Abstract
Metabolomic analysis of easily accessible biofluids has provided numerous biomarkers in urine and blood for biodosimetric purposes. In this pilot study we assessed saliva for its utility in biodosimetry using a mouse model. Mice were exposed to 0.5, 3 and 8 Gy total-body gamma irradiation and saliva was collected on day 1 and 7 postirradiation. Global metabolomic profiling was conducted through liquid chromatography mass spectrometry and metabolites were positively identified using tandem mass spectrometry. Multivariate data analysis revealed distinct metabolic profiles for all groups at day 1, whereas at day 7 the two lower dose profiles appeared to have minimal differences. Metabolites that were identified include amino acids and fatty acids, and intermediates of the nicotinate and nicotinamide metabolism. The specificity and sensitivity of the radiation signature, as expected, was higher for the 8 Gy dose at both time points, as determined through generation of receiver operating characteristic curves. To the best of our knowledge, this is the first metabolomics study in saliva of irradiated mice to demonstrate the utility of this biofluid as a potential matrix for identification of radiation and dose-specific biomarkers.
INTRODUCTION
In the event of a possible radiological or nuclear terrorist attack or accidental exposure, it will be necessary to rapidly assess a large number of individuals for their level of exposure, whether external or internal, to provide immediate and effective triage. While ionizing radiation exposure can raise the lifetime cancer and noncancer associated disease risk of an individual, the National Institute of Allergy and Infectious Diseases (NIAID) and Health and Human Services (HHS) have concentrated their efforts on the development of rapid and noninvasive biodosimetric methods to initially identify individuals exposed to higher doses that may lead to hematopoietic and/or gastrointestinal syndromes (1–3). These rapid high-throughput methods can further be followed by other techniques for the refinement of individual dose, such as classical cytogenetic assessment, and concurrent and lifetime monitoring by medical personnel.
In our laboratory we have concentrated our efforts on developing metabolomic methods to characterize small molecules in urine and blood and assess the utility of those metabolites as biomarkers of external radiation exposure (1). To date, we have explored the value of such markers in rodents, nonhuman primates, and total-body-irradiated humans with very encouraging results and significant cross-species validation (4–8). In this pilot study, we extended our studies to assess the utility of saliva as a biofluid for biodosimetry through metabolomics at day 1 and 7 postirradiation, time points that have been previously investigated with other biofluids (1, 4, 6, 7, 9–11). Since the salivary glands as well as oral mucosa are highly radiosensitive, this can be translated in products of radiation exposure directly present in saliva from cells undergoing cell death (12, 13). Reduction in saliva flow has been observed within the first week of radiation exposure and is a severe symptom that is associated in particular with radiotherapy patients undergoing treatment for head and neck cancers. Severe salivary gland dysfunction and xerostomia can lead to mucositis and dysphagia, with reduced quality of life and nutrient absorption (12).
The study results demonstrated significant changes in the metabolome at day 1 and 7 after total-body irradiation, which were more pronounced at the dose associated with the hematopoietic syndrome (8 Gy). Dose-specific changes and metabolites showing dose-response alterations were shown at both time points, which can be utilized for the construction of dose-specific signatures. This pilot study demonstrates the utility of saliva in radiation biodosimetry in addition to the currently established biofluids (urine, blood).
MATERIALS AND METHODS
Chemicals
All chemicals were of the highest purity and all reagents were of LC-MS grade. All chemicals utilized for validation of ions through tandem mass spectrometry (MS/MS) were acquired from Sigma-Aldrich® (St. Louis, MO), except 3-oxo-octadecanoic acid that was acquired from Cayman Chemical (Ann Arbor, MI).
Experimental Design, Irradiations and Sample Collection
Male wild-type C57BL/6 mice, age 8–10 weeks, were acquired from Charles River Laboratories, Inc. (Wilmington, MA). Mice were acclimated at Georgetown University for one week before irradiation and housed under a 12:12 h light-dark schedule and provided with food and water ad libitum (PicoLab® Rodent Diet 20 5053 irradiated). All experiments and animal handling were in accordance with approved IACUC protocols. Irradiations were conducted with a 137Cs source (n =5 per group: sham, 0.5, 3 and 8 Gy; 1.4 Gy/min). The doses were chosen to reflect low, sublethal and LD50/30 dose exposures, with the latter being the most relevant for the NIAID and requiring medical intervention. At day 1 and 7 postirradiation, 10–20 μl of saliva was collected from mice with a pipette tip. If saliva could not be immediately acquired, deionized water was pipetted in the mouth of mice to facilitate sample collection. Saliva samples were briefly centrifuged to precipitate cellular debris, the supernatant was transferred in fresh tubes and samples were stored at −80°C until analysis.
