Abstract
Introduction
Drug-metabolizing enzymes (DMEs) are primarily down-regulated during infectious and inflammatory diseases, leading to disruption in the metabolism of small molecule drugs (SMDs), which are increasingly being prescribed therapeutically in combination with biologics for a number of chronic diseases. The biologics may exert pro- or anti-inflammatory effect, which may in turn affect the expression/activity of DMEs. Thus, patients with infectious/inflammatory diseases undergoing biologic/SMD treatment can have complex changes in DMEs due to combined effects of the disease and treatment.
Areas covered
We will discuss clinical biologics-SMD interaction and regulation of DMEs during infection and inflammatory diseases. Mechanistic studies will be discussed and consequences on biologic-small molecule combination therapy on disease outcome due to changes in drug metabolism will be highlighted.
Expert opinion
The involvement of immunomodulatory mediators in biologic-SMDs is well known. Regulatory guidelines recommend appropriate in vitro or in vivo assessments for possible interactions. The role of cytokines in biologic-SMDs has been documented. However, the mechanisms of drug-drug interactions is much more complex, and is probably multi-factorial. Studies aimed at understanding the mechanism by which biologics effect the DMEs during inflammation/infection are clinically important.
Keywords: Biologics, small molecule drugs, drug-drug interaction, drug metabolizing enzymes, cytokines, inflammation, infection, Toll-like receptors, nuclear receptors, interferon
1. Introduction
Drug metabolism is known to be disrupted during infectious and inflammatory disease primarily due to down-regulation of the drug metabolizing enzymes (DMEs) at transcriptional/post-transcriptional level. Post-translational modifications of the DMEs are also reported to cause reductions in the DME activity, leading to impaired drug metabolism. Altered drug metabolism can lead to adverse drug reactions and drug-drug interactions in patients with infectious and inflammatory diseases.
In the recent decade, with the advent of newer sophisticated biotechnological methods, biologics are being used therapeutically for a range of disorders, including cancer, rheumatoid arthritis, gastro-enteric disorders, skin disorders, respiratory diseases, hormone deficiency disorders and other infections. Evaluate Pharma showed that the total sales of biologics in prescription and over the counter medicines grew from 12% in 2004 to 19% in 2011 and is estimated to reach 50% sales in 2020. [1]
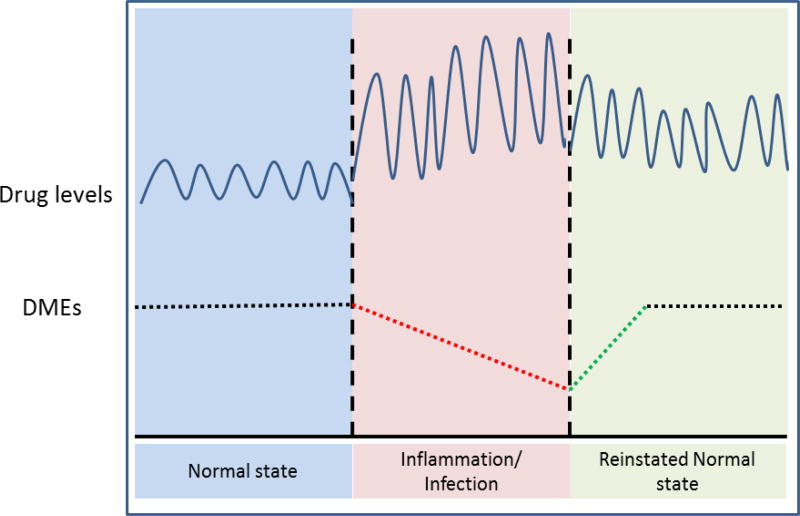
Since combination therapies offer the potential for improved therapeutic effect, biologics are combined with conventional small molecule drugs (SMDs) and/or with other biologics to attain greater benefits. However, there is a downside to biologics co-treatment. The SMDs undergo metabolism/clearance by DMEs and transporters. In patients with infectious and inflammatory diseases, down-regulation of DMEs can contribute to accumulation of SMDs in vivo due to their impaired metabolism and clearance (Fig. 1). Biologics given in combination with the SMD interferes with the inflammatory pathways. Thus, treatment with the biologic can further alter the expression and activity of the DMEs in these patients, causing further alterations in SMD metabolism and clearance. The impact of biologic treatment on DMEs and SMD metabolism will likely be dependent on the pro- or anti-inflammatory properties of the biologic. Thus, in patients with infectious and inflammatory diseases, biologic-drug interactions can impact the safety and effectiveness of the treatment.
Figure 1.
Schematic representation of correlation between disease-mediated alteration in drug metabolizing enzymes and small molecule drug levels
This review will provide insights into the mechanism of regulation of DMEs and the implications for biologic-small molecule drug interactions in patients with inflammatory and infectious diseases.
2. Methodology of Search strategy
Potentially relevant literature until August, 2016 was identified by performing searches in the following databases: Google Scholar, PubMed, FDA (Drug approvals and database). Literature was searched independently by the authors and finally screened according to their relevance to the topic undertaken in this review. Literature search was started using key words including “biologics”, “FDA approved biologics”, “biologics for inflammation and infection”, “regulation of drug metabolizing enzymes”. Thereafter, filters were used such as: “biologics drug interactions”, “altered regulation of drug metabolizing enzymes (DMEs) and transporters in infection & inflammation” and finally streamlined with drug interaction with small molecule drugs (SMDs). Selection criteria mainly focused on clinically studies that have reported effect of biologics on the pharmacokinetics/pharmacodynamics of SMDs, followed by in vivo and in vitro drug interaction studies with maximum weightage given to studies with emphasis on DMEs and transporters. Exclusion criteria included documents such as comments, letters without an experimental study, meta-analyses, and editorials despite being retrieved using the search terms.
3. Classification of biologics
U. S Food and Drug Administration (FDA) define biologics as “any virus, therapeutic serum, toxin, antitoxin, or analogous product applicable to the prevention, treatment or cure of diseases or injuries of human being”. The first-generation biologics were obtained from humans or animals, such as human blood, insulin, TNF-α or influenza vaccine. Second-generation biologics are complex proteins, manufactured using biotechnology and interact with surface receptors of target cells. Third generation biologics target different epitopes and are mostly designed for improved Fc-associated immune functions or half-life. For example, humanized type II CD20 antibody is less immunogenic than second generation monoclonal antibodies and with its glyco-engineered Fc, has superior cytotoxicity [2].
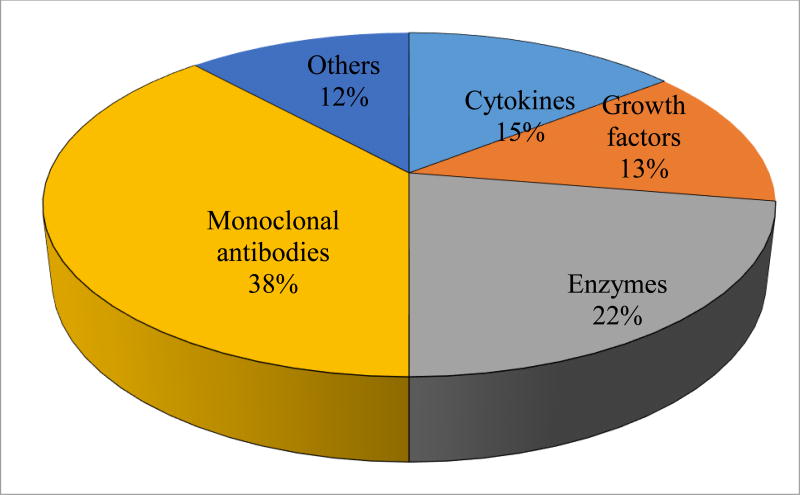
Biologics are categorized according to their molecular structure and have also been organized by Dimitrov et al [3–5] based on their mode of action as follows: (a) substitution of a defective/deficient protein, (b) modification of existing pathway, (c) offer novel function/activity, (d) meddling with molecules and (e) carrier of other compounds or proteins [5]. Benjamin et al have grouped biologics according to their molecular types as antibody-based drugs, Fc fusion proteins, anticoagulants, blood factors, bone morphogenetic proteins, engineered protein scaffolds, enzymes, growth factors, hormones, interferons, interleukins, and thrombolytics [6]. Among all categories of biologics, monoclonal antibodies represent a predominant class of biologics and accounts about 50% of the sales [3] (Fig. 2).
Figure 2.
Percentage of major classes of biologics approved by U.S FDA (2016)
4. Differences between biologics and small molecule drugs
Biologics are manufactured using biological processes while the conventional SMDs are synthesized chemically. Strict definition dictates that biologics has two critical traits; produced from living systems and relatively large molecules composed of more than 1,300 amino acids and can be as heavy as 150,000 g/mol (or 150 kDa). In contrast, SMDs are low molecular weight (<900 daltons) organic compounds that may help regulate a biological process. Due to their large size, protein folding and lack of long-term stability, biologics pose unique challenges for pharmacokinetic studies in comparison to SMDs (Table. 1) [6–10].
Table 1.
