Abstract
The localization and organization of mitochondria- and ribonucleoprotein granule-rich germ plasm is essential for many aspects of germ cell development. In Xenopus, germ plasm is maternally inherited and is required for the specification of primordial germ cells (PGCs). Germ plasm is aggregated into larger patches during egg activation and cleavage and is ultimately translocated perinuclearly during gastrulation. Although microtubule dynamics and a kinesin (Kif4a) have been implicated in Xenopus germ plasm localization, little is known about how germ plasm distribution is regulated. Here, we identify a role for maternal Xenopus Syntabulin in the aggregation of germ plasm following fertilization. We show that depletion of sybu mRNA using antisense oligonucleotides injected into oocytes results in defects in the aggregation and perinuclear transport of germ plasm and subsequently in reduced PGC numbers. Using live imaging analysis, we also characterize a novel role for Sybu in the collection of germ plasm in vegetal cleavage furrows by surface contraction waves. Additionally, we show that a localized kinesin-like protein, Kif3b, is also required for germ plasm aggregation and that Sybu functionally interacts with Kif3b and Kif4a in germ plasm aggregation. Overall, these data suggest multiple coordinate roles for kinesins and adaptor proteins in controlling the localization and distribution of a cytoplasmic determinant in early development.
Keywords: Xenopus, germ plasm, primordial germ cells, germline, mitochondrial transport
Introduction
Cell polarity and intracellular localization in the egg are important for many aspects of normal embryonic development, including overall body patterning and proper germ cell development. The role of a localized “germ plasm” in the formation of the germline has been particularly well studied. Germ plasm was first identified as a histologically distinct region localized within eggs and embryonic germline cells of various organisms and is composed mainly of cytoplasmic aggregates of mitochondria and other organelles, dense ribonucleoprotein germ line granules, and a collection of proteins and RNAs (reviewed in Beams and Kessel, 1974; Houston and King, 2000; Extavour and Akam, 2003; Aguero et al. 2017). The presence of germ plasm and certain localized RNAs is required for germline development in organisms such as Drosophila, Xenopus and zebrafish. Although maternal germ plasm per se is notably absent from the mammalian egg and early blastocyst (and also from salamander/newt eggs), the differentiating germ cells of these organisms are known to contain a “nuage” substance that is similar in ultrastructure to germ plasm (Eddy, 1975). Nuage also houses a similar set of localized proteins and RNAs and functions during later aspects of germ cell differentiation (Houston and King, 2000; Voronina et al. 2011). Whereas the overall role of the germ plasm in germ cell biology is well appreciated, the mechanisms controlling its assembly and localization in vertebrates are less well understood. In Xenopus eggs, germ plasm is found dispersed in small islands throughout the vegetal cortex (Czolowska, 1969; Whitington and Dixon, 1975). Germ plasm is thought to originate in the mitochondrial cloud/Balbiani body of pre-vitellogenic oocytes, which accumulates germ plasm-specific mRNAs, including nanos1, dazl, and vasa homologs. The mitochondrial cloud is fragmented during early oogenesis (stage II in Xenopus) and its material localizes to the future vegetal pole of the growing oocyte (Kloc and Etkin, 1995, 1998; Choo et al., 2005). Following oocyte maturation and subsequent fertilization, the germ plasm undergoes additional local clustering and further aggregates into larger masses at the vegetal pole. Aggregation continues during the cleavage stages, mediated in part by surface contraction waves (SCWs) in the egg and blastomeres undergoing mitosis, and is followed by “ingression” of germ plasm along the vegetal cleavage furrows (Ressom and Dixon, 1988; Savage and Danilchik, 1993). At these stages, germ plasm is asymmetrically inherited, resulting in an initial PGC population of about four cells. During gastrulation, the germ plasm transitions from a peripheral position to a perinuclear localization and is subsequently inherited symmetrically by daughter cells, presumptively increasing the number of cells specified toward the germline (reviewed in Houston and King, 2000). Previous inhibitor-based studies in Xenopus have indicated that both local and SCW-mediated germ plasm aggregation is microtubule dependent and likely independent of actin microfilaments (Ressom and Dixon, 1988; Savage and Danilchik, 1993). Correspondingly, the germ plasm is enriched in microtubules and depletion of maternal kif4a mRNA (Xklp1; Robb et al., 1996) inhibited local and SCW-mediated germ plasm aggregation, in the latter case by seemingly inhibiting SCW propagation. Depletion of kif15 (Xklp2) did not affect germ plasm aggregation (Robb et al., 1996), suggesting that different kinesin family members may play discrete roles in germ plasm localization. It is unclear whether Kif4a-mediated germ plasm aggregation is conserved in vertebrates. In zebrafish, germ plasm assembly appears to occur at the cleavage stages and involves a two-stage localization of RNAs to the blastodisc at the animal pole, followed by actin and microtubule mediated mechanisms acting at cleavage furrows (Theusch et al. 2006; Eno and Pelegri, 2016). It is thus likely that the germ plasm assembly process has been modified during evolution of the teleost egg/embryo morphology, and despite the similarity in overall composition and role of the cytoskeleton, the specific mechanisms regulating germ plasm aggregation in either organism are not well understood.
Another conserved cytoplasmic localization event occurring in fertilized eggs is the putative translocation of determinants by microtubule-mediated cortical rotation leading to dorsal axis formation (reviewed in Houston, 2013; Houston, 2017). Interestingly, several maternal genes involved in Xenopus axis formation exhibit at least partial mRNA localization to the germ plasm in eggs and embryos. These include trim36, dead end homolog 1 (dnd1) and plin2. Importantly, mRNA depletion of maternal stores of these molecules leads to loss of cortical rotation and to defects in dorsal axis formation (Chan et al. 2007; Cuykendall and Houston, 2009; Mei et al. 2013). In zebrafish, two maternal-effect mutants deficient in axis formation have been identified that encode mRNAs localized to the mitochondrial cloud and vegetal cortex of the oocyte and egg (tokkebi (tkk)/syntabulin (sybu), Nojima et al., 2010; hecate/grip2a, Ge et al., 2014). Both gene products are potentially involved in microtubule organization and/or trafficking. However, both Sybu and Grip2a protein and RNA fail to accumulate in the aggregating germ plasm following its transport to the forming blastodisc and thus do not localize to embryonic germ plasm. By contrast, in Xenopus, sybu and grip2a do associate with germ plasm in oocytes (Colozza and De Robertis, 2014; Kaneshiro et al. 2007; Tarbashevich et al. 2007) but also remain localized to the germ plasm throughout early embryogenesis. Depletion of Grip2a in fertilized embryos using morpholino antisense oligos in Xenopus can result in PGC migration defects (Kirilenko et al. 2008) but a role in axis formation was not specifically tested by depletion of maternal grip2a mRNA in oocytes. Maternal depletion of sybu by the host-transfer procedure does indeed result in axis deficient embryos (Colozza and De Robertis, 2014) but it remains unclear to what extent Sybu is acting similarly in fish and frogs. Nevertheless, the localization of sybu mRNA to the germ plasm in cleavage stage embryos prompted us to examine its roles in the germ plasm.
This work focuses on Sybu, previously characterized as a kinesin motor linker protein involved in the transport of mitochondria, syntaxin-associated vesicles and other cargoes in neuronal axons (Su et al., 2004; Cai et al., 2005; Cai et al., 2007; Frederick and Shaw 2007). We therefore hypothesized that Sybu would function in linking germ plasm to microtubule motors via mitochondria or other vesicles. Here we show that maternal sybu is required for PGC formation in Xenopus, likely by mediating kinesin-dependent aggregation in the early embryo. We also define the roles of additional kinesins in germ plasm aggregation and show that germ plasm aggregation at the vegetal pole is coupled to the surface contraction waves occurring after vegetal cleavage furrow formation.
Results
Syntabulin is associated with germ plasm in Xenopus
In Xenopus and zebrafish, sybu is expressed in the mitochondrial cloud/Balbiani body (Bb) of pre-vitellogenic oocytes and in the vegetal cortex of full-grown oocytes and eggs, congruent with the germ plasm (Colozza and De Robertis, 2014; Nojima et al., 2010). We verified this expression pattern in Xenopus, showing as previously reported that sybu localizes to the mitochondrial cloud of stage I oocytes and remains with the cloud as it fragments and disperses to the vegetal cortex during oogenesis (Colozza and De Robertis, 2014). In fertilized eggs, sybu is maintained in a germ plasm pattern, concentrating at the vegetal apex and at cleavage furrows, and later becoming restricted to a small group of vegetal cells in the gastrula (Fig. S1). Because a subset of other vegetally localized RNAs overlapping with the germ plasm have been shown to have roles in axis formation (reviewed in Houston, 2013), it was important to determine whether sybu transcript localization mirrored the localization of these other mRNAs. We therefore directly compared the expression of sybu to that of other localized RNAs involved in axis formation (trim36 and wnt11b), as well as to that of bona fide germ plasm-localized RNA (pgat/xpat; (Hudson and Woodland, 1998)), using oocytes from the same animal under identical conditions.
