Abstract
Carbohydrate metabolism is essential for cellular energy balance as well as for the biosynthesis of new cellular building blocks. As animal nutrient intake displays temporal fluctuations and each cell type within the animal possesses specific metabolic needs, elaborate regulatory systems are needed to coordinate carbohydrate metabolism in time and space. Carbohydrate metabolism is regulated locally through gene regulatory networks and signaling pathways, which receive inputs from nutrient sensors as well as other pathways, such as developmental signals. Superimposed on cell-intrinsic control, hormonal signaling mediates intertissue information to maintain organismal homeostasis. Misregulation of carbohydrate metabolism is causative for many human diseases, such as diabetes and cancer. Recent work in Drosophila melanogaster has uncovered new regulators of carbohydrate metabolism and introduced novel physiological roles for previously known pathways. Moreover, genetically tractable Drosophila models to study carbohydrate metabolism-related human diseases have provided new insight into the mechanisms of pathogenesis. Due to the high degree of conservation of relevant regulatory pathways, as well as vast possibilities for the analysis of gene–nutrient interactions and tissue-specific gene function, Drosophila is emerging as an important model system for research on carbohydrate metabolism.
Keywords: metabolism, insulin, glucose, gene regulation, nutrient sensing
Preface
CARBOHYDRATE metabolism is essential for all life, having profound implications for growth, reproduction, and organismal maintenance. As multicellular animals eat periodically and experience times of starvation, carbohydrate intake can undergo extreme fluctuations. Moreover, different cell types and developmental stages have their own metabolic requirements, which, together with the changing nutrient intake, pose the need for constant regulation of carbohydrate metabolism. Therefore, complex regulatory systems have evolved to integrate these functions. In recent years, Drosophila melanogaster (hereafter Drosophila) has been increasingly utilized to study the regulation of carbohydrate metabolism and new research fields have emerged around this topic. New insight has been gained into the regulatory pathways that respond to changes in carbohydrate intake and regulate metabolism to maintain homeostasis. These include gene regulatory networks and signaling pathways, which act locally in metabolically active peripheral tissues, as well as hormonal signals, which maintain organismal homeostasis through interorgan communication. Interesting cross-talk between carbohydrate metabolism and other physiological processes, such as circadian regulation and developmental transitions, have also been uncovered. Moreover, powerful Drosophila models to study carbohydrate metabolism-related human diseases have been established. The success of Drosophila research on providing new insights into carbohydrate metabolism has its foundation in the strengths of this model system. These include a high degree of conservation of the pathways controlling carbohydrate metabolism, the ease of using simple dietary schemes, which allow studies on interactions between genes and individual nutrients, as well as a powerful genetic toolkit, which is particularly advantageous in studies that address hormonal signaling between tissues. Here, we have highlighted the recent advances in Drosophila research on carbohydrate energy metabolism. For the sake of focus, we have excluded or only touched minimally upon some related themes, such as gustatory responses, the regulation of feeding behavior, lipid metabolism, and growth control.
Part I
Homeostatic control of carbohydrate metabolism through intracellular nutrient sensing
Carbohydrate-responsive gene regulation and signaling:
Fluctuations in nutrient intake pose constant requirements for homeostatic control of carbohydrate metabolism. Such regulation requires that cells are able to detect the levels of key carbohydrate-derived metabolites and consequently adjust the activity of regulatory pathways. An important layer of local regulation of carbohydrate homeostasis is mediated through so-called intracellular sugar sensing by a heterodimer of conserved basic helix-loop-helix transcription factors Mondo and Max-like protein X (Mlx, Bigmax) (Havula and Hietakangas 2012). In Drosophila larvae, Mondo-Mlx control the majority of the strongly sugar-responsive genes (Mattila et al. 2015).
Vertebrates have two Mondo paralogs, called MondoA (MLXIP) and ChREBP (Carbohydrate Response Element-Binding Protein, also known as MondoB, MLXIPL), both of which dimerize with Mlx (Havula and Hietakangas 2012). Studies in mammals have shown that the nuclear translocation and transcriptional activity of ChREBP/MondoA-Mlx are induced by glucose. The N-terminus of ChREBP and MondoA contains a so-called Glucose-Sensing-Module (GSM), which includes the low glucose inhibitory domain (LID) and the Glucose-Response Activation Conserved Element (GRACE), both of which are required for glucose sensing (Havula and Hietakangas 2012). It has been proposed that the GSM of the Mondo proteins contains a conserved motif, which resembles the glucose-6-phosphate (G-6-P)-binding site of metabolic enzymes. The binding of G-6-P to the GSM would prevent the intramolecular inhibition of GRACE imposed by LID (McFerrin and Atchley 2012). However, direct structural evidence about the interaction of G-6-P (and possibly other phosphorylated hexoses) with MondoA/ChREBP is still missing. The intracellular glucose sensing appears to be highly conserved. For example, the domain structure, glucose responsiveness, and the heterodimerization with Mlx are conserved in Drosophila Mondo (Li et al. 2006; Havula and Hietakangas 2012). Moreover, Drosophila Mondo contains a conserved LxxLL nuclear receptor box signature, which likely allows Mondo to interact with nuclear receptors (McFerrin and Atchley 2012). In mammals, the activity of ChREBP is further regulated through post-translational modifications, such as phosphorylation and O-GlcNAc (N-acetylglucosamine) modification, but the role of these modifications in Drosophila remains unclear [reviewed in Havula and Hietakangas (2012)].
The physiological importance of intracellular sugar sensing is reflected by the fact that Drosophila larvae deficient of Mondo-Mlx display lethality on any diet containing high levels of sucrose, glucose, or fructose (Havula et al. 2013). The sugar intolerance of mlx mutants manifests in a physiologically relevant range of dietary sugars, as mlx mutants are unable to develop on red grapes, which are naturally rich in sugars. Interestingly, mice lacking ChREBP also display impaired survival on a diet rich in simple carbohydrates (Iizuka et al. 2004). In Drosophila larvae, Mondo and Mlx display highest expression levels in the fat body, intestine, and Malpighian tubules (Havula et al. 2013). Moreover, both genes are upregulated upon sugar feeding (Zinke et al. 2002; Mattila et al. 2015). The sugar intolerance phenotype of mlx mutants can be rescued by fat body-specific transgenic expression. In addition to sugar tolerance, Mondo-Mlx also affects feeding behavior; knockdown of Mondo in the fat body decreases (Sassu et al. 2012), while neuronal knockdown increases feeding (Docherty et al. 2015). However, the underlying mechanisms of how Mondo-Mlx controls Drosophila feeding behavior remain unknown.
Mondo-Mlx regulates its target genes by binding to the so-called carbohydrate response element (ChoRE), which is composed of two imperfect E-boxes divided by five bases and is well-conserved in Drosophila (Shih et al. 1995; Jeong et al. 2011; Bartok et al. 2015; Mattila et al. 2015). In addition to direct regulation of metabolic target genes, Mondo-Mlx controls the expression of other transcription factors, namely Cabut and Sugarbabe (Bartok et al. 2015; Mattila et al. 2015) (Figure 1). Cabut is an ortholog of mammalian Krüppel-like factors 10 and 11 and is a transcriptional repressor with many physiological roles, including growth control as well as developmental, metabolic, and circadian regulation (Rodriguez 2011; Bartok et al. 2015; Ruiz-Romero et al. 2015). Mondo-Mlx binds directly to the promoter of the cabut gene and cabut expression is strongly downregulated in mlx mutants (Havula et al. 2013; Bartok et al. 2015). While Cabut is essential for development, partial knockdown of Cabut allows larval development on a low-sugar diet, although the larvae become sugar intolerant. Sugarbabe is also a direct target of Mondo-Mlx (Mattila et al. 2015). It is a homolog of mammalian Gli-similar transcription factors and it has been long known as one of the most strongly sugar-responsive genes in Drosophila (Zinke et al. 2002). In addition to transcriptional control, Sugarbabe is regulated by a nutrient-dependent miRNA, miR-14 (Varghese et al. 2010). Sugarbabe-deficient larvae display some intolerance toward a high-sugar diet, albeit to a lesser extent than mlx mutants (Mattila et al. 2015).
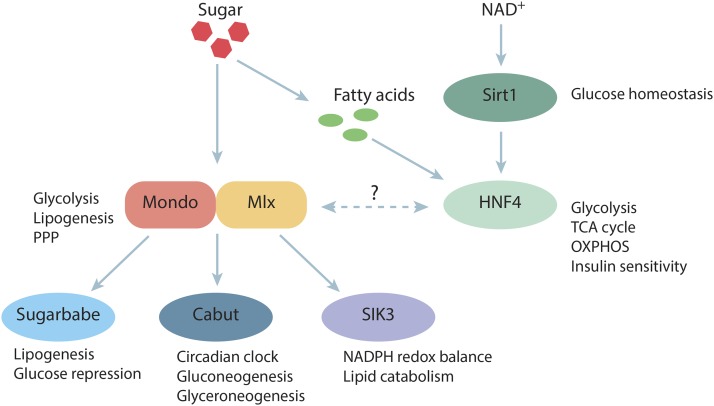
Figure 1.
Intracellular sugar-responsive gene regulatory network. The main regulators of sugar-responsive gene expression are the heterodimeric bHLH-Zip transcription factors Mondo and Mlx. Mondo-Mlx has a direct role in regulating gene expression programs, which are essential in glucose and fatty acid metabolism. In addition, Mondo-Mlx activates the transcription of a second tier of transcriptional regulators, including Sugarbabe and Cabut as well as other regulatory proteins such as protein kinase SIK3. In parallel, glucose regulates indirectly, through the generation of fatty acids and NAD+, transcription factor HNF4 and deacetylase Sirt1, respectively. The transcriptome regulated by Mondo-Mlx and HNF4 are partially overlapping. However, how these factors interact is yet unknown. bHLH, basic helix-loop-helix; FA, fatty acid; HNF4, hepatocyte nuclear factor 4; Mlx, Max-like protein X; NAD+, nicotinamide adenine dinucleotide; NADPH, nicotinamide adenine dinucleotide phosphate; OXPHOS, oxidative phosphorylation; PPP, pentose phosphate pathway; SIK3, salt-inducible kinase 3; Sirt1, Sirtuin 1; TCA, tricarboxylic acid cycle.
In addition to transcription factors, Mondo-Mlx controls other types of regulatory proteins, including protein kinase SIK3 (Salt-inducible kinase 3). SIK3 belongs to the family of AMP-activated protein kinase (AMPK)-related kinases and has been recently implicated in metabolic regulation downstream of insulin and glucagon signaling (Wang et al. 2011; Choi et al. 2015; Hirabayashi and Cagan 2015). Thus, SIK3 integrates signals from intracellular sugar sensing as well as hormonal control. SIK3-null mutants were originally identified as larval lethal, but recent data shows that on a low-sugar diet some pupae emerge, indicating that SIK3 loss-of-function leads to prominent sugar intolerance (Wang et al. 2011; Teesalu et al. 2017). Intracellular glucose sensing by Mondo-Mlx is also coupled with the systemic control of metabolism. Namely, sugar-inducible transforming growth factor β (TGF-β)/Activin ligand Dawdle (Daw) is a direct target of Mondo-Mlx. Daw signals through the Babo receptor and transcription factor SMAD2/Smox and is expressed in several peripheral tissues of larvae, displaying highest expression levels in the fat body (Mattila et al. 2015; Upadhyay et al. 2017). Similar to other regulatory genes that act downstream of Mondo-Mlx, dawdle is essential for sugar tolerance (Ghosh and O’Connor 2014; Mattila et al. 2015). Interestingly, Daw and Sugarbabe belong to a common regulatory pathway, since depletion of Daw and its downstream effector SMAD2/Smox prevents full activation of sugarbabe upon sugar feeding (Mattila et al. 2015). How Daw-dependent Activin signaling cooperates with Mondo-Mlx to control Sugarbabe expression is an interesting question for future studies. A more detailed description of TGF-β/Activin signaling will be presented later in this review.
Along with Mondo-Mlx and its downstream targets, other regulatory genes have been shown to be essential for sugar tolerance. One of them is a conserved nuclear receptor HNF4 (Hepatocyte nuclear factor 4), which can be activated in response to long-chain fatty acids (Palanker et al. 2009) (Figure 1). Mutants of HNF4 display normal larval development, but they fail to eclose on standard laboratory food. However, reduction of dietary sugar rescues the development of the majority of mutants into adulthood. Thus, the sugar intolerance of HNF4 mutants manifests at a later stage than that of Mondo-Mlx-deficient animals (Barry and Thummel 2016). Similar to mlx mutants, HNF4 mutants also display highly elevated circulating glucose and trehalose levels, and the circulating glucose responds strongly to the sugar content of the diet. HNF4 maintains glucose homeostasis both in the fat body as well as in the insulin-producing cells (IPCs). Considering the cooperative function between ChREBP and HNF4 in mammals (Adamson et al. 2006), and the conserved nuclear receptor box in Drosophila Mondo (McFerrin and Atchley 2012), studies examining the possible interplay between HNF4 and Mondo-Mlx in Drosophila are warranted.
Another nutrient sensor critical for carbohydrate homeostasis is the NAD+-dependent deacetylase Sirtuin 1 (Sirt1, Sir2). Loss of Sirt1 in the adult fat body leads to hyperglycemia, obesity, and insulin resistance, which are all age-progressive (Banerjee et al. 2012; Palu and Thummel 2016). Through the control of systemic free fatty acid levels and insulin signaling, the fat body-specific activity of Sirt1 is further reflected in other tissues, for example by affecting mitochondrial function in the muscle (Banerjee et al. 2013). Similar to adults, fat body Sirt1 negatively regulates triglyceride accumulation in larvae (Reis et al. 2010). Interestingly, Sirt1 mutants share many phenotypic features with the HNF4 mutants and display deregulation of an overlapping set of genes (Palu and Thummel 2016). Moreover, Sirt1 mutants display an age progressive reduction in HNF4 expression along with increased HNF4 acetylation, suggesting that Sirt1 maintains HNF4 stability through deacetylation (Palu and Thummel 2016). Since Sirt1 activity depends on the availability of the cofactor Nicotinamide adenine dinucleotide (NAD+), which in turn depends on carbohydrate metabolism, it is conceivable that the glucose-dependent cellular metabolic status is reflected in the regulation of HNF4 through Sirt1 (Figure 1).
O-GlcNAcylation, a link between carbohydrate metabolism and signaling:
In addition to direct glucose sensing through Mondo-Mlx, cells have evolved additional mechanisms to convey information about intracellular metabolic status to the regulation of cell physiology. One such mechanism is the post-translational modification of proteins through O-linked GlcNAcylation, where a nitrogen-containing nucleotide sugar GlcNAc is reversibly added to serine and/or threonine residues, altering target protein activity, stability, specificity, or localization. Protein O-GlcNAcylation has been shown to target several key regulators important for cellular energetics and growth. These include, for example, c-Myc, p53, calcium/calmodulin-dependent kinase IV (CamKIV), casein kinase 2 (CK2), AMPK, the cAMP response element-binding protein (CREB), Forkhead box subgroup O (FOXO1), and AKT [reviewed by Hardivillé and Hart (2014)]. Accordingly, a wealth of data indicate that deregulated O-linked GlcNAcylation is associated with metabolic disorders such as insulin resistance and cancer [reviewed by Bond and Hanover (2015); Ferrer et al. (2016)].
The substrate for O-linked GlcNAcylation, UDP-GlcNAc, is the end product of the hexosamine biosynthesis pathway (HBP), which integrates inputs from glucose (fructose-6-phosphate), amino acid (glutamine), fatty acid [acetyl-coenzyme A (CoA)], and nucleotide/energy (UDP) metabolism (Figure 2). The regulation of the HBP flux is not fully understood, but fructose-6-phosphate availability and negative feedback inhibition by UDP-GlcNAc likely play major roles (McKnight et al. 1992). The members of the HBP are well-conserved in Drosophila, with two rate-limiting enzymes, glucose-fructose amidotransferases (Gfat1/2) and the orthologs of glucosamine-phosphate N-acetyltransferase (CG1969), phosphoacetylglucosamine mutase (nst) and UDP-N-acetylglucosamine diphosphorylase (mmy) (Figure 2). The activity of the HBP is essential for fly development, since mutants for mmy and nst are lethal with various developmental defects. Notably, UDP-GlcNAc is also a substrate for the N-linked protein glycosylation necessary for appropriate protein folding, maturation, and membrane targeting, as well as chitin biosynthesis necessary for the production of apical extracellular matrices (Schimmelpfeng et al. 2006; Tonning et al. 2006). Therefore, the phenotypes of HBP pathway mutants may reflect defects in these functions.
