Abstract
The GTP hydrolysis activities of Rho GTPases are stimulated by GTPase-activating proteins (GAPs), which contain a RhoGAP domain equipped with a characteristic arginine finger and an auxiliary asparagine for catalysis. However, the auxiliary asparagine is missing in the RhoGAP domain of Myo9b (Myo9b-RhoGAP), a unique motorized RhoGAP that specifically targets RhoA for controlling cell motility. Here, we determined the structure of Myo9b-RhoGAP in complex with GDP-bound RhoA and magnesium fluoride. Unexpectedly, Myo9b-RhoGAP contains two arginine fingers at its catalytic site. The first arginine finger resembles the one within the canonical RhoGAP domains and inserts into the nucleotide-binding pocket of RhoA, whereas the second arginine finger anchors the Switch I loop of RhoA and interacts with the nucleotide, stabilizing the transition state of GTP hydrolysis and compensating for the lack of the asparagine. Mutating either of the two arginine fingers impaired the catalytic activity of Myo9b-RhoGAP and affected the Myo9b-mediated cell migration. Our data indicate that Myo9b-RhoGAP accelerates RhoA GTP hydrolysis by a previously unknown dual-arginine-finger mechanism, which may be shared by other noncanonical RhoGAP domains lacking the auxiliary asparagine.
Keywords: Rho GTPases, GTPase-activating proteins, RhoGAP domain, Myo9b, RhoA
Introduction
The Rho family of small GTPases, such as RhoA, Cdc42, and Rac1, is a master regulator of a range of cellular processes including cytoskeleton organization, cell adhesion and migration, cell polarity, and cell division [1–3]. As molecular switches, Rho GTPases can cycle between an active GTP-bound and an inactive GDP-bound form, which is regulated by guanine-nucleotide-exchange factors that facilitate GDP dissociation, GTPase-activating proteins (GAPs) that stimulate GTP hydrolysis, and guanine-nucleotide-dissociation inhibitors that sequester Rho GTPases [4,5]. The GAPs for Rho GTPases (RhoGAPs) often contain a characteristic RhoGAP domain that is capable of binding to Rho GTPases and promoting their GTP hydrolysis activities [5,6]. Elegant structural studies of the RhoGAP domains, in complex with their cognate Rho GTPases, have revealed that the canonical RhoGAP domain possesses a signature catalytic arginine residue (referred to as the arginine finger) that directly inserts into the nucleotide-binding pocket of Rho GTPases and interacts with the nucleotide to stabilize the transition state of GTP hydrolysis [5,7,8]. In the vicinity of the arginine finger, an essentially conserved asparagine residue in most, if not all, canonical RhoGAP domains interacts with and stabilizes the Switch I loop of Rho GTPases to assist the arginine finger in GTP hydrolysis [8,9] (Supplementary Fig. S1).
Myosin IXb (Myo9b), a member of class IX myosins in mammals, is a unique motorized Rho GAP that contains an N-terminal motor domain and a RhoGAP domain at the C-terminal tail (Fig. 1a). Myo9b has been detected in immune cells and controls cell shape and motility [10]. Myo9b-deficient macrophages fail to spread and remain in a round shape [11]. Additionally, Myo9b-depleted dendritic cells and T-cells are also defective in chemotactic migration [12,13]. It has been proposed that Myo9b acts as a single-headed myosin motor that is capable of moving along actin filaments [14,15]. The motor domain of Myo9b contains a large insertion sequence that can bind to actin filaments and provides the second tethering site for processive movement [16,17]. In addition to binding to actin filaments, Myo9b functions as a Rho GAP to inactivate Rho GTPases and thereby terminate Rho signaling [18–20]. Because Rho signaling is important for regulating the organization of actin filaments [3], it has been proposed that Myo9b might form some feedback loops in which it regulates actin tracks where it walks along [10]. Moreover, the motor activity of Myo9b is also responsible for the targeting of this motor–RhoGAP chimera to the leading edge of migrating cells [21]. Given the essential role of Myo9b in immune cells, it is not surprising that the defects of this Rho GAP are associated with immune disorders such as celiac disease and inflammatory bowel disease [22,23]. Interestingly, Myo9b has also been identified as a key player in Slit-Robo-mediated lung tumor suppression, and increased Myo9b expression is correlated with lung cancer progression and poor prognosis [24].
Fig. 1.
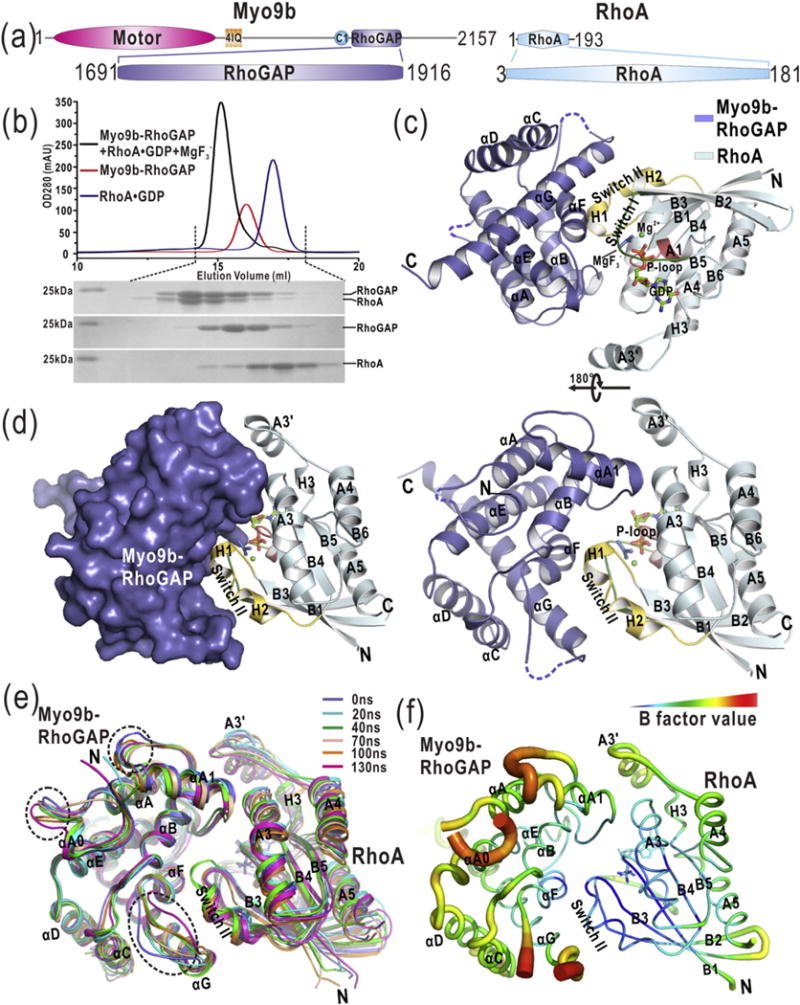
The overall structure of the Myo9b-RhoGAP/RhoA complex. (a) Domain organization of Myo9b and RhoA. Myo9b contains an N-terminal motor domain followed by four IQ motifs and a C1, and a RhoGAP domain at the C terminus. RhoA contains a Rho GTPase domain. (b) Analytical gel-filtration analysis of the formation of the Myo9b-RhoGAP/RhoA complex in the presence of . (c) A ribbon diagram of the structure of the Myo9b-RhoGAP/RhoA complex. Myo9b-RhoGAP and RhoA are colored in blue and cyan, respectively. The secondary structures of Myo9b-RhoGAP and RhoA are labeled and both N and C termini are also marked. The Switch I and II and P-loop of RhoA, colored in green, yellow, and pink, respectively, construct the nucleotide-binding pocket in which GDP, , and Mg2+ are located. (d) A combined surface and ribbon representation of the Myo9b-RhoGAP/RhoA complex structure. Myo9b-RhoGAP is in the surface representation (colored in blue), and RhoA is in the ribbon representation (colored in cyan). The figure shows that RhoA is cradled in the expanded concave target-binding groove of Myo9b-RhoGAP. (e) Superposition of the snapshots of a representative simulation of the Myo9b-RhoGAP/RhoA complex with simulation time indicated. The flexible loops of Myo9b-RhoGAP that exhibit dynamic conformations during the simulations are highlighted by dashed circles. (f) A diagrammatic representation of the complex structure with variable radii according to different B-factors. Consistent with data from the MD simulations, the flexible loops of Myo9b-RhoGAP have higher B-factors.
