Abstract
As the primary microtubule-organizing center, centrosomes play a key role in establishing mitotic bipolar spindles that secure correct transmission of genomic content. For the fidelity of cell division, centrosome number must be strictly controlled by duplicating only once per cell cycle. Proper levels of centrosome proteins are shown to be critical for normal centrosome number and function. Overexpressing core centrosome factors leads to extra centrosomes, while depleting these factors results in centrosome duplication failure. In this regard, protein turnover by the ubiquitin-proteasome system provides a vital mechanism for the regulation of centrosome protein levels. Here, we report that FZR-1, the Caenorhabditis elegans homolog of Cdh1/Hct1/Fzr, a coactivator of the anaphase promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase, functions as a negative regulator of centrosome duplication in the C. elegans embryo. During mitotic cell division in the early embryo, FZR-1 is associated with centrosomes and enriched at nuclei. Loss of fzr-1 function restores centrosome duplication and embryonic viability to the hypomorphic zyg-1(it25) mutant, in part, through elevated levels of SAS-5 at centrosomes. Our data suggest that the APC/CFZR-1 regulates SAS-5 levels by directly recognizing the conserved KEN-box motif, contributing to proper centrosome duplication. Together, our work shows that FZR-1 plays a conserved role in regulating centrosome duplication in C. elegans.
Keywords: FZR-1, C. elegans, Centrosome, E3 ubiquitin ligase, SAS-5
The centrosome is a small, nonmembranous organelle that serves as the primary microtubule-organizing center in animal cells. Each centrosome consists of a pair of barrel-shaped centrioles that are surrounded by a network of proteins called pericentriolar material (PCM). During mitosis, two centrosomes organize bipolar spindles that segregate genomic content equally into two daughter cells. Thus, tight control of centrosome number is vital for the maintenance of genomic integrity during cell division, by restricting centrosome duplication to once, and only once, per cell cycle. Erroneous centrosome duplication results in aberrant centrosome number that leads to chromosome missegregation and abnormal cell proliferation, and is associated with human disorders including cancers and microcephaly (Nigg and Stearns 2011; Gönczy 2015).
In the nematode Caenorhabditis elegans, extensive studies identified a set of core centrosome factors that are absolutely essential for centrosome duplication: the protein kinase ZYG-1 and the coiled-coil proteins SPD2, SAS-4, SAS-5, and SAS-6 (O’Connell et al. 2001; Kirkham et al. 2003; Leidel and Gönczy 2003; Dammermann et al. 2004; Delattre et al. 2004; Kemp et al. 2004; Pelletier et al. 2004; Leidel et al. 2005). SPD-2 and ZYG-1 localize early to the site of centriole formation and are required for the recruitment of the SAS-5/SAS-6 complex that sequentially recruits SAS-4 to the centriole (Delattre et al. 2006; Pelletier et al. 2006). These key factors are also present in other animal systems, suggesting an evolutionary conservation in centrosome duplication. For instance, the human genome contains homologs of the five centrosome factors found in C. elegans, Cep192/SPD-2 (Zhu et al. 2008), Plk4/ZYG-1 (Habedanck et al. 2005), STIL/SAS-5 (Arquint et al. 2012), HsSAS-6/SAS-6 (Leidel et al. 2005) and CPAP/SAS-4 (Kleylein-Sohn et al. 2007; Tang et al. 2009), and all these factors are shown to play a critical role in centrosome biogenesis (Fu et al. 2015; Gönczy 2015). However, the recently identified core centriole factor, SAS-7, that acts upstream of SPD-2 during C. elegans centriole duplication has not been found outside of nematodes (Sugioka et al. 2017).
Maintaining the proper levels of centrosome proteins is critical for normal centrosome number and function (Kleylein-Sohn et al. 2007; Strnad et al. 2007; Rogers et al. 2009; Tang et al. 2009, 2011; Holland et al. 2010; Brownlee et al. 2011; Puklowski et al. 2011; Song et al. 2011; Meghini et al. 2016; Levine et al. 2017). In light of this, protein turnover by proteolysis provides a key mechanism for regulating the abundance of centrosome factors. A mechanism regulating protein levels is their degradation by the 26S proteasome that catalyzes the proteolysis of polyubiquitinated substrates (Livneh et al. 2016). The anaphase promoting complex/cyclosome (APC/C) is a multi-subunit E3 ubiquitin ligase that targets substrates for degradation (Acquaviva and Pines 2006; Peters 2006; Chang and Barford 2014). The substrate specificity of the APC/C is directed through the sequential, cell-cycle-dependent activity of two coactivators, Cdc20/Fzy/FZY-1 (Hartwell and Smith 1985; Dawson et al. 1995; Kitagawa et al. 2002) and Cdh1/Fzr/Hct1/FZR-1 (Schwab et al. 1997; Sigrist and Lehner 1997; Visintin et al. 1997; Fay et al. 2002). During early mitosis, Cdc20 acts as coactivator of the APC/C, and Cdh1 functions as coactivator to modulate the APC/C-dependent events at late mitosis and in G1 (Irniger and Nasmyth 1997; Visintin et al. 1997; Fang et al. 1998; Prinz et al. 1998; Shirayama et al. 1998). Upregulated targets in Cdh1-deficient cells are shown to be associated with the genomic instability signature of human cancers, and show a high correlation with poor prognosis (Carter et al. 2006; García-Higuera et al. 2008). Furthermore, a mutation in SIL/STIL (a human homolog of SAS-5) linked to primary microcephaly (MCPH; Kumar et al. 2009) results in deletion of the Cdh1-dependent destruction motif (KEN-box), leading to deregulated accumulation of STIL protein and centrosome amplification (Arquint and Nigg 2014). In Drosophila, the APC/CFzr/Cdh1 directly interacts with Spd2 through KEN-box recognition and targets Spd2 for degradation (Meghini et al. 2016). Therefore, the APC/CCdh1/Fzr/Hct1 plays a critical role in regulating the levels of key centrosome duplication factors in mammalian cells and flies.
In C. elegans, FZR-1 has been shown to be required for fertility, cell cycle progression and cell proliferation during embryonic and postembryonic development via synthetic interaction with lin-35/Rb (Fay et al. 2002; The et al. 2015). However, the role of FZR-1 in centrosome assembly has not been described. In this study, we molecularly identified fzr-1 as a genetic suppressor of zyg-1. Our results suggest that APC/CFZR-1 negatively regulates centrosome duplication, in part, through proteasomal degradation of SAS-5 in a KEN-box dependent fashion. Therefore, FZR-1, the C. elegans homolog of Cdh1/Hct1/Fzr, plays a conserved role in centrosome duplication.
Materials and Methods
C. elegans strains and genetics
A full list of C. elegans strains used in this study is listed in Supplemental Material, Table S1 in File S1. All strains were derived from the wild-type Bristol N2 strain using standard genetic methods (Brenner 1974; Church et al. 1995).
Strains were maintained on MYOB plates seeded with Escherichia coli OP50 and grown at 19° unless otherwise indicated. The fzr-1::gfp::3xflag construct containing 21.6 Kbp of the fzr-1 5′UTR and 6 Kbp of the fzr-1 3′UTR was acquired from TransgenOme (construct number: 7127141463160758 F11, Sarov et al. 2012), which was used to generate the transgenic line, MTU10, expressing C-terminal GFP-tagged FZR-1. For the generation of N-terminal GFP-tagged FZR-1 (OC190), we used Gateway cloning (Invitrogen, Carlsbad, CA) to generate the construct. Coding sequence of fzr-1 was PCR amplified from the cDNA clone yk1338f2, and cloned into pDONR221 (Invitrogen) and then the resulting entry clone was recombined into pID3.01 (pMS9.3), which is driven by the pie-1 promotor. The transgenes were introduced into worms by standard particle bombardment (Praitis et al. 2001). For embryonic viability and brood size assays, individual L4 animals were transferred to clean plates, and allowed to self-fertilize for 24 hr at the temperatures indicated. For brood size assays, this was repeated until animals no longer produced embryos. Progeny were allowed at least 24 hr to complete embryogenesis before counting the number of progeny. The fzr-1(RNAi) experiments were performed by RNAi soaking (Song et al. 2008). To produce dsRNA for RNAi soaking, we amplified a DNA template from the cDNA clone yk1338f2 using the primers 5′-ATGGATGAGCAACCGCC-3′ and 5′-GCACTGTACGTAAAAAGTGATC-3′ that contained a T7 promoter sequence at their 5′ ends. In vitro transcription was performed using the T7-MEGAscript kit (Thermo-Fisher, Hanover park, IL). L4 animals were soaked overnight in M9 buffer containing either 0.1–0.4 mg dsRNA/ml or no dsRNA (control).
