Abstract
Transposons can impact the host genome by altering gene expression and participating in chromosome rearrangements. Therefore, organisms evolved different ways to minimize the level of transposition. In Saccharomyces cerevisiae and its close relative S. paradoxus, Ty1 copy number control (CNC) is mediated by the self-encoded restriction factor p22, which is derived from the GAG capsid gene and inhibits virus-like particle (VLP) assembly and function. Based on secondary screens of Ty1 cofactors, we identified LOC1, a RNA localization/ribosome biogenesis gene that affects Ty1 mobility predominantly in strains harboring Ty1 elements. Ribosomal protein mutants rps0bΔ and rpl7aΔ displayed similar CNC-specific phenotypes as loc1Δ, suggesting that ribosome biogenesis is critical for CNC. The level of Ty1 mRNA and Ty1 internal (Ty1i) transcripts encoding p22 was altered in these mutants, and displayed a trend where the level of Ty1i RNA increased relative to full-length Ty1 mRNA. The level of p22 increased in these mutants, and the half-life of p22 also increased in a loc1Δ mutant. Transcriptomic analyses revealed small changes in the level of Ty1 transcripts or efficiency of translation initiation in a loc1Δ mutant. Importantly, a loc1Δ mutant had defects in assembly of Gag complexes and packaging Ty1 RNA. Our results indicate that defective ribosome biogenesis enhances CNC by increasing the level of p22, and raise the possibility for versatile links between VLP assembly, its cytoplasmic environment, and a novel stress response.
Keywords: Ty1 retrotransposon, Saccharomyces, restriction factor, ribosome biogenesis, Loc1
Ty1 is the most abundant retrotransposon in the Saccharomyces cerevisiae reference strain at 32 copies per haploid genome, and is an effective model for understanding the mechanism and consequences of retrotransposition (Curcio et al. 2015). Ty1 shares many similarities with retroviruses, except that retrotransposition is not infectious. Ty1 long terminal repeats (LTRs) bracket two overlapping genes: GAG encodes the capsid protein of virus-like particles (VLPs), and POL encodes a polyprotein that contains protease (PR), integrase (IN), and reverse transcriptase (RT). Once transcribed by RNA polymerase II from the 5′ LTR to the 3′ LTR, Ty1 mRNA is exported to the cytoplasm and translated into two protein products, Gag-p49 and Gag-Pol-p199. Gag-Pol is generated by a programmed +1 frameshifting event near the end of GAG. Gag-p49 and Gag-Pol-p199 are present in a ratio of 20:1 (Kawakami et al. 1993). Ty1 mRNA and translated proteins form cytoplasmic granules called the retrosomes, or T-bodies, which are sites where VLPs assemble (Malagon and Jensen 2008; Checkley et al. 2010; Dutko et al. 2010). During or after Ty1 VLP assembly, Gag-p49 and Gag-Pol-p199 undergo maturation catalyzed by PR to form mature Gag-p45, PR, IN, and RT. Reverse transcription takes place using packaged dimeric Ty1 RNA as template, and a heterodimeric complex of IN and RT (Chapman et al. 1992; Wilhelm et al. 1994; Feng et al. 2000). Ty1 cDNA and IN likely form a preintegration complex or intasome, which is imported into the nucleus via a nuclear localization sequence present on IN (Kenna et al. 1998; Moore et al. 1998; McLane et al. 2008). Genomic regions upstream of genes transcribed by RNA polymerase III (such as tRNA genes) are favored sites for Ty1 integration, based on functional interactions between IN and subunits of RNA polymerase III (Devine and Boeke 1996; Baller et al. 2012; Bridier-Nahmias et al. 2015; Cheung et al. 2016).
While retrotransposons and retrovirus-like elements comprise almost 50% of the human genome, Ty1, Ty2, Ty3, Ty4, and Ty5 sequences make up ∼3% of the compact S. cerevisiae reference genome (Kim et al. 1998; Lander et al. 2001). Ty1 movement occurs at a low rate of ∼1 × 10−6 transposition events/Ty1 element/generation (Curcio and Garfinkel 1991), yet yeast has the capacity to harbor hundreds of additional Ty1 insertions when cells containing Ty1 fused to the GAL1 promoter undergo multiple rounds of induction (Scheifele et al. 2009). These observations suggest that cellular or element-encoded products prevent rampant Ty1 retrotransposition in S. cerevisiae. Interestingly, none of the silencing pathways used by other eukaryotes to minimize transposon movement, such as RNAi or DNA methylation, are present in the S. cerevisiae sensu stricto group (Drinnenberg et al. 2009, 2011; Levin and Moran 2011), implying there may be a novel pathway utilized by S. cerevisiae to modulate Ty1 retrotransposition. Copy-number dependent effects on Ty1 mobility, termed copy number control (CNC) (Garfinkel et al. 2003) were first addressed when a plasmid containing Ty1 tagged with the retromobility indicator his3-AI was expressed in a strain of S. paradoxus strain that lacks complete Ty1 elements (Curcio and Garfinkel 1991; Moore et al. 2004). A dramatically higher level of Ty1 mobility is observed in this Ty1-less strain when compared to the S. cerevisiae laboratory strain S288c, which carries 32 Ty1 elements, or when the Ty1-less strain is repopulated with Ty1. Interestingly, the minimal Ty1 sequence required for CNC is located within GAG and the 5′ LTR. In cells undergoing CNC, defective VLPs are formed that contain lower levels of mature RT and IN, and reverse transcription products (Matsuda and Garfinkel 2009; Purzycka et al. 2013).
Although early work suggested that Ty1 antisense transcripts confer CNC (Matsuda and Garfinkel 2009), extensive biochemical and genetic analyses of the CNC region revealed a self-encoded Ty1 protein, termed p22, that is necessary and sufficient for CNC (Saha et al. 2015). p22 is encoded by the 3′-half of GAG, and is produced from subgenomic, internally initiated, Ty1(i) transcripts. p22 is processed to p18 by PR cleavage at the same site utilized by the full-length Gag protein. p22/p18 confers CNC and a trans-dominant negative effect on Ty1 mobility. p22/p18 associates with VLPs, co-immunoprecipitates with Ty1 Gag, and colocalizes with Gag in the cytoplasm. p22/p18 disturbs retrosome formation and VLP assembly, and blocks maturation and reverse transcription within the VLPs. In addition, p18 interferes with the nucleic acid chaperone function of Gag-p45 (Nishida et al. 2015), and p22/p18 targets Gag and disrupts Gag–Gag interactions during VLP assembly (Tucker et al. 2015).
Like retroviruses, retrotransposons are capable of making only a few proteins critical for replication; therefore, a variety of host functions modulate their life cycle (Maxwell and Curcio 2007; Goff 2008). Host genes from diverse biological processes affect Ty1 transposition and can stimulate (cofactors) or inhibit (restriction factors) various aspects of the Ty1 life cycle. To date, ∼500 cofactor and 110 restriction factor genes have been identified by genome-wide screens using yeast gene deletion mutants (Griffith et al. 2003; Nyswaner et al. 2008; Dakshinamurthy et al. 2010; Risler et al. 2012) or transposon mutagenesis (Scholes et al. 2001). Ty1 cofactors are involved in processes such as transcription, chromatin modification, histone deacetylation, ribosome biogenesis and function, and mRNA turnover and transport (Griffith et al. 2003; Dakshinamurthy et al. 2010; Risler et al. 2012). In contrast, ∼80% of Ty1 restriction factors are involved in nuclear processes including DNA repair, transcription, chromatin structure and function, recombination, and the cell cycle (Scholes et al. 2001; Nyswaner et al. 2008; Risler et al. 2012). We set out to identify host modulators that influence the synthesis or activity of the newly discovered Ty1 restriction factor p22. Such host factors would greatly help us understand the mechanism of CNC.
We reasoned that CNC-specific host factors should stimulate or inhibit CNC in a Ty1 copy number-dependent manner. In other words, a host modulator displaying a CNC-specific phenotype would occur predominantly in CNC+ cells containing Ty1 elements, but not in CNC− cells lacking Ty1. To identify CNC-specific modulators, we analyzed previously identified Ty1 cofactor mutants involved in RNA metabolism, which is central to the process of Ty1 retrotransposition (Curcio et al. 2015), for those that displayed phenotypes strongly associated with CNC (Matsuda and Garfinkel 2009; Saha et al. 2015). We then analyzed candidate deletion mutants in congenic CNC− and CNC+ strains that differ only in Ty1 copy number. Surprisingly, we identified several genes involved in ribosome biogenesis that increase the level p22 and show CNC-specificity; Ty1 mobility decreased preferentially in CNC+ cells when compared to CNC− cells. In particular, LOC1, a gene required for asymmetric localization of ASH1 mRNA (Heym and Niessing 2012) and rRNA processing (Urbinati et al. 2006), facilitates Gag–Gag interactions and retrosome formation by modulating the level of p22.
Materials and Methods
Genetic techniques, media, strains, and plasmids
Strains are listed in Supplemental Material, Table S1. Standard yeast genetic and microbiological procedures were used in this work (Guthrie and Fink 1991). The S. cerevisiae CNC− strain DG3453 was derived from DJ12, which was isolated in Djibouti, Africa, and generously provided by Michel Aigle. A MATα ho ascospore (DJ12-1B) was subjected to 5-fluoroorotic acid (5-FOA) selection (Boeke et al. 1984), and a spontaneous ura3 mutant was isolated to generate DG3093. The results of Southern blot analysis (Garfinkel et al. 2003; Moore et al. 2004) suggest there are 10 Ty1 elements in DJ12-1B and DG3093. To eliminate the Ty1 elements from DG3093, we generated a Ty1:URA3 targeting fragment containing Ty1 sequences 1702–3945 (Boeke et al. 1988). The 2243-bp segment was amplified by PCR from DG3093 using primers 1702XhoBglF (5′-CCCGCTGCAGAGATCTGCTCATCACATACACTC-3′) and 3945BamR (5′-CCGCGGATCCTGCAATCAGGTGAATTCGT-3′) and subcloned into pSP70 (Promega, Madison, WI) digested with XhoI and BglII. URA3 bracketed by XhoI restriction sites on pB130 (kindly provided by Gerald Fink) was digested and cloned into the Ty1 SalI site (nucleotide 2173) of pSP70/Ty1. The resulting plasmid pBDG1374 was digested with XhoI and EcoRI to release the Ty1:URA3 targeting fragment. Ura+ transformants containing putative Ty1:URA3 recombination events with individual Ty1 elements were subjected to 5-FOA selection followed by Southern blot analysis to detect LTR-LTR recombinants (Winston et al. 1984). Changes in Ty1 hybridization patterns suggest that one Ty1 was eliminated with each round of transformation and 5-FOA selection. After removing nine of 10 Ty1 elements, HIS3 was deleted as described previously using pBDG652 (Garfinkel et al. 2003) to generate DG3453. The same Ty1 was present in independent elimination lineages, and could not be lost to generate a Ty1-less strain, raising the possibility that the remaining Ty1 insertion may be required for the function of an essential gene. Crosses between DG3093 containing 10 Ty1 elements and strains with a single Ty1, or crosses between the reference strain BY4741 (Brachmann et al. 1998) and the single Ty1 strains displayed 70% spore viability in 18–20 tetrads/cross. Ascosporal progeny displayed similar variations in growth and expected segregation of genetic markers. DG3453 was repopulated with ∼19 Ty1 elements by induction of a pGTy1 plasmid to generate DG3648, as described previously (Garfinkel et al. 2003). MATα deletion strains (Giaever et al. 2002) derived from BY4742 (Brachmann et al. 1998) were obtained from Invitrogen (Carlsbad, CA). Gene disruptions in DG3453 and DG3648 were carried out using the KanMX4 cassette (Wach et al. 1994). The KanMX disruption cassettes were PCR amplified using template DNA extracted from deletion mutants with gene-specific primers A and D from the Saccharomyces Genome Database (https://www.yeastgenome.org/). Correct gene disruptions by KanMX for all of the deletion mutant strains used in this study were confirmed by PCR using locus-specific primers. To analyze Ty1 mobility and protein levels, pBDG606 (pGTy1his3-AI/Cen/URA3) (Dakshinamurthy et al. 2010) was introduced to BY4742-, DG3453-, and DG3648-derived strains.
