Abstract
Tumor progression and metastasis are dependent on the intrinsic properties of tumor cells and the influence of microenvironment including the immune system. It would be important to identify target drug that can inhibit cancer cell and activate immune cells. Proteasome β subunits (PSMB) family, one component of the ubiquitin-proteasome system, has been demonstrated to play an important role in tumor cells and immune cells. Therefore, we used a bioinformatics approach to examine the potential role of PSMB family. Analysis of breast TCGA and METABRIC database revealed that high expression of PSMB5 was observed in breast cancer tissue and that high expression of PSMB5 predicted worse survival. In addition, high expression of PSMB5 was observed in M2 macrophages. Based on our bioinformatics analysis, we hypothesized that PSMB5 contained immunosuppressive and oncogenic characteristics. To study the effects of PSMB5 on the cancer cell and macrophage in vitro, we silenced PSMB5 expression with shRNA in THP-1 monocytes and MDA-MB-231 cells respectively. Knockdown of PSMB5 promoted human THP-1 monocyte differentiation into M1 macrophage. On the other hand, knockdown PSMB5 gene expression inhibited MDA-MB-231 cell growth and migration by colony formation assay and boyden chamber. Collectively, our data demonstrated that delivery of PSMB5 shRNA suppressed cell growth and activated defensive M1 macrophages in vitro. Furthermore, lentiviral delivery of PSMB5 shRNA significantly decreased tumor growth in a subcutaneous mouse model. In conclusion, our bioinformatics study and functional experiments revealed that PSMB5 served as novel cancer therapeutic targets. These results also demonstrated a novel translational approach to improve cancer immunotherapy.
Keywords: Breast cancer, proteasome, PSMB5, macrophage, bioinformatics
Introduction
According to a report of WHO, 2017, there is 8.8 million of deaths in 2015 caused by cancer in which top 5 cancer are breast, lung, liver, colorectal and stomach cancer. In the US, it is estimating of 252,710 malignant and 63,410 benign new cases of breast cancer (WHO, Fact Sheet, Feb 2017). As the growth of death rate due to breast cancer has increased steadily, studying of cancer biology, understanding of cancer mechanism, and developing therapeutic treatments and drugs became an important issue for breast cancer research.
Systemic therapy is used for patients who have multiple tumor metastases [1], a tumor with portal vein involvement, which limits the usage of locoregional therapies. Systemic therapies, including chemotherapy (doxorubicin, epirubicin, cisplatinum, etc.), hormonal compounds (anti-estrogens and anti-androgens) and immunotherapy (interferon). However, the negative side of traditional chemotherapeutic drugs is that they are toxic by nature and therefore influence the human body in adverse ways. For example, doxorubicin which is used as a potent chemotherapeutic agent has a wide range of toxicity such as cardiotoxicity at optimal therapeutic dosing [2]. A deeper understanding of cancer turned the focus of cancer research toward a more selective, targeted approach that would eventually produce anticancer therapies with fewer side effects and with improved efficacy. For instance, Bevacizumab (Avastin), a monoclonal anti-VEGF antibody [3], is another option for antiangiogenic therapy [4]. Bevacizumab showed a significant disease-stabilizing effect in HCC patients [5]. Nevertheless, Bevacizumab decreased white blood cells and attenuated immunity to diseases [6] and increased risk of bleeding as a potentially life-threatening complication limits the usage of bevacizumab [7]. In addition, possible side effects of some targeted therapy drugs are immune-attenuation, following an increased defenselessness to infections and decreased cancer immunosurveillance. On the other hand, STAT3 signaling in macrophages and neutrophils might negatively affect immune progress [8], and the STAT3 signaling induced tumor cell proliferation, survival, angiogenesis, and metastasis. Kortylewski’s group found that STAT3 antagonists can induce tumor cell apoptosis and modify the tumor microenvironment by reducing the immunosuppressive effects of STAT3 [9]. Therefore, it is important to design a target drug to boost the immune system and decrease the growth of tumor cells, functioning similarly to a STAT3 antagonist.
Macrophages present one of the most important cell types of the innate immune system for executing different functions including phagocytosis of the invading microorganisms, the foreign particles, or debris left behind after cell destruction. Moreover, macrophages were the sites for expression of different proteins or enzymes, the release of reactive oxygen species, chemokines, and pro-inflammatory or anti-inflammatory cytokines. In addition, macrophages are able to present antigen to T lymphocytes for activation of cellular immunity. In general, macrophages can be classified by M1 (classically activated macrophages) and M2 macrophages (alternatively activated macrophage). Classical immune activation of macrophages (M1) depends on the products of specifically activated T helper 1 (TH1)- type lymphocytes and natural killer (NK) cells- in particular, interferon-γ (IFN-γ). On the other hand, the alternative activation of macrophages (M2) depends on cytokines that are generally produced in TH2-type responses including interleukin-4 (IL-4), IL10 and IL-13 [10]. In addition to the differential dependence on cytokine, M1 and M2 macrophages have distinct biological functions in the immune system. M1 macrophages can activate TH1 immune response thus have stronger effects on viral or bacterial clearance and tumor-cell cytotoxicity (type I). M2 macrophages can induce TH2 response and the biological functions are prone to tissue repair thus somehow promote the growth of tumor cells. M2 macrophage was infiltrated to tumor tissue after chemotherapy, especially in breast cancer and glioblastoma [11-13]. Thus, target M2 macrophage could be a promising therapy for breast cancer patients. The predominance of STAT3 activation results in M2 macrophage polarization, associated with immune suppression and tumor progression. A similar M2 toward M1 switch has been reported for treating STAT3 inhibitor in breast cancer.
The ubiquitin-proteasome system (UPS) is an essential mechanism involved in the cellular processes such as degradation, antigen processing, cell cycle regulation, and signal transduction [14]. UPS functions in ubiquitin proteasome-dependent degradation of proteins and presentation of the pathogen peptide or oncoprotein peptide to T cell. The 26S proteasome is composed of two 19S regulatory particles (RP) and the barrel-shaped catalytic core particle (CP), termed the “20S proteasome”. These complexes are composed of four stacked rings which contained seven subunits either of the α (PSMA1-7) or the β type (PSMB1-7). Processing of protein antigens within inducible subunits of the proteasome is important in subsequent conjugation with the class I major histocompatibility complex (MHC) in antigen presenting cell (APC) such as dendritic cell (DCs) or macrophage [15]. A previous study reported that the mRNA expression of PSMB4 and PSMB7 were significantly increased in cancer tissue compared to normal tissues [16,17]. In addition, the PSMB8 is overexpressed in gastric cancer tissue [18] and PSMB2 was significantly associated with chronic myelogenous leukemia [19]. These studies suggest that PSMBs are involved in multiple human cancers, but a comprehensive analysis of these PSMB subunit genes, which might act as potential therapeutic targets or prognostic biomarkers is still absent.
Meanwhile, emerging evidence has indicated that these proteasome β subunits were implicated in regulating the biological progression of the immune system. For instance, matrix protein interferes with the immunoproteasome PSMB6 assembly and permits infected cells to escape detection and ejection by the immune system [20]. Zebrafish maintain much higher diversity in their antigen processing genes than other vertebrate species, especially MHC-linked antigen processing genes such as PSMB2 and PSMB3 [21]. Additionally, chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature (CANDLE) was caused by mutations in PSMB8 [22]. These evidences indicated that PSMBs subunits are involved in regulating immune-related disease.
