Abstract
Tumors from colorectal cancer (CRC) are generally immunogenic and commonly infiltrated with T lymphocytes. However, the details of the adaptive immune reaction to these tumors are poorly understood. We have accrued both colon tumor samples and adjacent healthy mucosal samples from 15 CRC patients to study lymphocytes infiltrating these tissues. We apply a method for detailed sequencing of T-cell receptor (TCR) sequences from tumor-infiltrating lymphocytes (TILs) in CRC tumors at high throughput to probe T-cell clones in comparison with the TCRs from adjacent healthy mucosal tissue. In parallel, we captured TIL counts using standard immunohistochemistry. The variation in diversity of the TIL repertoire was far wider than the variation of T-cell clones in the healthy mucosa, and the oligoclonality was higher on average in the tumors. However, the diversity of the T-cell repertoire in both CRC tumors and healthy mucosa was on average 100-fold lower than in peripheral blood. Using the TCR sequences to identify and track clones between mucosal and tumor samples, we determined that the immune response in the tumor is different than in the adjacent mucosal tissue, and the number of shared clones is not dependent on distance between the samples. Together, these data imply that CRC tumors induce a specific adaptive immune response, but that this response differs widely in strength and breadth between patients.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-013-1446-2) contains supplementary material, which is available to authorized users.
Keywords: Tumor-infiltrating lymphocytes, Immune repertoire sequencing, T-cell receptor β, Colorectal cancer
Introduction
Tumor immunogenicity is emerging as an important but incompletely understood contributor to cancer progression. A tumor’s ability to both evade and suppress the host’s T lymphocyte response likely affects survival, progression, and metastasis [1]. The presence of tumor-infiltrating lymphocytes (TILs) is often interpreted as evidence that the host’s immune system, at some level, is responding and attempting to eliminate the tumor [2]. This interpretation is supported by multiple observations that for several known immunogenic cancers, including colorectal cancer (CRC), the presence, density, and phenotype of TILs are correlated with outcomes including disease-free survival (DFS) and overall survival (OS) [3–5]. However, T-cell count is a very gross view of a T-cell population. Underneath the total number of infiltrating T cells is a complex population that may be characterized by a diverse or restricted T-cell receptor (TCR) repertoire. Capturing not just the total cell count, but the clonal lineages of T cells allows a new way to analyze and understand TIL populations. This project uses high-throughput sequencing of the TCR Beta (TCRB) repertoire to quantify the immune repertoire of infiltrating lymphocytes in CRC tumors and paired mucosal tissue. These sequencing data complement previously collected CD3+ immunohistochemistry data. These data allow us, as a vital first step, to characterize the variation in diversity and clonality of T cells infiltrating both normal mucosa and colon tumor.
While many cancers are considered immunogenic, data on CRC give strong evidence that anti-tumor immune response may be a clinically reliable prognostic indicator [3]. Multiple independent studies have indicated that the presence and density of TILs in CRC tumors are strongly correlated with DFS and OS. The methods and the type of T cells that were quantified varied by study; overall, presence and high density of CD3+ (total), CD8+ (cytotoxic), CD4+ (helper), and memory TILs were associated with better outcomes, while the presence and high density of Tregs were associated with poorer outcomes in CRC patients [1, 4]. In addition, higher expression of genes involved in the cytotoxic T-cell response (e.g. interferon gamma) is also correlated with lower risk of relapse. Pages et al. have reported that immune markers alone predicted outcome better than TNM staging [5]. Thus, CRC is an ideal cancer in which to study the abundance, phenotype, and clonal structure of tumor-infiltrating T lymphocytes.
While data on the prognostic significance of total count and phenotype of TILs are strong, less is known of the significance of the antigen receptor repertoire. Quantifying the antigen receptor repertoire would allow researchers to elucidate whether the population of T cells that infiltrates tumor cells is measurably different than the T cells that are associated with adjacent tissue. We apply this approach to investigate whether TIL populations harbor a specialized repertoire of T cells (presumably targeting specific tumor antigens) or whether the intratumoral T-cell repertoire is closely related to the repertoire of T cells infiltrating healthy adjacent tissue. Given the heterogeneity of patients’ immune response to tumors, the answer to this question could easily vary between patients.
However, quantifying the clonal structure of the TCRB repertoire is technically difficult as the cellular adaptive immune system generates a remarkable breadth of diversity in antigen-specific TCRs by combinatoric shuffling of gene segments in somatic cells. The TCR is composed of two peptide chains, encoded by the TCRA and TCRB genes, respectively. The antigenic specificity of αβ T lymphocytes is in large part determined by the amino acid sequence in the hypervariable complementarity-determining region 3 (CDR3) regions of TCRs [6]. The existence of multiple Vβ, Dβ, and Jβ gene segments in the TCRB locus permits a large combinatorial diversity in receptor composition, and template-independent deletion or insertion of nucleotides at the Vβ–Dβ, and Dβ–Jβ junctions further adds to the potential diversity of receptors that can be encoded.
Previous technologies permitted quantification of one component of the CDR3 chain diversity at a time (either V gene usage with Betamark or CDR3 length distribution with spectratyping). While betamark spectratyping combines these two measures, it is still an indirect measure of the TCRB repertoire diversity, both because betamark technology only assays some V gene segments and because V gene usage and CDR3 length together are still insufficient to generate a unique molecular tag for each T-cell clone. Herein, we apply a multiplex PCR method that amplifies rearranged TCRB CDR3 sequences [7] and exploits the capacity of high-throughput sequencing technology to sequence tens of thousands of TCRB CDR3 chains simultaneously. By directly sequencing the TCRB CDR3 chains, the assay captures the true diversity of the TCRB repertoire since the CDR3 region contains enough information for its full sequence to act as a unique molecular tag identifying each T-cell clone. We apply the multiplex PCR assay to amplify TCRB CDR3 chains from T cells infiltrating both colorectal tumor and matched mucosal tissue from 15 CRC patients.