Among the Rho family of small GTPases, RhoA is a specific target of the RhoGAP domain of Myo9b (Myo9b-RhoGAP) [18,24]. The selective inactivation of RhoA by Myo9b-RhoGAP is important for actin filament organization at the leading edge of cell protrusions and critical for migration [11]. We have determined the structure of Myo9b-RhoGAP and built a structural model of the Myo9b-RhoGAP/RhoA complex based on the homology modeling [24]. We have shown that Myo9b-RhoGAP contains a unique positively charged site within the target-binding groove for specific recognition of RhoA (rather than Cdc42 or Rac1) [24]. Mutations inside this positively charged region impaired the binding of Myo9b-RhoGAP to RhoA and compromised the specific catalytic activity of Myo9b-RhoGAP toward RhoA [24]. However, the high-resolution atomic structure of the Myo9b-RhoGAP/RhoA complex still remains to be determined. More interestingly, in stark contrast to those canonical RhoGAP domains, the highly conserved auxiliary asparagine is absent in Myo9b-RhoGAP; instead, the corresponding position is occupied by an alanine residue (Supplementary Fig. S1). These unique features make Myo9b-RhoGAP different from other canonical RhoGAP domains and suggest that Myo9b-RhoGAP may employ a distinct catalytic mechanism for RhoA GTP hydrolysis.
In this study, based on the previous structural studies of Myo9b-RhoGAP alone, we further determined the structure of Myo9b-RhoGAP in complex with GDP-bound RhoA and magnesium fluoride that mimics the transition state of GTP hydrolysis. Unexpectedly, in addition to the conventional arginine finger that inserts into the nucleotide-binding pocket of RhoA, Myo9b-RhoGAP possesses another arginine finger at the catalytic site that anchors the Switch I loop of RhoA and interacts with the nucleotide, which may compensate for the lack of the auxiliary asparagine. Mutations of either of the two arginine fingers disrupted the catalytic activity of Myo9b-RhoGAP and impaired the Myo9b-mediated cell migration. Together, our data demonstrate that Myo9b-RhoGAP accelerates RhoA GTP hydrolysis using a dual-arginine-finger mechanism that has not been reported previously.
Results
The overall structure of the Myo9b-RhoGAP/RhoA complex
Myo9b-RhoGAP has been demonstrated to exhibit the specific catalytic activity toward RhoA [18,24], although the conserved auxiliary asparagine is missing (Supplementary Fig. S1). To examine the catalytic mechanism of Myo9b-RhoGAP for RhoA GTP hydrolysis, we prepared the Myo9b-RhoGAP/RhoA complex for crystallization and structural determination. Magnesium fluoride has been found to mimic the transition state of GTP hydrolysis and stabilize the complexes formed between GDP-bound Rho GTPases and their cognate RhoGAP domains [25]. In the presence of magnesium fluoride, Myo9b-RhoGAP co-eluted with GDP-bound RhoA in the analytical gel-filtration analysis (Fig. 1b), indicating the formation of a stable Myo9b-RhoGAP/RhoA complex in solution. With this co-eluted fraction, we obtained the diffraction-quality crystals of the Myo9b-RhoGAP/RhoA complex for structural determination. The structure of Myo9b-RhoGAP in complex with GDP-bound RhoA and magnesium fluoride was solved by the molecular replacement method and was refined to 2.4 Å (Supplementary Table S1).
In this complex structure, Myo9b-RhoGAP adopts a classical RhoGAP fold with nine α-helices (αA0–αG) forming a cradle-shaped structure (Fig. 1c). Specifically, αA, αB, αE, and αF constitute the base of the cradle, while αA1 and αG (together with αC and αD) construct the two protruding wings, respectively (Fig. 1c). The expanded concave groove in this cradle (that binds to RhoA as the target-binding groove) is contributed by αA1, αB, αF, and αG of Myo9b-RhoGAP (Fig. 1c–d). On the other hand, RhoA forms a canonical GTPase fold with a central β-sheet (B1–B6) that is surrounded by five α-helices (A1–A5) and three 310 helices (H1–H3; Fig. 1c). The nucleotide-binding pocket of RhoA consists of Switch I (the A1/B2-loop) and II (H1 and H2), P-loop, H3, and the B6/A5-loop, in which GDP and magnesium fluoride ( that acts as the transition state analog of phosphoryl transfer [25]) reside (Fig. 1c–d). In this nucleotide-binding pocket, an additional Mg2+ coordinates GDP and . Overall, Switch I and II of RhoA are both embraced by the target-binding groove of Myo9b-RhoGAP, and the A3 helix of RhoA also contacts the protruding αA1 helix of Myo9b-RhoGAP (Fig. 1c–d).
Because Myo9b-RhoGAP adopts a classical RhoGAP fold, the prediction is that this RhoGAP domain possesses a target-binding mode similar to that of the canonical RhoGAP domains. To test this hypothesis, we compared the structure of the Myo9b-RhoGAP/RhoA complex with that of the canonical RhoGAP domain p50-RhoGAP/RhoA complex under the same condition (i.e., in complex with GDP-bound RhoA and ) [25]. As expected, RhoA binds to a similar target-binding groove in both Myo9b-RhoGAP and p50-RhoGAP (Supplementary Fig. S2a). The structures of the two complexes can be superimposed with the RMSD of ~0.77 Å for the backbone atoms (Supplementary Fig. S2a), indicating that the target recognition conformation of Myo9b-RhoGAP indeed resembles that of the canonical RhoGAP domains. Moreover, upon the superimposition of RhoA, Myo9b-RhoGAP moves more closely to grasp RhoA, with a shift of ~ 1.6 Å closer toward RhoA as compared to p50-RhoGAP (Supplementary Fig. S2b). This suggests that Myo9b-RhoGAP may bind to RhoA more strongly than p50-RhoGAP at the transition state (see below for details).
Molecular dynamics (MD) simulations of the Myo9b-RhoGAP/RhoA complex
In the structure of the Myo9b-RhoGAP/RhoA complex, the αA0/αA and αF/αG loops of Myo9b-RhoGAP could not be clearly defined due to their poor electron density maps (as indicated by the dashed lines in Fig. 1c), suggesting that these two loops may be flexible. To avoid the potential artifacts induced by crystal packing and evaluate the conformational properties of the missing loops, we performed the MD simulations of the complex structure in solution. As expected, the overall structure of the Myo9b-RhoGAP/RhoA complex underwent little conformational changes during the simulations (Fig. 1e), supporting the model that Myo9b-RhoGAP forms a compact and stable complex with GDP-bound RhoA in the presence of . However, in comparison to the other regions of Myo9b-RhoGAP, the αA0/αA and αF/αG loops exhibit more dynamic conformations (Fig. 1e), suggesting that these two loops within Myo9b-RhoGAP are intrinsically flexible. The conformation of the αA1/αB loop of Myo9b-RhoGAP is more dynamic as well. Consistently, all the flexible and dynamic regions of Myo9b-RhoGAP found in the MD simulations exhibit a higher value of crystallographic B-factors for the complex structure (Fig. 1f). On the other hand, the A3′ helix of RhoA that seems to be isolated from the complex structure and the regions of RhoA outside the interaction core for the complex formation also appear to be dynamic (Fig. 1c and e).
The interface between Myo9b-RhoGAP and RhoA in the complex structure
RhoA is intimately captured in the target-binding groove of Myo9b-RhoGAP (Fig. 1c). Based on the different regions of RhoA (i.e., Switch I and II and the A3 helix), the target-binding groove of Myo9b-RhoGAP and the interaction interface between Myo9b-RhoGAP and RhoA (buried with a surface area of ~1142 Å2) can be further divided into three sites (Site 1 to 3; Fig. 2a). At Site 1 (the interaction site for the A3 helix), R1742 and R1744 from αA1 of Myo9b-RhoGAP form the electrostatic and hydrogen-bonding interactions with D90, E93, and E97 from the A3 helix of RhoA (Fig. 2b and Supplementary Fig. S3). Although the side chain of R1742 points away from the A3 helix in the structure, the MD simulations demonstrated that it could flip back to favor by contacting the side chains of D90 and E93 in the A3 helix (Supplementary Fig. S4). The side chain of N94 from the A3 helix of RhoA forms a hydrogen bond with the main chain of A1740 from αA1 of Myo9b-RhoGAP (Fig. 2b and Supplementary Fig. S3), which would further stabilize this interaction site. At Site 2 (the interaction site for Switch II), K1772, Q1773, and R1776 from αB of Myo9b-RhoGAP form the electrostatic and hydrogen-bonding interactions with E64 and D65 from Switch II of RhoA (Fig. 2c and Supplementary Fig. S3). Additionally, P1852 from αF together with V1870 and M1867 from αG of Myo9b-RhoGAP also make the hydrophobic contacts with Y66 and L69 from Switch II of RhoA. At Site 3 (the interaction site for Switch I), I1848 from αF and V1870, L1871, and M1867 from αG of Myo9b-RhoGAP form the extensive hydrophobic interactions with V35, P36, V37, and F38 from Switch I of RhoA (Fig. 2d and Supplementary Fig. S3). Furthermore, Y34 from the Switch I region of RhoA is sandwiched between R1735 and R1840 from the αA/αA1-loop and αE/αF-loop of Myo9b-RhoGAP, respectively (Fig. 2d). It is likely that all these extensive electrostatic, hydrogen-bonding, and hydrophobic interactions between the target-binding groove of Myo9b-RhoGAP and RhoA contribute to the formation of this stable complex.
