Abstract
There are an increasing number of studies showing the order of drug presentation plays a critical role in achieving optimal combination therapy. Here, a nanoparticle design is presented using ion pairing and drug-polymer conjugate for the sequential delivery of gefitinib (Gi) and doxorubicin (Dox) targeting epidermal growth factor receptor (EGFR) signaling applicable for the treatment of triple negative breast cancers. To realize this nanoparticle design, Gi complexed with dioleoyl phosphatidic acid (DOPA) via ion paring was loaded onto the nanoparticle made of Dox-conjugated poly(L-lactide)-block-polyethylene glycol (PLA-b-PEG) and with an encapsulation efficiency of ~90%. The nanoparticle system exhibited a desired sequential release of Gi followed by Dox, as verified through release and cellular uptake studies. The nanoparticle system demonstrated approximate fourfold and threefold increases in anti-cancer efficacy compared to a control group of Dox-PLA-PEG conjugate against MDA-MB-468 and A549 cell lines in terms of half maximal inhibitory concentration (IC50), respectively. High tumor accumulation of the nanoparticle system was also substantiated for potential in vivo applicability by non-invasive fluorescent imaging.
Keywords: Nanoparticles, Sequential Delivery, Controlled Delivery, Combination therapy, EGFR inhibitor, Doxorubicin
Introduction
Breast cancer is one of the leading causes of cancer patient deaths in the U.S.1, 2 In particular, ~10–20% of breast cancer cells do not express the estrogen receptor, progesterone receptor, or the epidermal growth factor receptor 2, and are referred to as Triple Negative Breast Cancers (TNBC).3 Many conventional treatments for TNBC are reported to be ineffective, exhibiting an overall worse prognosis compared to other breast cancer subtypes.4, 5 Combination therapy, which utilizes multiple therapeutic agents for non-overlapping and synergistic mechanisms to block multiple oncogenic pathways, holds great potential in enhancing cancer treatments. Appropriately applied combination therapy has the potential to enhance therapeutic efficacy, overcome treatment resistance, and reduce adverse effects.6, 7
Overexpression of the epidermal growth factor receptor (EGFR), a member of the receptor tyrosine kinase (RTK) superfamily, correlates with a basal-like phenotype, and EGFR in TNBC is significantly associated with poor survival.3, 5, 8–10 Many large-scale genomic analyses have reported that ~80% of TNBCs show essential activation of members of EGFR–virus-induced rapidly accelerated fibrosarcoma (v-RAF) murine sarcoma viral oncogene homolog B signaling pathway.11–15 Several approved agents targeting EGFR including as cetuximab, panitumumab, gefitinib, and erlotinib have shown only limited efficacy in both preclinical and clinical settings.10, 16–18 Cetuximab treatment combined with either carboplatin or cisplatin showed modest improved results in two independent Phase II clinical trials.19, 20 However, it was reported that the basal B type of TNBC and non-small cell lung cancer (NSCLC) cells can remarkably be sensitized to the effects of DNA-damaging agents, doxorubicin (Dox), after EGFR signaling is suppressed.4, 21 Lung cancers containing high expressions of phosphorylated wild type or mutated EGFR were reported to respond to such time-delayed sequential treatment. The sensitization effect was derived from the rewiring of signaling networks by prolonged EGFR inhibition using EGFR inhibitors (EGFRi) including erlotinib, gefitinib, or lapatinib. This rewiring through EGFR inhibition showed the most effective DNA damaging efficacy for Dox after a 24-hour time delay when compared with synergistic combination chemotherapy.4 Other recent studies also reported that the order of drug presentation with a time delay in combination therapy is critical to achieving the enhanced therapeutic efficacy.22–24
To achieve the synergistic effects of combination therapy, there are two major factors in achieving an effective combination therapy, co-localization of drugs with effective doses and optimal drug ratio with a time course of drug presentation. However, different pharmacokinetics and biodistribution of each drug can cause difficulties realizing optimal conditions identified from in vitro studies for in vivo systemic delivery. In this study, we report a rational nanoparticle (NP) design using ion pairing and drug-polymer conjugate to address unmet needs of sequential co-delivery. Gi and Dox were co-loaded into the same NP carrier. To sequentially release Gi followed by Dox, Gi was physically encapsulated via ion pairing in a NP system made of Dox-conjugated poly(L-lactide)-block-polyethylene glycol (PLA-PEG). An ion-pairing approach was developed to maximize the Gi loading. The ion-paired Gi is amorphous, which increased the encapsulation efficiency compared to the original highly crystalline form of Gi. The sequential delivery of Gi and Dox was verified through in vitro drug release and cellular uptake studies. The efficacy of this NP system was examined for cytotoxicity against MDA-MB-468 of a TNBC cell line and A549 of a NSCLC cell line, respectively. Finally, an in vivo biodistribution using a syngeneic breast cancer model was also performed to evaluate the tumor accumulation capability of the NP system.
Materials and Methods
Materials
N,N-diisopropylethylamine (DIEA), N-hydroxysuccinimide (NHS), tetrahydrofuran (THF), dichloromethane (DCM), dimethyl sulfoxide (DMSO), acetonitrile (ACN), chloroform and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC·HCl) were purchased from Sigma-Aldrich (St. Louis, MO). Poly(L-lactide) (MW 20,000) with terminal carboxylate groups was purchased from Akina (West Lafayette, IN). Methoxy-poly(ethylene-glycol)-amine (mPEG-NH2 (MW 5,000) was purchased from Laysan Bio Inc. (Arab, AL). 1,2-dioleoyl-sn-glycero-3-phosphate (DOPA) and liposomal encapsulated doxorubicin (DOXIL) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). DiIC18(7) (1,1'-Dioctadecyl-3,3,3',3'-Tetramethylindotricarbocyanine Iodide, ‘DiR’)-labeled liposome with lipid compositions similar to DOXIL® was purchased from FormuMax Scientific, Inc. (Sunnyvale, CA). Doxorubicin hydrochloride (Dox), erlotinib (Ei), and gefitinib (Gi) were purchased from LC Laboratories (Woburn, MA). Dialysis membrane tubing (MWCO 100,000 Da) was purchased from Spectrum (Rancho Dominguez, CA). Amicon® Ultra-4 centrifugal filter (MWCO 100,000 Da) was purchased from Millipore (Billerica, MA). Acrodisc® syringe filters (0.45 µm and 0.2 µm) were purchased from Pall Corp. (Port Washington, NY). Cyanine 5.5 (Cy 5.5) NHS ester was purchased from Lumiprobe Corp. (Hallandale Beach, FL).