With these data we can describe the TCRB repertoire of colorectal tumor and adjacent mucosal tissue. Specifically, with these data we can assess (1) whether T cells infiltrating colorectal tumors are a different population than mucosal tissue-localized T cells, (2) whether the population-level characteristics of the intratumoral TCRB repertoire in colorectal tumors are different than those of mucosal tissue, and 3) whether such variations in T-cell repertoire are consistent across individuals.
Methods
Study population
This study is based on CRC patients who participate in the ColoCare study, Heidelberg, Germany. The ColoCare Consortium is a multicenter initiative establishing an international cohort of CRC patients for interdisciplinary studies of CRC prognosis and outcomes, with sites at the Fred Hutchinson Cancer Research Center, Seattle (Washington, USA), H. Lee Moffitt Cancer Center and Research Institute, Tampa (Florida, USA), and the National Center for Tumor Diseases, Heidelberg (Germany). This pilot study involves n = 15 CRC patients, recruited between October 2010 and November 2011 at the Division of Preventive Oncology, in Heidelberg, Germany (Table 1). Eligibility criteria for this study included: at least 18 years of age, first diagnosis of colon or rectal cancer (stages I-IV), and availability of both tumor and healthy mucosal tissue. The study was approved by the Institutional Review Board and all participants provided written informed consent.
Table 1.
Study population
| Variable | N |
|---|---|
| Sex | |
| Female | 7 (47 %) |
| Male | 8 (53 %) |
| Age | 62 (27–78) |
| Tumor stage | |
| I | 3 (20 %) |
| II | 5 (33 %) |
| III | 5 (33 %) |
| IV | 2 (13 %) |
| Tumor location | |
| Colon | 12 (80 %) |
| Rectum | 3 (20 %) |
| BMI | |
| ≤25 kg/m2 | 7 (47 %) |
| >25 kg/m2 | 8 (53 %) |
| MSI | |
| MSI-H | 2 (13 %) |
| MSS | 13 (87 %) |
Sample collection
Colorectal tumor tissue and healthy mucosal tissue were collected during surgery in collaboration with the University Clinic Section for Surgical Oncology, Heidelberg. Specimens were reviewed by pathologists to determine TNM (tumor, nodes and metastases) stage. Tissue samples were collected, processed and frozen within 45 min of excision (cold ischemia), placed into a storage cryovial pre-labeled with a unique specimen ID, immersed in liquid nitrogen, and stored at −80 °C until analysis. Additional sections of tumor were formalin-fixed and paraffin-embedded. DNA was extracted from ~50 mg each of frozen tumor and healthy mucosal tissue using the QIAGEN AllPrep DNA/RNA Mini Kit.
Microsatellite instability analysis
Tumors were classified based on microsatellite instability phenotype. Microsatellite instability (MSI) typing was performed using the marker panel CAT25, BAT25, and BAT26 [8]. Tumors showing microsatellite instability at two or more markers were scored as MSI-H; tumors were classified as microsatellite stable (MSS) if no instabilities were detected. The marker combination encompassing the quasi-monomorphic mononucleotide markers CAT25, BAT25, and BAT26 equals the sensitivity and specificity of the Bethesda marker panel and other five-nucleotide marker panels in colorectal cancer [8, 9].
TIL quantification
Tumor infiltration by CD3+ cells was analyzed immunohistochemically. Tumor sections were sliced into 4-μm-thick sections from FFPE tissue. Sections were deparaffinized, rehydrated, and placed on slides and then transferred to a fully automated staining facility (Leica BOND-MAX™, Leica Microsystems). To retrieve antigens, slides were boiled in 10 mM citrate buffer (pH 6) for 20 min and then endogenous peroxidase activity was quenched by 10 min incubation with 0.6 % H2O2 in methanol and blocked with 10 % normal goat serum (Vectastain® Elite ABC kit, Vector). Mouse monoclonal antibodies recognizing human CD3ε (1:50 dilution, clone PS1, Acris) were applied as primary antibody at room temperature (RT) for 30 min. To amplify the response, slides were then incubated with secondary antibody (rabbit anti-mouse IgG, Bond Refine Detection Kit, Leica) for 8 min at RT followed by incubation with a third antibody, conjugated with horseradish peroxidase and coupled to dextrane molecules, for 8 min at RT (mouse anti-rabbit IgG, Bond Refine Detection Kit, Leica). Revelation was performed by a colorimetric reaction with 3,3-di-amino-benzidine (DAB + chromogen, Leica). Sections were counterstained with hematoxylin (Leica) and mounted with Aquatex (Merck).
Positively stained immune cells were counted using a computerized image analysis system consisting of a NDP Nanozoomer (Hamamatsu Photonics) and a personal computer. The whole center of the tumor was analyzed—so in this analysis, the average CD3+ cell density across the measured region as well as the individual density of cells for each mm2 of tissue analyzed was used. All results are expressed as the mean of positively stained cells per mm2. Full sections were analyzed, with all evaluable tissue on the slide being used for quantification. Cell counts were performed with a dedicated software (VIS software suite, Visiopharm) to measure cell densities across a given region of interest as previously reported [10–12]. All evaluations were visually checked for consistency.