Ty1his3-AI mobility
Quantitative and qualitative Ty1his3-AI mobility assays were determined as described previously (Curcio and Garfinkel 1991; Garfinkel et al. 2003; Sharon et al. 1994) with minor modifications. For strains expressing pGTy1his3-AI (pBDG606), a single colony grown on SC-Ura at 30° was resuspended in 1 ml of SC-Ura + 2% raffinose, and incubated for 24 hr at 30° with aeration; 200 μl of the raffinose culture was centrifuged, and the cell pellet was resuspended in 1 ml of SC-Ura + 2% galactose in quadruplicate. The cultures were grown for 16 hr at 22°, washed, diluted, and spread onto SC-Ura and SC-His-Ura plates, which were incubated for 3–4 days at 30° until colonies formed. The frequency of Ty1his3-AI mobility was calculated by dividing the number of His+ Ura+ colonies by the number of Ura+ colonies. For qualitative Ty1his3-AI mobility assays, cells patched onto SC-Ura were incubated at 30° for 2 days. Cells were replica plated onto SC-Ura + 2% galactose medium plates and incubated at 22° for 16 hr. To detect Ty1HIS3 mobility events, galactose-induced cells were replica plated to SC-Ura-His followed by incubation for 2–3 days at 30°.
RNA isolation and northern blot analysis
For strains carrying pBDG606, 10 ml of SC-Ura + 2% raffinose was inoculated with a single colony, and the culture was grown at 30° to an OD600 of 2, or up to 24 hr, depending on the strain; 2 ml of the raffinose culture was centrifuged and cells were suspended in 10 ml of SC-Ura + 2% galactose and grown at 22° for 16 hr. To detect endogenous Ty1i RNA, a single colony was suspended in 5 ml YEPD and grown at 30° overnight. The overnight culture was diluted different amounts depending on the growth of each strain (1:10 for WT, 1:4 for loc1Δ, 1:5 for rps0bΔ, and 1:8 for rpl7aΔ) in a total of 10 ml fresh YEPD and grown at 22° for 8 hr. Total RNA was extracted using the MasterPure yeast RNA purification kit (Epicentre Biotechnologies, Madison, WI) with modifications as described previously (Saha et al. 2015). Poly(A)+ RNA was isolated from 250 μg total RNA using the NucleoTrap mRNA purification kit (Clontech, Mountain View, CA) following manufacturer’s protocol. Northern blot analysis using 32P-labeled riboprobes was performed as described previously (Saha et al. 2015). Hybridization signals were visualized and quantified using a STORM 840 phosphorimager and ImageQuant software (GE Healthcare).
RNA sequencing analyses
RNA sequencing (RNA-seq) and ribosome footprint profiling (Ribo-seq) datasets were created by S. Komili, D. Muzzey, and F.P. Roth, and downloaded from GEO (accession no. GSE34438). Three replicate RNA-seq or Ribo-seq libraries were analyzed from the wild-type parent BY4741 and the isogenic loc1Δ derivative (12 libraries in total). Note that deleting LOC1 confers a transposition defect in BY4741 or BY4742 (Risler et al. 2012), which are closely related MATa and MATα strains, respectively (Brachmann et al. 1998). Sequencing reads were analyzed as follows: reads were trimmed of adaptor sequences. For Ribo-seq, a size filter of 28 bp was imposed, as 28-bp Ribo-seq reads tended to have a high proportion of in-frame reads. As Ty1 is present in multiple copies in the genome, the Ty1-H3 reference element (GenBank M18706.1) was used in the initial round of mapping. Mapping was performed with STAR RNA-seq aligner (version 2.4.2a) allowing zero mismatches. The remaining (non-Ty1) reads were mapped to the S. cerevisiae genome (Release 85; Ensembl). Mapped reads were assigned to genes and plotted with custom scripts. Gene counts are the average of the three replicates for each library, with similar results observed for pairwise library comparisons.
Protein extraction and western blot analysis
Total protein was extracted from strains expressing pGTy1his3-AI as previously described (Braiterman et al. 1994). For detecting p22/p18, cells from 2 OD600 of culture were processed by trichloroacetic acid (TCA) extraction, and immunoblotted as described (Saha et al. 2015). Samples were separated on 7.5 or 8% (for detecting IN), 10% (for detecting Gag-isoforms) or 15% (for detecting Gag-isoforms and p22/p18) SDS-PAGE gels. Antibody dilutions were as follows: anti-p18 1:5000, anti-VLP 1:7000, anti-IN 1:2500, anti-Pgk1 1:40,000, and anti-TY tag (BB2; UAB Epitope Recognition and Immunoreagent Core, Birmingham, AL) 1:20,000 (Bastin et al. 1996). Immunoreactive protein signals were quantified using Quantity One software (Bio-Rad, Hercules, CA).
Stability of p22
p22 stability in wild type and loc1Δ strains was determined as described previously (Doh et al. 2014) with minor modifications; 5 ml of SC-Ura+2% raffinose medium was inoculated with a single colony and grown for ∼24 hr at 30°. The raffinose cultures (3 OD600 for HWA625 and 6 OD600 for HWA626) were added to 50 ml SC-Ura+2% galactose medium and grown at 20° for to an OD600 of 0.6–0.8. Thirty OD600 of cells per strain were harvested and washed three times in 5 ml of SC-Ura-Met+2% Gal. Click-iT L-HPG (Life Technologies) was added to 5 ml cell suspensions in SC-Ura-Met+2% Gal to a final concentration of 80 μM. Cultures were incubated at 20° for 30 min with shaking. Cells were washed twice with 5 ml of SC-Ura-Met+2% Gal and then resuspended in chase medium (SC-Ura-Met+2% Gal +50 mM l-methionine). Equal amounts of cells were harvested at 0, 1, 3, and 6 hr time points. Cell pellets were resuspended in 100 μl HB buffer (25 mM Tris, pH 7.5, 125 mM NaCl, 5 mM EDTA, 0.5% IPEGAL) with protease inhibitors (20 μl of 1 mg/ml Aprotonin, Leupeptin, Pepstatin, and 100 μl of 10 mg/ml PMSF) and vortexed at 4° for 5 min two times with 0.15 g of glass beads. Following addition of 5 μl of 10% SDS, the cell extract was boiled for 5 min and 500 μl ice-cold HB buffer was added. The extract was centrifuged for 20 min at 10,000 × g at 4° and precleared with 50 μl of Protein A agarose (Pierce) slurry (50% beads) washed with HB buffer and incubation at 4° for 1 hr; 10 μg of anti-p18 antibody was added to precleared lysate, and rotated at 4° for 16 hr, and then 50 μl of Protein A agarose slurry was added to each sample and rotated at 4° for 2 hr. Samples were washed three times with 500 μl HB buffer and proteins were eluted from the beads with 50 μl of 50 mM Glycine (pH 3.0) equilibrated in 200 μl of 50 mM Tris-Cl, pH 8.0, 1% SDS. Eluted proteins were precipitated by methanol/chloroform. Click-iT Cell Reaction Buffer Kit (Life Technologies) was used to conjugate TAMRA (Life Technologies) to HPG-labeled p22 according to the manufacturer’s protocol. Protein samples were separated on 15% SDS-PAGE gels. The fluorescent signal of TAMRA-conjugated p22 was detected using a Typhoon trio scanner with 580 BP30 filter, and p22 bands were quantified with ImageQuant Software (GE Healthcare). We assumed that p22 degradation follows first-order kinetics, and determined p22 half-life as described previously (Belle et al. 2006).
Fluorescence in situ hybridization/Immunofluorescence (FISH/IF)
Retrosomes were analyzed by FISH/IF as described previously (Checkley et al. 2010, 2013; Saha et al. 2015). To visualize endogenous retrosomes, 5 ml YEPD cultures were inoculated with a single colony and grown for 16 hr at 30°. The overnight cultures were diluted 40- (HWA215) or 500-fold (BY4742) into fresh YEPD, and grown at 20° for ∼24 hr to an OD600 of 0.8–1.0. To visualize pBDG606-induced retrosomes, 5 ml of SC-Ura+2% raffinose medium was inoculated with a single colony and grown for 24 hr at 30°. The raffinose cultures were diluted to 20- (HWA169) or 80-fold (HWA15) in SC-Ura+2% galactose medium and grown at 20° for ∼24 hr to an OD600 of 0.8–1.0. Formaldehyde was added directly to the culture to a final concentration of 4% and allowed to fix for 1 hr. Anti-VLP (rabbit polyclonal, 1:2000) and a cocktail of three GAG digoxigenin (DIG)-labeled antisense oligonucleotides (TyA_380, 5′-GCCTTCTCACATTCTTCTGTTTTGGAAGCTGAAACGTCTAACGGATCTTG-3′; TyA_444, 5′-TTCTCTGGAACAGCTGATGAAGCAGGTGTTGTTGTCTGTTGAGAGTTA-3′; TyA_545, 5′-CAACCAGATGGATTGGCTTGGTTTTGGGTCATCATGCACTGCTGTGGGTA-3′) were used to detect Gag and Ty1 mRNA, respectively. Secondary antibodies used here were anti-rabbit-AF594 (1:200; Life Technologies) and Fluorescein (FITC)-conjugated sheep anti-DIG Fab fragment (1:200; Roche Applied Science). Image acquisition was carried out using a ZeissAxio Observer microscope equipped with an AxioCam HSm camera, and images were analyzed with AxioVision v4.6 software (Carl Zeiss Microscopy, Thornwood, NY). The percentage of cells with retrosomes was calculated by dividing the number of cells with colocalized foci (Ty1 mRNA and Gag) by the total number of cells. At least 200 cells were analyzed for each strain.
Sucrose gradient sedimentation
Sedimentation of Gag complexes was analyzed as described (Tucker et al. 2015), with the following modifications. For strains BY4742 and HWA215, a 100-ml culture was grown as described for FISH/IF microscopy. Equal amounts of total protein (7–8 mg in 300–450 μl) were applied to a 7–47% continuous sucrose gradient and centrifuged at 25,000 rpm in a SW 41 swinging bucket rotor (Beckman Coulter, Brea, CA) for 3 hr at 4°. Nineteen 0.6-ml fractions were collected, and an equal volume of each fraction was subjected to western blot analysis to detect Ty1 Gag as described above.
Nuclease protection
Strain BY4742 and HWA215 were grown as described above for FISH/IF microscopy. Nuclease protection using benzonase (EMD Millipore, Billerica, MA) was performed as described previously (Nishida et al. 2015). Total RNA extraction and northern blot analysis was performed as described above.