Synthesis and characterization of sequential delivery NP
To form an ion-paired complex of Gi+DOPA, Gi and DOPA dissolved in THF were mixed at a predetermined molar ratio. The solubility values of Gi and Gi+DOPA complex in THF were determined from the individual saturated solutions. Undissolved Gi was separated from the solutions by centrifuging the samples at 16.1k × g for 30 minutes, and the concentrations of Gi in the saturated solutions were determined through HPLC analysis. To examine the crystallinity of Gi and Gi+DOPA complex, both samples were prepared, freeze dried, and analyzed using X-ray diffraction (XRD). The XRD measurement was carried out with an X’Pert Pro MPD X-ray diffractometer under CuK radiation (wavelength = 1.5406 Å). The powdered samples were loaded in an aluminum holder with a depth of 1 mm. The scanning range was from (2θ) 10 to 40° with a step size of 0.02° at a rate of 0.25 s/step. Proton nuclear magnetic resonance (1H NMR) spectroscopy was used to study the complexation of DOPA and Gi. For the 1H NMR analysis, DOPA, Gi, and DOPA/Gi were dissolved in CDCl3, DMSO-d6, and CDCl3/DMSO-d6 (1:1 by volume), respectively.
The nano-precipitation method was used to prepare the NPs.25–28 A di-block copolymer, PLA-b-mPEG, was made by conjugating PLA-COOH (15 kDa) with H2N-mPEG (5 kDa) through the carbodiimide-mediated coupling reaction.28–30 Dox-conjugated PLA-b-mPEG (Dox-PLA-PEG) was prepared by following published procedures.31–34 Unreacted Dox was removed by dialyzing Dox-PLA-PEG against water for 24 hrs (MWCO 3.5 kDa). The amount of conjugated Dox was determined to be 1.5 wt% by measuring UV absorbance at 485 nm. Then the Dox-PLA-PEG polymer was dissolved in THF to reach a polymer concentration of 5 mg/ml. To vary a Gi-to-Dox ratio, a different amount of the Gi+DOPA complex was added to the THF containing Dox-PLA-PEG. Then the solution was dropwise added to stirring water (water:THF =5:1 by volume) for NP formation. This mixture was stirred for 2 hours to remove THF by evaporation at room temperature. Then, the NPs were washed by ultrafiltration with water (3,000 g, 15 min, MWCO 100 kDa) to remove residual solvent, unencapsulated Dox, DOPA, and Gi. The NPs were then resuspended in phosphate-buffered saline (PBS) at pH 7.4.
In order to determine the amount of Gi encapsulated, the NPs were dissolved in acetonitrile, and the drug was quantified using HPLC (Agilent 1100 series). Dox was quantified by measuring the UV absorbance at 485 nm. The encapsulation efficiency (EE) was determined by dividing the amount of a drug loading by the amount of a drug initially added during the NP formation. Dynamic light scattering (DLS) was employed to measure the size of NPs at a concentration of 0.2 mg/ml.
The morphology of the NPs was investigated by FEI CM20 transmission electron microscopy (TEM). A 10 µl solution of 0.1 mg/ml freshly made NPs in PBS was dropped on a carbon-coated copper grid. Then an excess amount of the liquid was removed by filter paper. Next the grid was immersed in a staining solution (1%(w/v) uranyl acetate) for 2 min and then dried under the fume hood.35
The release kinetics of the NPs was studied by an indirect dialysis method. The NP solution was transferred to a dialysis membrane tubing (MWCO 100 kDa). Then the dialysis tubing was immersed in 500 ml of PBS solution at 37 °C. At predetermined times, a small fraction of the sample inside the dialysis tubing was collected. The individual amounts of Gi and Dox retained inside the NPs at each sampling time were determined using HPLC and UV plate reader, respectively, as described above.
In vitro cell tests for imaging and cytotoxicity
The Dulbecco's Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% penicillin-streptomycin (Invitrogen) was utilized as a cell culture medium for a basal B type TNBC cell line, MDA-MB-468 (ATCC) and a NSCLC cell line, A549 (ATCC). The cells were cultivated in a humidified environment at 37 °C with 5% CO2.
A fluorescence microscope study was performed to investigate the intracellular distribution of delivered Dox. The cells were cultured on cover slips in 12-well plates with 200,000 cells per well. Then the cells were incubated with Dox-conjugated PLA-b-mPEG NP (NP-D) and Dox, with a Dox concentration of 0.2 µg/ml for 30 min, 2 hours, and 12 hours. After the incubation, the cells were washed with PBS three times and fixed with 4% (wt/vol) formaldehyde in PBS for 15 min. The nuclei were stained with 4', 6-diamidino-2-phenylindole (DAPI). Finally, 10 µl of a mount medium (SlowFade® Gold Antifade Reagent, Life Technologies Corp., Grand Island, NY) was added onto glass slide, and the cover slip containing the cells treated with the samples was placed on the glass slide.
The MTS assay (Promega, Madison WI) was used to evaluate the cytotoxicity of different samples. The cells were plated in a 96-well plate with ~5,000 cells per well, and then the cells were treated with different NP formulations. The cytotoxicity of NPs was examined by the MTS assay following the protocol provided by the vendor.
In vivo non-invasive imaging
The R7 murine breast cancer cell line was derived from a mammary tumor from a transgenic MMTV-Ron mouse.36 For our in vivo experiments, 150,000 R7 cells were orthotopically injected into the inguinal mammary fat pads of syngeneic wild type FVB female mice following the protocols previously described.37 The tumor formation was examined by manual palpation. An alfalfa-free diet was fed to the mice upon the identification of a palpable tumor. To label PLA with Cy5.5, Cy5.5-NH2 was conjugated to PLA-COOH through the carbodiimide-mediated coupling reaction. Near infrared fluorescence (NIRF) nanoparticles were prepared by blending 5 wt% of PLA-Cy5.5 and 95 wt% Dox-PLA-PEG before nanoprecipitation. Free Cy5.5 was removed by washing the NIRF nanoparticles three times using centrifugal filter (MWCO 100 kDa). The DiR-labeled liposome was also used for a comparison. The NPs formed were then injected through the tail vein, and the mice were imaged 2 hours, 4 hours, and 24 hours after the injection using a fluorescent imaging system (Bruker In-Vivo MS FX PRO). All animal treatments and surgical procedures followed approved protocols and were performed in accordance with the Institutional Animal Care and Use Committee of the University of Cincinnati.