Mapping and molecular identification of szy-14
Both szy-14(bs31) and szy-14(bs38) suppressors were previously mapped between dpy-10 and unc-4 on chromosome II as described in Kemp et al. (2007). As szy-14(bs31) appears be a stronger suppressor (Figure 1B and Table 1, Kemp et al. 2007), we chose to use szy-14(bs31) for further mapping. Because szy-14 mutants show no embryonic lethality, we decided to use the suppression of the zyg-1(it25) embryonic lethality by the szy-14(bs31) mutation for phenotyping. For single-nucleotide polymorphism (SNP) mapping, we mated zyg-1(it25) dpy-10(e128) szy-14(bs31) unc-4(e120) hermaphrodites with Hawaiian CB4856 males as described in Song et al. (2008), and isolated a total of 104 independent Dpy-nonUnc recombinants from the F2 generation. After establishing homozygous recombinant lines, we screened for the fzr-1(bs31) presence using the suppression of the zyg-1(it25) lethality at 24° supplemented by reduced brood size phenotype (Fay et al. 2002). For each phenotyping, we used the following control strains in parallel to accurately score the suppression: zyg-1(it25), zyg-1(it25) dpy-10(e128), zyg-1(it25) dpy-10(e128) szy-14(bs31) unc-4(e120), zyg-1(it25) szy-14(bs31), and zyg-1(it25) szy-14(bs31) unc-4(e120). After careful phenotype examination, we determined 27 (out of 104) of the Dpy-nonUnc recombinants contain the fzr-1(bs31) mutation, which restores the zyg-1(it25) embryonic lethality, and that 77 (out of 104) of the Dpy-nonUnc recombinants do not contain the fzr-1(bs31) mutation, causing no suppression of the zyg-1 lethality. Through fine mapping, we narrowed down the szy-14 locus to a region of 57 kb between 9621265 and 9678204 on chromosome II. Then, we continued to molecularly screen for the szy-14 gene by sequencing several candidate genes (nos-3, kin-15, kin-16, wee-1.1, wee-1.3, and fzr-1) located within a 57-kb interval on chromosome II. For sequencing the fzr-1 gene, we used the following primers: forward 5′-TCTTGTTTCTGGTGGAGGT-3′ and reverse 5′-ACACGATACTGATGCCCAA-3′ for the bs31 suppressor, and forward 5′-ATGGATGAGCAGCAACCGCC-3′ and reverse 5′-CAAGCTTGAGCTGTTGG-3′ for the bs38 suppressor. Purified PCR amplicons were sequenced and aligned to the ORF, ZK1307.6 to identify the nucleotide substitution.
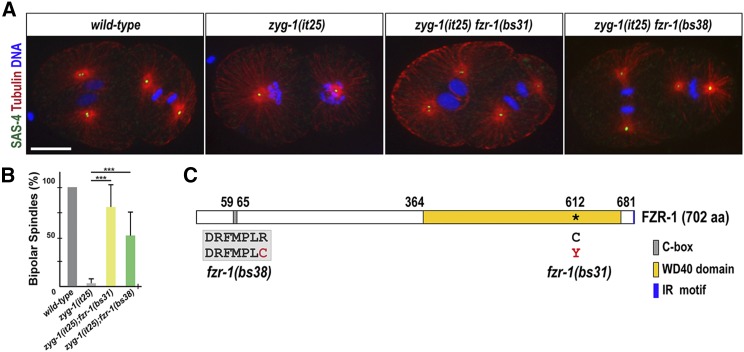
Figure 1.
fzr-1 mutations restore bipolar spindle formation to zyg-1(it25). (A) Embryos grown at 24° stained for centrioles (SAS-4), microtubules, and DNA, illustrating mitotic spindles at the second mitosis. While the wild-type embryo (N2) forms bipolar spindles, the zyg-1(it25) mutant embryo exhibits monopolar spindles. However, bipolar spindle formation is restored in zyg-1(it25) fzr-1(bs31) and zyg-1(it25) fzr-1(bs38) double-mutant embryos. Bar, 10 μm. (B) Quantification of bipolar spindle formation during the second cell cycle. At the restrictive temperature (24°), wild-type (N2) embryos invariably assemble bipolar spindles (100% bipolar spindles, n = 600), whereas a great majority of zyg-1(it25) mutant embryos form monopolar spindles (3.3 ± 4.4% bipolar spindles, n = 660). In contrast, bipolar spindle formation is restored in zyg-1(it25) fzr-1(bs31) (79.9 ± 22.0% bipolar spindles, n = 276, P < 0.001) and zyg-1(it25) fzr-1(bs38) (51.4 ± 24.4%, n = 404, P < 0.001) double mutants. Average values are presented. Error bars represent SD. n is the number of blastomeres. *** P < 0.001 (two-tailed t-test). (C) Schematic of FZR-1 protein structure illustrates functional domains and the location of the missense mutations: R65C within the C-box in the fzr-1(bs38) mutant, and C612Y within WD40 domain in the fzr-1(bs31) mutant allele.
Table 1. Genetic analysis.
| °C | % Embryonic Viability (Average ± SD) | n (Progeny) | |
|---|---|---|---|
| N2 | 24 | 99.4 ± 0.7 | 1500 |
| zyg-1(it25) | 0 ± 7.2 | 1350 | |
| fzr-1(bs31) | 96.8 ± 5.0 | 1200 | |
| zyg-1(it25) fzr-1(bs31) | 44.5 ± 7.3 | 1273 | |
| zyg-1(it25) fzr-1(bs38) | 28.6 ± 10.3 | 1004 | |
| zyg-1(it25) M9 buffer | 0 ± 0 | 600 | |
| zyg-1(it25) fzr-1(RNAi) | 10.7 ± 7.9 | 1045 | |
| zyg-1(or409) M9 buffer | 0 ± 0 | 466 | |
| zyg-1(or409) fzr-1(RNAi) | 2.2 ± 0 | 313 | |
| N2 | 24 | 100 ± 0 | 437 |
| zyg-1(it25) | 0 ± 0 | 1573 | |
| mat-3(or180) | 0 ± 0 | 636 | |
| zyg-1(it25); mat-3(or180) | 5.1 ± 1.2 | 1300 | |
| N2 | 24 | 100 ± 0 | 1143 |
| sas-5KEN-to-3A | 99 ± 1.1 | 1386 | |
| zyg-1(it25); sas-5KEN-to-KEN | 0 ± 0 | 1165 | |
| zyg-1(it25); sas-5KEN-to-3A | 0 ± 0 | 1216 | |
| N2 | 22.5 | 100 ± 0 | 400 |
| zyg-1(it25) | 4.0 ± 5.4 | 1159 | |
| emb-1(hc57) | 3.2 ± 2.1 | 1064 | |
| zyg-1(it25); emb-1(hc57) | 3.3 ± 4.3 | 1337 | |
| N2 | 22.5 | 0 ± 0 | 817 |
| sas-5KEN-to-3A | 0.1 ± 0.1 | 769 | |
| zyg-1(it25); sas-5KEN-to-KEN | 4.6 ± 4.0 | 1409 | |
| zyg-1(it25); sas-5KEN-to-3A | 35.3 ± 9.2 | 1341 |
CRISPR/CAS-9 mediated genome editing
For genome editing, we used the co-CRISPR technique as previously described in C. elegans (Arribere et al. 2014; Paix et al. 2015). In brief, we microinjected N2 and zyg-1(it25) animals using a mixture containing recombinant SpCas9 (Paix et al. 2015), crRNAs targeting sas-5 and dpy-10 at 0.4–0.8 μg/μl, tracrRNA at 12 μg/μl, and single-stranded DNA oligonucleotides to repair sas-5 and dpy-10 at 25–100 ng/μl. Microinjection was performed using the XenoWorks microinjector (Sutter Instruments, Novato, CA) with a continuous pulse setting at 400–800 hPa. All RNA and DNA oligonucleotides used in this study were synthesized by Integrated DNA Technologies (IDT, Coralville, IA) and are listed in Table S2 in File S1. As we were unable to engineer a silent mutation into the PAM sequence used by the sas-5 crRNA, we introduced six silent mutations to sas-5 (aa 201–206; Figure 5A) by mutating 8 out of 20 the nucleotides that comprise the sas-5 crRNA, in order to disrupt Cas9 recognition after homology-directed repair. After injection, animals were allowed to produce F1 progeny that were monitored for the presence of dpy-10(cn64)/+ rollers. To identify the sas-5KEN-to-3A mutation, we extracted genomic DNA from broods containing the highest frequency of F1 rollers. Using the primers, forward: 5′-TGCCCAAAATACGACAACG-3′ and reverse: 5′-TACACTACTCACGTCTGCT-3′, we amplified the region of sas-5 containing the KEN-box sequence. As the repair template for the sas-5KEN-to-3A mutation introduces an Hpy8I restriction enzyme (NEB, Ipswich, MA) cutting site, we used an Hpy8I enzyme digestion to test for the introduction of our targeted mutation. After isolating homozygotes based on the Hpy8I cutting, we confirmed the SAS-5KEN-to-3A mutation by genomic DNA sequencing. Sequencing revealed that several lines were homozygous for the SAS-5KEN-to-3A mutation (Figure 5A and Table S1 in File S1). However, the strain MTU14, contained all of the silent mutations that we designed to disrupt Cas9 recognition without affecting the KEN-box (Figure 5A and Table S1 in File S1). Thus, we used MTU14 as a control for our assays.
