Kazatskaya et al. uncover a role for the atypical MAP kinase MAPK15 in regulating primary cilium structure and morphology in both C. elegans and human..
Keywords: primary cilia, MAPK15, C. elegans, basal body
Abstract
Motile and immotile (or primary) cilia are microtubule-based structures that mediate multiple cellular functions, including the transduction of environmental cues, developmental signaling, cellular motility, and modulation of fluid flow. Although their core architectures are similar, motile and primary cilia exhibit marked structural differences that underlie distinct functional properties. However, the extent to which ciliogenesis mechanisms are shared between these different cilia types is not fully described. Here, we report that the atypical MAP kinase MAPK15 (ERK7/8), implicated in the formation of vertebrate motile cilia, also regulates the formation of primary cilia in Caenorhabditis elegans sensory neurons and human cells. We find that MAPK15 localizes to a basal body subdomain with the ciliopathy protein BBS7 and to cell–cell junctions. MAPK15 also regulates the localization of ciliary proteins involved in cilium structure, transport, and signaling. Our results describe a primary cilia-related role for this poorly studied member of the MAPK family in vivo, and indicate a broad requirement for MAPK15 in the formation of multiple ciliary classes across species.
CILIA are microtubule-based immotile or motile structures protruding from the surfaces of most eukaryotic cell types. These include most postmitotic vertebrate cells, which possess an immotile or primary cilium that senses and transduces environmental cues (Goetz et al. 2009; Drummond 2012; Ko 2012; Guemez-Gamboa et al. 2014). Defects in primary and motile cilia cause numerous human genetic disorders (ciliopathies) that affect the development of many tissues and organs (Waters and Beales 2011). While motile cilia can also serve sensory functions (Bloodgood 2010), most studies of these organelles focus on their role in mediating cellular movement and fluid flow (Lindemann and Lesich 2016; Meunier and Azimzadeh 2016; Spassky and Meunier 2017). Not surprisingly, motile and primary cilia exhibit significant differences in axoneme ultrastructures and mechanisms of biogenesis. For instance, motile cilia are typically present on multiciliated cells and nucleate from > 100 basal bodies per cell that are generated de novo, whereas a primary cilium arises from a basal body derived from a remodeled mother centriole (Kobayashi and Dynlacht 2011; Kim and Dynlacht 2013; Brooks and Wallingford 2014; Spassky and Meunier 2017). In addition, motile cilia contain specialized motility-associated components not found in primary cilia (Ishikawa 2017; Loreng and Smith 2017; Zhu et al. 2017). Despite such differences, many ciliogenic programs are shared by motile and primary cilia (Mitchell 2007; Carvalho-Santos et al. 2011).
The formation and function of all cilia rely on intracellular transport pathways that regulate protein trafficking to, from, and within the organelle. Microtubule-based intraflagellar transport (IFT) between the ciliary basal body and tip regulates ciliary protein delivery and export via kinesin-2 and IFT–dynein motors, together with cargo adaptor complexes (IFT-A, IFT-B, and BBSome) (Malicki and Avidor-Reiss 2014). The molecular composition of cilia is also regulated by gating mechanisms at the basal body and the proximal-most region of the axoneme (transition zone). For instance, ciliopathy-associated proteins facilitate diffusion barriers at the transition zone, and basal body distal appendage components regulate IFT/BBSome protein entry into cilia (Craige et al. 2010; Chih et al. 2011; Garcia-Gonzalo et al. 2011; Williams et al. 2011; Wei et al. 2013, 2016; Garcia-Gonzalo and Reiter 2016).
Mitogen-activated protein kinases (MAPKs) are a highly conserved family of enzymes that function in a plethora of cellular processes (Robinson and Cobb 1997; Chang and Karin 2001; Pearson et al. 2001). Classical MAPKs that act in the conventional tripartite MEKK-MEK-MAPK signaling cascade are known to regulate cilium length in mammalian cells, Caenorhabditis elegans, and Trypanosomatids, and ciliary beating in Schistosoma mansoni (Rotureau et al. 2009; Ressurreicao et al. 2011; Abdul-Majeed et al. 2012; van der Vaart et al. 2015). In contrast, the physiological roles of atypical MAPKs that appear to lack conventional MAPK cascade involvement are poorly understood (Coulombe and Meloche 2007). In cultured cells, MAPK15 (ERK7 or ERK8) regulates autophagy and protein trafficking/secretion, supports genomic integrity, and promotes tumorigenic properties (Abe et al. 2001, 2002; Groehler and Lannigan 2010; Xu et al. 2010; Zacharogianni et al. 2011; Colecchia et al. 2012). In the first description of an in vivo role, MAPK15 depletion in multiciliated Xenopus and mouse cells reduces motile cilia number and length, and disrupts basal body migration to the apical cell surface due to defects in the actin network (Miyatake et al. 2015). MAPK15 expression is controlled by the master regulator of motile cilia biogenesis, FoxJ1, and overexpressed MAPK15 localizes to basal bodies, cell–cell contacts, and weakly to ciliary axonemes (Choksi et al. 2014; Miyatake et al. 2015).
Here, we show that MAPK15 also regulates the formation of primary cilia. C. elegans only possess primary cilia, which extend from the dendritic endings of 60 sensory neurons (Ward et al. 1975; Perkins et al. 1986). C05D10.2, the C. elegans ortholog of MAPK15, is expressed exclusively in most, if not all, ciliated sensory neurons. Similar to observations for vertebrate multiciliated cells, MAPK15 localizes to the basal bodies and cell–cell junctions in C. elegans and human primary cilium-forming cells. In human cells, MAPK15 localizes to a discrete subdomain near the distal end of the basal body. Loss of nematode and human MAPK15 function causes defects in cilium length and morphology, and disrupts the localization of basal body and axonemal proteins important for cilium structure, trafficking, and ciliary gating. Together with previous work (Miyatake et al. 2015), our results demonstrate that the atypical MAP kinase, MAPK15, regulates the formation of motile and primary cilia across species, highlighting a conserved role for this protein in cilium biology.
Materials and Methods
C. elegans genetics
All C. elegans strains were grown at 20° on standard NGM plates seeded with Escherichia coli OP50 bacteria. Standard genetic crossing techniques were used to introduce transgenes into mutant genetic backgrounds. Transgenic animals were generated by microinjection of 5–10 ng/µl of plasmid(s) or 5–50 ng/µl linear DNA constructs with 30–50 ng/µl of co-injection marker (unc-122p::gfp, unc-122p::mCherry, or unc-122p::dsRed). Mutations were confirmed by PCR (genotyping) and sequencing (oligonucleotide primer sequences available on request). In all cases where expression of a transgene was compared between wild-type and mutant animals, the same transgenic array was examined in both backgrounds. Mutant alleles were provided by the Caenorhabditis Genetics Center, apart from the hyls-1(tm3067) allele, which was a gift from K. Oegema (University of California, San Diego). A list of all strains used in this work is provided in Supplemental Material, Table S1 in File S1.
Identification and cloning of mapk-15(oy112)
The oy112 allele was isolated in a genetic screen designed to identify suppressors of reduced str-1p::gfp expression in the AWB neurons in a kin-29(oy39) mutant background (Lanjuin and Sengupta 2002; van der Linden et al. 2007). The mutation was mapped to a 7.3-Mb region of chromosome III using standard mapping techniques (Davis et al. 2005). Whole-genome sequencing of oy112 mutants identified a premature stop codon (Q229*) in C05D10.2. Ciliary phenotypes of oy112 were rescued upon injection of C05D10.2 genomic DNA with endogenous upstream regulatory sequences at 50 ng/µl.
Dye-filling assay
Dye-filling (Dyf) assays were performed as previously described (Sanders et al. 2015). Briefly, animals were incubated in fluorescent lipophilic DiI (Life Technologies; diluted 1:200 or 1:1000 in M9 buffer for the standard or sensitized assays, respectively) for 30 min, and allowed to recover for 30 min on NGM plates containing bacterial food before imaging on a fluorescence microscope.
Molecular biology
C. elegans mapk-15 genomic sequences:
The mapk-15 genomic DNA used in Figure 1C was generated by PCR amplification of ∼1.6 kb of upstream regulatory sequence together with mapk-15 exonic and intronic sequences using iProof high-fidelity DNA polymerase (Bio-Rad, Hercules, CA). The mapk-15 genomic rescue fragment used in Figure 1B consists of 1.8 kb of upstream regulatory sequence, mapk-15 exonic/intronic sequences, and 461 bp of 3′-UTR sequences.
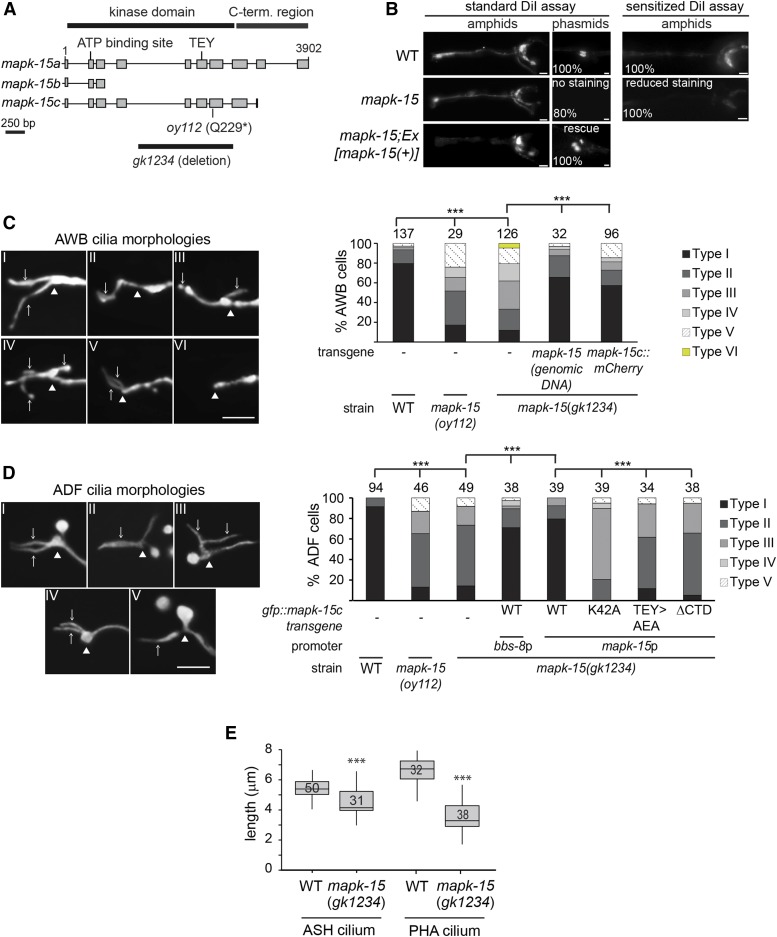
Figure 1.