Results and Discussion
Synthesis and Characterization of Sequential Co-Delivery Nanoparticles
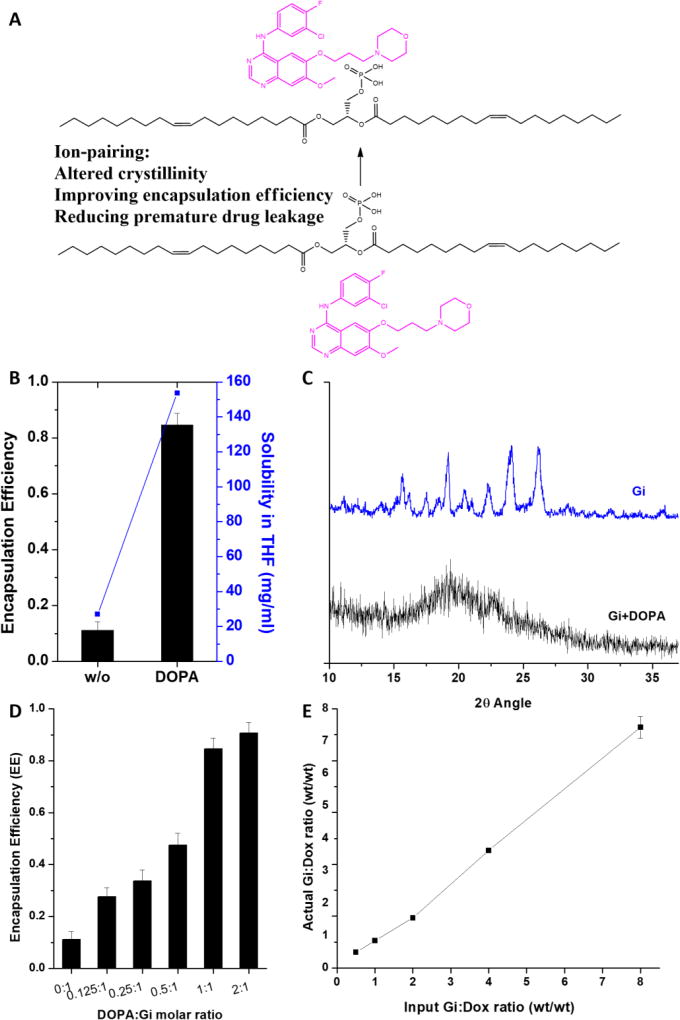
Combination therapy holds great promise in enhancing treatment outcomes. To achieve synergistic effects for combination therapy, tumor cells should be exposed to drugs with effective doses at optimal drug ratios. In addition, there are an increasing number of studies showing the order of drug presentations plays a critical role in achieving enhanced efficacy.4, 23, 38–41 However, due to diverse physicochemical properties of drugs, it is challenging to co-deliver effective doses of drugs to tumor sites at an optimal ratio and with a time delay in drug presentation. To address the sequential delivery of EGFRi and Dox for synergistic anti-cancer efficacy against the basal B type of TNBC and NSCLC, we developed a NP delivery system based on drug-polymer conjugation and ion pairing. Dox was chemically conjugated to PLA-PEG to ensure time delayed presentation to the TNBC cells. EGFRi was physically encapsulated by Dox-PLA-PEG, and is expected to be released faster than Dox. However, the encapsulation efficiencies of the two EGFRis of Ei and Gi were extremely low, ~10%. High initial drug inputs were also attempted to increase the loadings in the Dox-PLA-PEG NPs. However, the bulk precipitates of Ei and Gi were formed during the NP formation process. Precipitation was attributed to the highly crystalline nature of Ei and Gi, which is believed to lead to the recrystallization of Ei and Gi and the formation of lower energy macroscopic crystals in the aqueous phase during the NP formation step.42 Salt formation using ion pairing between a therapeutic drug and counterion is a widely used method in the pharmaceutical industry to adjust drug properties, and it has potential to alter the crystallinity of Ei and Gi (Fig. 1A).42–45 Dioleoyl phosphatidic acid (DOPA) was found to be extremely effective in altering the crystallinity of Gi and enhancing the solubility of Gi in THF, which is used as a solvent for nanoprecipitation (Fig. 1B). DOPA contains a phosphate group that can bind to the tertiary amine of Gi. The altered crystallinity was observed for Gi+DOPA complex through XRD (Fig. 1C). While the parent Gi exhibited the diffraction peaks showing its highly crystalline nature, the Gi+DOPA complex showed a broad peak indicating the characteristic of an amorphous compound. In addition, proton nuclear magnetic resonance (1H NMR) was used to characterize the complexation between Gi and DOPA (Fig. S1). The peaks assigned to the protons close to tertiary amine of Gi were shifted, indicating an electron density change resulting from the complexation (Fig. S2). We also found that a 1:1 molar ratio of DOPA:Gi was sufficient for full complexation and such complexation maximizes Gi encapsulation (Fig. 1D). It was also found that DOPA could significantly improve the encapsulation of other EGFRis of Ei and Li (data not shown). Due to the high encapsulation efficiency of Gi employing DOPA as a counterion, the ratio of Gi to Dox to be encapsulated inside the NPs can be precisely controlled by a simple method of varying initial drug inputs (Fig. 1E).
Figure 1.
A: schematic of ion-pairing; B: effects of DOPA complexation (DOPA:Gi molar ratio = 1:1) on enhancing Gi solubility in THF and improving encapsulation efficiency (EE) in nanoparticle; C: X-ray diffraction (XRD) spectra of parent drug (Gi) and DOPA complexed drug (Gi+DOPA); D: effect of DOPA:Gi molar ratio on encapsulation efficiency (EE); E: Control of Gi and Dox loadings with respect to different initial drug loadings.
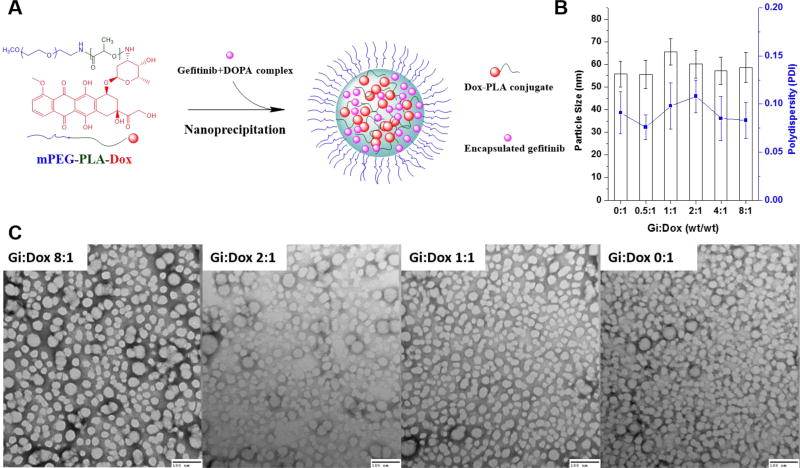
A schematic of the NP design using Gi+DOPA complexation and Dox-PLA-PEG conjugation is shown in Fig. 2A. In addition, any significant changes in the NP sizes (~50–60 nm) and polydispersity with respect to different Gi:Dox ratios ranging from 0:1 to 8:1 were not observed, as confirmed through DLS measurements (Fig. 2B). When the Dox-PLA-PEG and Gi+DOPA complex were first dissolved in THF and the mixture was then dropwise added to water, the NPs were formed as a result of the fast diffusion of THF in the aqueous phase and supersaturation of the Dox-PLA-PEG and Gi+DOPA complex. The TEM images showed consistent particle sizes of ~30–40 nm in a spherical shape (Fig. 2C), confirming that the encapsulation of different amounts of the Gi+DOPA complex did not change the size of the NPs formed. A difference in the NP sizes measured by TEM and DLS is attributed to the shrinkage of the NPs as TEM assessed the sample in a dried form as compared to in a wet form when DLS was used. Similar observations were previously reported.46–48
Figure 2.