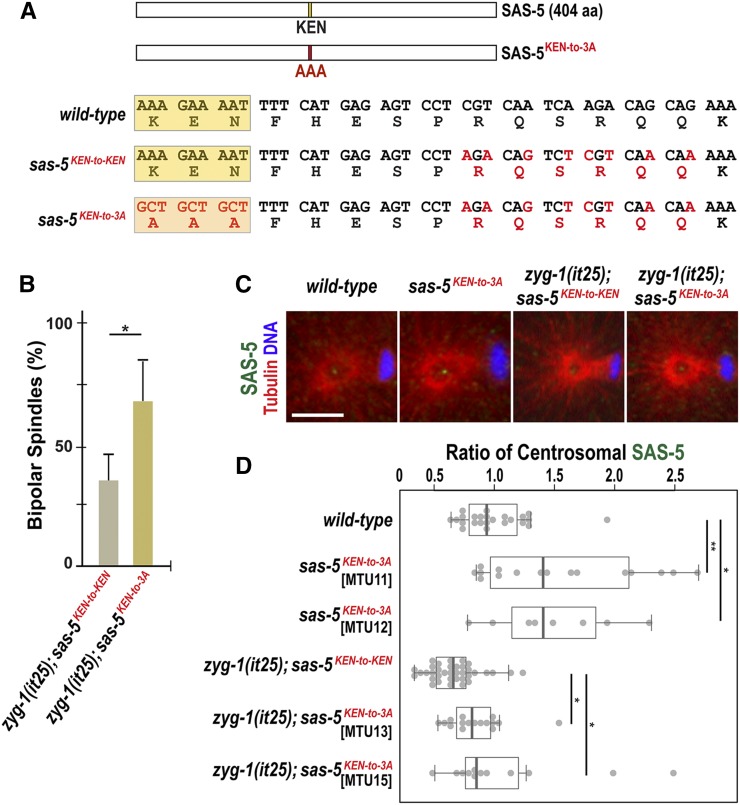
Figure 5.
Mutation of the SAS-5 KEN-box leads to increased SAS-5 levels at centrosomes and restores centrosome duplication to zyg-1(it25) mutants. (A) SAS-5 contains a KEN-box (aa 213–216) motif. Mutations (red) are introduced at multiple sites to make alanine substitutions (AAA; 3A) for the KEN-box and additional silent mutations for the CRISPR genome editing (see Materials and Methods). The KEN-box is highlighted in yellow. Note that the sas-5KEN-to-KEN mutation contains the wild-type SAS-5 protein. (B) Quantification of bipolar spindle formation during the second cell cycle in zyg-1(it25); sas-5KEN-to-KEN and zyg-1(it25); sas-5KEN-to-3A embryos at 22.5°. zyg-1(it25); sas-5KEN-to-3A double mutant embryos produce bipolar spindles at a higher rate (67.5 ± 16.3%, n = 124, P = 0.02) than zyg-1(it25); sas-5KEN-to-KEN controls (35.1 ± 10.7%, n = 164). n is the number of blastomeres. Average values are presented and error bars are SD. (C) Centrosomes stained for SAS-5 (green) during the first anaphase. Bar, 5 μm. (D) Quantification of centrosomal SAS-5 levels during the first anaphase. We used two independently generated sas-5KEN-to-3A mutant lines to quantify SAS-5 levels (MTU11 and 12, Table S1 in File S1). SAS-5 levels at centrosomes are normalized to the average fluorescence intensity in wild-type centrosmes. Mutating the SAS-5 KEN-box leads to increased levels of centrosomal SAS-5 in both MTU11 (1.54 ± 0.63-fold, n = 16; P = 0.04) and MTU12 (1.48 ± 0.50 fold, n = 8; P = 0.03), compared to wild type (1.00 ± 0.29-fold; n = 24). Consistently, there are a significant increase in centrosomal SAS-5 levels in both zyg-1(it25); sas-5KEN-to-3A double mutant lines (MTU13: 0.85 ± 0.24-fold, n = 16; P = 0.01 and MTU15: 1.09 ± 0.59-fold; n = 12; P = 0.03), compared to zyg-1(it25); sas-5KEN-to-KEN control that contains reduced levels of centrosomal SAS-5 (0.67 ± 0.20-fold; n = 36). n is the number of centrosomes. Each dot represents a centrosome. Box ranges from the first through third quartile of the data. Thick bar indicates the median. Solid gray line extends 1.5 times the interquartile range, or to the minimum and maximum data point. * P < 0.05, ** P < 0.01 (two-tailed t-test).
Cytological analysis
To perform immunostaining, the following antibodies were used at 1:2000–3000 dilutions: α-Tubulin (DM1a; Sigma, St-Louis, MO), α-GFP: IgG1κ (Roche, Indianapolis, IN), α-ZYG-1 (Stubenvoll et al. 2016), α-TBG-1(Stubenvoll et al. 2016), α-SAS-4 (Song et al. 2008), α-SAS-5 (Medley et al. 2017), and Alexa Fluor 488 and 561 (Invitrogen) as secondary antibodies. Confocal microscopy was performed as described (Stubenvoll et al. 2016) using a Nikon Eclipse Ti-U microscope equipped with a Plan Apo 60 × 1.4 NA lens, a Spinning Disk Confocal (CSU X1) and a Photometrics Evolve 512 camera. Images were acquired using MetaMorph software (Molecular Devices, Sunnyvale, CA). MetaMorph was used to draw and quantify regions of fluorescence intensity and Adobe Photoshop CS6 was used for image processing. To quantify centrosomal SAS-5 signals, the average intensity within an 8-pixel (1 pixel = 0.151 μm) diameter region was measured within an area centered on the centrosome and the focal plane with the highest average intensity was recorded. Centrosomal TBG-1 (γ-tubulin) levels were quantified in the same manner, except that a 25-pixel diameter region was used. For both SAS-5 and TBG-1 quantification, the average fluorescence intensity within a 25-pixel diameter region drawn outside of the embryo was used for background subtraction.
Immunoprecipitation (IP)
Embryos were collected from gravid worms using hypochlorite treatment (1:2:1 ratio of M9 buffer, 5.25% sodium hypochlorite, and 5 M NaCl), washed with M9 buffer five times and frozen in liquid nitrogen. Embryos were stored at −80° until use. IP experiment using α-GFP were performed following the protocol described previously (Stubenvoll et al. 2016); 20 μl of Mouse-α-GFP magnetic beads (MBL, Naka-ku, Nagoya, Japan) were used per reaction. The α-GFP beads were prepared by washing twice for 15 min in PBST (PBS; 0.1% Triton-X), followed by a third wash in 1× lysis buffer [50 mM HEPES, pH 7.4, 1 mM EDTA, 1 mM MgCl2, 200 mM KCl, and 10% glycerol (v/v)] (Cheeseman et al. 2004). Embryos were suspended in 1× lysis buffer supplemented with complete protease inhibitor cocktail (Roche) and MG132 (Tocris, Avonmouth, Bristol, UK). The embryos were then milled for 3 min at 30 Hz using a Retsch MM 400 mixer-mill (Verder Scientific, Newtown, PA). Lysates were sonicated for 3 min in ice water using an ultrasonic bath (Thermo-Fisher). Samples were spun at 45,000 rpm for 45 min using a Sorvall RC M120EX ultracentrifuge (Thermo-Fisher). The supernatant was transferred to clean microcentrifuge tubes. Protein concentration was quantified using a NanoDrop spectrophotometer (Thermo-Fisher) and equivalent amount of total proteins was used for each reaction. Samples and α-GFP beads were incubated and rotated for 1 hr at 4°, and then washed five times for 5 min using PBST (PBS + 0.1% Triton-X 100). Samples were resuspended in 20 μl of a solution containing 2× Laemmli Sample Buffer (Sigma) and 10% β-mercaptoethanol (v/v), then boiled for 5 min. For protein input, 5 μl of embryonic lysates were diluted using 15 μl of a solution containing 2× Laemmli Sample Buffer and 10% β-mercaptoethanol (v/v), and boiled for 5 min before fractionating on a 4–12% NuPAGE Bis-Tris gel (Invitrogen).
