Summary
A gradual restriction in lineage potential of multipotent stem/progenitor cells is a hallmark of adult hematopoiesis, but the underlying molecular events governing these processes remain incompletely understood. Here, we identified robust expression of the leukemia-associated transcription factor hepatic leukemia factor (Hlf) in normal multipotent hematopoietic progenitors, which was rapidly downregulated upon differentiation. Interference with its normal downregulation revealed Hlf as a strong negative regulator of lymphoid development, while remaining compatible with myeloid fates. Reciprocally, we observed rapid lymphoid commitment upon reduced Hlf activity. The arising phenotypes resulted from Hlf binding to active enhancers of myeloid-competent cells, transcriptional induction of myeloid, and ablation of lymphoid gene programs, with Hlf induction of nuclear factor I C (Nfic) as a functionally relevant target gene. Thereby, our studies establish Hlf as a key regulator of the earliest lineage-commitment events at the transition from multipotency to lineage-restricted progeny, with implications for both normal and malignant hematopoiesis.
Keywords: hematopoiesis, lineage commitment, gene regulation, transcription factor, lymphopoiesis, myelopoiesis
Graphical Abstract
Highlights
-
•
During hematopoiesis, Hlf is sharply downregulated upon exit from multipotency
-
•
Prolonged Hlf expression instructs GMLPs to adopt myeloid fates
-
•
Failure to downregulate Hlf is incompatible with appropriate B and T lymphopoiesis
-
•
Hlf governs molecular programs driving myelopoiesis and inhibiting lymphopoiesis
Regulators of early blood cell formation are important in both health and disease. Wahlestedt et al. identify abrupt downregulation of the transcription factor Hlf during hematopoietic differentiation. Failure to downregulate Hlf leads to a drastically skewed output of mature blood cells, positioning Hlf as a critical regulator of hematopoiesis.
Introduction
Blood cell formation is often viewed in a hierarchical manner, in which hematopoietic stem cells (HSCs) reside at the apex of the hematopoietic hierarchy (Figure 1A). Upon differentiation, HSCs produce intermediate progenitors with increasingly restricted lineage potential (Figure 1A). Although the extensive self-renewal that characterizes HSCs is lost in the initial stages of HSC differentiation, most existing data suggest that multipotency is retained. Thereafter, progenitors gradually lose multipotency through a branched differentiation scheme. Eventually, progenitors become locked to only one lineage, after which lineage-specific maturation events commence (Bryder et al., 2006, Mercer et al., 2011).
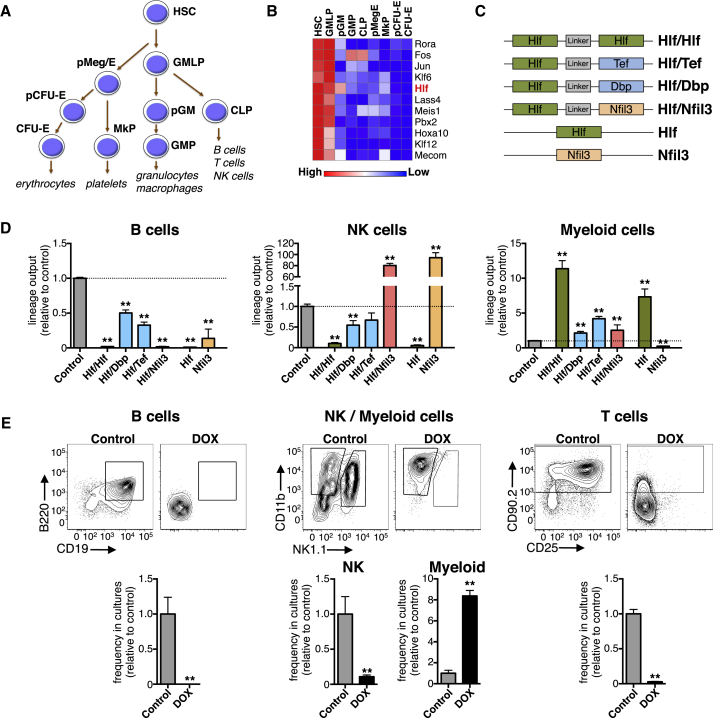
Figure 1.
Identification of Hlf as a Candidate Regulator of Early Hematopoiesis
(A) Schematic depiction of the progenitor subsets that underlie the early stages of hematopoiesis.
(B) Heatmap showing the expression levels of TFs displaying 2-fold or higher expression in HSCs and GMLPs compared with downstream progenitor cell types (mean values of three replicate arrays per cell type, except for HSCs and pMegEs, for which six and five replicate arrays were used, respectively). See also Table S1.
(C) Schematic depiction of the different lentiviral constructs used for OP9 stromal co-cultures.
(D) The lineage output relative to controls of B cells, NK cells, and myeloid cells produced from Hlf/Hlf, Hlf/Dbp, Hlf/Tef, Hlf/Nfil3, Hlf, Nfil3, and M33-Hlf transduced GMLPs in OP9 stroma co-cultures (n = 6, 6, 6, 6, 6, 3, 6, and 45 cultures, respectively, from two independent experiments).
(E) B, NK, and myeloid cell generation in OP9 co-culture experiments and T cell generation in OP9-DL1 co-cultures using Hlf-inducible GMLPs cultured in the absence or presence of DOX. Shown are representative FACS plots as well as bar graphs showing the frequency relative to untreated controls of CD19+ B cells, NK1.1+ NK cells, CD11b+ myeloid cells, and CD90+ T cells in the cultures from three independent experiments (n = 5 for the B, NK, and myeloid experiments, and n = 9 for the T cell experiments).
Error bars denote SEM. CFU-erythroid, colony-forming unit-erythroid; MkP, megakaryocyte progenitor; pCFU-erythroid, pre-colony-forming unit-erythroid; pGM, pre-granulocyte-monocyte progenitor; pMeg/erythroid, pre-megakaryocyte-erythroid progenitor. See also Figures S1 and S2.
The lymphoid lineages (B, T and natural killer [NK] cells) branch from a common lymphoid progenitor (CLP) (Kondo et al., 1997). The generation of CLPs from upstream multipotent progenitors involves upregulation of lymphoid specification factors, such as Tcf3, Gfi-1, and Ikaros (Mercer et al., 2011). CLPs give rise to committed B cell progenitors, a process guided by a network of transcription factors (TFs) that includes Foxo1, Ebf1, and Pax5 (Mercer et al., 2011). Following initial maturation in the bone marrow (BM), immature B cells home to secondary hematopoietic organs for further maturation (Hardy and Hayakawa, 2001). T cells represent the other major lineage of lymphopoiesis. Although a matter of dispute, much data support that the earliest T cell progenitors (ETPs) can be generated from CLPs, although alternative parental progenitors might also exist (Allman et al., 2003, Benz et al., 2008, Kondo et al., 1997, Yang et al., 2010). Regardless, alternative lineage options of ETPs can be overridden by Notch signaling in the thymus, which acts by locking cells to the T cell lineage (Yang et al., 2010). In the thymus, ETPs mature through a series of immature progenitor stages (DN1–DN4) to eventually give rise to more mature T cells that are exported from the thymus (Germain, 2002). Myelopoiesis (giving rise to granulocytes, macrophages, erythroid cells, and platelets) represents the other dominant branch of hematopoiesis. Work from others and us has provided insight into the first stages of specification for these lineages (Akashi et al., 2000, Pronk et al., 2007). For instance, granulocyte/monocyte/lymphoid progenitors (GMLPs) retain a combined lymphoid and granulocyte/monocyte potential but harbor little or no erythroid (E) or megakaryocytic (Meg) potential (Adolfsson et al., 2005, Arinobu et al., 2007). Instead, the Meg and erythroid lineages are generated from a common pre-MegE progenitor, whose closest upstream progenitors remain a matter of debate (Adolfsson et al., 2005, Arinobu et al., 2007, Forsberg et al., 2006, Pronk et al., 2007, Sanjuan-Pla et al., 2013, Yamamoto et al., 2013).
