Neel NF, Sai J, Ham AJ, Sobolik-Delmaire T, Mernaugh RL, Richmond A. IQGAP1 is a novel CXCR2-interacting protein and essential component of the “chemosynapse”. PLoS One. 2011;6:e23813. doi: 10.1371/journal.pone.0023813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 63.McCall MN, Jaffee HA, Zelisko SJ, Sinha N, Hooiveld G, Irizarry RA, Zilliox MJ. The Gene Expression Barcode 3.0: improved data processing and mining tools. Nucleic Acids Res. 2014;42:D938–43. doi: 10.1093/nar/gkt1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harton JA, Linhoff MW, Zhang J, Ting JP. Cutting edge: CATERPILLER: a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol. 2002;169:4088–93. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- 65.Ye Z, Lich JD, Moore CB, Duncan JA, Williams KL, Ting JP. ATP binding by monarch-1/NLRP12 is critical for its inhibitory function. Mol Cell Biol. 2008;28:1841–50. doi: 10.1128/MCB.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith JM, Hedman AC, Sacks DB. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 2015;25:171–84. doi: 10.1016/j.tcb.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kufer TA, Kremmer E, Banks DJ, Philpott DJ. Role for erbin in bacterial activation of Nod2. Infect Immun. 2006;74:3115–24. doi: 10.1128/IAI.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–6. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crow YJ. Type I interferonopathies: mendelian type I interferon up-regulation. Curr Opin Immunol. 2015;32:7–12. doi: 10.1016/j.coi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 70.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baetz A, Frey M, Heeg K, Dalpke AH. Suppressor of cytokine signaling (SOCS) proteins indirectly regulate toll-like receptor signaling in innate immune cells. J Biol Chem. 2004;279:54708–15. doi: 10.1074/jbc.M410992200. [DOI] [PubMed] [Google Scholar]
- 72.Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O’Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, Zhang Z, Arnott D, Dixit VM. DUBA: a deubiquitinase that regulates type I interferon production. Science. 2007;318:1628–32. doi: 10.1126/science.1145918. [DOI] [PubMed] [Google Scholar]
- 73.Surpris G, Poltorak A. The expanding regulatory network of STING-mediated signaling. Curr Opin Microbiol. 2016;32:144–50. doi: 10.1016/j.mib.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abel AM, Schuldt KM, Rajasekaran K, Hwang D, Riese MJ, Rao S, Thakar MS, Malarkannan S. IQGAP1: insights into the function of a molecular puppeteer. Mol Immunol. 2015;65:336–49. doi: 10.1016/j.molimm.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Owen D, Campbell LJ, Littlefield K, Evetts KA, Li Z, Sacks DB, Lowe PN, Mott HR. The IQGAP1-Rac1 and IQGAP1-Cdc42 interactions: interfaces differ between the complexes. J Biol Chem. 2008;283:1692–704. doi: 10.1074/jbc.M707257200. [DOI] [PubMed] [Google Scholar]
- 76.Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Okada M, Hozumi Y, Iwazaki K, Misaki K, Yanagida M, Araki Y, Watanabe T, Yagisawa H, Topham MK, Kaibuchi K, Goto K. DGKzeta is involved in LPS-activated phagocytosis through IQGAP1/Rac1 pathway. Biochem Biophys Res Commun. 2012;420:479–84. doi: 10.1016/j.bbrc.2012.03.057. [DOI] [PubMed] [Google Scholar]
- 78.Kim H, White CD, Li Z, Sacks DB. Salmonella enterica serotype Typhimurium usurps the scaffold protein IQGAP1 to manipulate Rac1 and MAPK signalling. Biochem J. 2011;440:309–18. doi: 10.1042/BJ20110419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mukai K, Konno H, Akiba T, Uemura T, Waguri S, Kobayashi T, Barber GN, Arai H, Taguchi T. Activation of STING requires palmitoylation at the Golgi. Nat Commun. 2016;7:11932. doi: 10.1038/ncomms11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Helgason E, Phung QT, Dueber EC. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: the emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 2013;587:1230–7. doi: 10.1016/j.febslet.2013.01.059. [DOI] [PubMed] [Google Scholar]
- 81.Suzuki T, Oshiumi H, Miyashita M, Aly HH, Matsumoto M, Seya T. Cell type-specific subcellular localization of phospho-TBK1 in response to cytoplasmic viral DNA. PLoS One. 2013;8:e83639. doi: 10.1371/journal.pone.0083639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McDonald C, Chen FF, Ollendorff V, Ogura Y, Marchetto S, Lecine P, Borg JP, Nunez G. A role for Erbin in the regulation of Nod2-dependent NF-kappaB signaling. J Biol Chem. 2005;280:40301–9. doi: 10.1074/jbc.M508538200. [DOI] [PubMed] [Google Scholar]
- 83.Kabi A, McDonald C. FRMBP2 directs NOD2 to the membrane. Proc Natl Acad Sci U S A. 2012;109:21188–9. doi: 10.1073/pnas.1219395110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakamura N, Lill JR, Phung Q, Jiang Z, Bakalarski C, de Maziere A, Klumperman J, Schlatter M, Delamarre L, Mellman I. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature. 2014;509:240–4. doi: 10.1038/nature13133. [DOI] [PubMed] [Google Scholar]
- 85.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.