Abstract
Excess consumption of energy-dense foods combined with a sedentary lifestyle is driving an obesity epidemic. Although obesity is closely associated with insulin resistance, most individuals meet the insulin demand by increasing their functional β-cell mass. Those who eventually develop type 2 diabetes are distinguished by a failure in this compensatory process. Although a causal role of insulin resistance in compensatory β-cell responses has received considerable experimental support, precisely how the β cell senses changes in the metabolic environment is still unknown. As metabolism of glucose, lipids and amino acids is profoundly altered in obesity, it is not surprising that these nutrients are conspicuous among the factors proposed to contribute. In this review we summarise our understanding of the role of nutrients, in particular glucose, fatty acids and amino acids in β-cell compensation with a particular emphasis on their relation to insulin resistance-induced factors and their underlying mechanism of action. Finally, we describe the concept of epigenetic programming and review recent studies illustrating how the status of the β cell epigenome is a product of its nutrient environment, and how metabolic programming of the β cell contributes to diabetes risk.
Keywords: β-cell proliferation, insulin secretion, insulin resistance, nutrients, epigenetics, metabolic programming
1. Introduction
The World Health Organization estimates that more than 600 million adults were obese in the world in 2014. The worldwide prevalence of obesity has more than doubled between 1980 and 2014. This can be explained in part by changes in dietary and physical activity patterns towards the consumption of energy-dense foods and an increase in sedentary lifestyles. Obesity is accompanied by a dramatic increase in the risk of diseases such as type 2 diabetes (T2D). In 2015, the International Diabetes Federation estimated that 415 million people suffered from diabetes in the world, among which 90% have T2D. T2D results from the inability of the pancreatic β cell to secrete sufficient amounts of insulin to maintain glucose homeostasis in the face of insulin resistance [1]. In non-diabetic individuals, for example in response to pregnancy or obesity-induced insulin resistance, the β cell has a remarkable capacity to adjust to the changing metabolic environment by increasing its functional mass. Euglycemia is thus maintained by elevating insulin levels through a combination of enhanced insulin secretion per cell and an increase in β-cell number [2]. However, in some individuals this compensatory response fails and T2D ensues. The underlying cause of β-cell failure is likely a combination of genetic, epigenetic, and environmental factors, but its precise nature is unknown. Thus, understanding the mechanisms underlying β-cell adaptation to its metabolic environment is critical to devise new strategies to prevent the progression to overt T2D. In adult rodents and possibly in humans, the increase in β-cell mass is mainly due to β-cell replication. B-cell proliferation can be modulated by diverse signals including hormones (e.g. insulin), growth factors (e.g. heparin-binding EGF-like growth factor), neurotransmitters (e.g. serotonin, GABA) and nutrients (e.g. glucose and fatty acids) [2]. However, the precise nature of the signals that trigger β-cell proliferation in response to a given metabolic situation remains elusive. Despite the recent identification of an effector of insulin resistance-induced β-cell proliferation [3], important questions remain. In this review, we summarize the current knowledge on the role of macronutrients (i.e. glucose, lipids, and amino acids) in rodent and human β-cell proliferation. As nutrients affect the epigenome, we finally discuss how epigenetic modifications lead to heritable changes in the capacity of the β cell to compensate under conditions of metabolic stress.
2. Glucose
Glucose is the key source of energy in the body and enters the blood stream by ingestion of carbohydrates or release from glycogen stores (glycogenolysis). In the β cell, glucose transport is not limiting and results in a rapid equilibrium between extra- and intracellular glucose levels. Once in the cell glucose is phosphorylated by the rate-limiting enzyme glucokinase (GK). GK acts as a glucose sensor whereby its low affinity for glucose allows for a significant variation in activity within the range of physiological glucose concentrations. Glucose metabolism couples glucose sensing to insulin release ensuring a precise regulation of insulin secretion to maintain blood glucose levels within a narrow physiological range.
Since the 1970s there has been an appreciation that glucose metabolism in the β cell not only regulates insulin secretion but also β-cell proliferation [4]. More recently, glucose-induced β-cell proliferation has been investigated in rodents infused intravenously with supraphysiological glucose concentrations [5–10]. In our latest study in rats, we adjusted the glucose infusion rate to maintain elevated blood glucose levels during 72 h which resulted in a 3-fold increase in β-cell proliferation with no change in apoptosis and a moderate expansion of β-cell mass [10]. Importantly, glucose infusion in streptozotocin-treated NOD-SCID mice transplanted with human islets also stimulates human β-cell proliferation [11] indicating that the proliferative effect of glucose is likely conserved between rodents and humans.
2.1 Insulin resistance and glucose sensing in β-cell proliferation
The mechanisms linking glucose to β-cell proliferation in vivo are complex and only beginning to be understood. Prominent among these, insulin resistance, which is elevated following glucose infusion, plays a major role. Indeed, β-cell proliferation largely parallels insulin resistance in glucose-infused rats [10]. A causal role of insulin resistance in β-cell proliferation was demonstrated by transplantation of wild-type islets into genetically insulin-resistant mice, which, in the absence of hyperglycemia, increased β-cell proliferation in the graft [12]. These results were later confirmed using conditional liver-specific insulin receptor knockout mice [13], studies which ultimately led to the identification of Serpin B1, a liver-derived secreted protein, as a regulator of β-cell proliferation in this context [3].
Although it is widely accepted that insulin resistance plays a central role, studies showing that β-cell proliferation occurs before the onset of insulin resistance in short-term high fat fed mice suggest that perturbations in nutrient levels might be directly sensed by the β cell [14, 15]. In support of this hypothesis, although mice with a heterozygous loss-of-function of the GK gene had normal β-cell mass when fed a regular chow diet, the compensatory increase following high fat feeding was severely compromised [16]. Then, Porat et al. used genetic and chemical means to alter GK activity in islets and showed that the rate of glucose metabolism controls β-cell proliferation [17]. These results have been corroborated in human studies correlating a GK gain-of-function mutation with increased β-cell replication and mass [18].