Key parameters determining ADME profiles of Biologics vs SMDs
| ADME-related factors |
Biologics | Small molecule drugs (SMDs) |
|---|---|---|
| Size | Large (mixture of related molecules), high molecular weight >>1000 Dalton | Small (single molecule), Low molecular weight <1000 Dalton |
| Source | Biologically produced or engineered | Chemically synthesized |
| Physical/chemical properties | Complex (heterogeneous), undergo post translational modification | Simple, well defined |
| Stability | Unstable, sensitive to external conditions | Stable |
| Absorption (Route of administration issues) | Parenteral (IV, SC, IM) (contribution of lymphatic absorption after subcutaneous injection) | All routes |
| Volume of distribution | Mainly limited to plasma or extracellular fluids (FcRn- and target-dependent mechanisms) | To any organs dictated by hydrophobicity and plasma and tissue protein binding |
| Half-life | Long (in days and weeks) | Short (in hours) |
| Elimination | Phagocytosis, endocytosis, proteolysis, formation of immune-complexes followed by complement- or Fc receptor-mediated clearance and importantly target mediated disposition | Biotransformation (oxidation andconjugation), renal |
| Immunogenicity | Immunogenic (immune system identify the molecule and initiates an immune response to clear away the biologics) | Mostly non-immunogenic (too small to be recognized by the immune system as “invaders) |
5. Biologics-small molecule drug interactions
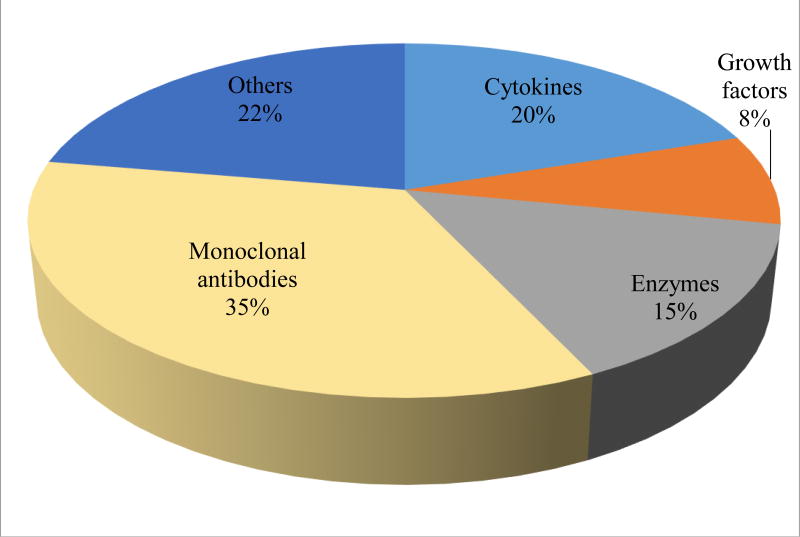
Combination therapies offer the potential for improved effectiveness, therefore to attain greater benefits, biologics are combined with conventional small molecule drugs and/or with other biologics. Until 2010, ~ 80 biologics were approved and very few biological license applications (BLAs) had information about biologic-drug interactions from clinical studies in the labelling [3]. Although later on many more biologics were approved by the FDA as therapeutics (Fig. 2), still information on biologic-drug interactions remain limited (Fig. 3). Unlike SMDs, there are only a few reported DDI studies for biologics. Since biologics don’t undergo hepatic metabolism via cytochrome P450s (CYP) enzymes or eliminated by biliary excretion, it was perceived that biologics have very low propensity for DDIs. Therapeutic monoclonal antibodies (mAbs), which are primarily used to treat moderate to severe and chronic diseases, are often prescribed with more than one concomitant medication, especially among geriatric patients. Therefore, potential DDIs with mAbs arise owing to the multiple medications and polypharmacy situations [11, 12]. There are reports of pharmacokinetics (PK) drug interaction studies with immunomodulators.
Figure 3.
Clinical biologic-DDIs included in labelling approved by FDA (2016)
Immunomodulators include both immunostimulator and immunosuppressor agents. To treat immune-mediated conditions, the desired therapeutic effect is to reduce or control inflammation. Over-production of pro-inflammatory cytokines is considered as underlying cause of progression of many inflammatory diseases like Rheumatoid arthritis and Crohn disease and also many infectious diseases including HIV, influenza H5N1 and malaria. Biologics used to reduce inflammation can be placed in one of three groups depending on their mechanism of action: (i) cytokine blockade, (ii) cell depletion and (iii) regulatory cell surface receptor blockade [6]. For treatment of many infectious diseases like hepatitis B and hepatitis C virus infection, immunostimulators capable of enhancing host defense are used to provide protection against infection. Drug interactions with immunomodulators have been reported for cytokines and antibodies (Table. 2) but hardly anything is known about DDIs of Growth Factors [13], Enzymes & Fusion Proteins. Therefore, here we discuss some of the interactions observed with cytokines and monoclonal antibodies.
Table 2.
Approved biologics and their reported clinical drug interactions
| Biologics | Indication | DDI information (FDA insert or clinical study) |
Ref |
|---|---|---|---|
| IFN- α2b (Intron A) | Hairy cell leukemia, malignant melanoma, follicular lymphoma, condylomata acuminata, AIDS- related kaposi’s sarcoma, hepatitis C, hepatitis B | IFN-α decreases theophylline clearance, resulting increase in theophylline serum levels | [14] |
| PEGylated IFN-α2a (Pegasys, Reiferon) | Chronic hepatitis C, hepatitis B |
|
[15] |
| PEGylated IFN- α2b (PegIntron) | Chronic Hepatitis C (CHC) in patients with compensated liver disease | PegIntron increased AUC of methadone clinically after 4 weeks of treatment. | [16] |
| Necitumumab (epidermal growth factor receptor antibody) | metastatic squamous NSCLC | Necitumumab increased area under curve (AUC) of gemcitabine by 22% and Cmax by 63% | [17] |
| Muromonab (CD3 receptor antibody) | acute, glucocorticoid-resistant rejection of allogeneic renal, heart and liver transplants | Increased concentration of Cyclosporine | [18, 19] |
| Trastuzumab (HER2 receptor antibody) | Treat breast cancer | Though not significant, paclitaxel peak plasma concentrations were ~ 25% lower | [20] |
| Adalimumab (TNF-α receptor antibody) | RA, psoriatic arthritis, ankylosing spondylitis, Crohn’s disease, ulcerative colitis | serum levels of duloxetine were decreased | [21] |
| Basiliximab | Indicated for prophylaxis of acute organ rejection in patients receiving renal transplantation (as part of an immunosuppressive regimen that includes cyclosporine, and corticosteroids) | Increased Tacrolimus blood trough levels (by 63%) | [93] |
| Tocilizumab (Anti IL-6) | Rheumatoid Arthritis, Polyarticular Juvenile Idiopathic Arthritis, Systemic Juvenile Idiopathic Arthritis | Increased metabolism of omeprazole and simvastatin and thus reduced AUC. | [94] |
5.1. Examples of cytokine-drug interactions
Over the last 20 years, successful development and clinical implementation of biologic strategies has been seen with respect to key cytokines in specific inflammatory/infectious diseases with efficacy, specificity, and toxicity profiles. The human body produces different types of cytokines such as colony stimulating factors, growth and differentiation factors, immunoregulatory and pro-inflammatory cytokines.
5.1.1. Interferons
Clinically, interferons are mainly used for the therapy of hepatitis B/C, multiple sclerosis and cancer. Williams et al. (1987) [15] reported that administration of IFN-α in chronic hepatitis B patients reduced the clearance of theophylline by 30–80% [14]. Similarly, theophylline clearance was significantly reduced in hepatitis C patients after IFN-β treatment, with a corresponding increase in terminal half-life by about 40% [22]. Serum theophylline concentrations were found to be significantly increased with a mean of 29.3 µg/mL (therapeutic range 10–20 µg/mL) in in children with influenza B outbreak [23, 24]. Additionally, IFN-α administration has also been shown to decrease erythromycin metabolism in patients with hepatitis B (15%), as determined by erythromycin breath test [25]. It has been reported that IFN-α2 also inhibits the metabolism of antipyrine [26]. Presently, PEGylated interferons, made by addition of poly-ethylene glycol side chain to the interferon, are being used in combination therapies for diseases such as hepatitis B and hepatitis C. As mentioned in the Table. 2, clinically significant drug interaction studies with PEGylated interferons have not been carried out to a large extent, however, studies have shown that its treatment was associated with increase in AUC of theophylline [27] and methadone [16].
5.1.2. Interleukins
Interleukins (ILs) are cytokines mainly synthesized by T lymphocytes, as well as monocytes, macrophages, and endothelial cells. In a clinical therapeutic protein- drug interaction study in rheumatoid arthritis patients, sirukumab (anti-IL-6 antibody) reduced the AUC of CYP probe substrates midazolam (CYP3A), omeprazole (CYP2C19), warfarin (CYP2C9) while caffeine AUC was increased [28]. To date, no clinical data have been published on the effect of IL-1 inhibitors on drugs that are substrate of CYP enzymes.
5.2. Examples of antibody-drug interactions
In the clinic, monoclonal antibodies (mAbs) are proving to be safe and effective for the treatment of a wide range of diseases including rheumatoid arthritis (RA), Crohn’s disease, spondyloarthropathies, psoriasis and allograft rejection. The clinical success of anti–TNF-α mAbs in human RA and psoriasis made these biologics as mainstream therapeutics. As of 2011, 35 mAbs have been approved by the FDA for use in humans. Table. 2 list some of the mAbs that have been introduced for immunologic indication.