Western blotting
For western blotting, samples were sonicated for 5 min and boiled in a solution of 2× Laemmli Sample Buffer and 10% β-mercaptoethanol before being fractionated on a 4–12% NuPAGE Bis-Tris gel (Invitrogen). The iBlot Gel Transfer system (Invitrogen) was then used to transfer samples to a nitrocellulose membrane. The following antibodies were used at 1:3000–10,000 dilutions: α-Tubulin: α-Tubulin (DM1a; Sigma), α-GFP: IgG1κ (Roche), α-SAS-5 (Song et al. 2011), and α-TBG-1 (Stubenvoll et al. 2016). IRDye secondary antibodies (LI-COR Biosciences, Lincoln, NE) were used at a 1:10,000 dilution. Blots were imaged using the Odyssey infrared scanner (LI-COR Biosciences), and analyzed using Image Studio software (LI-COR Biosciences).
Statistical analysis
All P-values were calculated using two-tailed t-tests assuming equal variance among sample groups. Statistics are presented as Average ± SD unless otherwise specified. Data were independently replicated at least three times for all experiments and subsequently analyzed for statistical significance.
Data availability
All strains used in this study are available upon request. File S1 contains the following: Figure S1, Centrosome-associated TBG-1 levels are unaffected in fzr-1(bs31) and sas-5KEN-to-3A mutant embryos; Figure S2, Brood size in sas-5KEN-to-3A and fzr-1(bs31) mutants; Figure S3, SAS-5 levels are increased in sas-5KEN-to-3A mutants; Table S1, List of strains used in this study; Table S2, List of oligonucleotides used for CRISPR/Cas9 genome editing.
Results and Discussion
The szy-14 mutation restores centrosome duplication to zyg-1(it25) mutants
Through a genetic suppressor screen, the szy-14 (suppressor of zyg-1) gene was originally identified that restores embryonic viability of the partial loss-of-function zyg-1(it25) mutant (Kemp et al. 2007). The zyg-1(it25) mutant embryo grown at the restrictive temperature (24°) fails to duplicate centrosomes during the first cell cycle, resulting in monopolar spindles at the second mitosis and 100% embryonic lethality (O’Connell et al. 2001). A complementation test identified two alleles, szy-14(bs31) and szy-14(bs38), of the szy-14 mutation that partially restore the embryonic viability of zyg-1(it25) but show slow growth phenotype without obvious cytological defects, indicating that the szy-14 gene is not essential for embryonic viability (Table 1, Kemp et al. 2007).
Given that ZYG-1 is essential for proper centrosome duplication (O’Connell et al. 2001), we speculated that the szy-14 mutation might suppress the embryonic lethality of zyg-1(it25) mutants via restoration of centrosome duplication. To examine centrosome duplication events, we quantified the percentage of bipolar spindles at the second mitosis, which indicates successful centrosome duplication during the first cell cycle (Figure 1, A and B). At the restrictive temperature 24°, both double mutant embryos, zyg-1(it25); szy-14(bs31) (79.9 ± 22.0%) and zyg-1(it25); szy-14(bs38) (51.4 ± 24.4%) produced bipolar spindles at a significantly higher rate, compared to zyg-1(it25) single mutant embryos (3.3 ± 4.4%) (Figure 1B). Our observation shows that the szy-14 mutation restores centrosome duplication in zyg-1(it25) embryos, thereby restoring embryonic viability to zyg-1(it25) mutants.
Molecular identification of szy-14
The szy-14 gene was initially mapped to the right arm of chromosome II between the morphological markers dpy-10 and unc-4 (Kemp et al. 2007). Using fine physical mapping, we located szy-14 to an interval of 57 kb (ChrII: 9621265.. 9678204; Wormbase.org) that contains several known cell cycle regulators. Based on the genetic map position of the szy-14 suppressor, we sequenced candidate genes within this interval to detect any mutations in szy-14 mutants. Sequencing revealed that szy-14(bs38) mutants contain a single substitution (C-to-T) in exon 2, and szy-14(bs31) mutants carry a mutation (G-to-A) in exon 5 of the ORF ZK1307.6 that corresponds to the fzr-1 gene. Consistently, inhibiting FZR-1 by RNAi soaking partially restores embryonic viability in both zyg-1(it25) and zyg-1(or409) mutant alleles (Table 1), indicating that loss-of-function of fzr-1 leads to the restoration of embryonic viability to the zyg-1 mutants. Together, we determined that the bs31 and bs38 mutations are alleles of the fzr-1 gene. Hereafter, we refer to szy-14(bs31) and szy-14(bs38) mutants as fzr-1(bs31) and fzr-1(bs38) mutants, respectively.
fzr-1 encodes a conserved coactivator of the anaphase promoting complex/cyclosome (APC/C), the C. elegans homolog of Cdh1/Hct1/Fzr (Schwab et al. 1997; Sigrist and Lehner 1997; Visintin et al. 1997; Fay et al. 2002). The APC/C is an E3 ubiquitin ligase that orchestrates the sequential degradation of key cell cycle regulators during mitosis and early interphase (Song and Rape 2008). As part of this process, specific activators modulate the APC/C activity in different phases of mitosis. Specifically, FZR-1/Cdh1 modulates the APC/C at late mitosis and events in G1 during the time when centrosome duplication occurs. In each of the fzr-1 mutant alleles, the single substitution leads to a missense mutation (Figure 1C). The fzr-1(bs31) mutation results in a missense mutation (C612Y) within the conserved WD40 repeat domain that is known to be involved in protein–protein interactions and is important for substrate recognition (Kraft et al. 2005; He et al. 2013). The fzr-1(bs38) mutation produces a missense mutation (R65C) at the conserved C-box of FZR-1. The C-box is known to be crucial for the physical interaction between FZR-1 and other APC/C subunits (Schwab et al. 2001; Thornton et al. 2006; Chang et al. 2015; Zhang et al. 2016). Thus, both fzr-1(bs31) and fzr-1(bs38) mutations appear to affect conserved domains that are critical for the function of the APC/C complex, suggesting that FZR-1 might regulate centrosome duplication through the APC/C complex.
FZR-1 localizes to nuclei and centrosomes during early cell division
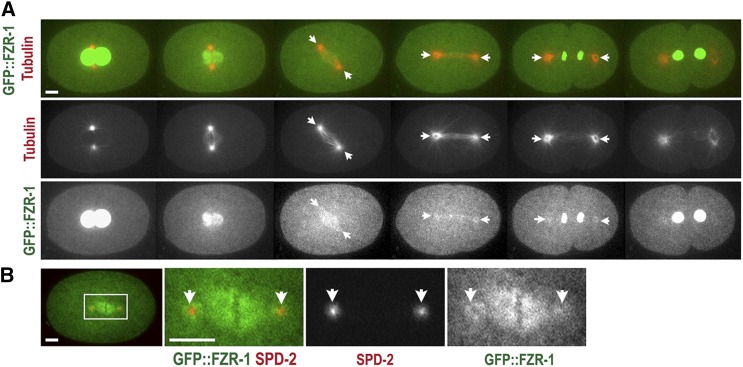
To determine where FZR-1 might function during the early cell cycle, we produced two independent transgenic strains that express FZR-1 tagged with GFP at the N- or C-terminus (see Materials and Methods). To label microtubules, we mated GFP-tagged FZR-1 transgenic animals with the mCherry::β-tubulin expressing line, and performed 4D time-lapse movies to observe subcellular localization of GFP::FZR-1 throughout the first cell cycle (Figure 2A). Confocal imaging illustrates that during interphase and early mitosis, GFP::FZR-1 is highly enriched at the nuclei. After the nuclear envelope breaks down (NEBD), GFP::FZR-1 diffuses to the cytoplasm and reappears to the nuclei at late mitosis when the nuclear envelop reforms. After NEBD, GFP::FZR-1 becomes apparent at spindle microtubules, and centrosomes that colocalize with SPD-2, a centrosome protein (Figure 2B). Both GFP-tagged FZR-1 transgenic embryos exhibit similar subcellular distributions, except a slight difference in fluorescent intensity (data not shown). While we do not exclude the possibility that FZR-1 functions in the cytoplasm to regulate cellular levels of centrosome factors, our observations suggest that C. elegans FZR-1 might direct APC/C activity at centrosomes during late mitosis in early embryos, which is consistent with the role of FZR-1 as the coactivator of the APC/C at late mitosis in other organisms (Raff et al. 2002; Zhou et al. 2003; Meghini et al. 2016).
Figure 2.