The need for insulin signaling in the β cell to maintain β-cell function and mass [13, 19] suggest that insulin might mediate the effect of glucose on β-cell proliferation. However, this view has been challenged on a number of grounds [20]. Stamateris et al. showed that exposure to insulin does not increase β-cell proliferation in either low or high glucose conditions in mouse islets ex vivo [21]. The authors also showed that mTOR signaling, a pivotal pathway controlling β-cell proliferation (see below), is strongly activated by glucose but not insulin. In our studies we demonstrated that exposure of rat islets ex vivo to insulin concentrations found in the plasma following nutrient infusion do not increase β-cell proliferation [10]. These results suggest that β-cell proliferation is likely to be independent of the autocrine/paracrine action of insulin. Instead, Sharma et al. suggest that insulin production, rather than secreted insulin per se, regulates β-cell proliferation by activating the endoplasmic reticulum (ER) stress-sensing unfolded protein response (UPR) pathway [22]. However, in an elegant genetic study Szabat et al. demonstrated that decreasing insulin production by conditional deletion of the Ins2 gene on an Ins1-null background increases mitogenic signaling and promotes cell-cycle progression in a cell-autonomous manner by mitigating ER stress [23]. The role of insulin production and ER stress in glucose-induced β-cell proliferation will need to be resolved in future studies.
2.2 Glucose signaling in the β cell
A number of pathways are activated in the β cell in response to glucose metabolism and their contribution to glucose-induced β-cell proliferation has been presented previously in a comprehensive series of reviews [24–26]. Here we summarise major pathways involving liver kinase B1 (LKB1), AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR); Ca2+-mediated signaling; and carbohydrate response element-binding protein (ChREBP) (Fig. 1).
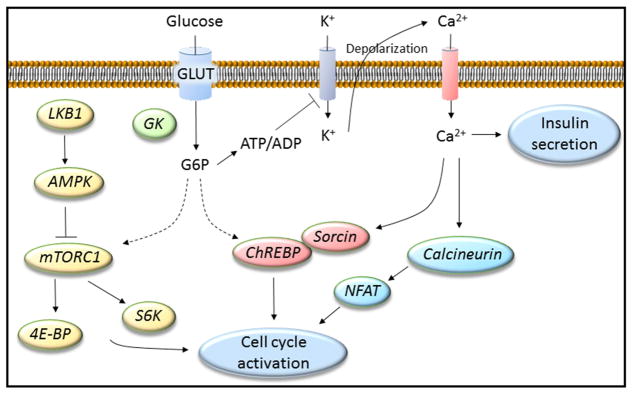
Figure 1. Glucose activates multiple pathways to control β-cell proliferation.
Glucose enters the β cell via GLUT transporters and is converted to glucose-6-phosphate (G6P) by glucokinase (GK), the first, committed step of glycolysis. Downstream, the stimulus-secretion coupling pathway, culminating in the accumulation of intracellular Ca2+, regulates insulin secretion but also contributes to β-cell proliferation via calcineurin/nuclear factor of activated T-cells (NFAT) and the carbohydrate-responsive element-binding protein (ChREBP). ChREBP is retained in the cytoplasm by Sorcin at low glucose concentrations but translocates to the nucleus upon glucose stimulation. In parallel, glucose metabolism regulates LKB1/AMPK/mTOR signaling to promote cell cycle activation.
LKB1/AMPK/mTOR
Conditional β-cell specific inactivation showed that LKB1 is a negative regulator of glucose-stimulated insulin secretion and β-cell proliferation, size and mass [27]. The effects of LKB1 on β-cell proliferation are mediated, in part, by its phosphorylation/activation of AMPK, which in turn inhibits mTOR signaling. Glucose metabolism promotes mTOR signaling and rapamycin exposure abrogates the β-cell proliferative response [21]. mTOR complexes (mTORC) include the rapamycin sensitive, mTORC1, and insensitive, mTORC2. mTORC1 is activated by nutrients and growth factors and controls cell growth and proliferation by directly modulating kinases, including S6 kinase (S6K) and 4E-BP.
Ca2+ signaling
Glucose metabolism generates multiple ATP molecules which increases the ATP/ADP ratio and promotes closure of ATP-sensitive K+ channels, depolarization of the plasma membrane and opening of voltage-dependent Ca2+ channels. The resulting increase in intracellular Ca2+ is not only involved in insulin exocytosis but is also linked to β-cell proliferation. Ca2+ activates the phosphatase calcineurin (via calmodulin) which in turn targets a number of substrates including nuclear factor of activated T cells (NFAT) to regulate cell cycle activator genes [28].
ChREBP
There are two isoforms of the ChREBP transcription factor. The alpha-isoform translocates to the nucleus in response to glucose metabolism, while the β-isoform is constitutively active. Both isoforms are necessary for glucose-induced β-cell proliferation and regulate the expression of cell cycle regulatory genes [29, 30]. ChREBPα nuclear translocation is coupled to Ca2+ signaling via the Ca2+-binding protein Sorcin which at low glucose concentrations retains ChREBPα in the cytoplasm [31].
3. Lipids
Lipids are a diverse group of molecules which include fatty acids and their derivatives, such as glycerolipids, phospholipids and sphingolipids, and sterol lipids such as cholesterol and its derivatives. Lipids exert a multitude of biological functions. Lipids serve an important structural role as the main component of membranes and are stored in the form of fat (triglycerides) which are hydrolysed to release fatty acids, an important source of energy. However, lipid also exert essential signaling roles controlling cell responses by activating G protein-coupled receptors (GPCR) at the cell surface and kinases, phosphatases, nuclear receptors and other effector proteins, intracellularly. In obesity, blood lipids in the form of triglycerides in lipoprotein complexes or fatty acids bound to albumin are elevated and have been extensively studied for their effect on β-cell functional mass.
