Abstract
A crucial step in plant breeding is the selection and combination of parents to form new crosses. Genome-based prediction guides the selection of high-performing parental lines in many crop breeding programs which ensures a high mean performance of progeny. To warrant maximum selection progress, a new cross should also provide a large progeny variance. The usefulness concept as measure of the gain that can be obtained from a specific cross accounts for variation in progeny variance. Here, it is shown that genetic gain can be considerably increased when crosses are selected based on their genomic usefulness criterion compared to selection based on mean genomic estimated breeding values. An efficient and improved method to predict the genetic variance of a cross based on Markov chain Monte Carlo samples of marker effects from a whole-genome regression model is suggested. In simulations representing selection procedures in crop breeding programs, the performance of this novel approach is compared with existing methods, like selection based on mean genomic estimated breeding values and optimal haploid values. In all cases, higher genetic gain was obtained compared with previously suggested methods. When 1% of progenies per cross were selected, the genetic gain based on the estimated usefulness criterion increased by 0.14 genetic standard deviation compared to a selection based on mean genomic estimated breeding values. Analytical derivations of the progeny genotypic variance-covariance matrix based on parental genotypes and genetic map information make simulations of progeny dispensable, and allow fast implementation in large-scale breeding programs.
Keywords: genomic selection, Bayesian statistics, plant breeding, progeny variance, usefulness criterion
IN plant breeding, superior inbred lines are developed either for direct cultivar release, as hybrid components, or as potential parents in population improvement. Generally, high yielding parental lines are crossed to secure a high mean performance of the progeny. To identify superior progeny, ensure genetic gain in the next selection cycle, and to maintain long-term selection gain, it is important that the cross also generates a high genetic variance. Following Schnell and Utz (1975), the “usefulness” of a cross is defined as the trait mean of a defined upper fraction of its progeny, and can be derived as the expected cross mean plus the expected selection gain as a function of the selection intensity, square-root of the trait heritability, and the genetic standard deviation of the cross. With decreasing genotyping costs, selection intensity can be increased by the use of genome-based prediction methods, and, consequently, the importance of considering the progeny variance when deciding about future crosses increases (Zhong and Jannink 2007). Several endeavors have been made in the past to predict the progeny variance. Earlier attempts used the phenotypic distance (Utz et al. 2001), and since the availability of markers the molecular distance of parental lines, to predict progeny variance but both with limited success (Bohn et al. 1999; Hung et al. 2012). Recently, the potential of genomic selection has been investigated for many species, and in major crops such as maize and wheat it has been fully integrated in commercial breeding programs. The possibility to get dense marker genotypes allows an integration of genomic prediction in many steps of line and hybrid improvement programs (Heslot et al. 2015). This has also led to the suggestion to use genomic estimated breeding values (GEBVs) to predict progeny variance (Endelman 2011; Bernardo 2014; Mohammadi et al. 2015). An R package developed by Mohammadi et al. (2015) predicts the progeny variance by using an appropriate training population of genotyped and phenotyped inbred lines and marker effects estimated with a whole-genome regression model like ridge-regression best linear unbiased prediction (RR-BLUP, Meuwissen et al. 2001). Subsequently, progenies from two genotyped parental lines are simulated in silico using genetic map information. In a third step, GEBVs of the simulated progenies are predicted using marker effect estimates from the training population. The progeny variance is finally estimated as sample variance of the GEBVs. Concerns about using the sample variance of GEBVs as estimate of the genetic variance were raised as GEBVs are shrunken toward zero, and thus the approach underestimates the true genetic variance (Lian et al. 2015). Lehermeier et al. (2017) showed that a fully Bayesian estimate as proposed by Sorensen et al. (2001) improves the estimation of the genetic variance explained by markers in a given population compared to estimation based on RR-BLUP variance components by taking linkage disequilibrium (LD) between quantitative trait loci (QTL) into account. Here, we suggest to integrate this fully Bayesian estimate of the genetic variance into the usefulness criterion (UC).
Following a different rationale than the UC, Daetwyler et al. (2015) proposed the concept of selecting heterozygous lines based on their optimal haploid value (OHV). The goal of OHV is to predict the best fully homozygous line that can be produced from a heterozygous line or a cross. The latter authors showed that selection based on OHV increases long-term genetic gain compared to standard genomic selection based on GEBVs.
We hypothesize that an increase in genetic gain can be obtained when crosses are selected based on their estimated UC compared to their mean GEBV or their OHV. In addition, genetic variance prediction is assumed to be more accurate with a fully Bayesian estimate based on Markov chain Monte Carlo (MCMC) samples compared to the sample variance of the GEBVs. We investigate our hypotheses in simulation studies based on genotypic maize data under varying selection intensities, trait heritabilities, training population sizes, and model complexities. We show that the genetic variance of progenies from a cross can be derived analytically from the parental genotypes and genetic map information without the need for in silico simulations. We provide formulas for calculating the expected genetic variance for a given type of population to be created from a biparental cross taking into account the expected frequency of recombinants under different levels of inbreeding.
Materials and Methods
Derivation of genetic variance among progenies
In this section and supporting Supplemental Material, File S1, we show how to derive the genetic variance of a cross under the assumption of biallelic QTL, known homozygous parental genotypes at QTL, known QTL allele substitution effects, known recombination frequencies between QTL, and absence of dominance and epistasis. We first concentrate on doubled haploid (DH) lines derived from the F1 generation of a biparental cross, and then extend formulas to general forms holding also for DH lines generated from higher selfing generations than F1 and to the genotypic variance of recombinant inbred lines (RILs).