Subcellular localization of FZR-1 during the first cell cycle. (A) Still images from time-lapse movie of an embryo expressing GFP::FZR-1 and mCherry::tubulin. Movie was acquired at 1-min intervals. GFP::FZR-1 localizes at nuclei, mitotic spindles, and centrosomes (arrows). Expression of mCherry::tubulin used as a subcellular land-marker. (B) Embryo expressing GFP::FZR-1 and mCherry::SPD-2 displays that GFP::FZR-1 localizes to mitotic spindles and centrosomes (arrows) that colocalize with mCherry-SPD-2, a centrosome marker. Bar, 5 μm.
FZR-1 might function as a part of the APC/C complex to regulate centrosome duplication
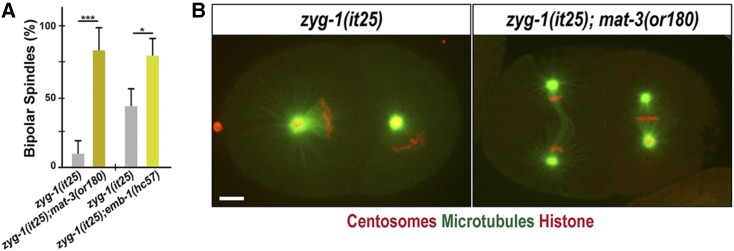
Given that FZR-1 is a conserved coactivator of the APC/C, an E3 ubiquitin ligase, we hypothesized that FZR-1 functions as a part of the APC/C complex in centrosome assembly. If so, depleting other APC/C subunits should have a similar effect that loss of FZR-1 had on the zyg-1(it25) mutant. To examine how other core subunits of the APC/C complex might affect zyg-1(it25) mutants, we mated the zyg-1(it25) strain with mat-3(or180) mutants for the core APC8/CDC23 subunit (Golden et al. 2000), and emb-1(hc57) mutants for the conserved subunit APC16 in the C. elegans APC/C complex (Kops et al. 2010; Green et al. 2011; Shakes et al. 2011). By generating double homozygote mutants, we assayed for bipolar spindle formation and embryonic viability in zyg-1(it25); mat-3(or180) and zyg-1(it25); emb-1(hc57) double homozygous mutants (Figure 3 and Table 1). At the restrictive temperature 24°, zyg-1(it25); mat-3(or180) double-mutant embryos exhibit a ninefold increase in bipolar spindle formation (81.8 ± 14.3%), compared to zyg-1(it25) single mutant embryos (9.1 ± 8.8%) during the second mitosis (Figure 3A). Consistently, 5% of zyg-1(it25); mat-3(or180) double mutants produce viable progeny while 100% of zyg-1(it25) or mat-3(or180) single mutant progeny die at 24° (Table 1). In support of our results, the mat-3(bs29) allele has been reported as a genetic suppressor of zyg-1 (Miller et al. 2016). This result also indicates that the zyg-1(it25) mutation partially restores embryonic viability of mat-3(or180) mutants, suggesting a mutual suppression between zyg-1 and mat-3. Furthermore, we observed that the emb-1 mutation suppresses the centrosome duplication phenotype of zyg-1(it25) mutants at the semi-restrictive temperature 22.5°. While 45.5 ± 11.9% of zyg-1(it25) embryos form bipolar spindles, 79.1 ± 12.4% of zyg-1(it25); emb-1(hc57) double-mutant embryos produce bipolar spindles (Figure 3A). We, however, observed no significant restoration of embryonic viability in zyg-1(it25); emb-1(hc57) double mutants (P = 0.691) compared to zyg-1(it25) single mutants (Table 1), presumably due to the strong embryonic lethality by the emb-1(hc57) mutation itself (Kops et al. 2010; Shakes et al. 2011). Our results indicate that loss of function mutations affecting the APC/C complex suppress the phenotype of zyg-1(it25) mutants. Therefore, FZR-1 might function as a component of the APC/C complex to regulate centrosome duplication in early C. elegans embryos.
Figure 3.
Inactivating the APC/C restores bipolar spindle formation to zyg-1(it25). (A) Quantification of bipolar spindle formation during the second cell cycle. At 24°, zyg-1(it25); mat-3(or180) double mutants assembled bipolar spindles at a significantly higher percentage (81.8 ± 14.3%, n = 78, P < 0.001) than zyg-1(it25) embryos (9.1 ± 8.8%, n = 144). At 22.5°, there is an increase in bipolar spindle formation in zyg-1(it25); emb-1(hc57) double mutants (79.1 ± 12.4%, n = 228, P = 0.03), compared to zyg-1(it25) single mutants (45.5 ± 11.9%, n = 238). n is the number of blastomeres. * P < 0.05, *** P < 0.001 (two-tailed t-test). (B) Still images of embryos expressing GFP::β-tubulin, mCherry::γ-tubulin (centrosome marker) and mCherry::histone raised at 24° illustrate monopolar spindle formation in the zyg-1(it25) embryo, and bipolar spindle formation in the zyg-1(it25); mat-3(or180) double-mutant embryo. Bar, 5 μm.
Loss of FZR-1 results in elevated SAS-5 levels
Next, we wanted to understand how FZR-1 contributes to centrosome duplication. Since FZR-1 appears to function through the APC/C complex in centrosome assembly, we hypothesized that the APC/CFZR-1 specifically targets one or more centrosome regulators for ubiquitin-mediated degradation. If that is the case, depleting FZR-1 should protect substrates from degradation leading to accumulation of target proteins. To identify a direct substrate of APC/CFZR-1 that regulates centrosome assembly, we utilized the conserved FZR-1 coactivator specific recognition motif, KEN-box, to screen for a potential substrate (Glotzer et al. 1991; Pfleger and Kirschner 2000; Song and Rape 2011). The KEN-box appears to be the major degron motif that APC/CFZR-1 recognizes in centrosome duplication (Strnad et al. 2007; Tang et al. 2009; Arquint and Nigg 2014; Meghini et al. 2016). In human cells, HsSAS-6, STIL/SAS-5, and CPAP/SAS-4 contain a KEN-box motif, and APC/CCdh1/FZR-1 targets these proteins for ubiquitin-mediated proteolysis, thereby preventing extra centrosomes (Strnad et al. 2007; Tang et al. 2009; Arquint et al. 2012; Arquint and Nigg 2014). The Drosophila APC/CFzr/Cdh1/FZR-1 is also shown to target Spd2 for destruction through direct interaction with a KEN-box (Meghini et al. 2016). Interestingly in C. elegans, a putative KEN-box motif is present in SAS-5, but not in SAS-4 and SAS-6, which indicates an evolutionary divergence between humans and nematodes.
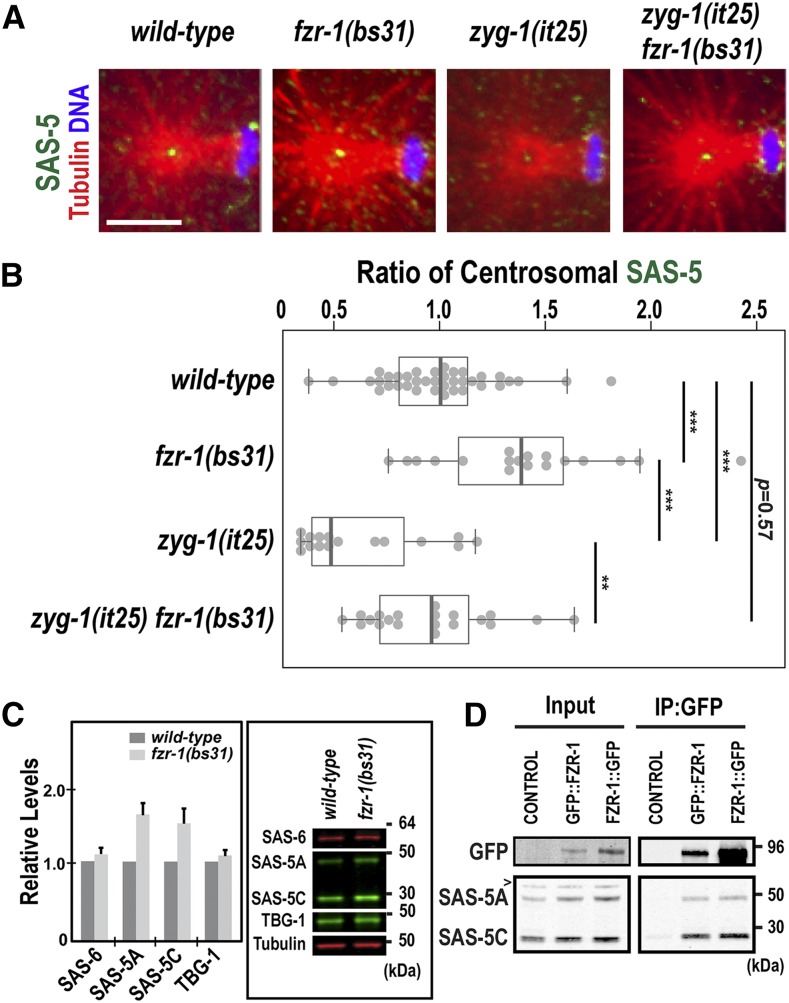
Protein stabilization by the fzr-1 mutation might lead to increased levels of a centrosome-associated substrate, which may compensate for impaired ZYG-1 function at the centrosome. In C. elegans, SAS-5 is the only core centrosome duplication factor containing a KEN-box, which suggests SAS-5 as a potential target of the APC/CFZR-1. If the APC/CFZR-1 targets SAS-5 directly through KEN-box for ubiquitin-mediated proteolysis, inhibiting FZR-1 should protect SAS-5 from degradation leading to SAS-5 accumulation. To examine how the fzr-1 mutation affected SAS-5 stability, we immunostained embryos with anti-SAS-5, and quantified the fluorescence intensity of centrosome-associated SAS-5 (Figure 4, A and B). As ZYG-1 is required for SAS-5 localization to centrosomes, hyper-accumulation of SAS-5 might compensate for partial loss-of-function of ZYG-1, thereby restoring centrosome duplication to zyg-1(it25) mutants. In fact, our quantitative immunofluorescence revealed that fzr-1(bs31) embryos exhibit a significant increase (1.41 ± 0.42 fold; P < 0.001) in centrosomal SAS-5 levels at the first anaphase, compared to wild-type (Figure 4B). Consistently, compared to zyg-1(it25) single mutants, zyg-1(it25); fzr-1(bs31) double mutant embryos exhibit a 1.48-fold increase (P < 0.001) in centrosome-associated SAS-5 levels (Figure 4B). Indeed, centrosomal SAS-5 are restored to near wild-type levels in zyg-1(it25); fzr-1(bs31) double mutants (0.95 ± 0.44 fold; P = 0.003). We, however, observed no significant changes in centrosomal TBG-1 (γ-tubulin) levels in fzr-1(bs31) mutants (Figure S1 in File S1).