Sybu localization was more restricted at the vegetal apex, closely resembling the germ plasm-like pattern represented by pgat rather than the more intermediate pattern of wnt11b and trim36 (Fig. 1A–D). The sybu pattern was however consistently broader than pgat, which may reflect the localization of pgat more exclusively to germinal granules (Kloc et al. 2002). The perdurance of sybu in the germ plasm of the dividing embryo in Xenopus differs from zebrafish in this regard; zebrafish sybu RNA remains in the vegetal yolk cell during early cleavage and is neither transported to the animal blastoderm cytoplasm nor incorporated into the embryonic germ plasm (Nojima et al., 2010). Also, zebrafish Sybu protein is degraded by 60-minutes post-fertilization, further suggesting that Sybu protein is not present in the early germ plasm. To determine whether Sybu protein might function in Xenopus germ plasm, we injected mRNA encoding a C-terminally-tagged Sybu-GFP fusion protein into oocytes. These were then matured and prick-activated to simulate fertilization and germ plasm aggregation. Sybu-GFP was localized to germ plasm aggregates in the egg, confirmed by co-staining with MitoTracker (Fig. 1E–J). And, Sybu-GFP was visible for several hours during germ plasm aggregation, suggesting that at least exogenous protein is not rapidly degraded after fertilization. Overall, these data show that Xenopus Sybu is localized to aggregating germ plasm in the embryo, although lack of specific antibodies precluded analysis of the endogenous protein localization.
Fig. 1.
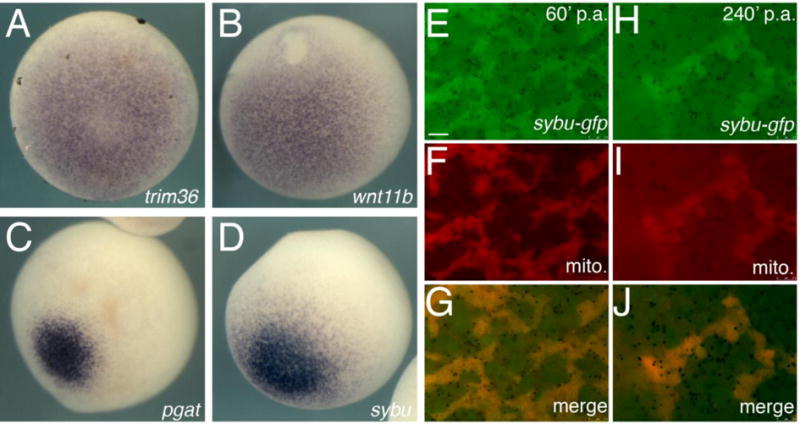
Germ plasm enrichment of Sybu mRNA and protein in oocytes and early embryos. (A–D) Representative images of whole-mount in situ hybridization on stage VI oocytes comparing expression of (A) trim36, (B) wnt11b, (C) pgat and (D) sybu. Vegetal views are shown. (E–J) Localization of Sybu-GFP to germ plasm. (E–G) Overlap of Sybu-GFP with clustered mitochondria (mito., MitoTracker) in pricked eggs 60-minutes post-activation (p.a.) and at 240-minutes post-activation (p.a.). Scale bars represent 7.5 μm.
Depletion of maternal syntabulin results in reduced primordial germ cell formation
To determine the extent that the germ plasm localization of maternal Sybu correlated with a role in normal germ cell development, we used an antisense oligo-mediated maternal mRNA depletion strategy (reviewed in Hulstrand et al., 2010). Although Colozza and De Robertis (2014) identified a role for Sybu in dorsal axis formation, our own oligo design identified several oligos that appeared to substantially deplete sybu RNA (Fig. 2A) although none of these resulted in ventralization (see below). X. laevis is an ancient allopolyploid with a genome comprising two “subgenomes,” L and S (corresponding to Long and Short homeologous chromosomes) derived from the hybridization of two ancestral species (Session et al. 2016). In many cases, transcripts are expressed from both homeologs and we suspected a potential incomplete targeting of one homeolog. Sybu is retained in both subgenomes, in contrast to typical core germ granule genes, which are preferentially retained within the L subgenome (Session et al., 2016). We therefore determined the extent of knockdown of sybu.L and sybu.S homeologs.
Fig. 2.
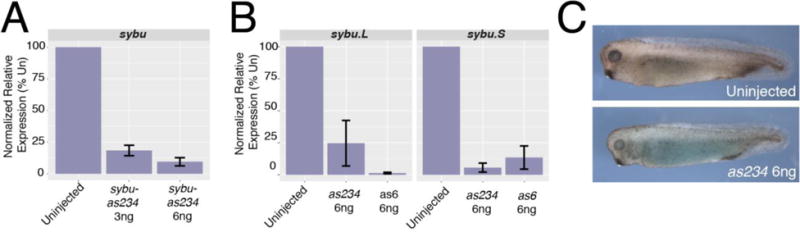
Antisense oligo-mediated depletion of sybu. (A) Real-time RT-PCR analysis of total sybu levels following injection of as234. (B) Differential depletion of sybu homeologs by as234 and as6 oligos. (C) Representative embryos obtained from host-transfer experiments using Uninjected control oocytes (upper panel) or sybu-depleted oocytes (lower panel). In (A–B), bars represent the average of two experiments; errors bars represent standard errors.
A comparison of candidate oligo sequences showed that our most effective oligo, as234, was 100% identical to sybu.S (the homeolog on chromosome (chr) 6S) but had one mismatch in the central region to sybu.L (chr6L). In contrast, the oligo identified by Colozza and De Robertis (2014; as6) was 100% identical to both L and S homeologs. Specific qRT-PCR primers and hydrolysis probes were used to detect the sybu.S and sybu.L homeologs in oligo-injected and control oocytes. Whereas both oligos were effective in reducing sybu.S to below ~ 12% of control levels, as234 was less effective at causing depletion of sybu.L, which remained at about 25% of control levels (Fig. 2B). However, overall sybu levels, as assessed by primers that amplify both homeologs, were nonetheless substantially depleted by oligo as234.
Because our pilot studies had suggested that embryos depleted of sybu using oligo as234 were not phenotypically ventralized, and because of sybu’s germ plasm localization, we took the opportunity to use this oligo to investigate whether Sybu might have an additional role in primordial germ cell (PGC) formation. This process cannot be effectively monitored in ventralized embryos owing to the involvement of dorsally derived structures. Embryos lacking maternal sybu were obtained by injecting cultured oocytes with oligo as234 and fertilizing these following host transfer into egg laying females. At doses of both 3.0 ng and 6.0 ng we recovered numerous fertilized eggs and these developed normally through gastrulation and formed normal swimming tadpoles (Fig. 2C). We assessed the extent of primordial germ cell development in sybu-depleted embryos by examining pgat expression in tailbud stage embryos, which identifies migrating PGCs at this stage (Hudson and Woodland, 1998). Control embryos derived from uninjected oocytes showed numerous pgat-expressing primordial germ cells in the endoderm, likely in the process of migrating to the future dorsal mesentery (Fig. 3A). In sybu-depleted embryos, these migrating PGCs were either absent or severely reduced in number (Fig. 3B). To demonstrate the specificity of this effect on PGCs, we reintroduced in vitro-transcribed sybu mRNA into depleted oocytes. PGC numbers were similar to control numbers in these rescued embryos, and the extent of migration also resembled the normal cases (Fig. 3C). These data suggested that in addition to possible functions in axis formation, in Xenopus Sybu likely has roles in the germ plasm to control either PGC specification and/or migration.
Fig. 3.
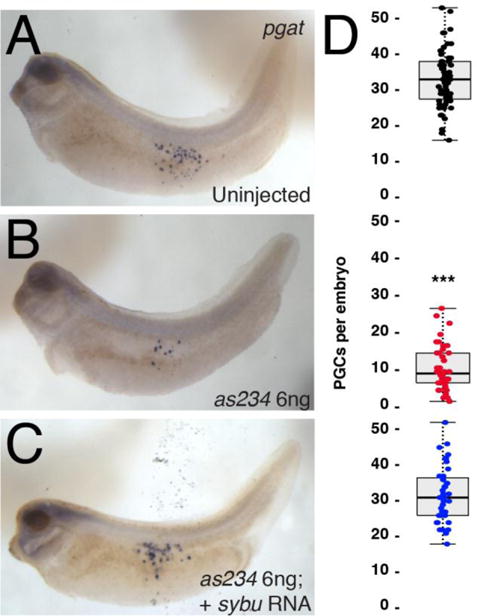
Reduction of PGCs in sybu-depleted embryos. (A–C) Whole-mount in situ hybridization for pgat mRNA. (A) control uninjected embryo, (B) sybu-depleted embryo, (C) depleted embryo rescued with injected sybu mRNA (5 pg). (D) Scatter plots of PGC numbers in experimental groups corresponding to the adjacent panel. Dots represent individual in which PGCs were counted (sum of three separate host-transfer experiments); dark line represents the median; shaded areas correspond to the limits of the first and third quartiles (25th/75th percentiles) and whiskers (dotted line) extends out to 1.5× the interquartile range. P < 0.001 by Kruskal-Wallis test; *** = P < 1.0e-11 by Dunn’s post hoc test w/2-sided Bonferroni adjustment.