MAPK-15 regulates the morphologies of sensory neuron cilia in C. elegans. (A) Genomic structure of C05D10.2 (mapk-15), showing three predicted isoforms (a, b, and c), the TEY activation motif, ATP-binding site, and the mutant alleles (gk1234 and oy112) employed in this study. Exons are denoted by gray rectangles. (B) Representative images of DiI uptake in the amphid (head) and phasmid (tail) neurons of wild-type (WT) and mapk-15(gk1234) worms. Left images from a standard dye-filling assay; right images from a sensitized assay (low concentration DiI). Transgene comprises WT mapk-15 genomic sequence (incorporating a, b, and c isoforms), together with endogenous upstream regulatory sequence. Numbers indicate the percentage of animals exhibiting the phenotype (n > 30 for each strain). Anterior is at left. Bar, 10 μm. (C and D) Representative images and quantification of AWB and ADF cilia defects in the indicated genetic backgrounds. AWB cilia morphologies categorized as: Type I, two normal cilia; Type II, one cilium missing; Type III, one misdirected cilium; Type IV, abnormally branched cilia; Type V, two short cilia; and Type VI, both cilia absent. ADF cilium morphologies categorized as: Type I, two normal cilia; Type II, collapsed cilia; Type III, one misdirected cilium; Type IV, two short cilia; and Type V, one missing cilium. White arrows denote cilia. White arrowheads denote the cilia base. Numbers at top indicate the number of scored neurons. AWB neurons visualized using str-1p::gfp; ADF neurons visualized using srh-142p::rfp. CTD; C-terminal domain. *** P < 0.001 (Generalized Linear Model test and Bonferroni corrections). Anterior is at left. Bar, 5 µm. (E) Box plots of ASH and PHA cilium lengths in WT and mapk-15(gk1234) worms. Horizontal lines denote 25, 50, and 75th percentiles; whiskers denote the full range. Numbers indicate the number of neurons quantified. ASH neurons visualized using sra-6p::gfp; PHA neurons visualized using flp-15p::gfp. *** P < 0.001 (Welch’s t-test vs. WT).
C. elegans transcriptional mapk-15 expression construct (mapk-15p::gfp):
To create the construct, 1811 bp of mapk-15 upstream regulatory sequence together with 11 bp of exon 1 (with the A of the ATG start codon modified to C) was fused to GFP sequences (1920 bp; amplified from pPD95.67; gift of A. Fire, Stanford University) (Figure 3A).
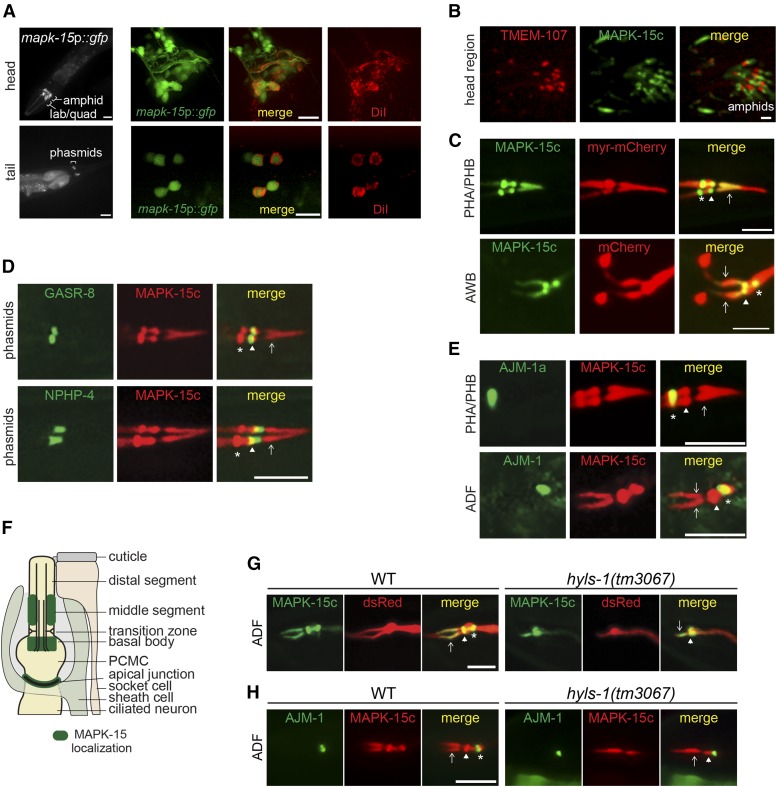
Figure 3.
MAPK-15 is expressed in ciliated sensory neurons and localizes to the ciliary axoneme, basal body, and apical junction. (A) Representative images of worms expressing GFP under the control of mapk-15 upstream regulatory sequences (mapk-15p). DiI costains six pairs of ciliated amphid head neurons and the pair of ciliated PHA/B phasmid tail neurons. Anterior is at left. Bar, 50 μm (left-most panels) and 15 μm (other panels). Lab, inner labial cells; quad, outer labial quadrant cells. (B) Representative images of head sensory neuronal cilia in worms expressing MAPK-15c::GFP (endogenous promoter) and the transition zone marker, TMEM-107::RFP. Anterior is at left. Bar, 2 μm. (C) Representative images of PHA/B and AWB cilia from worms expressing GFP::MAPK-15c (nphp-4 or str-1 promoter) and either mapk-15p::myr-mCherry (PHA/B) or str-1p::mCherry (AWB). Arrows, ciliary axoneme; arrowhead, basal body; asterisk, apical junction. Anterior is at left. Bar, 5 µm. (D) Representative images of phasmid cilia from worms expressing mCherry::MAPK-15c (bbs-8 promoter) and either the basal body marker, GFP::GASR-8 (nphp-4 promoter), or the transition zone marker, NPHP-4::GFP (endogenous promoter). Arrows, ciliary axoneme; arrowhead, basal body; asterisk, apical junction. Anterior is at left. Bar, 5 µm. (E) Representative images of PHA/B and ADF cilia from worms expressing mCherry::MAPK-15c (endogenous promoter for PHA/B; srh-142 promoter for ADF) and AJM-1a::GFP (nphp-4 promoter for PHA/B) or AJM-1::CFP (srh-142 promoter for ADF). Arrows, ciliary axoneme; arrowhead, basal body; asterisk, apical junction. Anterior is at left. Bar, 5 µm. (F) Cartoon representation of MAPK-15 localization (green) to the apical junction, basal body, and middle segments of amphid and phasmid channel cilia. Only one axoneme shown for simplicity. PCMC: periciliary membrane compartment. Socket and sheath cells are glial cells. (G) Representative images of MAPK-15c localization in ADF cilia of wild-type (WT) and mutant animals (n ≥ 16 each). Of hyls-1 mutants, 51% exhibit the shown phenotype. Arrows, ciliary axoneme; arrowhead, basal body; asterisk, apical junction. Anterior is at left. Bar, 5 µm. (H) Representative images of MAPK-15c and AJM-1 localization in ADF cilia of WT and hyls-1 mutant animals (n ≥ 20 each). Arrows, ciliary axoneme; arrowhead, basal body; asterisk, apical junction. Anterior is at left. Bar, 5 µm.
C. elegans translational reporter constructs (cDNA):
The mapk-15 (isoform c) and ajm-1 (isoform a) cDNAs were amplified from a mixed stage N2 cDNA library (Nechipurenko et al. 2016) using gene-specific primers. The nphp-4, dyf-19, ajm-1 [exons 2–9 minimal region from Koppen et al. (2001)], and jbts-14 cDNAs were gifts from M. Barr (Rutgers University), J. Hu (Mayo Clinic), M. Heiman (Harvard Medical School), and Inna Nechipurenko, respectively. All constructs were cloned into pPD95.77 or pMC10 (gift of M. Colosimo) C. elegans expression plasmids and confirmed by sequencing. N- and C-terminal reporter-tagged constructs were generated by subcloning fluorescent reporter sequences in-frame into expression vectors containing the gene of interest. mapk-15 mutant constructs were generated by PCR using Phusion polymerase (New England Biolabs, Beverly, MA). All mutations were confirmed by sequencing. Upstream regulatory sequences of the following genes were used to drive transgene expression in specific ciliated cells: srh-142 (ADF) (Troemel et al. 1997; Sagasti et al. 1999), str-1 (AWB) (Troemel et al. 1997), bbs-8 (all ciliated neurons) (gift from M. Leroux, Simon Fraser University), and nphp-4 (all ciliated neurons (gift from M. Barr, Rutgers University)). For mapk-15(cDNA) constructs driven by the endogenous promoter, ∼1.6 kb of upstream regulatory sequence of mapk-15 were used.
C. elegans translational reporter constructs (genomic DNA):
gasr-8 genomic DNA (gift of M. Leroux, Simon Fraser University) was cloned into pMC10 and confirmed by sequencing. All genomic mapk-15 reporter-tagged constructs were generated by fusion PCR as previously described (Hobert 2002). For the translational C-terminal-tagged GFP constructs (Figure 3B and Figure S2A in File S1), mapk-15 (a or c) exonic/intronic sequences together with 746 bp of upstream regulatory sequence were fused in-frame with GFP sequence (amplified from pPD95.77). For the translational N-terminal-tagged GFP construct (Figure S2B in File S1), GFP sequence was fused 3′ to 1811 bp of mapk-15 upstream regulatory sequence and 5′ to mapk-15a genomic sequence (3899 + 461 bp of 3′-UTR).
Human MAPK15 plasmid constructs:
Full-length cDNA encoding human MAPK15 was obtained from Promega (Madison, WI; clone FHC10717), amplified by PCR, and cloned into pEGFP-N1 (Clontech) to generate the expression construct for C-terminal-tagged MAPK15::GFP. Point mutations for the siRNA-resistant construct and kinase-dead mutant of MAPK15 were inserted by site-directed mutagenesis. All constructs were confirmed by DNA sequencing.
Cell culture and plasmid transfection
Human hTERT-immortalized renal proximal epithelial cells, RPTEC/TERT1 (gift from T. McMorrow, University College Dublin), were grown in low-glucose Dulbecco's Modified Eagle's Medium (DMEM)/F-12 supplemented with insulin, transferrin, sodium selenite (ITS), EGF, hydrocortisone, L-glutamine, and penicillin/streptomycin at 37°/5% CO2. For the analysis of basal body protein localization, RPTEC/TERT cells were maintained for at least 7 days after reaching confluence to allow cilia formation. Human hTERT-immortalized retinal pigmented epithelial cells, hTERT-RPE1 (gift from J. Simpson, University College Dublin), were maintained in DMEM/F12 medium supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine, and 0.348% sodium bicarbonate at 37°/5% CO2. Cilia formation in hTERT-RPE1 cells was induced by serum withdrawal for 24–48 hr. RPTEC/TERT and hTERT-RPE1 cells were transiently transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) or TransIT-LT1 (Mirus Bio), respectively. For generation of stable hTERT-RPE1 cell lines, hTERT-RPE1 cells were transfected with plasmid DNA using Lipofectamine 3000 (Invitrogen) followed by selection with 500 µg/ml G418 (Invitrogen) 24 hr after transfection.
RNA interference
For MAPK15 knockdown, hTERT-RPE1 cells were transfected with siRNAs using Lipofectamine 2000 (Invitrogen). Silencer Select siRNAs obtained from Ambion targeted the following sequences: MAPK15-si1 5′-AGAACGACAGGGACATTTA-3′ and MAPK15-si2 5′-TGAACGCAGTCATCCGGAA-3′. As negative control, nontargeting scrambled siRNA (5′-TAACGACGCGACGACGTAA-3′) was used. Efficiencies of mRNA depletion were measured by quantitative real-time PCR (qPCR). For this, total RNA was purified with a NucleoSpin RNA minikit (Macherey-Nagel) and reverse transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). qPCR analysis was performed using SYBR green detection in an Applied Biosystems 7500 system. GAPDH mRNA levels were used for normalization and results were obtained using the ΔΔCt method (Livak and Schmittgen 2001).