Schmitt et al. (2011), [29] reported that tocilizumab (Anti IL-6) administration increased metabolism of simvastatin and thus reduced AUC in rheumatoid arthritis patients. Both muromonab (CD3 receptor antibody) and basiliximab (IL-2 receptor antibody), when co-administered with cyclosporine, increases its serum concentrations [18, 19]. In a case report, adalimumab (TNF-α inhibitor) decreased serum levels of duloxetine [21]. In a Phase 1 pharmacokinetic study, co-administration of dalotuzumab (IGF-1 receptor antibody) with irinotecan did not affect the AUC of the parent drug but reduced the Cmax (from 16.8 to 13.0 ng/mL) and the AUC0–24h (by 13 %) of SN-38, the active metabolite of irinotecan [30]. Necitumumab, an IgG1 monoclonal antibody and an EFGR antagonist increased the geometric mean dose-normalized AUC and Cmax of gemcitabine [17]. Although not significant, trastuzumab (HER2 receptor antibody) lowered the Cmax of taxol in patients with advanced breast cancer [20]. A recent study in 2016 by Lee et al shows that sarilumab (which is a human monoclonal antibody blocking the IL-6-Rα) treatment resulted in reduced exposure of simvastatin (which is a CYP3A4 substrate) in patients with rheumatoid arthritis. This was attributed to the fact that sarilumab restores CYP3A4 activity, which results in decreased exposure of it’s sensitive substrate simvastatin [92].
5.3. Examples of Growth Factors, Enzymes & Fusion Protein- drug interactions
Not much is known about DDIs of Growth Factors [13], Enzymes & Fusion Protein. (i) Growth factors typically act as signaling molecules between cells, like cytokines and hormones which bind to specific receptors on the surface of their target cells and belong to different large families, including Epidermal growth factor (EGF), Granulocyte macrophage colony-stimulating factor (GMCSF), Fibroblast growth factor (FGF) and others. (ii) Enzymes as therapeutics are generally used for patients who lack certain enzymatic activities due to genetic defects. Thrombolytic therapy has been used for treatment of acute myocardial infarction, pulmonary embolism and various kinds of thrombosis. Several plasminogen activators (PA) have been developed to treat thrombotic diseases (Reteplase, Tenecteplase, Alteplase etc). (iii) Fusion proteins have an immunoglobin Fc domain directly linked to another peptide. Fc portions have powerful biological response and comprise both activating and inhibitory effects. Fc domain markedly prolongs therapeutic activity by increasing plasma half-life. Fc linked peptide partner has multiple function, as a ligand it gets activated upon interaction with a cell-surface receptor or acts as a decoy to distinguish binding partners assembled in a protein microarray. For these strong reasons, Fc-based fusion proteins are of most interest in infectious and inflammatory diseases including RA (abatacept) and psoriasis (alefacept).
6. Regulation of drug metabolizing enzymes during inflammation/infection: potential mechanisms of biologics–small molecule drug interactions
Alterations in drug metabolism are hugely detrimental and will have a very high potential for clinically relevant consequences if the parent drug is toxic and/or has a low safety margin. Clinical instances have shown that drug metabolism is disrupted during different pathophysiological conditions including diabetes, cancer, hepatitis, cirrhosis, endocrine disorders and other infections and inflammation. In patients with infections and inflammation, the expression and activity of the DMEs are primarily down-regulated due to the inflammatory micro-environment in vivo (Fig. 1). As SMDs undergo metabolism/clearance by DMEs, their in vivo concentration is higher in these patients, which may have major implications on safety and efficacy of SMDs with narrow therapeutic window. In 1978, the first case of altered drug metabolism was observed, when patients with influenza showed delayed theophylline elimination [31]. Further, the reduction in drug biotransformation capacity was shown to be paralleled by a decrease in total CYP content and CYP-associated enzyme activities. More recently, HIV infected patients demonstrated 18% lower hepatic CYP3A4 activity, 90% lower CYP2D6 activity and 53% lower N-acetyltransferase 2 activity [32].
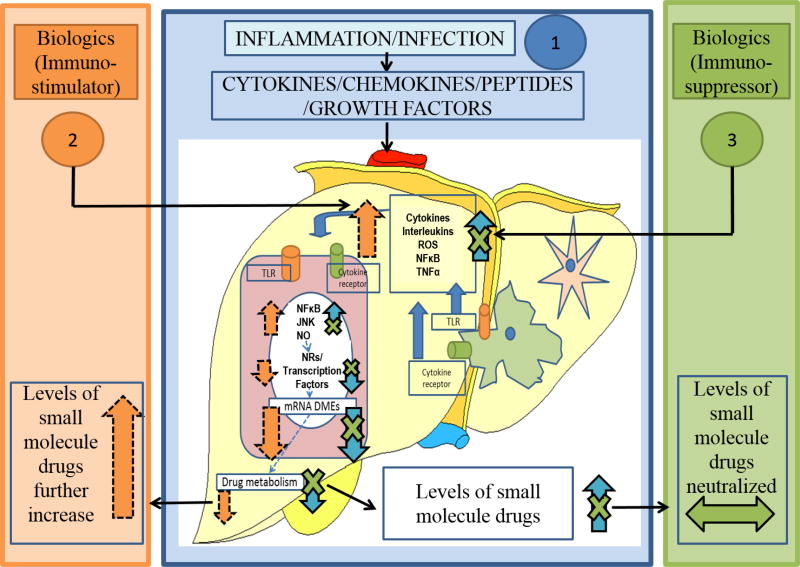
Patients with infectious and inflammatory diseases undergo treatments with a broad range of therapeutic options involving SMDs and biologics individually or in combination. Almost all biologic acts by targeting the inflammatory pathway, therefore treatment with the biologic can alter the expression and activity of the DMEs in these patients and have potential implications for biologic-SMD interactions. For e.g., in patients treated with anti-inflammatory agents can reverse the effect of disease-associated inflammation on DMEs (Fig. 4). As this area of research is still growing, limited information is available on effect of biologics on SMDs.
Figure 4.
Mechanism of Biologics-SMDs interaction. 1) In infection/inflammation disease states, plasma levels of cytokines/chemokines/peptides and growth factors are elevated. These factors act on the liver to activate kupffer cells to release pro-inflammatory cytokines. These intrinsic as well as the circulatory cytokines act on the hepatocytes through the cytokine receptors. Furthermore, the TLRs on the hepatocytes are activated, to induce cell-signaling pathways, leading to the down-regulation of basal transcription factors, NRs and DMEs. This leads to disruption of drug metabolism/clearance. 2) Upon treatment with biologics which are immunostimulators, cytokines in the liver are elevated further, thus potentiating the down-regulation of DMEs, causing more severe disruption of drug metabolism/clearance and increasing the levels of SMDs. 3) In contrast, biologics which are immunosuppressors, attenuate inflammatory disease-mediated elevation of cytokines and restore the expression/activity of DMEs and ultimately levels of SMDs are neutralized.
Mechanisms for biologic-SMD interactions are still poorly or partially understood, and these mechanisms are likely to be very complex. DDIs between two SMDs are different from that of biologics-SMDs primarily due to the differences in the clearance mechanisms of for SMDs and biologics. Like SMDs, clearance mechanisms for biologics don’t involve hepatic metabolism and/or biliary excretion. Therefore, biologics are unlikely to affect the hepatic or biliary elimination of SMDs by direct interactions with the SMDs. In biologics-SMDs interaction, there are three important elements: a perpetrator (biologics or SMD), a victim (biologics or SMD), and a mediator (disease state, immunogenicity, or target physiology). So far, observed effect of biologics on SMDs are indirect through modulation of cytokines. Biologics as immunostimulators will induce an overproduced state of cytokines during inflammatory conditions that will favor further negative regulation of DMEs and reduce metabolism of SMDs. In contrast, biologics that are immunosuppressors can restore/reverse cytokine-mediated suppression of DMEs in inflammatory diseases resulting in a “normalization” of DMEs activities. Such disease–drug interaction may result in risk of therapeutic failure, due to increased clearance of drugs metabolized by DMEs (Fig. 4).
The main mechanisms involved in alteration of hepatic DMEs in infectious and inflammatory states include the effect of inflammatory cytokines, oxidative stress, transcriptional regulation, post-transcriptional regulation, nitric oxide and protein phosphorylations.
6.1. Role of Cytokines
Majority of the studies on DMEs in infections and inflammation have focused on the alterations in CYP enzymes and in most cases, cytokines are mainly involved as mediators of gene expression of these CYP isoforms [33]. Pro-inflammatory cytokines are elevated in infectious/inflammatory diseases which upregulates transcription factors (as discussed below), ultimately leading to down regulated CYP enzymes. Similarly, during treatment with biologics for various cancers, infections and inflammatory disorders, cytokines are usually elevated, which may be responsible for down-regulation of CYP enzymes in a similar manner. Cytokines are released from immune cells and play an important role in cell signaling. Pro-inflammatory cytokines i.e. which promote systemic inflammation such as IL-1, IL-1 β, IL-4, IL-6, TNF-α, IFN-α, -β and -γ, transforming growth factor-b1, human hepatocyte growth factor, and lymphotoxin down-regulate major CYP enzymes with the specific effects on mRNA levels, protein expression, and enzyme activity. However, the extent to which each cytokine alters the expression of a subset of a CYP family may vary and sometimes these repressive effects of cytokines may be additive [34]. Studies employing cytokine or cytokine receptor knockouts have been carried out to better understand the role of cytokines in regulation of DMEs. For example, knock down of IL6, IL-1β genes had no effect on the downregulation of several CYP enzymes after treatment with the gram-negative bacterial endotoxin, lipopolysaccharide (LPS) [35, 36]. This suggests that there might be functional redundancy of various cytokines released during LPS-induced inflammation. Following are few studies that indicate involvement of cytokines in alteration of DMEs during infections and inflammation with focus on CYP enzymes.