Fig. 2.
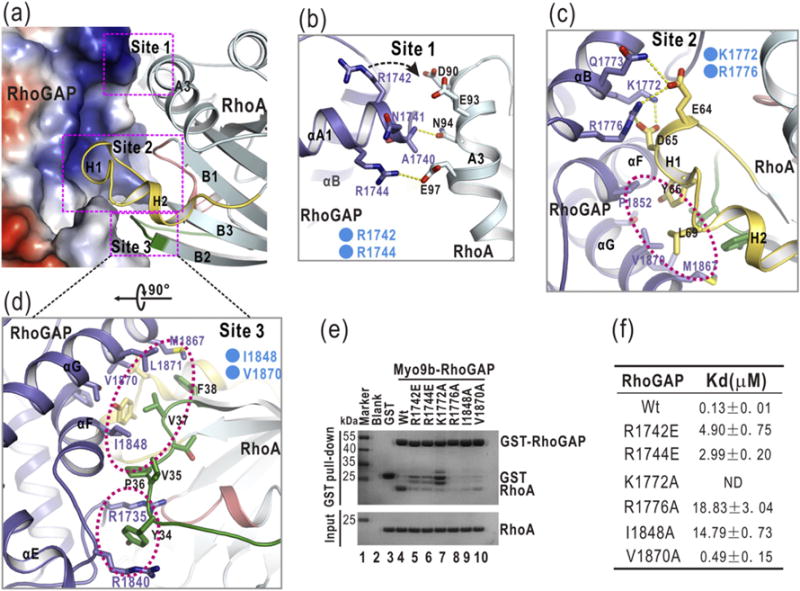
The interaction interface between Myo9b-RhoGAP and RhoA. (a) A combined surface-and-ribbon representation showing the interaction interface between Myo9b-RhoGAP and RhoA. RhoA is presented in the ribbon representation (colored in cyan), and Myo9b-RhoGAP is shown in the surface representation. In this surface drawing, the positive and negative charge potentials are colored in blue and red, respectively. Notably, the target-binding groove of Myo9b-RhoGAP is predominantly positively charged. The three interaction sites are also highlighted by dashed boxes. (b–d) A combined ribbon-and-stick model illustrates the three interaction sites (Site 1 to 3) between Myo9b-RhoGAP and RhoA. In this drawing, Myo9b-RhoGAP and RhoA are colored following the color patterns of Fig. 1c, and the side chains of the residues involved in the interaction are shown as sticks. Hydrogen bonds and salt bridges are indicated by dashed lines. Hydrophobic packing clusters are also highlighted by dashed circles. (e) GST pull-down analysis of the interactions between the Myo9b-RhoGAP mutants and RhoA. The GST protein alone was used as the negative control. As compared to the wild-type protein, all the mutations in Myo9b-RhoGAP severely impaired the binding between Myo9b-RhoGAP and RhoA. (f) A summary of the binding affinities between Myo9b-RhoGAP (or its mutants) and RhoA. Due to the poor behavior of the K1722A mutant in solution, the binding affinity of this mutant for RhoA could not be determined.
Our previous studies demonstrated that among the three aforementioned interaction sites, the positively charged residues of Myo9b-RhoGAP within Site 1 are the key components that determine the binding specificity for RhoA, which contains the negatively charged residues in the corresponding interaction region [24] (Fig. 2b). Consistently, the residues inside Myo9b-RhoGAP for the packing at Site 2 and Site 3 are highly conserved among different RhoGAP domains, whereas the residues for the contacts at Site 1 are largely non-conserved (Supplementary Fig. S1). However, in the p50-RhoGAP/RhoA complex structure, one of the positively charged residues (R1742) in Myo9b-RhoGAP is replaced by a valine (V289) in p50-RhoGAP (Supplementary Fig. S2b), which would likely weaken the interaction Site 1 in this complex. Thus, the cluster of the specific electrostatic interactions at Site 1 would also likely induce the closer distance of packing between Myo9b-RhoGAP and RhoA than that between p50-RhoGAP and RhoA (Supplementary Fig. S2b).
Upon forming the Myo9b-RhoGAP/RhoA complex, although the overall structural folds of Myo9b-RhoGAP and RhoA are similar to that of their free forms, both of them exhibit some local conformational changes in the interaction interface (Supplementary Fig. S2c). The αF/αG loop of Myo9b-RhoGAP in the complex structure undergoes an obvious shift toward Switch II of RhoA, and the C-terminal part of αG likely has an accompanying rotation (Supplementary Fig. S2c). On the other hand, the Switch I loop of RhoA in the complex structure also possesses some conformational changes induced by the insertion of the side chain of Y34 between R1735 and R1840 of Myo9b-RhoGAP (Fig. 2d and Supplementary Fig. S2c).
To test the essential role of the residues in the target-binding groove of Myo9b-RhoGAP for recognizing RhoA, we made specific point mutations inside Myo9b-RhoGAP to disrupt the three interaction sites, that is, R1742E and R1744E at the Site 1, K1772A and R1776A at the Site 2, and I1848A and V1870A at the Site 3 (Fig. 2b–d). We first used the glutathione S-transferase (GST) pull-down assay to evaluate the binding between Myo9b-RhoGAP and RhoA. In this assay, the purified GST-tagged Myo9b-RhoGAP (GST-RhoGAP) and various mutants were used to pull down the purified RhoA. As compared with the wild-type Myo9b-RhoGAP, the individual point mutations at each of these three interaction sites disrupted the interaction between Myo9b-RhoGAP and RhoA (Fig. 2e). To further examine the binding affinities between these Myo9b-RhoGAP mutants and RhoA, we performed the isothermal titration calorimetry (ITC) assay. Consistently, all the point mutations in the three interaction sites decreased the binding affinity between Myo9b-RhoGAP and RhoA (Fig. 2f and Supplementary Fig. S5). Taken together, these data indicate that all the three interaction sites within the target-binding groove of Myo9b-RhoGAP are essential for binding to RhoA.
The two arginine fingers in Myo9b-RhoGAP for RhoA GTP hydrolysis
In comparison with the canonical RhoGAP domain p50-RhoGAP [8,25], the structure of Myo9b-RhoGAP in complex with GDP-bound RhoA and is also analogous to the transition-state intermediate of RhoA GTP hydrolysis. To investigate how Myo9b-RhoGAP accelerates RhoA GTP hydrolysis, we dissected the catalytic site of the Myo9b-RhoGAP/RhoA complex structure by examining the interaction network around the nucleotide. The electron densities of GDP and were traced in the nucleotide-binding pocket of RhoA (Fig. 3a). Both GDP and are anchored at the catalytic site by the extensive interactions with Myo9b-RhoGAP and RhoA (Fig. 3b and the interaction network shown in Supplementary Fig. S6). Remarkably, Myo9b-RhoGAP contains two arginine fingers (R1735 and R1840) that directly interact with GDP and/or at the catalytic site (Fig. 3b). Similar to the canonical arginine finger (R282) in p50-RhoGAP (Fig. 3c), the first arginine finger (R1735) of Myo9b-RhoGAP inserts into the nucleotide-binding pocket of RhoA (Fig. 3b). Specifically, the backbone of R1735 forms a hydrogen bond with the side chain of Q63 from RhoA and reorientates it to interact with a water molecule and , and the bulky side chain of R1735 aligns well with the aromatic ring of Y34 from Switch I of RhoA and forms the extensive electrostatic and hydrogen-bonding interactions with the phosphogroups of GDP and (Fig. 3b). Thus, the first arginine finger of Myo9b-RhoGAP resembles the canonical catalytic arginine finger within p50-RhoGAP (Fig. 3b–c).
Fig. 3.