3.1 Divergent effects of fatty acids on β-cell proliferation
In the β cell, acute exposure to fatty acids potentiates glucose-stimulated insulin secretion whereas, chronically, elevated fatty acid levels in the background of hyperglycemia, as seen in T2D, promote β-cell dysfunction (a phenomenon referred to as glucolipotoxicity) [32, 33]. Nonetheless, a number of studies in rodents point to a propensity of fatty acids to promote compensatory β-cell responses that may serve to delay the progression to T2D in obese individuals. In a series of studies we showed that simultaneous infusion of a triglyceride emulsion (co-infused with heparin to raise circulating fatty acid levels) and glucose in rats leads to insulin resistance and a marked increase in β-cell proliferation and mass despite eventual β-cell dysfunction due to glucolipotoxicity [10, 34, 35]. Surprisingly, although infusion of glucose alone resulted in a comparable level of insulin resistance, the β-cell proliferative response was dramatically higher when co-infused with fatty acids [10]. Infusion of fatty acids alone did not have a significant effect. To determine whether circulating factors were involved we performed syngeneic islet transplantation and found that, similar to the endogenous pancreatic islets, β-cell proliferation was increased in transplanted islets in response to the co-infusion of glucose and fatty acids [10]. Then, we exposed rat islets ex vivo to infused rat serum and found that whereas serum from saline- or glucose-infused animals had no significant effect, serum from rats co-infused with glucose and fatty acids potentiated glucose-induced β-cell proliferation [10]. These studies exposed an indirect effect of nutrient infusion on the β cell, however, we also tested whether fatty acids directly promote β-cell proliferation by treating rat or human islets ex vivo with a mixture of long-chain fatty acids. Interestingly, in both low and high glucose conditions, these fatty acids potently stimulated β-cell proliferation [10]. Hence, although a background of insulin resistance appears to be necessary, these studies led us to suggest that fatty acids in synergy with glucose can bypass insulin resistance to stimulate β-cell proliferation.
The capacity of fatty acids to promote β-cell proliferation both in vitro and in vivo has been demonstrated in a number of rodent models. Hirose et al. showed that whereas both the Zucker fatty (ZF) and their diabetic counterparts, the Zucker diabetic fatty (ZDF) rats are hyperlipidemic and insulin resistant, ZF rats maintain normoglycemia thanks to a compensatory increase in β-cell function and mass [36]. Interestingly, exposing normal rat, but not ZDF rat, islets to fatty acids increased glucose responsiveness and β-cell hyperplasia to levels seen in ZF islets. Steil et al. found that infusion of fatty acids in rats increases β-cell proliferation and insulin resistance in the absence of hyperglycemia [6]. It is noteworthy that in these examples hyperlipidemia, as described above for hyperglycemia, correlates with insulin resistance possibly providing a parallel signal to promote β-cell compensation. In marked contrast, Pascoe et al. showed that whereas infusion of fatty acids in mice has no impact by itself, it blocked glucose-induced β-cell proliferation in vivo [9]. Although the underlying reason for these widely divergent effects has yet to be determined, it is notable that the type of fatty acid species infused differ considerably between studies and may account for the variable β-cell responses. Along these lines, depending on the degree of unsaturation, fatty acids have been shown to either inhibit or increase β cell proliferation in rodent and human islets ex vivo. Whereas saturated fatty acids such as palmitate are toxic to the β cell, monounsaturated fatty acids such as oleate promote β-cell proliferation, and oleate protects against palmitate-induced β-cell dysfunction [37, 38]. In the study by Pascoe et al. described above, palmitate inhibited glucose induced β-cell proliferation in mouse islets ex vivo whereas oleate was without effect [9]. Mechanistically, the detrimental effects of palmitate are coupled to de novo ceramide synthesis [37, 38], expression of the cyclin-dependent kinase 4 (INK4) family of cell cycle inhibitors p16 and p18 [9] and toll-like receptor 3 (TLR3) signaling [39].
3.2 Lipid signaling in the β cell
In the β cell, fatty acids and their derivatives signal via several GPCR, including the receptors for short-chain (FFA2 & FFA3) and long-chain (FFA1) fatty acids, 2-monoacyl-glycerol (Gpr119) and sphingosine-1-phosphate, to regulate β-cell function and survival. Fatty acids also cross the β cell plasma membrane by diffusion or via cell surface transporters (eg. CD36). Once inside, fatty acids are activated by fatty acyl-CoA synthetase to generate acyl-CoA which undergoes β-oxidation, enters the glycerolipid/free fatty acid cycle or participates in sphingolipid synthesis to generate an array of messenger metabolites. Fatty acid metabolism is tightly linked to glucose metabolism and under conditions of hyperglycemia, an increase in malonyl-CoA inhibits the enzyme carnitine palmitoyl transferase 1 (CPT1). This in turn blunts β-oxidation by preventing fatty acid entry into the mitochondria, thus favoring esterification and synthesis of fatty acid-derived signaling molecules. Although a role of these pathways in insulin secretion has been shown [40] (Fig. 2), the existing literature sheds little light on their contribution to β-cell proliferation. To begin to elucidate these processes we undertook a global analysis of the islet transcriptome and identified a number of genes whose expression was significantly altered by the co-infusion of glucose and fatty acids in rats [35]. In particular, we described an increase in the expression of the heparin binding epidermal growth factor-like (HB-EGF) gene and then showed that the surge in β-cell proliferation detected in this model was dependent on EGF receptor and mTOR-FoxM1 signaling. In future studies we will combine transcriptomic data with lipidomic/proteomic analyses to identify lipids and signaling pathways that mediate fatty acid-induced β-cell proliferation.
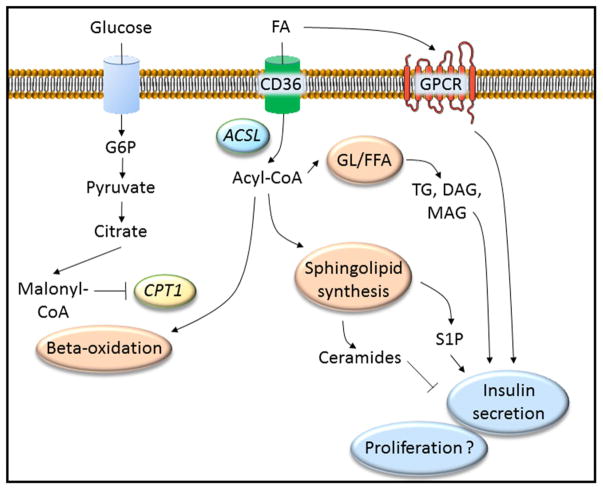
Figure 2. Lipid signaling in the β cell is mediated by multiple pathways.