A: schematic of preparing Dox-conjugated nanoparticle encapsulating Gi (NP(Gi)-D); B: particle sizes of NPs(Gi)-D with respect to different Gi:Dox ratios, determined by dynamic light scattering (DLS) measurements; C: Transmission electron microscopy (TEM) images of NPs(Gi)-D with respect to different Gi:Dox ratios. Samples were negatively stained using 1 wt% uranyl acetate. Scale bar shows 100 nm.
In vitro Evaluation of Sequential Co-Delivery Nanoparticles
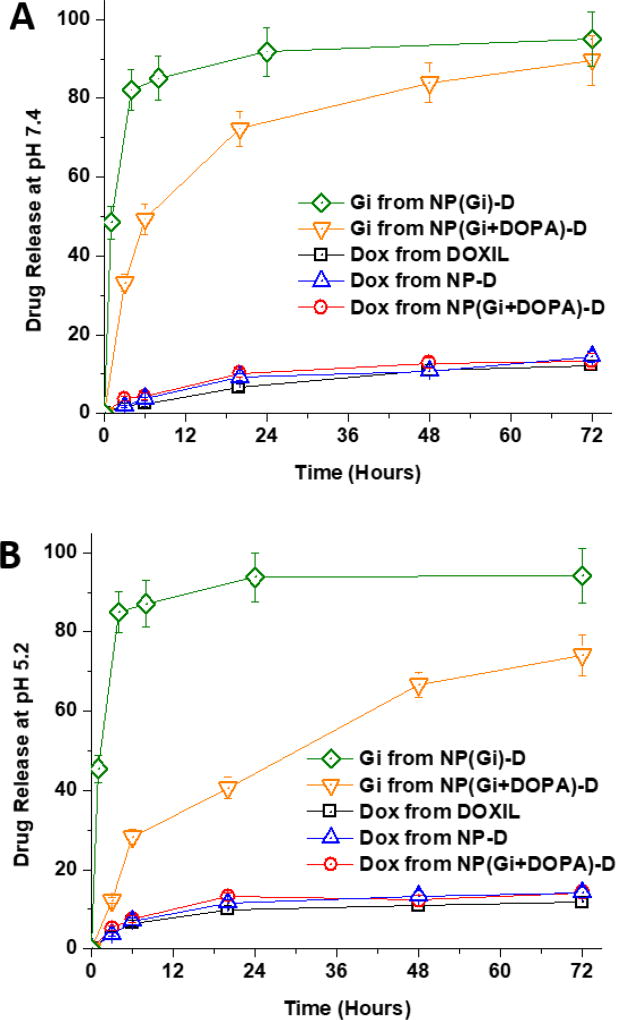
The in vitro release kinetics of Gi and Dox was investigated by dialysis against PBS at the physiological temperature (37 °C) and two pH conditions (pH 7.4 and pH 5.2) (Fig. 3). Gi was released at much slower rates when it was complexed with DOPA. Increased molar volume and hydrophobicity of drug-counterion complex relative to uncomplexed drug is presumed to be responsible for such altered drug release kinetics.49 The release kinetics of encapsulated drug has significant impacts on the efficacies of drug delivery systems.49, 50 Burst release would result in similar pharmacokinetics between encapsulated drug and a free form of drug, diminishing advantages of using NP as a drug carrier. Hence, the sustained release of Gi would benefit in vivo application.49, 50 The slow release kinetic profile obtained for the Gi+DOPA complex is highly expected to allow for sustained release during systemic circulation of the NPs. A desired sequential release of Gi and Dox was successfully achieved using the Gi+DOPA complex encapsulated in the Dox-conjugated PLA-PEG NP (NP(Gi+DOPA)-D). The Dox conjugation to PLA-PEG was used for a subsequent slow release of Dox.31, 33 Liposomal encapsulated doxorubicin (DOXIL) is known to release Dox slowly and was used as a control. The DOXIL and Dox-PLA-PEG conjugate released Dox at about the same rate, confirming a successful achievement of slow Dox release from NP(Gi+DOPA)-D. In addition, it was confirmed that Gi+DOPA complexation did not have impact on Dox release.
Figure 3.
Dox and gefitinib releases from liposomal encapsulated doxorubicin (DOXIL) and Dox-conjugated NP with DOPA complexation or without DOPA complexation at pH 7.4 (A) and pH 5.2 (B) and 37 °C.
To simulate the acidic condition around and inside the tumor sites, the releases of both drugs were also tested at pH 5.2 (Fig. 3B). It was found that pH did not affect the Dox release. However, Gi release was found to be further suppressed at pH 5.2, which is attributed to a larger difference in pKa (6.9 for Gi) and pH values. Although the slow release kinetics of Gi was successfully achieved using DOPA, the slow release kinetics of Ei was not achievable using the same DOPA (Fig. S3A). This is attributed to the low pKa value of Ei (i.e. 4.6), which unlikely keeps Ei at a protonated state required for ion pairing at the physiological pH 7 condition.42, 49 This speculation was validated from a much slower release kinetic profile obtained for the Ei+DOPA complex at pH 3, where the complex should be more stable than at pH 7 (Fig. S3B). Due to the burst release of Ei even with DOPA, Ei was not further pursued in this study.