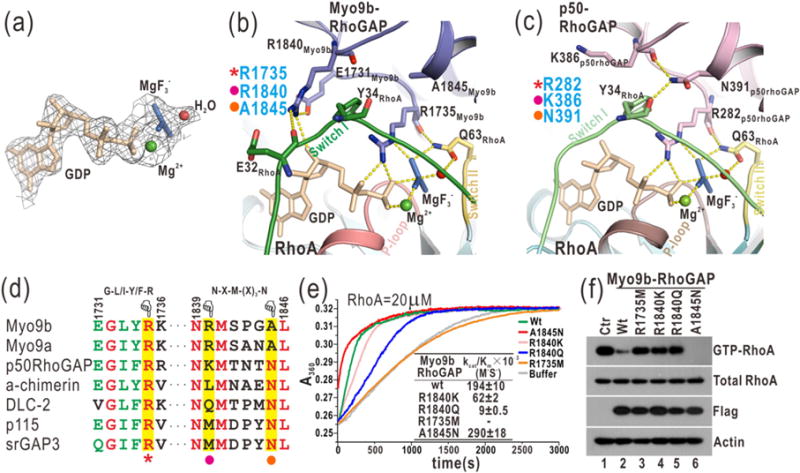
Myo9b-RhoGAP contains two arginine fingers for RhoA GTP hydrolysis. (a) A close-up view of GDP and in the complex structure by a stick model representation. The electron density map (2Fo-Fc map) of GDP, , Mg2+, and a putative nucleophilic water molecule is shown and contoured at 1.5 σ level. (b) A combined ribbon-and-stick model illustrates the catalytic site in the structure of the Myo9b-RhoGAP/RhoA complex. In this drawing, Myo9b-RhoGAP and the Switch I and II and P-loop of RhoA are colored in blue, green, yellow, and pink, respectively, and the residues essential for catalysis are shown as sticks. Hydrogen bonds and salt bridges are indicated by dashed lines. (c) A combined ribbon-and-stick model illustrates the catalytic site in the structure of the p50-RhoGAP/RhoA complex (PDB code: 1OW3). p50-RhoGAP is colored in violet. The color schemes of RhoA follow that shown in panel (b), and the residues essential for catalysis are shown as sticks. (d) Two selected key regions of the structure-based sequence alignment of the RhoGAP domains from different proteins. The identical and highly conserved residues are colored in red and green, respectively. The catalytic arginine finger, the auxiliary arginine finger, and the essential asparagine in the RhoGAP domains are highlighted with a red star, purple dot, and yellow dot, respectively, at the bottom. (e) Time courses of GTP hydrolysis for RhoA (20 μM) catalyzed by Myo9b-RhoGAP and its mutants (40 nM). The Michaelis–Menten kinetic parameters (kcat/Km) are summarized as an inset. Data shown are mean values ± SD from two independent experiments. (f) Biochemical pull-down analysis of active GTP-bound RhoA with Myo9b-RhoGAP and its mutants. The active RhoA levels were measured by GST pull-down analysis with GST-RBD and analyzed by Western blotting using a specific anti-RhoA antibody. The levels of total RhoA, Flag-tagged Myo9b-RhoGAP, and the actin (as an internal loading control) in the cell lysate were also shown. Consistently, mutations of the two arginine fingers impaired the catalytic activity of Myo9b-RhoGAP, while the A1845N mutation increased that of Myo9b-RhoGAP.
Myo9b-RhoGAP contains the second arginine finger (R1840) that is absent in p50-RhoGAP (Fig. 3c–d). This arginine finger directly interacts with Switch I of RhoA and GDP. Specifically, the side chain of R1840 stacks along the aromatic ring of Y34 from Switch I of RhoA and forms a hydrogen bond with the sugar ring of GDP (Fig. 3b). Interestingly, the second arginine finger is sandwiched between E32 from Switch I of RhoA and E1731 from its own αA/αA1-loop, which would form the additional electrostatic and hydrogen-bonding interactions to restrict the conformation of R1840 (Fig. 3b). Moreover, the side chain of R1840 tends to form a hydrogen bond with the backbone of E32 (Fig. 3b and Supplementary Fig. S4), further stabilizing the interaction between the second arginine finger and Switch I of RhoA. Consistently, the MD simulations demonstrated that the distances between the side chain of R1840 and these interacting residues are relatively stable (Supplementary Fig. S4). Thus, Myo9b-RhoGAP contains two parallel arginine fingers at the catalytic site for RhoA GTP hydrolysis (Fig. 3b–d), in stark contrast to a single arginine finger in the canonical RhoGAP domain p50-RhoGAP.
To examine the essential role of the two arginine fingers of Myo9b-RhoGAP for RhoA GTP hydrolysis, we generated point mutations in either arginine finger (i.e., R1735M, R1840K and R1840Q). We evaluated the in vitro catalytic activity of these Myo9b-RhoGAP mutants by the well-established GAP assay and measured the active RhoA levels in cells expressing Myo9b-RhoGAP using the biochemical pull-down experiment. In the in vitro GAP assay, the release of phospho-group accelerated by Myo9b-RhoGAP during RhoA GTP hydrolysis was measured by the phosphate assay kit. In the pull-down experiment, the purified GST-tagged rhotekin-binding domain (RBD) that selectively interacts with GTP-bound RhoA was used to pull down the active RhoA in cells expressing Flag-tagged Myo9b-RhoGAP. As expected, the wild-type Myo9b-RhoGAP accelerated RhoA GTP hydrolysis with high catalytic efficiency, and only a small fraction of GTP-bound RhoA was detected in the pull-down experiment (Fig. 3e–f). Consistent with the published data on Myo9b-RhoGAP [18], mutating the first arginine finger (R1735M) abolished the catalytic activity of Myo9b-RhoGAP (Fig. 3e–f), indicating that this arginine finger is the key catalytic determinant. On the other hand, the mutation of the second arginine finger to a positively charged lysine (R1840K) decreased the catalytic efficiency by ~3-fold, whereas mutating it to a hydrophilic glutamine (R1840Q) reduced the catalytic activity by ~20-fold (Fig. 3e–f). These data show that the second arginine finger also plays an essential but more auxiliary role as compared to the first one (R1735). As supported by the structural analysis (Fig. 3b–d), both of the two arginine fingers of Myo9b-RhoGAP are essential for RhoA GTP hydrolysis, with R1735 as a catalytic finger and R1840 likely acting as a stabilizing finger (see below).
The second arginine finger compensates for the lack of the auxiliary asparagine in Myo9b-RhoGAP
Myo9b-RhoGAP contains two arginine fingers at the catalytic site, whereas the canonical RhoGAP domain p50-RhoGAP has only one (Fig. 3b–c). However, in addition to the key catalytic arginine finger (R282), p50-RhoGAP contains a highly conserved asparagine (N391) in αF that forms the hydrogen-bonding interactions with the backbones of Y34 and K386 from Switch I of RhoA and its αE/αF loop, respectively (Fig. 3c and Supplementary Fig. S1), which would “zip” the two loops together and thus stabilize Switch I of RhoA at the transition state to aid the catalytic arginine finger. Mutating N391 to an alanine (N391A) dramatically decreased the catalytic activity of p50-RhoGAP toward its highly specific substrate Cdc42 [26,27] (Supplementary Fig. S7), indicating that this asparagine is essential for promoting GTP hydrolysis. Strikingly, the auxiliary asparagine found in p50-RhoGAP is absent in Myo9b-RhoGAP and is replaced by an alanine (A1845) at the corresponding position that shows no contact with Switch I of RhoA (Fig. 3b–d). Instead, the second arginine finger (close to the asparagine position) of Myo9b-RhoGAP interacts with both Switch I of RhoA and the nucleotide (Fig. 3b), apparently compensating for the lack of the asparagine residue in Myo9b-RhoGAP in stabilizing Switch I of RhoA. Thus, the second arginine finger (R1840) of Myo9b-RhoGAP likely plays a stabilizing role and functions as an auxiliary finger at the catalytic site, similar to the role of the essential asparagine in p50-RhoGAP.
Based on the structural comparison above, the auxiliary asparagine in p50-RhoGAP and the second arginine finger (R1840) in Myo9b-RhoGAP are both close to Switch I of RhoA and stabilize this loop (Fig. 3b–c). This would predict that the residues in these two positions in the RhoGAP domains could be altered to further enhance their catalytic activities. Consistent with this hypothesis, the conversion of the alanine back to asparagine in Myo9b-RhoGAP (A1845N) indeed increased its catalytic activity by ~1.5-fold (Fig. 3e–f). On the other hand, the corresponding position in p50-RhoGAP for the second arginine finger is occupied by a lysine residue (K386; Fig. 3c). The mutation of this K386 residue to an arginine (K386R) also increased the catalytic activity of p50-RhoGAP (Supplementary Fig. S7), suggesting that the catalytic activities of other RhoGAP domains may be enhanced if the corresponding residues are mutated to an arginine as the second arginine finger in Myo9b-RhoGAP.