Antibodies
Primary antibodies were obtained from the following sources: rabbit anti-MAPK15/ERK8 (BS2438; diluted 1:100) from BioWorld Technology; mouse anti-Ki67 (MA1-80199; diluted 1:200) from Pierce Chemical (Rockford, IL); mouse anti-polyglutamylated tubulin (AG-20B-0020-C100; diluted 1:1000) from Adipogen; rabbit anti-ZO-1 (40–2300; diluted 1:500) from Invitrogen; rabbit anti–γ-tubulin (T5192; diluted 1:500) from Sigma (Sigma Chemical, St. Louis, MO); and rabbit anti-ARL13B (17711-1-AP; diluted 1:3000), rabbit anti-FBF1 (11531-1-AP; diluted 1:500), rabbit anti-NPHP4 (13812-1-AP; diluted 1:50), and rabbit anti-BBS5 (14569-1-AP; diluted 1:100) from Proteintech. Rabbit anti-BBS7 (diluted 1:100) was a gift from P. Beales (University College London), and mouse anti-acetylated tubulin (diluted 1:250), guinea pig anti-CEP164 (diluted 1:2000), and guinea pig and rabbit anti-ODF2 antibodies (diluted 1:1000 and 1:500, respectively) were gifts from G. Pereira (University of Heidelberg) (Schmidt et al. 2012; Kuhns et al. 2013; Kurtulmus et al. 2016). Anti-mouse, rabbit, and guinea pig secondary antibodies conjugated to Alexa Fluor 488, 568, or 647 dyes were obtained from Life Technologies.
Immunofluorescence staining of human cells
Cells were grown on glass coverslips, washed with PBS and fixed with either 3% PFA in PBS for 10 min or ice-cold methanol for 5 min. PFA-fixed samples were permeabilized for 5 min in either 0.2% Triton X-100/PBS or ice-cold methanol. After blocking with 3% BSA in 0.2% Triton X-100/PBS for 30 min, cells were incubated with primary antibodies at room temperature for 2 hr. Antibodies were diluted in blocking solution. Samples were washed three times with PBS and incubated with appropriate secondary antibodies conjugated to Alexa Fluor dyes (diluted 1:500) for 1 hr at room temperature. DAPI (4’,6-Diamidino-2-phenylindole, Sigma) was included with secondary antibodies for DNA staining. Samples were washed three times in PBS, and coverslips (No 1.5 High precision, Marienfeld) were mounted on glass slides in Mowiol (Sigma). For stimulated emission depletion (STED) imaging, secondary antibodies were used at higher concentrations (1:100 dilution), DAPI staining was omitted, and coverslips were mounted on glass slides in ProLong Diamond (Life Technologies).
Transmission electron microscopy
Day 1 C. elegans adults were fixed, sectioned, and imaged as previously described (Sanders et al. 2015).
Light microscopy
C. elegans imaging:
Live worms were mounted onto 10% agarose pads and immobilized using 10 mM tetramisole (Sigma L9756) or microbeads (Polysciences, Warrington, PA; no. 00876-15). Animals were imaged on a Leica DM5000B compound fluorescence microscope, or inverted spinning disk imaging system [Nikon (Garden City, NY) Eclipse Ti/Andor Revolution or Zeiss ([Carl Zeiss], Tornwood, NY) Axiovert] fitted with a Yokogawa spinning disk confocal head. For confocal imaging, images were generated by collecting optical sections (z-stack) every 0.2–0.27 µm using Plan Apochromat 100×/1.40 NA or 63×/1.40 NA oil immersion objectives. Maximum intensity projections of z-stacks were generated using SlideBook 6.0 software (Intelligent Imaging Innovations, 3i) or Fiji/ImageJ [National Institutes of Health (NIH), Bethesda, MD). Protein localization and cilia structure were visualized and quantified using SlideBook 6.0 software (Intelligent Imaging Innovations, 3i) and ImageJ (NIH). Image quantification and analyses were performed on raw images. Control and test images were corrected for brightness, contrast, and directionality using ImageJ (NIH) and Photoshop (Adobe Systems) for presentation in figures. All animals were imaged on at least two different days for each data point. At least two independently generated lines were examined for each transgenic strain.
Imaging of human cells:
Widefield images were acquired as z-stacks at 0.25-µm intervals using a Zeiss AxioImager M1 microscope equipped with a Plan Apochromat 63×/1.40 NA oil immersion objective and an AxioCamHR camera. Confocal images were acquired either on a Zeiss LSM510 Meta microscope with a Plan Apochromat 63×/1.40 NA oil immersion objective or an Olympus Fluoview FV1000 microscope with a 60×/1.35 NA UPlanSApo oil immersion objective. Fluorescence intensity quantifications of basal body proteins were performed using Fiji/ImageJ (NIH) (Schindelin et al. 2012). The centriole was marked with polyglutamylated tubulin and the transition zone was defined by the gap in polyglutamylated tubulin staining between the centriole and the axoneme. For each cell, the mean intensity in a defined area of 100 pixels encompassing the signal at the centriole or the transition zone was measured. Background signals were measured in the near proximity of each centrosome and subtracted from the centrosomal measurement. For each experimental condition, 30–50 cells were analyzed. To determine the ciliary length in hTERT-RPE1 cells, ARL13B staining was used as a ciliary marker, and line measurements were performed using Fiji/ImageJ (NIH). For each experimental condition, 30–60 cilia were analyzed.
Super resolution (STED) imaging of human cells
STED imaging was performed on a Leica TCS SP8 STED microscope equipped with a white-light laser for excitation, two STED depletion lasers (592 nm, 660 nm), and a HCX PL APO 100×/1.40 NA oil objective. Images were acquired on Leica HyD detectors using time-gated detection. For single-color STED, Alexa 488 was excited at 499 nm and depleted at 592 nm. For dual-color STED, Alexa 488 and Alexa 568 were excited at 499 and 577 nm, respectively, and depletion was performed at 592 nm for Alexa 488 and at 660 nm for Alexa 565. To show the orientation of the ciliary axonemes, samples were stained with polyglutamylated tubulin antibodies (and detected with Alexa 647) and imaged under confocal conditions. Images were captured as z-stacks at 140-nm intervals and with a pixel size of ∼14 nm. Sequential acquisition (660–592) was employed to avoid cross talk and fluorophore damage by the 592 nm depletion laser.
Confocal and STED images were deconvolved with Huygens Professional software (Scientific Volume Imaging) using a theoretical point spread function calculated based on imaging parameters. Within the deconvolution wizard, manual background correction was applied to images, the signal to noise ratio set to 20 for confocal images and to seven for STED images, and deconvolution performed using the Classic Maximum Likelihood Estimation algorithm (CMLE).
Ring diameters at the ciliary base were determined in deconvolved STED images of RPTEC/TERT cells that showed the cilium in cross section. Images were analyzed using the Twin Slicer tool of the Huygens Professional software (Scientific Volume Imaging). Briefly, a line was drawn across the longest axis and the fluorescence intensity plotted. The resulting line intensity plot showed two peaks where the line crosses the ring signal, and the distance of the maxima of these two peaks was measured to provide the ring diameter.
Data availability
The authors state that all data necessary for confirmation of the conclusions discussed in the article are represented fully within the article. All worm strains (listed in Table S1 in File S1) and constructs are available upon request.
Results
MAPK-15 regulates the morphologies of multiple sensory cilia subtypes in C. elegans
We isolated the oy112 mutation in a forward genetic screen for animals exhibiting reduced sensory neuron function (see Materials and Methods) (van der Linden et al. 2007, 2008). Genetic mapping and complementation analyses showed that oy112 is an allele of the gene C05D10.2, which encodes the C. elegans ortholog (bidirectional best BLAST hit) of MAPK15 (Figure 1A and Figure S1, A and B in File S1). We henceforth refer to the gene product of C05D10.2 as mapk-15.
Three MAPK-15 isoforms are confirmed by cDNAs (www.wormbase.org): MAPK-15a (470 aa) and MAPK-15c (367 aa) contain the kinase domain and differ only in the length of their C-termini, whereas the MAPK-15b isoform encodes a highly truncated protein (86 aa) that lacks key features of the kinase domain (Figure 1A). Sequence analysis shows that oy112 represents a nonsense mutation predicted to encode a protein lacking part of the kinase domain (Figure 1A). An additional mapk-15 allele (gk1234) obtained from the C. elegans Knockout Consortium is an insertion/deletion allele resulting in a truncated protein lacking a large portion of the kinase domain, and is likely a null allele (Figure 1A).
To investigate the role of MAPK-15 in regulating neuronal properties, we first explored whether sensory neuron development is compromised in mapk-15 mutants. The DiI Dyf assay indirectly reports on the integrity of the dendrites and cilia of 16 sensory neurons (six and two pairs of head amphid and tail phasmid sensory neurons, respectively) (Figure 1B) (Perkins et al. 1986; Starich et al. 1995). We found that in adult mapk-15(gk1234) mutants, < 20% of PHA/B phasmid neurons fill with detectable levels of dye, whereas the amphid sensory neurons retain dye uptake, albeit at slightly reduced levels (Figure 1B). However, using a sensitized dye uptake assay (see Materials and Methods), adult mapk-15(gk1234) animals display a strong Dyf defect in the amphid neurons and a fully penetrant dye filling defect in the phasmid neurons (Figure 1B). The Dyf defect is rescued by wild-type mapk-15 genomic sequence driven under the endogenous promoter (Figure 1B).
We next examined the morphologies of individual sensory neurons in mapk-15 mutants using cell type-specific soluble fluorescent reporters. Both mapk-15(oy112) and mapk-15(gk1234) mutants exhibit a range of defects in the ciliary morphologies of the AWB amphid sensory neuron pair (Figure 1C). In addition, the rod-shaped ciliary axonemes of the ASH and ADF amphid head, and PHA/B phasmid tail, neuron pairs are significantly affected upon loss of mapk-15 function (Figure 1, D and E). In all cases, we observed severe structural defects in mapk-15(gk1234) cilia, including axonemes that are truncated, misdirected, collapsed, or missing (Figure 1, C–E). The AWB ciliary morphological defects in mapk-15(gk1234) animals are rescued by transgenic expression of the wild-type mapk-15 genomic sequence, and the AWB and ADF cilia phenotypes are rescued by the MAPK-15c isoform driven under the endogenous mapk-15 promoter (Figure 1, C and D). Transgenic expression of MAPK-15c driven under the ciliated neuron-specific promoter of bbs-8 also rescues the ADF cilium defects of mapk-15(gk1234) animals (Figure 1D), indicating that MAPK-15 functions in sensory neurons to regulate cilium structure. In contrast, transgenic expression of the kinase-dead mutants, MAPK-15c(K42A) and MAPK-15c(TEY > AEA) (Klevernic et al. 2006; see Figure 1A), or a MAPK-15c protein lacking the C-terminal 57 amino acids, fail to rescue the mapk-15(gk1234) ADF cilium defects (Figure 1D). These results indicate that the ciliary role of this protein is dependent on its kinase function.