High-throughput sequencing
Sequencing CDR3 regions
The TCRB CDR3 regions were amplified and sequenced from 600 ng of total extracted genomic DNA. Amplification and sequencing of TCRB CDR3 regions was carried out on the ImmunoSEQ platform (Adaptive Biotechnologies, Seattle, WA) at Adaptive Biotechnologies as previously described [7]. The sequences for both the TCRB CDR3 regions were delineated according to the definition established by the International ImMunoGeneTics collaboration [13]. Sequences that did not match CDR3 sequences were removed from the analysis. A standard algorithm was used to identify which V, D, and J segments contributed to each TCRB CDR3 sequence [13].
Sequence analysis and statistical analysis
In order to control for the types of CRC tumors, MSI-H and MSS, TCR data were analyzed using the entire dataset (15 samples) and excluding the two MSI-H samples (13 samples). To ascertain whether the sequencing data were consistent with IHC data, the correlation co-efficient was calculated between log-transformed values of CD3+ cells/mm2 and unique TCRB clones sequenced.
In order to test whether tumor and mucosal tissue samples had different variances with respect to the number of unique TCR clones observed, we used a nonparametric Fligner–Killeen [14] test for homogeneity of variances. To address the diverse versus oligoclonal nature of each sample’s TCR repertoire, we calculated the Shannon entropy [7] of the observed frequency distribution of TCR clones, then normalized this value by log2 (# of unique TCR clones observed), which produces a metric that represents the clonal nature of a sample while being insensitive to total sampling depth. To compare this metric between tumor and mucosal samples, we used a nonparametric two-tailed Wilcoxon signed-rank test. For Fig. 4, the proportion of TCR overlap between tumor and matched mucosal tissue samples was calculated as (# of TCR sequencing reads from clones found in both samples)/(# of total TCR sequencing reads from both samples).
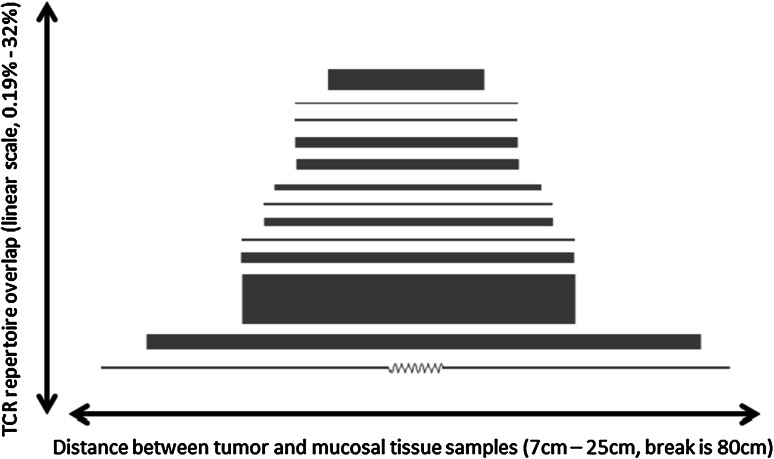
Fig. 4.
Relationship between spatial distance and TCR repertoire overlap. Above, we present each of 13 samples for which paired tumor and mucosal tissue samples were available along with information about the distance between the tumor site and the healthy mucosal tissue sample. The line connecting each pair of samples has a length proportional to the spatial distance between the two samples, and a width proportional to the proportion of TCR sequencing reads belonging to TCR clones shared in both samples. Samples are sorted by (distance/TCR overlaps). There is no significant evidence of a relationship between the distance between tumor and nearby healthy tissue and the degree of TCR repertoire overlap (Pearson’s r = 0.16)
Results
Our study cohort consisted of 15 patients diagnosed with colorectal cancer. Table 1 summarizes the characteristics of the study population, including stage, patient metadata, and tumor type. All tumor samples were CIMP negative. The paired primary tumor and nearby healthy mucosal tissue were analyzed using the ImmunoSEQ assay to generate a total of approximately 14 million sequencing reads distributed among about 32,000 unique TCRB rearrangements, summarized in Table 2. Using these data, we have attempted to characterize the intratumoral T-cell repertoire in colorectal tumors and compared it to nearby mucosal tissue.
Table 2.