Two fully homozygous parental lines, and , are assumed with known QTL genotypes and each a vector of length counting the number of favorable QTL alleles at each of QTL. We define the -dimensional vector of known allele substitution effects as The breeding values of and are then and Further, DH progenies generated from the F1 generation of a cross are considered. The genotypes of the progenies can be defined as a matrix with progeny as rows and the QTL genotypes as columns. The mean breeding value of the progenies can be derived as the mean of their parental lines’ breeding values:
| (1) |
The progeny variance can be derived as:
| (2) |
To obtain Bernardo (2014) and Mohammadi et al. (2015) suggested to simulate progenies in silico using the parental genotypes, and a genetic map in order to obtain If the approach is to be implemented in breeding programs to test the variance of a high number of potential crosses, the simulation of progeny becomes a computationally intensive task, as, per cross, a minimum of several 100 progenies need to be simulated for accurate variance estimation. In the following, we show how can be derived from the parental genotypes and the recombination frequencies without the need to simulate For fully inbred lines, the following holds:
| (3) |
where the j-th diagonal entry corresponds to the variance at the j-th QTL locus with inbreeding coefficient for fully inbred DH lines, and the allele frequency in the parental lines, which, by expectation, also holds for the progenies (). For DH lines, the variance at locus j is then either if the parental alleles differ at this locus, or 0 if both parental lines have the same allele, and progenies will not show segregation at this locus. The off-diagonal elements of show the disequilibrium covariances between two loci: where denotes the haplotype frequency. The disequilibrium parameter between loci j and l can be derived from the disequilibrium parameter among both parental lines and the expected frequency of recombinants between both loci as: Depending on the parental haplotypes, is either 0 if both parental lines show the same alleles at one or both loci, or 0.25 or −0.25 depending on the linkage phase of the parents. If DH lines are generated from a later generation than F1, it needs to be considered that the expected frequency of recombinants increases with increasing number of meioses. Depending on the generation k when DH lines are generated ( for DH from F1), the expected frequency of recombinants increases to
| (4) |
The general formula for DH lines generated from generation k is then:
| (5) |
We give a full derivation for arriving at and an adjustment that also holds for RILs after different numbers of selfing generations in File S1. Table 1 summarizes how the genotypic variance-covariance between two loci j and l can be derived for different populations, based on the LD parameter in the parental gametes and the expected frequency of recombinants between both loci in the first generation.
Table 1. Overview of genotypic covariance between loci j and l for different populations derived from two parental lines based on LD parameter in parental lines and expected frequency of recombinants in generation 1 .
| Population | Genotypic Variance-Covariance |
|---|---|
| DH F1 ()a | |
| DH generation k | |
| RILs generation kb | |
| DH and RILs generation |
DH lines derived from F1 generation.
RILs after selfing generations ( equals F2 population).
Estimation of progeny variance based on whole-genome regression
For estimating the variance of progeny, allele substitution effects need to be known. QTL and their effects cannot be observed, but, with high marker density, strong LD between markers and QTL can be exploited allowing to replace QTL with marker genotypes with only limited loss of information. We then can define the marker genotypes and their -dimensional vector of allele substitution effects accordingly. Following Bernardo (2014) and Mohammadi et al. (2015), we estimate marker effects in a phenotyped and genotyped training population with the linear regression model:
| (6) |
where is the vector of phenotypes of a training population, is an intercept, is a matrix of marker genotypes of the individuals in the training population, and and are the vectors of marker effects and residuals. We estimate marker effects in a fully Bayesian way, assigning independent and identical Gaussian prior distributions to the marker effects Residuals are also assumed to follow independent and identical Gaussian distributions: Scaled inverse- prior distributions are assigned to the residual and marker effect variance and Samples from the posterior distribution are created using a MCMC algorithm as implemented in the R package BGLR (Pérez and de los Campos 2014). Hyperparameters for the scaled inverse- prior distributions were chosen according to default rules in BGLR corresponding to relatively uninformative priors, and an a priori assumption of 50% of the phenotypic variance explained by markers. We used 20,000 iterations, where the first 5000 samples were discarded as burn-in. From the postburn-in samples, we saved only every fifth sample for posterior inference, corresponding to samples.
To estimate progeny variance based on the whole-genome regression model given in (6), two alternative methods were used. The first method—denoted as the “variance of posterior means” (VPM)—corresponds to calculating the sample variance of the GEBVs as described by Bernardo (2014) and Mohammadi et al. (2015), with
| (7) |
where is the vector of posterior means of marker effects obtained from model (6) using a training population.
Following Sorensen et al. (2001), the progeny variance can also be estimated by constructing a posterior distribution calculating in each MCMC sample the progeny variance as:
| (8) |
where is the s-th thinned postburn-in sample from the MCMC algorithm. By using the posterior mean from all samples, we obtain the estimate according to method M2 of Lehermeier et al. (2017), which we denote here as the “posterior mean variance” (PMV):
| (9) |
Equation (9) can also be formulated as:
| (10) |
where is the posterior variance-covariance matrix of marker effects, which is estimated based on L MCMC samples: with the -dimensional matrix of L samples of the marker effects centered by their posterior means. The second part of Equation (10) corresponds to (7), which can be interpreted as the variation among GEBVs. The first part of Equation (10) can be interpreted as variation of an individual’s GEBV originating from variation of marker effect estimates. If there is no uncertainty in marker effect estimates, and the first part is zero. In this case, estimates from PMV and VPM are equal, and should approach the true genetic variance.
Usefulness and optimal haploid value of a cross
As the interest in breeding is typically both increasing the mean of a population and identifying superior lines, crosses can be selected based on their UC (Schnell and Utz 1975) or their superior progeny value as defined by Zhong and Jannink (2007), which is the mean of the upper fraction of the selected lines. For a normally distributed trait, the mean of the genotypic values of selected progenies from a cross is:
| (11) |
where μ is the genotypic mean of the cross, i the selection intensity (Falconer and Mackay 1996), and the genetic standard deviation. Under absence of dominance and epistasis as assumed here, the genotypic value of a line equals its breeding value. We predicted the usefulness of crosses by estimating μ from the mean parental GEBVs and using the two alternative variance estimation methods VPM and PMV as described in the previous section. The genotypic variance-covariance matrix entering VPM and PMV was derived from parental genotypes and genetic map information using the formula given in Table 1, line 1.
For comparison, we also investigated the concept of optimal haploid value selection suggested by Daetwyler et al. (2015) to identify superior crosses. We predicted the optimal haploid value that can be generated from a cross by:
| (12) |
where is the number of segments into which the genome is split to calculate the OHV, defines a matrix with number of columns equal to the number of marker loci in segment w, and rows containing the four haplotype scores (0 or 1) of the two parental lines; and defines the vector containing the marker effects of segment w estimated in model (6) using a training population. Note, for fully homozygous parental lines, can be reduced to two rows by considering one gamete each. In our study, we split each chromosome into three segments corresponding to the default value as chosen by Daetwyler et al. (2015).