Previously, we established the genome-wide expression patterns of highly purified HSCs and a range of defined hematopoietic progenitor subsets (Norddahl et al., 2011, Pronk et al., 2007, Pronk et al., 2008, Wahlestedt et al., 2013, Weishaupt et al., 2010; Figure 1A). When interrogating these datasets for expression of TFs that associate with multipotency and rapid downregulation with differentiation (Figures 1B, S1, and S2), we identified hepatic leukemia factor (Hlf), a TF of the proline and acidic amino acid-rich basic leucine zipper family, previously studied for its role in circadian rhythm regulated neurotransmitter metabolism in the brain (Gachon et al., 2004, Mitsui et al., 2001), xenobiotic detoxification in the liver (Gachon et al., 2006), and renal function (Zuber et al., 2009). Although Hlf was originally proposed to not be expressed in BM (Inukai et al., 2005), later studies found its promoter differentially methylated in between multipotent progenitor cells and more committed cells (Ji et al., 2010), with enforced expression of Hlf linked to enhanced self-renewal (Gazit et al., 2013, Shojaei et al., 2005). More recently, Hlf was identified as a necessary factor to revert committed blood cells back into an HSC-like state (Riddell et al., 2014). Hlf has perhaps a more well established connection to leukemia as the fusion partner of TCF3/E2A resulting from the t(17;19) translocation, a rare but recurrent event associated with a highly aggressive subset of acute B-lymphoblastic leukemia (Fischer et al., 2015, Hunger et al., 1992, Inaba et al., 1992). However, little is known on how Hlf might influence on normal hematopoietic differentiation. Therefore, we set out to characterize the role of Hlf in normal hematopoietic lineage development.
Results
Prolonged Hlf Expression Critically Influences Multilineage Hematopoietic Differentiation
To begin to dissect the impact of Hlf on hematopoietic development, we first generated lentiviral constructs that included a single-copy Hlf overexpression vector and a construct in which Hlf is expressed as a forced homodimer (Hlf/Hlf; Figure 1C). We chose this complementary approach because HLH TFs generally target DNA as dimers. Although expression data suggested that Hlf is the only PAR bZIP family member with selectively high expression in immature blood cells, Hlf may affect early hematopoiesis by heterodimerizing with any of its other family members upon differentiation. We therefore also generated Hlf/Dbp, Hlf/Nfil3, and Hlf/Tef forced dimer constructs (Figure 1C) along with an Nfil3 overexpression vector, the latter because Nfil3 has been reported to act antagonistically to Hlf (Mitsui et al., 2001).
After confirmation of relevant Hlf expression induced by the different viral constructs (Figure S3A), transduced multipotent GMLPs were assessed using the OP9 stromal cell system (Nakano et al., 1994). These experiments revealed that Hlf and Hlf/Hlf overexpressing GMLPs displayed a strikingly reduced capacity to differentiate into CD19+ B cells and NK1.1+ NK cells, but with enhanced generation of Gr-1+/CD11b+ myeloid (Figure 1D). Overexpression of Nfil3 led to almost exclusive differentiation into NK cells (Figure 1D), reinforcing the importance of Nfil3 for NK cell development (Kamizono et al., 2009). A similar increase in NK cell development was observed from cells transduced with the Hlf/Nfil3 heterodimer construct (Figure 1D), suggesting that Nfil3 acts dominantly over Hlf. Finally, Hlf/Dbp and Hlf/Tef presented with an intermediate reduction in B and NK cell differentiation and an increase in myelopoiesis (Figure 1D).
To obtain a more controllable transgenic system, we next established a Tet-ON mouse model that upon doxycycline (DOX) administration allows conditional expression of Hlf (Figure S3B). Although systemic administration of DOX in these mice results in ubiquitous induction of transgene expression, its combination with cell isolation allows inducible expression of Hlf in any cell type of choice. To verify this model, we isolated Hlf-inducible GMLPs and subjected them to the OP9 co-culture system. In addition, we also investigated T lymphopoiesis by culturing transgenic GMLPs on OP9-DL1 stromal cells (Schmitt and Zúñiga-Pflücker, 2002). These experiments confirmed our previous findings using lentiviral-mediated transgenesis (Figure 1E), and OP9-DL1 co-culture experiments revealed that a failure to downregulate Hlf also inhibited T cell development (Figure 1E).
In Vivo Induction of Hlf Associates with Enhanced Myelopoiesis and Repressed Lymphopoiesis
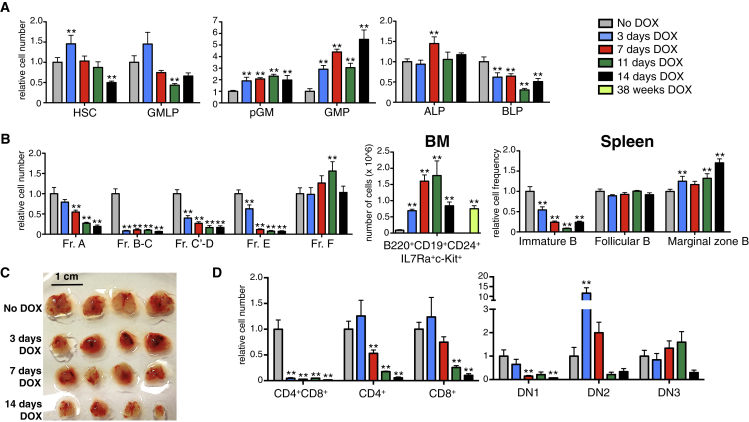
To investigate the roles of Hlf in vivo, we provided Hlf-inducible mice with DOX via their food. Analyses of BM from these mice revealed a slight but significant reduction in overall cellularity as early as 3 days following Hlf induction (Figure S4A). To investigate hematopoiesis in detail, we enumerated multipotent cell compartments (HSCs and GMLPs), early myeloid precursors, and different stages of B cell development (Figure S4B and Table S1). HSC numbers in the BM of Hlf-induced mice were mildly increased at 3 days and decreased at 14 days of Hlf induction (Figure 2A). GMLPs were unaltered at all time points except following 11 days of Hlf induction, when a slight decrease in their numbers was observed (Figure 2A). By contrast, we observed a robust and progressive increase of pre-granulocyte-monocyte progenitors (pGMs) and granulocyte-monocyte progenitors (GMPs) over time (Figure 2A). Investigations into early lymphoid development in the BM revealed a slightly decreased abundance of B cell-biased lymphoid progenitors (BLPs) (Inlay et al., 2009) (Figure 2A). A more dramatic depletion was observed in downstream B lymphocyte precursor populations, including Hardy’s fractions A, B-C, C′-D, and E (Hardy et al., 1991, Tung et al., 2004) (Figure 2B). By contrast, fraction F cells (mature recirculating B cells) (Hardy et al., 1991, Tung et al., 2004) were less affected (Figure 2B). These observations were accompanied by marked increases of a BM cell population with a Lin−B220+CD19+CD24+IgM−IgD−c-Kit+IL7Ra+ phenotype (Figure 2B). Retention of CD19 expression in vitro when cultured on both OP9 and OP9-DL1 stroma (data not shown), expression of several B cell-associated genes at levels comparable with fraction B-C cells, varying degrees of DJ and VDJ heavy-chain rearrangements, and their cell surface marker profile strongly suggested that they indeed represented a subset of early B cell progenitors (Figure S5). When Hlf was induced in fraction B-C cells for 48 hr, a large fraction of the cells (31.5 ± 8.1%, as opposed to 10.4 ± 3.6% of control cells) upregulated c-kit expression (Figure S4E), further emphasizing that the differentiation block in the B cell lineage caused by Hlf associates with a rapid induction of c-kit expression. In the spleen, the frequency of immature B cells was progressively decreased upon Hlf induction, whereas mature follicular B cells and marginal zone B cells were less affected (Figure 2B). The negative impact of Hlf on B lymphopoiesis therefore starts early and affects multiple progenitor stages, with little or no impact on more mature B cells.
Figure 2.
Hlf Induction In Vivo Negatively Influences Lymphopoiesis at the Expense of Enhanced Myelopoiesis
Hlf-inducible mice were given DOX via their food pellets for 0, 3, 7, 11, and 14 days (n = 7, 7, 7, 3, and 4 mice in each group, respectively, from two independent experiments) and 38 weeks (n = 5 mice, from one experiment).
(A) Bar charts showing the amount of HSCs, GMLPs, pGMs, GMPs, all lymphoid progenitors (ALPs), and B cell-biased lymphoid progenitors (BLPs) in the BM of the analyzed mice (relative to uninduced mice).