High-throughput technology such as RNA-sequence and microarray for determining global changes in gene expression is an important developmental paradigm that facilitates advances in functional genomics and systems biology. We have successfully used meta-analysis to predict the biological functions of ACSL, THBS, and VGCCs family genes [23-25], and many of these predictions have been confirmed by other research groups [26]. Meanwhile, we also used data mining strategy to explore the potential cancer therapeutic targets via bioinformatics approach and defined their characteristics for cancer development [27]. Since no systematic study has been performed to link PSMB member genes with both cancer and tumor immune system, we aimed to use a bioinformatics approach to screen PSMB member genes for a protein having dual roles for oncogenic and immune suppressive characteristics in public high-throughput database. We combined public NCBI GEO database, Oncomine [28], CCLE [29] as well as kmplot database to screen candidate genes which may have immunosuppressive and oncogenic character and used a variety of experimental approach for validating the hypothesis.
Materials and methods
ONCOMINE analysis
The mRNA levels of distinct PSMB subunits in different types of cancers were determined through analysis of ONCOMINE database, which is a publicly accessible tumor microarray database to facilitate discovery from genome-wide expression analyses. In current study, students’ t-test was used to calculate a p-value for comparison between cancer specimens and normal control datasets. The fold change was defined as 1.5 and p-value was set up at 0.05.
CCLE analysis
In order to investigate on PSMB subunits expression in different cancer cell lines, we obtained normalized mRNA expression and DNA copy-number data of human cancer cell lines in the Cancer Cell Line from Encyclopedia. Of the 947 human cancer cell lines in the CCLE database, gene expression data on subunits member across a panel of 947 cancer cell lines were downloaded and analyzed from the Cancer Cell Line Encyclopedia.
The Kaplan-Meier plotter survival analysis
Prognostic values of featured PSMB subunits with highly expressed in breast cancer samples were assessed by displaying the overall survival by using the Kaplan-Meier plotter database. Kaplan-Meier survival curve, log-rank P value and HR with 95% confidence intervals were calculated and plotted with the default setting.
Cells and cell culture
The human monocytic THP-1 cell line was obtained from Dr. Chiou-Feng Lin (National Cheng Kung University Medical College, Taiwan). THP-1 cells are referred to as THP-1 monocytes; for PMA-differentiated adhesion macrophage-like THP-1 cells are referred to as PMA-differentiated human THP-1 macrophages or human THP-1; for PMA (Sigma-Aldrich, MO, USA), LPS (Sigma-Aldrich, MO, USA) and IFN-γ (PeproTech, NJ, USA) differentiated macrophage are referred to as M1-polarized macrophages [30]. Before stimulation, 4 × 105 cells/ml cells were seeded into 6-well cell culture dishes in the presence of PMA (320 μM). Cells were incubated for another 6 hr before stimulation. After 24 hr stimulation, cells were collected. The human cancer cell lines were maintained in DMEM (Dulbecco’s Modified Eagle Medium) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin under 5% CO2 at 37°C.
Xenograft tumor model
Eight weeks Female NOD/SCID mice were obtained from the Laboratory Animal Center at National Cheng Kung University and housed in barrier facilities on twelve hours light and dark cycle. All experimental procedures were approved by the Animal Welfare Committee of National Cheng Kung University. The NOD/SCID mice were subcutaneously injected with MDA-MB-231 (1 × 106 cells) into the flank region and then shPSMB5 and luciferase control lentivirus were delivered into transplant tumor once a week (n=5 for each group). At eight-week post-transplantation, animals were sacrificed and tumors were collected for further analysis. Tumor sizes were measured with calipers every week and the tumor volume was calculated using the following formula: tumor volume = L × W2/2, where L means tumor length and W is the tumor width.
shRNA and antibody
For shRNA-mediated signaling, shRNAs targeting PSMB5 and luciferase were purchased from the National RNAi Core Facility (Academia Sinica, Taiwan; http://rnai.genmed.sinica.edu.tw) according to the accession number TRCN0000003920 and target sequence 5’-CCCATCCTCCATCCTATTTAT-3’. A control construct (pLKO.1 containing a luciferase non-silencing shRNA) was also purchased from National RNAi Core Facility as an expression control. Anti-CD68 FITC-conjugated and anti-TLR4 PE-conjugated antibodies were purchased from eBio-science (San Diego, CA, USA). Anti-GAPDH (GTX100118), Anti-HA (GTX115044) and Anti-PSMB5 (GTX104687) were purchased from GeneTex (Irvine, CA, USA).
RNA isolation and RT-PCR
Total RNA was isolated from the cells by using Trizol reagent (MDBIO, ROC) according to the manufacturers’ instructions. The concentration and purity were determined by measuring the absorbance at 260 and 280 nm, respectively, with a Nanodrop (ThermoFisher, MA). Total RNA was reverse transcribed into cDNA. The cDNA was synthesized using 2 μg of total RNA, 50 ng of primer, 0.5 mM dNTP mix, 1X RT buffer (Promega, USA) and 200 U of MMLV (Moloney Murine Leukemia Virus Reverse Transcriptase) RTase (Promega, USA) in a total volume of 20 μl. The reaction was carried out at 42°C for 60 min and terminated by deactivation of the enzyme at 70°C for 10 min. Control reactions, lacking either reverse transcriptase or template, were included to assess carryover of genomic DNA and RNA contamination, respectively. The RT-PCR were performed as previous described [31] and primer listed below. PSMB5 forward 5’-CCATACCTGCTAGGCACCAT-3’; PSMB5 reverse, 5’-GCACCTCCTGAGTAGGCATC-3’. GAPDH forward 5’-ACAACTTTGGTATCGTGGAAGG-3’; GAPDH reverse, 5’-GCCATCACGCCACAGTTTC-3’. MCP-1 forward 5’-CTGCTCATAGCAGCCACCTT-3’; MCP-1 reverse, 5’-GCACTGAGATCTTCCTATTGGTG-3’. IL-1β forward 5’-GAGGCACAAGGCACAACAG-3’; IL-1β reverse, 5’-CCATGGCTGCTTCAGACAC-3’. STAT1 forward 5’-AGAGGTCGTCTCGAGGTCAA-3’; STAT1 reverse, 5’-TTCAGAGCTCGTTTGTGGTG. STAT2 forward 5’-GAGGCCTCAACTCAGACCAG-3’; STAT2 reverse, 5’-GATTCGGGGATAGAGGAAGC-3’. STAT3 forward 5’-CAGCAGCTTGACACACGGTA-3’; STAT3 reverse, 5’-AAACACCAAAGTGGCATGTGA-3’. STAT5 forward 5’-GCAGAGTCCGTGACAGAGG-3’; STAT5 reverse, 5’-CCACAGGTAGGGACAGAGTCT-3’. STAT6 forward 5’-GTGAAAGCCTGGTGGACATT-3’; STAT6 reverse, 5’-GTTCTTGAACAGGGCAGAGC-3’. JAK3 forward 5’-TTGCCATCAACAAGCTCAAG-3’; JAK3 reverse, 5’-GCTGCTTCCAGGAATGACTC-3’. TGFβ-1 forward 5’-ACTACTACGCCAAGGAGGTCAC-3’; TGFβ-1 reverse, 5’-TGCTTGAACTTGTCATAGATTTCG-3’.
Western blot
Cells were lysed in modified RIPA buffer supplemented with protease inhibitors. Total cell lysates were separated using SDS-PAGE, and the proteins were transferred onto PVDF membranes (Millipore, Bedford, MA) using a Hoefer Semiphor Semi-Dry transfer unit (Amersham Pharmacia, San Francisco, CA). The membranes were incubated with the indicated primary antibody and followed by a horseradish peroxidase-conjugated antibody. The blots were developed using ECL western blotting detection reagents (Millipore, Bedford, MA) and detected using a BioSpectrum AC imaging system (UVP, CA).
Colony formation assay
PSMB5 and luciferase knockdown stable clone were harvested using trypsin/EDTA and the isolated cells were seeded in 6-well plate at a density of 2 × 104 cells per well. These cells were cultured for 10 days in the growth medium. To evaluate the number of high proliferative potential colony forming cells, the number of colonies more than 0.5 mm in diameter was also determined, and then the cells were fixing with Methanol and staining with Giemsa solution. The cells were calculated for each well by dividing the number of colonies by the number of colonies on the control plate.