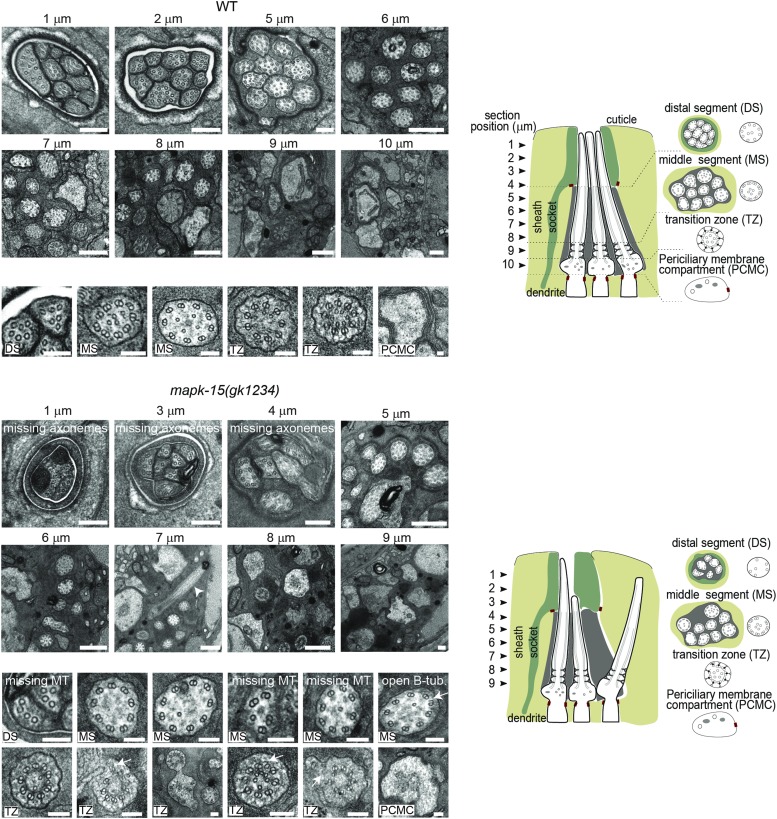
To examine the cilium structure defects of mapk-15 mutants in greater detail, we used transmission electron microscopy to investigate the amphid sensory pores, each of which contains 10 cilia from eight neurons (ADF and ADL are biciliated). In wild-type animals, each amphid channel axoneme consists of a modified basal body, a ∼0.8 μm long transition zone, followed by a ∼3 μm long middle (containing doublet microtubules) and ∼4 μm long distal segments (containing singlet microtubules) (Perkins et al. 1986; Doroquez et al. 2014). In mapk-15(gk1234) animals, 3–6 axonemes are absent from the distal pore regions, indicating that a subset of cilia are truncated in these mutants (Figure 2). We also observed axonemes that are misdirected, curving away from the main ciliary bundle (see image at 7 μm in mapk-15(gk1234) in Figure 2). In addition, a subset of cilia appear to lack the full complement of nine microtubule doublets in the middle segment and transition zone regions, and we occasionally observed misplaced doublet microtubules in the transition zones (Figure 2).
Figure 2.
Ultrastructure of amphid channel cilia in WT and mapk-15(gk1234) worms. Representative transmission election microscopy images from serial cross-sections of the amphid pore. Large panels show the entire pore; small panels show individual ciliary axonemes at higher magnifications. Schematics summarize the ultrastructure phenotypes (only 3 of the 10 axonemes are shown for simplicity). Section positions denoted above each image and in schematics. Arrowhead: misdirected ciliary axoneme. Arrows: mispositioned MTs or incomplete B-tubs. Bar, 200 nm (large panels) and 100 nm (small panels). B-tub, B-tubules; DS, distal segment; MS, middle segment; MT, microtubule; PCMC, periciliary membrane component; TZ transition zone.
MAPK-15 is expressed in ciliated sensory neurons
To determine which cells express mapk-15, we constructed a transcriptional gfp reporter (mapk-15p::gfp) under the control of ∼1.8 kb of mapk-15 upstream genomic regulatory sequence. mapk-15 constructs using the same promoter were sufficient to rescue the Dyf defects of mapk-15(gk1234) mutants (Figure 1B). Worms expressing mapk-15p::gfp display GFP expression in many, if not all, ciliated cells, including all amphid and phasmid neurons (Figure 3A). Little or no GFP expression was observed in nonciliated cells (Figure 3A), suggesting that MAPK-15 promoter activity is largely restricted to ciliated cells. It remains possible that regulatory elements present elsewhere in the genomic sequences may drive expression in other cell types.
Expression of many ciliary genes in C. elegans requires the regulatory factor X transcription factor, DAF-19, which binds a 14-bp X-box promoter motif (Swoboda et al. 2000). Indeed, a modestly-scoring X-box motif occurs ∼200-bp upstream of the mapk-15 start codon (Blacque et al. 2005) (Figure S2A in File S1). However, mapk-15p::gfp expression is not grossly affected in head ciliated neurons of daf-19(m86) loss-of-function mutants, although expression in tail neurons is reduced in this mutant background (Figure S2A in File S1). Consistent with only a weak or cell-specific effect of DAF-19 on mapk-15 regulation, expression profiling data has previously shown that mapk-15 transcription is not significantly altered in daf-19(m86) worms (Chen et al. 2006). Thus, ciliated neuron-specific expression of mapk-15 relies on DAF-19 activity only in a subset of sensory neurons.
C. elegans MAPK-15 localizes to the basal body, ciliary axoneme, and cell–cell junctions
We subsequently employed fluorescence protein-tagged MAPK-15 fusion proteins to examine the subcellular localization of MAPK-15 in ciliated cells. A functional MAPK-15c fusion protein (see Figure 1, C and D) localizes at the ciliary base and along the axonemes of most or all ciliated neurons (Figure 3, B–E and Figure S2B in File S1). The localization is similar in wild-type and mapk-15 mutants (Figure S2C in File S1), suggesting that the localization pattern is unlikely a consequence of overexpression. An identical localization pattern is also found for worms with a mapk-15a::gfp transgene expressed at low levels (Figure S2B in File S1). In cilia, MAPK-15c appears to localize preferentially to the middle segments in examined neurons (Figure 3, C–E and Figure S2, B and C in File S1). At the ciliary base, MAPK-15 is largely excluded from the transition zone marked with TMEM-107 and NPHP-4, but colocalizes with the basal body marker GASR-8 (Figure 3, B and D).
In addition to accumulating at the basal body and axoneme, we observed a prominent pool of MAPK-15 ∼1–2 μm proximal to the basal body, and in some images, the two pools appear to be connected by a strip of MAPK-15 signal (Figure 3, B–E and Figure S2, B and C in File S1). Based on its position, the proximal pool coincides with the proximal side of the periciliary membrane compartment (PCMC) where cell junctions (variably referred to as tight, belt, or apical junctions) connect the neuronal dendrites with a supporting sheath cell (Figure 3F) (Ware et al. 1975; Perkins et al. 1986; Doroquez et al. 2014). Indeed, MAPK-15c colocalizes with the junction marker AJM-1 in both ADF and PHA/B cells (Figure 3E). Together, these results indicate that MAPK-15 localizes to the ciliary axoneme, basal body, and apical junctions in sensory neurons (Figure 3F).
We subsequently asked whether molecules associated with structures at the ciliary base and/or involved in ciliary transport regulate MAPK-15 localization. We examined MAPK-15 distribution in loss-of-function alleles of IFT (osm-5/IFT88, che-11/IFT140, xbx-1/DLIC, klp-11/KIF3B, and osm-3/KIF17) and BBSome (bbs-8/BBS8) genes. We also investigated MAPK-15 localization in hyls-1 and dyf-5 mutants: HYLS-1 (Hydrolethalus syndrome protein 1) at the basal body regulates the formation of the transition fibers that dock IFT assemblies, and DYF-5 (MAK ortholog) is a MAP kinase-related protein that regulates kinesin-2 IFT motors (Burghoorn et al. 2007; Mohan et al. 2013; Wei et al. 2016). MAPK-15 retains grossly normal localization in most examined mutants despite the ciliary structural defects observed in many of these strains (Figure S3 in File S1). However, while MAPK-15 localization is retained at the axoneme and basal body in ∼51% of hyls-1 mutant animals, the accumulation of the fusion protein at the apical junction is fully abolished in these worms (Figure 3G). A cell junction marker (AJM-1::CFP) confirms that apical junction structure is retained in hyls-1 mutants (Figure 3H). Thus, the loss of MAPK-15 apical junction localization is not because these junctions are missing in hyls-1 worms. These results suggest that HYLS-1 plays a role in the correct localization of MAPK-15.
MAPK15 localizes to the distal end of the basal body in human cells
MAPK15 has previously been shown to localize to the basal bodies of motile cilia in vertebrates (Miyatake et al. 2015). Therefore, we asked whether MAPK15 also localizes to the base of primary cilia in human cells, similar to our findings in C. elegans sensory neurons. We found that overexpressed GFP-tagged MAPK15 preferentially accumulates at the basal bodies of hTERT-RPE1 and RPTEC/TERT primary cilia, and also decorates the daughter centriole in a subset of cells to a lesser extent (Figure 4, A and B and Figure S4E in File S1). Unlike in C. elegans, human MAPK15 is not found in the ciliary axonemes (Figure 4, A–D and Figure S4, A and E in File S1). However, reminiscent of MAPK-15 distribution to apical junctions in C. elegans, MAPK15::GFP colocalizes with the tight junction protein ZO-1 at RPTEC/TERT cell–cell junctions (Figure 4B).
Figure 4.
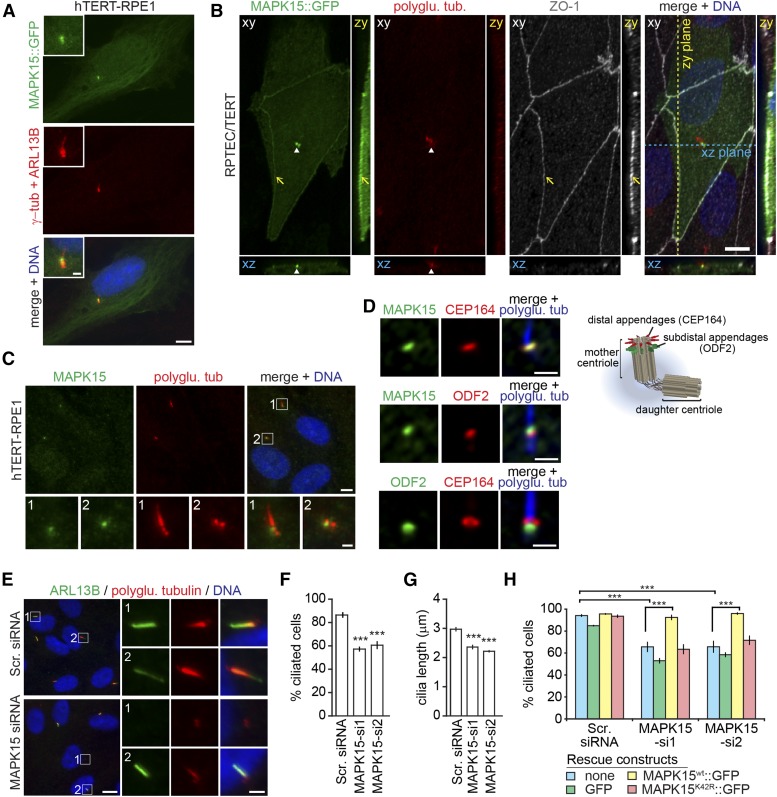
MAPK15 localizes to the ciliary base and regulates ciliogenesis in human cells. (A) Representative images of hTERT-RPE1 cells transiently transfected with MAPK15::GFP, serum starved for 24 hr ,and stained for γ-tubulin (centrosome), ARL13B (cilia), and DNA. Insets show higher magnification images of the centrosomal region. Bar, 5 µm (main images) and 1 µm (magnifications). (B) Representative images of RPTEC/TERT cells transiently transfected with MAPK15::GFP and stained for polyglutamylated tubulin (polyglu. tub, cilia), ZO-1 (tight junctions), and DNA. Blue and yellow dotted lines indicate positions of the XZ and YZ cross‐sections, respectively. Arrows, tight junctions; arrowheads, basal body. Bar, 5 µm. (C) Representative images of serum-starved hTERT-RPE1 stained for endogenous MAPK15, polyglutamylated tubulin (centrioles and cilia), and DNA. Regions within white boxes are shown at higher magnifications in the lower panels. Bar, 5 µm (main images) and 1 µm (magnifications). (D) Representative images of hTERT-RPE1 cells stained for the indicated endogenous proteins. Schematic of centrosome depicts CEP164 and ODF2 localization to distal and subdistal appendages, respectively. Bar, 1 µm. (E–G) Representative images of serum-starved hTERT-RPE1 cells transfected with scrambled siRNA (Scr. siRNA) or an siRNA targeting MAPK15 (MAPK15-si1). Cells stained for ARL13B, polyglutamylated tubulin, and DNA. Regions within white boxes shown at higher magnifications to the right. Bar, 5 µm (main images) and 1 µm (magnifications). ARL13B staining used to mark cilia and determine the percentages of ciliated cells (F) and cilia lengths (G). Data shown are means of five independent experiments. Error bars are ± SEM. For (F), 100–150 cells were assessed per experimental condition. For (G), 30–60 cilia were scored per experimental condition. *** P < 0.001 (ANOVA followed by Dunnett’s post hoc test; vs. Scr. siRNA). (H) hTERT-RPE1 cells stably expressing the indicated rescue constructs were treated as in (E) and percentages of ciliated cells determined using ARL13B as a cilia marker. Data points represented as mean ± SEM of three independent experiments. Per experimental condition, 100–150 cells were scored. *** P < 0.001 (ANOVA followed by Bonferroni post hoc test).