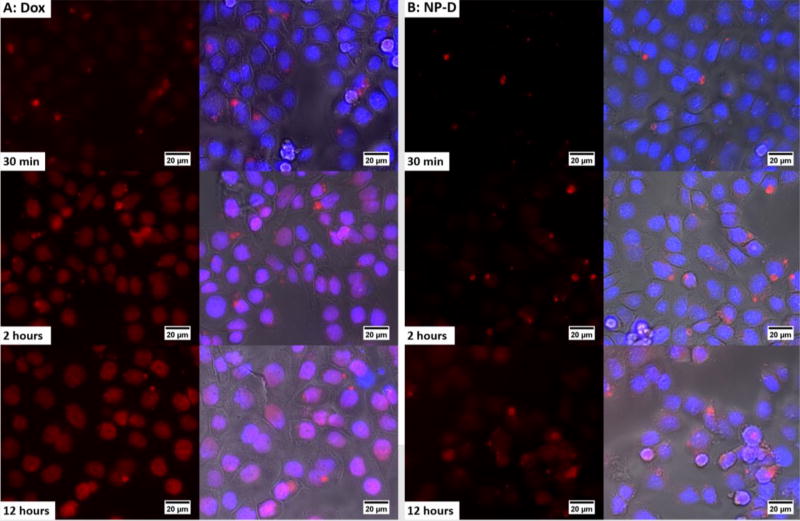
To investigate the uptake of the conjugated form of Dox (NP-D), a temporal fluorescence imaging study was carried out using MDA-MB-468 cells. Free form of Dox (Dox solution) was used as a positive control. Cells were incubated with NP-D and Dox with a Dox concentration of 0.2 µg/ml for 30 min, 2 hours, and 12 hours at 37 °C. DAPI was used to stain the nuclei and phase contrast images were taken to show the cell morphology. As shown in Fig. 4, in the case of free Dox, most of the red staining was derived from Dox present in the cytoplasm at 30 min and then started to enter the nuclei after 30 min. From 2 hours to 12 hours, the fluorescent response from Dox inside the nuclei was found to increase based on a color change from blue to pink, which comes from the overlap between red Dox and blue nuclei. This result indicates that Dox starts to accumulate inside the nuclei within 2 hours. On the other hand, in the case of the conjugated form of Dox, there was little Dox present inside the cells after 2 hours. Even after 12 hours of the incubation, most Dox still remained in the cytoplasm, indicating slow uptake of Dox from NP-D. In addition, liposomal encapsulated doxorubicin (DOXIL) is known to be stable and releases Dox very slowly, and was also tested as a reference (Fig. S4).51, 52 DOXIL showed a slow cellular uptake profile similar to that of NP-D. It is attributed to the comparable Dox release kinetics obtained from DOXIL and NP-D (Fig. 3). These results confirm that the slow Dox release can lead to the delayed presence of Dox inside the cells and Dox conjugation can significantly delay the permeation of Dox into the cells, validating the use of Dox-PLA-PEG for the sequential delivery.
Figure 4.
Cellular uptake of free form of Dox (A) and Dox-conjugated NP (NP-D) (B) in MDA-MB-468 cells. Nuclei were stained with 4', 6-diamidino-2-phenylindole (DAPI) (blue). The cells were incubated with NP (NP-D) and free Dox at a Dox concentration of 0.2 µg/ml for 30 min, 2 hours, and 12 hours at 37 °C.
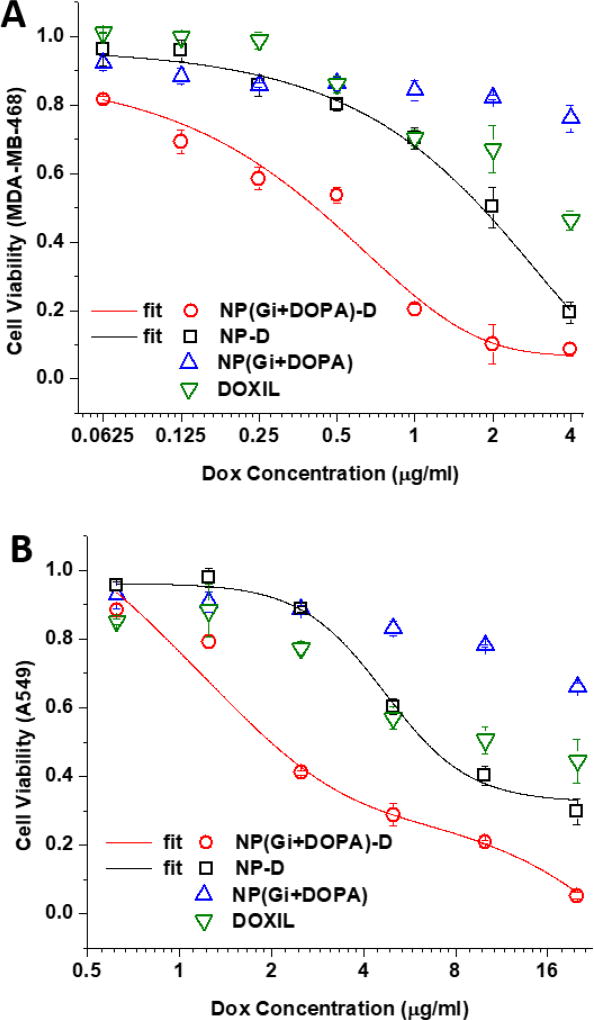
The cytotoxic efficacy of the PLA-PEG NP system for the sequential delivery was examined in vitro using MDA-MB-468 and A549 cell lines as model cell lines of the basal B type of TNBC and NSCLC, respectively (Fig. 5). The calculated half maximal inhibitory concentration (IC50) values are summarized in Table 1. First, it was confirmed that the cells have to be treated with Gi at least 4 hours prior to Dox application in order to obtain an enhanced therapeutic efficacy for MDA-MB-468 cells (Fig. S5). The simultaneous co-application of Dox and Gi only moderately improved the anti-cancer efficacy in comparison with the single application of Dox for both MDA-MB-468 and A549 cells. It was noted that A549 was more resistant to Dox than MDA-MB-468, which requires much higher doses of Dox to effectively inhibit the growth. Other studies also reported similar results.4, 53
Figure 5.
In vitro cytotoxicity in MDA-MB-468 cells (A) and A549 cells (B) after treatment with liposomal formulation of Dox (DOXIL®), NP encapsulating Gi+DOPA complex (NP(Gi+DOPA)), Dox-conjugated NP (NP-D), and Dox-conjugated NP encapsulating Gi+DOPA complex (NP(Gi+DOPA)-D) with Gi:Dox = 1:1 (wt:wt). Cell viability was determined using the MTS assay at 48 hours following treatment addition. Data represent mean ± standard deviation, n = 3.
Table 1.
IC50 values of MDA-MB-468 and A549 cells treated by various formulations after 2 days.
| IC50 (µg/ml) | Gi | DOXIL | NP(Gi+DOPA)-D | NP-D |
|---|---|---|---|---|
| MDA-MB-468 | \ | \ | 0.48 | 1.84 |
| A549 | \ | 9.92 | 2.02 | 6.37 |
In order to evaluate a combined strategy of the drug-polymer conjugation and counter-ion pairing designed for the sequential delivery, MDA-MB-468 and A549 cells were treated with NP-D and NP(Gi+DOPA)-D (Fig. 5). The conjugation of Dox to PLA-PEG (NP-D) showed cytotoxic efficacy similar to DOXIL, which is attributed to the comparable Dox release profiles. It should be noted that the Dox release kinetics from all the three delivery systems of NP-D, NP(Gi+DOPA)-D, and DOXIL® was almost the same over 72 hours (Fig. 3). However, when the Gi+DOPA complex was encapsulated in NP-D, the efficacy was greatly improved. The NP(Gi+DOPA)-D system demonstrated approximate fourfold and threefold increases in anti-cancer efficacy compared to a control group of Dox-PLA-PEG conjugate (NP-D) against MDA-MB-231 and A549 cell lines respectively, as shown by IC50. In combination therapy, there is an optimal or minimal ratio for synergistic effects. Other ratios of Gi to Dox, ranging from 0.5:1 to 8:1, were also examined for cytotoxicity against MDA-MB-468 cell line to find an optimal ratio (Fig. S6). It was found that a Gi-to-Dox ratio of 1:1 was sufficient to achieve an enhanced efficacy for the sequential delivery. Higher Gi-to-Dox ratios did not further improve the therapeutic effect. Such an enhanced cytotoxic efficacy is attributed to the desired order of drug presentation of Gi followed by Dox for Dox sensitization. These results successfully demonstrate that the proposed NP system can be used to achieve the enhanced cytotoxic efficacy of Dox.