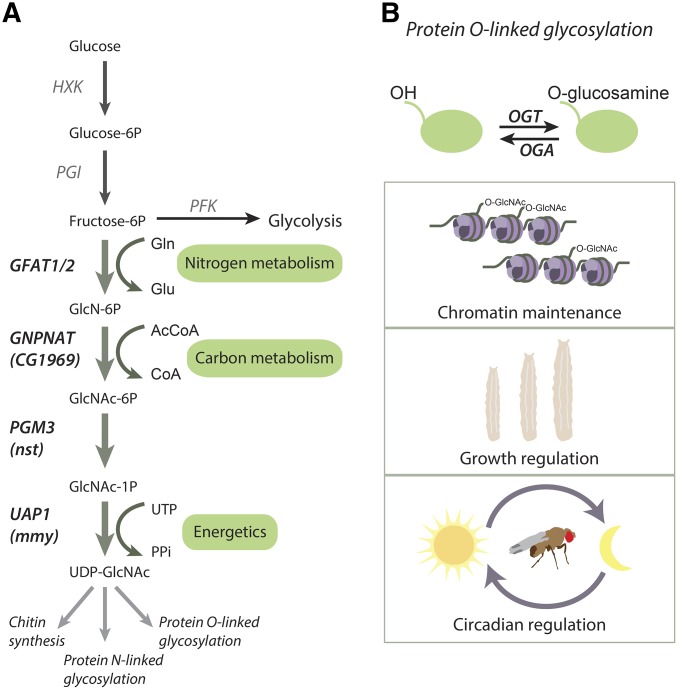
Figure 2.
The role of the HBP and protein O-linked GlcNAc conjugation in the regulation of Drosophila physiology. Schematic presentation of the HBP (A) and the known processes regulated by protein O-linked GlcNAc conjugation (B). (A) HBP competes for F-6-P with PFK, the rate-limiting enzyme of glycolysis. In the first and rate-limiting step of HBP, GFAT conjugates an amine group from glutamine to the F-6-P yielding GlcN-6-P and glutamate. In the following step, GNPNAT conjugates the acetyl group from acetyl-CoA to yield GlcNAc-6-P, which is then isomerized to GlcNAc-1-P by PGM3. Finally, UDP is conjugated to the GlcNAc-1-P by UAP1 to yield UDP-GlcNAc, which is a substrate for macromolecule glycosylation and chitin biosynthesis. Hence, HBP integrates inputs from glucose (G-6-P), amino acid (glutamine), fatty acid (acetyl-CoA) and energy (UDP) metabolism, making it a sensor of cellular nutrient and energy metabolism. N-linked glycosylation, covalent attachment of an oligosaccharide to asparagine residues, is a mechanism of protein maturation and trafficking between cell compartments. The UDP-GlcNAc polymer, also known as chitin, is the key component of Drosophila exoskeleton. (B) O-linked GlcNAcylation is a transient protein post-translational modification mechanism, which targets threonine and serine residues. O-linked GlcNAcylation is mediated by the activities of OGT and OGA to conjugate and deconjugate glucosamine, respectively. O-linked GlcNAcylation can compete, enhance, or attenuate protein phosphorylation, making it an important mechanism to regulate protein activity. In Drosophila, the activity of OGT is known to be involved in maintenance of chromatin state, in regulating larval growth through insulin signaling, and in regulating the maintenance of circadian rhythm. F-6-P, fructose-6-phosphate; G-6-P, glucose-6-phosphate; GFAT1/2, glutamine fructose-6-phosphate amidotransferase 1/2; GlcN-6-P, glucosamine-6-phosphate; GlcNAc, N-acetylglucosamine; GlcNAc-1-P, N-acetyl-D-glucosamine-1-phosphate; GlcNAc-6-P, N-acetyl-D-glucosamine-6-phosphate; GNPNAT, glucosamine-phosphate N-acetyltransferase; HBP, hexosamine biosynthesis pathway; HXK, hexokinase; OGA, O-GlcNAcase; OGT, O-GlcNAc transferase; PFK, phosphofructokinase; PGI, phosphoglucose isomerase; PGM3, phosphoglucomutase 3; UAP1, UDP-N-acetylglucosamine pyrophosphorylase; UTP, uridine triphosphate.
The dynamic regulation of O-linked GlcNAc conjugation/deconjugation is mediated by the activity of a conserved pair of enzymes, O-GlcNAc transferase (Ogt, encoded by the super sex combs gene) and O-GlcNAcase (Oga), which catalyze the addition and removal of O-GlcNAc, respectively. In Drosophila, Ogt was first identified as belonging to the Polycomb group genes (PcG) due to the characteristic homeotic transformations of the mutant animal (Ingham 1984). It was later shown that Ogt can modify other PcG proteins located at the Polycomb Repressor Element (PRE) loci, thereby affecting transcriptional repression during embryonic development (Gambetta et al. 2009; Sinclair et al. 2009). The role of Ogt as a transcriptional regulator was further extended by the finding that O-GlcNAc-modified proteins can bind to sites throughout the genome, and not only at loci containing PREs (Liu et al. 2017). However, it is not known how these modifications correlate with gene expression and which genomic loci are affected by changes in O-GlcNAcylation. Interestingly, in a recent study by Selvan et al. (2017), a catalytically inactive OGA mutant was used as a substrate trap to enrich O-GlcNAcylated proteins from Drosophila embryos. By this approach, > 2000 proteins were identified as substrates for O-GlcNAcylation, suggesting it to be a major mechanism of protein modification during development (Selvan et al. 2017).
Modulation of O-linked GlcNAc conjugation by inhibiting Ogt and Oga during larval stages attenuates or enhances larval growth, respectively (Park et al. 2011). In addition, Ogt knockdown during larval development leads to an increase in autophagy (Park et al. 2015). These results, together with the findings that AKT and FOXO are regulated through O-GlcNAcylation, have led to the proposal that nutritional status and Ogt activity contribute to the regulation of the insulin signaling pathway during larval growth (Park et al. 2011, 2015). However, the exact mechanism of this regulation remains to be elucidated. One possibility is that O-linked GlcNAcylation affects growth cell nonautonomously, through the regulation of insulin-like peptide (dILP) synthesis and release from the IPCs. Indeed, targeted RNA interference (RNAi) knockdown of Ogt and Oga in the IPCs has been shown to attenuate and elevate dilp expression, respectively (Sekine et al. 2010). Interestingly, mice lacking Ogt in pancreatic β-cells develop diabetes through ER stress-induced β-cell apoptosis (Alejandro et al. 2015). These findings suggest that the regulation of insulin synthesis through O-linked GlcNAc cycling is conserved through evolution.
In conclusion, O-linked GlcNAc modification is emerging as an important regulator of Drosophila growth, metabolism, and physiology. Furthermore, it is tempting to speculate that the role of the O-GlcNAcylation is emphasized in cells involved in nutrient sensing, as suggested by the data on O-GlcNAcylation in the IPCs. Further studies are needed to uncover the role of this post-translational modification on the major peripheral nutrient-sensing tissues, the fat body and the intestine.
Regulation of glycolysis and lipid metabolism upon sugar feeding:
Activation of glycolysis is critical for the elimination of excess glucose entering the circulating hemolymph and sugar feeding strongly activates the expression of genes encoding glycolytic enzymes (Mattila et al. 2015) (Figure 3). A key mediator of glycolytic gene activation is Mondo-Mlx, as sugar feeding fails to activate the expression of glycolytic genes in mlx mutant larvae. HNF4, which also controls glucose homeostasis upon high-sugar feeding, promotes the expression of the Glucokinase homolog Hexokinase C (Barry and Thummel 2016). Genetic inhibition of glycolytic gene activation prevents the clearance of circulating glucose and reduces survival on a high-sugar diet, possibly reflecting the toxicity of high circulating free glucose (Havula et al. 2013; Garrido et al. 2015).
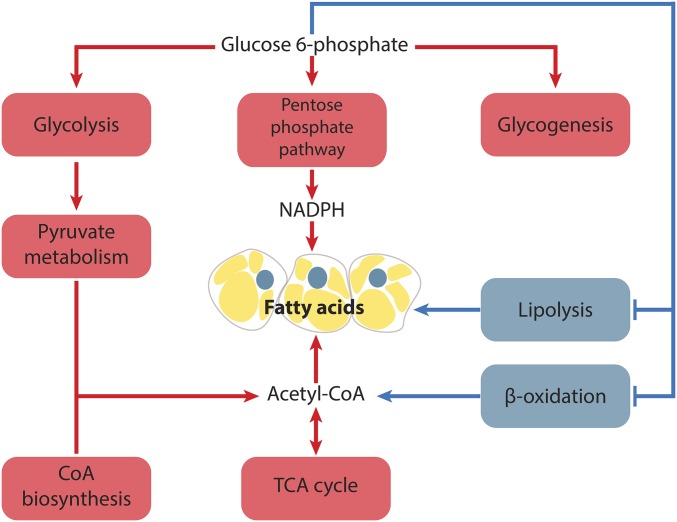
Figure 3.
The de novo synthesis of fatty acids and glycogenesis is coordinated in response to dietary sugars. The increase in cellular G-6-P levels leads to the orchestrated regulation of several metabolic processes important in the synthesis of fatty acids and glycogen and, as a result, clearance of intracellular sugars. The majority of G-6-P is channeled through glycolysis, resulting in elevated pyruvate and the production of acetyl-CoA. The process is coordinated with increased levels of CoA biosynthesis. Acetyl-CoA is utilized by the TCA cycle to produce intermediates of amino acid metabolism, ATP, NADH, and citrate. Citrate is further channeled to the fatty acid biosynthesis. The process of fatty acid synthesis is accompanied with the activity of the pentose phosphate pathway yielding the necessary reductive power in the form of NADPH. Parallel to the fatty acid synthesis, elevated levels of G-6-P shut down the process of lipid catabolism through lipolysis and generation of acetyl-CoA through β-oxidation. CoA, coenzyme A; G-6-P, glucose-6-phosphate; NADPH, nicotinamide adenine dinucleotide phosphate; TCA, tricarboxylic acid cycle.
Downstream of the glycolytic pathway, pyruvate needs to be transferred into mitochondria to be catabolized further in the tricarboxylic acid (TCA) cycle. Sugar feeding modestly activates the expression of the Drosophila mitochondrial pyruvate carrier (Mpc1) (Mattila et al. 2015). Inhibiting mitochondrial transport of pyruvate in Mpc1 mutants causes increased levels of glycolytic intermediates and high circulating glucose and trehalose. Mpc1 mutants also survive poorly on a carbohydrate-only diet (Bricker et al. 2012). HNF4 promotes the expression of genes encoding components of the TCA cycle and oxidative phosphorylation (OXPHOS) pathway. Strikingly, nearly all transcripts of the mitochondrial genome are significantly downregulated in HNF4 mutants and chromatin immunoprecipitation has revealed specific HNF4 enrichment in the mitochondrial DNA control region, which regulates the transcription of both mtDNA strands (Barry and Thummel 2016). Impaired OXPHOS leads to poor survival on a high-sugar diet, as evidenced by the mutant phenotype of the technical knockout (tko) gene (Kemppainen et al. 2016). Consistent with the gene expression changes, HNF4 mutants show elevated levels of G-6-P and dihydroxyacetone phosphate (Barry and Thummel 2016). Moreover, intermediates of the polyol pathway, namely sorbitol and fructose, are elevated in HNF4 mutants. This pathway is activated when normal homeostatic clearance of circulating glucose is impaired, such as in the case of untreated diabetes (Luo et al. 2016). Interestingly, elevated levels of sorbitol are also present in the mlx mutants, underlining the similarities of the metabolic phenotypes of HNF4 and mlx mutants (Teesalu et al. 2017). It will be interesting to learn whether the oxidative and osmotic stress caused by sorbitol synthesis and accumulation contributes to the sugar intolerance phenotype.
In addition to activating gene expression that promotes glucose catabolism, the counteracting flux of carbon from the TCA cycle toward gluco- and glyceroneogenesis needs to be inhibited. To this end, sugar feeding downregulates the expression of both cytoplasmic and mitochondrial isoforms of phosphoenolpyruvate carboxykinase (PEPCK) in adults (Bartok et al. 2015). The Mondo-Mlx target Cabut is upregulated upon sugar feeding and it directly binds to the promoter of the Pepck gene, repressing its activity.
Concomitantly, sugar feeding strongly activates the expression of lipogenic enzymes: ATP citrate lyase, acetyl-CoA carboxylase (ACC), and Fatty acid synthase 1 (FASN1), which convert citrate derived from the TCA cycle into fatty acids (Zinke et al. 2002; Sassu et al. 2012; Musselman et al. 2013; Mattila et al. 2015) (Figure 3). This response also depends on Mondo-Mlx, which directly binds to the promoters of at least FASN1 and ACC (Mattila et al. 2015). This highlights the evolutionary conservation of intracellular sugar sensing as FAS and ACC are well-established direct targets of mammalian ChREBP (Ishii et al. 2004). In Drosophila, lipogenic gene expression is also positively regulated by Sugarbabe, which constitutes a positive feed-forward loop to drive lipogenic gene expression during sustained sugar feeding (Mattila et al. 2015). Impaired fatty acid synthesis results in reduced survival on a high-sugar diet (Musselman et al. 2013; Garrido et al. 2015). Genetic inhibition of lipogenesis in mutants with defective FASN1 and FASN2 leads to developmental lethality, which can be partially rescued by dietary lipids. Under these lipid-rescued conditions, addition of sugar into the diet of FASN mutants causes lethality and an accumulation of advanced glycation end products (AGEs) (Garrido et al. 2015). The harmful effects of sugar feeding can be rescued by the overexpression of glyoxalase-1, which counteracts the toxicity of methyglyoxal, suggesting a causal link between sugar-induced toxicity and AGEs in this setting. Notably, the role of fatty acid synthesis on sugar tolerance is likely to depend on the other diet components, as on a yeast-based diet addition of sugar can give a growth advantage to larvae with reduced FASN1 expression (Havula et al. 2013). High lipogenesis in response to high dietary sugar poses an elevated need for CoA, which is a cosubstrate for FAS. Sugar feeding leads to strong activation of genes involved in CoA biosynthesis to compensate for the increased need for CoA (Palanker Musselman et al. 2016). On the other hand, genes encoding lipid catabolic enzymes, such as triacylglycerol lipases and acyl-CoA dehydrogenases, are downregulated by dietary sugar (Zinke et al. 2002; Mattila et al. 2015). Perilipin expression is also increased under these conditions, possibly suppressing basal levels of lipolysis (Beller et al. 2010; Mattila et al. 2015). In sum, high-sugar feeding promotes lipid biosynthesis and inhibits lipid catabolism to channel excess carbon derived from sugars into triacylglycerols (Figure 3).
Fatty acid biosynthesis requires the reductive power of NADPH. A key mechanism to reduce NADP+ into NADPH is via the activity of the oxidative branch of the pentose phosphate pathway (PPP). Sugar feeding strongly activates the expression of genes encoding PPP components, including the rate-limiting enzyme glucose-6-phosphate dehydrogenase [G-6-PD, encoded by the Zwischenferment (Zw) gene in Drosophila] (Zinke et al. 2002; Mattila et al. 2015). PPP gene expression is fully dependent on Mondo-Mlx (Mattila et al. 2015). In fact, the PPP constitutes one of the most highly enriched pathways among Mondo-Mlx targets. Moreover, HNF4 contributes to the expression of a subset of PPP genes (Barry and Thummel 2016). The PPP is also post-translationally activated through phosphorylation of G-6-PD by protein kinase SIK3 (Teesalu et al. 2017). Impaired activation of PPP in SIK3 and mlx mutants leads to elevated oxidative stress, which contributes to the sugar intolerance.
An important aspect of sugar-induced metabolism is the simultaneous regulation and coordination of multiple metabolic pathways (Figure 3). This encompasses the activation of glycolysis, lipogenesis, PPP, CoA synthesis, and lipid desaturation, along with inhibition of the counteracting catabolic routes. It is worth noting that sugar feeding also controls other metabolic pathways including the synthesis of nonessential amino acids serine and glutamine (Mattila et al. 2015). While the role of these pathways in sugar homeostasis is unclear, genetic inhibition of their activity is detrimental for survival and growth on a high-sugar diet, implying their physiological importance (Mattila et al. 2015).