Clinically, administration of IFN-α in chronic hepatitis B patients and during influenza, reduced the clearance and increased the concentration of CYP1A2 substrate, theophylline [15, 23]. Likewise, in healthy volunteers, PEG-IFN alfa-2a increased the AUC0-∞ of theophylline that paralleled with reduction in its clearance [27]. In vitro, interferon-α was found to down-regulate CYP1A2 gene, which probably explains the decreased theophylline clearance by interferon. In human primary hepatocytes, IL-6 down-regulated gene expression of CYP2C8, CYP3A4 and CYP2C9 and anti-IL-6 monoclonal antibody partially blocked the IL6-mediated suppression of CYP3A4 and CYP1A2 activities [37]. Therefore, it’s supposed that DDIs observed between sirukumab (anti-IL-6 antibody) and CYP probe substrates, may be due to reversed effect of sirukumab on IL-6 mediated suppression of CYP3A, CYP2C9 and CYP2C19 activities in rheumatoid arthritis patients [28]. Involvement of altered drug metabolism is suspected in increased cyclosporine serum concentrations by both muromonab-CD3 and basiliximab [18, 19]. Muromonab-CD3 releases cytokines such as TNF-α, interferon, and IL-2 [38]. IL-2 can suppress CYP3A activity which may explain the muromonab-CD3–cyclosporine interaction. Basiliximab is an immunosuppressive agent, it binds to the IL-2 receptor (IL-2R) on activated T cells and blocks IL-2 signaling [39]. It’s hypothesized that basiliximab displaces IL-2 which in turn alters CYP3A expression/activity [19]. Similarly, since TNF-α suppresses expression and activity of CYP enzymes, (including CYP3A), it is suggested that increased clearance of duloxetine by adalimumab, a TNF blocker, may be due to normalization of CYP activity.
6.2. Role of Toll like Receptors
In infection and inflammation, innate immune system is activated via pattern recognition receptors such as toll-like receptors (TLR) or nucleotide-binding oligomerization-domain protein (NOD) families. TLRs are present on the cell surface of various immune cells (the resident macrophages or Kupffer cells) as well as the hepatocytes. Similar to TLRs, NOD like receptors are expressed by antigen presenting cells and epithelial cells. They are composed of NOD1, NOD2 and NOD like family receptor proteins, NLRP3/NLRP4 etc. which recognize exogenous stimuli- pathogen associated molecular patterns (PAMPs) and endogenous signals of stress-danger associated molecular patterns (DAMPs) [40, 41]. Out of the 13 TLRs identified in mammals, TLR4 is activated by the bacterial gram-negative component, LPS, and TLR2 is activated by the bacterial gram-positive component, lipoteichoic acid (LTA) [41, 42]. Upon binding of TLR4 to LPS, adapter proteins such as myeloid differentiation factor 88 (MyD88), MyD88 adaptor-like protein (Mal, also known as TIRAP), TIR-containing adapter molecule (TRIF), and TRIF-related adapter molecule (TRAM) are activated. Downstream events in the activation of the MyD88- dependent pathway by LPS, leads to further activation of pro-inflammatory cytokines, NF-κB & MAPK pathways, which have been associated with down regulation of DMEs [43]. It was seen that down-regulation of Cyp3a11 in LPS-sensitive TLR4 wild type (TLR4-wt) (C3HeB/FeJ) mice could not be detected in TLR4-mutant (C3H/HeJ) mice, emphasizing the role of TLR4 [44]. In fact, down-regulation of gene expression of key hepatic phase I and phase II DMEs in TLR2+/+ mice by LTA was also blocked in TLR2−/− mice. Interestingly, our laboratory has shown that LPS or LTA can also directly down-regulate CYPs in primary mouse hepatocytes, independent of cytokines. TLR-mediated signaling is initiated by the down-stream adaptor protein, Toll-interleukin 1 receptor domain containing adaptor protein (TIRAP) and we showed that TIRAP was involved only in TLR2-mediated regulation of DME and transporter genes, and not by TLR4 [44]. Similarly, muramyl dipeptide (MDP) is a well-known NOD receptor ligand which either alone or in combination with TLR’s leads to activation of Kupffer cells and thereby pro-inflammatory cytokines [45]. However, a study conducted by Zidek et al demonstrated that MDP had no effect on the in vitro activity of both rat liver microsomal enzymes studied and similarly, no change was produced in specific rat cytochrome P-450 and b5 content [46].
6.3. Role of transcription factors and nuclear receptors
Transcription factors such as NF-κB, nuclear factor erythroid 2–related factor 2 (Nrf2), mitogen activated protein kinase (MAPK), c-Jun-N-terminal kinase (JNK), CCAAT enhancer binding protein, C/EBPβ etc. are also known to play a significant role in down-regulation of DMEs. It was observed that upon administration of LPS to mice, the mRNA levels of C/EBP α decreased, whereas C/EBPβ and γ mRNAs increased [47]. Also, expression of a truncated form of C/EBPβ lacking transactivation activity (LIP) is increased in hepatocytes after treatment with LPS [48] or pro-inflammatory cytokines [49]. Martinez-Jimenez et al [50] recently identified an enhancer site in the CYP3A4 gene, at which LAP (Liver enriched activator protein) binds and activates transcription, while LIP antagonizes LAP activity. Changes in the LAP:LIP ratio can influence the regulation of CYP3A4 by other well-characterized mechanisms such as rifampicin induction. A study by Jover et al [51] showed that IL-6 caused a marked increase in the translation of C/EBPβ-LIP. Recently, it has been shown that different forms of C/EBPβ were not significantly affected in rat liver 1 h after LPS injection and therefore changes in the LAP:LIP ratio are likely not involved in the suppression of CYP3A2, 2C11, or 2E1 at this early time point [52].
NF-κB is a transcription factor that can either indirectly regulate CYP gene expression through mutual repression between NF-κB and nuclear receptors, or can directly regulate CYP gene expression through binding to NF-κB response element in the promoter region of CYP genes. Interaction of NF-κB with nuclear receptors during pathophysiological conditions can alter expression of DMEs. Cross talk between nuclear receptor pregnane X- receptor (PXR) and NF-κB signaling pathways is believed to coordinately regulate hepatic gene expression during inflammatory response [53]. It was shown that NF-κB disrupts the binding of PXR:RXRα to its regulatory sites by inhibiting PXR-RXRα complex formation, thus downregulating CYP450 gene expression in HepG2 cells [54].
As mentioned earlier, NF-κB can either bind directly to regulatory elements on genes or affect the binding of other transcription factors such as nuclear receptors required for optimum induction of drug metabolizing enzymes. Reductions in mRNA levels of nuclear receptors such as PPARα, PXR, retinoid X receptor (RXR), and liver X receptor have been reported in liver and intestine of rodents treated with LPS [55–57]. However, mRNA and protein expression of several CYPs did not differ in PXR−/− or PPAR−/− mice treated with LPS. We have also previously shown that RNA levels of CAR were suppressed ~60% by LPS treatment of TLR4-wt mice, and this down-regulation was blocked in the TLR4-mutant mice. PXR RNA levels were not changed by LPS treatment of either the TLR4-wt or mutant mice and RXRα protein levels were significantly reduced in the nucleus of TLR4-wt mice, whereas no such reduction was detected in the TLR4-mutant mice [44]. Aryl hydrocarbon receptor (AhR), which is activated by polycyclic aromatic hydrocarbons such as dioxins, has also been known to be involved in acute inflammatory response and regulates CYP enzymes [58]. It has been shown recently that LPS administration leads to a significant reduction of CYP1A1 protein in the liver of AhR−/− mice compared to WT mice [59]. Apart from regulating gene expression of drug metabolizing enzymes, PXR activation has also been associated with suppression of inflammation and acute phase response by attenuating the activity of NF-κB signaling [60]. Further studies revealed that post-translational modifications by small-ubiquitin-related modifier (SUMO) to PXR was required to suppress hepatic acute phase response, specifically pro-inflammatory mediators (IL-1β, IL-6, and TNFα) at the level of transcription [61]. It might be possible that other post-translational modifications such as phosphorylation, acetylation of PXR may produce similar effects, but the exact mechanism still needs to be determined.