Lipid metabolites, including fatty acids (FA), activate cell surface G protein-coupled receptors (GPCR) such as Gpr40, Gpr120 and Gpr119, or enter the β cell via transporters (CD36). Once inside the cell FA are activated by fatty acid-CoA ligase (ACSL) to generate acyl-CoA. Acyl-CoA is a substrate for mitochondrial β-oxidation, however in the presence of high glucose concentrations malonyl-CoA inhibits the enzyme carnitine palmitoyl transferase 1 (CPT1) blunting β-oxidation and shunting acyl-CoA towards the glycerolipid/free fatty acid (GL/FFA) cycle, which generates triglycerides (TG) and mono- (MAG) and di-acyglycerols (DAG), or the sphingolipid synthesis pathway, which generates ceramides and sphingosine-1-phosphate (S1P) among other metabolites. Together these metabolites regulate insulin secretion, however, their role in β-cell proliferation has yet to be demonstrated.
4. Amino acids
Amino acids, especially branched-chain amino acids (BCAA), including leucine, isoleucine and valine, regulate diverse physiological and metabolic pathways including hormone secretion, lipolysis and glucose metabolism [41]. The study of BCAA is increasingly relevant in the context of T2D since increased levels of plasma BCAA have been reported to lead to insulin resistance, possibly via persistent activation of the mTOR signaling pathway or perturbation of mitochondrial function [41]. Although amino acids, in particular BCAA are well known stimulators of insulin secretion [42], very few studies suggest a role of amino acids in β-cell proliferation. Amino acids enter the cell via active transporters such as the system-L amino acid transporter 1 (LAT1), which is abundantly expressed in islets. siRNA-mediated LAT1 knockdown in clonal β cells and rat islet cultures inhibits leucine-stimulated mTORC1 activation, insulin secretion and islet cell proliferation [43]. However, whether the effect of leucine on β-cell proliferation is specific or merely reflects the absence of an aminoacid essential for global protein synthesis remains an open question. Furthermore, whether leucine-induced mTORC1 activation contributes to the proliferative response was not demonstrated. Mullooly et al. [44] found that long-term exposure of rat islets ex vivo to BCAA has little effect on insulin secretion, proliferation or apoptosis, whereas arginine was found to impair β-cell function and increase apoptosis through activation of the endoplasmic reticulum stress response. Indirectly, glutamine affects β-cell function and survival via the insulin-like growth factor (IGF) 2/IGF1R pathway. Biosynthesis of IGF2, an autocrine regulator of β-cell mass and function, is controlled by glutamine which also stimulates secretion of IGF2 and activates Akt phosphorylation [45]. Despite ample evidence that amino acids regulate the mTOR pathway [46], there is no direct proof that a change of amino acids levels alters β-cell proliferation.
5. Nutrient-induced epigenetic regulation of β-cell proliferation
Gene expression is regulated by interplay between the transcriptional machinery and chromatin. Chromatin, a higher order complex of DNA, RNA and histone proteins, is subject to a multitude of epigenetic modifications that are stable through subsequent cell divisions and even the germ line. DNA methylation and histone acetylation and methylation are the most widely studied epigenetic modifications, although other histone modifications including ubiquitylation, phosphorylation, SUMOylation, ribosylation and O-GlcNAcylation as well as microRNA and long non-coding RNA also play a role.
5.1 Nutrient control of epigenetic modifications
The epigenetic status of a cell is sensitive to nutrient availability and underlies cellular adaptation to the metabolic environment. Nutrient metabolism produces key intermediates, including S-adenosyl-methionine (SAM), acetyl-coenzyme A (CoA), nicotinamide adenine dinucleotide (NADH/NAD+), flavin adenine dinucleotide (FAD) and α-ketoglutarate among others, which act as co-substrates for the so called ‘writers’ and ‘erasers’ that modify chromatin and associated factors (Fig. 3). The actions of these chromatin modifiers and their regulation by nutrients have been discussed in detail in several recent reviews [47–49].
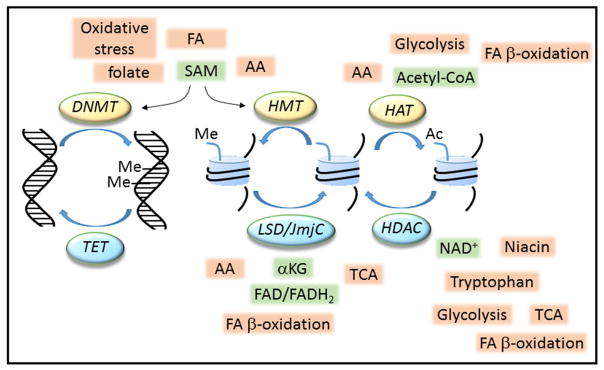
Figure 3. Nutrients regulate epigenetic modifications.
Carbohydrate, fatty acid (FA) and amino acid (AA) metabolism produces intermediates, including S-adenosyl-methionine (SAM), acetyl-coenzyme A (acetyl-CoA), nicotinamide adenine dinucleotide (NAD+), flavin adenine dinucleotide (FAD) and α-ketoglutarate (αKG), which act as co-substrates for epigenetic, chromatin modifying enzymes. DNA methyltrasferases (DNMT) methylate (Me) DNA, whereas histone methylation (Me) and acetylation (Ac) are mediated by methyltranseferases (HMT) and acetyltransferases (HAT), respectively. Histone demethylation and deacetylation are also subject to nutrient regulation via the action of demethylases (LSD/JmjC) and deacetylases (HDAC). Demethylation of DNA occurs passively or via enzymes of the Ten-eleven Translocation (TET) family. Together these modifications regulate heritable patterns of gene expression. TCA, tricarboxylic acid cycle.