Simulations
Our simulation study consists of two main parts. In the first part, we investigate the two variance estimation methods (VPM and PMV), and in the second, we assess the genetic gain from selection based on UC and OHV compared to selection based on mean GEBVs. For both, we simulated a training population based on genotypic data from 10 multi-parental populations of maize DH lines from the dent heterotic group, which were published by Bauer et al. (2013). The 841 DH lines were genotyped with the Illumina MaizeSNP50 BeadChip. After quality control and imputation of missing values as described by Lehermeier et al. (2014), 32,801 high-quality polymorphic SNPs were available and formed the genotypic data of our training population. A genetic consensus map of the 10 biparental families was constructed by Giraud et al. (2014), and is available at Maize GDB (http://maizegdb.org/cgi-bin/displayrefrecord.cgi?id=9024747). Based on this genetic map information, recombination frequencies between marker pairs were derived as with x being the map distance between two marker loci in Morgan (Haldane 1919).
Simulation part 1—investigation of variance prediction methods:
We randomly sampled loci from the marker data of the training population to be QTL, and randomly sampled QTL effects from independent and identical normal distributions with mean zero. True genotypic effects of the training population were then defined as with the QTL genotypes of the training population. To obtain phenotypic values, random error terms were sampled from a normal distribution with mean zero and variance equal to to obtain a heritability of Heritability values varied from 0.2 to 1, in steps of 0.2. To investigate different training population sizes, subsets varying in size from 100 to 600, in steps of 100, were sampled randomly from the full training population. An only-QTL scenario and an only-marker scenario were considered. In the only-QTL scenario, we exclusively included the 300 markers assigned a nonzero QTL effect in the whole-genome regression model. This can be considered as ideal situation. In the only-marker scenario, we excluded the 300 markers assigned QTL effects, and included a random subset of 3000 remaining markers in the whole-genome regression model. Model parameters were then estimated using the specified training population. We generated 200 crosses among randomly sampled lines from the training population. In each cross, we calculated the true genetic variance as in Equation (2) using the formula given in Table 1 for DH lines derived from the F1 generation. Further, variances were estimated using VPM and PMV as described above with their average bias calculated as true variance minus the estimated variance and standardized by the true variance. The predictive correlation of the variance estimates was calculated as correlation between true variance and estimated variance among the 200 crosses. The whole simulation approach was replicated 10 times and results of bias and predictive correlation were averaged over the 10 replications.
Simulation part 2—selection based on UC and OHV:
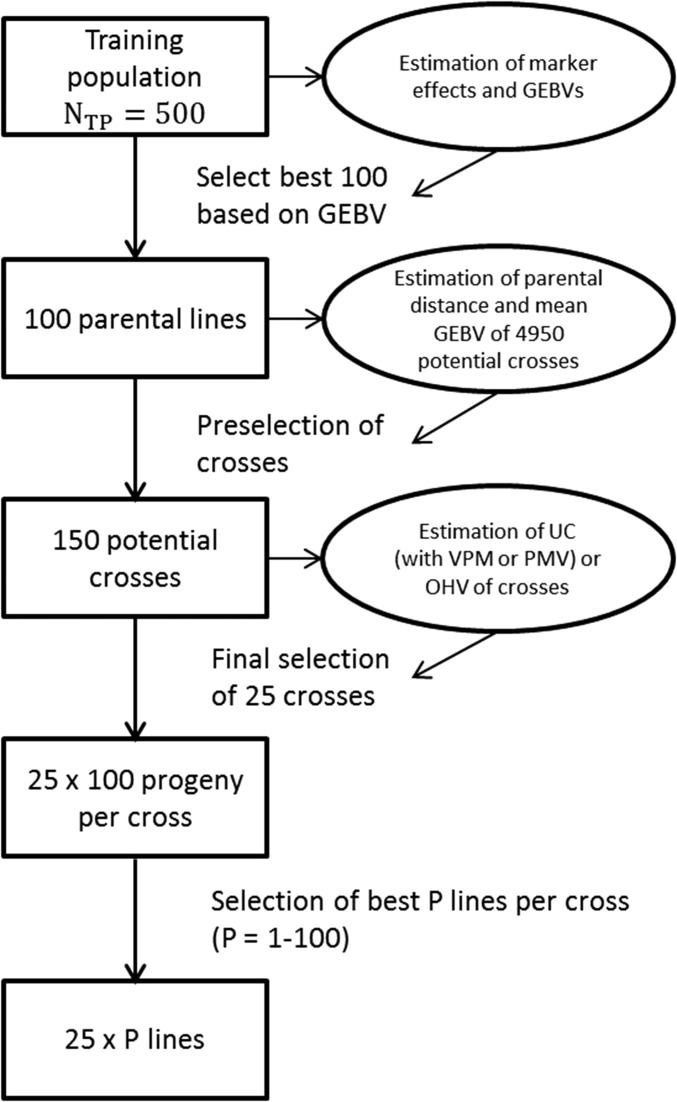
The simulation part 2 was based on a training population as simulated in part 1 with training population size of and two different heritability values ( and 0.6). Using only 3000 non-QTL markers, as in the only-marker scenario of simulation part 1, we fitted a whole-genome regression model as described in Equation (6). From the model, we obtained GEBVs for all lines in the training population. Using the GEBVs, we selected the 100 best lines (showing largest GEBVs) to form parental lines for new crosses. We calculated the mean of the GEBVs for all 4950 potential crosses of the 100 best lines. Further, we calculated the Rogers’ distance based on marker data between the parental lines of the 4950 crosses. To avoid crosses between closely related parents, and to ensure high means of the crosses, we selected those crosses where parental lines showed a minimum genetic distance of 0.2, and subsequently selected the 150 crosses with the highest mean parental GEBV. We used this approach of preselecting crosses on the one hand to reduce computing time for the further calculations, and, on the other hand, to best simulate a typical procedure in a breeding program. For comparison, we additionally show results where the 150 crosses were selected by mean parental GEBV alone without restriction on parental distance (minimum distance of 0.0). For the 150 crosses, we calculated the mean and the variance within each cross based on true QTL effects, and based on estimated marker effects. For each cross, we calculated the true and estimated UC with the two different variance estimation approaches. In addition, the OHV of each cross was estimated. The full simulation procedure was replicated 400 times. In each replication, we selected 25 crosses based either only on their mean GEBV, based on their true or estimated UC, or based on their estimated OHV. For each of the 25 crosses, we assumed a sample size of 100 progeny per cross, and, from those selected, the best lines per cross applying different selection intensities corresponding to a selection of 1–100 lines per cross in steps of 1. To assess the genetic gain of the different approaches, we calculated the mean true genotypic value of the selected lines from the different selection strategies. We report results for the difference in gain between selection based on estimated UC and mean GEBV, as well as between estimated OHV and mean GEBV in genetic standard deviations of the training population. An overview of the selection scheme applied in simulation part 2 is given in Figure 1.