Accumulation of reactive oxygen and nitrogen species generated by inflammatory cells leads to activation of another transcription factor Nrf2. LPS and other Toll-like receptor (TLR) agonists activate Nrf2 signaling and the activation is due to the reduction of Keap1, the key Nrf2 inhibitor [62]. The effect of activated Nrf2 on CYP genes was minimal, with only Cyp2a5, Cyp2c50, Cyp2c54, and Cyp2g1 increased, and Cyp2u1 decreased. However, Nrf2 increased mRNA of other phase-I enzymes in mice, such as aldo-keto reductases, carbonyl reductases, and phase-II enzymes such as glutathione S-transferases, UDP- glucuronosyltransferases, and UDP-glucuronic acid synthesis enzymes [63].
Pro-inflammatory cytokines/LPS/LTA are also known to activate mitogen-activated protein kinases (MAPKs) in the liver [45, 64, 65] and these MAPKs such as c-Jun N-terminal kinase (JNK), and extracellular signal activated kinase are involved in regulation of some DMEs [66]. It has been shown that activation of JNK by LPS or IL-1 results in modification and nuclear export of RXRα, and this may contribute to suppression of RXRα-dependent hepatic genes [57, 67]. Apart from RXRα, JNK also inhibits glucocorticoid receptor activity, leading to suppression of CAR gene expression [68]. Furthermore, LPS mediated down-regulation of DMEs was attenuated when primary mouse hepatocytes were treated with specific JNK inhibitor (SP600125, 10µM) [69].
6.4. Role of miRNAs
The kinetics of mRNA suppression suggest that inflammatory regulation may involve destabilization or stabilization of mRNA for some CYPs and this remains an under-studied area. MicroRNAs affect the stability of mRNA post-transcriptionally, either by translational repression or by mRNA degradation. MiRNAs are short RNAs (19–25 nucleotides) typically involved in the downregulation of gene expression. MiRNAs are also believed to play a key role in immune system development and regulation of innate immunity. In a study by Mi et al in 2007, the expression of over 270 mouse miRNAs was analyzed in LPS-treated mouse macrophage cell line RAW264.7. It was found that more than 25 miRNAs were differentially regulated in LPS-treated RAW264.7 cells compared to untreated cells, including the up-regulation of miR-155, miR-132, miR-22, miR-342 and miR-146 genes in LPS-treated cells and the down-regulation of miR-696, miR-805, miR-706, miR-710 and miR-214 genes [70]. Although the exact roles of these miRNAs on DMEs expression remains to be determined, other miRNAs such as miR148a, miR27b etc., have been implicated in the regulation of CYPs. miR27b, in particular, targets the 3’UTR of VDR and CYP3A4 [71]. A recent study suggests that miR-27b may target RXRα which is a necessary component for PXR and VDR to form functional heterodimers which ultimately regulate CYP3A4 transcriptional expression [72]. Lamba et al. (2014) recently identified miR34a as a strong predictor of mRNA levels of CYP3A4, CYP2C19, and several hepatic transcription factors (NR1I2, NR1I3, hepatocyte nuclear factor 4α) [73]. MiR34a has also been implicated in regulation of Nrf2 [74]. Given the observed association of miRNAs with expression levels of multiple CYPs and transcription factors (TFs) mediating inflammation, its upregulation could potentially lead to repression of DME levels in patients having infections and inflammation.
6.5. Role of oxidative stress
Pro-inflammatory cytokines induced during inflammation can also activate nitric-oxide synthase 2 to form nitric oxide (NO) in macrophages and hepatocytes. Although, this NO can lead to destabilization of DME protein structure, a large number of studies have now shown that the downregulation CYP proteins and mRNAs is not affected by deletion of the nitric oxide synthase 2 (NOS2) gene or by inhibitors of NOS enzymes. Pharmacologically derived NO is capable of inhibiting CYP2D6 reporter gene transcription by regulating the activity of transcription factors HNF4 and NFκB. In vitro studies show that IL-1β and TNF-α-mediated down-regulation of CYP protein was NO dependent, but not IL-6 mediated down-regulation [75].
7. Conclusion
The pharmacokinetics of co-administered SMDs can be altered by biologics. Interpretation of biologics-SMDs interaction from in vitro systems is difficult, so clinical cocktail studies are commonly used. Cytokine or cytokine modulators can act on expression and activity of DMEs. In vitro system utilizes only the potential of biologics to modulate cytokines as screening tool. Several different mechanisms can regulate DMEs during inflammation and infection, which suggests strategic inclusion of these mechanisms, will enable prospective prediction of biologics-SMDs interaction.
8. Expert Opinion
New therapeutic agents (SMDs and/or biologics) are being developed targeting inflammatory pathways, transcription factors, nuclear receptors, etc. It is conceivable, that the safety/efficacy of these agents can be compromised due to their impact on the regulation of DMEs in the inflammatory microenvironment associated with diseases states in vivo. Very few dedicated clinical DDIs studies between SMDs and biologics have been conducted and so far potential DDI mechanism suggested, is the involvement of pro-inflammatory cytokines. The critical weaknesses are the lack of understanding of the transcriptional/post-transcriptional mechanism of down-regulation of DMEs during inflammation/infection. Also, there is limited ability to predict clinical biologic-SMD interaction based on animal studies and/or in vitro data.
Recently, TLR agonists and antagonists are being developed as potential treatment for a broad array of diseases. For example, TLR9 agonist has shown substantial evidence of antitumor activity in human clinical trials. TLR9 detect the unmethylated CpG dinucleotides that are relatively common in bacterial and viral genomic DNAs [76]. HEPLISAV-B is combination of TLR9 agonist and hepatitis B surface antigen to elicit an efficient immune response after just two doses. It focuses the immune response to generate protective antibodies rapidly resulting in faster protection. Role of TLRs has also been indicated in atherosclerosis [77, 78]. Activation of TLR2, TLR4 and TLR9 impacts atherosclerotic lesion formation by contributing to foam cell formation via induced lipid uptake and disruption of cholesterol efflux mechanisms [79–81]. In TLR2−/− and TLR4−/− no lesion development occurred, making them attractive targets for treating atherosclerosis [82, 83]. OPN-305, monoclonal antibodies against TLR2 is being tested for the potential treatment of inflammatory diseases [77]. Considering role of TLRs in downregulation of DMEs, it is very much possible that TLR agonist will further downregulate DMEs that were already suppressed during infectious disease. In contrast, TLR antagonists may rebound the DME expression/activity to normal state that was initially altered due to inflammation. Thus, biologics targeting TLRs may affect SMD concentration in patients with inflammatory/infectious diseases.
Antibody-drug conjugates (ADCs) represent a modality of therapeutics in which antibodies are empowered with small molecules to produce a more profound effect on target cells [89]. Heterogeneous nature of conventional ADCs has generally precluded their uses in diseases outside of oncology. Recent progress in formulating very stable linkage, allows precise attachment of a drug at an optimal site on an antibody. Such advancements in technology show promise in development of ADCs for use in chronic diseases such as inflammation. After administration, the ADCs get unconjugated and the small molecule drug once released from ADCs may be metabolized by DMEs and subject to potential drug-drug interactions from P450 inhibitors or inducers.
Recombinant human erythropoietin (Darbepoetin alfa), human keratinocyte growth factor (KGF) (Palifermin), Granulocyte-macrophage colony stimulating factor receptor (GM-CSF-R-alpha or CSF2R) (Sargramostim) are biologics used for the treatment of anemia, oral mucositis and bone marrow transplant respectively [90]. They can bind to their respective receptors to activate the JAK-STAT signaling pathways within the cytosol [91]. Activated STAT (signal transducers and activators of transcription) proteins are translocated to the nucleus where they serve as transcription factors which regulate the activation of specific DME genes. Thus this can lead to potential exaggeration in disease-mediated altered DMEs expression/activity and result in biologic-SMD interaction.
With the discovery of miRNA dysregulation in inflammatory bowel diseases, cancers and viral infections, they emerge as potential drug therapies. Recent clinical trials with miRNA-122 antagonist, Miravirsen for treatment of HCV infection [84] has indicated their effective use. What makes miRNAs more attractive as therapy is their unique role in the regulation of genes involved in immune defense. Some miRNA are suppressor of TLR signaling, whereas others are activators. With LPS treatment, miR-146 & 155 are upregulated that negatively regulates mRNA levels of TNF receptor associated factor 6 (TRAF6), interleukin-1 receptor-associated kinase 1 (IRAK1) [85] and Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP-1), which is a negative regulator of NF-κB signaling [70, 86] respectively. Though miRNA come across as very promising biologics, ability of miRNA to regulate expression of DMEs as stated earlier, will pose potential caveat and risk for interaction with SMDs.
CCAAT/enhancer binding protein (C/EBP)-alpha transcription factors play important role in the transcriptional regulation of cytochromes P450 such as CYP2A6, CYP2B6, CYP2C9, CYP2D6, CYP3A4, CYP3A5 and CYP3A7 [87]. In hepatocytes, this regulation takes place in cooperation with HNFs and other transcription factors. C/EBP alpha and HNF-3 alpha regulate CYP3A4 gene expression probably by chromatin remodeling [88]. C/EBP-alpha has been indicated as potential therapeutic target in osteogenesis (In vitro C3H10T1/2 cells induced by BMP-2). Thus it can be well perceived that further development of C/EBP-alpha as agonist/antagonist for therapeutic proteins can alter DMEs post translationally and have implication on interaction with SMDs. HNF4α is potential targetable onco-protein in Gastric Cancer. Given that (C/EBP)-alpha along with HNF4α regulates post translational modification of DMEs, HNF4α agonists too have potential for DDI with SMDs.