Regulation of trehalose metabolism:
Trehalose is the most abundant circulating carbohydrate in insects. It is a disaccharide composed of two α-glucose molecules linked in a 1,1-glycosidic bond. Due to its nonreductive nature, it is nontoxic and therefore tolerated at high circulating levels (∼2000 mg/dl in third-instar larvae) (Ugrankar et al. 2015). This is in striking contrast with circulating glucose levels, which are typically maintained within a range of 5–30 mg/dl in larvae (Ugrankar et al. 2015). Interestingly, adult haemolymph contains 10–20-fold more glucose, suggesting a profound difference in carbohydrate metabolism and glucose tolerance between the life cycle stages (Tennessen et al. 2014b) (for discussion about differences in glucose sensing between larval and adult stages see Regulation of dILP expression and secretion by carbohydrates). The availability of high circulating trehalose has been considered critical to provide sufficient energy for insect flight muscle (Becker et al. 1996). Moreover, trehalose provides the energy needed for brain function. The Drosophila nervous system is surrounded by layers of glial cells, which maintain the blood-brain barrier (BBB). These BBB glial cells take up trehalose and metabolize it through the glycolytic pathway to secrete lactate and alanine to fuel neurons (Volkenhoff et al. 2015). Failures in BBB glial trehalose metabolism will lead to neuronal cell death. Trehalose was also suggested to contribute to the maintenance of neuroepithelial stem cells of the optic lobe (Chen et al. 2014), but this has been questioned in a recent study (Yasugi et al. 2017). In addition to its function as an energy source, trehalose has a role in protecting against environmental stresses, such as cold temperature and desiccation stress (Koštál et al. 2011; Thorat et al. 2016; Yoshida et al. 2016).
Trehalose is synthesized in the fat body from G-6-P and UDP-glucose by two enzymatic activities, Trehalose-6-phosphate synthase and Trehalose-6-phosphate phosphatase. These enzymatic activities are provided by the two catalytic domains of Drosophila Tps1 protein, both of which are essential for trehalose biosynthesis (Yoshida et al. 2016). Loss of Tps1 leads to trehalose-deficient animals (Matsuda et al. 2015). Surprisingly, trehalose is dispensable for larval development, as Tps1-deficient animals display lethality only at the late pupal stage. However, the trehalose-deficient larvae are sensitive to nutrient limitation, displaying rapid lethality upon starvation. The Drosophila genome encodes two putative trehalose transporters (Tret1-1 and Tret1-2). Tret1-1 has been shown to transport trehalose across the plasma membrane (Kanamori et al. 2010). Tret1-2 has emerged recently during evolution through a duplication event and is present only in D. melanogaster and its closest relatives (Volkenhoff et al. 2015). The Tret1-1 expression pattern suggests that it releases trehalose from the fat body into circulation and mediates the uptake of trehalose by other tissues (Kanamori et al. 2010; Volkenhoff et al. 2015). Trehalose is catabolized by the Trehalase enzyme. The Drosophila genome encodes two genes with putative trehalose-hydrolyzing catalytic activity, of which Treh displays ubiquitous expression and CG6262 is mainly expressed in the testis. Loss of trehalase activity prevents trehalose catabolism, leading to highly elevated circulating trehalose levels (Yoshida et al. 2016). Similarly to Tps1 mutants, Treh mutants are viable as larvae, but display pupal lethality and starvation sensitivity (Yoshida et al. 2016). Moreover, circulating glucose levels are significantly downregulated in Tps1 and Treh mutants, implying that trehalose turnover is needed to maintain systemic glucose levels (Matsuda et al. 2015; Yoshida et al. 2016).
In contrast to circulating glucose, trehalose levels do not respond strongly to dietary sugars (Ugrankar et al. 2015), although Tps1 expression is elevated by high-sugar feeding (Musselman et al. 2013). Moreover, many genes that impact circulating glucose levels do not affect trehalose levels, implying that glucose and trehalose levels are independently regulated (Ugrankar et al. 2015). However, trehalose levels display high variation during Drosophila development. Trehalose levels rise gradually during embryonic development and reach maximum levels in larvae (Matsuda et al. 2015). During metamorphosis, trehalose levels drop gradually, possibly reflecting high consumption or reduced synthesis of trehalose during pupal stages (Matsuda et al. 2015).
Regulation of glycogen metabolism:
Similar to other animals, glycogen is a key storage form of carbohydrates in Drosophila (Baker and Thummel 2007; Matsuda et al. 2015). In larvae, glycogen is predominantly stored in the body wall muscle while in the adult fly glycogen is abundant in the fat body and flight muscle (Wigglesworth 1949; Ruaud et al. 2011). Moreover, high levels of glycogen accumulate in the oocyte during late stages of oogenesis. Glycogen accumulation in oocytes occurs via remodeling of the electron transport chain into a respiratory quiescent mode, and is essential for the developmental competence of the oocyte (Sieber et al. 2016).
The expression of glycogen synthase (GlyS) is elevated by dietary sugar and depletion of GlyS from the larval fat body delays development on a high-sugar diet (Garrido et al. 2015). This suggests that glycogen synthesis needs to be dynamically controlled with respect to sugar intake. Two transcription factors, DHR38 and Mef2, have been demonstrated to regulate the expression of several glycogen biosynthesis genes (Ruaud et al. 2011; Clark et al. 2013). DHR38 is an orphan nuclear receptor homologous to the nuclear receptor 4A family in mammals. DHR38 is highly expressed in the larval body wall and gut, and its expression is induced by yeast feeding (Ruaud et al. 2011). DHR38 mutants show significantly reduced levels of glycogen in the body wall muscle, which is consistent with strongly reduced expression of Phosphoglucomutase, a critical enzyme of glycogen biosynthesis (Ruaud et al. 2011). Mef2 promotes glycogen synthesis in adults by activating the expression of several glycogen biosynthetic genes (Clark et al. 2013). Mef2 serves as a switch between metabolic and immune gene regulation. Once phosphorylated by S6K, Mef2 promotes glycogen and lipid biosynthesis. However, this phosphorylation is lost upon infection and activation of biosynthetic pathways is reduced, while immune response genes are upregulated. Glycogen stores are also regulated by the sugar sensor Mondo-Mlx, since mlx mutants possess strongly elevated glycogen stores (Havula et al. 2013). Whether this is due to direct regulation of glycogen metabolism or is an indirect consequence of impaired lipid biosynthesis remains to be explored. Supporting the latter, it has been observed that inhibition of fatty acid biosynthesis leads to elevated glycogen levels in larvae (Garrido et al. 2015). Low oxygen availability (hypoxia) has a strong impact on carbohydrate metabolism, including increased mobilization of glycogen. This response is prevented by loss of hypoxia-inducible factor (HIF) activity, but the underlying mechanisms remain to be explored (Y. Li et al. 2013).
Glycogen breakdown occurs through two parallel mechanisms: glycogenolysis and glycogen autophagy (Zirin et al. 2013). Genetic experiments in the larval skeletal muscle have shown that simultaneous inhibition of both autophagy and glycogenolysis fully prevents glycogen catabolism during starvation, and both pathways are needed for maximal efficiency of glycogen breakdown. Interestingly, GlyS interacts with Atg8, raising the possibility that GlyS acts as an adaptor between glycogen metabolism and the autophagy machinery. Another line of evidence linking glycogen synthesis and autophagy comes from the analysis of Rack1, a conserved guanine nucleotide-binding scaffold protein with a WD40-repeat. Loss of Rack1 leads to an attenuated autophagic response upon starvation and a dramatic > 10-fold reduction of glycogen stores in the larval fat body (Erdi et al. 2012). Furthermore, Rack1 colocalizes with glycogen particles as well as with Shaggy, the Drosophila ortholog of GlyS kinase 3β (GSK-3β) (Erdi et al. 2012), further suggesting that Rack1 might promote glycogen synthesis. Glycogen phosphorylase (GlyP), which catalyzes the rate-limiting step in glycogenolysis, is highly expressed in the carcass of the larva and in the fat body and carcass of the adult (FlyAtlas, Chintapalli et al. 2007). The expression of GlyP is elevated on a high-sugar diet, but the functional relevance of this regulation remains unknown (Musselman et al. 2011; Mattila et al. 2015). GlyP activity is essential to maintain Drosophila flight muscle function as GlyP mutants display severely reduced wing beat frequency (Eanes et al. 2006). Thus, glycogen autophagy is insufficient to compensate for the loss of glycogenolysis in the flight muscle.
Regulation of carbohydrate digestion through glucose repression:
Homeostatic control of metabolism affects not only channeling of metabolites into various end products, but also the enzymes involved in nutrient breakdown within the intestine. Specifically, it was recognized several decades ago that the presence of glucose in the Drosophila diet inhibits the activities of enzymes needed for the breakdown of polymeric carbohydrates, such as starch and oligosaccharides (Hickey and Benkel 1982; Benkel and Hickey 1986, 1987). Subsequent studies have shown that both at the larval and adult stages, various forms of sugar (i.e. sucrose, glucose, fructose, and trehalose) have a profound repressive effect on the expression of genes encoding enzymes that possess glycoside hydrolase activities, including α-amylases, maltases, and α-mannosidases (Zinke et al. 2002; Chng et al. 2014; Mattila et al. 2015). Collectively, the phenomenon of repressing the expression of enzymes and the digestion of carbohydrate polymers in the presence of a readily utilizable monosaccharide is termed “glucose repression” (Chng et al. 2014). Glucose repression might be a physiological response to suppress overload of systemic glucose under conditions where glucose catabolic pathways are close to saturation.
The mechanisms underlying glucose repression have been shown to involve several transcription factors. The maintenance of amylase expression is attributed to the nuclear receptors HNF4 and DHR38, whereas the repression by glucose is achieved through Mondo-Mlx, Sugarbabe, and the TGF-β/Activin target SMAD2 (Ruaud et al. 2011; Chng et al. 2014; Mattila et al. 2015; Barry and Thummel 2016). Mondo-Mlx resides high in the hierarchy of the sugar-responsive transcriptional network, which directly activates the expression of Sugarbabe and the TGF-β/Activin ligand Daw (Mattila et al. 2015). Interestingly, Daw expression is highest in the fat body where it functions as a secreted ligand to repress amylase expression in intestinal enterocytes through SMAD2 (Chng et al. 2014). In the intestine, Mondo-Mlx and SMAD2 converge to regulate sugarbabe expression, which is necessary and sufficient to repress the expression of amylases (Mattila et al. 2015). However, the details of the combinatorial function of Mondo-Mlx and TGF-β/activin signaling, as well as the significance of overlapping cell autonomous and noncell autonomous mechanisms, remain to be elucidated. The prevailing model suggests that while Mondo-Mlx monitors sugar uptake directly in the intestine, TGF-β/Activin signaling is needed to transmit information about the carbohydrate status of the fat body. Such a mechanism would coordinate the expression of amylases, utilization of carbohydrate polymers, and glucose uptake in the intestine according to the metabolic status of the animal.
Part II
Carbohydrate-responsive hormonal circuits: Insulin/glucagon axis and beyond
The dILP/glucagon circuit:
The maintenance of metabolic homeostasis during feeding and fasting periods requires constant communication between nutrient-storing and nutrient-consuming tissues. Several hormonal regulators have evolved for intertissue communication, which orchestrate the allocation of nutrients between growth, maintenance functions, and energy generation. The main hormonal system for maintaining carbohydrate homeostasis in metazoan organisms is the insulin–glucagon circuit, which responds to the levels of circulating glucose. In mammals, glucagon is released from the pancreatic α-cells upon low glucose concentration, promoting lipid and glycogen catabolism and gluconeogenesis to release glucose into the bloodstream. The rise in circulating glucose after feeding triggers insulin secretion from the pancreatic β-cells promoting anabolic metabolism such as lipogenesis and glycogenesis to clear excess glucose from circulation. In addition to the insulin/glucagon axis, several other hormones contribute to carbohydrate homeostasis. Some of them act in parallel to trigger independent responses, whereas many of the hormones regulating carbohydrate metabolism are interconnected to insulin/glucagon signaling, for example by modulating the secretion of these hormones or by influencing the sensitivity of the response in the signal-receiving tissue (Padmanabha and Baker 2014).
The insulin/glucagon axis is well-conserved in Drosophila. The Drosophila genome encodes eight dILPs (Brogiolo et al. 2001; Grönke et al. 2010; Colombani et al. 2012), and a single glucagon-like peptide, adipokinetic hormone (dAKH) (Schaffer et al. 1990). The regulation of the dILP–dAKH circuit varies between the larval and adult stages, reflecting the profound differences between these life cycle phases in terms of feeding behavior, nutritional demands, and growth (Figure 4). While larvae feed and grow constantly until they reach a critical size for pupariation, the adult life consists of periods of feeding and fasting regulated by the circadian clock and the availability of food. Thus, the framework of metabolic regulation in larvae and adult animals are very different.
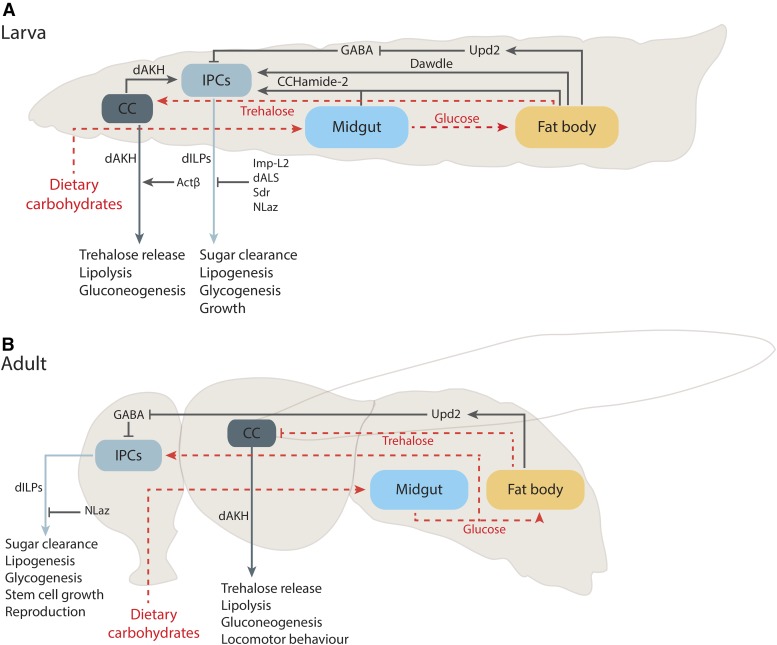
Figure 4.
Insulin-like peptide-glucagon circuit in Drosophila. Schematic presentation of larva (A) and adult fly (B), illustrating mechanisms that regulate the output of dILP and dAKH signaling in response to dietary carbohydrates. Dietary carbohydrates are digested in the midgut and glucose is taken up by the intestinal enterocytes. Glucose is converted into trehalose in the fat body and released into circulation. In the larva, trehalose has a biphasic effect to dAKH; low and high trehalose concentrations are shown to stimulate dAKH secretion. Whether such regulation also exists in adults is unknown. Larval IPCs are inherently insensitive to glucose, but carbohydrates regulate dILP secretion through remote mechanisms. These include dAKH from the CC as well as CCHamide-2, Dawdle, and Upd2 secreted from the fat body. Only Upd2 has been shown to function at the larval and adult stages. At the adult stage, glucose regulates IPCs directly by modulating the activity of KATP channels and cell depolarization leading to dILP secretion. The output of dAKH and dILP signaling is regulated through humoral factors, such as Activinβ, Imp-L2, dALS, Sdr, and NLaZ. Only NLaz has been shown to function at larval and Activinβ adult stage. Actβ, Activinβ; CC, corpora cardiaca; dAKH, Drosophila adipokinetic hormone; dALS, Drosophila acid-labile subunit; dILP, Drosophila insulin-like peptide; Imp-L2, imaginal morphogenesis protein-late 2; IPC, insulin producing cells; NLaz, Neural Lazarillo; Sdr, secreted decoy of InR; Upd2, unpaired 2.
dILPs:
Drosophila ILPs vary in terms of temporal and spatial expression patterns, suggesting the evolution of unique physiological functions and specific modes of regulation [reviewed by Nässel et al. (2015)]. The current interpretation is that the various dILPs have adopted specialized roles in maintaining growth and metabolic homeostasis in different nutritional conditions and stages of the life cycle. However, analyses of individual dILP mutants have revealed that none of the dILPs are essential during development, suggesting that they can act in a redundant and compensatory manner (Grönke et al. 2010). The main site for dILP synthesis is a cluster of median neurosecretory cells referred to as the IPCs of the fly brain, where dILPs 1, 2, 3, and 5 are expressed (Brogiolo et al. 2001; Ikeya et al. 2002; Rulifson et al. 2002; Broughton et al. 2005; Colombani et al. 2012; Liu et al. 2016). Loss of the IPCs by targeted induction of cell death during early larval stages causes profound consequences to larval growth and metabolic homeostasis. The animals are developmentally delayed, have impaired growth, and possess elevated hemolymph glucose concentration. In addition, the total levels of lipids, trehalose, and glycogen are elevated in these animals (Rulifson et al. 2002; Broughton et al. 2005). These phenotypes can be reversed by the expression of dILP2 (Rulifson et al. 2002). Adult flies that develop from IPC-ablated larvae, or larvae deficient for dILPs 2, 3, and 5, contain high hemolymph glucose concentrations and elevated stored trehalose, glycogen, and lipid (Ikeya et al. 2002; Broughton et al. 2005; Grönke et al. 2010). Collectively, these findings show that the dILPs emanating from the IPCs are critical regulators of glucose metabolism, and that the physiological function of the IPCs resembles that of mammalian pancreatic β-cells.