Figure 4.
Loss of FZR-1 results in elevated SAS-5 levels. (A) Images of centrosomes stained for SAS-5 (green) at the first anaphase. Bar, 5 μm. (B) Quantification of centrosome-associated SAS-5 levels at the first anaphase. SAS-5 levels are normalized to the average fluorescence intensity in wild-type centrosomes. fzr-1(bs31) embryos exhibit increased levels of centrosomal SAS-5 (1.41 ± 0.42 fold, n = 18; P < 0.001) relative to wild-type controls (1.00 ± 0.28 fold, n = 38). In zyg-1(it25) fzr-1(bs31) double mutants, centrosomal SAS-5 levels are restored to near wild-type levels (0.95 ± 0.44-fold, n = 20; P = 0.003), compared to zyg-1(it25) embryos that show decreased levels of centrosomal SAS-5 (0.64 ± 0.28-fold, n = 16). n is the number of centrosomes. Each dot represents a centrosome. Box ranges from the first through third quartile of the data. Thick bar indicates the median. Solid gray line extends 1.5 times the interquartile range or to the minimum and maximum data point. ** P < 0.01, *** P < 0.001 (two-tailed t-test). (C) Quantitative western blot analyses show that (left panel) fzr-1(bs31) mutant embryos possess increased levels of both SAS-5 isoforms, SAS-5A (1.56 ± 0.16-fold) and SAS-5C (1.48 ± 0.19-fold), compared to wild-type (N2) embryos. However, there were no significant differences in levels of either SAS-6 (1.09 ± 0.08-fold) or TBG-1 (1.08 ± 0.07-fold) between fzr-1(bs31) mutant and wild-type embryos. Four biological samples and eight technical replicates were used. Average values are presented and error bars are SD. Right panel: Representative western blot using embryonic lysates from fzr-1(bs31) mutants and N2 animals. Tubulin was used as a loading control. (D) IP using anti-GFP suggests that FZR-1 physically interacts with SAS-5. Both SAS-5 isoforms (SAS-5A, SAS-5C) coprecipitate with GFP::FZR-1 or FZR-1::GFP. Wild-type (N2) control embryos were used as a negative control of IP; ∼1% of total embryonic lysates was loaded in the input lanes. > indicates a nonspecific detection by the SAS-5 antibody.
Elevated protein levels might influence centrosome-associated SAS-5 levels in fzr-1(bs31) mutants. To determine how inhibition of the APC/CFZR-1 affected overall protein levels, we performed quantitative western blot analysis using embryonic protein lysates and antibodies against centrosome proteins (Figure 4C). Our data indicate that fzr-1(bs31) embryos possess increased SAS-5 levels (∼1.5-fold), relative to wild-type embryos, while the levels of SAS-6 and TBG-1 are not significantly affected in fzr-1(bs31) mutants (Figure 4C). Our observation on the SAS-6 levels in fzr-1(bs31) mutants is consistent with previous work by Miller et al. (2016), showing no increase in SAS-6 levels by the mat-3(bs29)/APC8 mutation that inhibits the APC/C function. These results suggest that C. elegans utilizes a different mechanism to control SAS-6 levels, unlike Human SAS-6, which is regulated by the APC/C-mediated proteolysis (Strnad et al. 2007). Furthermore, our immunoprecipitation suggests a physical interaction between SAS-5 and FZR-1 in C. elegans embryos (Figure 4D), supporting that SAS-5 might be a direct substrate of the APC/CFZR-1. Consistent with our results in this study, prior study has shown that inhibiting the 26S proteasome leads to increased levels of SAS-5 (Song et al. 2011). Thus, SAS-5 levels are likely to be controlled through the ubiquitin-proteasome system.
Collectively, our data show that the fzr-1 mutation leads to a significant increase in both cellular and centrosomal levels of SAS-5, suggesting that the APC/CFZR-1 might control SAS-5 levels via ubiquitin-mediated proteasomal degradation to regulate centrosome assembly in the C. elegans embryo.
Mutation of the KEN-box stabilizes SAS-5
If the APC/CFZR-1 directly targets substrates for destruction via the conserved KEN-box, mutating this motif should cause substantial resistance to ubiquitination-mediated degradation. To determine whether the APC/CFZR-1 targets SAS-5 through the KEN-box motif, we mutated the KEN-box at the endogenous sas-5 locus. By using CRISPR/CAS-9 mediated genome editing (Paix et al. 2015), we generated mutant lines (sas-5KEN-to-3A) carrying alanine substitutions of the SAS-5 KEN-box (Figure 5A). The sas-5KEN-to-3A mutant embryo exhibits no obvious cell cycle defects or embryonic lethality (Table 1), consistent with fzr-1 mutants (Kemp et al. 2007). sas-5KEN-to-3A animals exhibit a slightly reduced (∼80%) and irregular distribution of brood size within the population (Figure S2 in File S1). Reduced brood size and slow growth phenotypes were previously reported in fzr-1 mutant alleles (Fay et al. 2002; Kemp et al. 2007).