Sample Processing and Data Acquisition
Supernatant (5 μl) were diluted in 50:50 acetonitrile:water (1:20 dilution) with internal standards (4 μM debrisoquine sulfate, 30 μM 4-nitrobenzoic acid) and centrifuged for 20 min at 13,000g, 4°C and 2 μl were then injected into a Waters® Acquity UPLC® High Strength Silica, 1.8 μm, 50 mm column (Waters Corp., Milford, MA) with LC-MS conditions as described previously (5, 8). Data were acquired on a Waters UPLC coupled to a Xevo® G2 QTof, operated in both ESI+ and ESI−. Quality control (QC) samples consisting of a pooled sample were run every ten samples for instrument quality assessment and retention time drift. Additionally, 2 μl of each sample was assessed for total protein concentration with the micro BCA protein assay kit (Thermo Fisher Scientific™, Grand Island, NY) following the manufacturer’s instructions and statistical significance was assessed with analysis of variance (ANOVA).
Data Analysis and Validation
Data processing and deconvolution was performed with Marker-Lynx™ software (Waters Corp.) and each sample was normalized to its respective total protein concentration. Data analysis was performed with the in-house statistical software package MetaboLyzer (14), combining ESI+ and ESI− data. Complete presence of ions was set at 75% and analyzed using Welch’s t test, while partial presence data of <75% were analyzed with Barnard’s test. Statistical significance was considered at P < 0.05 for both tests. Putative identity assignment was performed with a parts per million (ppm) error of <10 through the databases HMDB and KEGG incorporated into MetaboLyzer (15–17). Multidimensional scaling (MDS) plots were constructed from the top 100 ranked ions through the machine-learning random forest algorithm. Validation of ions was performed through tandem mass spectrometry and fragmentation patterns of each candidate were matched against patterns of pure chemicals and/or through the METLIN metabolite database (18). Graphical representation and P values after identification and removal of outliers were all determined with Prism 6 (GraphPad Software Inc., LaJolla, CA). Receiver operating characteristic (ROC) curves for each ion assessed the sensitivity and specificity [area under the curve (AUC) value] of each ion and its significance, which was considered at P < 0.05.
RESULTS
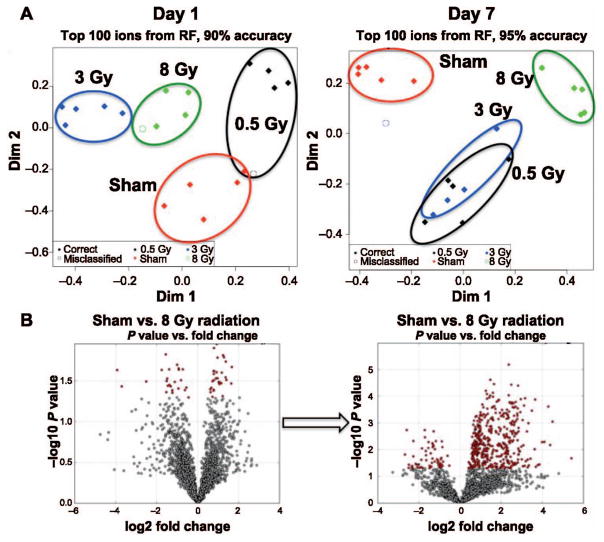
Protein quantification revealed no statistically significant changes within the groups at day 1 (P =0.5174) and 7 (P = 0.0987), and therefore normalization of the raw ion abundances was performed to total protein levels of each sample. To determine the existence of differences in the overall metabolic profiles, the top 100 ranked ions from the random forest algorithm were utilized to construct MDS plots (Fig. 1A). On day 1, tight clustering was observed in all groups (90% overall classification accuracy) with well-defined separation, indicative of substantially different metabolic profiles within each group. However, on day 7, while the classification accuracy remained high (95%), the metabolic profiles of the lower doses (0.5 and 3 Gy) were clustered more closely together, although they remained distinct from each other. Conversely, the 8 Gy irradiated group maintained its distinct profile. To further investigate the effect of time on the metabolic profiles, volcano plots for the highest dose mapping the fold change versus the P value of each ion in a given comparison revealed that while a number of ions were perturbed on day 1, the visible and significant differences were far more evident on day 7.