Data summary
| ColoCare_ID | Tumor | Mucosal | |||||
|---|---|---|---|---|---|---|---|
| MSI-status | CD3+ Cells/mm2 | % Stroma | TCRB CDR3 | TCRB CDR3 | |||
| Total | Unique | Total | Unique | ||||
| 400464 | MSS | 319.25 | 20 | 60,876 | 1,836 | 321,096 | 1,466 |
| 400480 | MSS | 52.01 | 15 | 130,921 | 2,432 | 15,556 | 1,773 |
| 400488 | MSI-H | 3,944.79 | 15 | 180,152 | 2,090 | 19,324 | 699 |
| 400600 | MSS | 59.09 | 0 | 19,366 | 862 | 15,573 | 984 |
| 400712 | MSS | 77.67 | 15 | 6,536 | 203 | 167,141 | 667 |
| 400728 | MSS | 18.00 | 15 | 1,115 | 41 | 29,676 | 1,110 |
| 401144 | MSS | 1,509.00 | 20 | 384,228 | 1,390 | 13,052 | 1,040 |
| 401176 | MSS | 261.44 | 15 | 41,096 | 723 | 65,571 | 883 |
| 401248 | MSS | 42.98 | 15 | 8,104 | 391 | 551,093 | 1,844 |
| 401256 | MSS | 262.61 | 20 | 37,093 | 1,711 | 13,572 | 1,068 |
| 401264 | MSS | 422.97 | 15 | 5,483,101 | 3,849 | 36,639 | 910 |
| 401304 | MSS | 244.68 | 20 | 1,962,748 | 1,659 | 21,987 | 1,612 |
| 401320 | MSS | 28.49 | 15 | 6,312,534 | 2,933 | 27,601 | 1,667 |
| 401336 | MSI-H | 120.88 | 15 | 257,195 | 988 | 18,932 | 1,228 |
TIL density
The average density of TILs varied by tumor, with the median at 120.1 cells/mm2. Average TIL count for seven tumor samples was <100 CD3+ cells/mm2; for six samples, it was between >100 and <500 CD3+ cells/mm2; and for two samples, it was > 1,000 CD3+ cells/mm2. While the sample with the highest TIL density (3,940 cells/mm2) was classified as MSI-H, the other sample classified as MSI-H was the median for TIL density (120.1 cells/mm2). Average TIL density was moderately correlated with unique TIL TCR sequences (r = 0.5, p = 0.04 by one-tailed normal approximation).
Distribution of T-cell clones infiltrating colorectal tumor and adjacent normal mucosal tissue
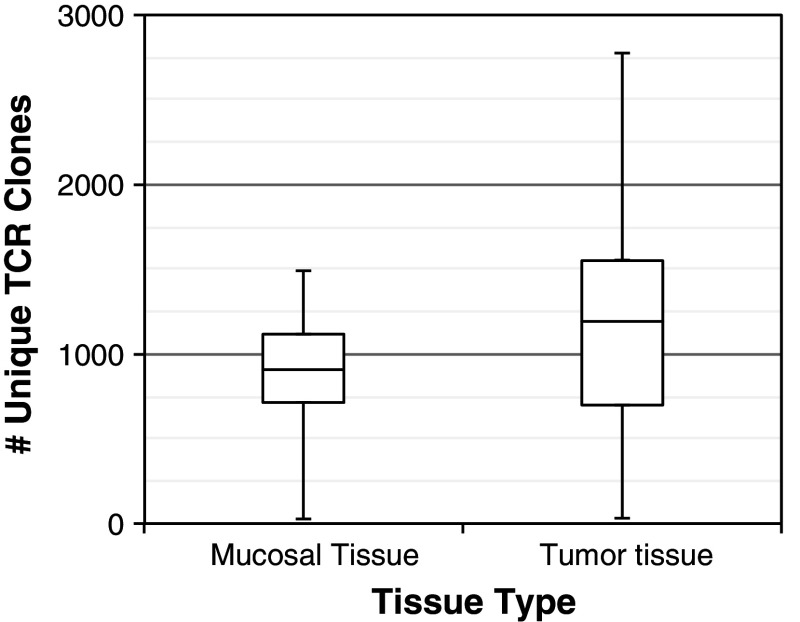
We used the junctional diversity that is present in CDR3 regions to identify T-cell clones and thus observe the number of unique clones as well as the distribution of clonal frequencies in each sample. We observed that the number of unique T-cell clones in each sample varied largely, ranging from a minimum of 26 T-cell clones to a maximum of 2,781 clones. Despite such a large variance across all samples, the number of T-cell clones we sequenced in tumors was similar to the number observed in healthy mucosal tissue samples (a median of 914 clones were observed in mucosal tissue v. 1195 in tumor samples, p = N/S). We observed no relationship between the number of T-cell clones detected in matched tumor and mucosal tissue samples, indicating the disparate nature of the T-cell repertoire in each tissue type.
Diversity of TILs in colorectal cancer is far more variable than in mucosa
While we observed similar numbers of T-cell clones in tumor and mucosal samples overall, there was a considerable discrepancy in the variance observed in the two tissue types: mucosal tissue samples had a standard deviation of 394 T-cell clones, versus 778 for tumor samples [p = 0.04 using a Fligner–Killeen test, Fig. 1; the same pattern is observed (p = 0.07; Supplemental Table 1) when ignoring the two MSI samples]. The higher variance observed in tumor samples is consistent with substantial diversity in the breadth of the intratumoral T-cell repertoire among patients.
Fig. 1.