Next, we asked how the sas-5KEN-to-3A mutation affected zyg-1(it25) mutants. If the APC/CFZR-1-mediated proteolysis of SAS-5 accounts for the suppression of zyg-1, sas-5KEN-to-3A mutants should mimic the fzr-1 mutation that suppresses zyg-1 mutants. By mating the sas-5KEN-to-3A mutant with zyg-1(it25) animals, we tested whether the sas-5KEN-to-3A mutation could genetically suppress zyg-1 mutants, by assaying for embryonic viability and centrosome duplication (Figure 5B and Table 1). For the zyg-1(it25) mutant control in this experiment, we used the strain MTU14 [zyg-1(it25); sas-5KEN-to-KEN, Table S1 in File S1] that contains the equivalent modifications, except the KEN-box, to the sas-5KEN-to-3A mutation (Figure 5A, see Materials and Methods). At the semirestrictive temperature 22.5°, zyg-1(it25); sas-5KEN-to-3A animals lead to a 7.7-fold increase in the frequency of viable progeny (35.3 ± 9.2%; P < 0.0001), compared to zyg-1(it25); sas-5KEN-to-KEN mutant controls (4.6 ± 4.0%) (Table 1). Consistently, zyg-1(it25); sas-5KEN-to-3A embryos exhibit successful bipolar spindle assembly at a significantly higher rate (67.5 ± 16.3%; P = 0.02) than zyg-1(it25); sas-5KEN-to-KEN embryos (35.1 ± 10.7%) at the two-cell stage (Figure 5B). These results suggest that the sas-5KEN-to-3A mutation does partially restore embryonic viability and centrosome duplication to zyg-1(it25) mutants at 22.5°. However, at the restrictive temperature (24°), where the fzr-1 mutation shows a strong suppression (Figure 1B and Table 1), both zyg-1(it25); sas-5KEN-to-3A double mutants and zyg-1(it25); sas-5KEN-to-KEN mutant animals result in 100% embryonic lethality (Table 1). zyg-1(it25); sas-5KEN-to-3A embryos (14.7% bipolar, n = 68) grown at 24° show only minor effect on centrosome duplication compared to zyg-1(it25); sas-5KEN-to-KEN control embryos (7.6% bipolar, n = 66). The data obtained at 24° reveal that the sas-5KEN-to-3A mutation results in much weaker suppression to zyg-1(it25) mutants than the fzr-1 mutation, suggesting that the SAS-5 KEN-box mutation does not generate the equivalent impact that results from the fzr-1 mutation. If SAS-5 is the only APC/CFZR-1 substrate that contributes to the suppression of zyg-1 mutants, the fzr-1 or KEN-box mutation might influence SAS-5 stability differently. In this scenario, FZR-1 might target SAS-5 through KEN-box and additional recognition motifs (e.g., D-box), causing a greater effect on SAS-5 stability than the KEN-box mutation alone. To examine how the KEN-box mutation affected SAS-5 stability, we measured the fluorescence intensity of SAS-5 at centrosomes by quantitative immunofluorescence (Figure 5, C and D). At 22.5°, where the sas-5KEN-to-3A mutation restores centrosome duplication and embryonic viability to zyg-1(it25), sas-5KEN-to-3A mutants exhibit a significant increase in centrosome-associated SAS-5 levels (∼1.5-fold, P < 0.001), compared to wild-type (Figure 5, C and D). Consistently, zyg-1(it25); sas-5KEN-to-3A embryos display ∼1.4-fold (P = 0.002) increased SAS-5 levels at centrosomes, compared to zyg-1(it25); sas-5KEN-to-KEN control embryos that contain reduced centrosomal SAS-5 levels (Figure 5D). Notably, zyg-1(it25); sas-5KEN-to-3A embryos exhibit centrosomal SAS-5 levels nearly equivalent (∼0.97-fold) to those of wild-type embryos (Figure 5D). As a control, we also quantified centrosomal TBG-1 levels, but saw no changes between sas-5KEN-to-3A mutants and the wild type (Figure S1 in File S1). Furthermore, we examined overall SAS-5 levels by quantitative western blot, finding that relative to wild-type embryos, sas-5KEN-to-3A mutant embryos possess ∼1.5-fold increased SAS-5 levels (Figure S3 in File S1). Together, our quantification data reveal that the sas-5KEN-to-3A or fzr-1 mutation leads to nearly equivalent fold change (∼1.5-fold) in both cellular and centrosome-associated SAS-5 levels (Figure 4, B and C, Figure 5D, and Figure S3 in File S1). Together, these results suggest that APC/CFZR-1 directly targets SAS-5 in a KEN-box-dependent manner to control SAS-5 turnover, and that SAS-5 stabilization by blocking proteolysis results in elevated SAS-5 levels at the centrosome, partially contributing to the suppression of the zyg-1(it25) mutation. In human cells, APC/CCdh1 recognizes a KEN-box to regulate the levels of STIL, the homolog of C. elegans SAS-5, and STIL deleted of the KEN-box leads to accumulation of STIL protein, and centrosome amplification (Arquint and Nigg 2014). While we do not observe extra centrosomes by the SAS-5 KEN-box mutation, our data show that that APC/CFZR-1 controls SAS-5 stability via the direct recognition of the conserved degron motif, KEN-box, to regulate centrosome duplication in C. elegans embryos, suggesting a conserved mechanism for regulating SAS-5 levels between humans and nematodes.
Interestingly, although either inhibiting FZR-1 or mutating KEN-box influences SAS-5 stability at a comparable level, we observe a notable difference in the suppression level by these two mutations. Weaker suppression by the sas-5KEN-to-3A mutation suggests that the APC/CFZR-1 might target additional substrates that cooperatively support the zyg-1 suppression. In this scenario, APC/CFZR-1 might target other centrosome proteins outside core duplication factors through the conserved degron motifs, such as destruction (D)-box and KEN-box (Glotzer et al. 1991; Pfleger and Kirschner 2000). Alternatively, APC/CFZR-1 might target additional core centrosome factors through other recognition motifs other than KEN-box, such as D-box (Glotzer et al. 1991) or unknown motif in the C. elegans system. In humans and flies, APC/CCdh1/Fzr has been shown to regulate the levels of STIL/SAS-5, Spd2, HsSAS-6, and CPAP/SAS-4 (Strnad et al. 2007; Tang et al. 2009; Arquint and Nigg 2014; Meghini et al. 2016). While C. elegans homologs of these factors, except SAS-5, lack a KEN-box, all five centrosome proteins contain at least one putative D-box. An intriguing possibility, given the strong genetic interaction observed between fzr-1 and zyg-1, is that ZYG-1 could be a novel substrate of APC/CFZR-1. Additional work will be required to understand the complete mechanism of APC/CFZR-1- dependent regulation of centrosome duplication in C. elegans. In summary, our study shows the APC/CFZR-1-dependent proteolysis of SAS-5 partially contributes to the suppression of the zyg-1 mutants, and we report that FZR-1 functions as a negative regulator of centrosome duplication in C. elegans.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.117.300260/-/DC1.
Acknowledgments
We thank members of the Song laboratory (Naomi Haque, Brittany Rettig, Karina Saiyad and Michael Stubenvoll) for technical support and Kevin O’Connell and Andy Golden for RNAi and worm stains. We especially thank WormBase and the Caenorhabditis Genetics Center (CGC). WormBase is supported by grant U41 HG002223 from the National Human Genome Research Institute at the United States National Institutes of Health, the United Kingdom (UK) Medical Research Council and the UK Biotechnology and Biological Sciences Research Council. The CGC (St. Paul, MN), is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by a grant [7R15GM11016-02 to M.H.S.] from the National Institute of General Medical Sciences, and Research Excellence Fund (to M.H.S.) from the Center for Biomedical Research at Oakland University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No competing interests are declared.
Author contributions: J.C.M. and M.H.S. designed the experiments and wrote the manuscript. J.C.M. and M.H.S. performed quantifications of confocal imaging and protein levels from western blots. J.C.M., L.E.D., M.M.K., and M.H.S. performed experiments and provided data.