DNA and histone methylation
DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B) use the methyl donor SAM to modify cytosine residues at CpG in DNA. Two-thirds of rodent and human genes have CpG islands in their proximal promoter regions. Promoter methylation interferes with transcription factor binding and promotes further chromatin modifications that lead to transcriptional repression. In contrast, methylation in the gene body is associated with highly transcribed genes. DNA demethylation takes place passively but DNA can be actively demethylated by the action of the Ten-eleven Translocation (TET) enzymes. Transfer of methyl groups from SAM to the ε-amino group of lysine by histone methyltransferases (HMTs) is the most prevalent form of histone methylation. Trimethylation of H3K4 and H3K36 at the promoter and in the gene body, respectively, is associated with actively transcribed genes. These modifications favor an ‘open’ chromatin conformation allowing the transcriptional machinery to access the DNA and promote gene transcription, In contrast, methylation of H3K9 or H3K27 leads to the formation of heterochromatin which represses gene transcription. Cellular SAM levels depend on vitamins such as folate, riboflavin, vitamin B12 and B6 and amino acids, such as methionine, cysteine, glycine and serine as well as dietary fats and oxidative stress. Levels of these methyl nutrients, and hence SAM, are affected in obese individuals.
Histone acetylation
Histone acetyltransferases (HATs) transfer the acetyl group from acetyl-CoA to the ε-amino group of lysine residues located in the N-terminal tail of histones. Acetylation removes the positive charge on histones, thereby decreasing the interaction with the negatively charged phosphate groups of DNA, which relaxes heterochromatin and increases gene transcription. Acetyl-CoA is a product of nutrient metabolism produced via glycolysis, fatty acid β-oxidation and amino acid degradation. Synthesized in the mitochondria Acetyl-CoA is shuttled to the cytoplasm as citrate where it is reconverted to acetyl-CoA by ATP-citrate lyase (ACL). Consumption of a high fat diet results in suppression of ACL levels in tissues leading to reduced acetyl-CoA.
Histone deacetylation
The acetylation status of histones is regulated by a balance between the activities of HATs and histone deacetylases (HDAC) of which the class III HDAC use NAD+ as a co-substrate. NAD+ is synthesized de novo from tryptophan, from niacin via the salvage pathway (vitamin B3) and is an important cofactor required by many enzymes involved in catabolic or oxidative pathways including glycolysis, the TCA cycle and fatty acid β-oxidation. Tissue NAD+ levels are increased during fasting and exercise whereas high fat diet and aging lead to a reduction.
Histone demethylation
The two major classes of histone demethylases, the lysine-specific (LSD) and the JmjC families, use FAD and α-ketoglutarate, respectively. FAD(H2) is a cofactor in fatty acid β-oxidation and oxidative phosphorylation, α-ketoglutarate is involved in amino-acid metabolism and both are products of the TCA cycle.
5.2 Epigenetic control of β-cell proliferation
Chromatin methylation has been mainly studied in the context of the age-dependent decline in the capacity of the β cell to proliferate, which is under the control of epigenetic events. p16/INK4a, p18/INK4c, p14/ARF, p27/KIP1 are cyclin-dependent kinase inhibitors (CDKI) whose expression is linked to the decline in β-cell proliferation. H3K4 and H3K27 methylation at the INK4a promoter are respectively, repressed and activated by Polycomb-Repressive Complexes, PRC1 and PRC2. In juvenile rodent and human β cells the PCR1 ring finger protein BMI1 and PRC2 EZH2, an HMT, are highly expressed leading to INK4a repression. In contrast, in adult β cells reduced BMI1 leads to recruitment of the HMT MLL1 increasing H3K4 methylation at the INK4a promoter. However, EZH2 is also reduced in adult β cells leading to loss of H3K27 methylation at the INK4a promoter. Together these histone marks increase INK4a gene expression down-regulating β-cell proliferation [50, 51]. Consistent with these findings, sustained expression of EZH2 in adult transgenic mice prevents the increase in INK4a gene expression and associated loss of β-cell proliferation [52]. Furthermore, β-cell specific deletion of the phosphatase and tensin homologue (PTEN), an inhibitor of PI3K/AKT signaling, prevents the decline in proliferation in aged β cells by up-regulating a cyclin D1/E2F/EZH2 pathway and repressing INK4a expression [53]. Alongside the age-dependent regulation of INK4a, the LIM-homeodomain transcription factor Islet-1 (ISL-1) promotes β-cell proliferation by recruiting the HMT SET7/9, which increases H3K4 methylation at the cyclin D1 promoter increasing its transcription, a pathway that is down-regulated in aging rats [54]. In a comprehensive study comparing the global methylation status of DNA from adolescent and adult mouse β cells an increase in de novo methylation and transcriptional repression of numerous genes involved in proliferation was revealed [55]. In this context it is surprising that β cell-specific deletion of DNMT3A, which is necessary for β cell functional maturation, does not affect β-cell proliferation [56] and suggest that compensation by other DNA methyltransferases may be responsible.
Modulating the activity of enzymes controlling histone acetylation also impacts β-cell proliferation. Mutation of serine 436 in CREB binding protein (CBP), an HAT, results in enhanced CREB-CBP interaction and activation of CBP-responsive genes. This mutation is associated with increased β-cell proliferation and mass but reduced glucose-stimulated insulin secretion [57]. Furthermore, exposure to butyrate, an HDAC inhibitor, increases β-cell proliferation and function and improves glucose homeostasis in diabetic rats by increasing histone H3/H4 acetylation [58]. Of note, the activity of HDAC is not limited to histones and decreasing SirT1 deacetylase activity, by exposure to GLP-1, leads to an increase in acetylation of the FoxO1 transcription factor down-regulating its activity and promoting β-cell proliferation [59].
Despite ample evidence that the status of the β cell epigenome affects proliferation, relatively few studies have addressed the role of epigenetic modifications in nutrient control of β-cell proliferation. Chronic exposure of islets ex vivo to high glucose concentrations or palmitate was found to alter patterns of expression and associated epigenetic marks at a number of genes important for β-cell function [60, 61]. Unfortunately, genes controlling proliferation were not investigated in these studies. However, Menin, a tumor suppressor protein participating in an HMT complex that mediates H3K4 methylation and transcriptional activation of CDKI, is inhibited following glucose treatment of rat islets ex vivo or glucose infusion in adult rats via the PI3K/AKT/FOXO1 pathway, releasing a brake on β-cell proliferation [62].