(B) Relative cell numbers of the analyzed B cell subsets in the BM and relative frequencies of the indicated splenic B cell fractions among all splenocytes in the analyzed mice (relative to uninduced mice). See also Figure S5.
(C) Photographs depicting thymi after 0, 3, 7, 11, and 14 days of enforced Hlf expression (four thymi per time point, representative of one of three experiments). The scale bar represents 1 cm.
(D) The amount of CD4+CD8+ double-positive, single-positive CD4+, single-positive CD8+ thymocytes, and DN1, DN2, and DN3 thymocytes following the different number of days of DOX administration (relative to uninduced mice). See also Table S1.
Error bars denote SEM. ALP, all lymphoid progenitor; BM, bone marrow; BLP, B cell biased lymphoid progenitor; DN, double negative. See also Figures S3 and S4.
We next asked whether Hlf might affect T cell development in vivo. Surprisingly, as early as 3 days after initiation of Hlf expression, we observed pronounced thymic atrophy that progressed rapidly (Figure 2C). Reasoning that this might be connected to thymic T cell development, we enumerated different T cell compartments (Figure S4C). After 3 days, CD4+ and CD8+ single-positive T cells were present at normal numbers, whereas CD4+CD8+ double-positive T cell numbers were dramatically decreased (Figure 2D). Analysis of CD4+CD8+ cells following brief Hlf induction in vitro revealed a massive induction of apoptosis (Figure S4D). Upon longer Hlf induction, the decrease in CD4+CD8+ cells persisted and single-positive subsets gradually decreased in numbers, such that by day 14, levels were only 5.7% (CD4+) and 10.2% (CD8+) of those observed in control mice (Figure 2D). When investigating more primitive T cell fractions, we observed a pronounced decrease in double-negative (DN) 1 cells (Figure 2D) from day 7 onward. DN2 cells were greatly expanded following 3 days of induction (∼7-fold; Figure 2D). However, this was attenuated 4 days later, and at 11 and 14 days, their levels displayed a decreasing trend compared with control mice (Figure 2D). Last, DN3 cells were present at normal numbers up to 11 days following Hlf induction but decreased by day 14 and at later time points (Figure 2D and data not shown).
Hlf Acts Intrinsically on Hematopoiesis
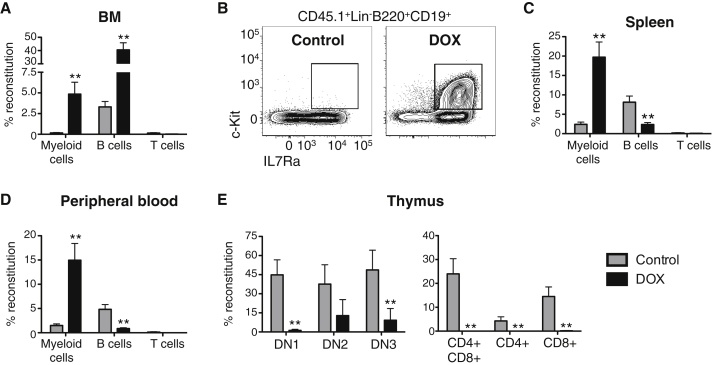
To exclude the possibility that Hlf mediates an indirect effect, we next transplanted Hlf-inducible GMLPs into sublethally irradiated wild-type (WT) mice under continuous Hlf induction. Twenty-one days post-transplantation, the Hlf-induced cells presented with a dramatic (28-fold) increase in myeloid cell reconstitution in the BM (Figure 3A). Somewhat surprisingly, we observed that CD19+ BM B cell reconstitution was increased 12-fold over the control (Figure 3A). More detailed examination revealed that these cells were almost exclusively restricted to the aforementioned population with a Lin−B220+CD19+c-Kit+IL7Ra+ phenotype (Figure 3B). Despite the dramatic expansion of this subset in the BM, B cell reconstitution was dramatically reduced in the spleen (3.4-fold) (Figure 3C) and in peripheral blood (PB) (5.7-fold) (Figure 3D). In agreement with the increased myeloid reconstitution in the BM, donor-derived myeloid cell abundance was increased 8.3-fold and 10.1-fold in the spleen and PB, respectively (Figures 3C and 3D). In the thymus, Hlf induction led to a total absence of donor-derived CD4+, CD8+, or CD4+CD8+ cells (Figure 3E). Although reconstitution of the more primitive DN1–DN3 cells was significantly lower than from control cells, their presence in induced mice suggested that initial homing to the thymus was at least to some degree functional (Figure 3E).
Figure 3.
Cell-Autonomous Alterations Underlie the Altered Hematopoiesis Following Enforced Hlf Expression In Vivo
Hlf-inducible GMLPs (CD45.1+) were transplanted into sublethally irradiated WT recipient hosts (CD45.2+) maintained on a DOX food diet (n = 6 and 7 mice for the control and DOX groups, respectively, from one of three experiments with similar results).
(A) Magnitudes of myeloid B, and T cell reconstitution in the BM of the recipient mice.
(B) FACS plots showing the appearance of CD45.1+c-Kit+IL7Ra+CD19+ cells in the BM of mice transplanted with Hlf-induced GMLPs.
(C and D) Donor-derived reconstitution of myeloid, B, and T cells in the spleen (C) and the PB (D).
(E) The thymi of recipient mice were assessed for donor-derived reconstitution of the indicated thymocyte subsets.
Error bars denote SEM. See also Figure S5.
Acute Loss and the Modulation of Hlf into a Transcriptional Repressor Enhance Lymphopoiesis from Multipotent Progenitors
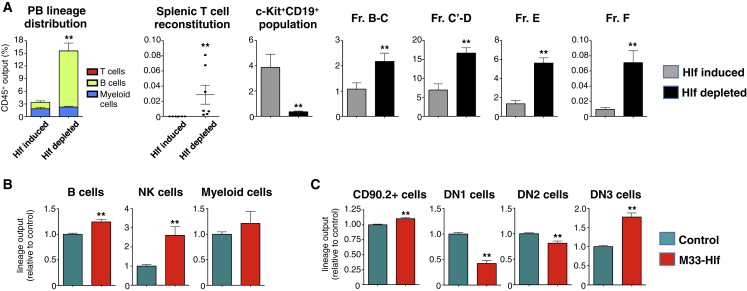
Next, we approached the roles of Hlf from a loss-of-function perspective. Because Hlf is rapidly downregulated upon normal differentiation, traditional knockout experiments provide little information. Therefore, we first transplanted mice with Hlf transgenic GMLPs under Hlf induction for 14 days. This was followed by removal of Hlf induction for 8 days to reduce Hlf levels. Compared with cells with continuous Hlf expression, Hlf depletion resulted in a rapid shift in PB lineage distribution, owing to an 8.7-fold increase in B cells (Figure 4A). Similarly, although continuous Hlf expression failed to associate with splenic T cell reconstitution, we detected low but consistent T cell generation following Hlf withdrawal (Figure 4A). Investigations of the BM revealed that the otherwise vastly expanded Lin−B220+CD19+c-Kit+IL7Ra+ population had greatly decreased in size (10.9-fold) following Hlf depletion, which coincided with strikingly elevated numbers at all stages of early B lymphopoiesis. Thus, the accumulation of this population represents a differentiation block that is released following DOX withdrawal (Figure 4A). In this setting, we failed to observe a decrease in myeloid cell output upon DOX withdrawal, which likely reflects the limited self-renewal potential of GMLPs and the fact that myeloid lineage commitment has already commenced during the induction period.
Figure 4.
Acute Hlf Depletion and Modulation of Hlf into a Transcriptional Repressor Lead to Enhanced Lymphopoiesis
Hlf-inducible GMLPs (CD45.1+) were transplanted into sublethally irradiated WT recipients (CD45.2+) on DOX food. After 14 days, one group of mice was shifted to normal food and kept for another 8 days prior to analysis (“Hlf depleted”), while control mice were treated with DOX throughout the experiment (“Hlf induced,” n = 7 and 6 mice, respectively, from one experiment).
(A) Data show the percentage of the indicated donor-derived cell types out of all CD45+ cells.