Regulation of dILP expression and secretion by carbohydrates:
The insulin signal emanating from the IPCs is subjected to several layers of regulation. These include cell-intrinsic regulation of transcription, protein processing, and protein secretion, as well as extrinsic factors such as neurotransmitters and hormonal signals from peripheral tissues [reviewed by Nässel and Broeck (2016); Alfa and Kim (2016)]. Below, we will review the mechanisms of IPC regulation by carbohydrates in the order of (1) direct regulation of secretion, (2) indirect regulation of secretion through hormones, and (3) regulation of dILP expression.
An important regulatory mechanism of the IPCs is direct glucose sensing from hemolymph, which differs significantly between the larval and adult stages. In mammalian pancreatic islets, glucose depolarizes β-cell membrane potential by shutting down the ATP-sensitive potassium channels (KATP), leading to action potential firing and the opening of voltage-dependent Ca2+ channels [reviewed by MacDonald et al. (2005)]. Interestingly, in contrast to the adult stage, larval IPCs lack the ability to respond to glucose directly, since they do not express the KATP complex necessary for cell depolarization (Kim and Rulifson 2004). Instead, carbohydrates regulate larval IPCs indirectly, through dAKH secreted by the corpora cardiaca (CC), and through other hormonal signals originating from the fat body and intestine (Rajan and Perrimon 2012; Ghosh and O’Connor 2014; Kim and Neufeld 2015; Sano et al. 2015). The larval IPC and CC neurons send axonal projections to each other and it is therefore likely that these cells interact in the process of nutrient sensing (Rulifson et al. 2002; Kim and Rulifson 2004; Lee and Park 2004). As an additional mechanism for nutrient sensing, larval IPCs employ the hexosamine synthesis pathway and O-linked GlcNAc conjugation (Sekine et al. 2010). RNAi knockdown of Ogt or Oga in the larval IPCs either increases or decreases ILP secretion, respectively. However, the targets for O-GlcNAcylation in the IPCs remain unknown.
In contrast to the larval stage, fully developed IPCs in adult flies have been shown to respond to glucose and secrete dILPs in a manner similar to mammalian β-cells. Glucose-uptake through GLUT1 stimulates mitochondrial ATP production, which shuts down KATP channels, leading to cell depolarization, potassium influx, and dILP release through exocytosis (Kréneisz et al. 2010; Park et al. 2014). Blocking ATP production by inhibiting pyruvate transport to mitochondria prevents dILP secretion in the adult fly, suggesting that the regulation of mitochondrial metabolism is a key step in the larval to adult IPC maturation (McCommis et al. 2016). A major regulator of this transition is the Drosophila ortholog of the nuclear receptor HNF4 (Hepatocyte nuclear factor 4). Loss-of-function of HNF4 in Drosophila has a profound impact on the regulation of glycolysis, mitochondrial respiration, and insulin signaling at the adult stage. In the IPCs, HNF4 coordinates gene expression to direct a metabolic switch toward OXPHOS and glucose-induced dILP secretion (Barry and Thummel 2016).
In addition to direct glucose sensing, the IPCs are subjected to nutrient regulation through multiple signals derived from the fat body and midgut (Figure 4). Here, we focus only on the mechanisms responding to carbohydrates, as the signals responding to amino acids are extensively reviewed elsewhere (Andersen et al. 2013; Droujinine and Perrimon 2016). The fat body conveys information about carbohydrates through at least three mechanisms. These include secretion of the cytokine Unpaired 2 (Upd2), secretion of the TGF-β/activin ligand Daw, and CCHamide-2 secretion (Rajan and Perrimon 2012; Ghosh and O’Connor 2014; Sano et al. 2015). upd2 expression is upregulated in the fat body of adult flies upon high-sugar and high-fat diet feeding, and knockdown of upd2 in the fat body leads to the hallmark phenotypes of IPC-deficient flies; small size and elevated hemolymph glucose concentration (Rajan and Perrimon 2012). Interestingly, Upd2 acts on IPCs indirectly, by silencing a set of intermediate γ-aminobutyric acid (GABA)ergic neurons. Knockdown of the JAK/STAT signaling components, the receptor Dome, or the transcription factor Stat92E, in the GABAergic neuron population inhibits dILP2 secretion and causes metabolic defects (Rajan and Perrimon 2012). The GABAergic neurons hence inhibit dILP release through synaptic firing to IPCs and JAK/STAT activation through Upd2 attenuates this effect. These results suggest that Upd2 functions analogously to the Leptin adipokine system in mammals. Indeed, overexpression of human Leptin in the fly fat body can rescue the phenotypes of upd2 mutant flies. While the exact mechanism of Upd2 activation in the fat body is unknown, the TGF-β/Activin ligand Daw responds to dietary sugars in a Mondo-Mlx-dependent manner (Ghosh and O’Connor 2014; Mattila et al. 2015). Loss-of-function mutants of daw are larval lethal and possess metabolic defects reminiscent of attenuated insulin signaling. Daw signals directly to the IPCs through the TGF-β/activin receptor Baboon, regulating the secretion of dILPs 2 and 5 (Ghosh and O’Connor 2014). A third mechanism by which carbohydrates remotely promote dILP expression and secretion from the IPCs is through CCHamide-2, which is synthesized by the fat body and midgut enteroendocrine cells in a sugar-inducible manner (see below for a further discussion of CCHamides) (Sano et al. 2015).
As discussed above, glucose sensing by the IPCs is accompanied by other nutrient-derived hormonal cues. However, the significance of these different nutritional signals and how they are integrated in the IPCs to elicit a physiological dILP response is still not fully understood. For example, a detailed study of dILP expression in the adult fly across a panel of isocaloric diets differing in their protein-to-carbohydrate ratios has shown that the expression of dILPs 2, 3, and 5 peak in response to different diets (Post and Tatar 2016). dilp2 mRNA is highest in response to diets low in protein whereas dilp5 expression peaks in response to high dietary protein content. On the other hand, dilp3 expression is enhanced in animals on a low-calorie diet, with a protein-to-carbohydrate ratio of 1:8. This suggests that the dILPs expressed in the IPCs are subjected to differential nutritional regulation. The expression of dILPs 3 and 5 respond to nutrient levels during larval stages, whereas dILP2 does not (Ikeya et al. 2002). Interestingly, only a few transcription factors are known to be involved in the nutrient-regulated dILP expressions. For example, these include Sugarbabe, which was shown to repress the expression of dILPs 3 and 5 (Varghese et al. 2010). Furthermore, it is not known how the differences in dILP expression correlate with circulating dILP proteins and signaling in the peripheral tissues. In fact, direct dILP visualization in the IPCs through immunofluorescence suggests that protein secretion is the key regulatory point regarding peripheral insulin signaling (Géminard et al. 2009). Under low nutritional conditions, dILPs accumulate in the neurosecretory cells and are rapidly released upon nutritional stimulus. The recent development of ELISA immunoassays that allow direct measurement of circulating dILP levels is likely to uncover the impact of dILP transcription and secretion more precisely (Pasco and Léopold 2012; Park et al. 2014; Post and Tatar 2016).
Regulation of insulin sensitivity and carbohydrate metabolism within insulin target tissues:
The insulin-induced signaling pathway (IIS) is an ancient signaling system to control animal growth, metabolism, and differentiation. In humans, the IIS is diversified into two branches, the insulin and insulin-like growth factor pathways, which control metabolism and growth, respectively (Saltiel and Kahn 2001; Chitnis et al. 2008). Drosophila ILPs signal through a sole ortholog of Insulin-like receptor (dInR) meaning that the regulation of growth and metabolism is achieved by the same downstream signaling events (Fernandez et al. 1995; Shingleton et al. 2005). An exception to this is the relaxin-like hormone dILP8, which acts through the leucine-rich repeat-containing G protein-coupled receptor (GPCR) 3 (Lgr3) to coordinate organ growth with the timing of larval maturation (Colombani et al. 2015; Garelli et al. 2015). The dInR signals through a well-known and conserved insulin receptor substrate (IRS)/phosphoinositide 3-kinase (PI3K)/AKT pathway (Teleman 2009; Nässel et al. 2015). Upon ligand binding, dInR is autophosphorylated and binds to the IRS Chico and to the SH2B family adapter protein Lnk (Böhni et al. 1999; Werz et al. 2009). Activation of the dInR leads to the phosphorylation of Chico, providing a binding site for the lipid kinase PI3K. Elevated levels of phosphatidylinositol-(3,4,5)-triphosphates causes recruitment of the AKT and PDK1 kinases at the plasma membrane via their lipid-binding pleckstrin homology domains. AKT is then phosphorylated and activated by PDK1 and TORC2 (Rintelen et al. 2001; Yang et al. 2006; Hietakangas and Cohen 2007). Activation of AKT is central to the growth-promoting and metabolic effects of IIS, having a large number of identified phosphorylation targets. These include, for example, the inhibitory regulation of GSK-3β (Shaggy), FOXO (dFOXO), and Tuberous sclerosis complex 2 (TSC2, Gigas) (Potter et al. 2002; Puig et al. 2003; Buttrick et al. 2008; Sieber et al. 2016). The best-known transcriptional mediator of IIS is dFOXO (Puig et al. 2003; Teleman et al. 2008; Alic et al. 2011). Upon activation, AKT phosphorylates dFOXO at three conserved sites (T44, S190, and S259), which leads to its cytoplasmic retention and inactivation (Puig et al. 2003). Hence, during low IIS, dFOXO is nuclear and binds to the promoter of genes involved in macromolecular catabolism, stress resistance, growth, apoptosis, and innate immunity (Gershman et al. 2007; Alic et al. 2011). In addition, dFOXO activates a second tier of transcriptional regulators through dMyc and the ETS-family transcription factors Anterior open (Aop) and Pointed (Pnt) (Teleman et al. 2008; Alic et al. 2014). Overexpression of a constitutively nuclear mutant of dFOXO in S2 cells, or in the wing imaginal disc, leads to a strong reduction of cell proliferation, partly through the transcriptional activation of d4EBP (Jünger et al. 2003; Puig et al. 2003). The role of dFOXO in glucose metabolism is less well understood. Transcriptional profiling experiments suggest that dFOXO has a major role in mitochondrial biogenesis through repression of PGC-1 (encoded by the spargel gene in Drosophila) and mitochondrial ribosome proteins (Gershman et al. 2007). In addition, dFOXO regulates gluconeogenesis through the activation of PEPCK expression, which could explain the increased hemolymph glucose levels observed in IPC-deficient larvae and adult flies (Rulifson et al. 2002; Broughton et al. 2005; Harvey et al. 2008). However, it is clear that additional players are involved in IIS-mediated carbohydrate homeostasis. This is exemplified by the finding that a direct target of dILPs, an α glucosidase encoded by the tobi gene, is regulated independently of dFOXO (Buch et al. 2008). One such factor could be dMyc, which is positively regulated by IIS through the TOR signaling branch and shares common transcriptional targets with dFOXO (Teleman et al. 2008; Li et al. 2010).
Perhaps the most prominent metabolic feature of IIS activation is the accumulation of stored triglyceride reserves manifested by the increase in lipid droplet number within fat body cells (Britton et al. 2002). One possible mechanism for IIS-induced lipogenesis is the AKT-mediated activation of the Sterol response element-binding protein (Porstmann et al. 2008). Yet surprisingly little is known about the direct mechanisms of IIS-induced lipogenesis in vivo. The IIS-induced transcriptional response has been studied by measuring gene expression from PI3K-overexpressing larvae (Li et al. 2010). Interestingly, only a few genes directly associated to the regulation of carbohydrate and fatty acid metabolism where found regulated in this data set. These included, for example, Hexokinase A (hex-A) and a long-chain fatty acid-CoA ligase (bgm) as well as the transcription factor Sugarbabe. These results suggest that the transcriptional program downstream of IIS is tissue-specific, and a more refined experimental strategy is needed to reveal the full spectrum of metabolic regulation by insulin-like signaling in the fly.
The physiological response to the activation of the IIS cascade is tightly regulated through negative feedback mechanisms, such as the inhibition of dAKT by TOR complex 1 (Kockel et al. 2010). Perturbations in these mechanisms might lead to insulin resistance, where even elevated levels of insulin are unable to activate the IIS cascade. For example, a high-sugar diet has been shown to lead to insulin resistance (Musselman et al. 2011; Pasco and Léopold 2012). This happens via a lipocalin-like protein Neural Lazarillo (NLaz) secreted from the fat body in a JNK-dependent manner (Hull-Thompson et al. 2009; Pasco and Léopold 2012). Mutant animals for NLaz contain less glucose, glycogen, and triglycerides, whereas overexpression of NLaz protein in the fat body results in higher glucose titers compared to control animals. In addition, mutant NLaz fat body cells have higher PI3K activity (Hull-Thompson et al. 2009). Taken together, the results suggest a model where upon organismal stress, such as high circulating sugars, NLaz expression is activated in the fat body through JNK, which leads to dampening of IIS cascade sensitivity. In addition, Drosophila IIS is modulated by secreted hormonal cues directly interacting with circulating dILPs. Some of these factors resemble the mammalian IGF-binding proteins (IGFBPs), which have several functions, ranging from carrier proteins to modulators of signaling activity (Duan and Xu 2005). An ortholog of the IGFBPs in Drosophila is the Imp-L2 protein, which is expressed in the fat body, CC, and IPCs. Imp-L2 binds to dILP2 and prevents downstream signaling under nutrient-deprived conditions (Honegger et al. 2008). Interestingly, the Imp-L2/dILP2 complex includes another IGFBP member, namely the Drosophila ortholog of Acid-labile subunit (ALS), dALS (Arquier et al. 2008). dALS is expressed in the fat body and IPCs, and its expression is strongly suppressed upon amino acid starvation. Overexpression or knockdown of dALS in the larval fat body reduces or increases adult body size, respectively. Furthermore, dALS overexpression in the fat body antagonizes the metabolic defects of dILP2 overexpression, including lowered circulating trehalose and higher total triglyceride levels. Interestingly, whereas dALS acts specifically on the Imp-L2/dILP2 complex, a glia-derived factor, referred to as Secreted Decoy of InR (SDR), binds with highest affinity to dILP3, independent of Imp-L2 or dALS (Okamoto et al. 2013). SDR antagonizes IIS under adverse dietary conditions.
dAKH, the Drosophila counterpart of glucagon:
Glucagon and AKHs are peptide hormones important for the maintenance of physiological levels of circulating sugars. AKH peptides were initially identified from a variety of insect species, mainly by immunochemical assays of crude extracts of CC, and were shown to be essential for energy regulation during insect flight [reviewed by Gäde (1990)]. dAKH is synthesized as a preprohormone containing a signal peptide, a single AKH of eight amino acids, and a C-terminal AKH precursor-related peptide (Schaffer et al. 1990; Noyes et al. 1995; Galikova et al. 2015). dAKH is enzymatically processed by proprotein convertases (PCs) before the active hormone is released into circulation. A mutant of the Drosophila PC-encoding gene amontillado (amon) phenocopies the loss of dAKH signaling, and is necessary and sufficient for dAKH-mediated regulation of carbohydrate metabolism. In addition, direct peptide profiling from CC cells has shown that amon mutants lack a mature dAKH peptide, linking it to dAKH maturation (Rhea et al. 2010).