Boyden chamber assay
Modified boyden chambers with polycarbonate membrane inserts (pore size 8 μm; Neuro Probes, Inc., Gaithersburg, MD) were used for cell migration assays. PSMB5 and luciferase knockdown stable clone were suspended in serum-free medium and placed in the upper chamber; 30 ul DMEM containing 10% FBS was then placed in the lower chamber. After 8 hr in culture, cells were fixed in methanol for 10 min and then stained with 10% Giemsa solution (Merck) for 1 hour. The number of cells on each membrane was counted under a microscope at a magnification of 20x (Olympus BX41, Japan), each sample was assayed in triplicate.
Statistical analysis
All statistical analyses by using GraphPad Prism (GraphPad Software, San Diego, CA, USA). This analysis was performed using unpaired t-test, one-way ANOVA for the analysis of the differences between experimental groups, respectively.
Results
High expression of PSMB subunits in breast cancer database
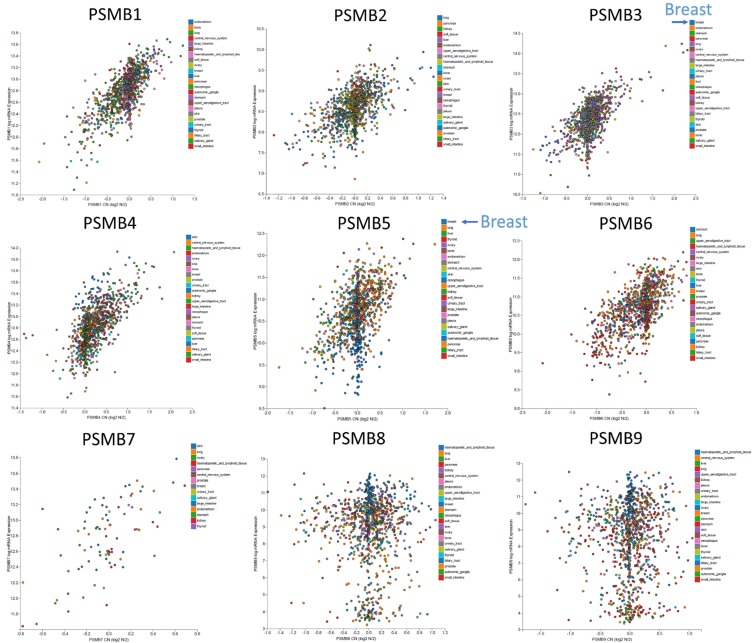
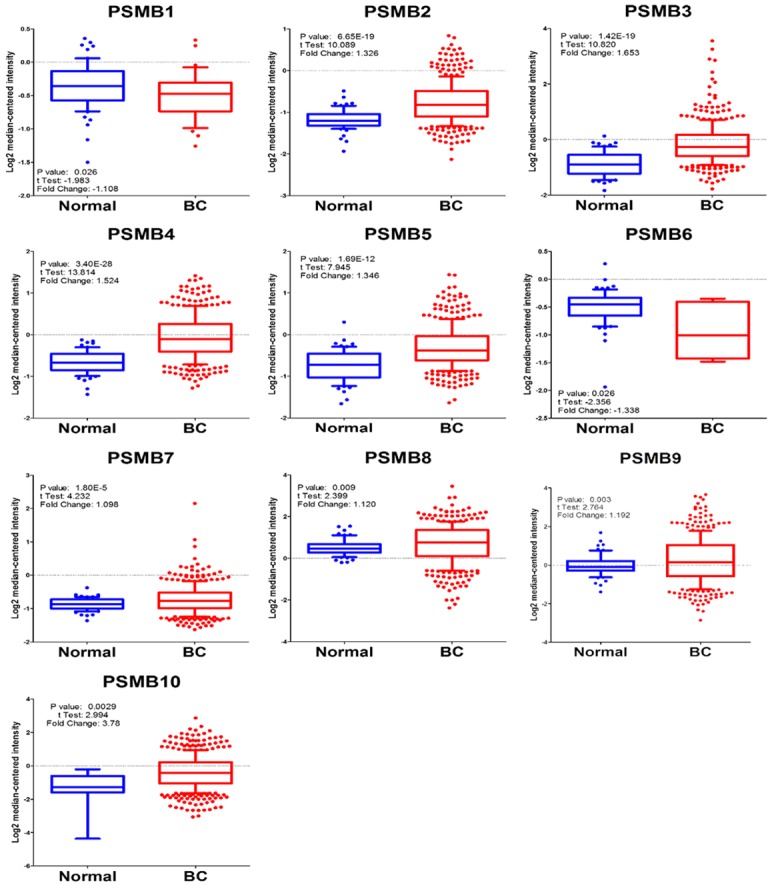
Ten PSMB subunits have been identified in human cancers, including hematological malignancies and solid tumors. In order to identify the PSMB subunits expression in breast cancer, a web-based ONCOMINE microarray database was employed (Supplementary Figure 1). We used TCGA (The Cancer Genome Atlas) database to evaluate PSMB subunits mRNA expression in breast cancer tissues and compare with normal tissues. Interesting, almost all the PSMB subunits had high expression in breast cancer tissue compared to normal group, except PSMB1 and PSMB6 (Figure 1). Notably, PSMB5 transcripts were 1.346 fold elevated in breast cancer samples as compared with normal tissues in a dataset with 593 samples that derived from TCGA database (Figure 1). We found that PSMB5 has high expression in triple negative breast cancer patients in comparison with normal tissue from TCGA database (Supplementary Figure 2A). Next, we studied the genomic alterations at the PSMB5 locus by using the cbioportal database, a resource designed to analyze the somatic mutation information. Interestingly, PSMB5 had high mutation rate in prostate cancer (NEPC, Neuroendocrine Prostate Cancer) or pancreas cancer but not for breast cancer patients. Strikingly, there was an extremely low frequency of alterations disrupting PSMB5 function in breast TCGA (n=825) or METABRIC (n=2509) database. Based on these information we suggested that PSMB5 mutations are tissue-specific. Overall, there are only a few PSMB5 mutations in breast cancer patients from both TCGA (n=825) and METABRIC (n=2509) database (Supplementary Figure 2B). Alterations in PSMB5 locus are unlikely to influence the breast cancer cell phenotype (Supplementary Figure 2C) based on the lack of large-scale genomic aberration and the non-synonymous mutation frequency of 0.1% (from breast TCGA database, n=817). The low frequency of genomic PSMB5 disruption led us to investigate whether increase of PSMB5 mRNA are favored during carcinogenesis. To address this question, we analyzed the transcription profile of the PSMB5 expression in public high-throughput database. In another dataset from Zhao’s study [32], PSMB5 was 2.121 fold elevated in breast cancer samples as compared with normal tissues (p=0.001). The dataset from Ma et al [33] also revealed consistent results, PSMB5 was 1.795 fold elevated in breast cancer samples as compared with normal tissues (p=5.23E-4). Similar results were obtained from METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) database [34]. PSMB1-10 mRNA expression except PSMB6 was significantly higher in breast cancer than normal samples across a wide variety of different cancer types. The P value from all PSMB subunits were from 7.12E-125 to 2.94E-25 in breast cancer samples as compared with normal tissues in a dataset with over 1700 clinical breast cancer patients (Figure 4A, 4B).
Figure 1.
Analysis of PSMB family subunits using breast cancer TCGA database (n=593). Comparison of the PSMB family subunits expression levels between the tumor and corresponding normal tissues obtained from 593 breast cancer patients. The breast TCGA dataset was obtained from Oncomine software which embedded in the TCGA database (https://tcga-data.nci.nih.gov/tcga). Box plots derived from gene expression data in breast cancer TCGA database demonstrating the differential expression of specific PSMB subunits in normal (left) and breast cancer tissues (right, BC), P value < 0.05 considered significant.