To determine the localization of endogenous MAPK15, we stained hTERT-RPE1 cells using a polyclonal antibody (S. W. Yang et al. 2013) and found a specific signal at the basal body/mother centriole in ciliated and nonciliated cells, with no signals at the daughter centriole (Figure 4C and Figure S4A in File S1). We confirmed the specificity of the staining by observing loss of signal in hTERT-RPE1 cells depleted for MAPK15 via RNA interference (Figure S4A in File S1). The ectopic recruitment of GFP-tagged MAPK15 to the daughter centriole may be explained by its overexpression, similar to what has been previously reported for other mother centriole-associated proteins such as ODF2 and TTBK2 (Soung et al. 2009; Cajanek and Nigg 2014).
To further characterize MAPK15 localization to the basal body, we costained hTERT-RPE1 cells for endogenous CEP164 and ODF2, which mark the distal and subdistal appendages of the mother centriole, respectively (Nakagawa et al. 2001; Graser et al. 2007) (Figure 4D). We found that the MAPK15 signal partially overlaps with both ODF2 and CEP164, and appears to be shifted distally to the ODF2 signal, suggesting association of MAPK15 with the distal end of the mother centriole (Figure 4D). These data indicate that, like in C. elegans, MAPK15 localizes to the basal body of primary cilia and cell–cell junctions in human cells.
MAPK15 regulates primary cilium formation in human cells
We next investigated a possible role for MAPK15 in the formation of human primary cilia by depleting MAPK15 in hTERT-RPE1 cells. Two independent siRNAs were used, both of which cause significant reductions in MAPK15 expression (Figure S4, A and B in File S1). Cilia formation was induced by serum withdrawal (48 hr), and cilia were visualized by staining for components of the ciliary membrane (ARL13B) and axoneme (polyglutamylated tubulin). Compared to a negative control (scrambled siRNA-treated), fewer MAPK15-depleted cells exhibit cilia (Figure 4, E and F). Moreover, cilia that are retained upon MAPK15 depletion are shorter than those in the control cells (Figure 4, E and G). Cilia loss is rescued by the expression of siRNA-resistant wild-type MAPK15 but not MAPK15(K42R) (kinase dead) (Figure 4H and Figure S4E in File S1), demonstrating that the cilium formation phenotype is not due to siRNA off-target effects and that the kinase activity of MAPK15 is crucial for its ciliogenesis function. We also confirmed by immunostaining with antibodies against the Ki-67 proliferation marker that the ciliogenesis phenotype in MAPK15-depleted cells is not an indirect consequence of defects in cell cycle arrest (Figure S4, C and D in File S1). Thus, as in C. elegans, human MAPK15 regulates primary cilium formation, which is dependent on its kinase activity.
MAPK15 is required for the localization of a subset of structural, ciliary signaling, and trafficking proteins in human cells and in C. elegans
The localization of MAPK15 to basal bodies, and the disruption of axonemal and transition zone ultrastructure in mapk-15 mutants, in C. elegans suggested that MAPK15 may facilitate ciliogenesis via regulation of basal body/transition zone structure and/or ciliary gating functions. To investigate this hypothesis, we examined the distribution of structural and ciliary proteins in mapk-15 mutant worms and in MAPK15-depleted human cells.
Basal body/transition zone/cell junction proteins:
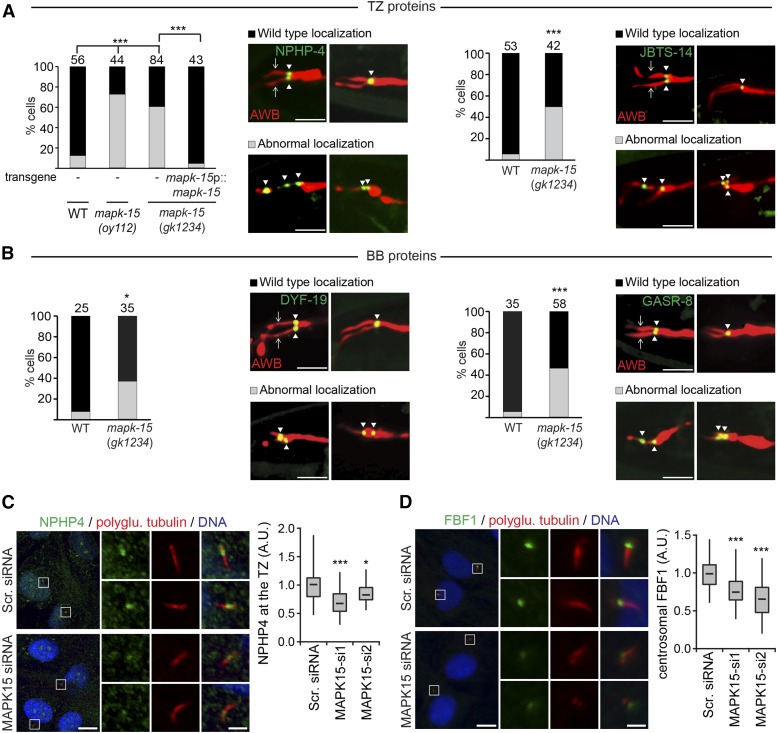
We observed mislocalization of the transition zone fusion proteins NPHP-4::GFP and JBTS-14::GFP, as well as the basal body-associated proteins DYF-19/FBF1::GFP and GFP::GASR-8 in AWB cilia in mapk-15 mutants (Figure 5, A and B). In all cases, we noted that the fusion proteins often accumulate at ectopic proximal locations in mapk-15 mutants (Figure 5, A and B). The NPHP-4::GFP mislocalization defects in AWB were fully rescued upon expression of wild-type mapk-15 sequence (Figure 5A). We also observed reduced levels of MKS-2::GFP and MKS-3::GFP at the transition zones of amphid neurons in mapk-15 mutants (Figure S5A in File S1). However, overall localizations of the MKS-5::GFP (transition zone) and AJM-1::CFP (apical junction) protein are largely unaltered in amphid and phasmid cilia in mapk-15 mutants (Figure S5, A and B in File S1). Similarly, in hTERT-RPE1 cells, MAPK15 depletion significantly reduces the levels of NPHP4 at the transition zone and FBF1 at the centrosome (Figure 5, C and D). Thus, MAPK15 regulates the localization of a subset of proteins associated with the ciliary base in both C. elegans and human cells.
Figure 5.
Localization of transition zone (TZ) and basal body (BB) proteins is disrupted in MAPK15-depleted human cells and MAPK-15 mutant worms. (A and B) Representative images and quantification of NPHP-4::GFP, JBTS-14::GFP, DYF-19::GFP, and GFP::GASR-8 localization in the AWB neurons of wild-type (WT) and mapk-15(gk1234) worms. AWB neurons visualized using str-1p::mCherry. GFP-tagged proteins expressed under the str-1 or srd-23 (GASR-8) promoter. Transgene used for rescue is mapk-15p::mapk-15c. Numbers of neurons examined are indicated above each data set. * P < 0.05, ** P < 0.01, and *** P < 0.001 (Generalized Linear Model test and Bonferroni corrections). Arrowheads denote GFP puncta. Arrows denote ciliary axonemes. Anterior is at left. Bar, 5 µm (all images identically scaled). (C and D) Representative images of hTERT-RPE1 cells transfected with nontargeting scrambled siRNA (Scr. siRNA) or an siRNA targeting MAPK15 (MAPK15-si1), serum starved for 48 hr, and stained for NPHP4 (C) or FBF1 (D), polyglutamylated (polyglu.) tubulin, and DNA. Regions within white boxes shown at higher magnification in smaller panels. Bar, 10 µm (large panels) and 2 µm (small panels). Box-and-whisker plots show the relative intensity of NPHP4 at the transition zone (TZ) (C), and FBF1 at the centrosome (D). Horizontal lines denote the 25, 50, and 75th percentiles; whiskers denote the full range. One representative experiment out of three is shown. Per experimental condition, 30–60 cells were scored.* P < 0.05 and *** P < 0.001 (ANOVA followed by Dunnett’s post hoc test; vs. Scr. siRNA). A.U.: arbitrary units.
Ciliary proteins:
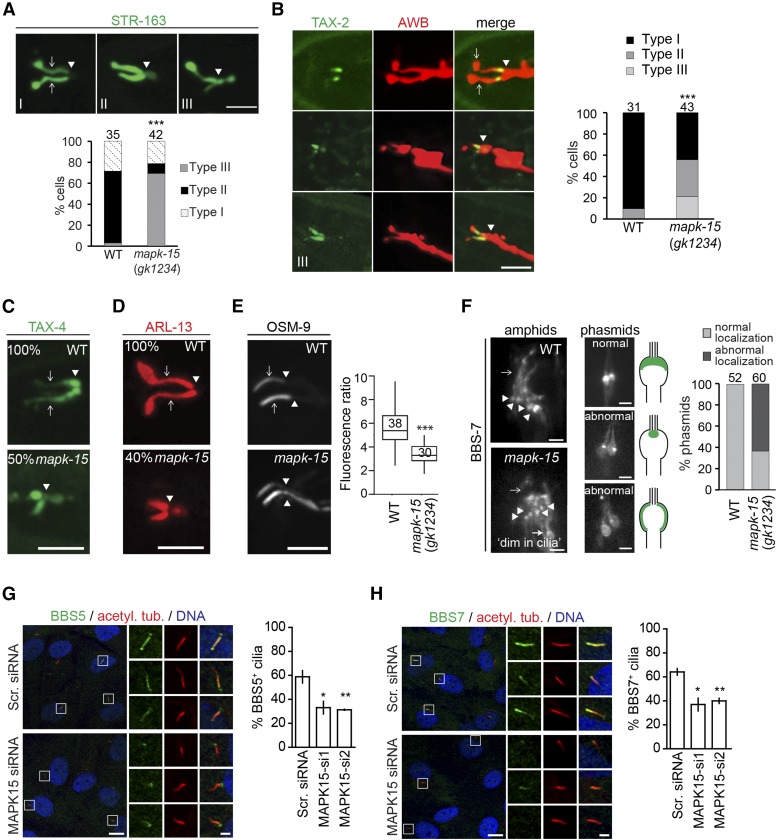
Since the transition zone acts as a ciliary gate, we explored whether altered distribution of transition zone proteins results in disrupted trafficking of ciliary molecules. We found that localization of the ciliary G protein-coupled receptor STR-163, cyclic nucleotide-gated channel subunits TAX-2 and TAX-4, and the membrane-associated small GTPase ARL-13, are affected in the AWB neurons of mapk-15 mutants (Figure 6, A–D). In all cases, accumulation of these GFP fusion proteins is observed at the distal dendritic tip, reminiscent of phenotypes observed in animals with defects in transition zone gating (Cevik et al. 2013; Brear et al. 2014). Consistent with the notion of a leaky ciliary gate in mapk-15 mutant worms, the transmembrane transient receptor potential vanilloid (TRVP) channel protein OSM-9 also shows abnormal accumulations in the PCMC and far distal dendrite region of OLQ cilia (Figure 6E). However, the examined IFT proteins (CHE-2/11/13, OSM-3/5/6, DYF-1/2/3/11/13, KAP-1, and XBX-1) display largely normal distributions in the amphid and phasmid channel cilia of mapk-15 mutants (Figure S5C in File S1). These observations indicate that loss of MAPK15 disrupts the localization of select ciliary proteins.