The two arginine fingers of Myo9b-RhoGAP are essential for Myo9b-mediated cell migration
The specific catalytic activity of Myo9b toward RhoA has been found to be important for the regulation of RhoA in migrating cells [11]. We have demonstrated that the catalytic activity of Myo9b is also essential for controlling lung cancer cell migration [24]. Since the two arginine fingers of Myo9b-RhoGAP are both essential for promoting RhoA GTP hydrolysis (Fig. 3e–f), we next tested whether they are important for Myo9b-mediated cell migration. We generated the point mutations of the two arginine fingers (R1735M, R1840K, and R1840Q) in the full-length Myo9b and evaluated the effects of these mutations on Myo9b-mediated cell migration using the cell-based transwell migration assay (Fig. 4a–b). Additionally, we also generated the A1845N mutation (that increases the catalytic activity of Myo9b-RhoGAP) in the full-length Myo9b. In this assay, cells were sealed in a well with the filter membrane; the more efficiently cells migrated, the more cells could be observed (Fig. 4b). We also probed the active RhoA levels in cells using the biochemical pull-down experiment coupled with Western blotting. Compared to the control, the knockdown of Myo9b by Myo9b-specific siRNA significantly impaired cell migration, and the active portion of RhoA was increased accordingly (Fig. 4a and c–d), indicating that Myo9b is indeed essential for cell migration. As expected, the transfection of the wild-type, full-length Myo9b largely rescued the cell migration defects induced by siRNA-mediated knockdown, whereas the transfection of the Myo9b mutants that carry the point mutations in either of the two arginine fingers did not (Fig. 4a and c), suggesting that the two arginine fingers within Myo9b-RhoGAP are both essential for Myo9b-mediated cell migration. Interestingly, the A1845N mutant could also rescue the cell migration defects even more efficiently than the wild-type protein (Fig. 4a and c), consistent with the higher catalytic activity of this mutant. Consistent with the data from the cell migration assay, the level of active RhoA was markedly decreased by the transfection of the wild-type Myo9b and A1845N mutant but not by the transfection of the mutants with impairment of the two arginine fingers (Fig. 4d). Taken together, our biochemical and cellular data demonstrated that the two arginine fingers of Myo9b-RhoGAP are both essential for regulating the RhoA activity and the subsequent Myo9b-mediated cell migration.
Fig. 4.
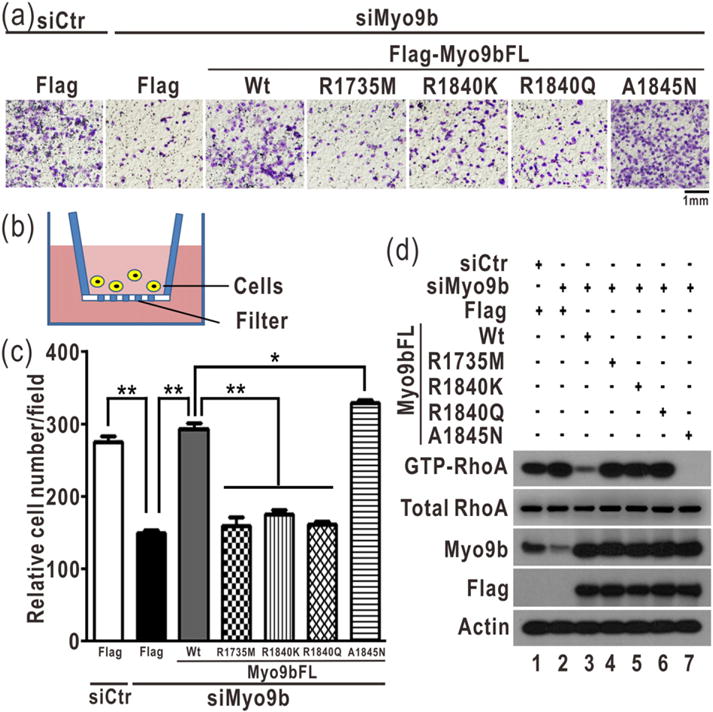
The two arginine fingers are essential for Myo9b-mediated cell migration. (a) Transwell assay of Myo9b-mediated cell migration with Myo9b and its mutants. In this assay, the cells were stained with crystal violet and the images were taken of the underside of a transwell, as shown in panel (b). The scale bar represents 1 mm. As expected, knockdown of Myo9b severely impaired cell migration. The transfection of the wild-type Myo9b largely rescued the cell migration defect induced by knockdown, whereas the transfection of the Myo9b mutants with the mutations of the two arginine fingers did not. The A1845N mutant could rescue the cell migration defect more efficiently than the wild-type protein. (b) A schematic diagram illustrates the transwell cell migration assay. In culture media, cells could penetrate into and migrate through the filter membrane in this assay. (c) Quantification of the transwell cell migration data shown in panel (a). The relative cell number of each field was quantified for each construct. Data were presented as the mean value ± SD from three independent experiments (five different fields for each experiment), **P < 0.01, *P < 0.05. (d) Biochemical pull-down analysis of active GTP-bound RhoA with the knockdown of Myo9b and transfection of the Flag-tagged, full-length Myo9b or its mutants. All the data were presented as shown in Fig. 3f. The level of Myo9b in the cell lysate was analyzed by Western blotting using the specific anti-Myo9b antibody. Consistently, the level of active GTP-bound RhoA was increased by down-regulating Myo9b, and the level of active RhoA was decreased by transfecting the wild-type Myo9b and A1845N mutant but not the Myo9b mutants with impairment of the two arginine fingers.
Discussion
Rho GTPases are the key regulators in a variety of cytoskeleton-related cellular processes and are inactivated by Rho GAPs that carry a RhoGAP domain for accelerating GTP hydrolysis [1,4,5]. The canonical RhoGAP domains are characterized by a unique single arginine finger for catalysis. In this study, we found that Myo9b-RhoGAP unexpectedly contains two arginine fingers at its catalytic site for RhoA GTP hydrolysis. This previously unknown dual-arginine-finger mechanism would broaden the catalytic versatility of the RhoGAP domains and might also be shared by other noncanonical RhoGAP domains.
The dual-arginine-finger mechanism for accelerating GTP hydrolysis
The RhoGAP domains contain a catalytic arginine finger that inserts into the nucleotide-binding pocket of Rho GTPases to facilitate the GTP hydrolysis process [4,5]. This key arginine finger interacts with the nucleotide and works together with the catalytic glutamine from Rho GTPases for the bond breakage (Fig. 3c). The Switch I loop of Rho GTPases wraps around both the arginine finger and the nucleotide to safeguard the catalytic center (Fig. 3c). During this catalytic process, the bond breakage has been indicated as the rate-limiting step, and the Switch I loop of Rho GTPases exhibits dynamic conformations [28]. Thus, in addition to the key arginine finger, the stabilization of the Switch I loop of Rho GTPases would also be critical for the highly efficient catalytic activities of the RhoGAP domains. Consistent with this hypothesis, the canonical RhoGAP domains often contain a highly conserved asparagine (close to the arginine finger) that forms the additional hydrogen-bonding interactions with the Switch I loop of Rho GTPases to stabilize this loop, thereby assisting the catalytic arginine finger in GTP hydrolysis (Figs. 3c and 5). Mutation of this auxiliary asparagine markedly decreased the catalytic activities of the canonical RhoGAP domains (Supplementary Fig. S7).
Fig. 5.
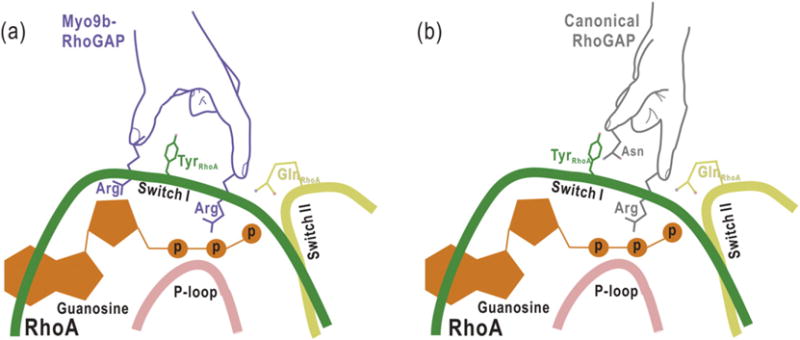
The proposed schematic models illustrating the catalytic mechanisms for Myo9b-RhoGAP and the canonical RhoGAP domain. Myo9b-RhoGAP contains two arginine fingers at the catalytic site (a), whereas the canonical RhoGAP domain has only one arginine finger together with an auxiliary asparagine (b). Similar to the catalytic arginine finger in canonical RhoGAP domain, the first arginine finger of Myo9b-RhoGAP inserts into the nucleotide-binding pocket of RhoA and interacts with the nucleotide for catalysis. In contrast, the second arginine finger of Myo9b-RhoGAP interacts with and stabilizes both the Switch I loop of RhoA and the nucleotide, which may compensate for the lack of the auxiliary asparagine residue in Myo9b-RhoGAP. Notably, a tyrosine residue from the Switch I loop of RhoA is sandwiched between the two arginine fingers of Myo9b-RhoGAP, further stabilizing the transition state of GTP hydrolysis.