Figure 4.
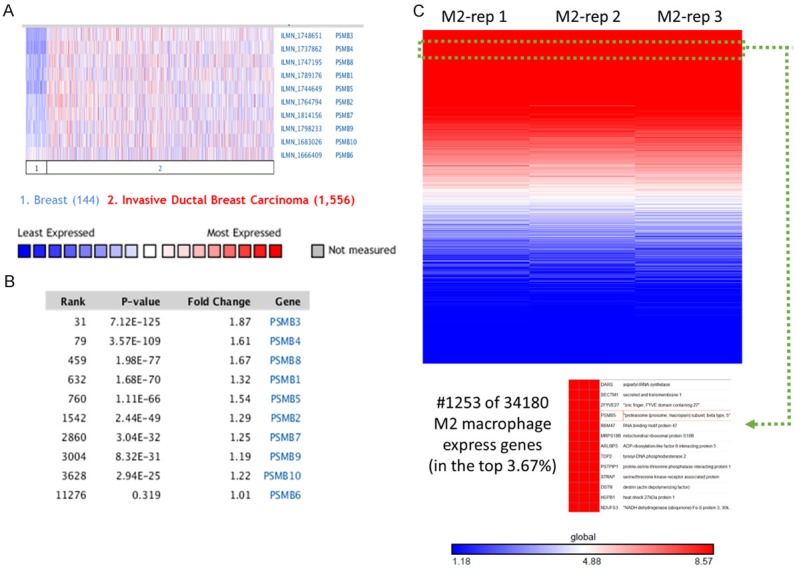
Identification of PSMB subunits mRNA expression in a variety of public database. A, B. Comparison of selected genes in METABRIC breast cancer database (n=1700). Over-expression of PSMB subunits in invasive ductal breast carcinoma vs. normal breast tissues (log2 median-centered intensity). C. Analysis of published microarray dataset (GSE34180) indicated that PSMB5 as highly expressed in M2 macrophage model. The heatmap shows genes rank ordered from highest to lowest for raw expression values across all M2 macrophage samples. The inset (bottom) highlights expression for PSMB5 ranked number 1253 out of over 34180 genes in the whole microarray dataset, which also means that PSMB5 had high expression in the top 3.67% gene ranking from these M2 macrophage samples.
We next aimed to establish the relationship of PSMB subunits mRNA expression and overall survival of breast cancer patients. We evaluated the clinical relevance of PSMB1-10 subunits in breast cancer patients by using an online kmplot software [35]. Clinical data revealed that higher expression levels of PSMB1-7 were significantly correlated with a poor survival rate, whereas high expression of PSMB8 and 9 were correlated with good survival rate (2898 patients, p < 0.05, Figure 2). In addition, the p-value for PSMB10 of prognosis curve was 0.14 which was not significant in these clinical breast cancer database (data not shown). Next, we used CCLE analysis [29] to confirm the bioinformatics data from TCGA, METABRIC, and kmplot database. Interestingly, CCLE analysis was consistent with these bioinformatics analysis, demonstrating that PSMB3 and PSMB5 were distinctively up-regulated in breast cancer cell lines, whereas other PSMB members were present at a low transcription level or absent in breast cancer cells (Figure 3).
Figure 2.
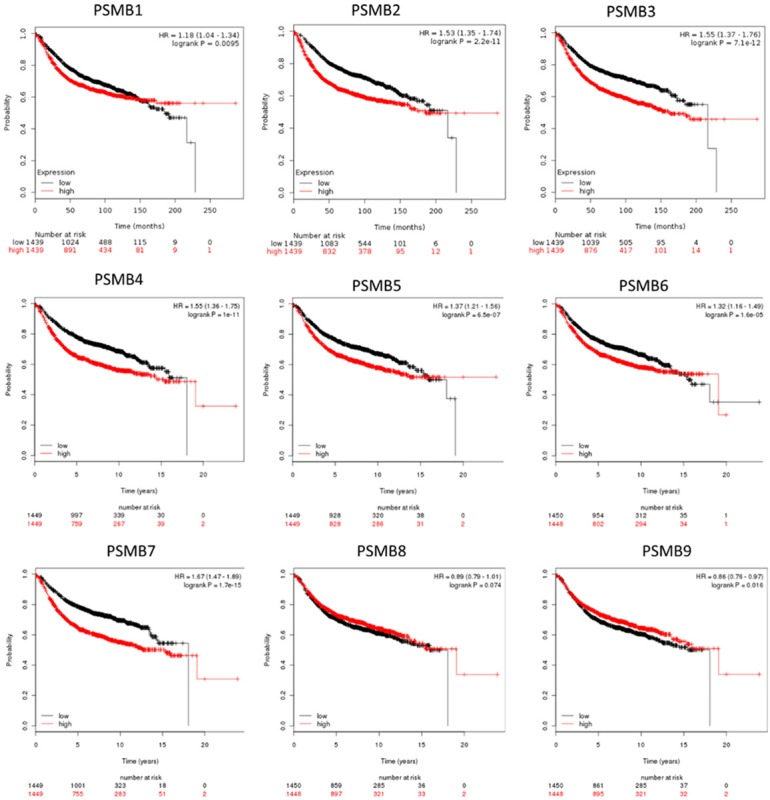
Relationship of PSMB1-9 expression and relapse-free survival of breast cancer patients (n=2898). Kaplan-Meier graphs show the relapse-free survival prognosis of breast cancer patients censored at twenty years, based on high or low PSMB family subunit’s mRNA expression. We used the median of expression as the cutoff, therefore the patients are divided into two groups based on the overexpression and under-expression for PSMB family subunit’s mRNA expression. Meanwhile, patients with expression above the median are shown in red line, whereas the patients with expression below the median in black line. High expression of PSMB1-7 was associated with poor survival, whereas high expression PSMB8 and 9 expressions were associated with better survival rate (total patient number=2898, P value < 0.05 consider significantly).
Figure 3.
PSMB3 and PSMB5 were highly expressed in breast cancer cell lines compared to other cancer types. Identification of genomic PSMB family subunits copy number variation (CNV) and correlation with gene expression in Cancer Cell Line Encyclopedia. DNA copy number was measured using Affymetrix high-density single nucleotide polymorphism arrays (Affymetrix SNP 6.0), and the mRNA expression levels were obtained for each cell line using Affymetrix U133 plus 2.0 arrays. Plots showing PSMB subunits mRNA expression z-scores vs. copy number value from 967 cell lines that were obtained through the CCLE. Each dot represents one cell line, and color coded by different tumor type or subtype.
Finally, we want to explore whether PSMB5 play an important role in the immune systems, we further analyzed published microarray dataset (GSE71253) and identified high expression of PSMB5 in M2 macrophage model [36]. The heatmap was made with GENE-E package in R environment as we previously described [23]. This data demonstrated that PSMB5 ranked number 1253 out of 34180 genes in the whole microarray dataset. It indicates that expression of PSMB5 is in the top 3.67% gene ranking from M2 macrophage samples (Figure 4C).
Collectively, our bioinformatics data on breast cancer and macrophage supported that PSMB5 in PSMB subunits may be a suitable candidate for our study on genes having mutual roles in tumor cells and tumor immune microenvironment. Furthermore, some of the proteasomes were immunoproteasome and play a critical role in the generation of MHC class I antigens. For example, PSMB1 (LMP2) knockout mice have a diminished or altered presentation of certain MHC class I antigens [42]. However, PSMB5 is a non-immunoproteasome and is suitable to be a drug target for immune activation without alteration of antigen presentation.