Crosslinking Gag complexes
HWA15 and HWA169 were grown as described above for FISH/IF microscopy. Formaldehyde (Fisher Scientific, Waltham, MA) was added directly to the culture medium to a final concentration of 1%. Fixed cells were incubated for 1 hr at 20°. Proteins were extracted as previously described (Braiterman and Boeke 1994) and separated on NuPAGE 3–8% Tris-Acetate gels (Invitrogen) for detecting Gag multimers and 10% SDS-PAGE gels for detecting Pgk1. An equal amount of protein extract (7.5 µg) was loaded per lane and electrophoresis was performed at 120 V for 2 hr. Proteins were transferred to PVDF membranes and probed with anti-TY tag at a 1:50,000 dilution. Gag1 quantification was performed as described above.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All strains and reagents are available upon request.
Results
Identifying CNC-specific Ty1 cofactors
To identify CNC-specific genes, 498 Ty1 cofactors were analyzed using DAVID (http://david.abcc.ncifcrf.gov/), which enables functional annotation of large gene lists (Huang et al. 2009). Functional annotation clustering was performed to determine the gene ontology (GO) terms enriched in Ty1 cofactor genes with reduced redundancy (for example, nucleic acid transport and RNA export were grouped in one cluster). Among the top four gene clusters with high enrichment scores (>4) (Table S2) were Ty1 modulators involved in post-transcriptional RNA biogenesis (RNA catabolic processes, RNA transport). To identify CNC-specific cofactors, 51 Ty1 cofactor mutants with defects in RNA metabolism were subjected to two secondary screens (Table 1). First, mutants involved in CNC should contain much less mature IN relative to Gag since CNC is associated with a block in the accumulation of mature Pol proteins (Garfinkel et al. 2003; Matsuda and Garfinkel 2009). Second, CNC-specific mutations should affect Ty1 mobility to a greater extent in CNC+ strains containing Ty1 elements vs. CNC− strains that do not.
Table 1. Ty1 cofactors with roles in post-transcriptional RNA biogenesis.
| Classification | Gene name |
|---|---|
| P-body components | DHH1, LSM1, LSM6, PAT1, PUB1, XRN1 |
| Nonsense-mediated decay | NAM7, NMD2, UPF3 |
| Exosome components | LRP1, RRP6, SKI8 |
| snRNA/snoRNA processing | BUD31, REF2, SNU66 |
| RNA splicing | DBR1, LEA1, MUD2, SNT309, SQS1 |
| CCR4-NOT complex | CCR4, MOT2, POP2 |
| TRAMP complex | PAP2, TRF5 |
| Nuclear cap-binding complex | CBC2, NPL3, STO1 |
| RNA export/transport | APQ12, BUD13, KAP123, LOC1, LOS1, MFT1, NUP120, NUP133, NUP170, NUP188, NUP84, PML39, SCP160, SOL1, SXM1, THO1, THP2, TOM1 |
| Miscellaneous | CTH1, MRT4, RPB4, SPT4, SSN3 |
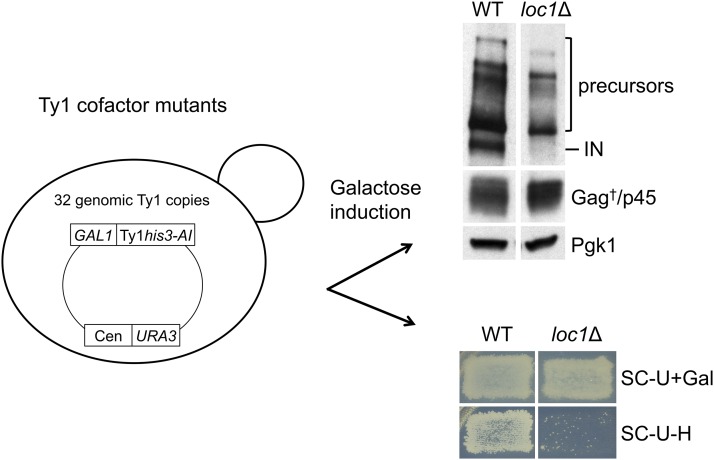
Selected Ty1 cofactor mutants were assessed for the level of Ty1 Gag and IN by western blot analysis using VLP and IN antisera (Figure S1A and Table S3). Transposition defects were monitored by a qualitative papillation assay for Ty1his3-AI mobility (Curcio and Garfinkel 1991) (Table S3). To reliably detect mature Ty1 IN but not overwhelm CNC by overexpressing Ty1 (Figure S1B) (Garfinkel et al. 2003), a low-copy centromere-based GAL1 expression plasmid pGTy1his3-AI plasmid (pBDG606) was introduced into the Ty1 cofactor deletion mutants (Giaever et al. 2002). For example, deletion of LOC1, a gene involved in biogenesis of 60S ribosomes and asymmetric localization of ASH1 RNA (Long et al. 2001; Urbinati et al. 2006), resulted in the preferential loss of mature IN relative to Gag following galactose induction of pBDG606 (Figure 1). Also, when wild type and loc1Δ cells were monitored qualitatively for Ty1his3-AI mobility after induction on SC-Ura + Galactose plates, the loc1Δ mutant harbored few Ty1HIS3 mobility events when compared to the wild-type parent. Two mutant classes were evident from the western blot analysis (Figure S1A) and qualitative mobility analyses (Table S3). We identified nine candidates (kap123∆, loc1∆, mot2∆, mrt4∆, nup170∆, pap2∆, ref2∆, ssn3∆, and xrn1∆) with considerably less mature IN relative to Gag and the Pgk1 loading control, and a low level of Ty1his3-AI mobility. Also, we identified mutants such as npl3Δ and sto1Δ that displayed a lower level of both Gag and IN. These mutants were not studied further as they may have less Ty1 mRNA or defects in Ty1 protein synthesis or stability.
Figure 1.
Identifying Ty1 cofactor mutants that phenocopy CNC. Selected Ty1 cofactor mutants containing pGTy1his3-AI (pBDG606) the BY4742 background were induced on SC-Uracil (U) + Galactose (Gal) solid and liquid media (Figure S1A). Results from loc1Δ are shown as an example. The level of mature IN and Gag isoforms was assessed by western blot analysis using IN and VLP antisera. IN and Pol precursors, mature Gag-p45 and Gag isoforms (†) that include Gag-p49 are noted alongside the blot. Pgk1 (3-phosphoglycerate kinase) served as a loading control. Cell patches from SC-U+Galactose were replicated to SC-U-Histidine (H) to detect Ty1HIS3 mobility events, as monitored by the formation of His+ papillae. Mutants showing a severe decrease in mature IN but not Gag and a decrease in Ty1 mobility were selected for further analysis.
We deleted nine CNC-specific candidate genes in congenic S. cerevisiae CNC− and CNC+ strains DG3453 (+1 Ty1) and DG3648 (+20 Ty1s), respectively. During strain construction, mutants showing CNC-independent Ty1 mobility defects (kap123∆), or no defect in Ty1 mobility (ref2∆, nup170∆), clonal variability in Ty1 mobility (pap2∆), or a severe growth defect (mot2∆) were not pursued further. The remaining four mutants (ssn3∆, mrt4∆, loc1∆, and xrn1∆) were assessed for Ty1his3-AI mobility (Figure S2A) and Ty1 protein level (Figure S2B) in the CNC− and CNC+ backgrounds. Mutants showing a decrease (ssn3∆) or possible increase (xrn1∆) of IN in the CNC− background or a relatively mild decrease of IN in the CNC+ background (mrt4∆) compared to wild type were not pursued further. Only loc1∆ displayed both phenotypes expected for a CNC-specific mutant: a severe decrease in mature IN relative to Gag, and a decrease in Ty1his3-AI mobility in CNC+ but not in CNC− strains. Therefore, we analyzed how LOC1 modulated Ty1 CNC in greater detail.
Ribosome biogenesis affects Ty1 CNC
LOC1 is important for ribosome biogenesis and asymmetric RNA localization (Urbinati et al. 2006; Heym and Niessing 2012). We hypothesized that, if ribosome biogenesis or asymmetric RNA localization were critical for CNC, disrupting additional genes in those pathways would phenocopy the loc1∆ mutant. For the ASH1 mRNA localization pathway, PUF6 and SHE2 were selected as representatives (Heym and Niessing 2012). For ribosome biogenesis, the Ty1 cofactors RPS0B and RPL7A as well as their paralogs RPS0A and RPL7B were analyzed for CNC-specificity (Dakshinamurthy et al. 2010; Risler et al. 2012; Palumbo et al. 2017). Deletion mutants were generated in S. cerevisiae CNC− and CNC+ backgrounds, and pBDG606-induced Ty1 mobility was determined (Table 2). The frequency of Ty1 mobility in CNC− and CNC+ strains was assessed individually, and also reported as a CNC−/CNC+ ratio, which estimates the increase in CNC observed in mutant strains when compared with wild type. The loc1Δ mutant displayed a 3.4- and a 117.6-fold decrease in Ty1his3-AI mobility in the CNC− and CNC+ backgrounds, respectively. The increase in CNC as estimated by the loc1Δ CNC−/CNC+ ratio was over 300-fold, which was ∼30-fold higher CNC than observed in the wild type. Interestingly, the ribosomal protein paralog mutants rps0aΔ and rps0bΔ, and rpl7aΔ and rpl7bΔ all showed an increase in CNC. However, the mutants identified as Ty1 cofactors (rps0bΔ and rpl7aΔ) had higher levels of CNC than their paralogs (rps0aΔ and rpl7bΔ), which were not recovered as cofactors. PUF6, previously identified as a Ty1 cofactor (Risler et al. 2012), displayed a CNC ratio of 69. In contrast, Ty1 mobility and CNC remained at wild-type levels in cells lacking SHE2, which neither modulates Ty1 transposition nor affects ribosome biogenesis. In addition, deleting other genes (myo4Δ, she3Δ, and scp160Δ) involved in asymmetric localization of ASH1 mRNA (Heym and Niessing 2012) did not affect Ty1his3-AI mobility in CNC− or CNC+ strains (Figure S3). Overall, our results show that defects in ribosome biogenesis trigger an increase in Ty1 CNC while defects in mRNA localization do not.
Table 2. Ty1his3-AI mobility in CNC− and CNC+ S. cerevisiae strains.
| Genotypea | Ty1his3-AI mobility, ×10−6 (SD) | Fold increase in CNC (CNC−/CNC+) | |
|---|---|---|---|
| CNC− | CNC+ | ||
| Wild typeb | 7481 (1805) | 823 (318) | 9 |
| loc1Δ | 2181 (534) | 7 (2) | 307 |
| puf6Δ | 1781 (748) | 26 (4) | 69 |
| rps0aΔ | 5002 (1585) | 59 (48) | 85 |
| rps0bΔ | 9592 (4285) | 67 (22) | 143 |
| rpl7aΔ | 5436 (1540) | 89 (22) | 61 |
| rpl7bΔ | 4212 (2308) | 166 (55) | 25 |
| she2Δ | 9370 (2038) | 750 (231) | 12 |
S. cerevisiae CNC− and isogenic CNC+ strains (WT and mutants) transformed with pBDG606 were induced with galactose. Cells were induced with galactose for 16 hr at 22°. Then cells were diluted, spread onto SC-Ura (for total number of colonies) and SC-Ura-His (for His+ colonies) plates, and incubated for 3–4 days at 30° until colonies formed.
Average of five trials.