FIG. 1.
Global metabolic profiling of saliva samples. Panel A: Construction of MDS plots using the random forest (RF) algorithm shows distinct clustering of each group on day 1 postirradiation. However, the profiles show a significant shift on day 7, with the lower doses clustering together while the higher dose of 8 Gy retains a distinct metabolic profile. Panel B: Volcano plots between sham-irradiated and 8 Gy irradiated groups depict the significant metabolic perturbations with increased fold changes and significant P values with significant increased levels of ions at one week postirradiation.
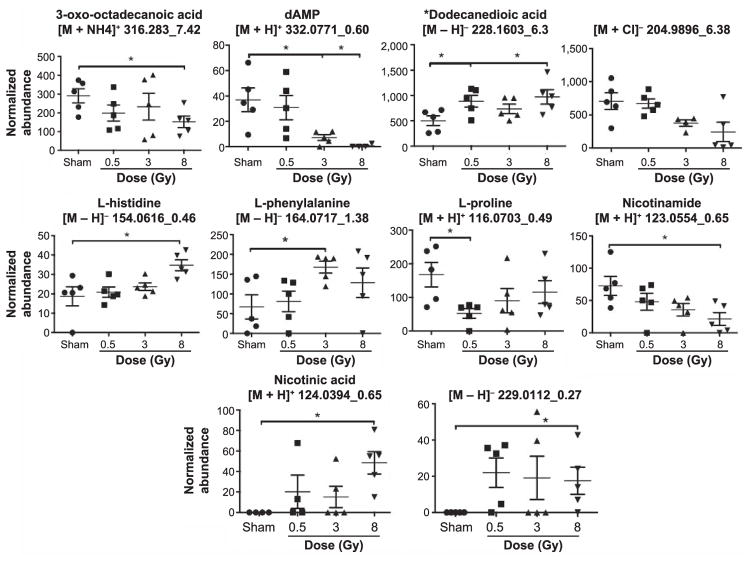
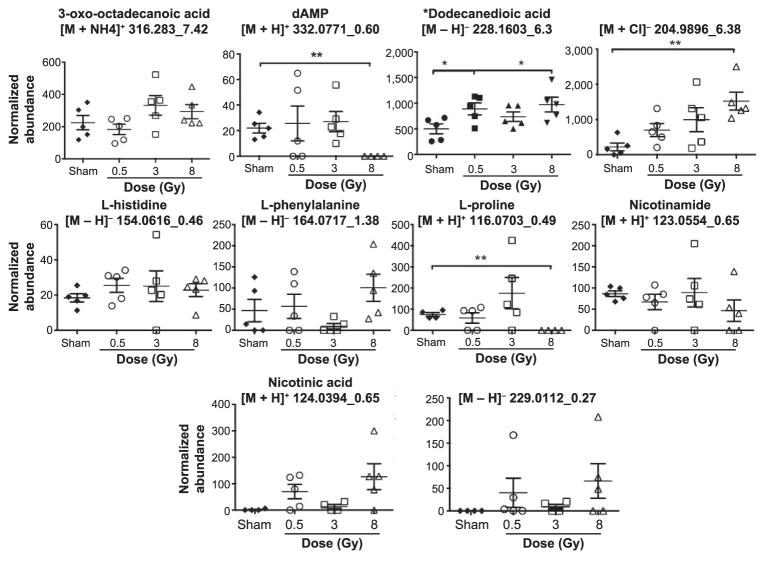
Fragments of ions in QC samples through MS/MS were compared to fragmentation patterns of pure chemicals. In all, 3-oxo-octadecanoic acid, dAMP, L-histidine, L-phenylalanine, L-proline, nicotinamide and nicotinic acid were positively identified as the metabolites in saliva samples. The fragments of the parent ion with [M – H]− = 228.1603 with retention time of 6.3 min were matched to the fragmentation profile of dodecanedioic acid through the METLIN MS/MS database. On the other hand, the ion [M + Cl]− 204.9896_6.38min was not positively validated, as it did not match against D-glyceraldehyde-3-phosphate. [M-H]− 229.0112_0.27min was tested against D-ribulose-5-phosphate, ribose-1-phosphate and ribose-5-phosphate and although the daughter ion matching to the phosphate group was present (m/z 96.9699), its identity could not be verified with certainty. The normalized abundance levels for each of these metabolites/ions were further graphed to depict the intragroup variation (error bars represent standard error of the mean). The results for day 1 are shown in Fig. 2 and for day 7 in Fig. 3. As expected, the most prominent responses are observed in the 8 Gy irradiated group, compared to the sham-irradiated group.