(B and C) The impact of Hlf target gene silencing on differentiation was studied by culturing control or M33-Hlf retrovirus infected WT GMLPs in B/NK and T cell permissive conditions (n = 6 replicates per group from two experiments). Shown is the lineage output relative to controls of indicated subsets in OP9 cultures (B) and OP9-DL1 cultures (C), respectively.
Error bars denote SEM.
We next generated an M33-Hlf construct, in which the strong repressive domain of the polycomb gene M33 (Cbx2) is predicted to turn Hlf from a transcriptional activator into a repressor (Argiropoulos et al., 2010). We reasoned that such an approach was highly relevant, given that Hlf associated mostly with transcriptionally active genes (see below). We introduced the M33-Hlf construct into WT GMLPs and subjected cells to OP9 and OP9-DL1 differentiation conditions. In OP9 cultures, expression of M33-Hlf led to an increased production of B cells and NK cells, with little influence on myeloid cell generation (Figure 4B). In OP9-DL1 co-cultures, the overall T cell production from M33-Hlf expressing GMLPs was enhanced, mainly because of an increase of DN3 cells at the expense of DN1 and DN2 cells (Figure 4C).
The Genome-wide Binding Characteristics of Hlf
We were next interested in identifying the genomic regions occupied by Hlf. For this, we generated a retroviral vector expressing a FLAG-tagged variant of Hlf and used this to infect Lin−Sca-1+c-Kit+ (LSK) cells. Although HSCs are included in the LSK fraction, the majority of cells in this population (>95%) represent multipotent progenitors (Bryder et al., 2006). Therefore, most observed binding represents Hlf activity in multipotent progenitors. Following 5 days of expansion, the infected cells were next used for chromatin immunoprecipitation sequencing (ChIP-seq) analysis using anti-FLAG or control antibodies. This revealed Hlf peaks scattered around different genomic regions; 7.4% of the sites were located at promoters, 1.8% in UTR regions, 45% at intragenic sites, and 45.8% in intergenic regions (Figure S6A). Investigations into a putative underlying Hlf-binding motif of the peaks revealed a predicted motif closely resembling that reported for human HLF (Inaba et al., 1994) (Figure S6B). Overrepresented gene ontology categories and mouse phenotypes on the basis of the predicted Hlf-binding peaks associated with blood cell development or immune system processes, further supporting the notion of Hlf as a regulator of blood cell formation (Figure S6C).
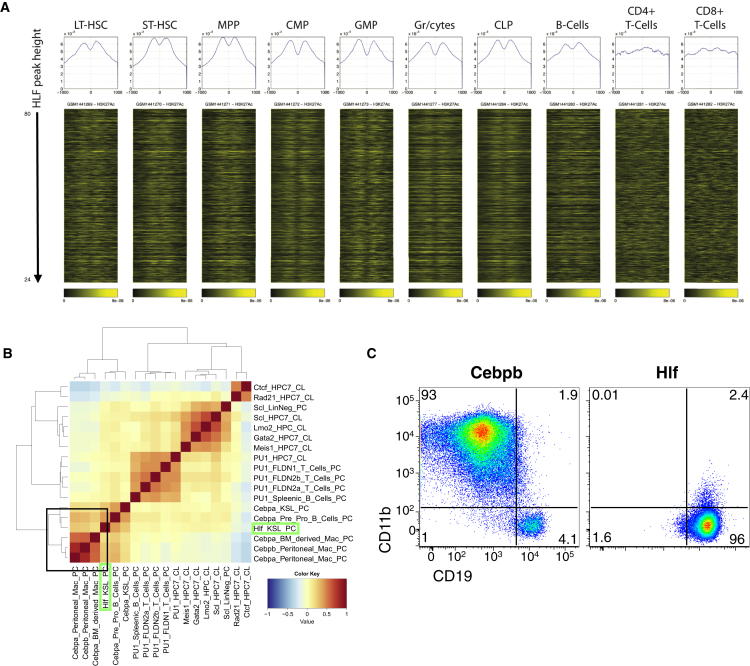
To investigate whether the intergenic peaks might overlap with known enhancer regions, we intersected them with enhancer regions recently identified in a variety of hematopoietic cell types on the basis of H3K27Ac occupancy (Lara-Astiaso et al., 2014) (Figure 5A). The elements bound by Hlf associated with a random distribution of H3K27Ac in mature B and T cells. In contrast, in long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), multipotent progenitor (MPP), common myeloid progenitor (CMP), GMP, mature granulocytes, and CLPs, Hlf peaks correlated with highly acetylated H3K27Ac positioned on either side of the Hlf peak summit, suggesting that Hlf may affect hematopoietic development by regulating transcription also via enhancer elements (Figure 5A). Further supporting this was the observation that identified binding motifs of several other well-known hematopoietic TFs, including c/EBP-like, Ets, Runx1, and AP1, co-associated with the identified Hlf peak areas (Figure S6D).
Figure 5.
The Genome-wide Binding Characteristics of Hlf
WT LSK cells were transduced with either a control retroviral vector or a vector encoding an N-terminal FLAG-tagged variant of Hlf/Hlf and were subjected to ChIP-seq analysis.
(A) The Hlf peaks were ranked in a decreasing peak height order, and H3K27Ac signal intensity (Lara-Astiaso et al., 2014) was plotted around the summit of the Hlf peaks and is shown as heatmaps. The histograms at the top of each heatmap depict the average signal intensity around the Hlf peaks for each cell type.
(B) Correlation analysis of the putative Hlf targets with those of other hematopoiesis associated TFs.
(C) BM resident B cells were subjected to transdifferentiation experiments by transduction with retroviral vectors encoding Cebpb or Hlf and cultured for 4 days on OP9 stromal cells prior to FACS analysis (representative FACS plots from two independent experiments).
ChIP, chromatin immunoprecipitation. See also Figure S6.
We thereafter compared the identified Hlf-bound regions with those of other hematopoietic TFs. This revealed a strong correlation between Hlf binding and c/EBP binding (Figure 5B), which has been proposed previously (Hunger et al., 1992), and suggested that Hlf might share c/EBP-binding sites during hematopoietic development. To functionally test a redundancy among these factors, we investigated if aberrant Hlf expression may lead to transdifferentiation of B cells into macrophages, as has previously been reported for C/EBP factors (Xie et al., 2004). However, although Cebpb efficiently transdifferentiated B cells into macrophages, Hlf was unable to do so (Figure 5C), demonstrating that although the underlying binding motif of Cebp and Hlf is similar, the functions of these factors are distinct.
Hlf Confers a Myeloid Fate on GMLPs that Is Accompanied by a Deregulation of Lymphoid and Myeloid Gene Expression Programs
Although our data suggested that Hlf acts as a strong lineage fate determinant in multipotent progenitors, it remained a possibility that one subset of GMLPs responded to Hlf by allowing enhanced myelopoiesis, while an alternative lymphoid-competent subset was selectively depleted. To investigate this, using fluorescence-activated cell sorting (FACS), we sorted individual Hlf-inducible GMLPs onto OP9 stromal cells under B and NK cell-promoting conditions and analyzed their progeny 14 days later. Forced Hlf induction significantly increased the cloning frequency (the ability of single-seeded GMLPs to promote growth of detectable progeny) by 50% (36.4% in the absence and 54.5% in the presence of DOX) (Figure S7A). WT GMLPs produced a variety of lineage outputs, in which lymphoid potential was dominant (Figure 6A). In stark contrast, forced Hlf expression caused most cells (96.5%) to produce only myeloid progeny (Figure 6A). These data strongly suggested that Hlf acts to divert lineage fate of multipotent GMLPs, rather than exercising differential effects on already committed progenitors. To investigate the genome-wide transcriptional consequences underlying this, we next conducted RNA sequencing (RNA-seq). We here used both viral-mediated overexpression in WT GMLPs and GMLPs from our inducible transgenic mouse strain (Figure S7B). We chose to use these complementary approaches to try to overcome limitations associated with each individual approach. By highly stringent analysis criteria, this combinatory approach revealed 43 and 41 up- and downregulated genes as a consequence of Hlf expression, respectively (Figure 6B; Table S2). In general, the most differentially expressed genes were the same with the two approaches (Figure 6C; Table S2). In addition, we found evidence for Hlf binding to 18 of the 43 upregulated genes, whereas only 4 of 41 downregulated genes presented as candidate Hlf targets (Figure 6C), perhaps suggesting that Hlf acts primarily to activate gene expression.