The dAKH gene is expressed exclusively in the CC neuroendocrine cells of larvae and adults (Isabel 2004; Kim and Rulifson 2004; Lee and Park 2004). In larvae, the AKHergic neurons make connections to the prothoracic gland, IPCs, and dorsal vessel (heart), where the peptide is released into the hemolymph (Kim and Rulifson 2004; Lee and Park 2004). In the adult, AKHergic neurons send axons to the brain (protocerebrum) and crop (Lee and Park 2004). Targeted ablation of the AKHergic neurons by overexpression of the proapoptotic gene reaper strongly reduces trehalose levels in the larval hemolymph (Isabel 2004; Kim and Rulifson 2004; Lee and Park 2004). This is consistent with the idea that dAKH functions like mammalian glucagon by releasing carbohydrates into circulation. CC-ablated adult flies are also more resistant to starvation and a show lack of starvation-induced hyperactivity (Isabel 2004; Lee and Park 2004). Analysis of dAKH mutants showed that AKH signaling is an essential mechanism in lipid catabolism and maintenance of normoglycemia in the adult fly, but is dispensable for larval energy metabolism. These findings suggest that larval CC ablation might have other, dAKH-independent consequences or that the role of AKH signaling is conditional and dependent on the larval nutrition uptake (Galikova et al. 2015).
dAKH release into the hemolymph is regulated through membrane depolarization by ATP-sensitive K+ -channels. These channels serve as intracellular AMP/ATP sensors that control membrane potential and hormone secretion. A rapid decrease in trehalose concentration triggers calcium influx into CC cells, which induces the release of dAKH into the hemolymph (Kim and Rulifson 2004). The inward flux of calcium into the CC cells, and subsequent dAKH secretion, is dependent on AMPK (Braco et al. 2012). Surprisingly, dAKH secretion is also triggered by high hemolymph trehalose concentration (Kim and Neufeld 2015). In addition, a recent study by Song et al. (2017) showed that larvae fed a high-sugar diet had higher dAKH signaling output in the fat body. The inhibition of this signal by knocking down dAKH downstream signaling components AKH receptor (AKHR), Ire1, Creb2, or CBP by RNAi significantly alleviates the high-sugar diet-promoted hyperglycemia (Song et al. 2017). Together, these observations suggest a model of biphasic regulation of dAKH, where its secretion is promoted by low and high hemolymph trehalose concentrations. Such regulation can be understood by the necessity to maintain constant hemolymph trehalose concentrations during the rapid larval growth phase, when insulin signaling is high, as well as during the wandering and pupal stages when feeding has ceased. In comparison, high glucose concentrations stimulate glucagon secretion from mouse pancreatic islets and glucagon, further promoting hyperglycemia in diabetic humans (Jiang and Zhang 2003; Salehi et al. 2006). Hence, it is possible that the Drosophila CC responds to sugars in a similar manner as the mammalian pancreatic α-cells. Further studies in well-defined nutritional regimes are required to uncover the elaborate regulation of dAKH.
Regulation of carbohydrate metabolism in dAKH target tissues:
The fly genome encodes one AKH-responsive GPCR, AKHR, which is expressed in the larval and adult fat body (Grönke et al. 2007; Bharucha et al. 2008). Consistent with the view of AKH being a lipolytic and glycolytic regulator, AKHR mutant animals display elevated triglyceride and glycogen levels compared to control animals, and are more resistant to starvation, probably due to changes in energy expenditure and reduced locomotor activity (Grönke et al. 2007; Bharucha et al. 2008). Overexpression of AKHR in the fat body leads to reduced triglyceride and glycogen stores (Grönke et al. 2007; Bharucha et al. 2008). The mechanism of GPCRs and the downstream intracellular events are well-documented in various model systems, as well as in humans (Pavlos and Friedman 2016). The downstream events following AKH binding to its receptor have also been studied in other insect species. For example, in adipocytes of the Lepidopteran Manduca sexta, AKHR activation leads to the cellular increase of classical second-messengers cAMP and Ca2+ (Arrese et al. 1999). In Drosophila, the GPCR signal transducers G protein α q subunit (Gαq), G protein γ1 (Gγ1), and Phospholipase C at 21C (Plc21C) control cellular and organismal fat storage downstream of AKHR (Baumbach et al. 2014). Genetic modulation of the GPCR signaling components leads to an impairment of intracellular Ca2+ through the inhibition of Store-Operated Calcium Entry (SOCE). As a consequence, lipid mobilization from the fat body is blocked through the regulation of Brummer lipase and diacylglycerol O-acyltransferase midway gene expression (Baumbach et al. 2014). Further details about AKHR signaling have been revealed in a recent study by Song et al. (2017), who showed that Drosophila AKHR employs an analogous signaling mechanism to mammalian glucagon, through the PKA-IRE-CREB2 pathway. Song et al. (2017) also elucidated a novel interaction between Activin and AKHR signaling in the fat body of chronically high-sugar-fed larvae. Activinβ derived from midgut enteroendocrine cells signals through the type I TGF-β receptor Babo and downstream transcription factor dSmad2 in the fat body to regulate AKHR expression, resulting to hyperglycemia. Transcriptomic analysis of fat bodies overexpressing dAKH has revealed a breadth of downstream metabolic processes, including the PPP, glycolysis, and gluconeogenesis. Interestingly, dAKH and IIS were also shown to interact under conditions of high circulating trehalose through the regulation of dILP3 (Kim and Neufeld 2015). As a response to trehalose and dAKH signaling, elevated levels of circulating dILP3 were shown to both activate mTOR signaling in the larval fat body and prevent autophagy.
Regulation of carbohydrate metabolism by transforming growth factor β/Activin signaling:
The Drosophila TGF-β family signaling pathway has two separate branches that utilize different ligands, namely, the bone morphogenetic proteins (BMPs) and Activins (Upadhyay et al. 2017). The BMP and Activin ligands signal through a separate set of receptors and downstream effectors. Recent studies have revealed that the Activin branch, which signals through the Babo receptor, has an important role in carbohydrate metabolism. This branch includes three ligands: Daw, Activinβ, and Myoglianin (Upadhyay et al. 2017). Daw is highly expressed in the fat body and muscles (Bai et al. 2013; Mattila et al. 2015). Its expression is strongly induced by sugar feeding and, at least in larvae, the majority of sugar-induced gene expression of Daw is mediated by Mondo-Mlx, which binds to the dawdle promoter (Mattila et al. 2015). Moreover, Daw is a direct target of FOXO, which negatively regulates its expression (Bai et al. 2013).
dawdle-null mutants display sugar intolerance similar to that of mlx mutants (Ghosh and O’Connor 2014; Mattila et al. 2015). On a carbohydrate-rich diet, most mutants die during larval stages and display delayed development, while the duration of larval development is normal and mutants pupariate in high numbers on a yeast diet. Moreover, dawdle mutants have high circulating trehalose and glucose as well as high glycogen and triacylglycerol levels (Ghosh and O’Connor 2014). Loss of dawdle also causes hemolymph acidification, possibly due to an accumulation of acidic TCA cycle intermediates. Daw seems to affect metabolic homeostasis through multiple mechanisms. It promotes secretion of dILPs, providing one of the many hormonal links between peripheral tissues and the IPCs (Ghosh and O’Connor 2014). Moreover, fat body-derived Daw influences signaling in the intestine, inhibiting expression of Amylases upon sugar feeding (Chng et al. 2014). Daw also contributes to the full activation of the sugar-responsive transcription factor Sugarbabe, possibly providing a feed-forward mechanism to the Mondo-Mlx-dependent activation of Sugarbabe (Mattila et al. 2015). Daw also maintains proteostasis in muscles, thereby extending life span (Bai et al. 2013). In addition to Daw, Activinβ was recently shown to contribute to carbohydrate metabolism. Chronic sugar feeding upregulates the expression of Activinβ from the enteroendocrine cells of the midgut (Song et al. 2017). It signals to the fat body, where it activates AKH signaling by upregulating AKHR expression, consequently causing hyperglycemia. In conclusion, Activin ligands are emerging as important carbohydrate-responsive signals that emanate from peripheral tissues.
CCHamides, emerging sugar-responsive hormones:
CCHamide is a short peptide hormone originally found in silkworms (Bombyx mori) (Roller et al. 2008). Subsequent work has led to the identification of two CCHamide genes, CCHamide-1 and -2, in Drosophila (Hansen et al. 2011). CCHamide-1 and -2 signal through their respective GPCRs, which are homologs of Bombesin Receptor Subtype 3 (BRS-3) in mammals. Mice lacking BRS-3 develop mild obesity and display impaired glucose metabolism (Ohki-Hamazaki et al. 1997). CCHamide-2 is expressed mainly in the fat body and gut endocrine cells and its expression is nutrient-dependent (S. Li et al. 2013; Sano et al. 2015). It is downregulated by starvation and activated by refeeding with nutritious sugars (Sano et al. 2015). CCHamide-2 Receptor (CCHamide-2 R) is expressed mainly in the CNS, displaying high levels in the IPCs (Sano et al. 2015). CCHamide-2 promotes secretion of dILP2 and dILP5 as well as expression of dILP5 in the IPCs. Consequently, mutants of CCHamide-2 R are growth impaired. Moreover, mutants of CCHamide-2 display strongly reduced feeding activity in both larvae and adults (Ren et al. 2015). In conclusion, CCHamide-2 is a carbohydrate-responsive hormone that mediates information from peripheral tissues to the CNS.
Part III
Physiological processes linked to carbohydrate metabolism
Circadian clock and carbohydrate metabolism:
Adult Drosophila feeding activity follows a circadian rhythm, with the highest feeding activity during the first few hours of daylight (Xu et al. 2008; Seay and Thummel 2011). This periodic feeding is reflected in the carbohydrate homeostasis of the animal, as circulating trehalose and glycogen levels increase a few hours after the highest feeding activity and are then gradually consumed during the remainder of the day (Seay and Thummel 2011). In contrast, triacylglycerol and protein levels do not display circadian oscillation (Seay and Thummel 2011). The circadian timekeeping system includes the central clock located in the brain and is composed of ∼150 clock-expressing neurons, as well as the peripheral clocks present in several peripheral tissues (Ito and Tomioka 2016). Cycling of the Drosophila feeding activity is controlled by the peripheral clock (Xu et al. 2008). In fact, a large number of metabolic genes, including Zw, display cyclic expression in the fat body, which depends on tissue autonomous clock activity (Xu et al. 2011). Moreover, flies lacking a functional clock in the fat body have significantly reduced glycogen storage along with starvation sensitivity, despite the fact that their total food consumption is higher than in control flies.
The interaction between feeding and the circadian clock is bidirectional, as the circadian clock can be reset by time-controlled feeding (Catterson et al. 2010; Xu et al. 2011). One mechanism mediating nutrient-dependent resetting of the circadian clock is through protein O-GlcNAcylation. O-GlcNAcylation is regulated in a circadian manner with inhibition of Drosophila Ogt in clock cells shortening the circadian period, while increased O-GlcNAcylation has the opposite effect (Kim et al. 2012). Central clock proteins, including Clock and Period, are modified by O-GlcNAc, which modulates their transcriptional activity (Kim et al. 2012; Kaasik et al. 2013). Protein O-GlcNAcylation is directly affected by the activity of the HBP, which is sensitive to glucose availability, providing a potential means for nutrient-dependent resetting of the circadian clock. Another point of interaction between nutrient sensing and circadian clock activity is through Mondo-Mlx-mediated intracellular sugar sensing. Mondo-Mlx directly regulates the expression of the Krüppel-like transcription factor Cabut (Havula et al. 2013; Bartok et al. 2015). The cabut promoter region is also bound by the circadian transcription factor CLK (Abruzzi et al. 2011), suggesting that sugar sensing and the circadian clock converge on Cabut regulation. Furthermore, Cabut overexpression leads to severe defects in circadian locomotor activity rhythms and deregulation of circadian cycling of metabolic targets, while having no effect on the core clock components (Bartok et al. 2015).
Regulation of carbohydrate metabolism upon developmental transitions:
During its life cycle, Drosophila undergoes different developmental stages with distinct metabolic needs. For example, during the larval stage, the body mass of Drosophila increases rapidly by ∼200-fold, which requires metabolic reprogramming into an anabolic mode. Temporal analysis of gene expression during the embryonic stage has revealed widespread changes in metabolic gene expression before the transition from an embryo to a larva, which is termed the embryonic metabolic transition (EmbMT) (Tennessen et al. 2014a). The genes activated during the EmbMT encode glycolytic enzymes, lactate dehydrogenase, as well as TCA cycle components and other mitochondrial metabolic enzymes. Along with gene expression changes, metabolite profiles of embryos change during embryogenesis (An et al. 2014; Tennessen et al. 2014a). During embryonic development, the animal consumes its triacylglycerol and glycogen stores and concomitantly accumulates glycerol-3-phosphate. At the onset of the EmbMT, uric acid levels increase dramatically, possibly reflecting catabolism of nitrogen-containing metabolites (Tennessen et al. 2014a). Concomitantly, levels of some amino acids, such as glutamate and aspartate, decline (An et al. 2014).
The EmbMT metabolically prepares the animal for the transition into larval development, when the animal starts feeding and growing. Interestingly, the metabolic profile of larvae includes high glycolytic activity and the production of lactate (Tennessen et al. 2011). Although the involvement of tissue-specific hypoxia has not been ruled out, the metabolic profile of growing larvae resembles the Warburg-type metabolism of malignant and other highly proliferative cells, which rely on high rates of biosynthesis. Furthermore, Drosophila larvae produce high levels of L-2-hydroxyglutarate (L-2HG), a metabolite found at high levels in cancers (Li et al. 2017). L-2HG is a product of lactate dehydrogenase activity, which is highly upregulated upon the EmbMT. L-2HG accumulation may have functional relevance since it inhibits 2-oxoglutarate-dependent dioxygenases, which are important epigenetic regulators.
A key regulator of the EmbMT is the Estrogen-Related Receptor (ERR) (Tennessen et al. 2011). ERR expression is upregulated during late embryogenesis. ERR mutants die during the second larval instar and display severe metabolic problems, for example low levels of ATP and high levels of both trehalose and sorbitol. At the gene expression level, ERR mutants fail to activate glycolytic gene expression and consequently have low levels of L-2HG (Y. Li et al. 2013). It will be interesting to learn how the developmental metabolic switch interacts with environmental signals influencing carbohydrate metabolism. In fact, it is already known that ERR binds to HIF and is essential for activating HIF-dependent gene expression upon hypoxia (Y. Li et al. 2013). Furthermore, ERR is essential for HIF-independent gene regulation upon hypoxia, including upregulation of glycolytic transcripts, but how ERR mediates these HIF-independent responses remains to be elucidated.
Another metabolic transition during Drosophila development occurs during metamorphosis when mitochondrial respiration is very low, and it is strongly activated at the onset of the adult stage (Merkey et al. 2011). This corresponds to a dramatic change in the animal’s mobility and feeding activity. Interestingly, the levels of HNF4 expression also strongly increase upon the transition to adulthood, which is followed by an upregulation of HNF4 downstream genes, including genes involved in glucose metabolism and mitochondrial OXPHOS (Barry and Thummel 2016). Notably, the effects of HNF4 on sugar tolerance are also observed during the late pupal stage or adult stage, in contrast to the larval phenotypes observed in mlx mutants (Havula et al. 2013). Thus, it is possible that carbohydrate metabolism is coordinated by distinct transcription factors during different developmental stages. As mentioned before, Drosophila IPCs undergo maturation during development. In the larval stage IPCs are nonresponsive to glucose, whereas in adults dILP secretion is glucose-responsive. Interestingly, IPC-specific knockdown of HNF4 prevents the transition to glucose responsiveness. Similarly, mutations in human HNF4a cause Mature-Onset Diabetes of the Young I (MODY I), with impaired pancreatic β-cell function (Fajans and Bell 2011). In conclusion, HNF4 appears to mediate the developmental switch to rewire carbohydrate metabolism into the mode of high mitochondrial respiration and trigger the maturation of IPCs into glucose responsiveness upon reaching adulthood.