FIG. 2.
Levels of ten ions, some validated through MS/MS, at day 1 postirradiation. Multiple ions are affected, some exhibiting a biodosimetric response. The more pronounced effects are evident in the highest dose group. *P < 0.05.
FIG. 3.
Levels of the same ten ions at day 7 postirradiation. A few ions exhibit the phenotypic dose response even at the later time point. The higher dose group shows the most significant differences, as was evident through the MDS plots as well. *P < 0.05; **P < 0.005.
Finally, the sensitivity and specificity of each of the metabolites and ions were assessed through construction of ROC curves and calculation of the AUC and P values associated with each one. An AUC value of ≥0.9 signifies an excellent marker, while the accuracy decreases with decreasing AUC values. The results are shown in Table 1. Boldface values with superscript a represent good or excellent markers that are also statistically significant. These markers can easily distinguish the higher dose from the rest in both day 1 and 7. However, while the lower doses do have significant markers associated with them, the responses are not as prevalent as in the 8 Gy irradiated group.
TABLE 1.
Receiver Operating Characteristic (ROC) Curve Results (Irradiated vs. Sham)
| Day 1
|
Day 7
|
|||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| 0.5 Gy | 3 Gy | 8 Gy | 0.5 Gy | 3 Gy | 8 Gy | |
| 3-Oxodecanoic acid | 0.76 | 0.52 | 0.92a | 0.64 | 0.8 | 0.72 |
| L-phenylalanine | 0.5 | 0.92a | 0.74 | 0.56 | 0.68 | 0.8 |
| dAMP | 0.6 | 0.92a | 1a | 0.6 | 0.56 | 1a |
| L-histidine | 0.56 | 0.68 | 0.96a | 0.72 | 0.72 | 0.72 |
| L-proline | 0.96a | 0.8 | 0.76 | 0.5 | 0.7 | 1a |
| Niacinamide | 0.68 | 0.84 | 0.92a | 0.6 | 0.56 | 0.8 |
| Nicotinic acid | 0.75 | 0.7 | 1a | 0.875 | 0.6875 | 0.875 |
| Dodecanedioic acid | 0.88a | 0.72 | 0.92a | 0.8 | 0.68 | 1a |
| 204.9896_6.38 | 0.56 | 0.85 | 0.84 | 0.88a | 0.88a | 1a |
| 229.0112_0.27 | 0.9a | 0.7 | 0.9 | 0.8 | 0.72 | 0.8 |
Significance considered at P < 0.05.
DISCUSSION
Efforts to utilize metabolomics for rapid biodosimetry of external exposure in easily accessible biofluids have been concentrated primarily in urine and blood. Here, we investigated saliva as a potential candidate and identified a number of metabolites and ions that can be used in the development of a biodosimetric radiation signature. To date, urinary and blood metabolomic studies have been limited in their ability to discern tissue-specific biomarkers. On the other hand, saliva metabolomics may be a better representative due to locally affected tissues (12, 13). To the best of our knowledge, this is the first study to investigate salivary radiation metabolomics, although different salivary parameters, such as electrolytes and amylase, have been investigated by others (13, 19). Salivary biomarkers have successfully discriminated patients with oral cancer, periodontal disease, breast cancer and pancreatic cancer from healthy controls (20). Other studies have been focused on saliva for autoimmune diseases and even dementia (21, 22). It is clear that this biofluid is an attractive candidate for use in screening and diagnostic purposes.