In vivo Biodistribution of Sequential Co-Delivery Nanoparticles
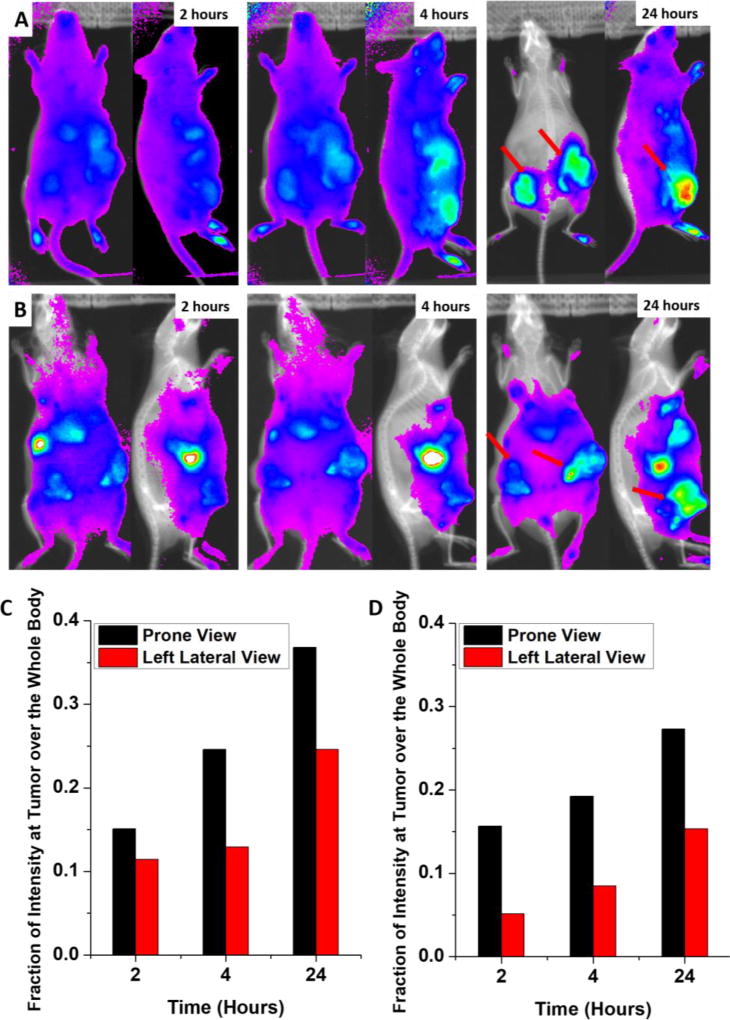
A bio-distribution study was conducted to examine the potential of this NP system for therapeutic applications (Fig. 6). For the real-time optical monitoring of in vivo distribution of NPs, time-dependent excretion profile, and tumor accumulation, Cy5.5 was labeled to the terminus of PLA (Fig. 6A). Then the Cy 5.5-labeled PLA (5 wt%) was mixed with Dox-PLA-PEG (95 wt%) during the formation of the NIRF NPs. A DiR-labeled liposome sample with lipid compositions similar to DOXIL® was also tested as a positive control for EPR effects (Fig. 6B). As expected, this liposome could passively accumulate at tumor sites. The Cy 5.5-labeled PLA-PEG NP started to accumulate in the tumors within 2 hours after NP injection and continued to accumulate for up to 24 hours (Fig. 6A). After 24 hours, the NIRF intensity at the tumor site was the highest throughout the whole body. The DiR-labeled liposome sample also showed a similar profile. In addition, the fluorescence intensities at the tumor site and the whole body were counted using ImageJ, and the fraction of the fluorescence intensities detected at the tumors were determined (Fig.6C and 6D). It is clear that the fluorescence intensities at the tumors steadily increased from 2 hours to 24 hours, showing NP accumulations at the tumor site. The fluorescence intensities of the PLA-PEG NP system was comparable to or slightly higher than that of DiR-labeled liposome. Such specific localization of the PLA-PEG NPs at the tumor site is anticipated to help enhance the therapeutic efficacy and reduce the systematic toxicity of the therapeutic agents by differentiating tumors and healthy tissues.
Figure 6.
In vivo non-invasive NIRF images of time-dependent prone and left lateral imaging of R7murine breast cancer tumor-bearing mice after 2, 4, and 24 hours of the i.v. injection of Cy5.5-NP (A) and DiR- labeled liposome (B); total NIRF intensity at the tumor site over the whole body after 2, 4, and 24 hours of the i.v. injection of Cy5.5-NP (C) and DiR-labeled liposome (D). Red arrows indicate tumors.
Conclusion
In summary, a rational NP design based on a combined strategy of Dox-polymer conjugation and counter-ion pairing was presented for the sequential delivery of Gi and Dox for enhanced therapeutic effects on basal B type TNBC and NSCLC cells. The Gi+DOPA complex was synthesized through ion paring, and was found to be suitable for the encapsulation in the Dox-PLA-PEG polymer. The ion pairing method increased the hydrophobicity and molar volume of Gi, leading to higher encapsulation efficiency and slower release kinetics compared to its parent form. The sequential delivery of Gi followed by Dox was validated through the drug release profiles of Gi and Dox as well as the delayed presence of Dox inside MDA-MB-468 cells. The sequential delivery of the drugs to the cells at a 1:1 ratio was found to be sufficient to achieve a maximum enhanced therapeutic efficacy. This nanoparticle design was also confirmed to exhibit tumor targeting through passive enhanced permeability and retention (EPR) effect. This NP system designed for sequential drug delivery is expected to maximize therapeutic efficacy while minimizing the amount of Dox and thus reducing cytotoxic and off-target side effects.