Given the potential of biologic-SMD interaction in clinical settings, it is critical for us to understand the mechanism of altered DME expression/activity during inflammation, and the role of biologics on DME regulation. Down-regulation of DMEs during inflammation may be mediated by signaling pathways activated by TLRs, kinases and other signaling pathways. Similarly, basal transcription factors and nuclear receptors may mediate the regulation of DMEs during inflammation. Post-transcriptional modification by miRNAs may regulate DMEs, and consequently drug disposition. To date, all the key regulators involved in alteration of DMEs during inflammation have not been identified. It is also plausible that specific regulators may be involved in modulation of specific phase I and phase II DMEs. Furthermore, a key limitation is our lack of understanding of human DME regulation in vivo, due to the limitations of experiments conducted in human cell-lines and in animal models. Therefore, it will be important to conduct exhaustive and comprehensive mechanistic studies using in vitro cell-culture models as well as in vivo humanized mice and patient tissue samples.
A thorough understanding of the mechanism will enable future predictions and interventions to prevent adverse effects due to biologic/SMD interactions in patients with infectious/inflammatory diseases undergoing biologic/SMD co-treatments.
highlights box.
Metabolism and clearance of small molecules may be disrupted during infectious and inflammatory diseases.
Biologics can alter metabolism/disposition of small molecule drugs (SMDs) directly, by altering their metabolism, or indirectly through modulation of cytokines.
Regulation of drug metabolizing enzymes (DMEs) by biologics may be mediated by modulation of transcription factors, nuclear receptors, TLRs, miRNA and oxidative stress.
Inflammatory modulators play key roles in controlling therapeutic efficacy and safety in patients undergoing treatments with biologics and small molecules in infectious/inflammatory diseases.
Acknowledgments
Funding
U.S. Department of Health and Human Services, National Institutes of Health, National Heart, Lung, and Blood Institute, HL-112516, HL129794; U.S. Department of Health and Human Services, National Institutes of Health, National Institute of Environmental Health Sciences, ES-009132, ES-019689; U.S. Department of Health and Human Services, National Institutes of Health, National Institute on Drug Abuse DA035751
Abbreviations
- DMEs
Drug metabolizing enzyme
- SMDs
small molecule drugs
- PXR
pregnane X receptor
- FDA
U. S Food and Drug Administration
- CYP450
Cytochrome P450s
- mAbs
monoclonal antibodies
- IFNs
interferons
- IL
Interleukins
- TNF
Tumor necrosis factors
- RA
Rheumatoid arthritis
- NAT
N-Acetyltransferase
- LPS
lipopolysaccharide
- TLR
toll-like receptors
- NOD
nucleotide-binding oligomerization-domain protein
- NLRP
NOD like family receptor proteins
- PAMPs
pathogen associated molecular patterns
- DAMPs
danger associated molecular patterns
- MyD88
myeloid differentiation factor 88
- TRIF
TIR-containing adapter molecule
- TRAM
TRIF-related adapter molecule
- TIRAP
Toll-interleukin 1 receptor domain containing adaptor protein
- MAPK
mitogen activated protein kinase
- JNK
c-Jun N-terminal kinase
- C/EBP
CCAAT/enhancer binding protein
- HNF
Hepatocyte nuclear factor
- PPAR
peroxisome proliferator-activated receptors
- RXR
retinoid X receptor
- AhR
Aryl hydrocarbon receptor
- Nrf2
Nuclear factor (erythroid-derived 2)-like 2
- NOS2
Nitric Oxide Synthase 2
- TRAF
TNF receptor associated factor
- IRAK1
Interleukin-1 receptor-associated kinase 1
- SHIP-1
Src homology-2 domain-containing inositol 5-phosphatase 1
- ADC
Antibody-drug conjugates
Footnotes
Declaration of interest
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Evaluate Pharma World Preview 2015, Outlook to 2020. Available at: http://info.evaluategroup.com/rs/evaluatepharmaltd/images/EP240614.pdf.
- 2.Alduaij W, Illidge TM. The future of anti-CD20 monoclonal antibodies: are we making progress? Blood. 2011 Mar 17;117:2993–3001. doi: 10.1182/blood-2010-07-298356. [DOI] [PubMed] [Google Scholar]
- 3.Lee JI, Zhang L, Men AY, et al. CYP-mediated therapeutic protein-drug interactions: clinical findings, proposed mechanisms and regulatory implications. Clinical pharmacokinetics. 2010 May;49:295–310. doi: 10.2165/11319980-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 4.Morrow T, Felcone LH. Defining the difference: What Makes Biologics Unique. Biotechnology healthcare. 2004 Sep;1:24–9. [PMC free article] [PubMed] [Google Scholar]
- 5.Dimitrov DS. Therapeutic proteins. Methods in molecular biology. 2012;899:1–26. doi: 10.1007/978-1-61779-921-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nature reviews Drug discovery. 2008 Jan;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 7.Vugmeyster Y, Xu X, Theil FP, et al. Pharmacokinetics and toxicology of therapeutic proteins: Advances and challenges. World journal of biological chemistry. 2012 Apr 26;3:73–92. doi: 10.4331/wjbc.v3.i4.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keizer RJ, Huitema AD, Schellens JH, et al. Clinical pharmacokinetics of therapeutic monoclonal antibodies. Clinical pharmacokinetics. 2010 Aug;49:493–507. doi: 10.2165/11531280-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 9.Brinks V, Jiskoot W, Schellekens H. Immunogenicity of therapeutic proteins: the use of animal models. Pharmaceutical research. 2011 Oct;28:2379–85. doi: 10.1007/s11095-011-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X, Vugmeyster Y. Challenges and opportunities in absorption, distribution, metabolism, and excretion studies of therapeutic biologics. The AAPS journal. 2012 Dec;14:781–91. doi: 10.1208/s12248-012-9388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seitz K, Zhou H. Pharmacokinetic drug-drug interaction potentials for therapeutic monoclonal antibodies: reality check. Journal of clinical pharmacology. 2007 Sep;47:1104–18. doi: 10.1177/0091270007306958. ● This review provides a systematic overview of current literature and offers some practical considerations while designing pharmacokinetic drug-drug interaction with monoclonal antibodies. [DOI] [PubMed] [Google Scholar]
- 12.Mahmood I, Green MD. Drug interaction studies of therapeutic proteins or monoclonal antibodies. Journal of clinical pharmacology. 2007 Dec;47:1540–54. doi: 10.1177/0091270007308616. [DOI] [PubMed] [Google Scholar]
- 13.Product Information GEM 21S (Growth-factor Enhanced Matrix) BioMimetic Pharmaceuticals, Inc
- 14.Product Information. IFNα2b (Intron A) Merck Sharp & Dohme Corp; [Google Scholar]
- 15.Williams SJ, Baird-Lambert JA, Farrell GC. Inhibition of theophylline metabolism by interferon. Lancet. 1987 Oct 24;2:939–41. doi: 10.1016/s0140-6736(87)91422-x. [DOI] [PubMed] [Google Scholar]
- 16.Product Information. PEGylated IFNα2b (PegIntron) Merck Sharp & Dohme Corp; [Google Scholar]
- 17.Product Information Poryrazza (Necitumumab) Eli Lilly and Company; [Google Scholar]
- 18.Vasquez EM, Pollak R. OKT3 therapy increases cyclosporine blood levels. Clinical transplantation. 1997 Feb;11:38–41. [PubMed] [Google Scholar]
- 19.Strehlau J, Pape L, Offner G, et al. Interleukin-2 receptor antibody-induced alterations of ciclosporin dose requirements in paediatric transplant recipients. Lancet. 2000 Oct 14;356:1327–8. doi: 10.1016/s0140-6736(00)02822-1. [DOI] [PubMed] [Google Scholar]
- 20.Furtlehner A, Schueller J, Jarisch I, et al. Disposition of paclitaxel (Taxol) and its metabolites in patients with advanced breast cancer (ABC) when combined with trastuzumab (Hercpetin) European journal of drug metabolism and pharmacokinetics. 2005 Jul-Sep;30:145–50. doi: 10.1007/BF03190613. [DOI] [PubMed] [Google Scholar]
- 21.Wu JJ, Fleming KF. Interaction between adalimumab with concurrent pregabalin and duloxetine administration in a psoriasis patient with diabetic peripheral neuropathy. Cutis. 2011 May;87:249–50. [PubMed] [Google Scholar]
- 22.Okuno H, Takasu M, Kano H, et al. Depression of drug-metabolizing activity in the human liver by interferon-beta. Hepatology. 1993 Jan;17:65–9. [PubMed] [Google Scholar]
- 23.Kraemer MJ, Furukawa CT, Koup JR, et al. Altered theophylline clearance during an influenza B outbreak. Pediatrics. 1982 Apr;69:476–80. [PubMed] [Google Scholar]
- 24.Prandota J. Important role of proinflammatory cytokines/other endogenous substances in drug-induced hepatotoxicity: depression of drug metabolism during infections/inflammation states, and genetic polymorphisms of drug-metabolizing enzymes/cytokines may markedly contribute to this pathology. American journal of therapeutics. 2005 May-Jun;12:254–61. [PubMed] [Google Scholar]
- 25.Craig PI, Tapner M, Farrell GC. Interferon suppresses erythromycin metabolism in rats and human subjects. Hepatology. 1993 Feb;17:230–5. [PubMed] [Google Scholar]