The overall metabolic differences between the irradiated and sham-irradiated groups in our study were very striking at day 1 postirradiation (see Fig. 1A). However, as expected only the higher dose, 8 Gy irradiated group maintained a consistently different metabolic fingerprint at day 7. As 8 Gy is considered a semi-lethal dose for male C57BL/6 mice (the LD50/30 dose in our studies) and an equivalent dose in humans can lead to acute radiation syndrome, it is of importance to investigate these effects at this dose in further detail. In a real-life scenario, a dose of >2 Gy to humans is currently considered the planned threshold for administration of cytokine therapy (3, 23). As our results demonstrated, a considerable number of metabolites show sensitivity and specificity that can distinguish individuals requiring immediate medical intervention.
Metabolites that were identified in this study belong primarily to nicotinate and nicotinamide metabolism, are amino acids and fatty acids, and also include one deoxyribonucleotide. Although products of direct DNA damage were not the predominant biomarkers, the inclusion of products associated with nicotinamide adenine dinucleotide (NAD) formation and general amino acid inclusion are suggestive of increased cell death, which is a characteristic after irradiation of the oral cavity (12). While it could be argued that the presence of these biomarkers can be attributed to diet, the results demonstrate a link with radiation exposure. A number of markers, such as dAMP, nicotinamide and 3-oxodecanoic acid are not present in the formulation of the chow, while others constitute a very low percentage of the ingredients. In addition, the intragroup variability is relatively low for the majority of the biomarkers, which is further minimized at day 7, making it unlikely that the levels are correlating with food intake. Future studies should consider the withholding of chow for 24 h postirradiation, although the effect of starvation in itself could further complicate metabolic processes. Regardless, this is a possibility in a real-life scenario and it should be investigated in further detail. Finally, it remains to be determined what the contribution of the oral microbiome is towards the radiation signature. Although some biomarkers could be mammalian specific, others may be more closely related to bacteria.
As we have demonstrated for the first time that saliva can be used for biomarker determination associated with external radiation exposure, we plan to expand its utility in assessing not only a larger radiation dose range, but also differences in radiation quality. Finally, while this proof-of-concept study in mice did provide valuable information on the utility of saliva as a biofluid for biodosimetry, the radiation signature should be further assessed in nonhuman primates and the human population, as has been done previously with urine (5, 8, 24).
Acknowledgments
This work was funded by the National Institutes of Health/National Institute of Allergy and Infectious Diseases, grant nos. U19AI067773 (DJB) and 1R01AI101798 (AJF), and the NIH/NCI grant no. P30 CA051008 (Louis Weiner). The content of the article is solely the responsibility of the authors and does not necessarily represent the official views of the NCI and NIH.
References
- 1.Coy SL, Cheema AK, Tyburski JB, Laiakis EC, Collins SP, Fornace A. Radiation metabolomics and its potential in biodosimetry. Int J Radiat Biol. 2011;87:802–23. doi: 10.3109/09553002.2011.556177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rios CI, Cassatt DR, Dicarlo AL, Macchiarini F, Ramakrishnan N, Norman MK, et al. Building the strategic national stockpile through the NIAID Radiation Nuclear Countermeasures Program. Drug Dev Res. 2014;75:23–8. doi: 10.1002/ddr.21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan JM, Prasanna PG, Grace MB, Wathen LK, Wallace RL, Koerner JF, et al. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 2013;105:540–54. doi: 10.1097/HP.0b013e31829cf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goudarzi M, Mak TD, Chen C, Smilenov LB, Brenner DJ, Fornace AJ. The effect of low dose rate on metabolomic response to radiation in mice. Radiat Environ Biophys. 