Syntabulin is required for proper germ plasm aggregation and localization in the embryo
We next wished to more precisely determine at which stage germline development becomes disrupted in the absence of Sybu. Maternal sybu-depleted embryos were examined over a time course of early germ plasm aggregation and relocalization. Germ plasm organization was initially assessed by staining for pgat RNA. In early cleavage-stage embryos derived from control uninjected oocytes, pgat RNA accumulates in aggregated germ plasm along vegetal cleavage furrows. In sybu-depleted embryos however, we saw reduced and uneven aggregation of germ plasm islands. These aggregates were smaller and dispersed away from vegetal cleavage furrows, in contrast to the rather compact pattern of germ plasm observed in controls (Fig. 4A–C). This pattern of mislocalization was also evident at the morula stage (32-cell, Fig. 4A′–C′) and into the gastrula stages, where germ plasm remained scattered in smaller islands throughout vegetal cells (not shown). Depletion using either oligo as234 or as6, which target different regions of sybu mRNA, showed similar germ plasm aggregation defects during cleavage stages. And, as with the PGC migration phenotype, reintroduction of sybu transcripts (5 pg per embryo) could rescue the effects of both oligos and restore normal aggregation (Fig. 4D, D′), thus confirming the specificity of the effects of oligo injection.
Fig. 4.
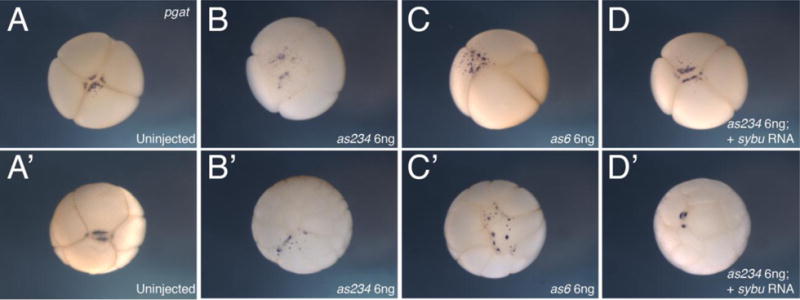
Abnormal germ plasm aggregation in sybu-depleted embryos. (A–D) Representative embryos at the 4-cell stage, probed for pgat to identify germ plasm, vegetal views. (A) uninjected control embryo, (B) sybu-depleted embryo, using as234, (C) sybu-depleted embryo using as6, (D) sybu-depleted embryo (as234) rescued by injection of sybu mRNA. (A′–D′) Representative embryos similarly analyzed at the 32-cell stage.
Relocalization of germ plasm from the cortex to the perinuclear region is an important hallmark of PGC specification. We therefore next sought to determine whether early germ plasm aggregation defects correlated with defects in perinuclear localization. Gastrula stage germ plasm is internal and not easily examined using in situ hybridization. We therefore briefly stained early embryos with the lipophilic dye DiOC6(3) to label the mitochondria-rich germ plasm (Savage and Danilchik, 1993; Venkatarama et al. 2010) and then dissected the vegetal mass from gastrula-stage embryos. This tissue was dissociated and cells examined by epifluorescence microscopy to identify germ plasm-containing cells. These presumptive PGCs could be readily identified and typically showed a bright cluster of germ plasm adjacent to the nucleus (Fig 5). DiOC6(3)-labelled cells were more difficult to find in sybu-depleted embryos, but in ten cases where the labelling was robust, the germ plasm was invariably located near the cortex or dispersed between the cortex and nucleus (Fig. 5B, 5B′). Overall, these data suggest that Sybu is necessary for germ plasm aggregation following fertilization, clustering a critical mass of germ plasm at the vegetal pole, and may also be required directly and/or indirectly for influencing the subsequent perinuclear accumulation of germ plasm for proper PGC specification.
Fig. 5.
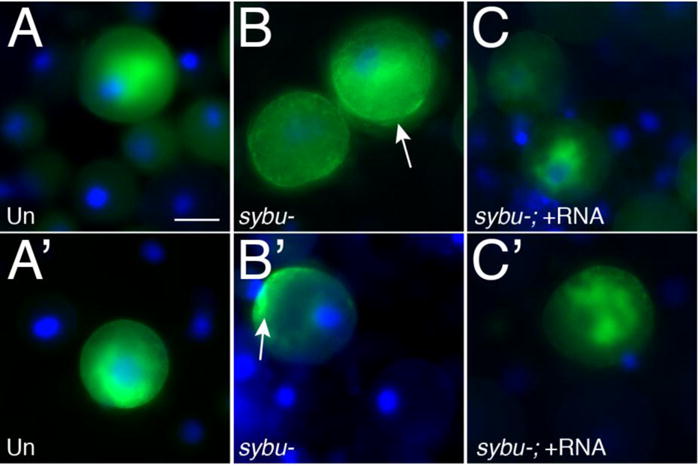
Failure of peri-nuclear germ plasm localization in sybu-depleted embryos. (A–C′) DiOC6(3) staining in isolated blastomeres (Green; DAPI in blue). Embryos were stained at the 4-cell stage and dissociated at the early gastrula stage. (A, A′) control uninjected isolated vegetal blastomeres, (B, B′) sybu-depleted blastomeres, (C, C′) blastomeres rescued by sybu-mRNA injection. arrows = cortical mitochondrial/germ plasm enrichment; scale bar = 30 μm.
Second SCW-driven germ plasm localization to vegetal pole cleavage furrows requires Sybu
Because our results indicated a defect in overall germ plasm aggregation in sybu-depleted embryos, we next sought to determine which aspects of this process might require Sybu function. Insofar as germ plasm aggregation has been investigated at the cellular and molecular level in Xenopus, aggregation requires microtubules (although not microfilaments) and the motor protein Kif4a, for generating and/or propagating SCWs that drive germ plasm clusters toward the vegetal cleavage furrows (Quaas and Wylie, 2002; Savage and Danilchik, 1993).
To further address this issue, we performed live imaging of control and sybu-depleted oocytes fertilized following host-transfer (Figs. 6 and 7; Supplementary Videos 1 and 2). Fertilizing eggs in small batches, we imaged eggs at low magnification using time-lapse epifluorescence microscopy (frame rate = 20 seconds), filming at least three eggs per sample per experiment over three experiments using eggs/oocytes from different donor females. Germ plasm was visualized by brief labeling with the lipophilic dye DiOC6(3) to preferentially stain mitochondria enriched in germ plasm (Ressom and Dixon, 1988; Savage and Danilchik, 1993; Quaas and Wylie, 2002). Movies were started at roughly eighty minutes after fertilization, to capture the end of cortical rotation (as an internal control for normal development), and then continued for ninety additional minutes (170 minutes post-fertilization total) to visualize the aggregation process.
Fig. 6.
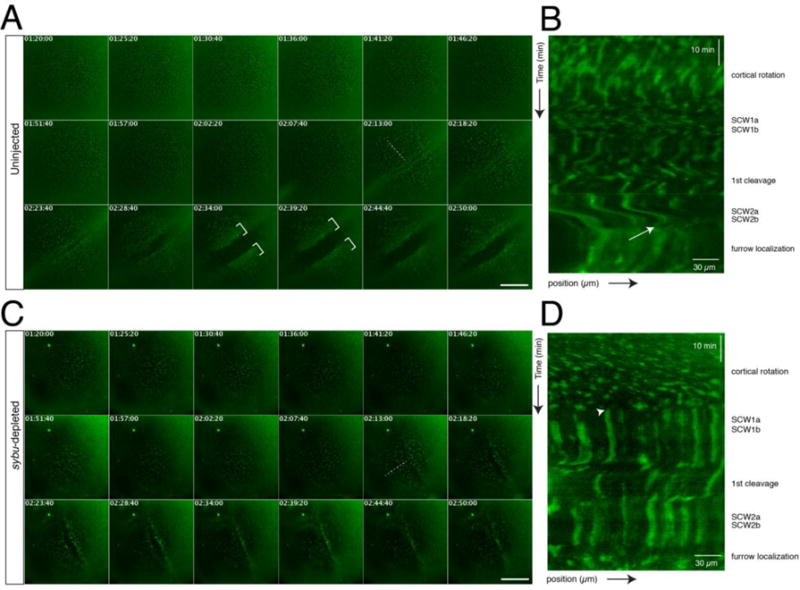
Time lapse imaging of germ plasm aggregation during early development in host-transfer fertilized eggs. (A) Representative time-lapse imaging of a zygote obtained by the host-transfer procedure, stained with DiOC6(3). Panels in (A) show selected frames at ~5-minute intervals from Supplementary Video 1 (170 minute time-lapse, frame rate = 20 seconds). In (A, C), elapsed time from in vitro fertilization is shown at the top of each frame (in hr:min:sec); dotted lines indicate the lines used for kymograph profiles; brackets indicate areas of clustered germ plasm; scale bars = 85 μm. (B) Representative kymograph analysis of an uninjected control embryo. The approximate timing of post-fertilization events is shown on the right. The white arrow indicates the beginning of aggregation into larger areas at the tail end of SCW2a/b. The position arrow indicates distance from the vegetal pole (bottom left corner). (C) Representative time-lapse imaging of a sybu-depleted zygote obtained by the host-transfer procedure, stained with DiOC6(3) (labels as in A). Panels show selected frames at ~5-minute intervals from Supplementary Video 2. (D) Representative kymograph analysis of a sybu-depleted embryo. Germ plasm is not disturbed by SCWs (arrowhead) and fails to form large continuous aggregates at the cleavage furrows.