5.3 Developmental programming of the β cell
Epigenetic modifications that affect an individual’s long-term metabolic health and response to diabetogenic insults are a product not only of the nascent nutrient environment but can be established early in life, in particular during fetal development and the perinatal period. Data from the Dutch famine (Winter 1944–45) has been a hotbed for researchers to decipher the long-term effects of malnutrition early in life on metabolic disease risk in adulthood. These studies have found that maternal malnutrition during gestation, but also moderate to severe malnutrition during the postnatal period, increases the risk of obesity and associated complications including T2D in adulthood [63, 64]. The effect of malnutrition during prenatal development on the β cell has been investigated in a rat model of intrauterine growth retardation (IUGR) through uterine artery ligation [65]. These rats develop diabetes in adulthood due to reduced β-cell function and mass. The authors then analyzed epigenetic marks at the Pdx1 transcription factor promoter, a gene critically involved in β-cell development, function and proliferation. These studies highlighted alterations in histone modifications and a role of HDAC1 in the transcriptional repression of Pdx1 during foetal and postnatal development. Following diabetes onset in adulthood the Pdx1 gene was found to be completely silenced due to promoter methylation. Interestingly, inhibition of HDAC in neonatal IUGR rats reversed the epigenetic changes and the reduction in Pdx1 expression suggesting that in some circumstances metabolic malprogramming can be reversed. Diabetes risk and β-cell function and mass are also affected in offspring exposed to maternal overnutrition and diabetes. Maternal exposure to high fat diet in mice predisposes male offspring to glucose intolerance, insulin resistance and decreased β-cell function and mass, characterized by reduced insulin content and Pdx1 gene expression [66]. In humans, offspring exposed to an intrauterine diabetic environment show comparable adiposity and insulin sensitive but a reduced insulin response to glucose compared to siblings born before their mothers were diagnosed with diabetes [67]. Similarly in rodents, Wistar rats exposed to a diabetic environment during foetal development, through embryo transfer into Goto Kakizaki (GK) mothers, have reduced β-cell mass and impaired glucose tolerance in adulthood compared to controls transferred to normal Wistar rats [68]. Beyond developmental programming, in a seminal study Ng et al. showed that paternal high fat feeding in rats predisposes their female offspring to impaired insulin secretion and glucose tolerance by altering the expression of hundreds of genes involved in β-cell function and mass [69].
6. Conclusion
Here, we presented the view that alterations in glucose and lipid homeostasis, arising in obesity, are directly sensed by the β cell and cooperate with insulin resistance-induced factors to promote functional β-cell mass expansion. We have also presented evidence suggesting that the nutrient environment impacts diabetes risk by altering the β cell epigenome. From this perspective, a number of issues need to be addressed. Firstly, although a causal link between hyperlipidemia and β-cell compensation was postulated many years ago, a thorough analysis of the underlying mechanisms has not been performed. The application of non-targeted lipidomic approaches to identify lipid species in plasma samples and islets associated with β-cell compensation in humans and rodents coupled with studies to elucidate lipid signals controlling β-cell proliferation may help to resolve this issue. Secondly, although a multitude of pathways, including LKB1/AMPK/mTOR, ChREBP and Ca2+ signalling described above, are recruited in the β cell and participate in glucose-induced proliferation, a unifying hypothesis integrating these signals with insulin demand-driven signals (eg. ER stress) as well as insulin resistance-induced factors (eg. serpin B1) and lipid signals is needed. Finally, knowledge of these pathways and their interaction with the β-cell epigenome will need to be harnessed to find novel ways to enhance β-cell compensation to prevent or delay diabetes onset.
Highlights.
In this review we discuss:
The role of macronutrients, glucose, lipids and amino acids in β-cell proliferation
Molecular mechanisms that couple nutrient metabolism to β-cell proliferation
The role of nutrients in epigenetic control of β-cell proliferation
Acknowledgments
Funding Sources
This work was supported by the CRCHUM (Postdoctoral Fellowship to VSM), the National Institutes of Health (grant R01-DK-58096 to VP) and the Canadian Institutes of Health Research (grant MOP 77686 to VP). VP holds the Canada Research Chair in Diabetes and Pancreatic Beta-Cell Function.
Footnotes
Conflict of Interest
The authors declared no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kahn SE. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of Type 2 diabetes. Diabetologia. 2003;46(1):3–19. doi: 10.1007/s00125-002-1009-0. [DOI] [PubMed] [Google Scholar]
- 2.Prentki M, Nolan CJ. Islet beta cell failure in type 2 diabetes. J Clin Invest. 2006;116(7):1802–12. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Ouaamari A, et al. SerpinB1 Promotes Pancreatic beta Cell Proliferation. Cell Metab. 2016;23(1):194–205. doi: 10.1016/j.cmet.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellerstrom C, Andersson A, Gunnarsson R. Regeneration of islet cells. Acta Endocrinol Suppl (Copenh) 1976;205:145–60. [PubMed] [Google Scholar]
- 5.Bonner-Weir S, et al. Compensatory growth of pancreatic beta-cells in adult rats after short-term glucose infusion. Diabetes. 1989;38(1):49–53. doi: 10.2337/diab.38.1.49. [DOI] [PubMed] [Google Scholar]
- 6.Steil GM, et al. Adaptation of beta-cell mass to substrate oversupply: enhanced function with normal gene expression. Am J Physiol Endocrinol Metab. 2001;280(5):E788–96. doi: 10.1152/ajpendo.2001.280.5.E788. [DOI] [PubMed] [Google Scholar]
- 7.Paris M, et al. Specific and combined effects of insulin and glucose on functional pancreatic beta-cell mass in vivo in adult rats. Endocrinology. 2003;144(6):2717–27. doi: 10.1210/en.2002-221112. [DOI] [PubMed] [Google Scholar]