Comparison of TCR diversity, colorectal tumor versus mucosal tissue. The range of TCR diversity (as measured by the number of unique TCR clones identified) is presented for 15 paired mucosal and tumor tissue samples. Values are given in quartiles, from min to max. There is no significant difference between the number of unique TCR clones identified in the two tissue hypes; however, the variance in TCR diversity is significantly higher in tumor than in mucosal tissue (p = 0.04 by a Fligner–Killeen test)
CRC TILs are more oligoclonal than T cells in mucosa
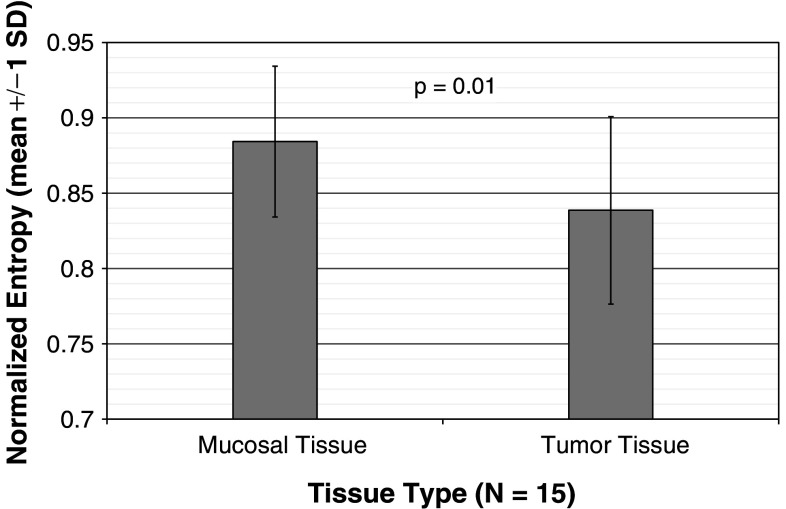
To quantitatively assess the distribution of T-cell clonal frequencies, we calculated the Shannon entropy of each tumor or mucosal tissue sample and normalized each sample’s entropy based on the number of T-cell clones observed (Fig. 2). This measure reflects the oligoclonal versus polyclonal nature of each sample’s T-cell repertoire independent of the total number of T cells that we sequenced. By this measure, the tumor samples tend toward oligoclonality compared to mucosal tissue samples [mean normalized entropy 0.88 for mucosal samples versus 0.84 for tumor samples; p = 0.01 by two-tailed Wilcoxon signed-rank test; the same pattern is observed (p = 0.006, Supplemental Table 1) when ignoring the two MSI samples]. We hypothesize that the more restricted T-cell repertoire observed in tumor samples may be indicative of a specific and oligoclonal T-cell response to antigens that are present in and restricted to the tumor environment.
Fig. 2.
Comparison of TCR distribution entropy, colorectal tumor versus mucosal tissue. Above is a comparison of Shannon entropy, normalized based on the number of unique TCR clones detected, between 15 mucosal and 15 tumor samples. The mean normalized entropy value is reported for each tissue type, with error bars representing one standard deviation. A two-tailed Wilcoxon signed-rank test (a nonparametric equivalent of the paired t test) demonstrates a significant trend toward lower entropy values (i.e., a more clonal TCR frequency distribution) in tumor samples (p = 0.01)
Repertoire-level characteristics are similar in CRC versus adjacent mucosal tissue
To determine whether this variance in the distribution of T-cell clones was matched by variation in the T-cell clones themselves, we characterized the high-level repertoire features of T cells sequenced from the fifteen tumor and matched healthy mucosal tissue samples by computing the following: usage frequency of the various TCRBV and TCRBJ gene segments; distribution of CDR3 length; number of unique T-cell clones detected by sequencing; and distribution of T-cell clonal frequencies. Figure 3 presents a summary of our results. As expected, V and J segment usage frequencies and CDR3 length distribution are quite similar among tumor and mucosal tissue samples, consistent with an intratumoral immune environment dominated by many specific interactions between TCRs, peptide antigens, and HLA proteins without a broad-based bias toward particular lengths or gene segments. Crude measurements of the T-cell repertoire such as antibody- or electrophoresis-based assays would be unlikely to identify any differences between tumor and healthy tissue samples.
Fig. 3.
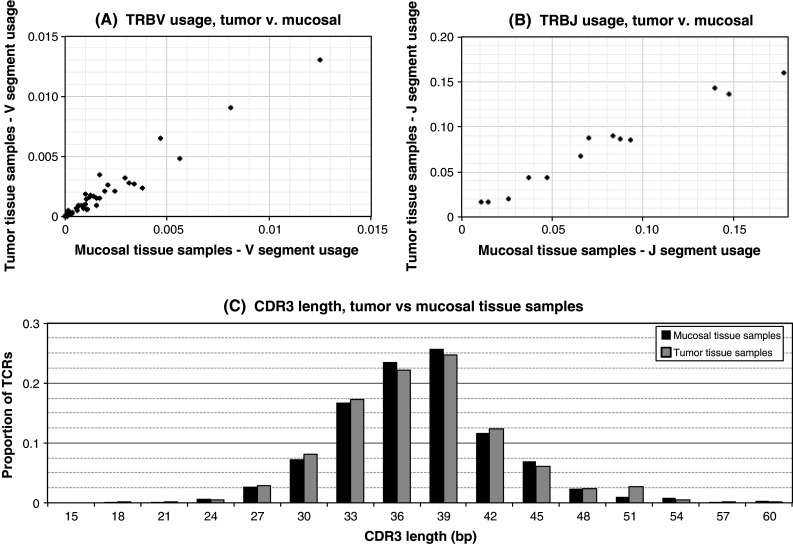
TCRβ sequence properties in tumor and mucosal tissue samples. a A comparison of TRBV gene segment usage in tumor and mucosal tissues (N = 15 samples of each tissue type). Each TRBV gene is graphed according to its frequency of usage in tumor and mucosal samples. TRBV gene usage is quite similar in the two tissue types. b A comparison of TRBJ gene segment usage in tumor and mucosal tissues. Each TRBV gene is graphed according to its frequency of usage in tumor and mucosal samples. TRBJ gene usage is very similar in both tissues. c A comparison of CDR3 region lengths between tumor and mucosal tissue samples. The proportion of TCR sequences with CDR3 regions of each length are presented in a histogram for each tissue type. CDR3 region length is similar in tumor and mucosal tissue samples
Overlap in cellular adaptive immune response in CRC versus adjacent mucosal tissue
In order to investigate the relationship between the immune environment in tumor compared to healthy mucosal tissue, we assessed the degree to which tumor samples and matched mucosal tissue samples shared common TCRB rearrangements, indicating the presence of clonally related T cells in both tissues (Fig. 4). By counting the number of TCRB CDR3 region sequencing reads (considering both the tumor and mucosal tissue samples) that originated from T-cell clones which were observed in both samples, we calculated a TCRB overlap index that ranges from 0 (if no clones are detected in both samples) to 1 (if an identical set of clones were detected in both samples). A wide range of overlap values are observed among the fifteen patients in this study: results ranged from 0.002, indicating that 0.2 % of T cells we sequenced were observed in both samples, to 0.38, indicating that 38 % of sequenced T cells were present in both the tumor and mucosal tissue samples. A median of 5 % overlap was observed—far higher than would be expected when comparing unrelated individuals, but low enough to strongly suggest that tumors have T-cell repertoires quite distinct from those of surrounding healthy tissue. Using these data and distance between mucosal and tumor tissue, we found no evidence for a relationship between the proportion of T-cell overlap and the distance between tumor samples and matched mucosal tissue samples at the time of collection (Fig. 4).