Ty1 transcript and protein levels in candidate CNC mutants
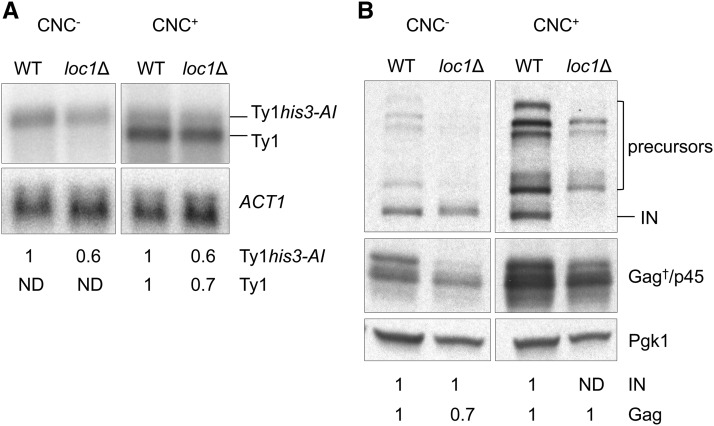
Total RNA and protein from wild-type and loc1Δ CNC− and CNC+ strains were subjected to northern (Figure 2A) and western blot (Figure 2B) analyses following pBDG606 induction. The level of Ty1his3-AI and Ty1 transcripts decreased modestly in wild type and loc1Δ CNC+ strains when hybridized with a 32P-labeled riboprobe derived from GAG and normalized to actin (ACT1) mRNA. The level of chromosomal Ty1 transcripts from the single copy CNC− strains was below the limit of detection, and the level of pGTy1-induced Ty1his3-AI mRNA decreased slightly in the CNC− loc1Δ mutant. Western blot analysis showed that the levels of IN precursors and mature IN remained about the same in the loc1Δ CNC− mutant. However, mature IN was not detected in the loc1Δ CNC+ mutant, which validates the results obtained in the BY4742 knockout strain (Figure 1 and Figure S1A). The level of Gag isoforms was also similar in the CNC− and CNC+ loc1Δ mutants. Together, our results suggest that the CNC-specific loss of mature IN is enhanced in a loc1Δ mutant while Ty1 mRNA level is comparable to wild type.
Figure 2.
Ty1 RNA and protein levels in congenic CNC− and CNC+ WT and loc1Δ strains expressing pBDG606. (A) Northern blot analysis of total RNA hybridized with a 32P-labeled riboprobe containing Ty1 nt 238–1702 to detect Ty1his3-AI and chromosomal Ty1 transcripts. ACT1 served as a loading control. Below are relative changes in RNA level compared to WT as determined by phosphorimage analysis. (B) Western blot analysis of Ty1 IN and Gag detected in total cell extracts (40 μg/lane) of CNC− and CNC+ loc1Δ mutant strains. Refer to Figure 1 legend for a description of Ty1 proteins. Pgk1 served as a loading control. Numbers below each panel indicate relative changes in protein level compared to WT as determined by quantification of protein bands by densitometry. ND, not detected.
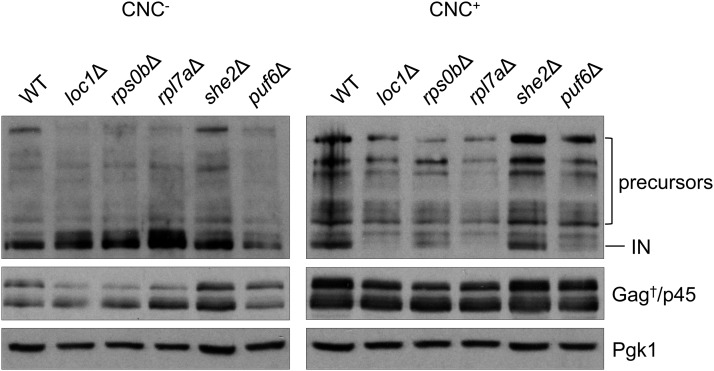
To determine if the level of Ty1 Gag and IN proteins was similar in strains defective in ribosome biogenesis or RNA localization, total protein from wild type, loc1Δ, rps0bΔ, rpl7aΔ, she2Δ, and puf6Δ strains expressing pBDG606 was subjected to western blot analysis (Figure 3). A trend emerged suggesting that mutants showing CNC-specificity (loc1Δ, rps0bΔ, and rpl7aΔ) (Table 2) contain less mature IN only in the CNC+ background, except the puf6Δ mutant showed a decrease of mature IN in both CNC− and CNC+ backgrounds. Normal levels of Gag and mature IN were present in a CNC− and CNC+ she2Δ mutant. Therefore, LOC1 likely modulates Ty1 transposition via its role in ribosome biogenesis and not through RNA localization.
Figure 3.
Ty1 IN and Gag levels in ribosome biogenesis and RNA localization mutants. Total cell extracts from CNC− and CNC+ wild type and loc1Δ, rps0bΔ, rpl7aΔ, she2Δ, and puf6Δ mutant strains expressing pBDG606 were subjected to western blot analysis using IN and VLP antisera. Pgk1 served as a loading control. Refer to Figure 1 for a description of Ty1 proteins.
Ty1i RNA and p22/p18 levels are altered in CNC-specific mutants
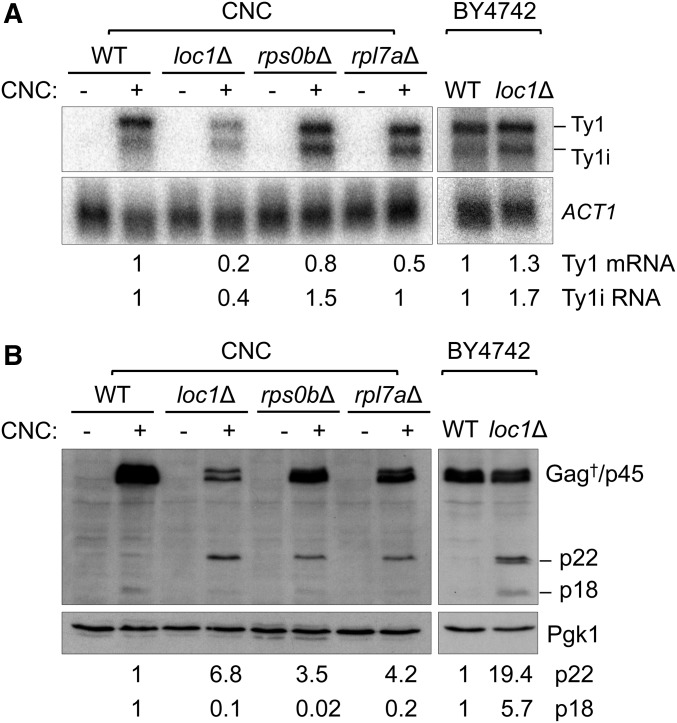
Suresh et al. (2015) reported that Ty1i RNA and p22/p18 levels increase in several ribosome biogenesis mutants including rps0bΔ. Therefore, endogenous levels of Ty1 mRNA, Ty1i RNA, Gag, and p22/p18 were determined in loc1Δ, rps0bΔ, and rpl7aΔ CNC mutants (Figure 4). To clearly distinguish Ty1i RNA from Ty1 mRNA (Saha et al. 2015; Suresh et al. 2015), poly(A)+ RNA was subjected to northern blot analysis using a 32P-labeled riboprobe containing Ty1 nucleotides 1266–1601 that hybridizes with Ty1 mRNA and Ty1i transcripts; ACT1 mRNA served as a loading control (Figure 4A). Ty1 transcripts were below the limit of detection in wild type or CNC− mutants since there is only one Ty1 element present in the CNC− background. The level of Ty1 mRNA decreased fivefold in the CNC+ loc1Δ background relative to wild type, whereas a similar decrease in Ty1 mRNA was not observed in a BY4742 loc1Δ mutant. We did not examine this strain-specific effect further. Ty1 mRNA levels also varied modestly in the CNC+ rps0bΔ and rpl7aΔ mutants. The level of poly(A)+ Ty1i RNA was altered in the CNC+ loc1Δ, rps0bΔ, and BY4742 loc1Δ mutants; slightly increased levels were detected in the CNC+ rps0bΔ and BY4742 loc1Δ mutants, and a decreased level was detected in the CNC+ loc1Δ mutant. The level of Ty1i RNA remained unchanged in a rpl7aΔ mutant. Although there was no clear pattern of changes for Ty1i and Ty1 poly(A)+ transcripts in the ribosome biogenesis mutants, the CNC-specific mutants displayed a trend where the ratio of Ty1i/Ty1 mRNA increased twofold. We also compared the ratio of Ty1i RNA and Ty1 mRNA in each lane without normalizing to the ACT1 mRNA loading control. In the CNC+ background, Ty1i/Ty1 mRNA ratio for wild type was 0.4, while the ratio for ribosome biogenesis mutants were 0.9, 0.8, and 0.8 for loc1Δ, rps0bΔ, and rpl7aΔ, respectively. In BY4742, the Ty1i/Ty1 mRNA ratio for wild type was 0.5, while the ratio for loc1Δ was 0.7. Taken together, the transcript ratios support the idea that the enhancement of CNC correlates with an increase in Ty1i RNA over Ty1 mRNA.
Figure 4.
Endogenous Ty1 transcript and p22 levels from CNC WT and ribosome biogenesis mutants, and BY4742 wild type and a loc1Δ mutant. (A) Northern blot analysis of poly(A)+ RNA hybridized with a 32P-labeled riboprobe containing Ty1 nt 1266–1601. This probe detects Ty1 mRNA, and Ty1 internally initiated (i) RNA that encodes p22/p18. ACT1 served as a loading control. The relative amount of Ty1 transcripts is shown below. (B) Western blot analysis of Ty1 and p22/p18 from total cell extracts using p18 antiserum, which detects Gag isoforms (†), p45, p22, and p18. Pgk1 served as a loading control. The relative amount of p22 and p18 is shown below.
To determine the level of endogenous Gag and p22/p18 in the CNC− and CNC+ strains, total protein was subjected to western blot analysis using an antiserum that detects both Gag and p22/p18 (Figure 4B) (Saha et al. 2015). In accordance with the levels of Ty1 RNAs (Figure 4A), full-length Gag and a low level of p22/18 were detected in CNC+ wild type but not in CNC− wild type strains. In the CNC+ loc1Δ mutant, we observed a decrease in full-length Gag and a 6.8-fold increase in p22 when compared to the CNC+ wild type. An increase in p22 of 3.5-fold, 4.2-fold, and 19.4-fold was observed in the CNC+ rps0bΔ and rpl7aΔ mutants, and the BY4742 loc1Δ mutant, respectively. Also, there was a 5- to 10-fold decrease in the level of p18 in the CNC+ ribosome biogenesis mutants. Some p18 was detected in the BY4742 loc1Δ mutant, although p22 was present in a much higher amount. Recent work suggests that PR-mediated cleavage of p22 to p18 takes place in VLPs (Saha et al. 2015; Tucker et al. 2015). Therefore, the lower level of p18 observed in the ribosome biogenesis mutants is consistent with a defect in VLP assembly or function.
Since Ty1 Gag stability decreased in several Ty1 cofactor mutants defective in ribosome biogenesis and translation (Doh et al. 2014), we examined p22 stability in wild type, and an isogenic loc1Δ mutant in the BY4742 background by pulse-chase immunoprecipitation (Figure S4). Due to the low level of endogenous p22 and p18 in BY4742, a previously characterized galactose-inducible p22 expression plasmid pBDG1565 (pAUG1p22/GAL1/2μ/URA3) (Nishida et al. 2015) was used to assess p22 stability. The half-life of p22 increased from 2.6 hr in wild-type cells to 4.1 hr in the isogenic loc1Δ mutant. Although a ∼60% increase in half-life suggests that p22 may be more stable, this result does not fully account for the increased level of p22 in the CNC+ or BY4742 loc1Δ mutants. We did not detect p18 in strains expressing pBDG1565, presumably because of the low-level of endogenous Ty1 protein processing by PR (Curcio and Garfinkel 1992).