Figure 6.
Hlf Instructs GMLPs to Adopt a Myeloid Fate by Upregulation of a Myeloid-Associated Transcriptional Program and a Concomitant Downregulation of Lymphoid-Affiliated Genes
(A) Single Hlf-inducible GMLPs were maintained on OP9 stromal cells for 14 days. The lineage potential of each seeded GMLP was determined using FACS, by scoring each well for the presence of B cells, NK cells, and myeloid cells. Shown is the distribution of the different colony types among the positive wells of the control and DOX treated groups.
(B) Venn diagrams depicting the number of shared up- and downregulated genes following Hlf induction using the two complementary approaches (compared with controls).
(C) Up- and downregulated genes, their fold changes compared with controls using both experimental strategies, and whether they were identified also using ChIP-seq. See also Table S2.
(D) GSEA in which the commonly up- and downregulated genes were used as gene sets and correlated against the whole transcriptomes of pGMs and CLPs.
Error bars denote SEM. GSEA, gene set enrichment analysis. See also Figure S7.
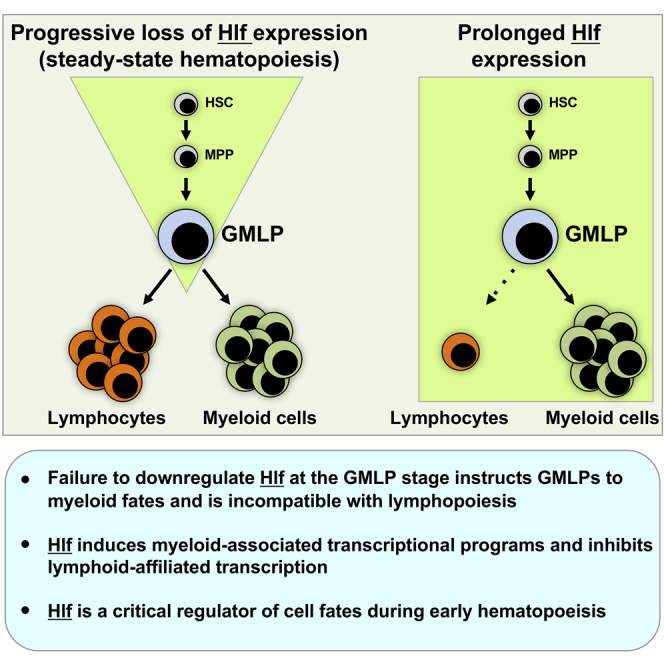
Next, we asked whether the up- and downregulated genes are part of larger transcriptional programs involved in early lympho- and myelopoiesis. Here, we used the up- and downregulated genes as gene sets for gene set enrichment analysis (GSEA) (Subramanian et al., 2005) and correlated the whole transcriptome of myeloid progenitors and CLPs against these genes (Figure 6D). Despite the very short induction period, this revealed striking associations of the upregulated genes with myeloid progenitors and downregulated genes to CLPs (Figure 6D), emphasizing Hlf downregulation as a critical requirement for early lymphoid development.
Identification of the TF Nfic as a Direct Target Gene of Hlf
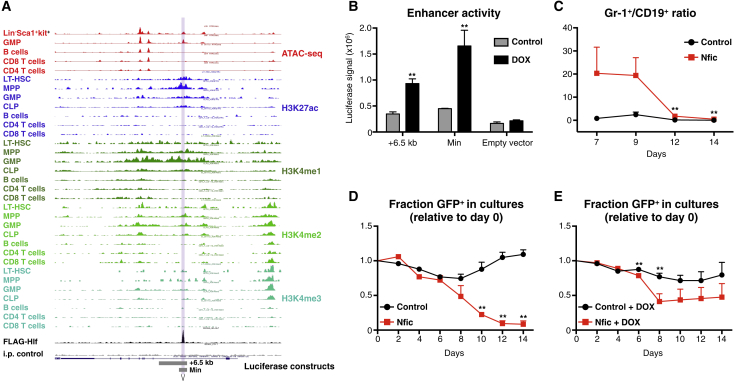
As a last part of our work, we explored whether we could identify any direct candidate target gene(s) of Hlf that might further detail its mechanism of action. For this, we generated retroviruses for 15 gene candidates selected on the basis of effect on transcription and chromatin binding (Figure 6C). Transduced cells were next assayed using the OP9 co-culture system. In preliminary analyses, the TF Nfic presented with both relative reductions in B cell and increases in myeloid output (Figure S7C). Closer interrogation of the chromatin environment around the Nfic-associated Hlf peak revealed a putative enhancer accessible in non-lymphoid cells (Figure 7A), and reporter assays in Hlf-inducible embryonic stem cells (ESCs) confirmed Hlf-dependent activation using both a larger domain covering the Hlf peak (+6.5 kb) and from a smaller domain (Min) restricted to encode the candidate Hlf-binding motifs (Figure 7B). However, although enforced Nfic expression had a strong and rapid negative effect on B cell output (Figure 7C), transgenic Nfic expression eventually also led to a reduction in myeloid cell output over time (Figure 7D), a feature not observed upon enforced Hlf expression alone (Figure 6A). Interestingly, when enforcing Hlf expression in the setting of Nfic overexpression, Hlf was able to counteract at least some of this reduction in cell output (Figure 7E), suggesting that the induction of Nfic alone cannot explain all the phenotypes observed upon persistent Hlf expression.
Figure 7.
Identification of Nfic as a Direct Hlf Target with Pronounced Functional Effects on Hematopoietic Differentiation
(A) UCSC Genome Browser track depicting the genomic context of Nfic with regard to accessible chromatin and histone modifications of indicated cell types (assay for transposase-accessible chromatin using sequencing [ATAC-seq] and histone modification data from Lara-Astiaso et al. (2014). The region occupied by Hlf is highlighted.
(B) Luciferase reporter assay of the candidate Nfic enhancer using Hlf-inducible ESCs. Bars denote the mean firefly luciferase signal (±SEM) after normalization against Renilla using the +6.5 kb, a minimal (Min) enhancer construct, or an empty reporter construct, with or without Hlf induction (triplicate cultures from one of three representative experiments).
(C and D) Frequency of myeloid and B cells generated as a consequence of enforced Nfic expression. Shown are the ratios of percentage GFP+ Gr1+ myeloid cells over GFP+CD19+ B cells for the indicated groups. Data are from one of three experiments with similar results (n = 6 cultures per group for all cultures except day 14 Nfic, for which five cultures were analyzed). Enforced Nfic expression leads to a general loss of hematopoietic cells over time. Hlf-inducible GMLPs were maintained in the absence (C) or presence (D) of DOX and were transduced with either a control (Migr1) or Nfic retroviral vector and were subsequently cultured on OP9 stroma cells with monitoring every 2 days for frequency of GFP+ cells (n = 1 replicate for days 0, 2, and 4 and n = 5 replicates for days 6, 8, 10, 12, and 14 for the groups in both C and D).
Error bars denote SEM. See also Figure S7.
Discussion
According to the prevailing view of hematopoiesis, appropriate blood cell formation is maintained in a hierarchical fashion and involves a range of progenitors with alternative differentiation and proliferation potentials. HSCs reside at the top of this hierarchy and have for decades been acknowledged as the primary cell type necessary for lifelong production of mature blood cells. Therefore, much focus has historically been aimed at isolation of near pure populations of HSCs. With this has come the realization that the HSC pool might not be as homogeneous as initially assumed. Rather, the pool of HSCs might be composed of clones with alternative blood-forming potentials in terms of both lineage potential and the magnitude of the mature cells they give rise to (Goodell et al., 2015). Such “lineage-biased” HSC clones have been sub-classified by the retrospective functional readout of single-cell transplanted mice and/or by differential marker expression prior to functional assessment. However, caveats of such work include the difficulty in distinguishing HSCs with declining differentiation and/or self-renewal potential from HSCs that are truly biased toward a given lineage. It is therefore conceivable that assessments of mechanisms governing multipotency are more easily approached by the study of cells that reside more proximal to the decision points to be studied, including the GMLPs, which was the main cell type studied here.