In contrast, Myo9b-RhoGAP does not possess this conserved asparagine residue, likely acting as a noncanonical RhoGAP domain (Fig. 3). Our structural analysis of the Myo9b-RhoGAP/RhoA complex has revealed that, in addition to the catalytic arginine finger (R1735), Myo9b-RhoGAP contains another arginine finger (R1840) at its catalytic site (Fig. 3). This additional arginine finger directly interacts with and stabilizes the Switch I loop of RhoA (Figs. 3b and 5), compensating for the lack of the prototypical asparagine residue in Myo9b-RhoGAP and stabilizing the transition state of GTP hydrolysis. Moreover, this additional arginine finger also interacts with the nucleotide (Figs. 3b and 5), which may further stabilize the nucleotide for the high catalytic efficiency of Myo9b-RhoGAP. Thus, the second arginine finger (R1840) of Myo9b-RhoGAP might play a more prominent role than the auxiliary asparagine in the canonical RhoGAP domains for GTP hydrolysis. Consistently, mutating the second arginine finger in Myo9b-RhoGAP attenuated its catalytic activity (Fig. 3e–f). Taken together, Myo9b-RhoGAP contains two arginine fingers at its catalytic site for RhoA GTP hydrolysis (Fig. 5), which is distinct from the catalytic mechanism utilized by the canonical RhoGAP domains.
It is interesting to note that, in addition to Myo9b-RhoGAP, a number of other RhoGAP domains also lack the auxiliary asparagine based on the structure-based sequence alignment (Supplementary Fig. S1), indicating that the absence of the auxiliary asparagine is not just a special case for Myo9b-RhoGAP. All of these RhoGAP domains lacking the asparagine may be grouped into the noncanonical RhoGAP domains, and some of them (such as the RhoGAP domains of ArhGAP12 and ArhGAP27) do contain an additional arginine residue at the position corresponding to the second arginine finger in Myo9b-RhoGAP and display the catalytic activity specific for Rho GTPases [29] (Supplementary Fig. S1). Thus, the dual-arginine-finger mechanism of Myo9b-RhoGAP for stimulating RhoA GTP hydrolysis presented in this study may represent a more general structural paradigm for these noncanonical RhoGAP domains.
Potential rational designs for highly efficient RhoGAP domains for Rho GTPases
In addition to the similar catalytic arginine finger, p50-RhoGAP and Myo9b-RhoGAP employ an auxiliary asparagine (analogously as an asparagine finger) and a second arginine finger to stabilize the Switch I loop of Rho GTPases, respectively (Fig. 5). Because the asparagine finger and the second arginine finger are both in close proximity to the Switch I loop of Rho GTPases, the combination of these two essential sites could enhance the catalytic activities of the RhoGAP domains. Consistent with this hypothesis, the introduction of the missing asparagine back into Myo9b-RhoGAP increased its catalytic activity, and similar was the case for the introduction of a second arginine finger into p50-RhoGAP (Fig. 3 and Supplementary Fig. S7). Based on the catalytic mechanisms of the canonical RhoGAP domains and noncanonical Myo9b-RhoGAP (Fig. 5), we predict that more efficient RhoGAP domains for Rho GTPases could be generated by rational design based on their structures. This might be useful in further designing the Rho-mediated modulators for various biological processes.
The potential second arginine finger hidden in other GAP domains
Similar to Rho GTPases, the inactivation of other families of small GTPases (such as Ras and Rab) is also regulated by their cognate GAP domains. Interestingly, most of these GAP domains possess a similar catalytic arginine finger for promoting GTP hydrolysis by small GTPases [4,5]. To investigate whether the “second arginine finger” found in Myo9b-RhoGAP may be present in these GAP domains, we have further analyzed the structures of several other GAP domains in complex with their cognate small GTPases. The structure of the RasGAP domain/Ras complex revealed that a similar arginine (R894) from the RasGAP domain is close to the Switch I loop of Ras and aligns well with Y32 from this loop (Supplementary Fig. S8). Moreover, the structure of the RabGAP TBC (Tre-2, Bub2, and Cdc16) domain/Rab complex showed that, in addition to the catalytic arginine finger (R343), the TBC domain also contains another arginine residue (R490) at its catalytic site (Supplementary Fig. S8). This arginine residue forms the hydrogen-bonding interactions with the backbone of A64 from the Switch I loop of Rab and interacts with D382 from the neighboring helix (Supplementary Fig. S8), which is reminiscent of the second arginine finger within Myo9b-RhoGAP (Fig. 3). Therefore, both the RasGAP and the RabGAP domains may also contain the “second arginine finger” that might be overlooked previously for stabilizing the Switch I loop of small GTPases. The biological and structural significance of the potential “second arginine finger” hidden in other GAP domains awaits future investigation.
Functional implications of the dual-arginine-finger mechanism for Myo9b-mediated cell migration
Rho-GTPase-mediated intricate organization of the cytoskeletons is critical for controlling cell migration [1,3]. Rho GTPases are spatially and temporally regulated within the restricted regions during the formation of cellular protrusions (such as neurites in neurons and invadopodia in cancer cells) and during migration of different cell types [30], which requires spatio-temporally precise and highly efficient activities of their regulators. Myo9b is localized at the leading edges of migrating cells, and its RhoGAP domain specifically targets RhoA for establishing the asymmetric RhoA activity for cell protrusion and migration [11]. As demonstrated in our previous work [24], the highly specific activity of Myo9b-RhoGAP toward RhoA may be determined by the specific interactions between Myo9b-RhoGAP and RhoA within Site 1 (Fig. 2). Moreover, the high efficiency of Myo9b-RhoGAP is likely to be accomplished by the dual-arginine-finger mechanism demonstrated in this study for RhoA GTP hydrolysis (Fig. 5). Mutating the two arginine fingers of Myo9b-RhoGAP decreased its catalytic efficiency and impaired Myo9b-mediated cell migration (Figs. 3 and 4), supporting that the dual-arginine-finger mechanism is essential for the highly efficient activity of Myo9b-RhoGAP for the control of cell migration. Finally, based on the comparison with the canonical RhoGAP domains (Fig. 5), the more efficient mutants of Myo9b-RhoGAP could be also generated to modulate the RhoA activity and to alter the role of Myo9b in modulating cell migration.
Materials and Methods
Protein expression and purification
The cDNA encoding the RhoGAP domain of human Myo9b (NCBI accession number: NM_004145.3; Myo9b-RhoGAP, residues 1691–1916) was cloned into a modified pET32a vector that contains an N-terminal Trx-His6 tag. Point mutations of Myo9b-RhoGAP were generated using the standard PCR-based mutagenesis method and confirmed by DNA sequencing. The recombinant Myo9b-RhoGAP protein and its mutants were expressed in Escherichia coli (BL21-CodonPlus) cells at 16 °C. The Trx-His6-tagged fusion proteins were purified by Ni2+-Sepharose 6 Fast Flow (GE Healthcare) affinity chromatography followed by size-exclusion chromatography (Superdex-200 26/60, GE Healthcare). After removing the Trx-tag, the resulting proteins were further purified by another round of size-exclusion chromatography in buffer [50 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM DTT, and 1 mM EDTA]. The human p50-RhoGAP protein (residues 237–439) and its mutants were expressed and purified using the same procedure for Myo9b-RhoGAP. The human RhoA protein (residues 3–181) containing the point mutation (F25N) to improve protein stability was also expressed and purified as a Trx-His6-tagged fusion protein. After removing the Trx-tag, the resulting protein was further purified by size-exclusion chromatography in buffer [50 mM Tris–HCl (pH 8.0), 100 mM NaCl, and 1 mM DTT]. The human Cdc42 protein (residues 1–179) was expressed and purified by using the same procedure as for RhoA.