Transcriptomic analyses in a loc1Δ mutant
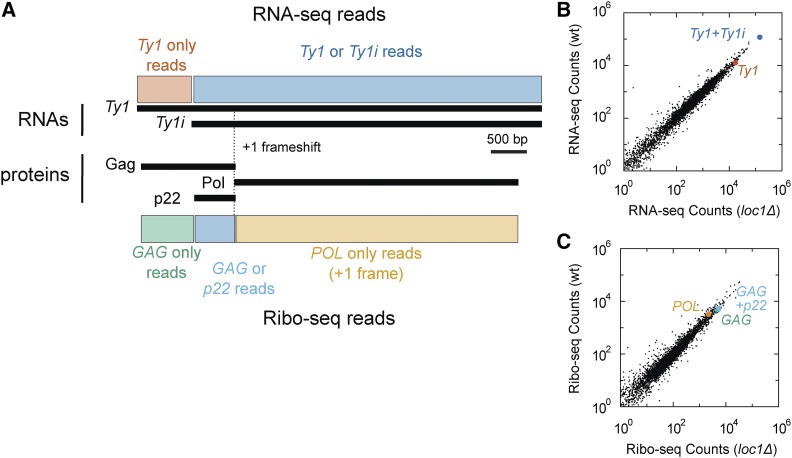
To determine if the changes in Ty1 transcript levels are part of a global response to a defect in ribosome biogenesis, we analyzed publicly available RNA-seq and ribosome profiling (Ribo-seq) datasets generated in BY4741 and a loc1Δ derivative (GEO accession no. GSE34438). First, RNA-seq datasets were analyzed for transcriptional changes. It is not possible to identify unique Ty1i reads from Ty1 as Ty1i transcripts overlap completely with full-length Ty1 mRNA (Saha et al. 2015). We split the RNA-seq counts into two regions: (1) Ty1 exclusive and (2) Ty1 or Ty1i-derived reads (Figure 5, A and B). We observed a slight decrease in counts assigned to both regions in the loc1Δ mutant. However, as Ty1 mRNA is more abundant than the Ty1i transcripts (Figure 4A) (Saha et al. 2015; Suresh et al. 2015), even a large fold change in Ty1i RNA might be undetectable by RNA-seq. In agreement with this idea, only a modest increase in Ty1i RNA relative to Ty1 mRNA was observed in the BY4742 loc1Δ mutant as determined by northern analysis (Figure 4A).
Figure 5.
Genomic analysis of RNA level and ribosome initiation in wild type and a loc1Δ mutant. (A) Transcripts and protein produced from Ty1. The element was divided into regions uniquely assignable to Ty1 (red), or Ty1 and Ty1i transcripts (blue). Three proteins are presented, two overlapping in the zero frame (Gag and p22), and a third produced after the +1 frameshift (Pol). These were assigned to Gag (green), Gag or p22 (light blue), or Pol (orange). (B) RNA-seq read counts for all genes (black dots) between wild type and loc1Δ. Each point is the average of three biological replicas for each strain. Highlighted are RNA read counts uniquely assignable to Ty1 (red), or Ty1 and Ty1i (blue). (C) Ribo-seq read counts for all genes (black dots) between wild type and loc1Δ. Analysis was restricted to reads 28 bp in length, and each point is the average of three biological replicas for each strain. Highlighted are read counts uniquely assignable to GAG in the zero frame (green), to either GAG or p22 in the zero frame (light blue), and reads from the POL region in the +1 frame (orange).
Previous work suggested that Loc1 represses translation of ASH1 mRNA (Komili et al. 2007). Therefore, we analyzed a Ribo-seq dataset generated in BY4741 and an isogenic loc1Δ mutant to determine if a specific increase in translation initiation can account for the increase of p22 in the loc1Δ mutant (Figure 5, A and C). The Ribo-seq reads were split according to region and frame: GAG (0-frame), GAG or p22 (0-frame), and POL (+1 frame). Ribo-seq coverage in these regions was not altered in the loc1Δ mutant compared to wild type. Thus, there are no remarkable changes in gene expression profiles to suggest a reason for observed CNC phenotype or increased p22 levels in the loc1Δ mutant.
There were 67 and 102 genes where RNA-seq transcript levels increased or decreased more than twofold in a loc1Δ mutant, respectively, with a false discovery rate (FDR)-adjusted P-value (padj) <0.05 (Table S4). Functional annotation clustering was performed to find which GO terms are enriched among these transcripts (Table S4). In general, expression of genes involved in growth-related processes such as cell wall structure, glycolysis/gluconeogenesis, ion transport, and amino acid biosynthesis were affected by the absence of LOC1. It remains to be determined whether these effects represent primary or secondary consequences of deleting LOC1.
Gag–Gag interactions and Ty1 mRNA packaging are altered in a loc1Δ mutant
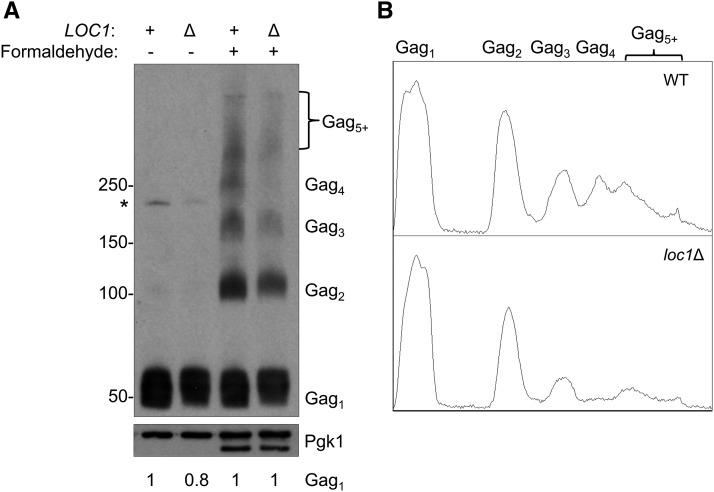
Formaldehyde cross-linking and the sedimentation pattern of Gag complexes was used to investigate the role of LOC1 in VLP assembly. Formaldehyde treatment of cells captures HIV-1 Gag assembly intermediates at the plasma membrane (Kutluay and Bieniasz 2010). However, our application is simplified since retrosome/VLP assembly takes place in the cytoplasm (Checkley et al. 2010; Dutko et al. 2010; Doh et al. 2014). To help ensure there are comparable levels of Gag prior to crosslinking, we induced pBDG606 expression in wild type BY4742 and an isogenic loc1Δ mutant (Figure 6). Following galactose induction, yeast cells were treated with formaldehyde, and total cell extracts were separated on a 3–8% gradient gel. Gag complexes and a Pgk1 loading control were detected by western blot analysis. In the absence of formaldehyde, Ty1 Gag was mostly present in a monomeric form (Gag1) (Figure 6A). We also detected a small amount of dimeric Gag (Gag2) and the 199 kDa Gag-Pol precursor (denoted by an asterisk). In wild-type cells treated with formaldehyde, there was an ordered accumulation of Gag multimers with Gag2 and trimeric Gag3 as the most prominent forms. When the loc1Δ mutant was treated with formaldehyde, a comparable amount of Gag1 was present when compared to wild type; however, there was a lower level of Gag2 and a marked decrease in higher order multimers, which is also evident in densitometric tracings (Figure 6B). There were also similar levels of Gag1 in wild type and loc1Δ cells when compared with Pgk1. Our results suggest that LOC1 facilitates early steps in VLP assembly as monitored by Gag multimerization.
Figure 6.
Gag multimerization in wild type and loc1Δ strains. (A) Total protein from pBDG606-induced WT and loc1Δ strains were cross-linked by formaldehyde treatment in vivo. Protein extracts from untreated (− formaldehyde) and treated (+ formaldehyde) cells were separated on a 3–8% Tris-Acetate gel and probed with Gag antiserum (TY-tag). Equal amounts of protein separated on a 10% SDS-PAGE gel were probed with Pgk1 antiserum, which served as a loading control. Estimated protein sizes are shown in kilodaltons. Extracts from untreated cells contain monomeric Gag (Gag1) and the Gag-Pol-p199 precursor (*), while cross-linked cells produce a ladder of Gag multimers. The gel conditions allow sufficient separation of Gag1–4, while larger multimers (Gag5+) are not resolved. An additional faster-migrating Pgk1 signal appeared in formaldehyde-treated samples, which likely results from cross-linking. Numbers below the panel indicate relative changes in Gag1 level compared to wild type with and without formaldehyde treatment, as determined by quantification of protein bands by densitometry. Both Pgk1 bands in formaldehyde treated cells were used in the quantification. (B) Densitometry traces of cross-linked Gag from WT and loc1Δ. The pixel intensity for each Gag-multimer per lane is shown along the Y-axis.
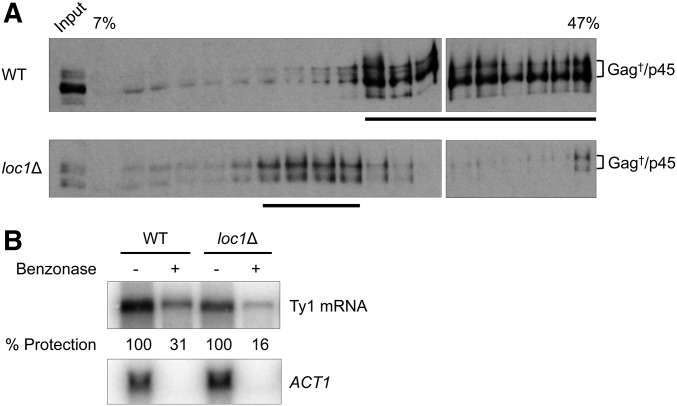
To visualize more complex assembly intermediates, total protein extracted from BY4742 wild type and loc1Δ cells was fractionated through 7–47% sucrose gradients, and the sedimentation pattern of endogenous Gag was monitored by western blot analysis (Figure 7A). The majority of Gag from wild type cells reproducibly sedimented as larger complexes (fractions 11–19) likely comprised of VLP assembly intermediates, since few endogenous VLPs are detected in the absence of pGTy1 expression (Checkley et al. 2010). In contrast, Gag from the loc1Δ mutant reproducibly sedimented as smaller complexes (fractions 7–10), suggesting that higher order Gag complex formation is also defective in a loc1Δ mutant.
Figure 7.
Gag sedimentation and nuclease protection of Ty1 mRNA in wild type and loc1Δ strains. (A) Total cell extracts from BY4742 WT or a loc1Δ mutant were centrifuged through a 7–47% continuous sucrose gradient. Ten micrograms of protein extract (input) and equal volumes of each fraction (15 μl/lane) were subjected to western blot analysis using Gag antiserum (TY-tag), which recognizes Gag isoforms and Gag-p45. Fractions containing the highest concentrations of Gag, as determined by densitometry tracing, are underlined. Sucrose gradient analysis was repeated three times and a representative experiment is shown here. (B) Ty1 mRNA packaging as monitored by sensitivity to the nuclease benzonase. Equal aliquots of whole cell extract from BY4742 WT and a loc1Δ mutant were incubated with (+) or without (−) benzonase. To detect Ty1 mRNA, total RNA extracted from these samples was analyzed by northern blot analysis. Nuclease protection assays were repeated three times and a representative experiment is shown. WT protection ranged from 26.5 to 31.4%, and loc1Δ protection ranged from 16.1 to 18.8%. ACT1 was used as a control to confirm RNA degradation in the benzonase treated samples.