Figure 1.
Scheme of simulation part 2.
Data availability
Simulations were based on genotypic maize data available under http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50558. The genetic map used can be downloaded from http://maizegdb.org/cgi-bin/displayrefrecord.cgi?id=9024747.
Results
Simulation part 1
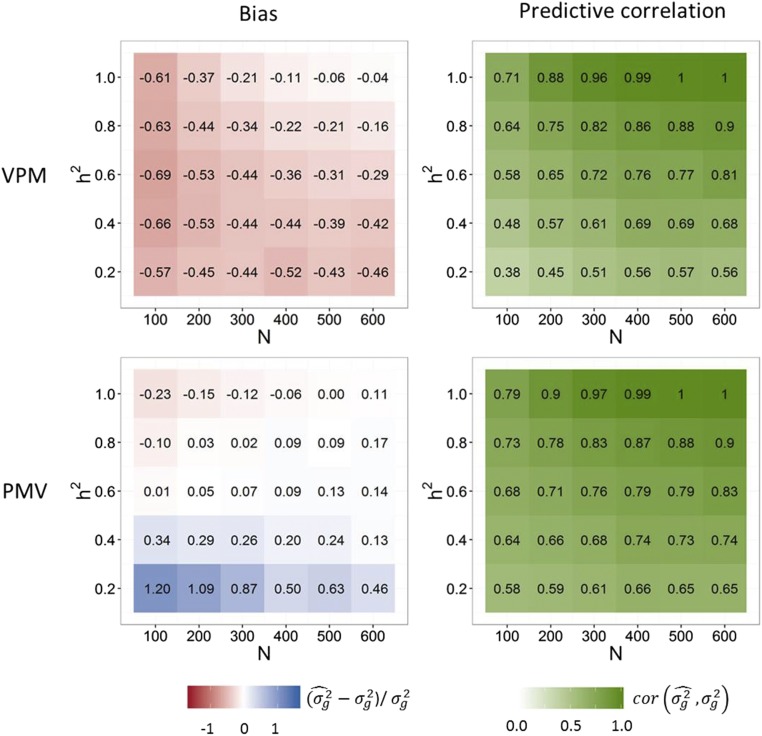
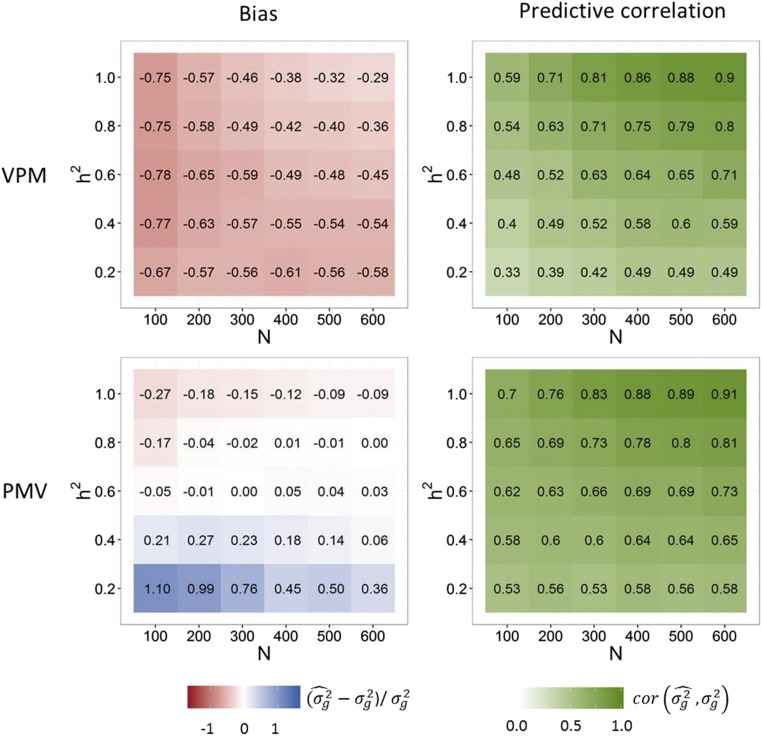
Figure 2 shows the bias and predictive correlation of VPM and PMV for prediction of the variance of new crosses in the only-QTL scenario. While VPM showed an underestimation of the true genetic variance with the bias approaching zero with increasing size of the training population (N) and heritability, PMV showed an overestimation with low N and heritability, and a slight underestimation with high heritability and low sample size. Except for a very low heritability of 0.2, PMV was considerably less biased than VPM. The correlation between true and estimated variance approached 1 with increasing N and heritability with estimation methods VPM and PMV, with PMV yielding a higher predictive correlation in all cases. Figure 3 shows the bias and predictive correlation for the scenario where only marker genotypes and no QTL were included in the whole-genome regression model. Compared to the only-QTL scenario, predictive correlations were reduced with both VPM and PMV, but only with VPM the bias increased. With heritability of 1 and training population size of 600, the predictive correlation was 0.91 when PMV was used and 0.90 when VPM was used. In all cases, PMV showed higher predictive correlations than VPM, and the superiority of PMV over VPM increased with decreasing heritability.
Figure 2.
Bias and predictive correlations of the genetic variance estimates for only-QTL scenario. Estimates for 200 randomly generated crosses were obtained with methods VPM and PMV for different and training population sizes N. Simulation scenario with 300 QTL genotypes coded in the marker matrix.
Figure 3.
Bias and predictive correlations of the genetic variance estimates for only-marker scenario. Estimates for 200 randomly generated crosses were obtained with methods VPM and PMV for different and training population sizes N. Simulation scenario with 3000 non-QTL marker genotypes coded in the marker matrix.