- 8.Alonso LC, et al. Glucose infusion in mice: a new model to induce beta-cell replication. Diabetes. 2007;56(7):1792–801. doi: 10.2337/db06-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pascoe J, et al. Free fatty acids block glucose-induced beta-cell proliferation in mice by inducing cell cycle inhibitors p16 and p18. Diabetes. 2012;61(3):632–41. doi: 10.2337/db11-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moulle VS, et al. Glucose and fatty acids synergistically and reversibly promote beta cell proliferation in rats. Diabetologia. 2017;60(5):879–888. doi: 10.1007/s00125-016-4197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levitt HE, et al. Glucose stimulates human beta cell replication in vivo in islets transplanted into NOD-severe combined immunodeficiency (SCID) mice. Diabetologia. 2011;54(3):572–82. doi: 10.1007/s00125-010-1919-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flier SN, Kulkarni RN, Kahn CR. Evidence for a circulating islet cell growth factor in insulin-resistant states. Proc Natl Acad Sci U S A. 2001;98(13):7475–80. doi: 10.1073/pnas.131192998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada T, et al. Insulin receptors in beta-cells are critical for islet compensatory growth response to insulin resistance. Proc Natl Acad Sci U S A. 2007;104(21):8977–82. doi: 10.1073/pnas.0608703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stamateris RE, et al. Adaptive beta-cell proliferation increases early in high-fat feeding in mice, concurrent with metabolic changes, with induction of islet cyclin D2 expression. Am J Physiol Endocrinol Metab. 2013;305(1):E149–59. doi: 10.1152/ajpendo.00040.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosser RE, et al. High-fat diet-induced beta-cell proliferation occurs prior to insulin resistance in C57Bl/6J male mice. Am J Physiol Endocrinol Metab. 2015;308(7):E573–82. doi: 10.1152/ajpendo.00460.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terauchi Y, et al. Glucokinase and IRS-2 are required for compensatory beta cell hyperplasia in response to high-fat diet-induced insulin resistance. J Clin Invest. 2007;117(1):246–57. doi: 10.1172/JCI17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porat S, et al. Control of pancreatic beta cell regeneration by glucose metabolism. Cell Metab. 2011;13(4):440–9. doi: 10.1016/j.cmet.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kassem S, et al. Large islets, beta-cell proliferation, and a glucokinase mutation. N Engl J Med. 2010;362(14):1348–50. doi: 10.1056/NEJMc0909845. [DOI] [PubMed] [Google Scholar]
- 19.Kulkarni RN, et al. Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell. 1999;96(3):329–39. doi: 10.1016/s0092-8674(00)80546-2. [DOI] [PubMed] [Google Scholar]
- 20.Rhodes CJ, et al. Direct autocrine action of insulin on beta-cells: does it make physiological sense? Diabetes. 2013;62(7):2157–63. doi: 10.2337/db13-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamateris RE, et al. Glucose Induces Mouse beta-Cell Proliferation via IRS2, MTOR, and Cyclin D2 but Not the Insulin Receptor. Diabetes. 2016;65(4):981–95. doi: 10.2337/db15-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma RB, et al. Insulin demand regulates beta cell number via the unfolded protein response. J Clin Invest. 2015;125(10):3831–46. doi: 10.1172/JCI79264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szabat M, et al. Reduced Insulin Production Relieves Endoplasmic Reticulum Stress and Induces beta Cell Proliferation. Cell Metab. 2016;23(1):179–93. doi: 10.1016/j.cmet.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 24.Kulkarni RN, et al. Human beta-cell proliferation and intracellular signaling: driving in the dark without a road map. Diabetes. 2012;61(9):2205–13. doi: 10.2337/db12-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernal-Mizrachi E, et al. Human beta-cell proliferation and intracellular signaling part 2: still driving in the dark without a road map. Diabetes. 2014;63(3):819–31. doi: 10.2337/db13-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart AF, et al. Human beta-cell proliferation and intracellular signaling: part 3. Diabetes. 2015;64(6):1872–85. doi: 10.2337/db14-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu A, et al. Loss of Lkb1 in adult beta cells increases beta cell mass and enhances glucose tolerance in mice. Cell Metab. 2009;10(4):285–95. doi: 10.1016/j.cmet.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Heit JJ, et al. Calcineurin/NFAT signalling regulates pancreatic beta-cell growth and function. Nature. 2006;443(7109):345–9. doi: 10.1038/nature05097. [DOI] [PubMed] [Google Scholar]
- 29.Metukuri MR, et al. ChREBP mediates glucose-stimulated pancreatic beta-cell proliferation. Diabetes. 2012;61(8):2004–15. doi: 10.2337/db11-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P, et al. Induction of the ChREBPbeta Isoform Is Essential for Glucose-Stimulated beta-Cell Proliferation. Diabetes. 2015;64(12):4158–70. doi: 10.2337/db15-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noordeen NA, et al. Glucose-induced nuclear shuttling of ChREBP is mediated by sorcin and Ca(2+) ions in pancreatic beta-cells. Diabetes. 2012;61(3):574–85. doi: 10.2337/db10-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poitout V, Robertson RP. Glucolipotoxicity: fuel excess and beta-cell dysfunction. Endocr Rev. 2008;29(3):351–66. doi: 10.1210/er.2007-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prentki M, Matschinsky FM, Madiraju SR. Metabolic signaling in fuel-induced insulin secretion. Cell Metab. 2013;18(2):162–85. doi: 10.1016/j.cmet.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 34.Fontes G, et al. Glucolipotoxicity age-dependently impairs beta cell function in rats despite a marked increase in beta cell mass. Diabetologia. 2010;53(11):2369–79. doi: 10.1007/s00125-010-1850-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarrouki B, et al. Epidermal growth factor receptor signaling promotes pancreatic beta-cell proliferation in response to nutrient excess in rats through mTOR and FOXM1. Diabetes. 2014;63(3):982–93. doi: 10.2337/db13-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirose H, et al. Defective fatty acid-mediated beta-cell compensation in Zucker diabetic fatty rats. Pathogenic implications for obesity-dependent diabetes. J Biol Chem. 1996;271(10):5633–7. doi: 10.1074/jbc.271.10.5633. [DOI] [PubMed] [Google Scholar]
- 37.Maedler K, et al. Distinct effects of saturated and monounsaturated fatty acids on beta-cell turnover and function. Diabetes. 2001;50(1):69–76. doi: 10.2337/diabetes.50.1.69. [DOI] [PubMed] [Google Scholar]