PMA, LPS and IFN-γ treatment promoted THP-1 cells differentiation into M1 macrophages
Macrophages are most abundant mononuclear phagocytes that reside within almost all tissues. They are responsible for host defense and regulation of inflammatory response. In order to mimic the human macrophages, we chose human THP-1 cell line as an experimental model. THP-1 cells can be differentiated into macrophage-like cells with treatment of PMA (320 nM) [37] for 6 hours followed by addition of LPS (100 ng/ml) and IFN-γ (20 ng/ml) for 24 hours [30]. To study M1 macrophage surface marker expression in M1-polarized THP-1 macrophages, the cells were analyzed by flow cytometry using CD68 and TLR4 as markers of macrophage differentiation and M1 macrophages, respectively. The M1-polarized THP-1 macrophages exhibited significant expression of macrophage differentiation marker (CD68) and M1 macrophage surface markers TLR4 (CD284) (Figure 5A, 5B) [38]. These data suggested that PMA, LPS and IFN-γ stimulation would promote THP-1 differentiation into M1 macrophage phenotype.
Figure 5.
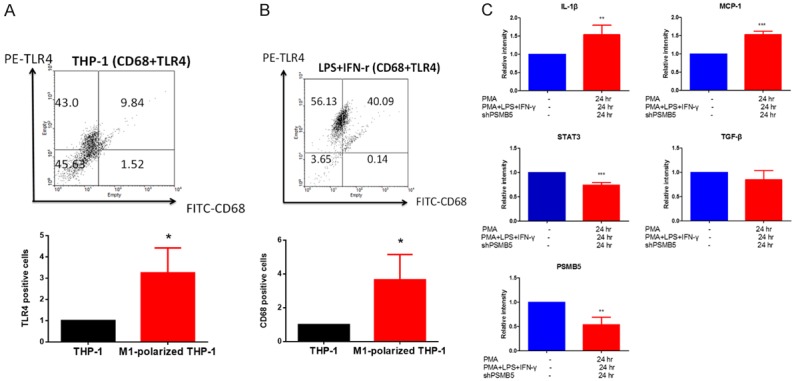
PMA, LPS and IFN-γ treatment promoted THP-1 cells differentiation to M1 macrophages. A, B. The effect of PMA, LPS and IFN-γ treatment on the number of M1 macrophages. THP-1 monocytes were treated with PMA (320 nM) for 6 hours and then cultured with PMA plus LPS (100 ng/ml) and IFN-γ (20 ng/ml) after 24 hours. TLR4 (M1 macrophage marker)-positive and CD68 (macrophage differentiated marker) positive fraction were analyzed using flow cytometry in THP-1 monocytes and M1-polarized THP-1 macrophages. Values are the average of assays performed in triplicate. Error bars represent SD (n=3), P value < 0.05 consider significantly. C. M1 Macrophage markers is expressed in PMA-treated THP-1 macrophage. MCP-1, IL-1β (a marker for M1 macrophage) and TGF-β (a marker for M2 macrophage) mRNA were measured in THP-1 cells and M1-polarized THP-1 macrophages. THP-1 monocytes were treated with PMA (320 nM) for 6 hours and then cultured with PMA plus LPS (100 ng/ml) and IFN-γ (20 ng/ml) after 24 hours. The mRNA levels were measured by RT-PCR and data were normalized according to GAPDH mRNA level and presented as a value relative to that for undifferentiated THP-1 monocytes. Values are the average of assays performed in triplicate. Error bars represent SD (n=3). *p < 0.05.
PSMB5 was downregulated in the M1-polarized THP-1 macrophage
To further confirm the phenotype of M1-polarized THP-1 macrophages, we evaluated the changes in the gene expression profiles of THP-1 and M1-polarized THP-1 macrophages. Both markers of the M1 macrophage, MCP-1, and IL-1β [39], were more abundantly expressed in M1-polarized THP-1 macrophages compared to undifferentiated THP-1 cells. In contrast, the expression of the M2 marker (TGF-β) [40] in M1-polarized THP-1 macrophages was not altered after PMA, LPS and IFN-γ treatment (Figure 5C).
Next, we used RT-PCR to analyze PSMB5 expression in M1-polarized THP-1 macrophages and undifferentiated THP-1 cells. These data demonstrated that PSMB5 was down-regulated in differentiated M1-polarized THP-1 macrophages (Figure 5C).
PSMB5 knockdown promoted differentiation of human THP-1 monocyte into M1 macrophage and decreased M1 macrophage markers including MCP-1 and IL-1β
To determine the knockdown efficiently of shRNA from RNAiCore on exogenous PSMB5, the HA-PSMB5 plasmid was co-transfected with PSMB5 shRNA plasmid in breast MDA-MB231 cell line. A significant reduction in the exogenous protein expression of HA-PSMB5 was observed upon transient co-transfection with a shPSMB5 plasmid (Figure 6A).
Figure 6.
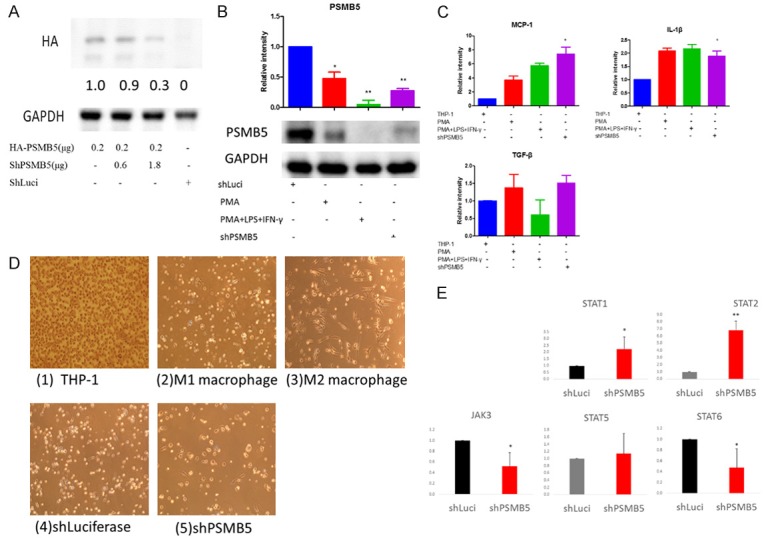
PSMB5 knockdown promoted differentiation of human THP-1 monocyte into M1 macrophage and decreased the transcription of M1 macrophage markers. A. Western blotting analysis of PSMB5 in MDA-MB-231 cells that were stably transfected with HA-PSMB5 and PSMB5 shRNA plasmids. Equal amounts (30 µg) of protein from whole-tissue lysates were analyzed for PSMB5 and GAPDH expression by Western blotting analysis. B. PSMB5 protein expression in PMA or PMA plus LPS (100 ng/ml) and IFN-γ (20 ng/ml) treated THP-1 cells or shPSMB5 knockdown cells were analyzed by western blot, Error bars represent SD (n=3). *p < 0.05. C. THP-1 cells were stimulated with PMA, LPS, IFN-γ or knockdown with shPSMB5 for 24 hr. Total RNA was prepared and analyzed by RT-PCR for M1 macrophage marker (MCP-1 and IL-1β) as well as M2 marker TGF-b. D. Phase-contrast 2D images of (1) Human THP-1 monocytes (2) THP-1 monocytes in the presence of PMA (320 nM), LPS (100 ng/ml) and IFN-γ (20 ng/ml) for differentiating into M1 macrophage. (3) THP-1 monocytes in the presence of IL-4 and IL-13 for differentiating into M2 macrophage. (4) Luciferase knockdown by pLKO-shLuc. (5) PSMB5 knockdown by shRNA in THP-1 cells. E. The involvement of PSMB5 in macrophage polarization signaling pathway such in PSMB knockdown macrophage treat with LPS for 24 hours and the analysis with RT-PCR.