Crystallization and data collection
To prepare the Myo9b-RhoGAP/RhoA complex for crystallization, we mixed the two purified proteins at a 1:1 M ratio together with NaF and MgCl2 to the final concentrations of 20 mM and 5 mM, respectively. The protein complex sample was rotated gently at 4 °C for 1 h and was further purified by size-exclusion chromatography (Superdex-200 10/300, GE Healthcare) in buffer containing 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM DTT, 20 mM NaF, and 5 mM MgCl2. The Myo9b-RhoGAP/RhoA complex sample was then concentrated to 15 mg/ml for crystallization. The crystal of the Myo9b-RhoGAP/RhoA complex was obtained at 16 °C using the vapor diffusion method (sitting drop) in 0.2 M sodium malonate (pH 7.0) and 20% (wt/vol) polyethylene glycol 3350. Before being flash-frozen in liquid nitrogen, the crystal was cryo-protected with the mother liquor supplemented with 10% ethylene glycol. Diffraction data were collected at the beamline BL17U of the Shanghai Synchrotron Radiation Facility with a wavelength of 0.979 Å at 100 K and were processed and scaled using iMOSFLM [31] and SCALA module in the CCP4 suite [32].
Structural determination
The structure of the Myo9b-RhoGAP/RhoA complex was solved by the molecular replacement method using the structure of the p50-RhoGAP/RhoA complex (PDB code: 1OW3) as the search model with PHASER [33]. Additional missing residues were manually modeled into the structure according to the 2Fo-Fc and Fo-Fc electron density maps. The structural model was further fitted and rebuilt with COOT [34] and refined with Phenix [35]. The overall quality of the final structural model of the complex was assessed by PROCHECK [36]. The protein structure figures were prepared using the program PyMOL‡. The statistics for the data collection and structural refinement were summarized in Supplementary Table S1.
MD simulations
The structure of Myo9b-RhoGAP in complex with GDP-bound RhoA and was used as the initial structural model for the MD simulations. The missing flexible loops in the structure were modeled with Modloop [37]. The complex structure was then soaked in an 83 × 92 × 96 Å3 water box, and the Na+ and Cl− ions were added to neutralize the system and maintain the ionic concentration of 150 mM. The MD simulations were performed using the NAMD software package [38] and CHARMM22 all-atom force field with CMAP corrections [39]. Under periodic boundary condition, a 12-Å cutoff was used for van der Waals interactions, and Particle Mesh Ewald summation was used to calculate the electrostatic interactions. Three independent simulations were performed. In each simulation, the energy of the resulting system was first carefully minimized in multisteps to avoid any possible clashes. The energy-minimized system was then equilibrated for 10 ns with temperature controlled at 310 K by Langevin dynamics and pressure controlled at 1 atm by Langevin piston method. With the equilibrated structure, the 130-ns simulation was further performed at constant temperature and pressure with 2-fs time step by rigid bond and SETTLE algorithm for each system. The simulation trajectories were analyzed with VMD [40].
GAP assay
Rho GTPases (RhoA and Cdc42) were loaded with GTP by incubating 2–3 mg of protein with a 10-fold molar excess of GTP (Sigma) at 37 °C for 1 min in 50 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM DTT, and 10 mM EDTA. Free nucleotide was removed with a D-salt column (GE Healthcare) pre-equilibrated with 50 mM Tris–HCl (pH 8.0), 100 mM NaCl, and 0.1 mM EDTA. The GAP assay was performed using the EnzChek Phosphate Assay Kit (Invitrogen). GTP-loaded Rho GTPases were mixed with solutions containing the assay reagents and RhoGAPs and then dispensed into 96-well microplates (Corning). The absor-bance at 360 nm was monitored using a Varioskan Flash apparatus (Thermo). The kinetics determination was carried out following the previously described procedure [41] using GraphPad Prism. Briefly, the data were analyzed and fitted to the pseudo-first-order Michaelis–Menten model function:
where kobs = kintr + (kcat/Km)[RhoGAP]. The catalytic efficiency (kcat/Km) and intrinsic rate constant for GTP hydrolysis (kintr) were treated as global parameters.
Cell culture and antibodies
The human lung cancer cell line H1299 was obtained from ATCC and cultured in a humidified 5%-CO2-containing atmosphere incubator at 37 °C (Thermo Scientific). The cell line was maintained in Dulbecco’s modified Eagle medium (DMEM; Gibco) supplemented with 10% (vol/vol) fetal bovine serum (Gibco), 100 U/ml of penicillin, and 100 μg/ml of streptomycin (P/S; Gibco).
Antibodies were purchased from respective companies: anti-Flag antibody from Sigma Aldrich, anti-RhoA antibody from Santa Cruz Biotechnology, anti-Cdc42 antibody from BD Biosciences Pharmingen, and anti-Myo9b together with anti-Actin antibodies from ProteinTech Group, Inc.
Plasmid constructs and Myo9b siRNAs
The full-length human Myo9b (residues 1–2157) and the RhoGAP domain of Myo9b (residues 1691–1916) were generated by PCR amplification. The RhoGAP domain of p50rhoGAP (residues 237–439) was amplified with the template cDNA reverse-transcribed from the total RNA of H1299 cells. All the fragments were inserted into a 3xFlag-pCMV 7.1 vector (Invitrogen) with the EcoR I (NEB) and Not I (NEB) sites. The plasmids containing mutations were constructed by overlap extension PCR. All the constructs were verified by DNA sequencing. The siRNA against the 3′UTR of Myo9b (5′-GUACGUGGUUUACGUAACUUU-3′) was purchased together with a control siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′) from Shanghai Gene Pharma Co., Ltd. Transient transfection with plasmid DNAs was carried out by using the Lipofectamine 2000™ (Invitrogen) according to the manufacturer’s instructions.
GST pull-down binding assay
GST-Myo9b-RhoGAP and its mutants were expressed in E. coli. The purified GST-fusion proteins (~2 μg) were incubated with the glutathione-Sepharose4B beads (GE Healthcare) for 2 h at 4 °C and then further incubated with the purified RhoA for another 2 h at 4 °C in the buffer [20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM DTT, 20 mM NaF, and 5 mM MgCl2]. The beads were washed three times with the same buffer, and the protein samples were resolved by SDS-PAGE and detected by Coomassie blue staining.
Measurement of levels of active Rho GTPases by the GST pull-down assay
The active form of RhoA or Cdc42 was determined by specific binding to GST-fusion protein containing the RhoA-binding domain of rhotekin (GST-RBD) [1] or GST-fusion protein containing the Cdc42-binding domain of p21-activated kinase (GST-PBD) [2]. The GST-RBD and GST-PBD proteins were expressed in E. coli. Plasmids expressing Flag-tagged Myo9b-RhoGAP or control vector were transfected into H1299 cells. The cells were lysed on ice in cell lysis buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 2 mM MgCl2, 1 mM DTT, 1% Nonidet P-40, 10% glycerol, and proteinase inhibitors (Roche)]. The cell lysates were incubated with either purified GST-RBD or GST-PBD coupled on the glutathione-Sepharose4B beads (GE Healthcare) for 2 h at 4 °C. The beads were washed three times with lysis buffer. The protein samples were resolved by SDS-PAGE and detected by Western blotting with each corresponding antibody.
ITC assay
The ITC assay was performed using a MicroCalorimeter ITC200 (Microcal LLC) at 25 °C. Prior to the experiment, the protein samples were dialyzed in the buffer containing 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM DTT, 20 mM NaF, and 5 mM MgCl2. Wild-type Myo9b-RhoGAP and its various mutants were each placed in the syringe of the instrument, and RhoA was in the sample cell. The Myo9b-RhoGAP protein sample (~0.7 mM) was sequentially injected into the stirred calorimeter cell initially containing RhoA (~0.05 mM) with the injection sequence of 19 × 2 μl at 2-min intervals. The heat of dilution obtained by the titration of Myo9b-RhoGAP into the buffer was subtracted. The integrated, corrected, and concentration-normalized peak areas of the raw data were finally fitted with a model of one binding site using ORIGIN 7.0 (Origin Lab).
Transwell cell migration assay
Cell migration was assayed using Costar cell culture chambers containing insert filters with 8.0-μm pores in 24-well plates (Costar). H1299 cells were transiently transfected with Myo9b-siRNA or control siRNA for 48 h and were then transfected with the plasmids encoding siRNA-resistant, full-length Myo9b or its mutants for 24 h (with ~90% transfection efficiency). The cells were suspended in serum-free DMEM at a density of 3 × 104 cells and plated in transwell insert. DMEM supplemented with 10% fetal bovine serum was placed at the bottom chamber of the plates. Cells that attached on membranes were fixed in methanol and stained with 0.05% crystal violet 13 h after seeding. The cells migrating through the pores to the underside of the filters were photographed under a microscope in five different fields from triplicate wells, and the cell migration index was quantified by measuring the number of cells on the underside of the filter membrane. Comparisons were performed using a two-tailed Student’s t-test, and P < 0.05 was considered as statistically significant. All statistical calculations were carried out using GraphPad Prism.