Our gene expression profiling revealed Hlf to be highly expressed in multipotent GMLPs. A failure to downregulate Hlf at this developmental stage instructed GMLPs to adopt a myeloid fate and was incompatible with B, T, and NK cell development. When artificially turning Hlf into a transcriptional repressor by its fusion to the polycomb member M33, we observed a reciprocal phenotype, despite the otherwise highly lymphoid-permissive conditions that associates with the OP9 and OP9-DL1 differentiation systems. The failure to downregulate Hlf in GMLPs coincided with repression of lymphoid-affiliated genes and the concomitant induction of myeloid-associated genes, with many of the latter identified as candidate Hlf targets also by ChIP-seq.
An unexpected but to us highly interesting finding, given the involvement of Hlf in E2A-HLF driven acute B cell leukemia, was the prominent accumulation of a c-kit+CD19+ population in the BM of Hlf-induced mice. Our approaches to characterize these cells collectively suggested that they represent committed B cell progenitors that are blocked in differentiation at a fraction B-C stage, which was further supported by a rapid gain of c-Kit expression of these cells upon Hlf induction. These cells also accumulated in the BM of WT mice transplanted with Hlf-inducible GMLPs, which coincided with a vastly decreased B cell reconstitution in the periphery. Given the genomic context of the E2A-HLF fusion oncogene, one outcome due to the more extended expression pattern of E2A should be aberrant expression of HLF in cell types normally devoid of HLF. However, as we have failed to observe leukemia upon Hlf induction, even following extensive time periods, additional mechanisms other than aberrant activation of Hlf target genes seem crucial for E2A-HLF-mediated transformation. At the same time, our studies appear in line with the observed phenotypes of murine transgenic models of the E2A-HLF fusion that manifest with myelo- rather than lymphoproliferation (Duque-Afonso et al., 2015).
By searching for candidate target genes of Hlf that could help explain the actions of Hlf on lineage commitment, we identified prominent Hlf binding to an intronic enhancer of Nfic and robust increases in Nfic mRNA expression upon Hlf induction. Although little is known about the role of Nfic in hematopoiesis, the Nfi family member Nfix was recently identified as a critical modulator of hematopoietic lineage choice by repressing B lymphoid formation while maintaining myeloid activity (O’Connor et al., 2015), suggesting redundancy among Nfi family members with respect to this activity. On the other hand, Nfic does not appear to explain all effects of Hlf, underscored not the least by a general loss of cells upon enforced Nfic expression. Although outside the scope of the current work, perhaps gene dosage and/or expression of alternative isoforms might underlie the differences observed between Nfic induced by Hlf and the retroviral approach we used, which does not exclude the likelihood that other target genes collaborate with Nfic to mediate a combined effect. When interrogating our Hlf ChIP-seq data (see Experimental Procedures, Data and Software Availability), we find Hlf binding to a recently identified distal c-Kit enhancer (chr5: 75,729,914–75,732,243) (Aranda-Orgilles et al., 2016). This might be relevant for our work, given the upregulation of c-Kit we see in the B cell lineage, the anti-apoptotic effect of c-kit signaling, and its downregulation in both B and T cell development (Godfrey et al., 1992, Rico-Vargas et al., 1994). Another potential candidate is the TF Meis1, whose promoter is occupied by Hlf (chr11: 18,741,886–19,157,511). In previous work from Kumar and colleagues (Roychoudhury et al., 2015), Meis1 was suggested to regulate Hlf expression, with Meis1 loss leading to pronounced apoptosis. Our observations extend these findings to include Hlf regulation of Meis1, indicative of a feedback loop between these factors.
As it is becoming increasingly apparent that multipotent progenitor cells other than HSCs are critical for steady-state hematopoiesis even over extended time periods (Busch et al., 2015, Sun et al., 2014), we believe that the ability of Hlf to strongly direct differentiation of an otherwise multipotent progenitor toward specific lineages is most relevant. A deeper understanding of the regulatory circuits governed by Hlf will not only be important to understand physiological blood cell formation but will likely also lead to an increased understanding of the leukemic processes that are characterized by involvement of Hlf.
Experimental Procedures
Further details and an outline of resources used in this work can be found in Supplemental Experimental Procedures.
Generation of Viral Vectors
Viral constructs were generated using PCR-mediated Gibson Assembly and T4 DNA ligase cloning (New England Biolabs). Lentiviruses were produced by transient transfection of 293T producer cells and retroviruses by transient transfection into Plat-erythroid packaging cells.
Generation of Transgenic Mice and In Vivo Hlf Transgenic Strategy
DOX-inducible Hlf transgenic mice were generated by cloning the coding sequence and the 5′ and 3′ UTR of the murine Hlf gene into the pBS31 targeting vector (Beard et al., 2006). Next, the pBS31-Hlf construct was used to target KH2 ESCs by Flpase-mediated recombination (Beard et al., 2006). See Figure S3B for a schematic of the model. Following successful targeting, engineered ESCs were used to generate transgenic mice, which were backcrossed to CD45.1+ C57BL/6 mice and bred to homozygosity for the modified loci. Transgenic Hlf expression in vivo was achieved by administering DOX-containing chow ad libitum (2 g/kg; Bio-Serv). All animal experiments were performed with consent from a local ethics committee.
Immunophenotypic Analysis and Cell Sorting
For isolation and analysis of HSCs, the indicated progenitor populations and mature cells by FACS were performed as previously described (Inlay et al., 2009, Pronk and Bryder, 2011, Pronk et al., 2007, Tung et al., 2004).
In Vitro Evaluation of NK, B, and T Cell Potential by OP9/OP9-DL1 Co-culture
To determine the differentiation potential of Hlf-inducible GMLPs, cells were sorted directly into 48-well plates (bulk cultures) or 96-well plates (single-cell cultures) pre-plated with OP9 or OP9-DL1 stromal cells. In some experiments, WT GMLPs were instead first transduced viral vectors before stromal cell co-culture. B/NK cell permissive cultures were maintained on OP9 stroma cells in medium containing 20 ng/mL interleukin-15 (IL-15), 40 ng/mL IL-2, 10 ng/mL stem cell factor (SCF), 10 ng/mL fms-like tyrosine kinase 3 ligand (Flt3L), and 10 ng/mL IL-7, while T cells were generated on OP9-DL1 stroma supplemented with 10 ng/mL Flt3L and 10 ng/mL IL-7. Hlf-inducible cultures were maintained in the presence or absence of 1 μg/mL DOX (Sigma-Aldrich). Cultures were evaluated at the indicated time points by cell counting and FACS analysis.
Evaluation of Apoptosis
To investigate whether Hlf expression resulted in increased levels of apoptosis in B and T lymphoid progenitors, thymic CD4+CD8+ and BM-derived fraction B-C cells were FACS-sorted from Hlf-inducible mice and cultured on OP9-DL1 and OP9, respectively, with or without DOX (1 μg/mL) for 48 hr. Next, fraction B-C cultures were stained with a c-Kit antibody (2B8; eBioscience), and both culture types were thereafter incubated with annexin V conjugated to Cy5 and propidium iodide for 15 min before immediate FACS analysis.
Reconstitution Experiments
Eight thousand five hundred to 15,000 Hlf-inducible GMLPs (CD45.1+) were transplanted into sublethally irradiated (500 rad) WT CD45.2+ C57BL/6 recipient mice preconditioned for 5 days prior to transplantation by allowing mice to eat DOX-containing food. The mice were kept on a DOX pellet diet for 21 days (when investigating the effects of prolonged Hlf expression) or 22 days and 14 days followed by 8 days with a normal diet (when investigating the effect of DOX withdrawal on the established phenotypes) post-transplantation; BM, spleen, thymus, and PB of the recipient mice were analyzed for donor reconstitution.
Affymetrix Gene Expression Analysis and qRT-PCR
Microarray data were analyzed using dChip (Li and Hung Wong, 2001). For qRT-PCR experiments, the indicated populations were FACS-sorted directly into RLT lysis buffer and purified using the RNeasy Micro mRNA purification kit (QIAGEN), followed by first-strand cDNA synthesis as previously described (Norddahl et al., 2011). In some cases, the cDNA was amplified using KAPA HiFi Hotstart Readymix (Kapa Biosystems). qRT-PCRs were run with SYBR GreenER (Invitrogen) or EvaGreen (Bio-Rad).