Then we determined whether PSMB5 shRNA was able to silence “endogenous” PSMB5 gene expression. THP-1 cells exhibited a dramatic decrease in endogenous PSMB5 after transfection with shPSMB5 (Figure 6B). As a positive control, THP-1 monocytes were treated with PMA (320 nM) for 6 hours and then cultured with PMA plus LPS (100 ng/ml) and IFN-γ (20 ng/ml) after 24 hours. The protein level of PSMB5 was down-regulated in M1-polarized THP-1 macrophages (Figure 6B), which was consistent with our previous data (Figure 5C).
In order to study whether PSMB5 knockdown resulted in the differentiation of THP-1 cells into M1 macrophages, we examined cell morphology and the M1 macrophage surface marker mRNA expression. MCP-1 and IL-1β were M1 macrophage marker and were significantly elevated after shPSMB5 transfection. In contrast, the TGF-β (M2 macrophage marker) was not altered (Figure 6C).
The 2D morphology data demonstrated that shPSMB5 promoted THP-1 to form large, round cells which were similar to differentiated M1 macrophages (Figure 6D). In contrast, IL-4 and IL-13 would differentiate THP-1 cells into M2 macrophages, which displayed predominantly long, spindle-shaped cells and only a few round cells. These data suggested that knockdown of PSMB5 with shRNA promoted THP-1 differentiation into M1 macrophage.
Next, we wanted to explore the involvement of PSMB5 in macrophage polarization signaling pathway [41,42]. PSMB5-knockdown macrophage was treated with LPS for 24 hours and STAT1, 2, 5 6 and JAK3 were analyzed with RT-PCR. The increase of STAT1 and STAT2 were observed in PSMB5 knockdown macrophage, whereas JAK3 and STAT6 were decreased. These data suggested that downregulation of PSMB5 would promote M1 macrophage polarization (Figure 6E).
Downregulation of PSMB5 inhibits growth, migration, and tumor progression of MDA-MB-231 cell
Since tumorigenesis consists of multiple processes including proliferation, migration, and anchorage-independent growth, we investigated whether modulation of PSMB5 expression affected these oncogenic processes. First, we investigated the anti-proliferation effect of PSMB5 shRNA in human breast MDA-MB-231 cancer cell. Cells were seeded and grown for ten days, the colonies were detected by methyl blue staining, and the number of colonies was counted for statistical analysis. Decreased cell growth was observed in the PSMB5 shRNA stable transfectants of MDA-MB-231 cells. (Figure 7A, 7C). Next, we compared the migration ability of MDA-MB231 cells and PSMB5 transfectants with Boyden chamber migration assay. Downregulation of PSMB5 inhibited migration ability (Figure 7B, 7D). These results indicated that PSMB5 regulated cellular migration as well as proliferation in cancer progression and development. Finally, in order to determine whether shRNA PSMB5 inhibited tumor growth in vivo, NOD-SCID mice were subcutaneously implanted with MDA-MB231 cells and then treated with shPSMB5 lentivirus once a week. It’s very interesting that we found tumor volume were different in the beginning of the shPSMB5 treatment group, however, not caused a significantly therapeutic effect in the following weeks. Therefore, we supposed that that lentiviral delivery of shPSMB5 significantly decrease early-stage tumor size but not for the late stage tumors (Figure 7E).
Figure 7.
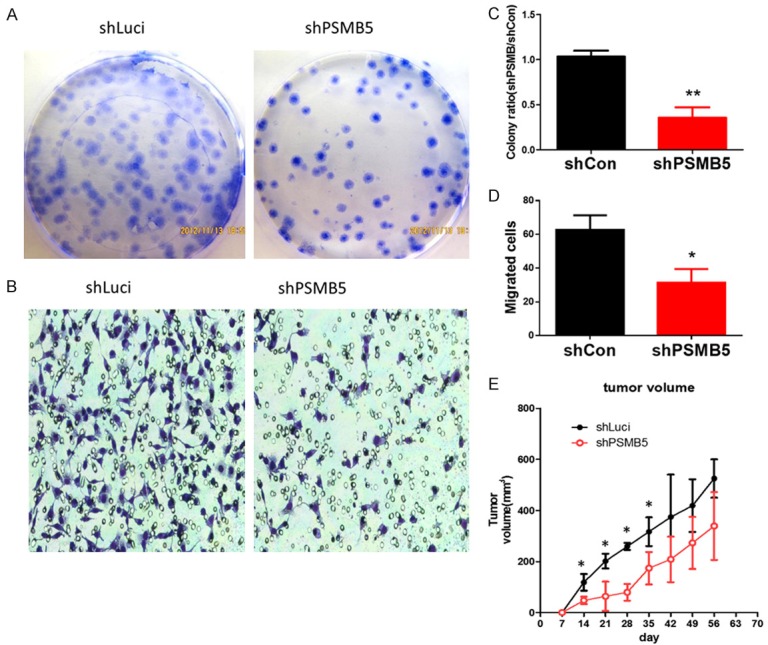
PSMB5 knockdown by shRNA significantly attenuated MDA-MB231 breast cancer proliferation and migration in vitro and in vivo. (A) The proliferation rates of the MDA-MB231 luciferase control cell lines and stable shPSMB5 knockdown MDA-MB231 cell line were determined by colony formation assay (B) the static data of colony formation assay. Values are the average of assays performed in triplicate. Error bars represent SD (n=3). *p < 0.05. (C) Motility of shPSMB5 knockdown as well as luciferase control MDA-MB231 cells were evaluated by Boyden chamber migration assays. (D) Quantification of random motility to PSMB5 knockdown and luciferase control MDA-MB231 cells with boyden chamber assay Error bars represent SD (n=3). *p < 0.05. (E) 1 × 106 MDA-MB231 cells were subcutaneous injected into NOD-SCID mice and the mice were treated with shPSMB5 or luciferase shRNA lentivirus once a week, tumor volume was calculated using the following formula: tumor volume (millimeters cubed) = L × W2/2, where L is the length, and W is the width (n=5 for each group, P=0.016).
Discussion
In this report, we demonstrate that PSMB5 plays an oncogenic role as well as an immune suppressor in cancer progression. Bioinformatics analysis reveals that overexpression of PSMB5 predicts a poor prognosis in breast cancer patients and PSMB5 is highly expressed in M2 type macrophage. Targeting PSMB5 using shRNA can attenuate tumor-cell proliferation, inhibit cell migration and modify the tumor microenvironment by alleviating the immunosuppressive effects of PSMB5. Meanwhile, blocking PSMB5 in immune cells can generate potent anti-tumor immunity by increasing the number of M1 macrophages and enhance their activity. Therefore, PSMB5 is a promising target for enhancing cancer immunotherapy.
Some of these PSMB subunits has been reported to be associated with cancer progression, as well as survival status [18,43,44]; however, little is known about the expression and function of these PSMB subunits in breast cancer development. Moreover, our multi-database-based meta-analysis may provide a more precise effect for the prediction of potential cancer therapeutic target. Our approach to bioinformatics analysis utilized the integration and validation of multiple high-throughput databases, therefore, the most potential PSMB family subunit could be identified for further investigation. Identifying novel targets of PSMB family subunits and classifying different subtypes of cancers on the basis of either microarray or high-throughput sequence data may promote the accuracy for development of new cancer therapy drugs. Besides, this study provides a comprehensive overview on the mediator subunits expressed in clinical breast cancer tissue. Ultimately, by performing a transcriptome analysis of the Oncomine database, CCLE, kmplot and NCBI GEO database, we identified distinct PSMB5 subunits, which show tumor-specific profiles and may influence the tumor and immune systems (Supplementary Figure 1).