- 26.Williams SJ, Farrell GC. Inhibition of antipyrine metabolism by interferon. British journal of clinical pharmacology. 1986 Nov;22:610–2. doi: 10.1111/j.1365-2125.1986.tb02943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennan BJ, Xu ZX, Grippo JF. Effect of peginterferon alfa-2a (40KD) on cytochrome P450 isoenzyme activity. British journal of clinical pharmacology. 2013 Feb;75:497–506. doi: 10.1111/j.1365-2125.2012.04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhuang Y, de Vries DE, Xu Z, et al. Evaluation of disease-mediated therapeutic protein-drug interactions between an anti-interleukin-6 monoclonal antibody (sirukumab) and cytochrome P450 activities in a phase 1 study in patients with rheumatoid arthritis using a cocktail approach. Journal of clinical pharmacology. 2015 Dec;55:1386–94. doi: 10.1002/jcph.561. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt C, Kuhn B, Zhang X, et al. Disease-drug-drug interaction involving tocilizumab and simvastatin in patients with rheumatoid arthritis. Clinical pharmacology and therapeutics. 2011 May;89:735–40. doi: 10.1038/clpt.2011.35. [DOI] [PubMed] [Google Scholar]
- 30.Doi T, Muro K, Yoshino T, et al. Phase 1 pharmacokinetic study of MK-0646 (dalotuzumab), an anti-insulin-like growth factor-1 receptor monoclonal antibody, in combination with cetuximab and irinotecan in Japanese patients with advanced colorectal cancer. Cancer chemotherapy and pharmacology. 2013 Sep;72:643–52. doi: 10.1007/s00280-013-2240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang KC, Bell TD, Lauer BA, et al. Altered theophylline pharmacokinetics during acute respiratory viral illness. Lancet. 1978 May 27;1:1132–3. doi: 10.1016/S0140-6736(78)90305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones AE, Brown KC, Werner RE, et al. Variability in drug metabolizing enzyme activity in HIV-infected patients. European journal of clinical pharmacology. 2010 May;66:475–85. doi: 10.1007/s00228-009-0777-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aitken AE, Richardson TA, Morgan ET. Regulation of drug-metabolizing enzymes and transporters in inflammation. Annual review of pharmacology and toxicology. 2006;46:123–49. doi: 10.1146/annurev.pharmtox.46.120604.141059. ●● This review presents good overview of experimental findings that indicates role of inflammatory markers in regulation of DMEs during inflammation and infection. [DOI] [PubMed] [Google Scholar]
- 34.Zidek Z, Anzenbacher P, Kmonickova E. Current status and challenges of cytokine pharmacology. British journal of pharmacology. 2009 Jun;157:342–61. doi: 10.1111/j.1476-5381.2009.00206.x. ● This is a good review that is tailored to cytokine’s potential as immunotherapeutic interventions in major disease areas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siewert E, Bort R, Kluge R, et al. Hepatic cytochrome P450 down-regulation during aseptic inflammation in the mouse is interleukin 6 dependent. Hepatology. 2000 Jul;32:49–55. doi: 10.1053/jhep.2000.8532. [DOI] [PubMed] [Google Scholar]
- 36.Ashino T, Oguro T, Shioda S, et al. Involvement of interleukin-6 and tumor necrosis factor alpha in CYP3A11 and 2C29 down-regulation by Bacillus Calmette-Guerin and lipopolysaccharide in mouse liver. Drug metabolism and disposition: the biological fate of chemicals. 2004 Jul;32:707–14. doi: 10.1124/dmd.32.7.707. [DOI] [PubMed] [Google Scholar]
- 37.Dickmann LJ, Patel SK, Rock DA, et al. Effects of interleukin-6 (IL-6) and an anti-IL-6 monoclonal antibody on drug-metabolizing enzymes in human hepatocyte culture. Drug metabolism and disposition: the biological fate of chemicals. 2011 Aug;39:1415–22. doi: 10.1124/dmd.111.038679. [DOI] [PubMed] [Google Scholar]
- 38.Chatenoud L, Ferran C, Legendre C, et al. In vivo cell activation following OKT3 administration. Systemic cytokine release and modulation by corticosteroids. Transplantation. 1990 Apr;49:697–702. doi: 10.1097/00007890-199004000-00009. [DOI] [PubMed] [Google Scholar]
- 39.Product Information. Simulect (Basiliximab) Novartis Pharmaceutical Corp
- 40.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006 Feb 24;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi O, Hoshino K, Kawai T, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999 Oct;11:443–51. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 42.Aliprantis AO, Yang RB, Mark MR, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999 Jul 30;285:736–9. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 43.Ghose R, Guo T, Vallejo JG, et al. Differential role of Toll-interleukin 1 receptor domain-containing adaptor protein in Toll-like receptor 2-mediated regulation of gene expression of hepatic cytokines and drug-metabolizing enzymes. Drug metabolism and disposition: the biological fate of chemicals. 2011 May;39:874–81. doi: 10.1124/dmd.110.037382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghose R, White D, Guo T, et al. Regulation of hepatic drug-metabolizing enzyme genes by Toll-like receptor 4 signaling is independent of Toll-interleukin 1 receptor domain-containing adaptor protein. Drug metabolism and disposition: the biological fate of chemicals. 2008 Jan;36:95–101. doi: 10.1124/dmd.107.018051. ●● This article shows TLR as novel mechanism involved in alteration of drug metabolism through regulation of DMEs and transporters. [DOI] [PubMed] [Google Scholar]
- 45.Geddes K, Magalhaes JG, Girardin SE. Unleashing the therapeutic potential of NOD-like receptors. Nature reviews Drug discovery. 2009 Jun;8:465–79. doi: 10.1038/nrd2783. [DOI] [PubMed] [Google Scholar]
- 46.Zidek Z, Kamenikova L, Buchar E, et al. Biotransformation of drugs in rats treated with a synthetic muramyl dipeptide, N-acetylmuramyl-L-alanyl-D-isoglutamine (MDP) International journal of immunopharmacology. 1983;5:151–5. doi: 10.1016/0192-0561(83)90007-3. [DOI] [PubMed] [Google Scholar]
- 47.Cantwell CA, Sterneck E, Johnson PF. Interleukin-6-specific activation of the C/EBPdelta gene in hepatocytes is mediated by Stat3 and Sp1. Molecular and cellular biology. 1998 Apr;18:2108–17. doi: 10.1128/mcb.18.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.An MR, Hsieh CC, Reisner PD, et al. Evidence for posttranscriptional regulation of C/EBPalpha and C/EBPbeta isoform expression during the lipopolysaccharide-mediated acute-phase response. Molecular and cellular biology. 1996 May;16:2295–306. doi: 10.1128/mcb.16.5.2295. ● Good article indicating the role of transcription factors in the regulation of drug metabolizing enzymes during inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baumann H, Morella KK, Campos SP, et al. Role of CAAT-enhancer binding protein isoforms in the cytokine regulation of acute-phase plasma protein genes. The Journal of biological chemistry. 1992 Sep 25;267:19744–51. [PubMed] [Google Scholar]
- 50.Jover R, Bort R, Gomez-Lechon MJ, et al. Down-regulation of human CYP3A4 by the inflammatory signal interleukin-6: molecular mechanism and transcription factors involved. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2002 Nov;16:1799–801. doi: 10.1096/fj.02-0195fje. [DOI] [PubMed] [Google Scholar]
- 51.Martinez-Jimenez CP, Gomez-Lechon MJ, Castell JV, et al. Transcriptional regulation of the human hepatic CYP3A4: identification of a new distal enhancer region responsive to CCAAT/enhancer-binding protein beta isoforms (liver activating protein and liver inhibitory protein) Molecular pharmacology. 2005 Jun;67:2088–101. doi: 10.1124/mol.104.008169. [DOI] [PubMed] [Google Scholar]
- 52.Cheng PY, Wang M, Morgan ET. Rapid transcriptional suppression of rat cytochrome P450 genes by endotoxin treatment and its inhibition by curcumin. The Journal of pharmacology and experimental therapeutics. 2003 Dec;307:1205–12. doi: 10.1124/jpet.103.057174. [DOI] [PubMed] [Google Scholar]
- 53.Zhou C, Tabb MM, Nelson EL, et al. Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. The Journal of clinical investigation. 2006 Aug;116:2280–89. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gu X, Ke S, Liu D, et al. Role of NF-kappaB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. The Journal of biological chemistry. 2006 Jun 30;281:17882–9. doi: 10.1074/jbc.M601302200. ● Good article illustrating the mechanism of NF-kappaB-mediated transcriptional regulation of DMEs by physiological and pathological stimuli. [DOI] [PubMed] [Google Scholar]
- 55.Beigneux AP, Moser AH, Shigenaga JK, et al. The acute phase response is associated with retinoid X receptor repression in rodent liver. The Journal of biological chemistry. 2000 May 26;275:16390–9. doi: 10.1074/jbc.M000953200. [DOI] [PubMed] [Google Scholar]
- 56.Kalitsky-Szirtes J, Shayeganpour A, Brocks DR, et al. Suppression of drug-metabolizing enzymes and efflux transporters in the intestine of endotoxin-treated rats. Drug metabolism and disposition: the biological fate of chemicals. 2004 Jan;32:20–7. doi: 10.1124/dmd.32.1.20. [DOI] [PubMed] [Google Scholar]