Figure 6.
Localization of ciliary membrane and BBSome proteins in C. elegans and human cells. (A and B) Representative images and quantification of ciliary transmembrane STR-163::GFP (seven transmembrane receptor) and TAX-2::GFP (cyclic nucleotide-gated ion channel subunit) localization in the AWB neurons of wild-type (WT) and mapk-15(gk1234) worms. AWB neurons visualized using an str-1p::mCherry reporter. STR-163 localization categorized as: Type I, signal restricted to cilia; Type II, additional diffuse localization in the PCMC; and Type III, additional localization in the distal dendrite. TAX-2 localization was categorized as: Type I, restricted to ciliary middle segment; Type II, additional localization in the distal dendrite; and Type III, localization throughout the entire ciliary axonemes. N-values indicated above each data set. Only cells retaining both ciliary axonemes were analyzed. Arrowheads denote cilium base. Arrows denote ciliary axoneme (shown only for Type I). Anterior is at left. Bar, 5 µm. (C and D) Representative images and quantification of ciliary transmembrane TAX-4::GFP (cyclic nucleotide-gated ion channel subunit) and membrane-associated ARL-13::tagRFP localization in the AWB neurons of WT and mapk-15(gk1234) worms. Panels show the percentage of worms with indicated pattern of TAX-4 and ARL-13 localization (n ≥ 22 for WT and mapk-15 worms). Only cells retaining both ciliary axonemes were analyzed. Arrowheads denote cilium base. Arrows denote ciliary axonemes (shown only for WT images). Anterior is at left. Bar, 5 µm. (E) Representative images and quantification of OSM-9::GFP (TRPV channel) localization in OLQ neurons. Box plots show the ratio of GFP signal (maximum fluorescence) at the distal ciliary region compared with the proximal ciliary region (and distal dendrite). Horizontal lines denote 25, 50, and 75th percentile; whiskers denote the full range. N-values indicated above each data set. Arrows denote ciliary axonemes (shown only for WT). *** P < 0.001 (Welch’s t-test vs. WT). Anterior is at left. Bar, 5 µm. (F) Representative images and quantification of BBS-7::GFP localization in amphid and PHA/B (phasmid) cilia of WT and mapk-15(gk1234) worms. Schematics show normal and abnormal BBS-7 localizations at the ciliary base. For phasmids, normal and abnormal localization phenotypes also shown in schematics. n > 50 for each data set in graph. Arrowheads denote cilium base. Arrows denote ciliary axonemes (shown only for WT images). Anterior is at top in amphid images and at bottom in phasmid images. Bar, 2 µm. (G and H) Representative images of hTERT-RPE1 cells transfected with nontargeting scrambled siRNA (Scr. siRNA) or an siRNA targeting MAPK15 (MAPK15-si1), serum starved for 48 hr and stained for BBS5 (G) or BBS7 (H), acetylated tubulin (acetyl. tub), and DNA. Regions within white boxes shown at higher magnifications in smaller image panels. Bar, 10 µm (large panels) and 2 µm (small panels). Bar graphs show percentages of cilia positive for BBS5 (G) or BBS7 (H). Data points represented as mean ± SEM of three independent experiments. Per experimental condition, 100–150 cells were scored. * P < 0.05 and ** P < 0.001 (ANOVA followed by Dunnett’s post hoc test; vs. Scr. siRNA).
BBSome proteins:
The BBSome acts as an adaptor for the trafficking of a range of ciliary signaling proteins (Berbari et al. 2008; Lechtreck et al. 2009; Jin et al. 2010; Domire et al. 2011; Cevik et al. 2013; Loktev and Jackson 2013; Xu et al. 2015). The altered localization of ciliary transmembrane proteins in mapk-15 mutants raised the possibility that MAPK15 regulates BBSome localization and/or function. To test this hypothesis, we examined the localization of BBS proteins in C. elegans mapk-15 mutant cilia and MAPK15-depleted human cells. In mapk-15 mutants, significantly reduced and altered BBS-7::GFP signals are found at the basal body of amphid and phasmid neurons, and in some worms, BBS-7 localization within the ciliary axonemes is also diminished (Figure 6F). Similarly, we observed a reduced number of BBS5- and BBS7-positive cilia in MAPK15-depleted hTERT-RPE1 cells as compared to controls (Figure 6, G and H). These findings suggest that MAPK15 may regulate the trafficking of ciliary proteins by impacting BBSome localization in C. elegans and human ciliated cells.
MAPK15 and BBS7 occupy a distinct subcompartment near the distal end of the basal body
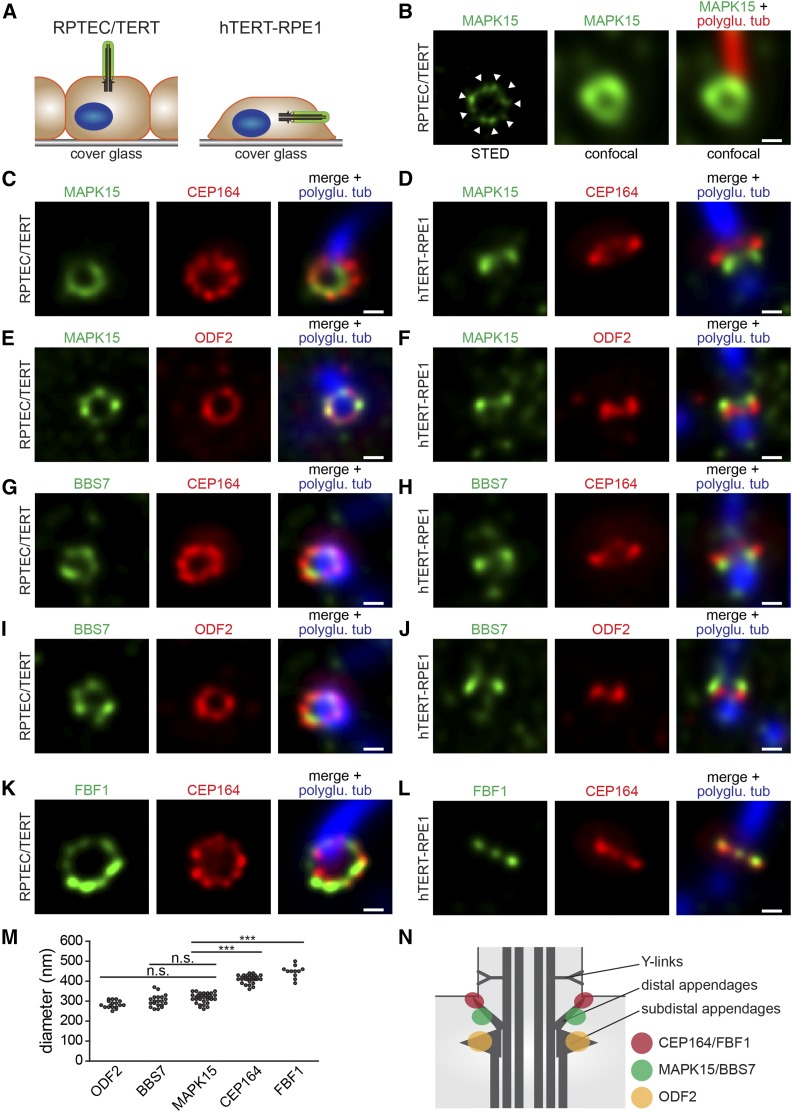
A conserved requirement for MAPK15 at the basal body in recruiting ciliary transport regulators (BBS7 and FBF1) prompted us to employ STED (superresolution) microscopy in human cells to investigate the precise spatial organization of basal body proteins in relation to MAPK15. First, we used single-color STED microscopy to image endogenous MAPK15 in RPTEC/TERT1 cells. In these cells, the cilium lies perpendicular to the imaging plane and is imaged in cross section (Figure 7A). Compared to confocal images, the improved resolution of STED reveals clusters of MAPK15 signal arranged as a ring at the ciliary base (Figure 7B). In most rings, six to eight or possibly nine individual clusters of signal can be resolved (Figure 7B), which suggests that MAPK15 associates with a specific basal body substructure. A similar distribution pattern has been described previously for CEP164 at the distal appendages (Lau et al. 2012).
Figure 7.
MAPK15 and BBS7 localize to a distinct subdomain near the distal end of the basal body in human cells. (A) Schematic depicting primary cilium orientation relative to the imaging plane for RPTEC/TERT and hTERT-RPE1 cells. Basal bodies and cilia imaged in cross-section orientation for RPTEC/TERT cells and longitudinal orientation for hTERT-RPE1 cells. (B) Representative single-color stimulated emission depletion (STED) image of endogenous MAPK15 at the basal body in RPTEC/TERT cells. Corresponding confocal images of MAPK15 and polyglutamylated tubulin staining shown to the right. Arrowheads indicate MAPK15 clusters. Bar, 200 nm. (C–L) Representative dual-color STED fluorescence images of RPTEC/TERT (C, E, G, I, and K) and hTERT-RPE1 (D, F, H, J, and L) cells costained with the indicated antibodies. Right-hand panels show merges of both STED images with corresponding confocal image of polyglutamylated tubulin (polyglu. tub) staining. Bar, 200 nm. (M) Scatter plots showing the distribution of ring diameters for ODF2, BBS7, MAPK15, CEP164, and FBF1 signals at the distal end of the basal body. *** P < 0.001 (Kruskel–Wallis followed by Dunnett’s post hoc test). (N) Cartoon summarizing the observed localization patterns. MAPK15/BBS7 are proposed to occupy a more proximal (and narrower) domain than CEP164/FBF1 at the distal appendages.
Next, we employed dual-color STED imaging to compare the localization of endogenous MAPK15 with that of CEP164, ODF2, BBS7, and FBF1. In RPTEC/TERT1 cells, we found that all these proteins form rings at the ciliary base (Figure 7, C, E, G, I, and K). We noted that the diameters of the CEP164 and FBF1 rings are larger (∼410–450 nm) than the BBS7, ODF2, and MAPK15 rings (∼280–320 nm each) (Figure 7M). Since it was difficult to precisely determine the spatial organization of these proteins along the axial plane in RPTEC/TERT1 cells due to the cilium being perpendicular to the imaging plane, we instead imaged hTERT-RPE1 cells where cilia lie parallel to the imaging plane (Figure 7A). We found that while CEP164 colocalizes with FBF1 (Figure 7L), MAPK15 and BBS7 associate with a more proximal domain (Figure 7, D and H). In contrast, the MAPK15 and BBS7 signals lie immediately distal to the ODF2 domain (Figure 7, F and J). Together, our results indicate that MAPK15 and BBS7 share a distinct basal body subdomain sandwiched between CEP164/FBF1 (distal appendages) and ODF2 (subdistal appendages) subdomains (Figure 7N). A shared localization domain for MAPK15 and BBS7 is also consistent with our observation that MAPK15 regulates BBSome protein localization in worms and human cells.