ChIP-Seq Experiments
WT LSK cells were infected with a pMX-GFP control or a pMX-3xFLAG-Hlf/Hlf-IRES-GFP virus and grown for 5 days in basal OP9 medium supplemented with 50 ng/mL SCF, 10 ng/mL IL-7, 10 ng/mL Flt3L, and 5 ng/mL IL-3 (all from Peprotech). Next, 2 × 107 cells were cross-linked in 1% formaldehyde, and nuclei were prepared and snap-frozen in a dry ice/isopropanol bath. The frozen nuclei were lysed, and chromatin was sonicated in a Bioruptor (Diagenode) before immunoprecipitating with a FLAG-tag antibody (F3165; Sigma-Aldrich). Next, cross-links were reversed, and DNA was purified using QIAGEN PCR clean-up columns. Sequencing libraries were prepared using the Illumina TruSeq ChIP sample preparation kit (IP-202-1012; Illumina) and sequenced on an Illumina HiSeq 2500. Further details along with data-processing regimens can be found in Supplemental Experimental Procedures.
RNA-Seq
WT GMLPs were transduced with pHAGE2-Hlf/Hlf-IRES-ZsGreen or a pHAGE2 control virus, and Hlf-inducible GMLPs maintained in the presence or absence of DOX were cultured on OP9 stroma in B/NK cell conditions for 4 days. Next, ZsGreen+/CD45+ cells were FACS-sorted. Following RNA purification, libraries were prepared by the SMARTer Ultra Low Input RNA Kit for Sequencing (Clontech), and RNA-seq analysis was performed using an Illumina HiSeq 2500 platform by the Genome Technology Access Center (GTAC; Washington University School of Medicine). Data-processing details can be found in Supplemental Experimental Procedures.
B Cell Transdifferentiation Experiments
B cell-to-macrophage transdifferentiation experiments were performed in essence as previously described (Xie et al., 2004). In brief, 100,000 BM-resident B220+CD19+ B cells were isolated by FACS and transduced on retronectin-coated plates with a MigR1 control virus, MigR1-Hlf, and a MigR1-Cebpb virus and cultured on OP9 stroma for 4 days in basal OP9 medium supplemented with SCF, M-CSF, IL-3, IL-7, and Flt3L (all at 10 ng/mL) prior to FACS analysis.
VDJ Rearrangement PCR
The PCR reactions were run using 10,000 cells of each indicated population exactly as previously described (Schlissel et al., 1991).
Luciferase Assays
A 1 kb fragment containing the Hlf-bound region of the Nfic gene or a more specific 204 bp region containing three candidate HLF-binding sites was inserted into the pGL2 vector upstream of a SV40 promoter sequence and firefly luciferase reporter. The resulting vectors, or an empty pGL2 vector, were used to co-transfect Hlf-inducible ESCs maintained with or without DOX using Lipofectamine LTX reagent (Invitrogen) together with a Renilla luciferase reporter gene plasmid. Luciferase reporter signals were determined 24 hr post-transfection using the Dual-Luciferase Reporter Assay System kit (Promega).
Statistics
Data analysis was performed using Microsoft Excel and GraphPad Prism (GraphPad Software). All FACS data were analyzed with FlowJo (Tree Star). The heatmap in Figure 1B was prepared using GEN-E (http://www.broadinstitute.org/cancer/software/GENE-E/index.html). Venn diagrams were generated using Venny (Oliveros, 2007–2015). Significance values, with the exception of Figure 4A (splenic T cell reconstitution), for which a Mann-Whitney test was used, were calculated using Student’s two-tailed t test, and a p value of <0.05 was considered to indicate statistical significance.
Author Contributions
M.W. and D.B. designed the study. M.W., V.L., I.H., P.S., H.W., M.D.-P. and G.L.N. performed experiments. M.M. provided intellectual input. V.L., M.S.C., R.H., and B.G. were involved in the design, interpretation, and analysis of the ChIP-seq experiment. D.B. conceived and supervised the study and wrote the paper together with M.W.
Acknowledgments
This work was generously supported by project grants to D.B. from the Swedish Cancer Society, the Swedish Medical Research Council, the Swedish Pediatric Leukemia Foundation, the Knut and Alice Wallenberg Foundation, and an European Research Council (ERC) consolidator grant (615068). Work in the Gottgens lab is supported by the Wellcome Trust, CRUK, Bloodwise, MRC, NIH-NIDDK, and core support funding by the Wellcome Trust and MRC to the Cambridge Stem Cell Institute. We would like to acknowledge Tom Serwold, Ewa Sitnicka, and Mikael Sigvardsson for valuable scientific discussions and Eva Erlandsson and Gerd Sten for expert technical assistance. The GTAC, Department of Genetics, Washington University School of Medicine, assisted with genomic analysis and is partially supported by National Cancer Institute (NCI) Cancer Center Support Grant P30 CA91842 to the Siteman Cancer Center, Institute of Clinical and Translational Sciences (ICTS)/Clinical and Translational Science Award (CTSA) Grant UL1TR000448 from the National Center for Research Resources (NCRR; a component of the NIH), and the NIH Roadmap for Medical Research.
Published: November 21, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2017.10.112.
Data and Software Availability
The accession numbers for the microarray data reported in this paper are GEO: GSE44923 and GEO: GSE27686 (HSCs); GEO: GSE8407 (Pre Meg/erythroid, Pre CFU-erythroid, CFU-erythroid, MkP, pGM and CLP); GEO: GSE18734 (GMLP); and GEO: GSE14833 (GMP). The accession number for the generated ChIP-seq data reported in this paper is GEO: GSE69817. Our ChIP data can also be visualized using a custom University of California, Santa Cruz (UCSC), Genome Browser session (http://genome-euro.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=promufa&hgS_otherUserSessionName=Lund_160719) together with previously established ChIP data (Lara-Astiaso et al., 2014). The accession number for the RNA-seq data reported in this paper is GEO: GSE69858.