Supplementary Material
Acknowledgments
This study was supported by the Marlene Harris-Ride Cincinnati Breast Cancer Foundation, and the authors appreciate their support.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR, Palacios J, Rakha EA, Richardson AL, Schmitt FC, Tan PH, Tse GM, Weigelt B, Ellis IO, Reis-Filho JS. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24(2):157–67. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 4.Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149(4):780–94. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13(15 Pt 1):4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 6.Greco F, Vicent MJ. Combination therapy: opportunities and challenges for polymer-drug conjugates as anticancer nanomedicines. Adv Drug Deliv Rev. 2009;61(13):1203–13. doi: 10.1016/j.addr.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Broxterman HJ, Georgopapadakou NH. Anticancer therapeutics: "Addictive" targets, multi-targeted drugs, new drug combinations. Drug Resist Updat. 2005;8(4):183–97. doi: 10.1016/j.drup.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Corkery B, Crown J, Clynes M, O'Donovan N. Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Annals of Oncology. 2009;20(5):862–867. doi: 10.1093/annonc/mdn710. [DOI] [PubMed] [Google Scholar]
- 9.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. The New England journal of medicine. 2010;363(20):1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 10.Kalimutho M, Parsons K, Mittal D, Lopez JA, Srihari S, Khanna KK. Targeted Therapies for Triple-Negative Breast Cancer: Combating a Stubborn Disease. Trends Pharmacol Sci. 2015;36(12):822–46. doi: 10.1016/j.tips.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Network CGA. Comprehensive molecular portraits of human breast tumors. Nature. 2012;490(7418):61. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amin DN, Sergina N, Ahuja D, McMahon M, Blair JA, Wang D, Hann B, Koch KM, Shokat KM, Moasser MM. Resiliency and vulnerability in the HER2-HER3 tumorigenic driver. Science translational medicine. 2010;2(16):16ra7–16ra7. doi: 10.1126/scitranslmed.3000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T, North-East Japan Study G. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. The New England journal of medicine. 2010;362(25):2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 18.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, Porta R, Cobo M, Garrido P, Longo F, Moran T, Insa A, De Marinis F, Corre R, Bover I, Illiano A, Dansin E, de Castro J, Milella M, Reguart N, Altavilla G, Jimenez U, Provencio M, Moreno MA, Terrasa J, Munoz-Langa J, Valdivia J, Isla D, Domine M, Molinier O, Mazieres J, Baize N, Garcia-Campelo R, Robinet G, Rodriguez-Abreu D, Lopez-Vivanco G, Gebbia V, Ferrera-Delgado L, Bombaron P, Bernabe R, Bearz A, Artal A, Cortesi E, Rolfo C, Sanchez-Ronco M, Drozdowskyj A, Queralt C, de Aguirre I, Ramirez JL, Sanchez JJ, Molina MA, Taron M, Paz-Ares L, Pneumocancerologie GF, Toracica AIO. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 19.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero-Torres A. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. Journal of Clinical Oncology. 2012;30(21):2615–2623. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baselga J, Gómez P, Greil R, Braga S, Climent MA, Wardley AM, Kaufman B, Stemmer SM, Pêgo A, Chan A. Randomized phase II study of the anti–epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. Journal of clinical oncology. 2013;31(20):2586–2592. doi: 10.1200/JCO.2012.46.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mok TS, Wu YL, Yu CJ, Zhou C, Chen YM, Zhang L, Ignacio J, Liao M, Srimuninnimit V, Boyer MJ, Chua-Tan M, Sriuranpong V, Sudoyo AW, Jin K, Johnston M, Chui W, Lee JS. Randomized, placebo-controlled, phase II study of sequential erlotinib and chemotherapy as first-line treatment for advanced non-small-cell lung cancer. J Clin Oncol. 2009;27(30):5080–7. doi: 10.1200/JCO.2008.21.5541. [DOI] [PubMed] [Google Scholar]
- 22.Westhoff MA, Faham N, Marx D, Nonnenmacher L, Jennewein C, Enzenmuller S, Gonzalez P, Fulda S, Debatin KM. Sequential dosing in chemosensitization: targeting the PI3K/Akt/mTOR pathway in neuroblastoma. PLoS One. 2013;8(12):e83128. doi: 10.1371/journal.pone.0083128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seino M, Okada M, Sakaki H, Takeda H, Watarai H, Suzuki S, Seino S, Kuramoto K, Ohta T, Nagase S, Kurachi H, Kitanaka C. Time-staggered inhibition of JNK effectively sensitizes chemoresistant ovarian cancer cells to cisplatin and paclitaxel. Oncol Rep. 2016;35(1):593–601. doi: 10.3892/or.2015.4377. [DOI] [PubMed] [Google Scholar]
- 24.Ubezio P, Falcetta F, Carrassa L, Lupi M. Integrated experimental and simulation study of the response to sequential treatment with erlotinib and gemcitabine in pancreatic cancer. Oncotarget. 2016;7(13):15492–506. doi: 10.18632/oncotarget.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legrand P, Lesieur S, Bochot A, Gref R, Raatjes W, Barratt G, Vauthier C. Influence of polymer behaviour in organic solution on the production of polylactide nanoparticles by nanoprecipitation. Int J Pharm. 2007;344(1–2):33–43. doi: 10.1016/j.ijpharm.2007.05.054. [DOI] [PubMed] [Google Scholar]
- 26.Fessi H, Puisieux F, Devissaguet JP, Ammoury N, Benita S. Nanocapsule Formation by Interfacial Polymer Deposition Following Solvent Displacement. International Journal of Pharmaceutics. 1989;55(1):R1–R4. [Google Scholar]
- 27.Quintanar-Guerrero D, Allemann E, Fessi H, Doelker E. Preparation techniques and mechanisms of formation of biodegradable nanoparticles from preformed polymers. Drug Dev Ind Pharm. 1998;24(12):1113–28. doi: 10.3109/03639049809108571. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Z, Badkas A, Stevenson M, Lee JY, Leung YK. Herceptin conjugated PLGA-PHis-PEG pH sensitive nanoparticles for targeted and controlled drug delivery. Int J Pharm. 2015;487(1–2):81–90. doi: 10.1016/j.ijpharm.2015.03.081. [DOI] [PubMed] [Google Scholar]