Simulation part 2
The efficiency of selection based on the different criteria (UC, OHV, GEBV) depends on the variation of the genetic means and genetic standard deviations between crosses. Table 2 shows the sample average and variance of the genetic mean, genetic standard deviation, and UC of the 150 crosses, preselected by parental GEBVs based on simulated heritabilities of 0.2 and 0.6, and with a minimum distance between parents of 0.2 and 0.0. The average genetic mean, and, consequently, the average UC increased with higher heritability as the preselection by GEBVs selected high performing lines more reliably. The average within-cross genetic standard deviation was only marginally affected by the heritability. Similarly, the sample variance among-cross genetic means and the UC were smaller with higher heritability, while the sample variance of genetic standard deviations remained constant. Without restriction on parental distance, the average genetic mean increased, but the average genetic standard deviation and UC decreased. The sample variance of the genetic standard deviations and UC increased compared to the fraction of crosses preselected based on a minimal parental distance of 0.2.
Table 2. Average genetic mean (), standard deviation (), and usefulness criterion () as well as the sample variance of genetic means (), standard deviations (), and usefulness criterions () among the 150 potential crosses of simulation part 2.
Minimum Rogers’ distance between parents.
Simulated heritability.
Means standard deviations across the 400 simulation runs.
Figure 4 shows the additional genetic gain that can be obtained when selection of crosses is based on the predicted UC or OHV compared to selection based on the mean GEBV of the parents as a function of the selection intensity within crosses for heritabilities of 0.2 (Figure 4A) and 0.6 (Figure 4B) when crosses were restricted to a minimum parental distance of 0.2. The additional gain of selection based on the UC increased with higher selection intensities. For all heritabilities and selection intensities, predicting UC based on PMV yielded the highest genetic gain. When selecting only 1% of lines per cross, and with heritability 0.6, the genetic gain increased up to With heritability of 0.6, estimating the variance with method PMV yielded up to more gain compared to using the VPM as estimate. With low heritability of 0.2, the superiority of PMV over VPM increased up to Selecting crosses based on their OHV led to an increase in genetic gain compared to selection based on the mean GEBV for high selection intensity. When >10% progenies per cross were selected, selection based on OHV was inferior to selection based on mean GEBV. In general, selection based on estimated UC was greatly superior to selection based on estimated OHV. With training population heritability of 0.6, the additional genetic gain of selecting crosses based on the UC compared to the mean GEBV was higher than for a training population heritability of 0.2. The additional genetic gain of selection based on UC compared to selection based on mean GEBVs increased considerably when crosses were not restricted to a minimum parental distance of 0.2, and reached a maximum increase of for heritability 0.2 (Figure 4C) and 0.24 for heritability 0.6 (Figure 4D).
Figure 4.
Additional gain of selecting crosses based on estimated UC and OHV. Gain is given as the difference in mean genotypic values of the selected lines from selection based on UC or OHV compared to selecting based on mean GEBV alone, standardized by the true genotypic standard deviation in the training population. Different variance estimates were used to estimate the UC (VPM and PMV). Gain is given for different fractions of selected lines per cross. Results are shown for preselected crosses based on minimum Rogers’ distance between parents of 0.2 and a heritability of 0.2 (A) and 0.6 (B), as well as for crosses not preselected by parental distance, and a heritability of 0.2 (C) and 0.6 (D).
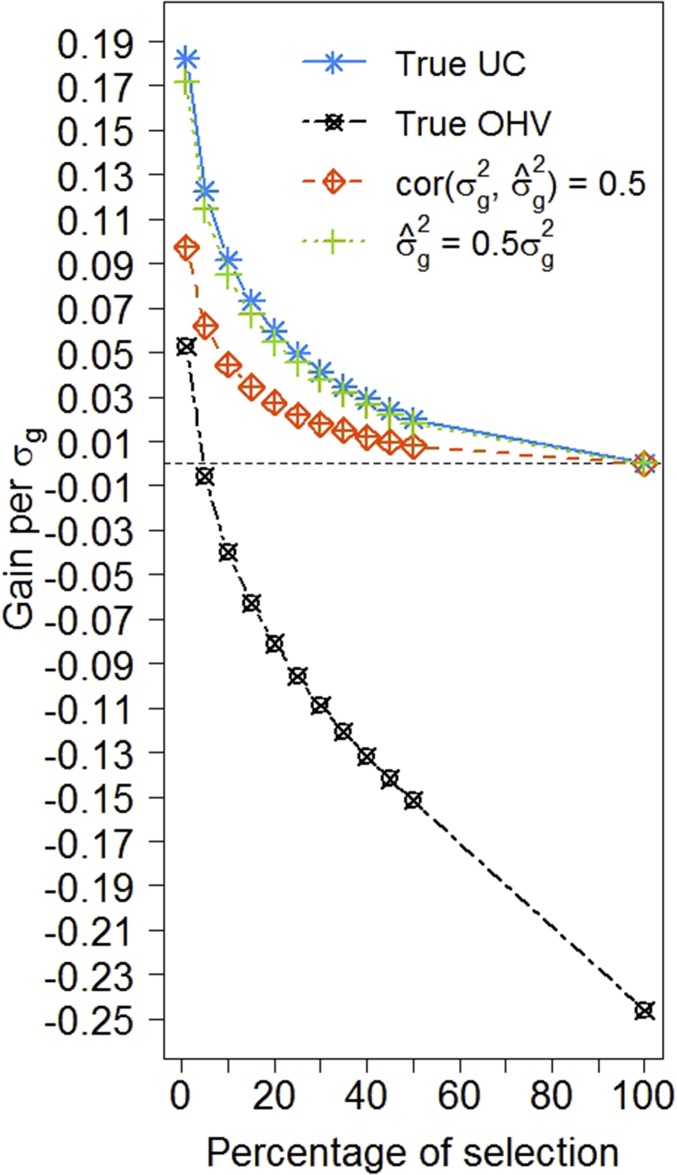
Figure 5 shows the genetic gain when true UC and true OHV were used for selection of crosses compared to selection based on true mean genotypic values of parental lines considering selection among 150 potential crosses preselected by minimum genetic distance of 0.2 and heritability of 0.6 (comparable to Figure 4B). Here, QTL effects were assumed to be known to calculate the UC and OHV. With true effects the genetic gain from selection with UC increased up to Similarly as under the use of estimated marker effects, selection based on true OHV resulted in reduced gain compared to selection based on true UC and was only superior to selection based on mean genotypic values when keeping <10% of progenies per cross. In addition to selection based on true UC, Figure 5 shows selection based on artificially biased UC. For this the true genetic variance entering the UC was either divided by two to simulate a variance estimate that is biased, but has a predictive correlation of 1; or a random error was added to the true genetic variance to simulate a variance estimate that is unbiased, but shows predictive correlation of 0.5. A biased genetic variance in the UC led to a small decrease in genetic gain of while a genetic variance with a predictive correlation of 0.5 decreased the additional genetic gain by around one half.