Fig. 7.
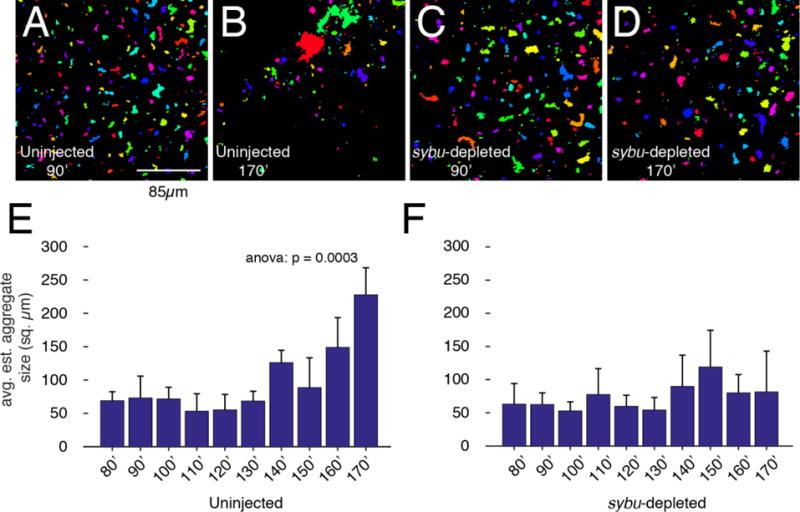
Quantification of germ plasm aggregation in early development. (A–D) Representative images from time-lapse videos (Videos 1, 2) from uninjected and sybu-depleted embryos, showing frames analyzed at ~90 minutes (A, C) and ~170 minutes (B, D) after fertilization. Vegetal views. Thresholded areas were randomly pseudocolored to verify that they could be distinguished and measured as separate clusters. (E–F) Graphs showing mean aggregate size at the indicated time points; error bars indicate standard errors. N=3 host-transferred eggs; >100 germ plasm aggregates (three experiments). The p-value from one-way ANOVA is indicated at the top. Scale bar in A–D = 85 μm.
In control eggs (n=10/10), DiOC6(3)-labelled germ plasm is seen translocating with the vegetal yolk mass during cortical rotation until the appearance of the first mitotic SCWs, at which point cortical rotation ceases. In agreement with previous studies, germ plasm did not substantially aggregate during cortical rotation (Savage and Danilchik, 1993). Aggregate size did not visibly increase following the passage of the first SCWs, suggesting little aggregation, whereas a greater amount of aggregation was seen in response to cytoplasmic perturbations created by first cleavage furrow (Fig. 6A, Supplementary Video 1). Subsequently, prior to the second cleavage, germ plasm clusters grew slightly larger following the passage of the SCW of the second mitosis (~15 minutes pre-furrow). At the end of this set of SCWs, or the final continuation of it, a subset of germ plasm clusters became visible at the egg surface and were then rapidly swept toward the vegetal pole, accumulating in a large patch along the furrow. A similar process was repeated after the formation second cleavage furrow, resulting in the typical four “island” pattern typical of germ plasm. The events in these movies were analyzed using kymographs, in which SCWs were evident as lateral waves (Fig. 6B). The collection along furrow at the end of the second SCW appears as an interruption in the wave pattern and as a hazy area in the kymograph, as the particles become grouped in larger areas (bottom, Fig. 6B).
In sybu-depleted embryos, DiOC6(3)-labelled germ plasm translocated normally during the period of cortical rotation (Fig. 6C, Supplementary Video 2), however there was no noticeable increase in aggregation following first cleavage. Further, the formation of dispersed particles and their collection along the vegetal cleavage furrows was much diminished or absent (n=9/10) and kymographs did not show evidence of perturbation by SCWs or of furrow aggregation (Fig. 6D). To validate the specificity of this aspect of the sybu depletion phenotype, we also made movies of rescued embryos (Supplementary Video 3) and found that the appearance of rapid translocation vegetally was restored (n=3/3).
To better quantify the effect of sybu depletion on germ plasm aggregates, we estimated DiOC6(3)-labelled aggregate size in frames at 10-minute intervals from three movies of control and sybu-depleted eggs. We consider these only estimated sizes owing to loss of information through imaging of a round egg and because of the thresholding involved in image processing. Frames were extracted from movies and exported using Fiji/ImageJ, followed by thresholding and measurement using a custom Matlab script. Examples of the processed images and the average germ plasm aggregate area in control and experimental eggs are shown in Fig. 7. As evident from the movies, these measurements show little change in aggregate size during cortical rotation and in response to the first SCWs (80–90 minutes), as well as during the period up to the first cleavage furrow formation (140′, Fig. 7E), at which time there is a slight but significant increase. Consistent with the lack of aggregation in the absence of sybu, the average germ plasm aggregate size remained unchanged during the period of analysis (Fig. 7F).
Taken together, these observations highlight several aspects of Sybu-dependent germ plasm aggregation: 1) as reported, little to no germ plasm aggregation happens during cortical rotation (Quaas and Wylie, 2002; Savage and Danilchik, 1993); 2) SCWs in general do not drive aggregation at the furrows – the 1st SCWs promote some local aggregation whereas the 2nd (and subsequent) SCWs promote large-scale collection of germ plasm at the cleavage furrows; 3) noticeable aggregation occurs after the passage of cleavage furrows, and 4) only a subset of clusters and/or germ plasm within a cluster become enriched at the furrow.
Sybu functionally interacts with multiple Kinesin-related proteins in germ plasm aggregation
In the context of both neuronal transport and early zebrafish development, Sybu has been suggested to serve as a linker for Kif5b/KinesinI (Su et al. 2004; Nojima et al., 2010). The extent that Sybu might interact with different kinesins to mediate germ plasm aggregation in the Xenopus egg is unknown. Kif4a has been implicated, likely acting through the generation and or maintenance of SCWs, although its protein is also enriched in germ plasm (Robb et al. 1996; Quaas and Wylie 2002). Prior maternal depletion experiments ruled out a role for Kif15/Xklp2 (Robb et al. 1996). Kif5b has not been explicitly tested in Xenopus germ plasm aggregation, although inhibitory antibody studies suggest it is dispensable for other microtubule-dependent events including cortical rotation and melanosome dispersal. Interestingly, we also noted from the literature that kif3b mRNA and protein is localized to the germ plasm in Xenopus oocytes (Betley et al. 2004), resembling sybu.
To begin to determine whether Sybu might act with these kinesins in germ plasm aggregation, we determined the extent of functional interaction between Sybu and kinesins Kif4a and Kif3b. We designed antisense oligos against kif3b and used a published oligo sequence against kif4a to deplete these mRNAs from oocytes (Fig. S2A). These were injected into oocytes and germ plasm aggregation was assessed by prick-activating these after progesterone-induced maturation (Fig. 8A). Pricking mimics fertilization and these oocytes will undergo normal cellular events like cortical rotation and germ plasm aggregation but not cleavage. For analyzing and comparing larger numbers of samples, this experimental design is often preferable to using host-transferred eggs. Consistent with our results presented above and with published reports (Robb et al., 1996; Quaas and Wylie 2002), we found that germ plasm aggregation (here labelled using Mitotracker) was specifically reduced in sybu- and kif4a-depleted eggs 3–4 hours after prick activation. (Fig. 8B–F). Germ plasm aggregation was also significantly reduced to a similar extent in kif3b-depleted eggs (Fig. 8F), with all eggs examined lacking large aggregates (p < 0.001; n=20). Furthermore, germ plasm aggregation was reduced in kif3b-depleted eggs fertilized by host transfer and analyzed by live imaging (Supplementary Video 3, Fig. S3, n=5/5). Although these embryos cleaved normally, all but a few were arrested in gastrulation and did not survive. This precluded an analysis of germ cell formation but suggested a role for maternally-supplied Kif3b in possibly establishing cell polarity during convergent extension gastrulation movements, as hypothesized by Shindo et al. (2008).
Fig. 8.