Footnotes
Communicating editor: D. Fay
Literature Cited
- Acquaviva C., Pines J., 2006. The anaphase-promoting complex/cyclosome: APC/C. J. Cell Sci. 119: 2401–2404. [DOI] [PubMed] [Google Scholar]
- Arquint C., Nigg E. A., 2014. STIL microcephaly mutations interfere with APC/C-mediated degradation and cause centriole amplification. Curr. Biol. 24: 351–360. [DOI] [PubMed] [Google Scholar]
- Arquint C., Sonnen K. F., Stierhof Y. D., Nigg E. A., 2012. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J. Cell Sci. 125: 1342–1352. [DOI] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee C. W., Klebba J. E., Buster D. W., Rogers G. C., 2011. The protein phosphatase 2A regulatory subunit twins stabilizes Plk4 to induce centriole amplification. J. Cell Biol. 195: 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S. L., Eklund A. C., Kohane I. S., Harris L. N., Szallasi Z., 2006. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38: 1043–1048. [DOI] [PubMed] [Google Scholar]
- Chang L., Barford D., 2014. Insights into the anaphase-promoting complex: a molecular machine that regulates mitosis. Curr. Opin. Struct. Biol. 29: 1–9. [DOI] [PubMed] [Google Scholar]
- Chang L., Zhang Z., Yang J., McLaughlin S. H., Barford D., 2015. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 522: 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J. R., III, et al. , 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18: 2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church D. L., Guan K. L., Lambie E. J., 1995. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 121: 2525–2535. [DOI] [PubMed] [Google Scholar]
- Dammermann A., Müller-Reichert T., Pelletier L., Habermann B., Desai A., et al. , 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7: 815–829. [DOI] [PubMed] [Google Scholar]
- Dawson I. A., Roth S., Artavanis-Tsakonas S., 1995. The Drosophila cell cycle gene fizzy is required for normal degradation of cyclins A and B during mitosis and has homology to the CDC20 gene of Saccharomyces cerevisiae. J. Cell Biol. 129: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M., Leidel S., Wani K., Baumer K., Bamat J., et al. , 2004. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6: 656–664. [DOI] [PubMed] [Google Scholar]
- Delattre M., Canard C., Gönczy P., 2006. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16: 1844–1849. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu H., Kirschner M. W., 1998. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell 2: 163–171. [DOI] [PubMed] [Google Scholar]
- Fay D. S., Keenan S., Han M., 2002. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 16: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Hagan I. M., Glover D. M., 2015. The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 7: a015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Higuera I., Manchado E., Dubus P., Cañamero M., Méndez J., et al. , 2008. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 10: 802–811. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W., Kirschner M. W., 1991. Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138. [DOI] [PubMed] [Google Scholar]
- Golden A., Sadler P. L., Wallenfang M. R., Schumacher J. M., Hamill D. R., et al. , 2000. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J. Cell Biol. 151: 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., 2015. Centrosomes and cancer: revisiting a long-standing relationship. Nat. Rev. Cancer 15: 639–652. [DOI] [PubMed] [Google Scholar]
- Green R. A., Kao H. L., Audhya A., Arur S., Mayers J. R., et al. , 2011. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145: 470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y. D., Wilkinson C. J., Nigg E. A., 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7: 1140–1146. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D., 1985. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 110: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Chao W. C., Zhang Z., Yang J., Cronin N., et al. , 2013. Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol. Cell 50: 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A. J., Lan W., Niessen S., Hoover H., Cleveland D. W., 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S., Nasmyth K., 1997. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J. Cell Sci. 110: 1523–1531. [DOI] [PubMed] [Google Scholar]
- Kemp C. A., Kopish K. R., Zipperlen P., Ahringer J., O’Connell K. F., 2004. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6: 511–523. [DOI] [PubMed] [Google Scholar]
- Kemp C. A., Song M. H., Addepalli M. K., Hunter G., O’Connell K., 2007. Suppressors of zyg-1 define regulators of centrosome duplication and nuclear association in Caenorhabditis elegans. Genetics 176: 95–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M., Müller-Reichert T., Oegema K., Grill S., Hyman A. A., 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112: 575–587. [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Law E., Tang L., Rose A. M., 2002. The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr. Biol. 12: 2118–2123. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y. D., et al. , 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell 13: 190–202. [DOI] [PubMed] [Google Scholar]
- Kops G. J., van der Voet M., Manak M. S., van Osch M. H., Naini S. M., et al. , 2010. APC16 is a conserved subunit of the anaphase-promoting complex/cyclosome. J. Cell Sci. 123: 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Vodermaier H. C., Maurer-Stroh S., Eisenhaber F., Peters J. M., 2005. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell 18: 543–553. [DOI] [PubMed] [Google Scholar]
- Kumar A., Girimaji S. C., Duvvari M. R., Blanton S. H., 2009. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am. J. Hum. Genet. 84: 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Gönczy P., 2003. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4: 431–439. [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gönczy P., 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7: 115–125. [DOI] [PubMed] [Google Scholar]
- Levine M. S., Bakker B., Boeckx B., Moyett J., Lu J., et al. , 2017. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell 40: 313–322.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh I., Cohen-Kaplan V., Cohen-Rosenzweig C., Avni N., Ciechanover A., 2016. The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. Cell Res. 26: 869–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medley J. C., Kabara M. M., Stubenvoll M. D., DeMeyer L. E., Song M. H., 2017. Casein kinase II is required for proper cell division and acts as a negative regulator of centrosome duplication in Caenorhabditis elegans embryos. Biol. Open 6: 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meghini F., Martins T., Tait X., Fujimitsu K., Yamano H., et al. , 2016. Targeting of Fzr/Cdh1 for timely activation of the APC/C at the centrosome during mitotic exit. Nat. Commun. 7: 12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. G., Liu Y., Williams C. W., Smith H. E., O’Connell K. F., 2016. The E2F–DP1 transcription factor complex regulates centriole duplication in Caenorhabditis elegans. G3 6: 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E. A., Stearns T., 2011. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13: 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell K. F., Caron C., Kopish K. R., Hurd D. D., Kemphues K. J., et al. , 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105: 547–558. [DOI] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D., Seydoux G., 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., Ozlü N., Hannak E., Cowan C., Habermann B., et al. , 2004. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14: 863–873. [DOI] [PubMed] [Google Scholar]
- Pelletier L., O’Toole E., Schwager A., Hyman A. A., Müller-Reichert T., 2006. Centriole assembly in Caenorhabditis elegans. Nature 444: 619–623. [DOI] [PubMed] [Google Scholar]
- Peters J. M., 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7: 644–656. [DOI] [PubMed] [Google Scholar]
- Pfleger C. M., Kirschner M. W., 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14: 655–665. [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J., 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S., Hwang E. S., Visintin R., Amon A., 1998. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr. Biol. 8: 750–760. [DOI] [PubMed] [Google Scholar]
- Puklowski A., Homsi Y., Keller D., May M., Chauhan S., et al. , 2011. The SCF-FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat. Cell Biol. 13: 1004–1009. [DOI] [PubMed] [Google Scholar]
- Raff J. W., Jeffers K., Huang J. Y., 2002. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J. Cell Biol. 157: 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. C., Rusan N. M., Roberts D. M., Peifer M., Rogers S. L., 2009. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M., Murray J. I., Schanze K., Pozniakovski A., Niu W., et al. , 2012. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell 150: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Lutum A. S., Seufert W., 1997. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90: 683–693. [DOI] [PubMed] [Google Scholar]
- Schwab M., Neutzner M., Mocker D., Seufert W., 2001. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 20: 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes D. C., Allen A. K., Albert K. M., Golden A., 2011. emb-1 encodes the APC16 subunit of the Caenorhabditis elegans anaphase-promoting complex. Genetics 189: 549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Zachariae W., Ciosk R., Nasmyth K., 1998. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17: 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist S. J., Lehner C. F., 1997. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 90: 671–681. [DOI] [PubMed] [Google Scholar]
- Song L., Rape M., 2008. Reverse the curse–the role of deubiquitination in cell cycle control. Curr. Opin. Cell Biol. 20: 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Rape M., 2011. Substrate-specific regulation of ubiquitination by the anaphase-promoting complex. Cell Cycle 10: 52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. H., Aravind L., Müller-Reichert T., O’Connell K. F., 2008. The conserved protein SZY-20 opposes the Plk4-related kinase ZYG-1 to limit centrosome size. Dev. Cell 15: 901–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. H., Liu Y., Anderson D. E., Jahng W. J., O’Connell K. F., 2011. Protein phosphatase 2A-SUR-6/B55 regulates centriole duplication in C. elegans by controlling the levels of centriole assembly factors. Dev. Cell 20: 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., et al. , 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell 13: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubenvoll M. D., Medley J. C., Irwin M., Song M. H., 2016. ATX-2, the C. elegans ortholog of human Ataxin-2, regulates centrosome size and microtubule dynamics. PLoS Genet. 12: e1006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugioka K., Hamill D. R., Lowry J. B., McNeely M. E., Enrick M., et al. , 2017. Centriolar SAS-7 acts upstream of SPD-2 to regulate centriole assembly and pericentriolar material formation. eLife 6: e20353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C. J., Fu R. H., Wu K. S., Hsu W. B., Tang T. K., 2009. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 11: 825–831. [DOI] [PubMed] [Google Scholar]
- Tang C. J., Lin S. Y., Hsu W. B., Lin Y. N., Wu C. T., et al. , 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 30: 4790–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The I., Ruijtenberg S., Bouchet B. P., Cristobal A., Prinsen M. B., et al. , 2015. Rb and FZR1/Cdh1 determine CDK4/6-cyclin D requirement in C. elegans and human cancer cells. Nat. Commun. 6: 5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton B. R., Ng T. M., Matyskiela M. E., Carroll C. W., Morgan D. O., et al. , 2006. An architectural map of the anaphase-promoting complex. Genes Dev. 20: 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Prinz S., Amon A., 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278: 460–463. [DOI] [PubMed] [Google Scholar]
- Zhang S., Chang L., Alfieri C., Zhang Z., Yang J., et al. , 2016. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature 533: 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Ching Y. P., Ng R. W., Jin D. Y., 2003. Differential expression, localization and activity of two alternatively spliced isoforms of human APC regulator CDH1. Biochem. J. 374: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Lawo S., Bird A., Pinchev D., Ralph A., et al. , 2008. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr. Biol. 18: 136–141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains used in this study are available upon request. File S1 contains the following: Figure S1, Centrosome-associated TBG-1 levels are unaffected in fzr-1(bs31) and sas-5KEN-to-3A mutant embryos; Figure S2, Brood size in sas-5KEN-to-3A and fzr-1(bs31) mutants; Figure S3, SAS-5 levels are increased in sas-5KEN-to-3A mutants; Table S1, List of strains used in this study; Table S2, List of oligonucleotides used for CRISPR/Cas9 genome editing.