Shared or public intertumoral immune response between CRC patients
To investigate whether there were public TCRs that respond to colorectal tumors, we investigated the presence of T-cell receptor sequences that were observed in multiple patients independently. Table 2 compiles all the TCR sequences seen in at least 2 separate tumor samples and provides the V and J gene segments used as well as the CDR3 sequence. To avoid any possibility of contamination from the sequencing process, we only counted a TCR protein sequence as having been observed twice if the amino acid sequence was coded by different DNA sequences in the two samples. In total, we observed 29 TCR sequences in two separate tumor samples, and a further 3 TCR sequences that were present in two patients and had been independently rearranged twice in one of those patients (for a total of 3 AA-identical TCRs). While potential public T-cell responses to antigens specific to colorectal cancers are expected to be HLA-restricted and will require a much larger sample size to study thoroughly, the ability to sequence TCRs opens up the possibility of such studies, and the presence of many TCR sequences observed in multiple patients suggests that such public responses may exist.
Discussion
There is no measurable difference between the V gene usage and CDR3 length of the TCRB chains sequenced from colorectal tumor and mucosal tissue. Previously used technologies, specifically Betamark and spectratyping, would be unable to differentiate the two tissues’ TCRB repertoire [15]. However, by using the TCRB CDR3 sequences, it is apparent that the TCRB repertoire of the two tissues is markedly different. Tumor tissue and mucosal tissue diverged in two ways: the TCRB repertoires of the tumor tissues are more oligoclonal than their matched tumor tissues and the infiltrating T-cell clones differed between matched tumor and mucosal tissue based on TCRB CDR3 sequences—independent of the physical distance between the two tissues (Fig. 4).
Although the gastrointestinal system has a large population of T cells, like peripheral blood, the mucosal T-cell population is estimated to have a different TCRB repertoire with the mucosal tissue estimated to have a more oligoclonal population of T cells [16–18]. In our dataset, we find that measures of TCRB diversity in T cells from mucosal tissue and its distribution are similar across individuals. In contrast, the TCRB repertoire diversity and distribution of tumor-infiltrating lymphocytes is not similar across individuals (Fig. 3). Within individuals, the TCRB repertoire, as measured by unique TCRB CDR3 sequences, has a different distribution between mucosal and tumor tissue. Individuals’ tumor samples have a measurably more clonal TCRB repertoire (Fig. 1). This matches observations that a targeted immune response to a tumor is dominated by a few clones [19–21]. While clinical outcome data are not yet available for this dataset, the variation we see between individuals could be representative of the variation in the immune response to the tumor. While abundance of TILs is correlated with better prognosis, it is currently not understood how TIL TCR diversity impacts prognosis. When clinical data are available, we will test this hypothesis.
Given the variation in the TCRB repertoire diversity between mucosal tissue and matched tumor tissue, we tested to see whether other measures of TCRB repertoire were divergent between the two tissue types. Like Ochsenreither et al. [22], we find that the V gene usage and CDR3 length are conserved between these two tissue types (Fig. 3). However, when we used TCRB sequence data to test whether the same T-cell clones infiltrated the two tissues, we find that the two tissue types are divergent. Based on shared CDR3 sequences, mucosal and tumor tissue have very different T-cell populations. This highlights that the immune response to the tumor is likely unique and separate from the standard immune repertoire of the adjacent mucosal tissue. In addition, these data highlight how repertoire sequencing permits greater granularity of tracking the immune response in the colorectal tumor.
Given that the tumor tissue has a unique repertoire relative to the mucosal tissue, we asked whether there were public, i.e., TCRB CDR3 chains shared by individuals, colorectal tumor T-cell clones. Previous TCRB repertoire sequencing data found that the peripheral repertoires of unrelated individuals had an unexpectedly high number of shared TCRB CDR3 chains, >10,000 shared TCRB amino acid sequences in the CD8+ memory compartment [23]. Later TCRB sequencing studies identified that a known EBV specific clone is public. If colorectal tumor cells present similar antigens, it is possible that we could find public T-cell receptors. We searched for shared TCRB CDR3 amino acid chains between individuals and found that several individuals have a few shared TCRB chains, including a chain with the same amino acid sequence but many different underlying DNA sequences (Table 3). While the size of this study precludes the ability to identify true public CRC tumor clones, further experiments with a larger sample size would permit this search.