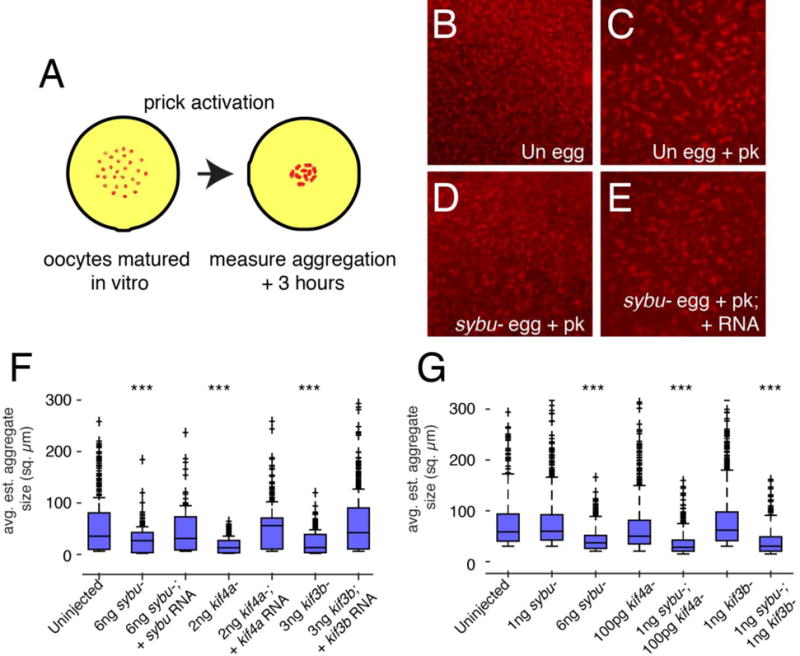
Functional interaction of Sybu with kinesins Kif3b and Kif4a in germ plasm aggregation. (A) Model of the experimental design. Oocytes are stained with MitoTracker and imaged following prick-activation. (B–E) Representative images control eggs before and after aggregation (B, Uninjected (Un) egg; C, Un egg + prick activation (pk)) and of sybu-depleted and rescued prick-activated eggs (D, sybu- egg +pk; E, sybu- egg +pk + RNA). (F–G) Average germ plasm aggregate size in depleted and rescued groups (F) and in different combinations of sub-effective doses of oligos (G). The dark bar in each box indicates the median size per group, lower and upper edges of the colored boxes indicate 25th and 75th percentiles respectively, whiskers represent maximum and minimum adjacent values. Outliers (>1.5× interquartile range) are indicated by crosses (+). N=20 oocytes, > 500 total islands. ***P<0.001 by Kruskal-Wallis test/Dunn’s post-hoc.
To provide insight into whether Sybu might be working with these kinesins to mediate germ plasm aggregation, we performed functional interaction studies using combinations of sub-effective oligo doses to partially deplete each component. Whereas the individual lower oligo-injected groups showed normal aggregation, the combinations of low-dose sybu oligos with either low-dose kif4a or kif3b oligos reduced germ plasm aggregation to a significant extent (Fig. 8G; p < 0.001; n=20). Taken together, these data demonstrate that Sybu likely mediates germ plasm aggregation by interacting with at least two kinesin motors, Kif4a and Kif3b.
Discussion
The role of cytoplasmic inheritance of a localized “germ plasm” during the specification of the germline has long-been appreciated but the mechanisms that assemble and localize the germ plasm (and the degree that these mechanisms are conserved) are little understood. Here we show a role for maternal syntabulin, which is localized to the germ plasm, in the aggregation of germ plasm during the cleavage stages of early Xenopus development and subsequent specification of PGCs. Interestingly, we noted that normal germ plasm aggregation likely involves localization towards cleavage furrows by surface contraction waves in the dividing egg. Syntabulin has been implicated as a protein linking kinesins to various cargo, including mitochondria, Syntaxin-containing vesicles and (in Xenopus and zebrafish) putative dorsal determinants (reviewed in Hirokawa et al. 2009; Houston, 2017). Our work shows that Sybu functionally interacts with a wider range of kinesins and has adopted an additional role in germ plasm aggregation in Xenopus.
We used antisense-based loss-of-function experiments to elucidate the role of maternal sybu in Xenopus. Prior studies have shown embryos derived from tokkaebi mutant female zebrafish are variably ventralized in a manner sensitive to genetic background (Nojima et al. 2010) and Xenopus embryos derived from oocytes injected with sybu antisense oligos are also ventralized to some extent (Colozza and De Robertis, 2014). Interestingly, we designed several antisense oligos that substantially depleted maternal sybu mRNA in Xenopus oocytes but did not generate ventralized embryos. Although extensive transcriptome and genome sequence data has been available for the diploid Xenopus tropicalis (Hellsten et al. 2010) for some time, this has only recently become accessible for Xenopus laevis (James-Zorn et al. 2015; Karpinka et al. 2015; Session et al. 2016). Upon retrospective inspection of our oligo sequences, we found our most effective oligo (as234) contained a one base mismatch with one of the homeologs (sybu.L) and depleted this homolog to a lesser extent. Therefore, it may either not deplete enough of the sybu.L homeolog mRNA or of overall sybu to elicit ventralization, whereas the role of Sybu in the germ plasm may be more sensitive to depletion. Regardless, embryos derived from oocytes injected with this oligo develop reproducible and rescuable defects in primordial germ cell formation and germ plasm aggregation.
It is unclear to what extent these homeologs have become subfunctionalized in Xenopus. The sybu.S-homeolog is expressed at twice the level of sybu.L (RNAseq data from Session et al. 2016 accessed on Xenbase; (James-Zorn et al. 2015; Karpinka et al. 2015; Session et al. 2016)). And, the rescue construct used by ourselves and Colozza and De Robertis (2014) encodes the S-homeolog. There is little amino acid difference between the two forms that might account for the different functions. It is thus unclear whether a suboptimal depletion of a minority species (the L-homeolog) would be responsible for the lack of ventralization in our hands. Similar to the zebrafish, there may be strain-specific requirements for Sybu in axis formation in different outbred populations Xenopus. It would be useful to repeat some of these experiments in several of the inbred Xenopus strains that have recently become available. Regardless of the role of Sybu in axis formation, our data show that Sybu also functions in germ plasm aggregation, resulting in loss of PGCs by the tailbud stage. We show this with both the as234 oligo and as6 oligo (Colozza and De Robertis, 2014), and the effects on aggregation can be rescued by injection of sybu.
We see several defects in PGC specification in embryos depleted of maternal sybu. First, germ plasm fails to aggregate into large “islands” at the vegetal pole, and during gastrulation, the germ plasm fails to become perinuclear. Therefore, one proximal cause of fewer PGCs in sybu-depleted embryos is likely to be this lack of perinuclear recruitment. Perinuclear localization has long been thought to be indicative of germ plasm “activity” (e.g., Blackler, 1958) but its mechanistic function is unclear. Our data do not distinguish between a direct role for maternally supplied Sybu in transporting germ plasm perinuclearly or whether this is a consequence of earlier aggregation defects or lack of a critical mass of germ plasm. In either case, presumptive PGCs receiving sybu-depleted germ plasm are likely not specified properly and do not initiate migration.
In examining the dynamics of germ plasm aggregation, we noted a correlation of cleavage furrow formation and the accumulation of germ plasm into large vegetal islands. Current models of germ plasm aggregation suggest this occurs through local Brownian aggregation, augmented by surface contraction waves (Quaas and Wylie, 2002). Our observations suggest several refinements to this model. First, we find that not all SCWs are involved in aggregation. We saw little aggregation in response to the first set of SCWs preceding first cleavage. These occur at the end of cortical rotation, during which we and others have found that no aggregation occurs. Also, in our hands local aggregation is enhanced by cleavage furrow formation, after which individual clusters appear larger. Importantly we show that end of the second set of SCWs, after first cleavage and prior to 2nd cleavage, germ plasm is robustly driven towards the vegetal cleavage furrow. Similar processes occur after the 2nd division, resulting in the typical “four-island” appearance of germ plasm.
These results are interesting because they suggest that aggregation occurs by a more dynamic process than previously thought. Many germ plasm clusters remain unaggregated after SCW passage, indicating that this is not simply a physical response to SCWs. This aspect could be indicative of an underlying cytoskeletal regulation, likely involving transport on microtubules by Kinesin-Sybu and other motor complexes. Organized microtubules are present near the cleavage furrows, termed the furrow microtubule array (FMA; Danilchik et al. 2003) and it is possible these microtubules mediate germ plasm transport.
Our work identifies several kinesins likely to mediate Sybu-mediated germ plasm transport. Using functional interaction in RNA depletion studies, this work suggests that germ plasm aggregation is mediated by Kif4a and Kif3b interacting with Sybu. Kif4a was previously shown to be required for germ plasm aggregation but its role remains unclear. Kif4a depleted embryos fail to undergo normal mitosis and thus do not generate SCWs, secondarily impacting germ plasm aggregation. Kif4a protein is also enriched in germ plasm and local aggregation is affected (Robb et al. 1996), suggesting a dual role. Because SCWs are normal in Sybu-depleted embryos, and Sybu is also enriched in the germ plasm, we infer that Sybu could mediate the local aggregation function of Kif4a. Kif4a has been implicated in vesicle transport as well as spindle regulation, although mitochondria have not been specifically examined (reviewed in Hirokawa et al. 2009). Human KIF4A has been implicated in synaptic dysregulation, which likely involves presynaptic mitochondrial localization (Willemsen et al. 2014), suggesting a possible role for Sybu-Kif4a complexes in this process.
Prior work also indicated localization of Kif3b to the germ plasm and implicated its function in RNA localization and/or germ plasm localization during oogenesis (Betley et al. 2004; Messitt et al. 2008). Our work shows that depletion of Kif3b generally mimics Sybu depletion with regard to germ plasm aggregation. Thus, Kif3b may be a key interacting partner for Sybu in germ plasm aggregation, mediating both local and FMA-mediated aggregation. Given the prominent role of Kif5b in multiple transport events in the egg and early embryo, it is likely that this motor is also involved in germ plasm aggregation at some level, although in general is little understood how the action of different kinesins and other motors is integrated and regulated for different transport substrates.