To examine if Ty1 mRNA packaging into nuclease-resistant Gag assemblies is defective (Purzycka et al. 2013; Nishida et al. 2015), cell extracts from BY4742 wild type and loc1Δ were treated with the nuclease benzonase followed by northern blot analysis (Figure 7B). Hybridization signals from the benzonase treated samples were normalized to untreated controls, and ACT1 mRNA served as a control for RNA integrity and benzonase activity. Ty1 mRNA level reproducibly decreased about threefold when extracts from wild type cells were treated with benzonase, whereas Ty1 mRNA decreased more than sixfold when extracts from the loc1Δ mutant were treated with the nuclease. These results suggest that less Ty1 mRNA is packaged into nuclease resistant Gag complexes in the absence of LOC1.
Retrosomes are not detected in loc1Δ cells
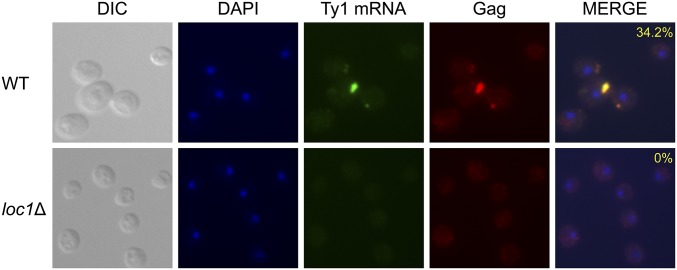
BY4742 wild type and loc1Δ cells were subjected to FISH/IF to visualize endogenous (Figure 8) or pBDG606 induced retrosomes (Figure S5). VLP antiserum was used to detect Gag, and a cocktail of DIG-labeled oligonucleotide GAG probes was used to detect full-length Ty1 mRNA (Saha et al. 2015). About 34% of wild type cells contained retrosomes, which are defined as distinct foci containing Gag and Ty1 mRNA (Malagon and Jensen 2008; Checkley et al. 2010). Retrosomes were not detected in the loc1Δ mutant. We also failed to observe nondistinct punctate staining for both Ty1 mRNA and Gag, which is present in other Ty1 cofactor mutants (Checkley et al. 2010; Dutko et al. 2010). Instead, diffuse cytoplasmic staining was detected in the loc1Δ mutant. Furthermore, retrosomes were also absent when pBDG606 was expressed in loc1Δ cells (Figure S5). Taken together, our results indicate that steps leading to assembly of functional VLPs are disrupted in a loc1Δ mutant.
Figure 8.
Retrosome formation in wild type and loc1Δ cells. BY4742 WT and loc1Δ mutant strains were visualized by DIC and fluorescence microscopy. DNA was stained with DAPI, Ty1 mRNA was detected using a cocktail of GAG-DIG probes, and Gag was detected with VLP antiserum. The percentage of cells containing retrosomes, defined as having at least one colocalized Ty1 mRNA/Gag foci, is shown in the DAPI/Ty1 mRNA/Gag merge. The experiment was repeated twice and >200 cells were analyzed for each strain.
Discussion
Genome-wide screens have revealed cellular modulators of Ty1 retrotransposition, and some are conserved with cellular factors involved in retroviral replication (Maxwell and Curcio 2007; Goff 2008). Since all the genetic screens for Ty1 cofactors (Griffith et al. 2003; Dakshinamurthy et al. 2010; Risler et al. 2012) were performed in CNC+ strains containing genomic Ty1 elements and producing the p22 restriction factor (Saha et al. 2015), a subset of previously identified Ty1 cofactors may affect the level or activity of p22. RNA metabolism is central to the process of retrotransposition; therefore, cellular processes affecting the Ty1 life cycle between RNA transcription and VLP assembly (Ty1 RNA export, translation, and retrosome formation) may contribute to CNC. Here, we utilized a candidate gene approach to identify CNC-specific modulators based on informative secondary screens that monitor preferential loss of mature IN and Ty1 mobility in a CNC+ strain compared to an isogenic CNC− strain. One weakness of a candidate gene approach is that not all possible causative genes may be queried (Zhu and Zhao 2007). However, the hundreds of known Ty1 modulators (Scholes et al. 2001; Griffith et al. 2003; Nyswaner et al. 2008; Dakshinamurthy et al. 2010; Risler et al. 2012) provide a rich source of interactions between Ty1 and its host that can be mined by additional phenotypic analyses. Our analysis of CNC-specific modulators revealed an unexpected connection between Ty1 CNC and a large group of host cofactors involved in ribosome biogenesis and function (Suresh et al. 2015).
The secondary screens identified LOC1 as a CNC-specific Ty1 cofactor since the level of mature Ty1 IN and retromobility decreased much more in CNC+ than in CNC− strains. LOC1 encodes a nucleolar protein that binds RNA (Long et al. 2001), associates with the pre-60S ribosome complex (Harnpicharnchai et al. 2001), and is necessary for rRNA processing, 60S assembly and nuclear export of pre-60S subunits (Urbinati et al. 2006). Loc1 also helps localize ASH1 mRNA to the distal tip of anaphase cells (Long et al. 2001) via a direct interaction with She2 (Niedner et al. 2013). To determine whether LOC1 modulated Ty1 transposition by altering RNA localization or ribosome biogenesis, we analyzed two other genes implicated in ASH1 mRNA dynamics, PUF6 and SHE2, for their ability to phenocopy the alteration of CNC observed with LOC1. Disrupting SHE2 leads to complete loss of ASH1 mRNA localization, and Puf6 interacts with Loc1 in the nucleolus, and has a role in translational repression of ASH1 mRNA (Heym and Niessing 2012). Our data suggest that the ASH1 mRNA localization pathway is not involved in CNC because deleting a key component of the pathway, SHE2, does not affect Ty1 mobility, confer a CNC-specific transposition defect, or result in loss of IN. Furthermore, several other genes implicated in ASH1 mRNA localization (MYO4, SHE3, and SCP160) do not affect Ty1 CNC (Figure S3).
Like LOC1, PUF6 is involved in both ASH1 mRNA localization as well as 60S ribosome biogenesis (Li et al. 2009). The puf6Δ mutant showed a decrease of IN in both CNC− and CNC+ strains, but its transposition defect was CNC-specific. Perhaps PUF6 is required for the process of Ty1 retrotransposition, and is also involved in CNC. Interestingly, Loc1 function may be compromised in the absence of Puf6 since these proteins interact (Shen et al. 2009; Yang et al. 2016). While their interaction is important for ASH1 mRNA localization and 60S ribosome assembly, previous studies suggest that Loc1 and Puf6 carry out distinct functions. For example, puf6Δ, loc1Δ, and a puf6Δ loc1Δ double mutant each display different phenotypes with respect to the localization of ribosomal protein paralogs, and assembly of Rpl43 in the 60S ribosome subunit (Komili et al. 2007; Yang et al. 2016).
Analysis of LOC1 focused our attention on how ribosome biogenesis affects Ty1 transposition and CNC. Remarkably, 71 of 458 host genes identified in multiple genetic screens for Ty1 modulators (Scholes et al. 2001; Griffith et al. 2003; Nyswaner et al. 2008; Dakshinamurthy et al. 2010; Risler et al. 2012) encode ribosomal subunits, ribosome biogenesis factors, and translation factors. Recent work suggests that these genes affect multiple steps during transposition, including translation initiation, programmed frameshifting, protein stability, and subcellular protein localization (Suresh et al. 2015). A rare tRNA-Arg(CCU) gene HSX1 specifically involved in programmed frameshifting is also required for Ty1 mobility (Kawakami et al. 1993). rpl21aΔ, rpl27aΔ, rpl39Δ, and rps0b mutants all show transcriptional patterns where Ty1i RNA increases relative to Ty1 mRNA. Additionally, Ty1 mRNA and Gag localization in retrosomes is absent in a rpl7aΔ mutant, further implicating ribosome biogenesis in Ty1 retrotransposition (Doh et al. 2014).
Ribosome biogenesis and nucleolar functions influence the replication of retroviruses and other retrotransposons. In addition to its primary function in ribosome biogenesis, the nucleolus is involved in ribonucleoprotein remodeling, cell cycle progression, and stress response (Boisvert et al. 2007). Cellular stress and viral infection change the organization, composition, and morphology of the nucleolus (Boulon et al. 2010). The nucleolus is the target for viral infection by RNA viruses (Hiscox 2007; Salvetti and Greco 2014), and nucleolar trafficking of viral proteins occurs with retroviruses such as Rous Sarcoma Virus (RSV), Mouse Mammary Tumor Virus (MMTV), and HIV (Jarboui et al. 2012; Beyer et al. 2013; Lochmann et al. 2013). Ty3 and L1 retrotransposon proteins (Lin et al. 2001; Goodier et al. 2004) and Alu RNA (Caudron-Herger et al. 2015) display nucleolar localization. Moreover, ribosomal protein Rpl7 affects HIV-1 Gag nucleic acid chaperone activity (Mekdad et al. 2016), and Rpl9 helps traffic MMTV Gag proteins (Beyer et al. 2013; Lee et al. 2013; Mekdad et al. 2016). Taken together, our work provides additional evidence linking ribosome biogenesis and retroelement replication.
We expanded the CNC analysis to include the ribosomal protein gene paralogs RPS0A and RPS0B, and RPL7A and RPL7B. As is the case for other paralogs, only RPS0B and RPL7A were recovered as Ty1 cofactors (Dakshinamurthy et al. 2010; Risler et al. 2012). rps0bΔ and rpl7aΔ mutants showed loss of IN and a lower level of Ty1 mobility in CNC+ but not in CNC− strains, demonstrating that these mutants are CNC-specific (Table 2). However, the rpl7bΔ and rps0aΔ paralogs also displayed CNC-specificity but at a lower level than rpl7aΔ and rps0bΔ. Expression of these ribosomal protein genes may account for these results. The level of RPL7A mRNA is fourfold higher than RPL7B, and RPS0B mRNA is 1.5-fold higher when compared to RPS0A (Komili et al. 2007), which correlates with CNC−/CNC+ ratios of Ty1his3-AI mobility (Table 2). Based on mRNA expression, Rpl7a may be the predominant ribosomal protein in the cell. Therefore, the absence of Rpl7a caused a more severe CNC-specific transposition defect. However, losing Rpl7b does not affect CNC because there is still a substantial amount of Rpl7a present in a cell. For Rps0a and Rps0b, comparable amounts of protein may be expressed from each gene. The absence of one RPS0 paralog could impact CNC, as suggested by the mobility defect observed in both CNC+ rps0aΔ and rps0bΔ mutants. Our results also agree with recent work regarding dosage-dependent rather than paralog-specific function of Rpl7a and Rpl7b on different cellular phenotypes, including Ty1 life cycle stages (Palumbo et al. 2017). Taken together, any paralog-specific effects of ribosomal proteins on CNC is probably due to expression levels of the duplicated ribosomal protein genes.