The tumor microenvironment consists of diverse components including blood vessels, immune cells, fibroblasts, inflammatory cells, lymphocytes, cytokine, chemokine and the extracellular matrix. In the tumor microenvironment, tumor-associated macrophages (TAMs) play a crucial role in cancer progression and metastasis. M1/M2 macrophage conversion can switch the macrophage from anti-tumor to pro-tumor phenotype. Meanwhile, M2-like macrophage produces immunosuppressive molecules such as HLA-G, IL-10, and TGF-β to suppress the immune system. Many studies have demonstrated a relationship between macrophages and immunosuppression [45,46]. A recent promising Phase II clinical trials indicated that a systemically non-toxic M2-to-M1 macrophage stimulating agent named RRx-001 has underscored the essential role of the immunotherapy [47]. Thus, developing the novel drug or inhibitor to target M2-like macrophage became important issues for cancer research.
The 26S proteasome is composed of two 19S regulatory particles (RP) and the barrel-shaped catalytic core particle (CP), termed the “20S proteasome”. These complexes are composed of four stacked rings each composed of seven subunits either of the α (PSMA1-7) or the β type (PSMB1-7), which share α and β sandwich fold. A previous study reported that the mRNA expression of PSMB subunits was significantly increased in cancer tissue compared to normal tissues [16,17,43]. Overexpression of PSMB4 was associated with ovarian cancer [44] and myeloma growth [44,48]. Meanwhile, the matrix protein interferes with the immunoproteasome PSMB6 assembly and permits infected cells to escape detection and ejection by the immune system [20]. This evidence indicated that PSMBs subunits are clearly involved in regulating immune-related disease. However, a detailed understanding of how the PSMB subunits act as a driver of carcinogenesis as well as immunosuppress requires further studies for a more detailed understanding of the complexity of this proteasome complex.
The proteasome inhibitor bortezomib is a FDA-approved drug which has been widely used to treat multiple myeloma patients, however, the application of bortezomib in breast cancer patients still largely unknown. In other respects, most myeloma patients show primary or secondary resistance to bortezomib, thereby limiting its clinical efficacy [49-51]. In addition, some articles also demonstrated that overexpression of the PSMB5 gene contributes to bortezomib resistance in T-lymphoblastic lymphoma [52,53]. Coincidentally, the other report suggested that combined CSF1R inhibitor as well as a PSMB5 inhibitor-bortezomib displayed an additive therapeutic efficacy against myeloma-associated macrophages (MAMs) [54]. However, surprisingly, Beyar-Katz et al recently found that bortezomib-treated mice could promote multiple myeloma cell migration and proliferation by stimulating a host inflammatory response via IL-16 signaling pathway [55]. Since the most studies demonstrated that bortezomib could attenuate cancer growth, it’s very important to understand why bortezomib could also promote myeloma cell become aggressiveness? In addition to IL-16 interfered response, the second plausible explanation for these opposed data could be that bortezomib interacts with PSMB subunits 1, 2, and 5 and some documented evidence of polymorphisms in these subunits such as PSMB6, 7, and 8 [56-59]. In other words, bortezomib was not specific target PSMB5 but also target for other PSMB subunits. Therefore, instead of using not-specific proteasome inhibitor, in the present study we try to use specific shRNA to target PSMB5 and explore the potential cancer immunotherapeutic mechanism for breast cancer.
In the current study, we extended the research field to breast cancers based on large databases, with the purpose of determining the expression pattern of PSMB subunits in breast cancer and normal tissues. Next, we explore the correlations between PSMB subunits with characteristic molecular markers, as well as their corresponding prognostic values in breast cancer. Collectively, we found that PSMB5 is overexpressed in breast cancer patient in TCGA human breast cancer database, CCLE, and kmplot database. In the experimental approach, PSMB5 were down-regulated in M1-polarized THP-1 macrophages, which conforms to the general definition of immunosuppressive genes. Knockdown of PSMB5 with shRNA promotes THP-1 differentiation into the M1 macrophage. Downregulation of PSMB5 expression by shRNA attenuates cell growth in vitro, meanwhile, down-regulation of PSMB5 also leads to a lower migration ability. Finally, lentiviral delivery of PSMB5 shRNA inhibits tumor growth in vivo. In the future, it would be worthwhile to investigate the migration and growth related mechanism in the PSMB5 knockdown stable clone.
In conclusion, PSMB5 not only play an immunosuppressive function in M1-polarized-THP-1 macrophages, but also exert tumorigenic activities by modulating cancer cell growth and migration. Most of all, using shRNA to knockdown PSMB5 gene expression would decrease tumor-cell growth, migration and modify the tumor microenvironment by active M1 macrophage to increase the efficacy of cancer immunotherapy (Figure 8). Further investigations are still required, but our new approach to discovering anti-cancer and upregulate-immune system inhibitor may shed the new insight of the treatment with cancer.
Figure 8.
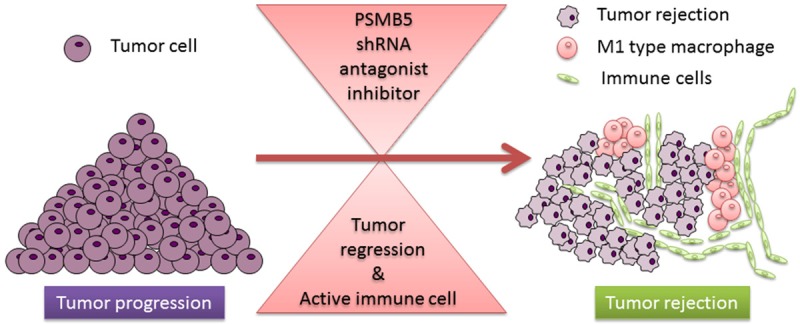
Crosstalk between cancer and immune cells: the role of PSMB5 in the tumor microenvironment. Targeting PSMB5 using shRNA can attenuate tumor-cell proliferation, inhibit cell migration and modify the tumor microenvironment by alleviating the immunosuppressive effects of PSMB5. Blocking PSMB5 in immune cells can generate potent anti-tumor immunity by decreasing the number of M2 macrophages and enhance M1 macrophage activity. Thereupon, PSMB5 has emerged as a promising target for advancing cancer immunotherapy.