Discussion
A conserved role for MAPK15 in primary cilium formation
We have found that loss of C. elegans MAPK-15 disrupts sensory neuron cilium length, morphology, and position, as well as microtubule number and integrity. Similarly, human MAPK15 depletion in hTERT-RPE1 epithelial cells abrogates ciliogenesis and reduces ciliary length. Together, these observations indicate that MAPK15 plays a conserved role in regulating primary cilium formation and structure. Our findings are consistent with, and expand upon, a role for MAPK15 in regulating the number and length of motile cilia in Xenopus embryos and mouse multiciliated cells (Miyatake et al. 2015). We conclude that MAPK15 regulates the formation of both motile and immotile cilia.
MAPK15 regulates ciliary protein localization
We observed partially disrupted localization of multiple ciliary proteins in MAPK15-depleted human cells and C. elegans mapk-15 mutants. Although the effects on protein distribution are partly distinct in worm and human ciliated cells, the affected proteins in both systems include ciliary base and transition zone molecules that regulate ciliary protein entry/exit, transition fiber formation, transition zone integrity, and basal body stability (Huang et al. 2011; Williams et al. 2011; Mohan et al. 2013; Wei et al. 2013, 2016). MAPK15 is also required for correct localization of ciliary axonemal proteins including ciliogenic trafficking regulators (BBS7 and ARL-13), and subsets of sensory signaling molecules. In addition, MAPK-15 has very recently been shown to regulate the ciliary localization of the PKD-2 polycystin transmembrane protein in males (Piasecki et al. 2017). Thus, MAPK15 regulates the distribution of a wide range of ciliary transport-associated proteins linked to cilium formation and function.
A potential mechanism by which MAPK15 influences the localization of a seemingly diverse set of proteins may be modulation of ciliary gating. Indeed, a subset of our data for MAPK15 correlates with findings for HYLS-1 and DYF-19, which are key components of the ciliary gate that also accumulate at the distal end of the basal body (discussed further below) (Wei et al. 2013, 2016). Like MAPK15, HYLS-1 regulates basal body recruitment of DYF-19 and GASR-8 (Wei et al. 2016), and we report here that HYLS-1 also regulates localization of MAPK-15. Similar to MAPK15 (C. elegans and human), DYF-19 and/or HYLS-1 regulate the basal body levels and ciliary entry of the BBSome IFT cargo adaptor, as well as the ciliary restriction of OSM-9 (Wei et al. 2013, 2016). A gating function for MAPK15 is also consistent with our observation that MAPK15 facilitates ciliary restriction of ARL-13 (worms) and the localization of several ciliopathy-associated transition zone proteins implicated in diffusion barrier control (worms and human cells) (Williams et al. 2011; Cevik et al. 2013). Based on these observations, we speculate that MAPK15 may function as a ciliary gate regulator at the basal body and transition zone, together with HYLS-1 and DYF-19/FBF1. However, compared to known ciliary gate gene mutants, the “gating” phenotype of mapk-15 null mutants is less penetrant, suggesting that MAPK15 facilitates gating efficiency rather than being a core component of the gating machinery. It also remains possible that MAPK15 has additional cilia-related roles, and that the gating defects are a secondary consequence of loss of MAPK15 function in other ciliary pathways. We note that in a recent study, MAPK-15 is implicated in synaptic trafficking of the dopamine transporter protein in C. elegans dopaminergic neurons (Bermingham et al. 2017), suggesting a role for MAPK-15 in regulating protein transport at multiple sites in C. elegans ciliated sensory neurons.
Dual localization of MAPK15 to the basal body and cell–cell junctions
We found that MAPK15 localizes to the basal body of primary cilia in C. elegans and human cells, in agreement with findings for MAPK15 in Xenopus multiciliated cells (Miyatake et al. 2015). We refined the basal body distribution of MAPK15 using superresolution (STED) imaging and found that this kinase forms clusters, approaching nine in number, arranged as a ring toward the distal end of the basal body. In addition, dual-color STED imaging reveals that MAPK15 and BBS7 occupy a discrete axial subdomain of the basal body that lies between CEP164/FBF1 (distal appendages) and ODF2 (subdistal appendages) subdomains. These findings suggest that MAPK15/BBS7 associate with distinct basal body structural elements, with one possibility being that CEP164 and FBF1 decorate the tips of the distal appendages, whereas MAPK15/BBS7 localize at more proximal regions of the same appendages (Figure 7N). Indeed, this model agrees with various ultrastructural studies using electron microscopy, indicating that distal appendages taper out from the centriolar microtubules at an oblique angle (Anderson 1972; Ibrahim et al. 2009). An appendage association for the IFT cargo adaptor protein BBS7 is also consistent with the reported localization of IFT machinery at these sites (Deane et al. 2001; T. T. Yang et al. 2013). Furthermore, a shared MAPK15/BBS7 basal body subdomain correlates with the requirement of MAPK15 for BBS7 ciliary localization in worms and human cells. However, MAPK-15 is also associated with the axoneme middle segments in C. elegans but not in human cells, suggesting that MAPK-15 may have additional cilia-associated functions in worms.
We also observed highly enriched pools of MAPK15 at cell junctions in C. elegans and human cells, which is again consistent with findings in Xenopus embryos (Miyatake et al. 2015). In worms, these cell junctions occur between the dendritic endings and the supporting glial cells (Perkins et al. 1986), and may play a role in anchoring basal bodies (Nechipurenko et al. 2016). A functional connection between the C. elegans basal body and cell junction is also supported by our observation that HYLS-1 at the ciliary base controls MAPK-15 localization at the cell junction. While, to our knowledge, MAPK-15 is the first protein for which a dual basal body/junction localization has been described in C. elegans, similar dual distribution has been reported for several NPHP proteins in cultured mammalian cells (Delous et al. 2009; Sang et al. 2011). Although these NPHP proteins have also been shown to regulate junction formation (Delous et al. 2009; Sang et al. 2011), MAPK-15 does not appear to modulate the localization of the apical junction protein AJM-1 in C. elegans sensory neurons. However, it is possible that the altered axoneme positioning observed in mapk-15 mutants reflects defects in apical junction structure/function. Therefore, we suggest that MAPK15 plays dual roles at the basal body and associated cell junctions.
Conclusions
In multiciliated cells, MAPK15 has been shown to regulate the apical actin network and basal body migration in part via phosphorylation of the actin regulator CapZIP (Miyatake et al. 2015). Our current data do not support a role for MAPK15 in the migration and docking of basal bodies in cells containing primary cilia. Instead, MAPK15 appears to organize the basal body region to ensure proper localization of ciliary proteins. Moreover, the C. elegans genome does not encode a CapZIP ortholog, suggesting that the targets of MAPK15 function may be different for ciliogenesis of motile and primary cilia. In the future, identification of MAPK15 substrates in cells with primary cilia will allow us to further characterize the shared and divergent molecular mechanisms that underlie the generation and function of different cilia types across species.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300383/-/DC1.
Acknowledgments
We are grateful to M. Barr, J. Hu, M. Leroux, M. Heiman, T. McMorrow, J. Simpson, V. Hernandez-Hernandez, P. Beales, and G. Pereira for reagents, and K. Oegema and the Caenorhabditis Genetics Center for worm strains. We thank Pin-Hao Chao for technical assistance, Dimitri Scholz, Tiina O’Neill (both University College Dublin Conway Institute Imaging Facility), and Una Prendergast [Dublin City University (DCU) Nano Research Facility] for microscopy assistance, and members of the Sengupta and Blacque laboratories for discussion and advice. Stimulated Emission Depletion imaging was carried out at the Nano Research Facility at DCU, which was funded under the Programme for Research in Third Level Institutions (PRTLI) Cycle 5. The PRTLI is cofunded through the European Regional Development Fund, part of the European Union Structural Funds Programme 2011–2015. This work was funded in part by the National Institutes of Health (R37 GM-56223 and R35 GM-22463 to P.S. and T32 GM-7122 to A.K.), Science Foundation Ireland (11/PI/1037 to O.E.B.), and the European Community’s Seventh Framework Programme FP7/2009 (SYSCILIA grant agreement 241955 to O.E.B.).
Footnotes
Communicating editor: D. Greenstein
Literature Cited
- Abdul-Majeed S., Moloney B. C., Nauli S. M., 2012. Mechanisms regulating cilia growth and cilia function in endothelial cells. Cell. Mol. Life Sci. 69: 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe M. K., Kahle K. T., Saelzler M. P., Orth K., Dixon J. E., et al. , 2001. ERK7 is an autoactivated member of the MAPK family. J. Biol. Chem. 276: 21272–21279. [DOI] [PubMed] [Google Scholar]
- Abe M. K., Saelzler M. P., Espinosa R., III, Kahle K. T., Hershenson M. B., et al. , 2002. ERK8, a new member of the mitogen-activated protein kinase family. J. Biol. Chem. 277: 16733–16743. [DOI] [PubMed] [Google Scholar]
- Anderson R. G., 1972. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J. Cell Biol. 54: 246–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari N. F., Lewis J. S., Bishop G. A., Askwith C. C., Mykytyn K., 2008. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc. Natl. Acad. Sci. USA 105: 4242–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham D. P., Hardaway J. A., Refai O., Marks C. R., Snider S. L., et al. , 2017. The atypical MAP kinase SWIP-13/ERK8 regulates dopamine transporters through a Rho-dependent mechanism. J. Neurosci. 37: 9288–9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O. E., Perens E. A., Boroevich K. A., Inglis P. N., Li C., et al. , 2005. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 15: 935–941. [DOI] [PubMed] [Google Scholar]
- Bloodgood R. A., 2010. Sensory reception is an attribute of both primary cilia and motile cilia. J. Cell Sci. 123: 505–509. [DOI] [PubMed] [Google Scholar]
- Brear A. G., Yoon J., Wojtyniak M., Sengupta P., 2014. Diverse cell type-specific mechanisms localize G protein-coupled receptors to Caenorhabditis elegans sensory cilia. Genetics 197: 667–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks E. R., Wallingford J. B., 2014. Multiciliated cells. Curr. Biol. 24: R973–R982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghoorn J., Dekkers M., Rademakers S., de Jong T., Willemsen R., et al. , 2007. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104: 7157–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajanek L., Nigg E. A., 2014. Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc. Natl. Acad. Sci. USA 111: E2841–E2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J. B., Bettencourt-Dias M., 2011. Evolution: tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik S., Sanders A. A., Van Wijk E., Boldt K., Clarke L., et al. , 2013. Active transport and diffusion barriers restrict Joubert syndrome-associated ARL13B/ARL-13 to an Inv-like ciliary membrane subdomain. PLoS Genet. 9: e1003977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Karin M., 2001. Mammalian MAP kinase signalling cascades. Nature 410: 37–40. [DOI] [PubMed] [Google Scholar]
- Chen N., Mah A., Blacque O. E., Chu J., Phgora K., et al. , 2006. Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol. 7: R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B., Liu P., Chinn Y., Chalouni C., Komuves L. G., et al. , 2011. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 14: 61–72. [DOI] [PubMed] [Google Scholar]
- Choksi S. P., Lauter G., Swoboda P., Roy S., 2014. Switching on cilia: transcriptional networks regulating ciliogenesis. Development 141: 1427–1441. [DOI] [PubMed] [Google Scholar]
- Colecchia D., Strambi A., Sanzone S., Iavarone C., Rossi M., et al. , 2012. MAPK15/ERK8 stimulates autophagy by interacting with LC3 and GABARAP proteins. Autophagy 8: 1724–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe P., Meloche S., 2007. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta 1773: 1376–1387. [DOI] [PubMed] [Google Scholar]
- Craige B., Tsao C. C., Diener D. R., Hou Y., Lechtreck K. F., et al. , 2010. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell Biol. 190: 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. W., Hammarlund M., Harrach T., Hullett P., Olsen S., et al. , 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane J. A., Cole D. G., Seeley E. S., Diener D. R., Rosenbaum J. L., 2001. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 11: 1586–1590. [DOI] [PubMed] [Google Scholar]