During developmental morphogenesis, the growth of individual cells is also closely regulated. Notch signaling promotes cell proliferation in specific developmental contexts, including the wing imaginal disc (Djiane et al. 2013). In this setting, Notch signaling activates the expression of genes involved in glucose uptake and glycolysis through its downstream effector, Suppressor of Hairless (Slaninova et al. 2016). On the other hand, Notch signaling inhibits the expression of genes of the TCA cycle by activating the transcriptional repressor Hairy (Slaninova et al. 2016). Through these gene expression changes, Notch signaling reprograms cellular metabolism to match the needs of proliferative cells.
Part IV
Drosophila as a model of carbohydrate metabolism-related pathophysiologies
Pathophysiologies of deregulated glycogen metabolism:
In addition to understanding normal carbohydrate physiology, Drosophila has been increasingly used to model pathophysiologies related to ones observed in humans. Defects in glycogen autophagy lead to myopathies in humans. These myopathies can be hereditary or caused by drugs, such as chloroquine, which is used for the treatment of malaria. Interestingly, chloroquine feeding to Drosophila larvae leads to a dramatic accumulation of glycogen in autophagic vesicles upon starvation (Zirin et al. 2013). In addition, muscle sarcomere structure is impaired and locomotor function is reduced, suggesting that chloroquine-treated larvae can be used as a genetically tractable model to study myopathies caused by defective glycogen autophagy (Zirin et al. 2013). The accumulation of glycogen into autophagosomes can be prevented by inhibition of the autophagy machinery as well as activation of TOR signaling, which is known to regulate starvation-induced autophagy. Moreover, inhibition of GlyS in the muscle inhibits glycogen accumulation in chloroquine-treated larvae. Thus, GlyS function, either by driving glycogen synthesis or by acting as a scaffold between glycogen and the autophagy machinery, is essential for the myopathy phenotype (Zirin et al. 2013).
Glycogen synthesis needs to be kept under strict tissue-specific control. Aberrant accumulation of glycogen in neurons coincides with aggressive neurodegeneration in humans, as observed in Lafora disease. In Drosophila, deregulated glycogen synthesis leads to neuronal loss, reduced locomotion, and short life span (Duran et al. 2012), demonstrating the causal relationship between neuronal glycogen and neurodegeneration. Also during the physiological aging process, glycogen clusters accumulate in neuronal processes. Inhibition of Drosophila GlyS expression in neurons improves neurological function with age and extends life span (Sinadinos et al. 2014). In conclusion, Drosophila has emerged as an important model to understand the regulation of glycogen metabolism and the underlying pathophysiologies.
Modeling diabetes and its complications in Drosophila:
While Drosophila displays a high degree of flexibility with respect to macronutrient content, chronic feeding of superphysiological concentrations (1 M) of sugar has been shown to cause a phenotype that resembles human obesity and diabetes. Larvae grown on a diet with high sucrose, but not on high fat or high protein diets, display high hemolymph glucose and high triacylglycerol levels (Musselman et al. 2011). This suggests that the physiological mechanism to cope with high-sugar intake and maintain glucose homeostasis has been saturated in this setting. Furthermore, high-sugar-fed larvae show signs of insulin resistance, including reduced phosphorylation of protein kinase AKT, a component of the IIS pathway. In adult animals, high-sugar feeding increases triglyceride storage while dietary sugar levels > 10% also shorten life span and reduce fecundity (Skorupa et al. 2008). Similar to larvae, signs of insulin resistance are observed in high-sugar-fed adults (Morris et al. 2012).
Stable isotope tracer experiments have unraveled differences in the metabolic profiles of larvae fed a high-sugar diet. High-sugar diet feeding leads to reduced fatty acid chain length and increased levels of desaturation, which are correlated with elevated expression of stearoyl-CoA desaturase 1 (Musselman et al. 2013), a target of Mondo-Mlx (Havula et al. 2013). Interestingly, loss-of-function of king tubby, a homolog of the mammalian obesity-associated gene tubby (Coleman and Eicher 1990; Shiri-Sverdlov et al. 2006), increase triglyceride storage and concomitantly lead to lower circulating glucose levels. Similar observations have been made following overexpression of Sugarbabe, a lipogenic Gli-similar transcription factor (Mattila et al. 2015). Thus, the lipogenic capacity of the fat body appears to be positively reflected in the ability to maintain the homeostasis of hemolymph glucose. High-sugar diet feeding also increases the levels of nonesterified fatty acids (NEFAs) (Musselman et al. 2013). Biosynthetic genes of CoA, a cosubstrate for lipogenesis, are activated following high-sugar diet feeding, and the inhibition of CoA biosynthesis leads to increased levels of NEFAs (Palanker Musselman et al. 2016). On the other hand, dietary supplementation of the CoA precursor pantothenate facilitates triglyceride biosynthesis, consequently lowering the levels of NEFAs and circulating glucose. Notably, however, this dietary intervention does not improve insulin sensitivity, suggesting that hemolymph glucose and lipid homeostasis can be uncoupled from insulin sensitivity (Palanker Musselman et al. 2016). One candidate for mediating the development of insulin resistance upon high-sugar feeding is the lipocalin NLaz. Lipocalins are secreted proteins, which bind small hydrophobic ligands and are known to promote insulin resistance in mammals (Yan et al. 2007). NLaz is under the control of the stress-activated JNK signaling pathway (Hull-Thompson et al. 2009), and it displays strongly elevated expression following high-sugar diet feeding (Pasco and Léopold 2012). Loss of NLaz expression suppresses insulin resistance in the fat body and improves the clearance of circulating glucose (Pasco and Léopold 2012).
Other approaches to use Drosophila as a model for understanding diabetes and other metabolic pathophysiologies include genetic screens with type 2 diabetes-associated genes as well as screens to identify novel genes that affect circulating glucose levels. While genome-wide association (GWAS) studies have been powerful in identifying genomic regions associated with metabolic disorders in human populations, the causative gene often remains uncertain. Complementing GWAS studies with functional experiments in Drosophila allows systematic discovery of candidate genes with an in vivo phenotype. Pendse and co-workers analyzed the sugar intolerance phenotypes of 83 Drosophila genes, which were homologs of human genes associated with type 2 diabetes or related metabolic traits (Pendse et al. 2013). This approach led to the identification of several new genes essential for sugar tolerance, including the homeobox transcription factor HHEX, mediating its function via activity in the intestine (Pendse et al. 2013). Another strategy utilizing Drosophila to identify human disease-associated candidate genes involves comparative analysis of gene expression data. Variants of the sugar sensing transcription factor ChREBP (MLXIPL, the mammalian ortholog of Drosophila Mondo) are strongly associated with circulating triglyceride levels in human (Kooner et al. 2008). Interestingly, human homologs of Drosophila Mondo-Mlx target genes are significantly overrepresented in the vicinity of triglyceride-associated SNPs, suggesting that novel candidate genes can be discovered through such comparative approaches (Mattila et al. 2015). RNAi screening for novel genes that affect circulating glucose levels have also uncovered > 150 candidate genes involved in circulating glucose homeostasis (Ugrankar et al. 2015). These were further divided into muscle and fat body-specific genes, which displayed substantial overlap. Moreover, most genes that affect circulating glucose levels do not have a similar effect on trehalose, implying that these two forms of circulating sugars are independently regulated. This emphasizes the need to measure levels of both circulating metabolites when characterizing the metabolic phenotype of Drosophila mutants. Roughly half of the identified genes had been implicated in diabetes before, but several novel regulators of glucose homeostasis were observed, including the protein kinase-encoding gene Ck1α (Ugrankar et al. 2015). Ck1α displays a conserved role in maintaining glucose homeostasis, as adipose tissue-specific loss of the Ck1α homolog (CSNK1a1) leads to hyperglycemia in mice.
Untreated diabetes leads to several secondary problems, including kidney failure and diabetic nephropathy (Sharma et al. 2017). The Drosophila nephrocyte has anatomical, molecular, and functional similarities to the glomerular podocyte of vertebrates, a cell that forms the main size-selective barrier in the kidney (Weavers et al. 2009). A high-sucrose diet, which elevates circulating glucose, damages the nephrocytes, suggesting that Drosophila is a valid model for diabetic nephropathy (Na et al. 2015). A high-sucrose diet leads to loss of the Nephrin-like protein Sns, which is mediated by a glucosamine-dependent pathway (Na et al. 2015). This suggests that inhibition of the HBP might be a strategy to attenuate diabetic nephropathy. A sugar-rich diet and elevated circulating glucose levels also increase the risk of heart disease. In Drosophila, high dietary sugars increase cardiac arrhythmias and lead to structural deterioration of heart tissue (Na et al. 2013). Similar to the nephrocyte, this sugar-dependent toxicity can be alleviated by inhibition of the hexosamine biosynthesis pathway. These studies underline the possibility of using the fly as a model to test strategies for selective inhibition of the HBP and/or O-GlcNAc modification in treating diet-derived organ deterioration.
Mechanisms of dietary sugar-induced tumor growth:
An unbalanced diet, obesity, and diabetes increase the risk of cancer (Khandekar et al. 2011). In Drosophila, specific combinations of oncogenes will produce tumor-like uncontrolled overgrowth, which allows the identification of genetic and dietary modifiers of tumor growth and metastasis (Gonzalez 2013). For example, a high-sugar diet strikingly increases the growth of Ras/Src-transformed tumors (Hirabayashi et al. 2013). Furthermore, sugar feeding converts local tumors into metastatic ones, thus significantly increasing their aggressiveness. High-sugar diet feeding activates salt-inducible kinases, which inhibit the growth-suppressing Hippo signaling pathway (Wehr et al. 2013; Hirabayashi and Cagan 2015). Consequent activation of the transcriptional cofactor Yorkie, which increases Wingless signaling in addition to driving the expression of growth-promoting and antiapoptotic genes, drives tumorigenesis (Hirabayashi and Cagan 2015). Wingless, in turn, promotes insulin receptor gene expression to promote insulin sensitivity of the tumor tissue, leading to a feed-forward cycle that enhances the malignant phenotypes of the tumors (Hirabayashi et al. 2013). While it remains uncertain which aspects of the sugar-induced growth of Drosophila tumors can be applied to human cancer, the fact that nutrient sensing and growth control pathways are highly conserved implies that this topic deserves a deeper look. However, the novel concepts emerging from Drosophila research should be actively tested in human tumor models.
Concluding Remarks and Future Prospects
Recent years have brought to light a number of new mechanisms that control Drosophila carbohydrate metabolism. These include discoveries of regulatory networks, both intracellular and systemic, which control organismal carbohydrate homeostasis by directing specific metabolic responses in both an organ and cell type-specific manner. Drosophila developmental stages have unique metabolic needs, which has enabled research on metabolic reprogramming, for example the shift from the larval biosynthetic growth phase into energy-intensive high OXPHOS metabolism of the adult. Considering the importance of metabolic reprogramming in stem cells and cancer, an increased understanding of such mechanisms may have relevance much beyond Drosophila development. High-sugar intake and impaired carbohydrate metabolism are implicated in an increasing number of health conditions within human populations. Key advantages of Drosophila disease models are their amenability to a wide range of defined diets, which allows discovery of interactions between genes and nutrients, as well as the powerful genetic toolkit, which can be utilized in unbiased screens.
While research along the aforementioned topics will undoubtedly continue, new questions are likely to emerge. These include specific roles for different dietary sugars. In fact, some existing literature already indicates disparate physiological outcomes for different dietary sugars (Rovenko et al. 2015). As carbohydrate metabolism is connected to the whole carbon metabolism of the animal, future studies will likely address the complex interactions between carbohydrate metabolism and other types of nutrients, including macro- and micronutrients. For such questions, the use of fully defined “holidic” Drosophila diets will be an asset (Lee and Micchelli 2013; Piper et al. 2014). In addition, an important factor affecting animal metabolism is the intestinal microbiota. Interestingly, recent evidence shows that chronic high-sugar feeding influences Drosophila microbiota, promoting uracil-secreting bacteria (Whon et al. 2017). Although normally considered as pathogenic, these bacteria protect the host against the deleterious effects of a high-sugar diet. This underlines the need for a more careful look into the interactions between Drosophila dietary carbohydrates and microbiota. Finally, it should be recognized that the Drosophila genus includes a wealth of different species, each metabolically adapted to distinct nutritional conditions. In addition to insight into evolution, understanding the molecular mechanisms that underlie the natural variation of Drosophila species will provide a new understanding of the complex regulation of carbohydrate metabolism.
Acknowledgments
We thank members of the Hietakangas group for valuable feedback, Michael Boutros for feedback and support, and Paula Jouhten for help with the figures. Research on carbohydrate metabolism in the V.H. group is supported by the Academy of Finland (grant no. 286767), the Sigrid Juselius Foundation, the Novo Nordisk Foundation (NNF16OC0021460), and The Diabetes Research Foundation. The work of J.M. is supported by the Sigrid Juselius Foundation.