Figure 5.
Additional gain of selecting crosses based on true UC, true OHV, or biased UC. For the biased UC, either an underestimated variance () or a variance estimate with predictive correlation of was considered. Gain is given in comparison to the genetic gain of selection based on mean parental true genotypic values, standardized by the genetic standard deviation of the training population.
Discussion
Here, we showed that increased genetic gain can be obtained when selection decisions are based on the estimated progeny variance in addition to the estimated mean of a cross. For a typical scenario of a selected proportion of 10% per cross and a heritability of 0.6, selection gain increased by when the estimated UC with PMV was used for selection decisions. We assumed 100 derived progenies per selected cross. Selection intensities within crosses will be smaller, with considerably fewer derived progenies per cross, and, consequently, the additional genetic gain that can be obtained from selection based on UC will decrease. However, with decreasing genotyping costs and the implementation of genomic selection, selection intensities are likely to increase, which will make selection of crosses based on UC more advantageous. Selection gain also depends on the level of variation in the genetic means and standard deviations of crosses. The additional genetic gain of selection based on UC compared to selection based on mean GEBVs was higher for a heritability of 0.6 compared to 0.2, as due to a more precise preselection of high performing parental lines based on GEBVs the variation between the genetic means of the 150 potential crosses was lower, leading to a more favorable ratio of sample variance of genetic standard deviations () over means (). Without a restriction on parental distance, the 150 potential crosses showed a larger variation in genetic standard deviations, and, consequently, the additional genetic gain of selection based on the UC considerably increased.
Considering, as parental lines, RILs derived from one cross, Zhong and Jannink (2007) concluded that the impact of the progeny variance of crosses on the superior progeny value decreases rapidly with increasing number of QTL. We simulated 300 QTL to warrant normally distributed genotypic values, and did not see a decrease in gain when the number of QTL was increased (results not shown). The reduced selection gain when using VPM compared to PMV as variance estimate in the UC can mainly be explained from a lower predictive correlation, as a correct ranking of the crosses based on their variances is most important (Figure 5). However, also an underestimated variance with perfect predictive correlation slightly decreases the genetic gain, as less weight is given to the variance part in the UC and selection becomes similar to selecting based on the mean of the crosses alone. An unbiased estimate of the genetic variance is clearly advantageous, as it can be directly used for selecting among different crosses based on their UC. Thus, the methods suggested here are clearly superior to variance prediction methods based on phenotypic or genetic distance between parents that can, at best, rank the variance of different crosses (Lian et al. 2015).
Other approaches independent of the UC have been proposed to guide mating decisions. Daetwyler et al. (2015) suggested selecting crosses or individuals based on their optimal haploid value—a concept that has been proposed in animal breeding for investigating selection limits (Cole and VanRaden 2011). Selecting crosses based on their OHV corresponds to summing over the best haploid segments present in both parental lines. In our simulation, selection based on the estimated OHV gave an increase in genetic gain compared to selection based on mean GEBVs for high selection intensities. This is expected as the OHV potentially identifies the best line that can be derived from a cross, assuming an infinite number of progeny per cross and an infinite selection intensity. With decreasing selection intensity, the genetic mean of the selected fraction moves away from the optimal value and approaches the mean GEBVs of the parents. Consequently, selection based on mean GEBVs was superior to selection based on OHV when >10% of lines per cross were selected. The OHV can be adapted by varying the number of segments in which absence of recombination is assumed. Thus, one might argue that a small number of segments per chromosome should be chosen if selection intensity is low and the focus is on short-term genetic gain (Goiffon et al. 2017). We observed very similar genetic gain with one segment per chromosome compared to using three segments for estimating the OHV (results not shown). As neither selection intensity nor recombination frequencies directly enter into the OHV, selection based on OHV does not take into account the probability of realizing a specific OHV. To alleviate this shortcoming, Han et al. (2017) suggested the predicted cross value for selection, which is defined as the probability that a gamete produced from a cross will only consist of desirable alleles. However, this approach does not differentiate between large and small allelic effects, and, similar to other approaches, would rely on knowledge or precise estimation of desirable alleles for use in practice. In contrast, selection based on UC and PMV takes into account all available information, including LD between loci as well as uncertainty of effect estimates, and provides an increase in genetic gain compared to selection based on mean GEBV for the entire range of selection intensities.
We showed that the variance of genotypes from a new cross can be derived analytically from parental genotypes and genetic map information, and, thus, no in silico simulations are needed for predicting the genetic variance of progenies under the formulated assumptions. The theoretical derivation corresponds to a simulation of an infinite number of progenies per cross, and is most precise (see supporting File S1). The simulation of progenies can become computationally intense if the variance of several thousands of crosses needs to be predicted, so we consider our approach highly advantageous for application in a breeding program. One limitation of our approach is that, for the derivation of the progeny variance, assumptions regarding the expected recombination frequency need to be made. Our results are based on the assumption of known recombination frequencies by assuming the given genetic map as true, and the absence of interference (Haldane 1919). In practice, precision of estimated recombination frequencies might vary between species, depending on available mapping information and the presence of interference. Furthermore, recombination rates might vary among crosses (Bauer et al. 2013), which might reduce the accuracy of variance prediction, and, consequently, the superiority of the UC. All these limitations equally apply to both the analytical derivation of progeny variance as well as in silico progeny simulations. The expected genetic variance depends on the population type derived from a cross. Our results are focused on DH lines derived from the F1 generation of a cross, but we give comprehensive formulas also for different generations of RILs and DH lines considering the respective expected frequency of recombinants and different levels of inbreeding of the derived population. The specific formulas quantify if there is a gain in genetic variance of a cross when DH lines are derived from a later generation than F1 (Sleper and Bernardo 2016), without the need of additional time-consuming computations.