- Delous M., Hellman N. E., Gaude H. M., Silbermann F., Le Bivic A., et al. , 2009. Nephrocystin-1 and nephrocystin-4 are required for epithelial morphogenesis and associate with PALS1/PATJ and Par6. Hum. Mol. Genet. 18: 4711–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domire J. S., Green J. A., Lee K. G., Johnson A. D., Askwith C. C., et al. , 2011. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell. Mol. Life Sci. 68: 2951–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroquez D. B., Berciu C., Anderson J. R., Sengupta P., Nicastro D., 2014. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. Elife 3: e01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond I., 2012. Cilia functions in development. Curr. Opin. Cell Biol. 24: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F. R., Reiter J. F., 2017. Open sesame: how transition fibers and the transition zone control ciliary composition. Cold Spring Harb. Perspect. Biol. 9: a028134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F. R., Corbit K. C., Sirerol-Piquer M. S., Ramaswami G., Otto E. A., et al. , 2011. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43: 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S. C., Ocbina P. J., Anderson K. V., 2009. The primary cilium as a Hedgehog signal transduction machine. Methods Cell Biol. 94: 199–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y. D., Lavoie S. B., Gassner O. S., Lamla S., et al. , 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groehler A. L., Lannigan D. A., 2010. A chromatin-bound kinase, ERK8, protects genomic integrity by inhibiting HDM2-mediated degradation of the DNA clamp PCNA. J. Cell Biol. 190: 575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guemez-Gamboa A., Coufal N. G., Gleeson J. G., 2014. Primary cilia in the developing and mature brain. Neuron 82: 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730. [DOI] [PubMed] [Google Scholar]
- Huang L., Szymanska K., Jensen V. L., Janecke A. R., Innes A. M., et al. , 2011. TMEM237 is mutated in individuals with a Joubert syndrome related disorder and expands the role of the TMEM family at the ciliary transition zone. Am. J. Hum. Genet. 89: 713–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim R., Messaoudi C., Chichon F. J., Celati C., Marco S., 2009. Electron tomography study of isolated human centrioles. Microsc. Res. Tech. 72: 42–48. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., 2017. Axoneme structure from motile cilia. Cold Spring Harb. Perspect. Biol. 9: a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., White S. R., Shida T., Schulz S., Aguiar M., et al. , 2010. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 141: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Dynlacht B. D., 2013. Assembling a primary cilium. Curr. Opin. Cell Biol. 25: 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevernic I. V., Stafford M. J., Morrice N., Peggie M., Morton S., et al. , 2006. Characterization of the reversible phosphorylation and activation of ERK8. Biochem. J. 394: 365–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko H. W., 2012. The primary cilium as a multiple cellular signaling scaffold in development and disease. BMB Rep. 45: 427–432. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Dynlacht B. D., 2011. Regulating the transition from centriole to basal body. J. Cell Biol. 193: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M., Simske J. S., Sims P. A., Firestein B. L., Hall D. H., et al. , 2001. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat. Cell Biol. 3: 983–991. [DOI] [PubMed] [Google Scholar]
- Kuhns S., Schmidt K. N., Reymann J., Gilbert D. F., Neuner A., et al. , 2013. The microtubule affinity regulating kinase MARK4 promotes axoneme extension during early ciliogenesis. J. Cell Biol. 200: 505–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtulmus B., Wang W., Ruppert T., Neuner A., Cerikan B., et al. , 2016. WDR8 is a centriolar satellite and centriole-associated protein that promotes ciliary vesicle docking during ciliogenesis. J. Cell Sci. 129: 621–636. [DOI] [PubMed] [Google Scholar]
- Lanjuin A., Sengupta P., 2002. Regulation of chemosensory receptor expression and sensory signaling by the KIN-29 Ser/Thr kinase. Neuron 33: 369–381. [DOI] [PubMed] [Google Scholar]
- Lau L., Lee Y. L., Sahl S. J., Stearns T., Moerner W. E., 2012. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys. J. 102: 2926–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.-F., Johnson E., Sakai T., Cochran D., Ballif B., et al. , 2009. The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187: 1117–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann C. B., Lesich K. A., 2016. Functional anatomy of the mammalian sperm flagellum. Cytoskeleton (Hoboken) 73: 652–669. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Loktev A. V., Jackson P. K., 2013. Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep. 5: 1316–1329. [DOI] [PubMed] [Google Scholar]
- Loreng T. D., Smith E. F., 2017. The central apparatus of cilia and eukaryotic flagella. Cold Spring Harb. Perspect. Biol. 9: a028118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicki J., Avidor-Reiss T., 2014. From the cytoplasm into the cilium: bon voyage. Organogenesis 10: 138–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier A., Azimzadeh J., 2016. Multiciliated cells in animals. Cold Spring Harb. Perspect. Biol. 8: a028233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. R., 2007. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv. Exp. Med. Biol. 607: 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatake K., Kusakabe M., Takahashi C., Nishida E., 2015. ERK7 regulates ciliogenesis by phosphorylating the actin regulator CapZIP in cooperation with Dishevelled. Nat. Commun. 6: 6666. [DOI] [PubMed] [Google Scholar]
- Mohan S., Timbers T. A., Kennedy J., Blacque O. E., Leroux M. R., 2013. Striated rootlet and nonfilamentous forms of rootletin maintain ciliary function. Curr. Biol. 23: 2016–2022. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y., Yamane Y., Okanoue T., Tsukita S., Tsukita S., 2001. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol. Biol. Cell 12: 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechipurenko I. V., Olivier-Mason A., Kazatskaya A., Kennedy J., McLachlan I. G., et al. , 2016. A conserved role for Girdin in basal body positioning and ciliogenesis. Dev. Cell 38: 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson G., Robinson F., Beers Gibson T., Xu B. E., Karandikar M., et al. , 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22: 153–183. [DOI] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N., Culotti J. G., 1986. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117: 456–487. [DOI] [PubMed] [Google Scholar]
- Piasecki B. P., Sasani T. A., Lessenger A. T., Huth N., Farrell S., 2017. MAPK-15 is a ciliary protein required for PKD-2 localization and male mating behavior in Caenorhabditis elegans. Cytoskeleton (Hoboken) 74: 390–402. [DOI] [PubMed] [Google Scholar]
- Ressurreicao M., Rollinson D., Emery A. M., Walker A. J., 2011. A role for p38 MAPK in the regulation of ciliary motion in a eukaryote. BMC Cell Biol. 12: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. J., Cobb M. H., 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9: 180–186. [DOI] [PubMed] [Google Scholar]
- Rotureau B., Morales M. A., Bastin P., Spath G. F., 2009. The flagellum-mitogen-activated protein kinase connection in Trypanosomatids: a key sensory role in parasite signalling and development? Cell Microbiol. 11: 710–718. [DOI] [PubMed] [Google Scholar]
- Sagasti A., Hobert O., Troemel E. R., Ruvkun G., Bargmann C. I., 1999. Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes Dev. 13: 1794–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A. A., Kennedy J., Blacque O. E., 2015. Image analysis of Caenorhabditis elegans ciliary transition zone structure, ultrastructure, molecular composition, and function. Methods Cell Biol. 127: 323–347. [DOI] [PubMed] [Google Scholar]
- Sang L., Miller J. J., Corbit K. C., Giles R. H., Brauer M. J., et al. , 2011. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell 145: 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K. N., Kuhns S., Neuner A., Hub B., Zentgraf H., et al. , 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199: 1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung N. K., Park J. E., Yu L. R., Lee K. H., Lee J. M., et al. , 2009. Plk1-dependent and -independent roles of an ODF2 splice variant, hCenexin1, at the centrosome of somatic cells. Dev. Cell 16: 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N., Meunier A., 2017. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 18: 423–436. [DOI] [PubMed] [Google Scholar]
- Starich T. A., Herman R. K., Kari C. K., Yeh W. H., Schackwitz W. S., et al. , 1995. Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139: 171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P., Adler H. T., Thomas J. H., 2000. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell 5: 411–421. [DOI] [PubMed] [Google Scholar]
- Troemel E. R., Kimmel B. E., Bargmann C. I., 1997. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91: 161–169. [DOI] [PubMed] [Google Scholar]
- van der Linden A. M., Nolan K. M., Sengupta P., 2007. KIN-29 SIK regulates chemoreceptor gene expression via an MEF2 transcription factor and a class II HDAC. EMBO J. 26: 358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden A. M., Wiener S., You Y. J., Kim K., Avery L., et al. , 2008. The EGL-4 PKG acts with KIN-29 salt-inducible kinase and protein kinase A to regulate chemoreceptor gene expression and sensory behaviors in Caenorhabditis elegans. Genetics 180: 1475–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart A., Rademakers S., Jansen G., 2015. DLK-1/p38 MAP kinase signaling controls cilium length by regulating RAB-5 mediated endocytosis in Caenorhabditis elegans. PLoS Genet. 11: e1005733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Thomson N., White J. G., Brenner S., 1975. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J. Comp. Neurol. 160: 313–337. [DOI] [PubMed] [Google Scholar]
- Ware R. W., Clark D., Crossland K., Russell R. L., 1975. The nerve ring of the nematode Caenorhabditis elegans: sensory input and motor output. J. Comp. Neurol. 162: 71–110. [Google Scholar]
- Waters A. M., Beales P. L., 2011. Ciliopathies: an expanding disease spectrum. Pediatr. Nephrol. 26: 1039–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Xu Q., Zhang Y., Li Y., Zhang Q., et al. , 2013. Transition fibre protein FBF1 is required for the ciliary entry of assembled intraflagellar transport complexes. Nat. Commun. 4: 2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Zhang Y., Schouteden C., Zhang Y., Zhang Q., et al. , 2016. The hydrolethalus syndrome protein HYLS-1 regulates formation of the ciliary gate. Nat. Commun. 7: 12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. L., Li C., Kida K., Inglis P. N., Mohan S., et al. , 2011. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192: 1023–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Zhang Y., Wei Q., Huang Y., Li Y., et al. , 2015. BBS4 and BBS5 show functional redundancy in the BBSome to regulate the degradative sorting of ciliary sensory receptors. Sci. Rep. 5: 11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. M., Zhu F., Cho Y. Y., Carper A., Peng C., et al. , 2010. Extracellular signal-regulated kinase 8-mediated c-Jun phosphorylation increases tumorigenesis of human colon cancer. Cancer Res. 70: 3218–3227. [DOI] [PubMed] [Google Scholar]
- Yang S. W., Huang H., Gao C., Chen L., Qi S. T., et al. , 2013. The distribution and possible role of ERK8 in mouse oocyte meiotic maturation and early embryo cleavage. Microsc. Microanal. 19: 190–200. [DOI] [PubMed] [Google Scholar]
- Yang T. T., Hampilos P. J., Nathwani B., Miller C. H., Sutaria N. D., et al. , 2013. Superresolution STED microscopy reveals differential localization in primary cilia. Cytoskeleton (Hoboken) 70: 54–65. [DOI] [PubMed] [Google Scholar]
- Zacharogianni M., Kondylis V., Tang Y., Farhan H., Xanthakis D., et al. , 2011. ERK7 is a negative regulator of protein secretion in response to amino-acid starvation by modulating Sec16 membrane association. EMBO J. 30: 3684–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Liu Y., Yang P., 2017. Radial spokes-a snapshot of the motility regulation, assembly, and evolution of cilia and flagella. Cold Spring Harb. Perspect. Biol. 9: a028126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirmation of the conclusions discussed in the article are represented fully within the article. All worm strains (listed in Table S1 in File S1) and constructs are available upon request.