- 57.Ghose R, Zimmerman TL, Thevananther S, et al. Endotoxin leads to rapid subcellular re-localization of hepatic RXRalpha: A novel mechanism for reduced hepatic gene expression in inflammation. Nuclear receptor. 2004 Aug 16;2:4. doi: 10.1186/1478-1336-2-4. ● This article provides a novel molecular mechanism for regulation of drug metabolizing enzymes (DMEs) in inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimura A, Naka T, Nakahama T, et al. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. The Journal of experimental medicine. 2009 Aug 31;206:2027–35. doi: 10.1084/jem.20090560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu D, Li W, Lok P, et al. AhR deficiency impairs expression of LPS-induced inflammatory genes in mice. Biochemical and biophysical research communications. 2011 Jul 1;410:358–63. doi: 10.1016/j.bbrc.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shah YM, Ma X, Morimura K, et al. Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-kappaB target gene expression. American journal of physiology Gastrointestinal and liver physiology. 2007 Apr;292:G1114–22. doi: 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- 61.Sun M, Cui W, Woody SK, et al. Pregnane X receptor modulates the inflammatory response in primary cultures of hepatocytes. Drug metabolism and disposition: the biological fate of chemicals. 2015 Mar;43:335–43. doi: 10.1124/dmd.114.062307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin S, Cao W. Toll-Like Receptor Signaling Induces Nrf2 Pathway Activation through p62-Triggered Keap1 Degradation. Molecular and cellular biology. 2015 Aug;35:2673–83. doi: 10.1128/MCB.00105-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu KC, Cui JY, Klaassen CD. Effect of graded Nrf2 activation on phase-I and -II drug metabolizing enzymes and transporters in mouse liver. PloS one. 2012;7:e39006. doi: 10.1371/journal.pone.0039006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moriguchi T, Toyoshima F, Masuyama N, et al. A novel SAPK/JNK kinase, MKK7, stimulated by TNFalpha and cellular stresses. The EMBO journal. 1997 Dec 1;16:7045–53. doi: 10.1093/emboj/16.23.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghose R, Guo T, Haque N. Regulation of gene expression of hepatic drug metabolizing enzymes and transporters by the Toll-like receptor 2 ligand, lipoteichoic acid. Archives of biochemistry and biophysics. 2009 Jan 1;481:123–30. doi: 10.1016/j.abb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu R, Lei W, Mandlekar S, et al. Role of a mitogen-activated protein kinase pathway in the induction of phase II detoxifying enzymes by chemicals. The Journal of biological chemistry. 1999 Sep 24;274:27545–52. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- 67.Li D, Zimmerman TL, Thevananther S, et al. Interleukin-1 beta-mediated suppression of RXR:RAR transactivation of the Ntcp promoter is JNK-dependent. The Journal of biological chemistry. 2002 Aug 30;277:31416–22. doi: 10.1074/jbc.M204818200. [DOI] [PubMed] [Google Scholar]
- 68.Pascussi JM, Dvorak Z, Gerbal-Chaloin S, et al. Pathophysiological factors affecting CAR gene expression. Drug metabolism reviews. 2003 Nov;35:255–68. doi: 10.1081/dmr-120026394. [DOI] [PubMed] [Google Scholar]
- 69.Shah P, Omoluabi O, Moorthy B, et al. Role of Adaptor Protein Toll-Like Interleukin Domain Containing Adaptor Inducing Interferon beta in Toll-Like Receptor 3- and 4-Mediated Regulation of Hepatic Drug Metabolizing Enzyme and Transporter Genes. Drug metabolism and disposition: the biological fate of chemicals. 2016 Jan;44:61–7. doi: 10.1124/dmd.115.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tili E, Michaille JJ, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. Journal of immunology. 2007 Oct 15;179:5082–9. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 71.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug metabolism and disposition: the biological fate of chemicals. 2009 Oct;37:2112–7. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ji J, Zhang J, Huang G, et al. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS letters. 2009 Feb 18;583:759–66. doi: 10.1016/j.febslet.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 73.Lamba V, Ghodke Y, Guan W, et al. microRNA-34a is associated with expression of key hepatic transcription factors and cytochromes P450. Biochemical and biophysical research communications. 2014 Mar 7;445:404–11. doi: 10.1016/j.bbrc.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell. 2003 Dec 26;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 75.Carlson TJ, Billings RE. Role of nitric oxide in the cytokine-mediated regulation of cytochrome P-450. Molecular pharmacology. 1996 May;49:796–801. [PubMed] [Google Scholar]
- 76.Hemmi H, Takeuchi O, Kawai T, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000 Dec 7;408:740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 77.Hennessy EJ, Parker AE, O’Neill LA. Targeting Toll-like receptors: emerging therapeutics? Nature reviews Drug discovery. 2010 Apr;9:293–307. doi: 10.1038/nrd3203. ● This review very nicely highlights the promising role of Toll-like receptors (TLRs) in treatment of inflammatory diseases. [DOI] [PubMed] [Google Scholar]
- 78.Cole JE, Mitra AT, Monaco C. Treating atherosclerosis: the potential of Toll-like receptors as therapeutic targets. Expert review of cardiovascular therapy. 2010 Nov;8:1619–35. doi: 10.1586/erc.10.149. [DOI] [PubMed] [Google Scholar]
- 79.Funk JL, Feingold KR, Moser AH, et al. Lipopolysaccharide stimulation of RAW 264.7 macrophages induces lipid accumulation and foam cell formation. Atherosclerosis. 1993 Jan 4;98:67–82. doi: 10.1016/0021-9150(93)90224-i. [DOI] [PubMed] [Google Scholar]
- 80.Oiknine J, Aviram M. Increased susceptibility to activation and increased uptake of low density lipoprotein by cholesterol-loaded macrophages. Arteriosclerosis and thrombosis : a journal of vascular biology / American Heart Association. 1992 Jun;12:745–53. doi: 10.1161/01.atv.12.6.745. [DOI] [PubMed] [Google Scholar]
- 81.Choi SH, Harkewicz R, Lee JH, et al. Lipoprotein accumulation in macrophages via toll-like receptor-4-dependent fluid phase uptake. Circulation research. 2009 Jun 19;104:1355–63. doi: 10.1161/CIRCRESAHA.108.192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mullick AE, Tobias PS, Curtiss LK. Modulation of atherosclerosis in mice by Toll-like receptor 2. The Journal of clinical investigation. 2005 Nov;115:3149–56. doi: 10.1172/JCI25482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Michelsen KS, Wong MH, Shah PK, et al. Lack of Toll-like receptor 4 or myeloid differentiation factor 88 reduces atherosclerosis and alters plaque phenotype in mice deficient in apolipoprotein E. Proceedings of the National Academy of Sciences of the United States of America. 2004 Jul 20;101:10679–84. doi: 10.1073/pnas.0403249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. The New England journal of medicine. 2013 May 2;368:1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 85.Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006 Aug 15;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Connell RM, Chaudhuri AA, Rao DS, et al. Inositol phosphatase SHIP1 is a primary target of miR-155. Proceedings of the National Academy of Sciences of the United States of America. 2009 Apr 28;106:7113–8. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pitarque M, Rodriguez-Antona C, Oscarson M, et al. Transcriptional regulation of the human CYP2A6 gene. The Journal of pharmacology and experimental therapeutics. 2005 May;313:814–22. doi: 10.1124/jpet.104.081570. [DOI] [PubMed] [Google Scholar]
- 88.Rodriguez-Antona C, Bort R, Jover R, et al. Transcriptional regulation of human CYP3A4 basal expression by CCAAT enhancer-binding protein alpha and hepatocyte nuclear factor-3 gamma. Molecular pharmacology. 2003 May;63:1180–9. doi: 10.1124/mol.63.5.1180. [DOI] [PubMed] [Google Scholar]
- 89.Bouchard H, Viskov C, Garcia-Echeverria C. Antibody-drug conjugates-a new wave of cancer drugs. Bioorganic & medicinal chemistry letters. 2014 Dec 1;24:5357–63. doi: 10.1016/j.bmcl.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 90.Product Information. Leukine (sargramostim) Sanofi-aventis US LLC; [Google Scholar]
- 91.Fortin CF, Larbi A, Dupuis G, et al. GM-CSF activates the Jak/STAT pathway to rescue polymorphonuclear neutrophils from spontaneous apoptosis in young but not elderly individuals. Biogerontology. 2007 Apr;8:173–87. doi: 10.1007/s10522-006-9067-1. [DOI] [PubMed] [Google Scholar]
- 92.Lee EB, Daskalakis N, Xu C, et al. Disease-Drug Interaction of Sarilumab and Simvastatin in Patients with Rheumatoid Arthritis. Clin Pharmacokinet. 2016 doi: 10.1007/s40262-016-0462-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Product Information. Simulect (basiliximab) Novartis Pharmaceutical Corp; [Google Scholar]
- 94.Product Information. ACTEMRA (tocilizumab) Genentech; [Google Scholar]