Footnotes
Communicating editor: C. Thummel
Literature Cited
- Abruzzi K. C., Rodriguez J., Menet J. S., Desrochers J., Zadina A., et al. , 2011. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 25: 2374–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson A. W., Suchankova G., Rufo C., Nakamura M. T., Teran-Garcia M., et al. , 2006. Hepatocyte nuclear factor-4alpha contributes to carbohydrate-induced transcriptional activation of hepatic fatty acid synthase. Biochem. J. 399: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejandro E. U., Bozadjieva N., Kumusoglu D., Abdulhamid S., Levine H., et al. , 2015. Disruption of O-linked N-Acetylglucosamine signaling induces ER stress and β cell failure. Cell Rep. 13: 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfa R. W., Kim S. K., 2016. Using Drosophila to discover mechanisms underlying type 2 diabetes. Dis. Model. Mech. 9: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic N., Andrews T. D., Giannakou M. E., Papatheodorou I., Slack C., et al. , 2011. Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol. Syst. Biol. 7: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic N., Giannakou M. E., Papatheodorou I., Hoddinott M. P., Andrews T. D., et al. , 2014. Interplay of dFOXO and two ETS-family transcription factors determines lifespan in Drosophila melanogaster. PLoS Genet. 10: e1004619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P. N., Yamaguchi M., Bamba T., Fukusaki E., 2014. Metabolome analysis of Drosophila melanogaster during embryogenesis. PLoS One 9: e99519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen D. S., Colombani J., Léopold P., 2013. Coordination of organ growth: principles and outstanding questions from the world of insects. Trends Cell Biol. 23: 336–344. [DOI] [PubMed] [Google Scholar]
- Arquier N., Géminard C., Bourouis M., Jarretou G., Honegger B., et al. , 2008. Drosophila ALS regulates growth and metabolism through functional interaction with insulin-like peptides. Cell Metab. 7: 333–338. [DOI] [PubMed] [Google Scholar]
- Arrese E. L., Flowers M. T., Gazard J. L., Wells M. A., 1999. Calcium and cAMP are second messengers in the adipokinetic hormone-induced lipolysis of triacylglycerols in Manduca sexta fat body. J. Lipid Res. 40: 556–564. [PubMed] [Google Scholar]
- Bai H., Kang P., Hernandez A. M., Tatar M., 2013. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet. 9: e1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K. D., Thummel C. S., 2007. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 6: 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K. K., Ayyub C., Sengupta S., Kolthur-Seetharam U., 2012. dSir2 deficiency in the fatbody, but not muscles, affects systemic insulin signaling, fat mobilization and starvation survival in flies. Aging 4: 206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee K. K., Ayyub C., Sengupta S., Kolthur-Seetharam U., 2013. Fat body dSir2 regulates muscle mitochondrial physiology and energy homeostasis nonautonomously and mimics the autonomous functions of dSir2 in muscles. Mol. Cell. Biol. 33: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry W. E., Thummel C. S., 2016. The Drosophila HNF4 nuclear receptor promotes glucose-stimulated insulin secretion and mitochondrial function in adults. Elife 5: e11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartok O., Teesalu M., Ashwall-Fluss R., Pandey V., Hanan M., et al. , 2015. The transcription factor Cabut coordinates energy metabolism and the circadian clock in response to sugar sensing. EMBO J. 34: 1538–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach J., Xu Y., Hehlert P., Kühnlein R. P., 2014. Gαq, Gγ1 and Plc21C control Drosophila body fat storage. J. Genet. Genomics 41: 283–292. [DOI] [PubMed] [Google Scholar]
- Becker A., Schlöder P., Steele J. E., Wegener G., 1996. The regulation of trehalose metabolism in insects. Experientia 52: 433–439. [DOI] [PubMed] [Google Scholar]
- Beller M., Bulankina A. V., Hsiao H.-H., Urlaub H., Jäckle H., et al. , 2010. PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 12: 521–532. [DOI] [PubMed] [Google Scholar]
- Benkel B. F., Hickey D. A., 1986. Glucose repression of amylase gene expression in DROSOPHILA MELANOGASTER. Genetics 114: 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkel B. F., Hickey D. A., 1987. A Drosophila gene is subject to glucose repression. Proc. Natl. Acad. Sci. USA 84: 1337–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha K. N., Tarr P., Zipursky S. L., 2008. A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. J. Exp. Biol. 211: 3103–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhni R., Riesgo-Escovar J., Oldham S., Brogiolo W., Stocker H., et al. , 1999. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell 97: 865–875. [DOI] [PubMed] [Google Scholar]
- Bond M. R., Hanover J. A., 2015. A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 208: 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braco J. T., Gillespie E. L., Alberto G. E., Brenman J. E., Johnson E. C., 2012. Energy-dependent modulation of glucagon-like signaling in Drosophila via the AMP-activated protein kinase. Genetics 192: 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker D. K., Taylor E. B., Schell J. C., Orsak T., Boutron A., et al. , 2012. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science 337: 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton J. S., Lockwood W. K., Li L., Cohen S. M., Edgar B. A., 2002. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2: 239–249. [DOI] [PubMed] [Google Scholar]
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., et al. , 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. CB 11: 213–221. [DOI] [PubMed] [Google Scholar]
- Broughton S. J., Piper M. D. W., Ikeya T., Bass T. M., Jacobson J., et al. , 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA 102: 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch S., Melcher C., Bauer M., Katzenberger J., Pankratz M. J., 2008. Opposing effects of dietary protein and sugar regulate a transcriptional target of Drosophila insulin-like peptide signaling. Cell Metab. 7: 321–332. [DOI] [PubMed] [Google Scholar]
- Buttrick G. J., Beaumont L. M. A., Leitch J., Yau C., Hughes J. R., et al. , 2008. Akt regulates centrosome migration and spindle orientation in the early Drosophila melanogaster embryo. J. Cell Biol. 180: 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterson J. H., Knowles-Barley S., James K., Heck M. M. S., Harmar A. J., et al. , 2010. Dietary modulation of Drosophila sleep-wake behaviour. PLoS One 5: e12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Quan Y., Wang H., Luo H., 2014. Trehalase regulates neuroepithelial stem cell maintenance and differentiation in the Drosophila optic lobe. PLoS One 9: e101433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J., Dow J. A. T., 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39: 715–720. [DOI] [PubMed] [Google Scholar]
- Chitnis M. M., Yuen J. S. P., Protheroe A. S., Pollak M., Macaulay V. M., 2008. The type 1 insulin-like growth factor receptor pathway. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 14: 6364–6370. [DOI] [PubMed] [Google Scholar]
- Chng W. A., Sleiman M. S. B., Schüpfer F., Lemaitre B., 2014. Transforming growth factor β/Activin signaling functions as a sugar-sensing feedback loop to regulate digestive enzyme expression. Cell Rep. 9: 336–348. [DOI] [PubMed] [Google Scholar]
- Choi S., Lim D. S., Chung J., 2015. Feeding and fasting signals converge on the LKB1–SIK3 pathway to regulate lipid metabolism in Drosophila. PLoS Genet. 11: e1005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. I., Tan S. W. S., Péan C. B., Roostalu U., Vivancos V., et al. , 2013. MEF2 is an in vivo immune-metabolic switch. Cell 155: 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman D. L., Eicher E. M., 1990. Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J. Hered. 81: 424–427. [DOI] [PubMed] [Google Scholar]
- Colombani J., Andersen D. S., Léopold P., 2012. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336: 582–585. [DOI] [PubMed] [Google Scholar]
- Colombani J., Andersen D. S., Boulan L., Boone E., Romero N., et al. , 2015. Drosophila Lgr3 couples organ growth with maturation and ensures developmental stability. Curr. Biol. 25: 2723–2729. [DOI] [PubMed] [Google Scholar]
- Djiane A., Krejci A., Bernard F., Fexova S., Millen K., et al. , 2013. Dissecting the mechanisms of Notch induced hyperplasia. EMBO J. 32: 60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docherty J. E. B., Manno J. E., McDermott J. E., DiAngelo J. R., 2015. Mio acts in the Drosophila brain to control nutrient storage and feeding. Gene 568: 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droujinine I. A., Perrimon N., 2016. Interorgan communication pathways in physiology: focus on Drosophila. Annu. Rev. Genet. 50: 539–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C., Xu Q., 2005. Roles of insulin-like growth factor (IGF) binding proteins in regulating IGF actions. Gen. Comp. Endocrinol. 142: 44–52. [DOI] [PubMed] [Google Scholar]
- Duran J., Tevy M. F., Garcia-Rocha M., Calbó J., Milán M., et al. , 2012. Deleterious effects of neuronal accumulation of glycogen in flies and mice. EMBO Mol. Med. 4: 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eanes W. F., Merritt T. J. S., Flowers J. M., Kumagai S., Sezgin E., et al. , 2006. Flux control and excess capacity in the enzymes of glycolysis and their relationship to flight metabolism in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103: 19413–19418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdi B., Nagy P., Zvara A., Varga A., Pircs K., et al. , 2012. Loss of the starvation-induced gene Rack1 leads to glycogen deficiency and impaired autophagic responses in Drosophila. Autophagy 8: 1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajans S. S., Bell G. I., 2011. MODY: history, genetics, pathophysiology, and clinical decision making. Diabetes Care 34: 1878–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez R., Tabarini D., Azpiazu N., Frasch M., Schlessinger J., 1995. The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 14: 3373–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer C. M., Sodi V. L., Reginato M. J., 2016. O-GlcNAcylation in cancer biology: linking metabolism and signaling. J. Mol. Biol. 428: 3282–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gäde G., 1990. The adipokinetic hormone/red pigment-concentrating hormone peptide family: structures, interrelationships and functions. J. Insect Physiol. 36: 1–12. [Google Scholar]
- Galikova M., Diesner M., Klepsatel P., Hehlert P., Xu Y., et al. , 2015. Energy homeostasis control in Drosophila adipokinetic hormone mutants. Genetics 201: 665–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta M. C., Oktaba K., Müller J., 2009. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325: 93–96. [DOI] [PubMed] [Google Scholar]
- Garelli A., Heredia F., Casimiro A. P., Macedo A., Nunes C., et al. , 2015. Dilp8 requires the neuronal relaxin receptor Lgr3 to couple growth to developmental timing. Nat. Commun. 6: 8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D., Rubin T., Poidevin M., Maroni B., Le Rouzic A., et al. , 2015. Fatty acid synthase cooperates with glyoxalase 1 to protect against sugar toxicity. PLoS Genet. 11: e1004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géminard C., Rulifson E. J., Léopold P., 2009. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10: 199–207. [DOI] [PubMed] [Google Scholar]
- Gershman B., Puig O., Hang L., Peitzsch R. M., Tatar M., et al. , 2007. High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol. Genomics 29: 24–34. [DOI] [PubMed] [Google Scholar]
- Ghosh A. C., O’Connor M. B., 2014. Systemic Activin signaling independently regulates sugar homeostasis, cellular metabolism, and pH balance in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 111: 5729–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C., 2013. Drosophila melanogaster: a model and a tool to investigate malignancy and identify new therapeutics. Nat. Rev. Cancer 13: 172–183. [DOI] [PubMed] [Google Scholar]
- Grönke S., Müller G., Hirsch J., Fellert S., Andreou A., et al. , 2007. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 5: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S., Clarke D. F., Broughton S., Andrews T. D., Partridge L., 2010. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6: e1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K. K., Hauser F., Williamson M., Weber S. B., Grimmelikhuijzen C. J. P., 2011. The Drosophila genes CG14593 and CG30106 code for G-protein-coupled receptors specifically activated by the neuropeptides CCHamide-1 and CCHamide-2. Biochem. Biophys. Res. Commun. 404: 184–189. [DOI] [PubMed] [Google Scholar]
- Hardivillé S., Hart G. W., 2014. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 20: 208–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey K. F., Mattila J., Sofer A., Bennett F. C., Ramsey M. R., et al. , 2008. FOXO-regulated transcription restricts overgrowth of Tsc mutant organs. J. Cell Biol. 180: 691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havula E., Hietakangas V., 2012. Glucose sensing by ChREBP/MondoA–Mlx transcription factors. Semin. Cell Dev. Biol. 23: 640–647. [DOI] [PubMed] [Google Scholar]
- Havula E., Teesalu M., Hyötyläinen T., Seppälä H., Hasygar K., et al. , 2013. Mondo/ChREBP-Mlx-regulated transcriptional network is essential for dietary sugar tolerance in Drosophila. PLoS Genet. 9: e1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey D. A., Benkel B., 1982. Regulation of amylase activity in drosophila melanogaster: effects of dietary carbohydrate. Biochem. Genet. 20: 1117–1129. [DOI] [PubMed] [Google Scholar]
- Hietakangas V., Cohen S. M., 2007. Re-evaluating AKT regulation: role of TOR complex 2 in tissue growth. Genes Dev. 21: 632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S., Cagan R. L., 2015. Salt-inducible kinases mediate nutrient-sensing to link dietary sugar and tumorigenesis in Drosophila. Elife 4: e08501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S., Baranski T. J., Cagan R. L., 2013. Transformed Drosophila cells evade diet-mediated insulin resistance through wingless signaling. Cell 154: 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger B., Galic M., Köhler K., Wittwer F., Brogiolo W., et al. , 2008. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J. Biol. 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull-Thompson J., Muffat J., Sanchez D., Walker D. W., Benzer S., et al. , 2009. Control of metabolic homeostasis by stress signaling is mediated by the lipocalin NLaz. PLoS Genet. 5: e1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka K., Bruick R. K., Liang G., Horton J. D., Uyeda K., 2004. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. USA 101: 7281–7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya T., Galic M., Belawat P., Nairz K., Hafen E., 2002. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. CB 12: 1293–1300. [DOI] [PubMed] [Google Scholar]
- Ingham P. W., 1984. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell 37: 815–823. [DOI] [PubMed] [Google Scholar]
- Isabel G., 2004. AKH-producing neuroendocrine cell ablation decreases trehalose and induces behavioral changes in Drosophila. AJP Regul. Integr. Comp. Physiol. 288: R531–R538. [DOI] [PubMed] [Google Scholar]
- Ishii S., Iizuka K., Miller B. C., Uyeda K., 2004. Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc. Natl. Acad. Sci. USA 101: 15597–15602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito C., Tomioka K., 2016. Heterogeneity of the peripheral circadian systems in Drosophila melanogaster: a review. Front. Physiol. 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y.-S., Kim D., Lee Y. S., Kim H.-J., Han J.-Y., et al. , 2011. Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. PLoS One 6: e22544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Zhang B. B., 2003. Glucagon and regulation of glucose metabolism. Am. J. Physiol. Endocrinol. Metab. 284: E671–E678. [DOI] [PubMed] [Google Scholar]
- Jünger M. A., Rintelen F., Stocker H., Wasserman J. D., Végh M., et al. , 2003. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaasik K., Kivimäe S., Allen J. J., Chalkley R. J., Huang Y., et al. , 2013. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17: 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori Y., Saito A., Hagiwara-Komoda Y., Tanaka D., Mitsumasu K., et al. , 2010. The trehalose transporter 1 gene sequence is conserved in insects and encodes proteins with different kinetic properties involved in trehalose import into peripheral tissues. Insect Biochem. Mol. Biol. 40: 30–37. [DOI] [PubMed] [Google Scholar]
- Kemppainen E., George J., Garipler G., Tuomela T., Kiviranta E., et al. , 2016. Mitochondrial dysfunction plus high-sugar diet provokes a metabolic crisis that inhibits growth. PLoS One 11: e0145836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandekar M. J., Cohen P., Spiegelman B. M., 2011. Molecular mechanisms of cancer development in obesity. Nat. Rev. Cancer 11: 886–895. [DOI] [PubMed] [Google Scholar]
- Kim E. Y., Jeong E. H., Park S., Jeong H.-J., Edery I., et al. , 2012. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 26: 490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Neufeld T. P., 2015. Dietary sugar promotes systemic TOR activation in Drosophila through AKH-dependent selective secretion of Dilp3. Nat. Commun. 6: 6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. K., Rulifson E. J., 2004. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature 431: 316–320. [DOI] [PubMed] [Google Scholar]
- Kockel L., Kerr K. S., Melnick M., Brückner K., Hebrok M., et al. , 2010. Dynamic switch of negative feedback regulation in Drosophila Akt–TOR signaling. PLoS Genet. 6: e1000990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooner J. S., Chambers J. C., Aguilar-Salinas C. A., Hinds D. A., Hyde C. L., et al. , 2008. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat. Genet. 40: 149–151. [DOI] [PubMed] [Google Scholar]
- Koštál V., Korbelová J., Rozsypal J., Zahradníčková H., Cimlová J., et al. , 2011. Long-term cold acclimation extends survival time at 0°C and modifies the metabolomic profiles of the larvae of the fruit fly Drosophila melanogaster. PLoS One 6: e25025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kréneisz O., Chen X., Fridell Y.-W. C., Mulkey D. K., 2010. Glucose increases activity and Ca2+ in insulin-producing cells of adult Drosophila. Neuroreport 21: 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Park J. H., 2004. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics 167: 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.-C., Micchelli C. A., 2013. Development and characterization of a chemically defined food for Drosophila. PLoS One 8: e67308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chawla G., Hurlburt A. J., Sterrett M. C., Zaslaver O., et al. , 2017. Drosophila larvae synthesize the putative oncometabolite L-2-hydroxyglutarate during normal developmental growth. Proc. Natl. Acad. Sci. USA 114: 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Edgar B. A., Grewal S. S., 2010. Nutritional control of gene expression in Drosophila larvae via TOR, Myc and a novel cis-regulatory element. BMC Cell Biol. 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. V., Chang B., Imamura M., Poungvarin N., Chan L., 2006. Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module. Diabetes 55: 1179–1189. [DOI] [PubMed] [Google Scholar]