We investigated two different methods for predicting the genetic variance of newly generated crosses. Method VPM, the sample variance of the GEBVs, has been used for this and similar purposes by other authors (Bernardo 2014; Segelke et al. 2014; Mohammadi et al. 2015; Tiede et al. 2015; Wittenburg et al. 2016). As VPM is known to underestimate the true genetic variance as it is based on shrunken marker effect estimates (Cole and VanRaden 2011; Lian et al. 2015), we investigated its performance for obtaining accurate and precise progeny variance estimates under different training population properties. In our study, VPM largely underestimated the true genetic variance with incomplete LD between markers and QTL (only-marker scenario). Only under ideal scenarios where markers were in perfect LD with the QTL, the training population size largely exceeded the number of markers in the model, and when heritability was high, did VPM provide a nearly unbiased estimate. The underestimation of VPM originates from the fact that uncertainty of the marker effect estimates is not taken into account. The posterior mean of the genetic variance calculated from MCMC samples (PMV) takes this uncertainty into account, and, consequently, provided an improved variance estimator. As shown in Equation (10), the estimated variance obtained from PMV can be split into the estimated variance obtained from VPM and a part that originates from the marker effect variances. Accordingly, it yielded consistently larger variance estimates than VPM, and showed only a slight deviation from the true genetic variance for heritability values >0.2 in the only-QTL and only-marker scenario. An overestimation was observed for very low heritability, which can be explained by model overfitting and large Monte Carlo errors due to the low signal-to-noise ratio in the training population data. As expected from theoretical considerations, variance estimates obtained from VPM and PMV converged with increasing heritability and training population size, and both yielded a predictive correlation of 1 in the more ideal scenarios (only QTL in the model, ). Zhong and Jannink (2007) made a similar observation, and found that a fully Bayesian treatment of the superior progeny value was better than using an approach based on the posterior means of marker effects (comparable to VPM). It has been shown that, for a given number of markers, increasing training population size and heritability increases the accuracy of marker effect estimates (Wimmer et al. 2013). Accordingly, as variance estimates obtained with VPM and PMV are based on marker effect estimates, they became more accurate with increasing heritability and training population size. In agreement with the results of the predictive correlations, using PMV always led to more genetic gain than using VPM, and this superiority increased with decreasing heritability.
In our study, we used an MCMC algorithm for obtaining both variance estimates (VPM and PMV). For VPM, MCMC would not have been necessary, and variance component estimates could have been obtained with restricted maximum likelihood (REML). For PMV, an estimate for the variance-covariance matrix of the marker effects is needed. A closed form estimate for this variance-covariance matrix can be derived only under an RR-BLUP model with known variance components and and thus, given shrinkage parameter which is then However, when variance components are unknown, no closed form exists and MCMC can be used to obtain an estimate. Alternatively, estimated variance components (e.g., from REML) might be plugged in to obtain but then the uncertainty of the variance component estimates is not taken into account, which might underestimate (Sorensen et al. 2001). This approach would provide an alternative if one wants to avoid using MCMC to save computing time. As expected, in our simulations this approach was superior considering predictive correlation and bias compared to VPM, but inferior compared to PMV (results not shown).
This study is based on simulations because a large number of crosses and a large number of progenies per cross are needed for inference. Such large numbers of unselected progenies are rarely available in experimental breeding programs. Further, true genetic variances are unknown in experimental data, and proper estimation requires replicated field trials to take genotype-by-environment interaction effects, which are typically large in maize, into account (Acosta-Pech et al. 2017). Our results might represent an upper limit for the application of selecting crosses based on UC in practical breeding. Nonadditive or multi-allelic QTL effects, which might affect the accuracy of variance prediction, were not considered. We conjecture that the prediction of progeny variance is affected by nonadditive and population-specific effects similar to the prediction of breeding values where accuracies of methods are very similar irrespective if nonadditive effects are accounted for or not. In cases where nonadditive effects are considered important, the whole-genome regression model to predict the genetic variance could be readily extended to include epistatic (Jiang and Reif 2015) or population-specific effects (de los Campos et al. 2015; Lehermeier et al. 2015). In addition, Bayesian whole-genome regression models, including marker-specific shrinkage priors like Bayesian Lasso or BayesB, could be used if large QTL effects are assumed to be segregating for the traits under study.
Predictions of the genetic variance are not only of interest for selection of crosses in plant breeding but have also been studied in animal breeding for mating decisions (Segelke et al. 2014; Bonk et al. 2016). We concentrated here on improving a single trait in a directional selection approach. In general, knowledge of the genetic mean and variance of normally distributed breeding values allows estimating the probability that an offspring exceeds a specific threshold, or that it is within a specific range. Instead of increasing the genetic variance for fast selection progress, in specific situations, the goal might lie in a large mean combined with a low genetic variance, for example, in animal breeding to obtain a homogeneous population (Cole and VanRaden 2011; Segelke et al. 2014). For such inferences and subsequent mating optimizations, an unbiased and precise variance prediction as can be provided by PMV is important. The prediction of genetic variance can also be extended to the prediction of genetic covariances and correlations among multiple traits. To estimate genetic correlations with PMV, genetic sample correlations among breeding values of two traits can be calculated in each postburn-in MCMC sample, and, from those, posterior means can be formed. Further, the single-trait whole-genome regression model could be extended to a multi-trait model to profit from genetic correlations for the estimation of marker effects (Jia and Jannink 2012). We conjecture that a multivariate extension of PMV also provides a superior prediction of genetic correlations between traits compared to VPM, which warrants further investigations. Knowledge of the genetic variance of single traits and genetic correlations among multiple traits of future crosses allows breeders to optimize their allocation of resources. Further, by applying formulas for different generations of inbreeding, how selection gain changes with additional selfing steps can be deduced. Here, our work provides a good starting point for the optimization of a genome-based prediction guided breeding program.
Supplementary Material
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.117.300403/-/DC1.