Table 3.
TCR sequences observed in multiple tumor samples
| CDR3 sequence | No. patients | No. rearrangements | TRBV | TRBJ |
|---|---|---|---|---|
| CASSPYQETQYF | 2 | 3 | TRBV18 | TRBJ2-5 |
| CASSLGGRDGYTF | 2 | 3 | TRBV5-4 | TRBJ1-2 |
| CASSLGPIQETQYF | 2 | 3 | TRBV7-9 | TRBJ2-5 |
| CASSPGGYEQYF | 2 | 2 | TRBV18 | TRBJ2-7 |
| CASSPSPNTEAFF | 2 | 2 | TRBV18 | TRBJ1-1 |
| CASRGQGNQPQHF | 2 | 2 | TRBV19 | TRBJ1-5 |
| CASRLAGETQYF | 2 | 2 | TRBV19 | TRBJ2-5 |
| CASSLGTGNNEKLFF | 2 | 2 | TRBV19 | TRBJ1-4 |
| CASSRTGGNTEAFF | 2 | 2 | TRBV19 | TRBJ1-1 |
| CASSTGQGALSGANVLTF | 2 | 2 | TRBV19 | TRBJ2-6 |
| CSAELQETQYF | 2 | 2 | TRBV20-1 | TRBJ2-5 |
| CSARDLEAGEFNEQFF | 2 | 2 | TRBV20-1 | TRBJ2-1 |
| CSARDRVYEQYF | 2 | 2 | TRBV20-1 | TRBJ2-7 |
| CAWSPGLNTEAFF | 2 | 2 | TRBV30 | TRBJ1-1 |
| CASSEEPSSGNTIYF | 2 | 2 | TRBV6-1 | TRBJ1-3 |
| CASSELAGGPNEQFF | 2 | 2 | TRBV6-1 | TRBJ2-1 |
| CASSELAGGQETQYF | 2 | 2 | TRBV6-1 | TRBJ2-5 |
| CASSELAGGYNEQFF | 2 | 2 | TRBV6-1 | TRBJ2-1 |
| CASRISYEQYF | 2 | 2 | TRBV6-2/TRBV6-3 | TRBJ2-7 |
| CASSSGRDTGELFF | 2 | 2 | TRBV6-2/TRBV6-3 | TRBJ2-2 |
| CASSYSSGGAETQYF | 2 | 2 | TRBV6-2/TRBV6-3 | TRBJ2-5 |
| CASSDSTSGGADTQYF | 2 | 2 | TRBV6-4 | TRBJ2-3 |
| CASSLAGGPTDTQYF | 2 | 2 | TRBV7-2 | TRBJ2-3 |
| CASSLEAEGTQYF | 2 | 2 | TRBV7-2 | TRBJ2-3 |
| CASSSSGTWGETQYF | 2 | 2 | TRBV7-3 | TRBJ2-5 |
| CASSADRGNTGELFF | 2 | 2 | TRBV7-8 | TRBJ2-2 |
| CASSLGREAQETQYF | 2 | 2 | TRBV7-8 | TRBJ2-5 |
| CASSLVPGASYNEQFF | 2 | 2 | TRBV7-8 | TRBJ2-1 |
| CASSLDDSPLHF | 2 | 2 | TRBV7-9 | TRBJ1-6 |
| CASSLDGTGANEQFF | 2 | 2 | TRBV7-9 | TRBJ2-1 |
| CASSLGAEQFF | 2 | 2 | TRBV7-9 | TRBJ2-1 |
| CASSLGGSSYEQYF | 2 | 2 | TRBV7-9 | TRBJ2-7 |
This is the first study to sequence at high-throughput the TCRB repertoire of tumor tissue. While other technologies exist to survey the immune repertoire of tissue, like immunohistochemistry, spectratyping, and V beta spectratyping (Betamark), this study demonstrates the limitations of these tools. Both TCRB sequencing and V beta spectratyping find that the TCRB repertoire of colon tissue is oligoclonal [16–18]. However, we find that mucosal tissue and CRC tumor tissue have substantially different populations of infiltrating T cells based on their TCRB sequences (Figs. 1, 2). Yet, the CDR3 length (assessed by spectratyping) and V gene usage (Betamark) was indistinguishable between the two tissue types (Fig. 3). V beta spectratyping would be unable to detect the variation between these two tissue types. While we recognize the limitations of the present study, namely the small sample size for this first investigation, the need for clinical outcomes that have not yet been ascertained, and to test for tumor heterogeneity, we identified characteristics of TIL populations that were not measurable using previous tools.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Conflict of interest
Anna Sherwood, Ryan Emerson, and Cindy Desmarais have employment and stock options at Adaptive Biotechnologies. Harlan Robins has consults for Adaptive Biotechnologies and owns stock. Dominique Scherer, Nina Habermann, Katharina Buck, Jürgen Staffa, Niels Halama, Dirk Jaeger, Peter Schirmacher, Esther Herpel, Matthias Kloor, Alexis Ulrich, Martin Schneider, Cornelia M Ulrich declare that they do not have any conflict of interest.
Footnotes
Anna M. Sherwood, Ryan O. Emerson, and Dominique Scherer contributed equally to this work.
Cornelia M. Ulrich and Harlan Robins contributed equally to this work.