- Li S., Torre-Muruzabal T., Søgaard K. C., Ren G. R., Hauser F., et al. , 2013. Expression patterns of the Drosophila neuropeptide CCHamide-2 and its receptor may suggest hormonal signaling from the gut to the brain. PLoS One 8: e76131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Padmanabha D., Gentile L. B., Dumur C. I., Beckstead R. B., et al. , 2013. HIF- and non-HIF-regulated hypoxic responses require the estrogen-related receptor in Drosophila melanogaster. PLoS Genet. 9: e1003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.-W., Myschyshyn M., Sinclair D. A., Cecioni S., Beja K., et al. , 2017. Genome-wide chemical mapping of O-GlcNAcylated proteins in Drosophila melanogaster. Nat. Chem. Biol. 13: 161–167. [DOI] [PubMed] [Google Scholar]
- Liu Y., Liao S., Veenstra J. A., Nässel D. R., 2016. Drosophila insulin-like peptide 1 (DILP1) is transiently expressed during non-feeding stages and reproductive dormancy. Sci. Rep. 6: 26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Wu J., Jing S., Yan L.-J., 2016. Hyperglycemic stress and carbon stress in diabetic glucotoxicity. Aging Dis. 7: 90–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald P. E., Joseph J. W., Rorsman P., 2005. Glucose-sensing mechanisms in pancreatic -cells. Philos. Trans. R. Soc. B Biol. Sci. 360: 2211–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda H., Yamada T., Yoshida M., Nishimura T., 2015. Flies without trehalose. J. Biol. Chem. 290: 1244–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila J., Havula E., Suominen E., Teesalu M., Surakka I., et al. , 2015. Mondo-Mlx mediates organismal sugar sensing through the Gli-Similar transcription factor Sugarbabe. Cell Rep. 13: 350–364. [DOI] [PubMed] [Google Scholar]
- McCommis K. S., Hodges W. T., Bricker D. K., Wisidagama D. R., Compan V., et al. , 2016. An ancestral role for the mitochondrial pyruvate carrier in glucose-stimulated insulin secretion. Mol. Metab. 5: 602–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFerrin L. G., Atchley W. R., 2012. A novel N-terminal domain may dictate the glucose response of Mondo proteins. PLoS One 7: e34803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight G. L., Mudri S. L., Mathewes S. L., Traxinger R. R., Marshall S., et al. , 1992. Molecular cloning, cDNA sequence, and bacterial expression of human glutamine:fructose-6-phosphate amidotransferase. J. Biol. Chem. 267: 25208–25212. [PubMed] [Google Scholar]
- Merkey A. B., Wong C. K., Hoshizaki D. K., Gibbs A. G., 2011. Energetics of metamorphosis in Drosophila melanogaster. J. Insect Physiol. 57: 1437–1445. [DOI] [PubMed] [Google Scholar]
- Morris S. N. S., Coogan C., Chamseddin K., Fernandez-Kim S. O., Kolli S., et al. , 2012. Development of diet-induced insulin resistance in adult Drosophila melanogaster. Biochim. Biophys. Acta 1822: 1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman L. P., Fink J. L., Narzinski K., Ramachandran P. V., Hathiramani S. S., et al. , 2011. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis. Model. Mech. 4: 842–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman L. P., Fink J. L., Ramachandran P. V., Patterson B. W., Okunade A. L., et al. , 2013. Role of fat body lipogenesis in protection against the effects of caloric overload in Drosophila. J. Biol. Chem. 288: 8028–8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na J., Musselman L. P., Pendse J., Baranski T. J., Bodmer R., et al. , 2013. A Drosophila model of high sugar diet-induced Cardiomyopathy. PLoS Genet. 9: e1003175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na J., Sweetwyne M. T., Park A. S. D., Susztak K., Cagan R. L., 2015. Diet-induced podocyte dysfunction in Drosophila and mammals. Cell Rep. 12: 636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel D. R., Broeck J. V., 2016. Insulin/IGF signaling in Drosophila and other insects: factors that regulate production, release and post-release action of the insulin-like peptides. Cell. Mol. Life Sci. 73: 271–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel D. R., Liu Y., Luo J., 2015. Insulin/IGF signaling and its regulation in Drosophila. Gen. Comp. Endocrinol. 221: 255–266. [DOI] [PubMed] [Google Scholar]
- Noyes B. E., Katz F. N., Schaffer M. H., 1995. Identification and expression of the Drosophila adipokinetic hormone gene. Mol. Cell. Endocrinol. 109: 133–141. [DOI] [PubMed] [Google Scholar]
- Ohki-Hamazaki H., Watase K., Yamamoto K., Ogura H., Yamano M., et al. , 1997. Mice lacking bombesin receptor subtype-3 develop metabolic defects and obesity. Nature 390: 165–169. [DOI] [PubMed] [Google Scholar]
- Okamoto N., Nakamori R., Murai T., Yamauchi Y., Masuda A., et al. , 2013. A secreted decoy of InR antagonizes insulin/IGF signaling to restrict body growth in Drosophila. Genes Dev. 27: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabha D., Baker K. D., 2014. Drosophila gains traction as a repurposed tool to investigate metabolism. Trends Endocrinol. Metab. TEM 25: 518–527. [DOI] [PubMed] [Google Scholar]
- Palanker L., Tennessen J. M., Lam G., Thummel C. S., 2009. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 9: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker Musselman L., Fink J. L., Baranski T. J., 2016. CoA protects against the deleterious effects of caloric overload in Drosophila. J. Lipid Res. 57: 380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palu R. A., Thummel C. S., 2016. Sir2 acts through Hepatocyte nuclear factor 4 to maintain insulin signaling and metabolic homeostasis in Drosophila. PLoS Genet. 12: e1005978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Park S.-H., Baek J. Y., Jy Y. J., Kim K. S., et al. , 2011. Protein O-GlcNAcylation regulates Drosophila growth through the insulin signaling pathway. Cell. Mol. Life Sci. 68: 3377–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Alfa R. W., Topper S. M., Kim G. E. S., Kockel L., et al. , 2014. A genetic strategy to measure circulating Drosophila insulin reveals genes regulating insulin production and secretion. PLoS Genet. 10: e1004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee Y., Pak J. W., Kim H., Choi H., et al. , 2015. O-GlcNAc modification is essential for the regulation of autophagy in Drosophila melanogaster. Cell. Mol. Life Sci. CMLS 72: 3173–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasco M. Y., Léopold P., 2012. High sugar-induced insulin resistance in Drosophila relies on the lipocalin Neural Lazarillo. PLoS One 7: e36583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlos, N. J., and P. A. Friedman, 2017 GPCR Signaling and Trafficking: The Long and Short of It. Trends Endocrinol. Metab. 28: 213–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendse J., Ramachandran P. V., Na J., Narisu N., Fink J. L., et al. , 2013. A Drosophila functional evaluation of candidates from human genome-wide association studies of type 2 diabetes and related metabolic traits identifies tissue-specific roles for dHHEX. BMC Genomics 14: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M. D. W., Blanc E., Leitão-Gonçalves R., Yang M., He X., et al. , 2014. A holidic medium for Drosophila melanogaster. Nat. Methods 11: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstmann T., Santos C. R., Griffiths B., Cully M., Wu M., et al. , 2008. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 8: 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post S., Tatar M., 2016. Nutritional geometric profiles of insulin/IGF expression in Drosophila melanogaster. PLoS One 11: e0155628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C. J., Pedraza L. G., Xu T., 2002. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4: 658–665. [DOI] [PubMed] [Google Scholar]
- Puig O., Marr M. T., Ruhf M. L., Tjian R., 2003. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A., Perrimon N., 2012. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151: 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis T., Van Gilst M. R., Hariharan I. K., 2010. A buoyancy-based screen of Drosophila larvae for fat-storage mutants reveals a role for Sir2 in coupling fat storage to nutrient availability. PLoS Genet. 6: e1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G. R., Hauser F., Rewitz K. F., Kondo S., Engelbrecht A. F., et al. , 2015. CCHamide-2 is an Orexigenic Brain-Gut peptide in Drosophila. PLoS One 10: e0133017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhea J. M., Wegener C., Bender M., 2010. The proprotein convertase encoded by amontillado (amon) is required in Drosophila Corpora Cardiaca endocrine cells producing the glucose regulatory hormone AKH. PLoS Genet. 6: e1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintelen F., Stocker H., Thomas G., Hafen E., 2001. PDK1 regulates growth through Akt and S6K in Drosophila. Proc. Natl. Acad. Sci. USA 98: 15020–15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez I., 2011. Drosophila TIEG is a modulator of different signalling pathways involved in wing patterning and cell proliferation. PLoS One 6: e18418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller L., Yamanaka N., Watanabe K., Daubnerová I., Zitnan D., et al. , 2008. The unique evolution of neuropeptide genes in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38: 1147–1157. [DOI] [PubMed] [Google Scholar]
- Rovenko B. M., Perkhulyn N. V., Gospodaryov D. V., Sanz A., Lushchak O. V., et al. , 2015. High consumption of fructose rather than glucose promotes a diet-induced obese phenotype in Drosophila melanogaster. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 180: 75–85. [DOI] [PubMed] [Google Scholar]
- Ruaud A.-F., Lam G., Thummel C. S., 2011. The Drosophila NR4A nuclear receptor DHR38 regulates carbohydrate metabolism and glycogen storage. Mol. Endocrinol. 25: 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Romero M., Blanco E., Paricio N., Serras F., Corominas M., 2015. Cabut/dTIEG associates with the transcription factor Yorkie for growth control. EMBO Rep. 16: 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K., Nusse R., 2002. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296: 1118–1120. [DOI] [PubMed] [Google Scholar]
- Salehi A., Vieira E., Gylfe E., 2006. Paradoxical stimulation of glucagon secretion by high glucose concentrations. Diabetes 55: 2318–2323. [DOI] [PubMed] [Google Scholar]
- Saltiel A. R., Kahn C. R., 2001. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414: 799–806. [DOI] [PubMed] [Google Scholar]
- Sano H., Nakamura A., Texada M. J., Truman J. W., Ishimoto H., et al. , 2015. The nutrient-responsive hormone CCHamide-2 controls growth by regulating insulin-like peptides in the brain of Drosophila melanogaster. PLoS Genet. 11: e1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassu E. D., McDermott J. E., Keys B. J., Esmaeili M., Keene A. C., et al. , 2012. Mio/dChREBP coordinately increases fat mass by regulating lipid synthesis and feeding behavior in Drosophila. Biochem. Biophys. Res. Commun. 426: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer M. H., Noyes B. E., Slaughter C. A., Thorne G. C., Gaskell S. J., 1990. The fruitfly Drosophila melanogaster contains a novel charged adipokinetic-hormone-family peptide. Biochem. J. 269: 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmelpfeng K., Strunk M., Stork T., Klämbt C., 2006. Mummy encodes an UDP-N-acetylglucosamine-dipohosphorylase and is required during Drosophila dorsal closure and nervous system development. Mech. Dev. 123: 487–499. [DOI] [PubMed] [Google Scholar]
- Seay D. J., Thummel C. S., 2011. The circadian clock, light, and cryptochrome regulate feeding and metabolism in Drosophila. J. Biol. Rhythms 26: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine O., Love D. C., Rubenstein D. S., Hanover J. A., 2010. Blocking O-Linked GlcNAc Cycling in Drosophila insulin-producing cells perturbs glucose-insulin homeostasis. J. Biol. Chem. 285: 38684–38691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvan N., Williamson R., Mariappa D., Campbell D. G., Gourlay R., et al. , 2017. A mutant O-GlcNAcase enriches Drosophila developmental regulators. Nat. Chem. Biol. DOI: 10.1038/nchembio.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Bhattacharya P., Kalia K., Tiwari V., 2017. Diabetic nephropathy: new insights into established therapeutic paradigms and novel molecular targets. Diabetes Res. Clin. Pract. 128: 91–108. [DOI] [PubMed] [Google Scholar]
- Shih H. M., Liu Z., Towle H. C., 1995. Two CACGTG motifs with proper spacing dictate the carbohydrate regulation of hepatic gene transcription. J. Biol. Chem. 270: 21991–21997. [DOI] [PubMed] [Google Scholar]
- Shingleton A. W., Das J., Vinicius L., Stern D. L., 2005. The temporal requirements for insulin signaling during development in Drosophila. PLoS Biol. 3: e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiri-Sverdlov R., Custers A., van Vliet-Ostaptchouk J. V., van Gorp P. J. J., Lindsey P. J., et al. , 2006. Identification of TUB as a novel candidate gene influencing body weight in humans. Diabetes 55: 385–389. [DOI] [PubMed] [Google Scholar]
- Sieber M. H., Thomsen M. B., Spradling A. C., 2016. Electron transport chain remodeling by GSK3 during oogenesis connects nutrient state to reproduction. Cell 164: 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinadinos C., Valles-Ortega J., Boulan L., Solsona E., Tevy M. F., et al. , 2014. Neuronal glycogen synthesis contributes to physiological aging. Aging Cell 13: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair D. A. R., Syrzycka M., Macauley M. S., Rastgardani T., Komljenovic I., et al. , 2009. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. USA 106: 13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupa D. A., Dervisefendic A., Zwiener J., Pletcher S. D., 2008. Dietary composition specifies consumption, obesity, and lifespan in Drosophila melanogaster. Aging Cell 7: 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaninova V., Krafcikova M., Perez-Gomez R., Steffal P., Trantirek L., et al. , 2016. Notch stimulates growth by direct regulation of genes involved in the control of glycolysis and the tricarboxylic acid cycle. Open Biol. 6: 150155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Cheng D., Hong S., Sappe B., Hu Y., et al. , 2017. Midgut-derived Activin regulates glucagon-like action in the fat body and Glycemic control. Cell Metab. 25: 386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teesalu M., Rovenko B. M., Hietakangas V., 2017. Salt-inducible kinase 3 provides sugar tolerance by regulating NADPH/NADP+ redox balance. Curr. Biol. 27: 458–464. [DOI] [PubMed] [Google Scholar]
- Teleman A. A., 2009. Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 425: 13–26. [DOI] [PubMed] [Google Scholar]
- Teleman A. A., Hietakangas V., Sayadian A. C., Cohen S. M., 2008. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 7: 21–32. [DOI] [PubMed] [Google Scholar]
- Tennessen J. M., Baker K. D., Lam G., Evans J., Thummel C. S., 2011. The Drosophila Estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 13: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Bertagnolli N. M., Evans J., Sieber M. H., Cox J., et al. , 2014a Coordinated metabolic transitions during Drosophila Embryogenesis and the onset of aerobic glycolysis. G3 (Bethesda) 4: 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Barry W. E., Cox J., Thummel C. S., 2014b Methods for studying metabolism in Drosophila. Methods 68: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorat L., Mani K.-P., Thangaraj P., Chatterjee S., Nath B. B., 2016. Downregulation of dTps1 in Drosophila melanogaster larvae confirms involvement of trehalose in redox regulation following desiccation. Cell Stress Chaperones 21: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonning A., Helms S., Schwarz H., Uv A. E., Moussian B., 2006. Hormonal regulation of mummy is needed for apical extracellular matrix formation and epithelial morphogenesis in Drosophila. Development 133: 331–341. [DOI] [PubMed] [Google Scholar]
- Ugrankar R., Berglund E., Akdemir F., Tran C., Kim M. S., et al. , 2015. Drosophila glucome screening identifies Ck1alpha as a regulator of mammalian glucose metabolism. Nat. Commun. 6: 7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay A., Moss-Taylor L., Kim M. J., Ghosh A. C., O’Connor M. B., 2017. TGF-β family signaling in Drosophila. Cold Spring Harb. Perspect. Biol. DOI: 10.1101/cshperspect.a022152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J., Lim S. F., Cohen S. M., 2010. Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes Dev. 24: 2748–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkenhoff A., Weiler A., Letzel M., Stehling M., Klämbt C., et al. , 2015. Glial glycolysis is essential for neuronal survival in Drosophila. Cell Metab. 22: 437–447. [DOI] [PubMed] [Google Scholar]
- Wang B., Moya N., Niessen S., Hoover H., Mihaylova M. M., et al. , 2011. A hormone-dependent module regulating energy balance. Cell 145: 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weavers H., Prieto-Sánchez S., Grawe F., Garcia-López A., Artero R., et al. , 2009. The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature 457: 322–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr M. C., Holder M. V., Gailite I., Saunders R. E., Maile T. M., et al. , 2013. Salt-inducible kinases regulate growth through the Hippo signalling pathway in Drosophila. Nat. Cell Biol. 15: 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werz C., Köhler K., Hafen E., Stocker H., 2009. The Drosophila SH2B family adaptor Lnk acts in parallel to chico in the insulin signaling pathway. PLoS Genet. 5: e1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whon T. W., Shin N.-R., Jung M.-J., Hyun D.-W., Kim H. S., et al. , 2017. Conditionally pathogenic gut microbes promote larval growth by increasing redox-dependent fat storage in high sugar diet-fed Drosophila. Antioxid. Redox Signal. DOI: 10.1089/ars.2016.6790. [DOI] [PubMed] [Google Scholar]
- Wigglesworth V. B., 1949. The utilization of reserve substances in Drosophila during flight. J. Exp. Biol. 26: 150–163. [DOI] [PubMed] [Google Scholar]
- Xu K., Zheng X., Sehgal A., 2008. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 8: 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., DiAngelo J. R., Hughes M. E., Hogenesch J. B., Sehgal A., 2011. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 13: 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q.-W., Yang Q., Mody N., Graham T. E., Hsu C.-H., et al. , 2007. The adipokine lipocalin 2 is regulated by obesity and promotes insulin resistance. Diabetes 56: 2533–2540. [DOI] [PubMed] [Google Scholar]
- Yang Q., Inoki K., Ikenoue T., Guan K.-L., 2006. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20: 2820–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T., Yamada T., Nishimura T., 2017. Adaptation to dietary conditions by trehalose metabolism in Drosophila. Sci. Rep. 7: 1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Matsuda H., Kubo H., Nishimura T., 2016. Molecular characterization of Tps1 and Treh genes in Drosophila and their role in body water homeostasis. Sci. Rep. 6: 30582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I., Schütz C. S., Katzenberger J. D., Bauer M., Pankratz M. J., 2002. Nutrient control of gene expression in Drosophila: microarray analysis of starvation and sugar-dependent response. EMBO J. 21: 6162–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J., Nieuwenhuis J., Perrimon N., 2013. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol. 11: e1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]