Our results did not identify a step during ribosome biogenesis that impacts Ty1 CNC. Although LOC1 was analyzed in greatest detail as a loc1Δ mutation conferred robust CNC-specificity, both small and large subunit ribosomal protein mutations as well as a ribosome assembly factor mutation also affect CNC to varying degrees. Instead of responding to specific steps in ribosome biogenesis, Ty1 CNC may be modulated by ribosome availability. Polysome profiles of several mutants analyzed for Ty1 CNC suggest there is an imbalance of 40S and 60S subunits. A decrease in 40S and an increase in 60S subunits is observed in a rps0bΔ mutant (Suresh et al. 2015), a decrease of both 40S and 60S subunits occurs in a loc1Δ mutant (Yang et al. 2016), and a decrease in 60S subunits, 80S monosomes, and polysomes is detected in a rpl7aΔ mutant (Palumbo et al. 2017). Defects in functional ribosome production affect cell growth and proliferation (Woolford and Baserga 2013). Interestingly, there is also a correlation between ribosome availability, cell growth, and Ty1 transposition. loc1Δ and puf6Δ mutants grow slowly, contain decreased amounts of 40S and 60S subunits (Yang et al. 2016), and display a high CNC−/CNC+ ratio (Table 2). Growth of RPL7 paralog mutants also show a similar relationship with Ty1 CNC. An rpl7aΔ mutant grows more slowly, and impacts Ty1 CNC more than a rpl7bΔ mutant, which contains more Rpl7a due to the higher expression of RPL7A (Palumbo et al. 2017). In addition, the slow growth of ribosomal protein mutants is related to other cellular phenotypes such as enhanced resistance to endoplasmic reticulum stress (Steffen et al. 2012). Although the magnitude of CNC may correlate with the level of functional ribosomes, other CNC phenotypes may not be directly related. For example, retrosomes are not detected in loc1Δ (Figure 8) and rpl7aΔ mutants (Doh et al. 2014); however, a more severe transposition defect is conferred by a loc1Δ mutant (Table 2). Together, our data support the idea that different steps in the Ty1 life cycle have different thresholds for ribosome levels.
The observed CNC-specific phenotypes in loc1Δ and two ribosomal protein mutants likely results from an increased amount of p22 when compared to the isogenic wild type strain (Figure 4B). We examined steps in Ty1 gene expression and protein stability to determine what might account for the increase in p22 observed in several ribosome biogenesis mutants. Our data permit the following conclusions:
The level of poly(A)+ Ty1 mRNA and Ty1i RNA is altered in the absence of LOC1, RPS0B, and RPL7A, and displays a trend suggesting that more Ty1i RNA is present relative to Ty1 mRNA (Figure 4A). However, the decrease in the level of poly(A)+ Ty1i RNA in the loc1Δ and rpl7aΔ mutants or the increase in the level of Ty1i RNA observed in other mutants fails to explain the 3.5 to 19.4-fold increase in the level of p22. Perhaps nonpolyadenylated Ty1i transcripts can also be used to synthesize p22 in the ribosome biogenesis mutants (Saha et al. 2015; Suresh et al. 2015).
Loc1 is reported to function as a translational repressor (Komili et al. 2007); therefore, loss of Loc1 may enhance the initiation of p22 translation. However, Ribo-seq analysis of p22 in a loc1Δ mutant is comparable to other Ty1 proteins as well as cellular proteins (Figure 5C). Although this result raises the possibility that initiation of translation is not enhanced in a loc1Δ mutant, it is unclear why the Ribo-seq analysis does not account for the increased level of p22 observed in loc1Δ as well as the rps0bΔ and rpl7aΔ mutants (Figure 4B). Possible reasons for this discrepancy include: (a) the effects on translation initiation are obscured by the overlapping nature of RNAs and proteins in this region, (b) the experimental conditions or strains used for the libraries differ from those used in our work, or (c) Loc1 may exert its effects through some non-RNA or nontranslational mechanism.
Loc1 enhances p22 turnover. Pulse-chase immunoprecipitation analysis suggests that p22 stability increases in a loc1Δ mutant (Figure S4). However, a ∼60% increase in p22 stability does not completely explain the increase in p22 level (Figure 4B).
What could be causing the transcriptional changes of Ty1 mRNA and Ty1i RNA in the ribosome biogenesis mutants? Perhaps defects in ribosome biogenesis cause a form of stress that leads to changes in Ty1 transcript level. A nucleolar stress response pathway activates p53-dependent as well as p53-independent downstream pathways in mammalian cells (James et al. 2014). Nucleolar stress may also occur in yeast (Gomez-Herreros et al. 2013; Thapa et al. 2013), though the signaling pathway has not been defined. We considered the possibility that the environmental stress response (ESR), which alters expression of a core set of genes under diverse environmental changes (Gasch and Werner-Washburne 2002) is activated in a loc1Δ mutant. However, analysis of the loc1Δ and ESR transcriptomes (Gasch et al. 2000) reveals little overlap with the ESR (Table S4). Interestingly, stress-responsive transcription factors such as Msn2, Tye7, and Gcn4 affect Ty1 mRNA expression may also be involved in Ty1i RNA transcription. Msn2 binds to Ty1 elements in vivo and is capable of acting as a transcriptional activator as well as a repressor (Schmitt and McEntee 1996; Elfving et al. 2014). Tye7 is a bHLH transcription factor that activates Ty1-mediated gene expression and regulates Ty1 mRNA and antisense RNAs expression upon adenine starvation (Lohning and Ciriacy 1994; Servant et al. 2012). Gcn4 is a transcription factor that responds to amino acid starvation via the TOR pathway, and can activate Ty1 transcription by recruiting chromatin remodelers Swi/Snf and SAGA (Hinnebusch and Natarajan 2002; Morillon et al. 2002). Further work will be required to understand the relationship between factors responsible for Ty1 mRNA and Ty1i RNA transcription.
Several Ty1 defects in cells lacking LOC1 or ribosomal proteins can be reconciled by an increase in the level of p22 (Saha et al. 2015). These include a marked decrease in mature IN relative to Gag (Figure 2B and Figure 3), altered Gag–Gag interactions (Figure 6 and Figure 7), and formation of endogenous (Figure 8) or pGTy1-induced retrosomes (Figure S5). Previous work (Saha et al. 2015), along with data presented here, led us to consider retrosome formation in CNC− and CNC+ cells. In CNC− cells, retrosomes may not form, or are undetectable, due to low levels of Ty1 products, including p22. The Ty1 copy number dependence of CNC, however, suggests the low level of p22 present in cells containing just a few elements may be less inhibitory (Garfinkel et al. 2003, 2016). In CNC+ cells, the increase in retrosome abundance may sensitize Ty1 retrotransposition to small changes in p22 levels and disturbances in the retrosome environment or network of interactions.
Recent work on biomolecular condensates (Banani et al. 2017; Riback et al. 2017), also known as membrane-less organelles or quinary structures, have influenced our view of retrosomes and their interactions with other cellular components. We postulate that Ty1 retrosomes are biomolecular condensates as they contain Ty1 RNA and proteins, and likely cellular proteins in an undefined assembly, no fixed membranous structure, and are highly dynamic. Phase separation caused by multivalent macromolecular interactions of nucleic acids and proteins drive the formation of molecular condensates. Indeed, P-bodies and stress granules are biomolecular condensates and required for Ty1 transposition. Retrosomes disassemble in P-body component as well as stress granule component mutants (Checkley et al. 2010; Dutko et al. 2010; Doh et al. 2014). The highly dynamic nature of retrosomes is also revealed by their disassembly under conditions, such as glucose deprivation, that causes P-bodies to form (Checkley et al. 2010). Our work favors the idea that normal ribosome biogenesis and functional ribosomes contribute to the retrosome environment required to maintain these condensates in a phase-separated or demixed state. The continuum from retrosome formation to VLP assembly may be similar to the formation of condensates such as stress granules (Riback et al. 2017) or transcriptional super-enhancers (Hnisz et al. 2017) from ribonucleoprotein complexes or “normal” enhancers, respectively. As various chemical and environmental factors or stress conditions influence the structure and function of biomolecular condensates (Mitrea and Kriwacki 2016), changes in the cellular environment resulting from defects in ribosome biogenesis in combination with increased p22 levels contribute to altered retrosome dynamics and defects in Ty1 mobility.
In summary, our study demonstrates that ribosome biogenesis is critical for Ty1 CNC. The diverse defects in ribosome biogenesis that enhance Ty1 CNC raise the possibility of a cellular response that leads to an increase in the amount of p22 relative to its target Gag. Further work will be required to define a stress response in yeast that affects p22 abundance and retrosome formation, characterize the retrosome interactome both genetically and biochemically, and determine what triggers VLP assembly from retrosomes.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300388/-/DC1.
Acknowledgments
We thank Katherine Nyswaner and Karen Stafanisko for technical help generating the S. cerevisiae CNC− strain. We thank Jeremy Thorner (Pgk1), Stephen Hajduk (TY tag), and Alan Kingsman (Ty1 VLP) for providing antisera, Gerald Fink for plasmid pB130, and Michel Aigle for yeast strains. We thank Claiborne Glover, Steven Hajduk, William Lanzilotta, Anne Summers, and Michael Terns for sharing equipment. We also thank Agniva Saha, Yuri Nishida, and Wioletta Czaja for helpful discussions. This work was supported by National Institutes of Health grant GM095622 (D.J.G.) and funds from University of Georgia Research Foundation (D.J.G). Early portions of this work were funded by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (D.J.G.).