- 38.Maedler K, et al. Monounsaturated fatty acids prevent the deleterious effects of palmitate and high glucose on human pancreatic beta-cell turnover and function. Diabetes. 2003;52(3):726–33. doi: 10.2337/diabetes.52.3.726. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, et al. Elevated toll-like receptor 3 inhibits pancreatic beta-cell proliferation through G1 phase cell cycle arrest. Mol Cell Endocrinol. 2013;377(1–2):112–22. doi: 10.1016/j.mce.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Nolan CJ, et al. Fatty acid signaling in the beta-cell and insulin secretion. Diabetes. 2006;55(Suppl 2):S16–23. doi: 10.2337/db06-s003. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S, et al. Novel metabolic and physiological functions of branched chain amino acids: a review. J Anim Sci Biotechnol. 2017;8:10. doi: 10.1186/s40104-016-0139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair KS, Short KR. Hormonal and signaling role of branched-chain amino acids. J Nutr. 2005;135(6 Suppl):1547s–52s. doi: 10.1093/jn/135.6.1547S. [DOI] [PubMed] [Google Scholar]
- 43.Cheng Q, et al. System-L amino acid transporters play a key role in pancreatic beta-cell signalling and function. J Mol Endocrinol. 2016;56(3):175–87. doi: 10.1530/JME-15-0212. [DOI] [PubMed] [Google Scholar]
- 44.Mullooly N, et al. Elevated levels of branched-chain amino acids have little effect on pancreatic islet cells, but L-arginine impairs function through activation of the endoplasmic reticulum stress response. Exp Physiol. 2014;99(3):538–51. doi: 10.1113/expphysiol.2013.077495. [DOI] [PubMed] [Google Scholar]
- 45.Modi H, Cornu M, Thorens B. Glutamine stimulates biosynthesis and secretion of insulin-like growth factor 2 (IGF2), an autocrine regulator of beta cell mass and function. J Biol Chem. 2014;289(46):31972–82. doi: 10.1074/jbc.M114.587733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDaniel ML, et al. Metabolic and autocrine regulation of the mammalian target of rapamycin by pancreatic beta-cells. Diabetes. 2002;51(10):2877–85. doi: 10.2337/diabetes.51.10.2877. [DOI] [PubMed] [Google Scholar]
- 47.Fan J, et al. Metabolic regulation of histone post-translational modifications. ACS Chem Biol. 2015;10(1):95–108. doi: 10.1021/cb500846u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janke R, Dodson AE, Rine J. Metabolism and epigenetics. Annu Rev Cell Dev Biol. 2015;31:473–496. doi: 10.1146/annurev-cellbio-100814-125544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su X, Wellen KE, Rabinowitz JD. Metabolic control of methylation and acetylation. Curr Opin Chem Biol. 2016;30:52–60. doi: 10.1016/j.cbpa.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H, et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23(8):975–85. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dhawan S, Tschen SI, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes Dev. 2009;23(8):906–11. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou JX, et al. Combined modulation of polycomb and trithorax genes rejuvenates beta cell replication. J Clin Invest. 2013;123(11):4849–58. doi: 10.1172/JCI69468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng N, et al. PTEN controls beta-cell regeneration in aged mice by regulating cell cycle inhibitor p16ink4a. Aging Cell. 2013;12(6):1000–11. doi: 10.1111/acel.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, et al. ISL-1 promotes pancreatic islet cell proliferation by forming an ISL-1/Set7/9/PDX-1 complex. Cell Cycle. 2015;14(24):3820–9. doi: 10.1080/15384101.2015.1069926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Avrahami D, et al. Aging-Dependent Demethylation of Regulatory Elements Correlates with Chromatin State and Improved beta Cell Function. Cell Metab. 2015;22(4):619–32. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dhawan S, et al. DNA methylation directs functional maturation of pancreatic beta cells. J Clin Invest. 2015;125(7):2851–60. doi: 10.1172/JCI79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hussain MA, et al. Increased pancreatic beta-cell proliferation mediated by CREB binding protein gene activation. Mol Cell Biol. 2006;26(20):7747–59. doi: 10.1128/MCB.02353-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khan S, Jena GB. Protective role of sodium butyrate, a HDAC inhibitor on beta-cell proliferation, function and glucose homeostasis through modulation of p38/ERK MAPK and apoptotic pathways: study in juvenile diabetic rat. Chem Biol Interact. 2014;213:1–12. doi: 10.1016/j.cbi.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Bastien-Dionne PO, et al. Glucagon-like peptide 1 inhibits the sirtuin deacetylase SirT1 to stimulate pancreatic beta-cell mass expansion. Diabetes. 2011;60(12):3217–22. doi: 10.2337/db11-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hall E, et al. Effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets. BMC Med. 2014;12:103. doi: 10.1186/1741-7015-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishikawa K, et al. Long-term pancreatic beta cell exposure to high levels of glucose but not palmitate induces DNA methylation within the insulin gene promoter and represses transcriptional activity. PLoS One. 2015;10(2):e0115350. doi: 10.1371/journal.pone.0115350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H, et al. Glucose-mediated repression of menin promotes pancreatic beta-cell proliferation. Endocrinology. 2012;153(2):602–11. doi: 10.1210/en.2011-1460. [DOI] [PubMed] [Google Scholar]
- 63.Ravelli AC, et al. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351(9097):173–7. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- 64.van Abeelen AF, et al. Famine exposure in the young and the risk of type 2 diabetes in adulthood. Diabetes. 2012;61(9):2255–60. doi: 10.2337/db11-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park JH, et al. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118(6):2316–24. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yokomizo H, et al. Maternal high-fat diet induces insulin resistance and deterioration of pancreatic beta-cell function in adult offspring with sex differences in mice. Am J Physiol Endocrinol Metab. 2014;306(10):E1163–75. doi: 10.1152/ajpendo.00688.2013. [DOI] [PubMed] [Google Scholar]
- 67.Gautier JF, et al. Low acute insulin secretory responses in adult offspring of people with early onset type 2 diabetes. Diabetes. 2001;50(8):1828–33. doi: 10.2337/diabetes.50.8.1828. [DOI] [PubMed] [Google Scholar]
- 68.Portha B, et al. The GK rat: a prototype for the study of non-overweight type 2 diabetes. Methods Mol Biol. 2012;933:125–59. doi: 10.1007/978-1-62703-068-7_9. [DOI] [PubMed] [Google Scholar]
- 69.Ng SF, et al. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467(7318):963–6. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]