Footnotes
Communicating editor: F. Eeuwijk
Literature Cited
- Acosta-Pech R., Crossa J., de los Campos G., Teyssèdre S., Claustres B., et al. , 2017. Genomic models with genotype x environment interaction for predicting hybrid performance: an application in maize hybrids. Theor. Appl. Genet. 130: 1431. [DOI] [PubMed] [Google Scholar]
- Bauer E., Falque M., Walter H., Bauland C., Camisan C., et al. , 2013. Intraspecific variation of recombination rate in maize. Genome Biol. 14: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo R., 2014. Genomewide selection of parental inbreds: classes of loci and virtual biparental populations. Crop Sci. 54: 2586–2595. [Google Scholar]
- Bohn M., Utz H. F., Melchinger A. E., 1999. Genetic similarities among winter wheat cultivars determined on the basis of RFLPs, AFLPs, and SSRs and their use for predicting progeny variance. Crop Sci. 39: 228–237. [Google Scholar]
- Bonk S., Reichelt M., Teuscher F., Segelke D., Reinsch N., 2016. Mendelian sampling covariability of marker effects and genetic values. Genet. Sel. Evol. 48: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. B., VanRaden P. M., 2011. Use of haplotypes to estimate Mendelian sampling effects and selection limits. J. Anim. Breed. Genet. 128: 446–455. [DOI] [PubMed] [Google Scholar]
- Daetwyler H. D., Hayden M. J., Spangenberg G. C., Hayes B. J., 2015. Selection on optimal haploid value increases genetic gain and preserves more genetic diversity relative to genomic selection. Genetics 200: 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Veturi Y., Vazquez A. I., Lehermeier C., Pérez-Rodríguez P., 2015. Incorporating genetic heterogeneity in whole-genome regressions using interactions. J. Agric. Biol. Environ. Stat. 20: 467–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J. B., 2011. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4: 250–255. [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Longman, Essex, England. [Google Scholar]
- Giraud H., Lehermeier C., Bauer E., Falque M., Segura V., et al. , 2014. Linkage disequilibrium with linkage analysis of multiline crosses reveals different multiallelic QTL for hybrid performance in the flint and dent heterotic groups of maize. Genetics 198: 1717–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goiffon M., Kusmec A., Wang L., Hu G., Schnable P., 2017. Improving response in genomic selection with a population-based Selection strategy: optimal population value selection. Genetics 206: 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J., 1919. The combination of linkage values and the calculation of distances between the loci of linked factors. J. Genet. 8: 299–309. [Google Scholar]
- Han Y., Cameron J. N., Wang L., Beavis W. D., 2017. The predicted cross value for genetic introgression of multiple alleles. Genetics 205: 1409–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslot N., Jannink J.-L., Sorrells M. E., 2015. Perspectives for genomic selection applications and research in plants. Crop Sci. 55: 1–12. [Google Scholar]
- Hung H.-Y., Browne C., Guill K., Coles N., Eller M., et al. , 2012. The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity 108: 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Jannink J.-L., 2012. Multiple-trait genomic selection methods increase genetic value prediction accuracy. Genetics 192: 1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Reif J. C., 2015. Modeling epistasis in genomic selection. Genetics 201: 759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehermeier C., Krӓmer N., Bauer E., Bauland C., Camisan C., et al. , 2014. Usefulness of multiparental populations of maize (Zea mays L.) for genome-based prediction. Genetics 198: 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehermeier C., Schön C.-C., de los Campos G., 2015. Assessment of genetic heterogeneity in structured plant populations using multivariate whole-genome regression models. Genetics 201: 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehermeier C., de los Campos G., Wimmer V., Schön C.-C., 2017. Genomic variance estimates: with or without disequilibrium covariances? J. Anim. Breed. Genet. 134: 232–241. [DOI] [PubMed] [Google Scholar]
- Lian L., Jacobson A., Zhong S., Bernardo R., 2015. Prediction of genetic variance in biparental maize populations: genomewide marker effects vs. mean genetic variance in prior populations. Crop Sci. 55: 1181–1188. [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E., 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157: 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M., Tiede T., Smith K. P., 2015. PopVar: a genome-wide procedure for predicting genetic variance and correlated response in biparental breeding populations. Crop Sci. 55: 2068–2077. [Google Scholar]
- Pérez P., de los Campos G., 2014. Genome-wide regression & prediction with the BGLR statistical package. Genetics 198: 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell F. W., Utz H. F., 1975. F1-Leistung und Elternwahl in der Züchtung von Selbstbefruchtern, pp. 234–258 in Bericht über die Arbeitstagung der Vereinigung Österreichischer Pflanzenzüchter. Gumpenstein, Österreich. [Google Scholar]
- Segelke D., Reinhardt F., Liu Z., Thaller G., 2014. Prediction of expected genetic variation within groups of offspring for innovative mating schemes. Genet. Sel. Evol. 46: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleper J. A., Bernardo R., 2016. Recombination and genetic variance among maize doubled haploids induced from F1 and F2 plants. Theor. Appl. Genet. 129: 2429–2436. [DOI] [PubMed] [Google Scholar]
- Sorensen D., Fernando R., Gianola D., 2001. Inferring the trajectory of genetic variance in the course of artificial selection. Genet. Res. 77: 83–94. [DOI] [PubMed] [Google Scholar]
- Tiede T., Kumar L., Mohammadi M., Smith K. P., 2015. Predicting genetic variance in bi-parental breeding populations is more accurate when explicitly modeling the segregation of informative genomewide markers. Mol. Breed. 35: 199. [Google Scholar]
- Utz H. F., Bohn M., Melchinger A. E., 2001. Predicting progeny means and variances of winter wheat crosses from phenotypic values of their parents. Crop Sci. 41: 1470–1478. [Google Scholar]
- Wimmer V., Lehermeier C., Albrecht T., Auinger H.-J., Wang Y., et al. , 2013. Genome-wide prediction of traits with different genetic architecture through efficient variable selection. Genetics 195: 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenburg D., Teuscher F., Klosa J., Reinsch N., 2016. Covariance between genotypic effects and its use for genomic inference in half-sib families. G3 6: 2761–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Jannink J.-L., 2007. Using quantitative trait loci results to discriminate among crosses on the basis of their progeny mean and variance. Genetics 177: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Simulations were based on genotypic maize data available under http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50558. The genetic map used can be downloaded from http://maizegdb.org/cgi-bin/displayrefrecord.cgi?id=9024747.