Sybu has been implicated in the kinesin-based anterograde transport of mitochondria in neurons (Su et al. 2004). Other adaptor proteins involved in anterograde or other aspects of mitochondrial transport include Milton/Oip106/Trak1, Rhot1/Miro, and RanBP2, primarily identified from studies in cultured neurons or in Drosophila (Frederick and Shaw 2007). A recent study in Xenopus using a dominant-inhibitory construct implicated Rhot1/Miro in germ plasm aggregation, displaying results strikingly similar to Sybu depletion (Tada et al. 2016). Thus, germ plasm aggregation is likely to entail complex interactions between multiple kinesins and adaptor proteins, with unique and overlapping functions, and dynamic interaction with an also dynamic cytoskeleton.
In general, the mechanisms controlling the localization of organelles and supra-organellar structures such as germ plasm are little understood. Our works lends some insight into this process, showing that localization of mitochondria by Sybu-Kinesin complexes is likely important for different aspects of mitochondrial dynamics in dividing cells and gastrulae. Interestingly, Sybu is also thought to mediate the transport of so-called “dorsal determinants” in Xenopus and zebrafish. Our studies and those in fish show there is some variability in this requirement, which could suggest that Sybu is not the only adaptor protein involved. Also, since Sybu inhibition is achieved by acute depletion in Xenopus oocytes and by maternal genetic mutation in zebrafish, this comparison may not be exactly commensurable. There could also be a further requirement for Sybu-mediated localization during oocyte development. Indeed, rescue of the tkk dorsal axis mutant phenotype requires expression of a sybu transgene during oogenesis (Nojima et al. 2010). Additionally, the nature of sybu expression during early development and the fertility of maternal-zygotic tkk mutant fish would seem to rule out a role for Sybu in germ plasm aggregation and PGC specification in fish. Indeed, sybu RNA and protein are no longer detectable when germ plasm is transported and re-assembled at distal cleavage furrows in the zebrafish zygote.
This idea fits with the proposed model for germ plasm evolution, which suggests that maternally-localized germ plasm evolved independently in different animal lineages (Extavour and Akam, 2003) and germ plasm aggregation proceeds quite differently in zebrafish and in Xenopus. It is also interesting to note that another maternal mutation implicated in zebrafish dorsal axis formation, hecate/grip2a, is also apparently dispensable for PGC development, whereas its Xenopus homolog is involved in germ plasm function and likely dispensable for axis formation (Kirilenko et al. 2008; Ge et al. 2014). It may be that the functions of Sybu and Grip2a related to the germ plasm were lost concomitant with evolution of meroblastic cleavage (i.e., loss of cleavage at the vegetal pole) and preferentially retained transport of axis signaling components, whereas the converse may have predominated in Xenopus.
Materials and Methods
Plasmids
The coding region of sybu (sybu.S) was amplified from a full-length cDNA obtained commercially (GenBank Accession: BC079762; GE/Dharmacon) and cloned into the pCR8/GW/TOPO vector (Invitrogen). Sequence verified clones were recombined into custom pCS2+ Gateway vectors for untagged and for C-terminal HA- and GFP-tagged versions (custom vector conversion kit; Invitrogen). sybu-pCS2+ and tagged derivatives were linearized with NotI and capped mRNA was synthesized using a SP6 mMessage mMachine kit (Ambion).
Antisense oligos and host transfer
Antisense oligodeoxynucleotides (oligos) are phosphorothioate-phosphodiester chimeric oligos obtained from Integrated DNA Technologies (Coralville, Iowa) (* represent phosphorothioate bonds): sybu234, 5′-A*A*G*GAGATTCCGAAG*A*C*G-3′ (new); sybu_as6, 5′-G*C*TACTGGCAGATGA*A*A*C-3′ (Colozza and de Robertis, 2014); kif3b, G*A*G*CTCTTAGACTTGGA*C*A*T (new); kif4a, A*T*G*CCCTCATCCTTCCC*C*A*T (Robb et al. 1996).
Stage VI oocytes were manually defolliculated and injected with the appropriate sybu antisense oligos at the vegetal pole. After culturing for 24 hours at 18°C in oocyte culture medium [OCM; 70% Leibovitz L-15, 0.045% polyvinyl alcohol, 1× penicillin-streptomycin, pH7.6–7.8] (modified from Heasman et al., 1991) oocytes were matured in 2 μM Progesterone. Rescue injections with 5 pg sybu mRNA were done right before oocyte maturation. Host transfer was carried out as described by Olson et al. (2012).
RT-PCR
Total RNA was isolated from flash frozen matured oocytes homogenized in cell lysis buffer (50 mM Tris pH7.5, 50 mM NaCl, 5 mM EDTA, 0.5% SDS, 250 mg/mL Proteinase K) incubated at 37°C for 1 hr. The mixture was further supplemented with 0.2 volumes 5M ammonium acetate and 20 μg glycogen, extracted with phenol/chloroform/isoamyl alcohol and precipitated with 100% isopropanol. RNA was resuspended in nuclease free water, and treated with DNase I (10U; Roche Applied Science) for 30 min at 37°C and precipitated with lithium chloride at −20°C. Resuspended RNAs were primed with random hexamer primers and cDNA was generated with M-MLV reverse transcriptase (Invitrogen) for 30min at 37°C.
Levels of gene expression were quantified using real time PCR with the LightCycler480 System (Roche Applied Sciences) using either SYBR Green I Master Mix and/or Universal Probe Library hydrolysis probes as previously described (Houston and Wylie, 2005; Schneider et al. 2011). All samples were normalized to ornithine decarboxylase (odc) and levels of expression were calculated against a standard curve of diluted uninjected oocyte cDNA. Individual primers, probes, and PCR conditions are available upon request.
Whole mount in-situ hybridization
Whole mount in-situ hybridization was performed as previously described (Sive et al., 2000; Kerr et al., 2008). The xpat/pgat template was linearized with EcoRI and transcribed with T7 polymerase (Promega) to synthesize the antisense probe with digoxygenin-11-UTPs (Roche). sybu/pCMV-SPORT6.1 was linearized with SmaI and transcribed with T7.
Germ plasm aggregation
For in vitro assays, oocytes were chemically defolliculated with collagenase and cultured overnight in OCM prior to injections of 6 ng sybu234 or sybu_as6 oligos at the vegetal pole. Oocytes were then incubated for 24 hrs and subsequently matured with 2 μM Progesterone for 20 hrs at 18°C. Germ plasm islands were stained in 100 nM MitoTracker™ (ThermoFisher) for 15 minutes at room temperature and washed several times over 30 minutes in OCM. Oocytes were prick-activated in the animal pole, mimicking fertilization, and germ plasm aggregation was monitored by epifluorescence over 3 hours. For in vivo assays, manually defolliculated oocytes were fertilized by the host-transfer procedure. Resulting embryos were dejellyed with 2% l-cysteine (Sigma) about 1 hour post fertilization and stained with DiOC6(3) (1:1000, ThermoFisher) for 10 minutes, with gentle rocking. Embryos were washed several times in 0.1X MMR before the first cell division and imaged, starting about 80 minutes post-fertilization.
Microscopy and time lapse imaging
Microscopy was performed at room temperature on an inverted, wide-field epifluorescence microscope (DMI4000B, Leica) using a 5× objective. Images were acquired using a monochrome digital camera (DFC3000G camera, Leica) and Leica AF6000 (64 bit) software. The pixel size was 1.07 × 1.07 × 0.200 μm and image size was 512 × 512 pixels. Time lapse images were collected at twenty-second intervals. Exposure times were approximately 0.30 seconds on the FITC channel. Samples were kept in either OCM or 0.1× MMR solution for imaging. Embryos were placed in custom aluminum chamber slides cut to the size of a standard microscope slide. Cover slips were attached to the bottom of the wells (1 cm diameter) with nail polish. Images were taken at or near the vegetal pole. Activated oocytes were placed into a mesh grid and imaged through the mesh in serial fashion.
Image processing and quantitation
Movies were initially processed in Fiji/ImageJ v1.51n (64-bit). Images were cropped to the relevant germ plasm regions, adjusted for threshold manually and exported for analysis. Montage construction and kymograph analysis were performed in ImageJ using the respective plugins, respectively. Germ plasm area measurements were made manually in Fiji/ImageJ or using a custom script in MATLAB_R2015a (available on request). Parameters were statistically analyzed using one-way ANOVA or Kruskal-Wallis tests with appropriate post-hoc tests using MATLAB. Images were prepared for publication in Fiji/ImageJ and Adobe Photoshop/Illustrator; only minor level and contrast adjustments were made and applied uniformly to the entire image. Other modifications included resizing and changes to stroke/fill weights and colors and the addition of annotation overlays.
Supplementary Material
Fig. S1 shows in situ hybridization of sybu during oogenesis and embryonic development (related to Fig. 1). Videos 1 and 2 show representative time-lapse imaging movies of germ plasm aggregation in control and sybu-depleted host-transferred embryos, respectively (related to Figs. 6–7). Video 3 is a movie of germ plasm aggregation in a sybu rescued embryo (related to Figs. 6–7). Video 4 is a movie of a kif3b-depleted host-transferred embryo (related to Fig. 8). Fig. S2 shows RT-PCR analysis of kif3b and kif4a mRNA levels in antisense oligo injected oocytes and kif3b-depleted gastrulae (related to Fig. 8). Fig. S3 shows a montage of frames from Video 4 and a kymograph analysis of mitochondrial movement from that sample (related to Fig. 8, Video 4).