Supplementary Material
Acknowledgments
We thank the team at the beamline BL17U of the Shanghai Synchrotron Radiation Facility for the beam time. This work was supported by grants from the National Major Basic Research Program of China (2014CB910202 and 2011CB910503), and the National Natural Science Foundation of China (31190062, 31470746, 31300611, 31200561, 31501133, and 31200577). J.Y.W. was supported by NIH (RO1CA175360). R.K. was supported by China Postdoctoral Science Foundation (20110490615). We are grateful to Dr. Warren McGee and the members of the Feng and Wu teams for their suggestions and help during the study and manuscript preparation.
Abbreviations used
- GAPs
GTPase-activating proteins
- RhoGAPs
GAPs for Rho GTPases
- Myo9b
Myosin IXb
- Myo9b-RhoGAP
RhoGAP domain of Myo9b
- MD
molecular dynamics
- GST
glutathione S-transferase
- ITC
isothermal titration calorimetry
- RBD
rhotekin-binding domain
- DMEM
Dulbecco’s modified Eagle medium
- GST-RBD
GST-fusion protein containing the RhoA-binding domain of rhotekin
- GST-PBD
GST-fusion protein containing the Cdc42-binding domain of p21-activated kinase
Appendix A. Supplementary Data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2016.06.014.
Footnotes
Edited by James Sellers
Accession numbers
The atomic coordinate of the Myo9b-RhoGAP/RhoA complex has been deposited in the Protein Data Bank with the accession number 5HPY.
References
- 1.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 2.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 3.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 4.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 5.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 7.Rittinger K, Walker PA, Eccleston JF, Nurmahomed K, Owen D, Laue E, et al. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
- 8.Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389:758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 9.Hakoshima T, Shimizu T, Maesaki R. Structural basis of the Rho GTPase signaling. J Biochem. 2003;134:327–331. doi: 10.1093/jb/mvg149. [DOI] [PubMed] [Google Scholar]
- 10.Bahler M, Elfrink K, Hanley PJ, Thelen S, Xu Y. Cellular functions of class IX myosins in epithelia and immune cells. Biochem Soc Trans. 2011;39:1166–1168. doi: 10.1042/BST0391166. [DOI] [PubMed] [Google Scholar]
- 11.Hanley PJ, Xu Y, Kronlage M, Grobe K, Schon P, Song J, et al. Motorized RhoGAP myosin IXb (Myo9b) controls cell shape and motility. Proc Natl Acad Sci U S A. 2010;107:12,145–12,150. doi: 10.1073/pnas.0911986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Pektor S, Balkow S, Hemkemeyer SA, Liu Z, Grobe K, et al. Dendritic cell motility and T cell activation requires regulation of Rho-cofilin signaling by the Rho-GTPase activating protein myosin IXb. J Immunol. 2014;192:3559–3568. doi: 10.4049/jimmunol.1300695. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Xu Y, Zhang X, Song J, Sorokin L, Bahler M. The motorized RhoGAP myosin IXb (Myo9b) in leukocytes regulates experimental autoimmune encephalomyelitis induction and recovery. J Neuroimmunol. 2015;282:25–32. doi: 10.1016/j.jneuroim.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Inoue A, Saito J, Ikebe R, Ikebe M. Myosin IXb is a single-headed minus-end-directed processive motor. Nat Cell Biol. 2002;4:302–306. doi: 10.1038/ncb774. [DOI] [PubMed] [Google Scholar]
- 15.O’Connell CB, Mooseker MS. Native myosin-IXb is a plus-, not a minus-end-directed motor. Nat Cell Biol. 2003;5:171–172. doi: 10.1038/ncb924. [DOI] [PubMed] [Google Scholar]
- 16.Nalavadi V, Nyitrai M, Bertolini C, Adamek N, Geeves MA, Bahler M. Kinetic mechanism of myosin IXB and the contributions of two class IX-specific regions. J Biol Chem. 2005;280:38,957–38,968. doi: 10.1074/jbc.M507161200. [DOI] [PubMed] [Google Scholar]
- 17.Struchholz S, Elfrink K, Pieper U, Kalhammer G, Honnert U, Grutzner A, et al. Functional role of the extended loop 2 in the myosin 9b head for binding F-actin. J Biol Chem. 2009;284:3663–3671. doi: 10.1074/jbc.M808338200. [DOI] [PubMed] [Google Scholar]
- 18.Muller RT, Honnert U, Reinhard J, Bahler M. The rat myosin myr 5 is a GTPase-activating protein for Rho in vivo: essential role of arginine 1695. Mol Biol Cell. 1997;8:2039–2053. doi: 10.1091/mbc.8.10.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirth JA, Jensen KA, Post PL, Bement WM, Mooseker MS. Human myosin-IXb, an unconventional myosin with a chimerin-like rho/rac GTPase-activating protein domain in its tail. J Cell Sci. 1996;109(Pt 3):653–661. doi: 10.1242/jcs.109.3.653. [DOI] [PubMed] [Google Scholar]
- 20.Reinhard J, Scheel AA, Diekmann D, Hall A, Ruppert C, Bahler M. A novel type of myosin implicated in signalling by rho family GTPases. EMBO J. 1995;14:697–704. doi: 10.1002/j.1460-2075.1995.tb07048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Boom F, Dussmann H, Uhlenbrock K, Abouhamed M, Bahler M. The myosin IXb motor activity targets the myosin IXb RhoGAP domain as cargo to sites of actin polymerization. Mol Biol Cell. 2007;18:1507–1518. doi: 10.1091/mbc.E06-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monsuur AJ, de Bakker PI, Alizadeh BZ, Zhernakova A, Bevova MR, Strengman E, et al. Myosin IXB variant increases the risk of celiac disease and points toward a primary intestinal barrier defect. Nat Genet. 2005;37:1341–1344. doi: 10.1038/ng1680. [DOI] [PubMed] [Google Scholar]
- 23.Nunez C, Oliver J, Mendoza JL, Gomez-Garcia M, Pinero A, Taxonera C, et al. MYO9B polymorphisms in patients with inflammatory bowel disease. Gut. 2007;56:1321–1322. doi: 10.1136/gut.2007.121905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong R, Yi F, Wen P, Liu J, Chen X, Ren J, et al. Myo9b is a key player in SLIT/ROBO-mediated lung tumor suppression. J Clin Invest. 2015;125:4407–4420. doi: 10.1172/JCI81673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham DL, Lowe PN, Grime GW, Marsh M, Rittinger K, Smerdon SJ, et al. MgF(3)(−) as a transition state analog of phosphoryl transfer. Chem Biol. 2002;9:375–381. doi: 10.1016/s1074-5521(02)00112-6. [DOI] [PubMed] [Google Scholar]
- 26.Lancaster CA, Taylor-Harris PM, Self AJ, Brill S, van Erp HE, Hall A. Characterization of rhoGAP. A GTPase-activating protein for rho-related small GTPases. J Biol Chem. 1994;269:1137–1142. [PubMed] [Google Scholar]
- 27.Barfod ET, Zheng Y, Kuang WJ, Hart MJ, Evans T, Cerione RA, et al. Cloning and expression of a human CDC42 GTPase-activating protein reveals a functional SH3-binding domain. J Biol Chem. 1993;268:26,059–26,062. [PubMed] [Google Scholar]
- 28.Kotting C, Gerwert K. The dynamics of the catalytic site in small GTPases, variations on a common motif. FEBS Lett. 2013;587:2025–2027. doi: 10.1016/j.febslet.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 29.Gentile A, D’Alessandro L, Lazzari L, Martinoglio B, Bertotti A, Mira A, et al. Met-driven invasive growth involves transcriptional regulation of Arhgap12. Oncogene. 2008;27:5590–5598. doi: 10.1038/onc.2008.173. [DOI] [PubMed] [Google Scholar]
- 30.Pertz O. Spatio-temporal Rho GTPase signaling—where are we now? J Cell Sci. 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- 31.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM, Acta Crystallogr. D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dodson EJ, Winn M, Ralph A. Collaborative computational project, number 4: providing programs for protein crystallography. Methods Enzymol. 1997;277:620–633. doi: 10.1016/s0076-6879(97)77034-4. [DOI] [PubMed] [Google Scholar]
- 33.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 37.Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 38.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 41.Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