- 29.Kamaly N, Fredman G, Subramanian M, Gadde S, Pesic A, Cheung L, Fayad ZA, Langer R, Tabas I, Farokhzad OC. Development and in vivo efficacy of targeted polymeric inflammation-resolving nanoparticles. Proc Natl Acad Sci U S A. 2013;110(16):6506–11. doi: 10.1073/pnas.1303377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Z, Kennell C, Lee JY, Leung YK, Tarapore P. Calcium phosphate-polymer hybrid nanoparticles for enhanced triple negative breast cancer treatment via co-delivery of paclitaxel and miR-221/222 inhibitors. Nanomedicine. 2017;13(2):403–410. doi: 10.1016/j.nano.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 31.Yoo HS, Park TG. Biodegradable polymeric micelles composed of doxorubicin conjugated PLGA–PEG block copolymer. Journal of Controlled Release. 2001;70(1–2):63–70. doi: 10.1016/s0168-3659(00)00340-0. [DOI] [PubMed] [Google Scholar]
- 32.Yoo HS, Oh JE, Lee KH, Park TG. Biodegradable nanoparticles containing doxorubicin-PLGA conjugate for sustained release. Pharm Res. 1999;16(7):1114–8. doi: 10.1023/a:1018908421434. [DOI] [PubMed] [Google Scholar]
- 33.Yoo HS, Lee KH, Oh JE, Park TG. In vitro and in vivo anti-tumor activities of nanoparticles based on doxorubicin–PLGA conjugates. Journal of Controlled Release. 2000;68(3):419–431. doi: 10.1016/s0168-3659(00)00280-7. [DOI] [PubMed] [Google Scholar]
- 34.Sengupta S, Eavarone D, Capila I, Zhao G, Watson N, Kiziltepe T, Sasisekharan R. Temporal targeting of tumour cells and neovasculature with a nanoscale delivery system. Nature. 2005;436(7050):568–72. doi: 10.1038/nature03794. [DOI] [PubMed] [Google Scholar]
- 35.Booth DS, Avila-Sakar A, Cheng Y. Visualizing proteins and macromolecular complexes by negative stain EM: from grid preparation to image acquisition. J Vis Exp. 2011;(58):e3227. doi: 10.3791/3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zinser GM, Leonis MA, Toney K, Pathrose P, Thobe M, Kader SA, Peace BE, Beauman SR, Collins MH, Waltz SE. Mammary-specific Ron receptor overexpression induces highly metastatic mammary tumors associated with beta-catenin activation. Cancer Res. 2006;66(24):11967–74. doi: 10.1158/0008-5472.CAN-06-2473. [DOI] [PubMed] [Google Scholar]
- 37.Wagh PK, Gray JK, Zinser GM, Vasiliauskas J, James L, Monga SP, Waltz SE. [beta]-Catenin is required for Ron receptor-induced mammary tumorigenesis. Oncogene. 2011;30(34):3694–3704. doi: 10.1038/onc.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian X, Ren Y, Shi Z, Long L, Pu P, Sheng J, Yuan X, Kang C. Sequence-dependent synergistic inhibition of human glioma cell lines by combined temozolomide and miR-21 inhibitor gene therapy. Mol Pharm. 2012;9(9):2636–45. doi: 10.1021/mp3002039. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy N. CELL DEATH 1+1 not equal 2. Nature Reviews Cancer. 2012;12(7):450–450. doi: 10.1038/nrc3301. [DOI] [PubMed] [Google Scholar]
- 40.Li YT, Qian XJ, Yu Y, Li ZH, Wu RY, Ji J, Jiao L, Li X, Kong PF, Chen WD, Feng GK, Deng R, Zhu XF. EGFR tyrosine kinase inhibitors promote pro-caspase-8 dimerization that sensitizes cancer cells to DNA-damaging therapy. Oncotarget. 2015;6(19):17491–17500. doi: 10.18632/oncotarget.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Lu Z, Zhao Q, Huang J, Shen H, Zhang Z. Enhanced chemotherapy efficacy by sequential delivery of siRNA and anticancer drugs using PEI-grafted graphene oxide. Small. 2011;7(4):460–4. doi: 10.1002/smll.201001522. [DOI] [PubMed] [Google Scholar]
- 42.Pinkerton NM, Grandeury A, Fisch A, Brozio J, Riebesehl BU, Prud'homme RK. Formation of stable nanocarriers by in situ ion pairing during block-copolymer-directed rapid precipitation. Mol Pharm. 2013;10(1):319–28. doi: 10.1021/mp300452g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adjei A, Rao S, Garren J, Menon G, Vadnere M. Effect of ion-pairing on 1-octanol-water partitioning of peptide drugs. I: The nonapeptide leuprolide acetate. International Journal of Pharmaceutics. 1993;90(2):141–149. [Google Scholar]
- 44.Meyer JD, Manning MC. Hydrophobic ion pairing: altering the solubility properties of biomolecules. Pharm Res. 1998;15(2):188–93. doi: 10.1023/a:1011998014474. [DOI] [PubMed] [Google Scholar]
- 45.Stahl PH, Wermuth CG. Handbook of Pharmaceutical salts properties, selection, and use. John Wiley & Sons; 2008. [Google Scholar]
- 46.Tang MH, Dou HJ, Sun K. One-step synthesis of dextran-based stable nanoparticles assisted by self-assembly. Polymer. 2006;47(2):728–734. [Google Scholar]
- 47.Liu M, Zhou ZM, Wang XF, Xu J, Yang K, Cui Q, Chen X, Cao MY, Weng J, Zhang QQ. Formation of poly(L,D-lactide) spheres with controlled size by direct dialysis. Polymer. 2007;48(19):5767–5779. [Google Scholar]
- 48.Zhang YW, Jiang M, Zhao JX, Zhou J, Chen DY. Hollow spheres from shell cross-linked, noncovalently connected micelles of carboxyl-terminated polybutadiene and poly(vinyl alcohol) in water. Macromolecules. 2004;37(4):1537–1543. [Google Scholar]
- 49.Song YH, Shin E, Wang H, Nolan J, Low S, Parsons D, Zale S, Ashton S, Ashford M, Ali M, Thrasher D, Boylan N, Troiano G. A novel in situ hydrophobic ion paring (HIP) formulation strategy for clinical product selection of a nanoparticle drug delivery system. J Control Release. 2016;229:106–19. doi: 10.1016/j.jconrel.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 50.Ashton S, Song YH, Nolan J, Cadogan E, Murray J, Odedra R, Foster J, Hall PA, Low S, Taylor P, Ellston R, Polanska UM, Wilson J, Howes C, Smith A, Goodwin RJ, Swales JG, Strittmatter N, Takats Z, Nilsson A, Andren P, Trueman D, Walker M, Reimer CL, Troiano G, Parsons D, De Witt D, Ashford M, Hrkach J, Zale S, Jewsbury PJ, Barry ST. Aurora kinase inhibitor nanoparticles target tumors with favorable therapeutic index in vivo. Sci Transl Med. 2016;8(325):325ra17. doi: 10.1126/scitranslmed.aad2355. [DOI] [PubMed] [Google Scholar]
- 51.YechezkelChezy B, Doxil? Handbook of Harnessing Biomaterials in Nanomedicine. Pan Stanford Publishing; 2012. The First FDA-Approved Nano-Drug; pp. 335–398. [Google Scholar]
- 52.Barenholz Y. Doxil(R)--the first FDA-approved nano-drug: lessons learned. J Control Release. 2012;160(2):117–34. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 53.Morton SW, Lee MJ, Deng ZJ, Dreaden EC, Siouve E, Shopsowitz KE, Shah NJ, Yaffe MB, Hammond PT. A nanoparticle-based combination chemotherapy delivery system for enhanced tumor killing by dynamic rewiring of signaling pathways. Sci Signal. 2014;7(325):ra44. doi: 10.1126/scisignal.2005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.