References
- 1.Mlecnik B, Bindea G, Pages F, Galon J. Tumor immunosurveillance in human cancers. Cancer Metastasis Rev. 2011;30:5–12. doi: 10.1007/s10555-011-9270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnet FM. The concept of immunological surveillance. Prog Exp Tumor Res. 1970;13:1–27. doi: 10.1159/000386035. [DOI] [PubMed] [Google Scholar]
- 3.Gooden MJ, Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer. 2011;105:93–103. doi: 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mlecnik B, Tosolini M, Kirilovsky A, Berchuck A, Bindea G, et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol. 2011;29:610–618. doi: 10.1200/JCO.2010.30.5425. [DOI] [PubMed] [Google Scholar]
- 5.Pages F, Galon J, Fridman WH. The essential role of the in situ immune reaction in human colorectal cancer. J Leukoc Biol. 2008;84:981–987. doi: 10.1189/jlb.1107773. [DOI] [PubMed] [Google Scholar]
- 6.Arstila TP, Casrouge A, Baron V, Even J, Kanellopoulos J, et al. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 7.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Findeisen P, Kloor M, Merx S, Sutter C, Woerner SM, et al. T25 repeat in the 3′ untranslated region of the CASP2 gene: a sensitive and specific marker for microsatellite instability in colorectal cancer. Cancer Res. 2005;65:8072–8078. doi: 10.1158/0008-5472.CAN-04-4146. [DOI] [PubMed] [Google Scholar]
- 9.Tikidzhieva A, Benner A, Michel S, Formentini A, Link K-H, et al. Microsatellite instability and beta2-microglobulin mutations as prognostic markers in colon cancer: results of the FOGT-4 trial. Br J Cancer. 2012;106:1239–1245. doi: 10.1038/bjc.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halama N, Zoernig I, Spille A, Michel S, Kloor M, Grauling-Halama S, et al. Quantification of prognostic immune cell markers in colorectal cancer using whole slide imaging tumor maps. Anal Quant Cytol Histol. 2010;32:333–340. [PubMed] [Google Scholar]
- 11.Halama N, Michel S, Kloor M, Zoernig I, Benner A, et al. Localization and density of immune cells in the invasive margin of human colorectal cancer liver metastases are prognostic for response to chemotherapy. Cancer Res. 2011;71:5670–5677. doi: 10.1158/0008-5472.CAN-11-0268. [DOI] [PubMed] [Google Scholar]
- 12.Halama N, Zoernig I, Spille A, Westphal K, Schirmacher P, Jaeger D, et al. Estimation of immune cell densities in immune cell conglomerates: an approach for high-throughput quantification. PLos One. 2009;4:e7847. doi: 10.1371/journal.pone.0007847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yousfi Monod M, Giudicelli V, Chaume D, Lefranc MP. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004;20(Suppl 1):i379–i385. doi: 10.1093/bioinformatics/bth945. [DOI] [PubMed] [Google Scholar]
- 14.Conover WJ, Johnson ME, Johnson MM. A comparative study of tests for homogeneity of variances, with applications to the outer continental shelf bidding data. Technometrics. 1981;23:351–361. doi: 10.1080/00401706.1981.10487680. [DOI] [Google Scholar]
- 15.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 16.Akolkar PN, Gulwani-Akolkar B, McKinley M, Fisher SE, Silver J. Comparisons of T cell receptor (TCR) V beta repertoires of lamina propria and peripheral blood lymphocytes with respect to frequency and oligoclonality. Clin Immunopathol. 1995;76:155–163. doi: 10.1006/clin.1995.1110. [DOI] [PubMed] [Google Scholar]
- 17.Gulwani-Akolkar B, Akolkar PN, McKinley M, Fisher SE, Silver J. Crohn’s disease is accompanied by changes in the CD4+, but not CD8+, T cell. Clin Immunol Immunopatholal. 1995;1:95–106. doi: 10.1016/0090-1229(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 18.May E, Lambert C, Holtmeier W, Hennemann A, Zeitz M, et al. Regional variation of the α/β T cell repertoire in the colon of healthy individuals and patients with Crohn’s disease. Hum Immunol. 2002;65:467–480. doi: 10.1016/S0198-8859(02)00378-6. [DOI] [PubMed] [Google Scholar]
- 19.Stumpf M, Hasenburg A, Riener MO, Jutting U, Wang C, et al. Intraepithelial CD8-positive T lymphocytes predict survival for patients with serous stage III ovarian carcinomas: relevance of clonal selection of T lymphocytes. Br J Cancer. 2009;101:1513–1521. doi: 10.1038/sj.bjc.6605274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiech T, Nikolopoulos E, Hausmann M, Walch A, Werner M, et al. A case of heterogeneous breast cancer with clonally expanded T-Cells in the HER2+ Breast J. 2008;14:487–491. doi: 10.1111/j.1524-4741.2008.00627.x. [DOI] [PubMed] [Google Scholar]
- 21.Goodell V, Waisman J, Salazar LG, de la Rosa C, Link J, et al. Level of HER-2/neu protein expression in breast cancer may affect the development. Mol Cancer Ther. 2008;7:449–454. doi: 10.1158/1535-7163.MCT-07-0386. [DOI] [PubMed] [Google Scholar]
- 22.Ochsenreither S, Fusi A, Wojtke S, Busse A, Nussler NC, et al. Comparison of T-cell receptor repertoire restriction in blood and tumor tissue of colorectal cancer patients. Journal Transl Med. 2010;8:35. doi: 10.1186/1479-5876-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robins H, Srivastava S, Campregher P, Turtle C, Andriesen J, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2:ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.