Footnotes
Communicating editor: A. Mitchell
Literature Cited
- Baller J. A., Gao J., Stamenova R., Curcio M. J., Voytas D. F., 2012. A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Res. 22: 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18: 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P., Bagherzadeh Z., Matthews K. R., Gull K., 1996. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77: 235–239. [DOI] [PubMed] [Google Scholar]
- Belle A., Tanay A., Bitincka L., Shamir R., O’Shea E. K., 2006. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl. Acad. Sci. USA 103: 13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A. R., Bann D. V., Rice B., Pultz I. S., Kane M., et al. , 2013. Nucleolar trafficking of the mouse mammary tumor virus gag protein induced by interaction with ribosomal protein L9. J. Virol. 87: 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R., 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197: 345–346. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Eichinger D., Castrillon D., Fink G. R., 1988. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol. Cell. Biol. 8: 1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F. M., van Koningsbruggen S., Navascues J., Lamond A. I., 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8: 574–585. [DOI] [PubMed] [Google Scholar]
- Boulon S., Westman B. J., Hutten S., Boisvert F. M., Lamond A. I., 2010. The nucleolus under stress. Mol. Cell 40: 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288c: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Braiterman L. T., Boeke J. D., 1994. In vitro integration of retrotransposon Ty1: a direct physical assay. Mol. Cell. Biol. 14: 5719–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braiterman L. T., Monokian G. M., Eichinger D. J., Merbs S. L., Gabriel A., et al. , 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: phenotypic analysis. Gene 139: 19–26. [DOI] [PubMed] [Google Scholar]
- Bridier-Nahmias A., Tchalikian-Cosson A., Baller J. A., Menouni R., Fayol H., et al. , 2015. Retrotransposons. An RNA polymerase III subunit determines sites of retrotransposon integration. Science 348: 585–588. [DOI] [PubMed] [Google Scholar]
- Caudron-Herger M., Pankert T., Seiler J., Nemeth A., Voit R., et al. , 2015. Alu element-containing RNAs maintain nucleolar structure and function. EMBO J. 34: 2758–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K. B., Bystrom A. S., Boeke J. D., 1992. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc. Natl. Acad. Sci. USA 89: 3236–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley M. A., Nagashima K., Lockett S. J., Nyswaner K. M., Garfinkel D. J., 2010. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol. Cell. Biol. 30: 382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley M. A., Mitchell J. A., Eizenstat L. D., Lockett S. J., Garfinkel D. J., 2013. Ty1 Gag enhances the stability and nuclear export of Ty1 mRNA. Traffic 14: 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung S., Ma L., Chan P. H., Hu H. L., Mayor T., et al. , 2016. Ty1 integrase interacts with RNA polymerase III-specific subcomplexes to promote insertion of Ty1 elements upstream of polymerase (Pol) III-transcribed genes. J. Biol. Chem. 291: 6396–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M. J., Garfinkel D. J., 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88: 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M. J., Garfinkel D. J., 1992. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol. Cell. Biol. 12: 2813–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M. J., Lutz S., Lesage P., 2015. The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae. Microbiol. Spectr. 3: 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakshinamurthy A., Nyswaner K. M., Farabaugh P. J., Garfinkel D. J., 2010. BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae. Genetics 185: 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine S. E., Boeke J. D., 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10: 620–633. [DOI] [PubMed] [Google Scholar]
- Doh J. H., Lutz S., Curcio M. J., 2014. Co-translational localization of an LTR-retrotransposon RNA to the endoplasmic reticulum nucleates virus-like particle assembly sites. PLoS Genet. 10: e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I. A., Weinberg D. E., Xie K. T., Mower J. P., Wolfe K. H., et al. , 2009. RNAi in budding yeast. Science 326: 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinnenberg I. A., Fink G. R., Bartel D. P., 2011. Compatibility with killer explains the rise of RNAi-deficient fungi. Science 333: 1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutko J. A., Kenny A. E., Gamache E. R., Curcio M. J., 2010. 5′ to 3′ mRNA decay factors colocalize with Ty1 Gag and human APOBEC3G and promote Ty1 retrotransposition. J. Virol. 84: 5052–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfving N., Chereji R. V., Bharatula V., Bjorklund S., Morozov A. V., et al. , 2014. A dynamic interplay of nucleosome and Msn2 binding regulates kinetics of gene activation and repression following stress. Nucleic Acids Res. 42: 5468–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. X., Moore S. P., Garfinkel D. J., Rein A., 2000. The genomic RNA in Ty1 virus-like particles is dimeric. J. Virol. 74: 10819–10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Nyswaner K., Wang J., Cho J. Y., 2003. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 165: 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel D. J., Tucker J. M., Saha A., Nishida Y., Pachulska-Wieczorek K., et al. , 2016. A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces. Curr. Genet. 62: 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P., Werner-Washburne M., 2002. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2: 181–192. [DOI] [PubMed] [Google Scholar]
- Gasch A. P., Spellman P. T., Kao C. M., Carmel-Harel O., Eisen M. B., et al. , 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Goff S. P., 2008. Knockdown screens to knockout HIV-1. Cell 135: 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Herreros F., Rodriguez-Galan O., Morillo-Huesca M., Maya D., Arista-Romero M., et al. , 2013. Balanced production of ribosome components is required for proper G1/S transition in Saccharomyces cerevisiae. J. Biol. Chem. 288: 31689–31700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J. L., Ostertag E. M., Engleka K. A., Seleme M. C., Kazazian H. H., Jr., 2004. A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 13: 1041–1048. [DOI] [PubMed] [Google Scholar]
- Griffith J. L., Coleman L. E., Raymond A. S., Goodson S. G., Pittard W. S., et al. , 2003. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R., 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194: 1–863. [PubMed] [Google Scholar]
- Harnpicharnchai P., Jakovljevic J., Horsey E., Miles T., Roman J., et al. , 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8: 505–515. [DOI] [PubMed] [Google Scholar]
- Heym R. G., Niessing D., 2012. Principles of mRNA transport in yeast. Cell. Mol. Life Sci. 69: 1843–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Natarajan K., 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1: 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox J. A., 2007. RNA viruses: hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., Sharp P. A., 2017. A phase separation model for transcriptional control. Cell 169: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- James A., Wang Y., Raje H., Rosby R., DiMario P., 2014. Nucleolar stress with and without p53. Nucleus 5: 402–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarboui M. A., Bidoia C., Woods E., Roe B., Wynne K., et al. , 2012. Nucleolar protein trafficking in response to HIV-1 Tat: rewiring the nucleolus. PLoS One 7: e48702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., Pande S., Faiola B., Moore D. P., Boeke J. D., et al. , 1993. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 135: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna M. A., Brachmann C. B., Devine S. E., Boeke J. D., 1998. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol. Cell. Biol. 18: 1115–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., Vanguri S., Boeke J. D., Gabriel A., Voytas D. F., 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8: 464–478. [DOI] [PubMed] [Google Scholar]
- Komili S., Farny N. G., Roth F. P., Silver P. A., 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell 131: 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay S. B., Bieniasz P. D., 2010. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 6: e1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., et al. , 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lee A. S., Burdeinick-Kerr R., Whelan S. P., 2013. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc. Natl. Acad. Sci. USA 110: 324–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin H. L., Moran J. V., 2011. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 12: 615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Lee I., Moradi E., Hung N. J., Johnson A. W., et al. , 2009. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 7: e1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. S., Nymark-McMahon M. H., Yieh L., Sandmeyer S. B., 2001. Integrase mediates nuclear localization of Ty3. Mol. Cell. Biol. 21: 7826–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochmann T. L., Bann D. V., Ryan E. P., Beyer A. R., Mao A., et al. , 2013. NC-mediated nucleolar localization of retroviral gag proteins. Virus Res. 171: 304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohning C., Ciriacy M., 1994. The TYE7 gene of Saccharomyces cerevisiae encodes a putative bHLH-LZ transcription factor required for Ty1-mediated gene expression. Yeast 10: 1329–1339. [DOI] [PubMed] [Google Scholar]
- Long R. M., Gu W., Meng X., Gonsalvez G., Singer R. H., et al. , 2001. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol. 153: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagon F., Jensen T. H., 2008. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol. Cell. Biol. 28: 6022–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda E., Garfinkel D. J., 2009. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc. Natl. Acad. Sci. USA 106: 15657–15662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P. H., Curcio M. J., 2007. Host factors that control long terminal repeat retrotransposons in Saccharomyces cerevisiae: implications for regulation of mammalian retroviruses. Eukaryot. Cell 6: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane L. M., Pulliam K. F., Devine S. E., Corbett A. H., 2008. The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus. Nucleic Acids Res. 36: 4317–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekdad H. E., Boutant E., Karnib H., Biedma M. E., Sharma K. K., et al. , 2016. Characterization of the interaction between the HIV-1 Gag structural polyprotein and the cellular ribosomal protein L7 and its implication in viral nucleic acid remodeling. Retrovirology 13: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrea D. M., Kriwacki R. W., 2016. Phase separation in biology; functional organization of a higher order. Cell Commun. Signal. 14: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. P., Rinckel L. A., Garfinkel D. J., 1998. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol. Cell. Biol. 18: 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. P., Liti G., Stefanisko K. M., Nyswaner K. M., Chang C., et al. , 2004. Analysis of a Ty1-less variant of Saccharomyces paradoxus: the gain and loss of Ty1 elements. Yeast 21: 649–660. [DOI] [PubMed] [Google Scholar]
- Morillon A., Benard L., Springer M., Lesage P., 2002. Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons. Mol. Cell. Biol. 22: 2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedner A., Muller M., Moorthy B. T., Jansen R. P., Niessing D., 2013. Role of Loc1p in assembly and reorganization of nuclear ASH1 messenger ribonucleoprotein particles in yeast. Proc. Natl. Acad. Sci. USA 110: E5049–E5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y., Pachulska-Wieczorek K., Blaszczyk L., Saha A., Gumna J., et al. , 2015. Ty1 retrovirus-like element Gag contains overlapping restriction factor and nucleic acid chaperone functions. Nucleic Acids Res. 43: 7414–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyswaner K. M., Checkley M. A., Yi M., Stephens R. M., Garfinkel D. J., 2008. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178: 197–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo R. J., Fuchs G., Lutz S., Curcio M. J., 2017. Paralog-specific functions of RPL7A and RPL7B mediated by ribosomal protein or snoRNA dosage in Saccharomyces cerevisiae. G3 7: 591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purzycka K. J., Legiewicz M., Matsuda E., Eizentstat L. D., Lusvarghi S., et al. , 2013. Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res. 41: 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback J. A., Katanski C. D., Kear-Scott J. L., Pilipenko E. V., Rojek A. E., et al. , 2017. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168: 1028–1040.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risler J. K., Kenny A. E., Palumbo R. J., Gamache E. R., Curcio M. J., 2012. Host co-factors of the retrovirus-like transposon Ty1. Mob. DNA 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A., Mitchell J. A., Nishida Y., Hildreth J. E., Ariberre J. A., et al. , 2015. A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J. Virol. 89: 3922–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti A., Greco A., 2014. Viruses and the nucleolus: the fatal attraction. Biochim. Biophys. Acta 1842: 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheifele L. Z., Cost G. J., Zupancic M. L., Caputo E. M., Boeke J. D., 2009. Retrotransposon overdose and genome integrity. Proc. Natl. Acad. Sci. USA 106: 13927–13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A. P., McEntee K., 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93: 5777–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes D. T., Banerjee M., Bowen B., Curcio M. J., 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G., Pinson B., Tchalikian-Cosson A., Coulpier F., Lemoine S., et al. , 2012. Tye7 regulates yeast Ty1 retrotransposon sense and antisense transcription in response to adenylic nucleotides stress. Nucleic Acids Res. 40: 5271–5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G., Burkett T. J., Garfinkel D. J., 1994. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol. Cell. Biol. 14: 6540–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Paquin N., Forget A., Chartrand P., 2009. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol. Biol. Cell 20: 2265–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen K. K., McCormick M. A., Pham K. M., MacKay V. L., Delaney J. R., et al. , 2012. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh S., Ahn H. W., Joshi K., Dakshinamurthy A., Kananganat A., et al. , 2015. Ribosomal protein and biogenesis factors affect multiple steps during movement of the Saccharomyces cerevisiae Ty1 retrotransposon. Mob. DNA 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa M., Bommakanti A., Shamsuzzaman M., Gregory B., Samsel L., et al. , 2013. Repressed synthesis of ribosomal proteins generates protein-specific cell cycle and morphological phenotypes. Mol. Biol. Cell 24: 3620–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker J. M., Larango M. E., Wachsmuth L. P., Kannan N., Garfinkel D. J., 2015. The Ty1 retrotransposon restriction factor p22 targets Gag. PLoS Genet. 11: e1005571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbinati C. R., Gonsalvez G. B., Aris J. P., Long R. M., 2006. Loc1p is required for efficient assembly and nuclear export of the 60S ribosomal subunit. Mol. Genet. Genomics 276: 369–377. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P., 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wilhelm M., Wilhelm F. X., Keith G., Agoutin B., Heyman T., 1994. Yeast Ty1 retrotransposon: the minus-strand primer binding site and a cis-acting domain of the Ty1 RNA are both important for packaging of primer tRNA inside virus-like particles. Nucleic Acids Res. 22: 4560–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Chaleff D. T., Valent B., Fink G. R., 1984. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107: 179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J. L., Baserga S. J., 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. T., Ting Y. H., Liang K. J., Lo K. Y., 2016. The roles of Puf6 and Loc1 in 60S biogenesis are interdependent, and both are required for efficient accommodation of Rpl43. J. Biol. Chem. 291: 19312–19323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Zhao S., 2007. Candidate gene identification approach: progress and challenges. Int. J. Biol. Sci. 3: 420–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All strains and reagents are available upon request.