2014;53:645–57. doi: 10.1007/s00411-014-0558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laiakis EC, Mak TD, Anizan S, Amundson SA, Barker CA, Wolden SL, et al. Development of a metabolomic radiation signature in urine from patients undergoing total body irradiation. Radiat Res. 2014;181:350–61. doi: 10.1667/RR13567.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laiakis EC, Strassburg K, Bogumil R, Lai S, Vreeken RJ, Hankemeier T, et al. Metabolic phenotyping reveals a lipid mediator response to ionizing radiation. J Proteome Res. 2014;13:4143–54. doi: 10.1021/pr5005295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laiakis EC, Pannkuk EL, Diaz-Rubio ME, Wang YW, Mak TD, Simbulan-Rosenthal CM, et al. Implications of genotypic differences in the generation of a urinary metabolomics radiation signature. Mutat Res. 2016 doi: 10.1016/j.mrfmmm.2016.03.003. (Epub ahead of print.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pannkuk EL, Laiakis EC, Authier S, Wong K, Fornace AJ. Global metabolomic identification of long-term dose-dependent urinary biomarkers in nonhuman primates exposed to ionizing radiation. Radiat Res. 2015;184:121–33. doi: 10.1667/rr14091.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones JW, Tudor G, Li F, Tong Y, Katz B, Farese AM, et al. Citrulline as a biomarker in the murine total-body irradiation model: correlation of circulating and tissue citrulline to small intestine epithelial histopathology. Health Phys. 2015;109:452–65. doi: 10.1097/HP.0000000000000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mak TD, Tyburski JB, Krausz KW, Kalinich JF, Gonzalez FJ, Fornace AJ. Exposure to ionizing radiation reveals global dose-and time-dependent changes in the urinary metabolome of rat. Metabolomics. 2015;11:1082–94. doi: 10.1007/s11306-014-0765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menon SS, Uppal M, Randhawa S, Cheema MS, Aghdam N, Usala RL, et al. Radiation metabolomics: current status and future directions. Front Oncol. 2016;6:20. doi: 10.3389/fonc.2016.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grundmann O, Mitchell GC, Limesand KH. Sensitivity of salivary glands to radiation: from animal models to therapies. J Dent Res. 2009;88:894–903. doi: 10.1177/0022034509343143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pernot E, Cardis E, Badie C. Usefulness of saliva samples for biomarker studies in radiation research. Cancer Epidemiol Biomarkers Prev. 2014;23:2673–80. doi: 10.1158/1055-9965.EPI-14-0588. [DOI] [PubMed] [Google Scholar]
- 14.Mak TD, Laiakis EC, Goudarzi M, Fornace AJ. MetaboLyzer: a novel statistical workflow for analyzing postprocessed LC-MS metabolomics data. Anal Chem. 2014;86:506–13. doi: 10.1021/ac402477z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3. 0–The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 19.Soni S, Agrawal P, Kumar N, Mittal G, Nishad DK, Chaudhury NK, et al. Salivary biochemical markers as potential acute toxicity parameters for acute radiation injury: a study on small experimental animals. Hum Exp Toxicol. 2016;35:221–8. doi: 10.1177/0960327115579433. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Sun J, Lin C-C, Abemayor E, Wang MB, Wong DT. The emerging landscape of salivary diagnostics. Oral Health Dent Manag. 2014;13:200–10. [PubMed] [Google Scholar]
- 21.Mikkonen JJW, Herrala M, Soininen P, Lappalainen R, Tjäderhane L, Seitsalo H, et al. Metabolic profiling of saliva in patients with primary Sjogren’s syndrome. Metabolomics: Open Access. 2013;3:128. ( http://bit.ly/1qJiTZ5) [Google Scholar]
- 22.Tsuruoka M, Hara J, Hirayama A, Sugimoto M, Soga T, Shankle WR, et al. Capillary electrophoresis mass spectrometry based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis. 2013;34:2865–72. doi: 10.1002/elps.201300019. [DOI] [PubMed] [Google Scholar]
- 23.Dicarlo AL, Maher C, Hick JL, Hanfling D, Dainiak N, Chao N, et al. Radiation injury after a nuclear detonation: medical consequences and the need for scarce resources allocation. Disaster Med Public Health Prep. 2011;5:S32–44. doi: 10.1001/dmp.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson CH, Patterson AD, Krausz KW, Kalinich JF, Tyburski JB, Kang DW, et al. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat Res. 2012;178:328–40. doi: 10.1667/rr2950.1. [DOI] [PMC free article] [PubMed] [Google Scholar]