Supplementary Video 1 (related toFigs. 6–7). Representative time-lapse video (20s frame rate, 170 minutes) of a control embryo fertilized by host-transfer, stained with DiOC6(3), vegetal view.
Supplementary Video 2 (related toFigs. 6–7). Representative time-lapse video (20s frame rate, 170 minutes) of a sybu-depleted embryo fertilized by host-transfer, stained with DiOC6(3), vegetal view.
Supplementary Video 3 (related toFigs. 6–7). Representative time-lapse video (20s frame rate, 170 minutes) of a sybu-depleted embryo, rescued with 5 pg sybu mRNA, fertilized by host-transfer, stained with DiOC6(3), vegetal view.
Supplementary Video 4 (related toFig. 8). Representative time-lapse video (20s frame rate, 170 minutes) of a kif3b-depleted embryo fertilized by host-transfer, stained with DiOC6(3), vegetal view.
Supplementary Fig. S1. In situ hybridization of sybu expression in (A) oocytes (stages I–IV upper row; stages V–VI lower row), (B) 4-cell embryos, and (C) gastrulae. Vegetal views.
Supplementary Fig. S2. Antisense oligo-mediated depletion of kif3b and kif4a. (A) Real-time RT-PCR analysis of total kif3b and kif4a levels following injection of corresponding oligos. (B–C) Images of control uninjected (B) and kif3b-depleted gastrulae (C), vegetal views.
Supplementary Fig. S3. Time lapse imaging of germ plasm aggregation during early development in kif3b-deplted, host-transfered eggs. (A) Representative time-lapse imaging stained with DiOC6(3). Panels in (A) show selected frames at ~5-minute intervals from Supplementary Video 4 (170 minute time-lapse, frame rate = 20 seconds). In (A), elapsed time from in vitro fertilization is shown at the top of each frame (in hr:min:sec); the dotted line indicates the line used for the kymograph profile; scale bar = 85 μm. (B) Representative kymograph analysis of kif3b-depleted embryo. The approximate timing of post-fertilization events is shown on the right. The position arrow indicates distance from the vegetal pole (bottom left corner).
Highlights.
Maternal syntabulin (sybu) RNA is depleted using antisense oligos.
Germ plasm aggregation and subsequent PGC specification are abnormal in sybu-depleted embryos.
Sybu mediates accumulation of vegetal accumulation of germ plasm in response to surface contraction waves.
Depletion of maternal kif3b also affects normal germ plasm aggregation.
Multiple coordinate roles for kinesins and adaptor proteins in controlling germ plasm localization are proposed.
Acknowledgments
The authors would like to thank members of the Houston lab for discussions and for critical reading of the manuscript. This work was supported by NIH R01 GM083999 to DWH. DO received support through a graduate fellowship from the Developmental Studies Hybridoma Bank (DSHB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguero T, Kassmer S, Alberio R, Johnson A, King ML. Vertebrate Development. Springer; 2017. Mechanisms of Vertebrate Germ Cell Determination; pp. 383–440. [DOI] [PubMed] [Google Scholar]
- Beams HW, Kessel RG. The problem of germ cell determinants. Int Rev Cytol. 1974;39:413–479. doi: 10.1016/s0074-7696(08)60944-4. [DOI] [PubMed] [Google Scholar]
- Betley JN, Heinrich B, Vernos I, Sardet C, Prodon F, Deshler JO. Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Current Biology. 2004;14:219–224. doi: 10.1016/j.cub.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Bowes JB, Snyder KA, Segerdell E, Jarabek CJ, Azam K, Zorn AM, Vize PD. Xenbase: gene expression and improved integration. Nucleic Acids Res. 2009;38:D607–D612. doi: 10.1093/nar/gkp953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Gerwin C, Sheng ZH. Syntabulin-mediated anterograde transport of mitochondria along neuronal processes. J Cell Biol. 2005;170:959–969. doi: 10.1083/jcb.200506042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Pan PY, Sheng ZH. Syntabulin-kinesin-1 family member 5B-mediated axonal transport contributes to activity-dependent presynaptic assembly. J Neurosci. 2007;27:7284–7296. doi: 10.1523/JNEUROSCI.0731-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AP, Kloc M, Larabell CA, LeGros M, Etkin LD. The maternally localized RNA fatvg is required for cortical rotation and germ cell formation. Mech Dev. 2007;124:350–363. doi: 10.1016/j.mod.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo S, Heinrich B, Betley JN, Chen Z, Deshler JO. Evidence for common machinery utilized by the early and late RNA localization pathways in Xenopus oocytes. Dev Biol. 2005;278:103–117. doi: 10.1016/j.ydbio.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Colozza G, De Robertis EM. Maternal syntabulin is required for dorsal axis formation and is a germ plasm component in Xenopus. Differentiation. 2014 doi: 10.1016/j.diff.2014.03.002. [DOI] [PubMed] [Google Scholar]
- Cuykendall TN, Houston DW. Vegetally localized Xenopus trim36 regulates cortical rotation and dorsal axis formation. Development. 2009;136:3057–3065. doi: 10.1242/dev.036855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czołowska R. The fine structure of the “germinal cytoplasm” in the egg ofXenopus laevis. Dev Genes Evol. 1972 doi: 10.1007/BF00580253. [DOI] [PubMed] [Google Scholar]
- Eddy E. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- Eno C, Pelegri F. Germ Cell Determinant Transmission, Segregation, and Function in the Zebrafish Embryo. InTech. 2016 doi: 10.5772/62207. [DOI] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Grotjahn D, Welch E, Lyman Gingerich J, Holguin C, Dimitrova E, Abrams EW, Gupta T, Marlow FL, Yabe T, Adler A, Mullins MC, Pelegri F. Hecate/Grip2a acts to reorganize the cytoskeleton in the symmetry-breaking event of embryonic axis induction. PLoS Genet. 2014;10:e1004422. doi: 10.1371/journal.pgen.1004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Holwill S, Wylie CC. Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. Methods Mol Biol. 1991;36:213–230. doi: 10.1016/s0091-679x(08)60279-4. [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, Blitz IL, Blumberg B, Dichmann DS, Dubchak I, Amaya E, Detter JC, Fletcher R, Gerhard DS, Goodstein D, Graves T, Grigoriev IV, Grimwood J, Kawashima T, Lindquist E, Lucas SM, Mead PE, Mitros T, Ogino H, Ohta Y, Poliakov AV, Pollet N, Robert J, Salamov A, Sater AK, Schmutz J, Terry A, Vize PD, Warren WC, Wells D, Wills A, Wilson RK, Zimmerman LB, Zorn AM, Grainger R, Grammer T, Khokha MK, Richardson PM, Rokhsar DS. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Houston DW, King ML. Germ plasm and molecular determinants of germ cell fate. Curr Top Dev Biol. 2000;50:155–181. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- Houston DW. Regulation of cell polarity and RNA localization in vertebrate oocytes. Int Rev Cell Mol Biol. 2013;306:127–185. doi: 10.1016/B978-0-12-407694-5.00004-3. [DOI] [PubMed] [Google Scholar]
- Hudson C, Woodland HR. Xpat, a gene expressed specifically in germ plasm and primordial germ cells of Xenopus laevis. Mech Dev. 1998;73:159–168. doi: 10.1016/s0925-4773(98)00047-1. [DOI] [PubMed] [Google Scholar]
- Hulstrand AM, Schneider PN, Houston DW. The use of antisense oligonucleotides in Xenopus oocytes. Methods. 2010;51:75–81. doi: 10.1016/j.ymeth.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingold AL, Cohn SA, Scholey JM. Inhibition of kinesin-driven microtubule motility by monoclonal antibodies to kinesin heavy chains. J Cell Biol. 1988;107:2657–2667. doi: 10.1083/jcb.107.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James-Zorn C, Ponferrada VG, Burns KA, Fortriede JD, Lotay VS, Liu Y, Brad Karpinka J, Karimi K, Zorn AM, Vize PD. Xenbase: Core features, data acquisition, and data processing. Genesis. 2015;53:486–497. doi: 10.1002/dvg.22873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneshiro K, Miyauchi M, Tanigawa Y, Ikenishi K, Komiya T. The mRNA coding for Xenopus glutamate receptor interacting protein 2 (XGRIP2) is maternally transcribed, transported through the late pathway and localized to the germ plasm. Biochem Biophys Res Commun. 2007;355:902–906. doi: 10.1016/j.bbrc.2007.02.059. [DOI] [PubMed] [Google Scholar]
- Karpinka JB, Fortriede JD, Burns KA, James-Zorn C, Ponferrada VG, Lee J, Karimi K, Zorn AM, Vize PD. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res. 2015;43:D756–D763. doi: 10.1093/nar/gku956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirilenko P, Weierud FK, Zorn AM, Woodland HR. The efficiency of Xenopus primordial germ cell migration depends on the germplasm mRNA encoding the PDZ domain protein Grip2. Differentiation. 2008;76:392–403. doi: 10.1111/j.1432-0436.2007.00229.x. [DOI] [PubMed] [Google Scholar]