Supplemental Information
References
- Adolfsson J., Månsson R., Buza-Vidas N., Hultquist A., Liuba K., Jensen C.T., Bryder D., Yang L., Borge O.J., Thoren L.A. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Akashi K., Traver D., Miyamoto T., Weissman I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Allman D., Sambandam A., Kim S., Miller J.P., Pagan A., Well D., Meraz A., Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- Aranda-Orgilles B., Saldaña-Meyer R., Wang E., Trompouki E., Fassl A., Lau S., Mullenders J., Rocha P.P., Raviram R., Guillamot M. MED12 regulates HSC-specific enhancers independently of mediator kinase activity to control hematopoiesis. Cell Stem Cell. 2016;19:784–799. doi: 10.1016/j.stem.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiropoulos B., Yung E., Xiang P., Lo C.Y., Kuchenbauer F., Palmqvist L., Reindl C., Heuser M., Sekulovic S., Rosten P. Linkage of the potent leukemogenic activity of Meis1 to cell-cycle entry and transcriptional regulation of cyclin D3. Blood. 2010;115:4071–4082. doi: 10.1182/blood-2009-06-225573. [DOI] [PubMed] [Google Scholar]
- Arinobu Y., Mizuno S., Chong Y., Shigematsu H., Iino T., Iwasaki H., Graf T., Mayfield R., Chan S., Kastner P., Akashi K. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Benz C., Martins V.C., Radtke F., Bleul C.C. The stream of precursors that colonizes the thymus proceeds selectively through the early T lineage precursor stage of T cell development. J. Exp. Med. 2008;205:1187–1199. doi: 10.1084/jem.20072168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryder D., Rossi D.J., Weissman I.L. Hematopoietic stem cells: the paradigmatic tissue-specific stem cell. Am. J. Pathol. 2006;169:338–346. doi: 10.2353/ajpath.2006.060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K., Klapproth K., Barile M., Flossdorf M., Holland-Letz T., Schlenner S.M., Reth M., Höfer T., Rodewald H.R. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- Duque-Afonso J., Smith K.S., Cleary M.L. Conditional expression of E2A-HLF induces B-cell precursor death and myeloproliferative-like disease in knock-in mice. PLoS ONE. 2015;10:e0143216. doi: 10.1371/journal.pone.0143216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Forster M., Rinaldi A., Risch T., Sungalee S., Warnatz H.J., Bornhauser B., Gombert M., Kratsch C., Stütz A.M. Genomics and drug profiling of fatal TCF3-HLF-positive acute lymphoblastic leukemia identifies recurrent mutation patterns and therapeutic options. Nat. Genet. 2015;47:1020–1029. doi: 10.1038/ng.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg E.C., Serwold T., Kogan S., Weissman I.L., Passegué E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- Gachon F., Fonjallaz P., Damiola F., Gos P., Kodama T., Zakany J., Duboule D., Petit B., Tafti M., Schibler U. The loss of circadian PAR bZip transcription factors results in epilepsy. Genes Dev. 2004;18:1397–1412. doi: 10.1101/gad.301404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F., Olela F.F., Schaad O., Descombes P., Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Gazit R., Garrison B.S., Rao T.N., Shay T., Costello J., Ericson J., Kim F., Collins J.J., Regev A., Wagers A.J., Rossi D.J., Immunological Genome Project Consortium Transcriptome analysis identifies regulators of hematopoietic stem and progenitor cells. Stem Cell Reports. 2013;1:266–280. doi: 10.1016/j.stemcr.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain R.N. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2002;2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., Zlotnik A., Suda T. Phenotypic and functional characterization of c-kit expression during intrathymic T cell development. J. Immunol. 1992;149:2281–2285. [PubMed] [Google Scholar]
- Goodell M.A., Nguyen H., Shroyer N. Somatic stem cell heterogeneity: diversity in the blood, skin and intestinal stem cell compartments. Nat. Rev. Mol. Cell Biol. 2015;16:299–309. doi: 10.1038/nrm3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.R., Hayakawa K. B cell development pathways. Annu. Rev. Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Carmack C.E., Shinton S.A., Kemp J.D., Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger S.P., Ohyashiki K., Toyama K., Cleary M.L. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 1992;6:1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- Inaba T., Roberts W.M., Shapiro L.H., Jolly K.W., Raimondi S.C., Smith S.D., Look A.T. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science. 1992;257:531–534. doi: 10.1126/science.1386162. [DOI] [PubMed] [Google Scholar]
- Inaba T., Shapiro L.H., Funabiki T., Sinclair A.E., Jones B.G., Ashmun R.A., Look A.T. DNA-binding specificity and trans-activating potential of the leukemia-associated E2A-hepatic leukemia factor fusion protein. Mol. Cell. Biol. 1994;14:3403–3413. doi: 10.1128/mcb.14.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlay M.A., Bhattacharya D., Sahoo D., Serwold T., Seita J., Karsunky H., Plevritis S.K., Dill D.L., Weissman I.L. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai T., Inaba T., Dang J., Kuribara R., Ozawa K., Miyajima A., Wu W., Look A.T., Arinobu Y., Iwasaki H. TEF, an antiapoptotic bZIP transcription factor related to the oncogenic E2A-HLF chimera, inhibits cell growth by down-regulating expression of the common beta chain of cytokine receptors. Blood. 2005;105:4437–4444. doi: 10.1182/blood-2004-08-2976. [DOI] [PubMed] [Google Scholar]
- Ji H., Ehrlich L.I., Seita J., Murakami P., Doi A., Lindau P., Lee H., Aryee M.J., Irizarry R.A., Kim K. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizono S., Duncan G.S., Seidel M.G., Morimoto A., Hamada K., Grosveld G., Akashi K., Lind E.F., Haight J.P., Ohashi P.S. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer E.M., Lin Y.C., Murre C. Factors and networks that underpin early hematopoiesis. Semin. Immunol. 2011;23:317–325. doi: 10.1016/j.smim.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui S., Yamaguchi S., Matsuo T., Ishida Y., Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Kodama H., Honjo T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- Norddahl G.L., Pronk C.J., Wahlestedt M., Sten G., Nygren J.M., Ugale A., Sigvardsson M., Bryder D. Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell. 2011;8:499–510. doi: 10.1016/j.stem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- O’Connor C., Campos J., Osinski J.M., Gronostajski R.M., Michie A.M., Keeshan K. Nfix expression critically modulates early B lymphopoiesis and myelopoiesis. PLoS ONE. 2015;10:e0120102. doi: 10.1371/journal.pone.0120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros, J.C. (2007–2015). Venny 2.1. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Pronk C.J., Bryder D. Flow cytometry-based identification of immature myeloerythroid development. Methods Mol. Biol. 2011;699:275–293. doi: 10.1007/978-1-61737-950-5_13. [DOI] [PubMed] [Google Scholar]
- Pronk C.J., Rossi D.J., Månsson R., Attema J.L., Norddahl G.L., Chan C.K., Sigvardsson M., Weissman I.L., Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Pronk C.J., Attema J., Rossi D.J., Sigvardsson M., Bryder D. Deciphering developmental stages of adult myelopoiesis. Cell Cycle. 2008;7:706–713. doi: 10.4161/cc.7.6.5565. [DOI] [PubMed] [Google Scholar]
- Rico-Vargas S.A., Weiskopf B., Nishikawa S., Osmond D.G. c-kit expression by B cell precursors in mouse bone marrow. Stimulation of B cell genesis by in vivo treatment with anti-c-kit antibody. J. Immunol. 1994;152:2845–2852. [PubMed] [Google Scholar]
- Riddell J., Gazit R., Garrison B.S., Guo G., Saadatpour A., Mandal P.K., Ebina W., Volchkov P., Yuan G.C., Orkin S.H., Rossi D.J. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychoudhury J., Clark J.P., Gracia-Maldonado G., Unnisa Z., Wunderlich M., Link K.A., Dasgupta N., Aronow B., Huang G., Mulloy J.C., Kumar A.R. MEIS1 regulates an HLF-oxidative stress axis in MLL-fusion gene leukemia. Blood. 2015;125:2544–2552. doi: 10.1182/blood-2014-09-599258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan-Pla A., Macaulay I.C., Jensen C.T., Woll P.S., Luis T.C., Mead A., Moore S., Carella C., Matsuoka S., Bouriez Jones T. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502:232–236. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- Schlissel M.S., Corcoran L.M., Baltimore D. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 1991;173:711–720. doi: 10.1084/jem.173.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt T.M., Zúñiga-Pflücker J.C. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- Shojaei F., Trowbridge J., Gallacher L., Yuefei L., Goodale D., Karanu F., Levac K., Bhatia M. Hierarchical and ontogenic positions serve to define the molecular basis of human hematopoietic stem cell behavior. Dev. Cell. 2005;8:651–663. doi: 10.1016/j.devcel.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Ramos A., Chapman B., Johnnidis J.B., Le L., Ho Y.J., Klein A., Hofmann O., Camargo F.D. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J.W., Parks D.R., Moore W.A., Herzenberg L.A., Herzenberg L.A. Identification of B-cell subsets: an exposition of 11-color (Hi-D) FACS methods. Methods Mol. Biol. 2004;271:37–58. doi: 10.1385/1-59259-796-3:037. [DOI] [PubMed] [Google Scholar]
- Wahlestedt M., Norddahl G.L., Sten G., Ugale A., Frisk M.A., Mattsson R., Deierborg T., Sigvardsson M., Bryder D. An epigenetic component of hematopoietic stem cell aging amenable to reprogramming into a young state. Blood. 2013;121:4257–4264. doi: 10.1182/blood-2012-11-469080. [DOI] [PubMed] [Google Scholar]
- Weishaupt H., Sigvardsson M., Attema J.L. Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood. 2010;115:247–256. doi: 10.1182/blood-2009-07-235176. [DOI] [PubMed] [Google Scholar]
- Xie H., Ye M., Feng R., Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yamamoto R., Morita Y., Ooehara J., Hamanaka S., Onodera M., Rudolph K.L., Ema H., Nakauchi H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Yang Q., Jeremiah Bell J., Bhandoola A. T-cell lineage determination. Immunol. Rev. 2010;238:12–22. doi: 10.1111/j.1600-065X.2010.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber A.M., Centeno G., Pradervand S., Nikolaeva S., Maquelin L., Cardinaux L., Bonny O., Firsov D. Molecular clock is involved in predictive circadian adjustment of renal function. Proc. Natl. Acad. Sci. U S A. 2009;106:16523–16528. doi: 10.1073/pnas.0904890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.