- Kloc M, Dougherty MT, Bilinski S, Chan AP, Brey E, King ML, Patrick CW, Etkin LD. Three-dimensional ultrastructural analysis of RNA distribution within germinal granules of Xenopus. Dev Biol. 2002;241:79–93. doi: 10.1006/dbio.2001.0488. [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development. 1995;121:287–297. doi: 10.1242/dev.121.2.287. [DOI] [PubMed] [Google Scholar]
- Kloc M, Larabell C, Chan AP, Etkin LD. Contribution of METRO pathway localized molecules to the organization of the germ cell lineage. Mech Dev. 1998;75:81–93. doi: 10.1016/s0925-4773(98)00086-0. [DOI] [PubMed] [Google Scholar]
- Mei W, Jin Z, Lai F, Schwend T, Houston DW, King ML, Yang J. Maternal Dead-End1 is required for vegetal cortical microtubule assembly during Xenopus axis specification. Development. 2013;140:2334–2344. doi: 10.1242/dev.094748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messitt TJ, Gagnon JA, Kreiling JA, Pratt CA, Yoon YJ, Mowry KL. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell. 2008;15:426–436. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H, Rothhamel S, Shimizu T, Kim CH, Yonemura S, Marlow FL, Hibi M. Syntabulin, a motor protein linker, controls dorsal determination. Development. 2010;137:923–933. doi: 10.1242/dev.046425. [DOI] [PubMed] [Google Scholar]
- Olson DJ, Hulstrand AM, Houston DW. Maternal mRNA Knock-down Studies: Antisense Experiments Using the Host-Transfer Technique in Xenopus laevis and Xenopus tropicalis. Methods Mol Biol. 2012;917:167–182. doi: 10.1007/978-1-61779-992-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaas J, Wylie CC. Surface contraction waves (SCWs) in the Xenopus egg are required for the localization of the germ plasm and are dependent upon maternal stores of the kinesin-like protein Xklp1. Dev Biol. 2002;243:272–280. doi: 10.1006/dbio.2001.0564. [DOI] [PubMed] [Google Scholar]
- Ressom R, Dixon K. Relocation and reorganization of germ plasm in Xenopus embryos after fertilization. Development. 1988;103:507–518. doi: 10.1242/dev.103.3.507. [DOI] [PubMed] [Google Scholar]
- Savage RM, Danilchik MV. Dynamics of germ plasm localization and its inhibition by ultraviolet irradiation in early cleavage Xenopus embryos. Dev Biol. 1993;157:371–382. doi: 10.1006/dbio.1993.1142. [DOI] [PubMed] [Google Scholar]
- Robb DL, Heasman J, Raats J, Wylie CC. A kinesin-like protein is required for germ plasm aggregation in Xenopus. Cell. 1996;87:823–831. doi: 10.1016/s0092-8674(00)81990-x. [DOI] [PubMed] [Google Scholar]
- Schneider PN, Slusarski DC, Houston DW. Differential Role of Axin RGS Domain Function in Wnt Signaling during Anteroposterior Patterning and Maternal Axis Formation. PLoS ONE. 2012;7:e44096. doi: 10.1371/journal.pone.0044096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, van Heeringen SJ, Quigley I, Heinz S, Ogino H, Ochi H, Hellsten U, Lyons JB, Simakov O, Putnam N, Stites J, Kuroki Y, Tanaka T, Michiue T, Watanabe M, Bogdanovic O, Lister R, Georgiou G, Paranjpe SS, van Kruijsbergen I, Shu S, Carlson J, Kinoshita T, Ohta Y, Mawaribuchi S, Jenkins J, Grimwood J, Schmutz J, Mitros T, Mozaffari SV, Suzuki Y, Haramoto Y, Yamamoto TS, Takagi C, Heald R, Miller K, Haudenschild C, Kitzman J, Nakayama T, Izutsu Y, Robert J, Fortriede J, Burns K, Lotay V, Karimi K, Yasuoka Y, Dichmann DS, Flajnik MF, Houston DW, Shendure J, DuPasquier L, Vize PD, Zorn AM, Ito M, Marcotte EM, Wallingford JB, Ito Y, Asashima M, Ueno N, Matsuda Y, Veenstra GJC, Fujiyama A, Harland RM, Taira M, Rokhsar DS. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo A, Yamamoto TS, Ueno N. Coordination of cell polarity during Xenopus gastrulation. PLoS ONE. 2008;3:e1600. doi: 10.1371/journal.pone.0001600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Cai Q, Gerwin C, Smith CL, Sheng ZH. Syntabulin is a microtubule-associated protein implicated in syntaxin transport in neurons. Nat Cell Biol. 2004;6:941–953. doi: 10.1038/ncb1169. [DOI] [PubMed] [Google Scholar]
- Tarbashevich K, Koebernick K, Pieler T. XGRIP2.1 is encoded by a vegetally localizing, maternal mRNA and functions in germ cell development and anteroposterior PGC positioning in Xenopus laevis. Dev Biol. 2007;311:554–565. doi: 10.1016/j.ydbio.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Theusch EV, Brown KJ, Pelegri F. Separate pathways of RNA recruitment lead to the compartmentalization of the zebrafish germ plasm. Dev Biol. 2006;292:129–141. doi: 10.1016/j.ydbio.2005.12.045. [DOI] [PubMed] [Google Scholar]
- Venkatarama T, Lai F, Luo X, Zhou Y, Newman K, King ML. Repression of zygotic gene expression in the Xenopus germline. Development. 2010;137:651–660. doi: 10.1242/dev.038554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. RNA Granules in Germ Cells. Cold Spring Harbor Perspectives in Biology. 2011;3 doi: 10.1101/cshperspect.a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitington P, Dixon K. Quantitative studies of germ plasm and germ cells during early embryogenesis of Xenopus laevis. J Embryol Exp Morph. 1975;33:57–74. [PubMed] [Google Scholar]
- Willemsen MH, Ba W, Wissink-Lindhout WM, de Brouwer APM, Haas SA, Bienek M, Hu H, Vissers LELM, van Bokhoven H, Kalscheuer V, Nadif Kasri N, Kleefstra T. Involvement of the kinesin family members KIF4Aand KIF5Cin intellectual disability and synaptic function. J Med Genet. 2014;51:487–494. doi: 10.1136/jmedgenet-2013-102182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 shows in situ hybridization of sybu during oogenesis and embryonic development (related to Fig. 1). Videos 1 and 2 show representative time-lapse imaging movies of germ plasm aggregation in control and sybu-depleted host-transferred embryos, respectively (related to Figs. 6–7). Video 3 is a movie of germ plasm aggregation in a sybu rescued embryo (related to Figs. 6–7). Video 4 is a movie of a kif3b-depleted host-transferred embryo (related to Fig. 8). Fig. S2 shows RT-PCR analysis of kif3b and kif4a mRNA levels in antisense oligo injected oocytes and kif3b-depleted gastrulae (related to Fig. 8). Fig. S3 shows a montage of frames from Video 4 and a kymograph analysis of mitochondrial movement from that sample (related to Fig. 8, Video 4).
Supplementary Video 1 (related toFigs. 6–7). Representative time-lapse video (20s frame rate, 170 minutes) of a control embryo fertilized by host-transfer, stained with DiOC6(3), vegetal view.
Supplementary Video 2 (related toFigs. 6–7). Representative time-lapse video (20s frame rate, 170 minutes) of a sybu-depleted embryo fertilized by host-transfer, stained with DiOC6(3), vegetal view.
Supplementary Video 3 (related toFigs. 6–7). Representative time-lapse video (20s frame rate, 170 minutes) of a sybu-depleted embryo, rescued with 5 pg sybu mRNA, fertilized by host-transfer, stained with DiOC6(3), vegetal view.
Supplementary Video 4 (related toFig. 8). Representative time-lapse video (20s frame rate, 170 minutes) of a kif3b-depleted embryo fertilized by host-transfer, stained with DiOC6(3), vegetal view.
Supplementary Fig. S1. In situ hybridization of sybu expression in (A) oocytes (stages I–IV upper row; stages V–VI lower row), (B) 4-cell embryos, and (C) gastrulae. Vegetal views.
Supplementary Fig. S2. Antisense oligo-mediated depletion of kif3b and kif4a. (A) Real-time RT-PCR analysis of total kif3b and kif4a levels following injection of corresponding oligos. (B–C) Images of control uninjected (B) and kif3b-depleted gastrulae (C), vegetal views.
Supplementary Fig. S3. Time lapse imaging of germ plasm aggregation during early development in kif3b-deplted, host-transfered eggs. (A) Representative time-lapse imaging stained with DiOC6(3). Panels in (A) show selected frames at ~5-minute intervals from Supplementary Video 4 (170 minute time-lapse, frame rate = 20 seconds). In (A), elapsed time from in vitro fertilization is shown at the top of each frame (in hr:min:sec); the dotted line indicates the line used for the kymograph profile; scale bar = 85 μm. (B) Representative kymograph analysis of kif3b-depleted embryo. The approximate timing of post-fertilization events is shown on the right. The position arrow indicates distance from the vegetal pole (bottom left corner).