Acknowledgements
Computational analyses and data mining were performed using the system provided by the Bioinformatics Core at the National Cheng Kung University (Tainan, Taiwan), supported by the MOST in Taiwan. The study was supported by the Ministry of Science and Technology (MOST) for the grants MOST103-2325-B006-012 (to MDL) and 104-2917-I-006-002 (to CYW).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Nowak AK, Chow PK, Findlay M. Systemic therapy for advanced hepatocellular carcinoma: a review. Eur J Cancer. 2004;40:1474–1484. doi: 10.1016/j.ejca.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 2.Gabizon A, Shmeeda H, Barenholz Y. Pharmacokinetics of pegylated liposomal Doxorubicin: review of animal and human studies. Clin Pharmacokinet. 2003;42:419–436. doi: 10.2165/00003088-200342050-00002. [DOI] [PubMed] [Google Scholar]
- 3.Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, Morin PJ, Buetow KH, Strausberg RL, De Souza SJ, Riggins GJ. An anatomy of normal and malignant gene expression. Proc Natl Acad Sci U S A. 2002;99:11287–11292. doi: 10.1073/pnas.152324199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoeben A, Landuyt B, Highley MS, Wildiers H, Van Oosterom AT, De Bruijn EA. Vascular endothelial growth factor and angiogenesis. Pharmacol Rev. 2004;56:549–580. doi: 10.1124/pr.56.4.3. [DOI] [PubMed] [Google Scholar]
- 5.Siegel AB, Cohen EI, Ocean A, Lehrer D, Goldenberg A, Knox JJ, Chen H, Clark-Garvey S, Weinberg A, Mandeli J, Christos P, Mazumdar M, Popa E, Brown RS Jr, Rafii S, Schwartz JD. Phase II trial evaluating the clinical and biologic effects of bevacizumab in unresectable hepatocellular carcinoma. J. Clin. Oncol. 2008;26:2992–2998. doi: 10.1200/JCO.2007.15.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samaras P, Bauer S, Stenner-Liewen F, Steiner R, Zweifel M, Renner C, Knuth A. Treatment of POEMS syndrome with bevacizumab. Haematologica. 2007;92:1438–1439. doi: 10.3324/haematol.11315. [DOI] [PubMed] [Google Scholar]
- 7.Mailliez A, Baldini C, Van JT, Servent V, Mallet Y, Bonneterre J. Nasal septum perforation: a side effect of bevacizumab chemotherapy in breast cancer patients. Br J Cancer. 2010;103:772–775. doi: 10.1038/sj.bjc.6605828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, Akira S. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 9.Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mule J, Kerr WG, Jove R, Pardoll D, Yu H. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 10.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Shree T, Olson OC, Elie BT, Kester JC, Garfall AL, Simpson K, Bell-McGuinn KM, Zabor EC, Brogi E, Joyce JA. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011;25:2465–2479. doi: 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stafford JH, Hirai T, Deng L, Chernikova SB, Urata K, West BL, Brown JM. Colony stimulating factor 1 receptor inhibition delays recurrence of glioblastoma after radiation by altering myeloid cell recruitment and polarization. Neuro Oncol. 2016;18:797–806. doi: 10.1093/neuonc/nov272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kioi M, Vogel H, Schultz G, Hoffman RM, Harsh GR, Brown JM. Inhibition of vasculogenesis, but not angiogenesis, prevents the recurrence of glioblastoma after irradiation in mice. J Clin Invest. 2010;120:694–705. doi: 10.1172/JCI40283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Ponz-Sarvise M, Chang SS, Chang WC, Chen CH, Hsu JL, Hung MC. Proteasome inhibition enhances the killing effect of BikDD gene therapy. Am J Transl Res. 2015;7:319–327. [PMC free article] [PubMed] [Google Scholar]
- 15.Kloetzel PM. Antigen processing by the proteasome. Nat Rev Mol Cell Biol. 2001;2:179–187. doi: 10.1038/35056572. [DOI] [PubMed] [Google Scholar]
- 16.Munkacsy G, Abdul-Ghani R, Mihaly Z, Tegze B, Tchernitsa O, Surowiak P, Schafer R, Gyorffy B. PSMB7 is associated with anthracycline resistance and is a prognostic biomarker in breast cancer. Br J Cancer. 2010;102:361–368. doi: 10.1038/sj.bjc.6605478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu R, Lu SM, Deng Y, Yang SY, He S, Cai J, Qiang FL, Chen C, Zhang WW, Zhao SY, Qian L, Mao GX, Wang YY. PSMB4 expression associates with epithelial ovarian cancer growth and poor prognosis. Arch Gynecol Obstet. 2016;293:1297–1307. doi: 10.1007/s00404-015-3904-x. [DOI] [PubMed] [Google Scholar]
- 18.Kwon CH, Park HJ, Choi YR, Kim A, Kim HW, Choi JH, Hwang CS, Lee SJ, Choi CI, Jeon TY, Kim DH, Kim GH, Park DY. PSMB8 and PBK as potential gastric cancer subtype-specific biomarkers associated with prognosis. Oncotarget. 2016;7:21454–21468. doi: 10.18632/oncotarget.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruzzoni-Giovanelli H, Gonzalez JR, Sigaux F, Villoutreix BO, Cayuela JM, Guilhot J, Preudhomme C, Guilhot F, Poyet JL, Rousselot P. Genetic polymorphisms associated with increased risk of developing chronic myelogenous leukemia. Oncotarget. 2015;6:36269–36277. doi: 10.18632/oncotarget.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beilstein F, Obiang L, Raux H, Gaudin Y. Characterization of the interaction between the matrix protein of vesicular stomatitis virus and the immunoproteasome subunit LMP2. J Viro. 2015;89:11019–11029. doi: 10.1128/JVI.01753-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McConnell SC, Hernandez KM, Wcisel DJ, Kettleborough RN, Stemple DL, Yoder JA, Andrade J, de Jong JL. Alternative haplotypes of antigen processing genes in zebrafish diverged early in vertebrate evolution. Proc Natl Acad Sci U S A. 2016;113:E5014–E5023. doi: 10.1073/pnas.1607602113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunimoto K, Kimura A, Uede K, Okuda M, Aoyagi N, Furukawa F, Kanazawa N. A new infant case of Nakajo-Nishimura syndrome with a genetic mutation in the immunoproteasome subunit: an overlapping entity with JMP and CANDLE syndrome related to PSMB8 mutations. Dermatology. 2013;227:26–30. doi: 10.1159/000351323. [DOI] [PubMed] [Google Scholar]
- 23.Chen WC, Wang CY, Hung YH, Weng TY, Yen MC, Lai MD. Systematic analysis of gene expression alterations and clinical outcomes for long-chain acyl-coenzyme a synthetase family in cancer. PLoS One. 2016;11:e0155660. doi: 10.1371/journal.pone.0155660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang CY, Lai MD, Phan NN, Sun ZD, Lin YC. Meta-analysis of public microarray datasets reveals voltage-gated calcium gene signatures in clinical cancer patients. PLoS One. 2015;10:e0125766. doi: 10.1371/journal.pone.0125766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weng TY, Wang CY, Hung YH, Chen WC, Chen YL, Lai MD. Differential expression pattern of THBS1 and THBS2 in lung cancer: clinical outcome and a systematic-analysis of microarray databases. PLoS One. 2016;11:e0161007. doi: 10.1371/journal.pone.0161007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacquemet G, Baghirov H, Georgiadou M, Sihto H, Peuhu E, Cettour-Janet P, He T, Perala M, Kronqvist P, Joensuu H, Ivaska J. L-type calcium channels regulate filopodia stability and cancer cell invasion downstream of integrin signalling. Nat Commun. 2016;7:13297. doi: 10.1038/ncomms13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho CY, Lee KT, Chen WC, Wang CY, Chang YS, Huang HL, Hsu HP, Yen MC, Lai MZ, Lai MD. MST3 promotes proliferation and tumorigenicity through the VAV2/Rac1 signal axis in breast cancer. Oncotarget. 2016;7:14586–14604. doi: 10.18632/oncotarget.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu JJ, Aspesi P, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li NX, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity (vol 483, pg 603, 2012) Nature. 2012;492:290–290. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tjiu JW, Chen JS, Shun CT, Lin SJ, Liao YH, Chu CY, Tsai TF, Chiu HC, Dai YS, Inoue H, Yang PC, Kuo ML, Jee SH. Tumor-associated macrophage-induced invasion and angiogenesis of human basal cell carcinoma cells by cyclooxygenase-2 induction. J Invest Dermatol. 2009;129:1016–1025. doi: 10.1038/jid.2008.310. [DOI] [PubMed] [Google Scholar]
- 31.Huang HL, Hsu HP, Shieh SC, Chang YS, Chen WC, Cho CY, Teng CF, Su IJ, Hung WC, Lai MD. Attenuation of argininosuccinate lyase inhibits cancer growth via cyclin A2 and nitric oxide. Mol Cancer Ther. 2013;12:2505–2516. doi: 10.1158/1535-7163.MCT-12-0863. [DOI] [PubMed] [Google Scholar]
- 32.Zhao HJ, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Karesen R, Botstein D, Borresen-Dale AL, Jeffrey SS. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma XJ, Dahiya S, Richardson E, Erlander M, Sgroi DC. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan YY, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S, Grp M. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 36.Municio C, Palacios BS, Estrada-Capetillo L, Benguria A, Dopazo A, Garcia-Lorenzo E, Fernandez-Arroyo S, Joven J, Miranda-Carus ME, Gonzalez-Alvaro I, Puig-Kroger A. Methotrexate selectively targets human proinflammatory macrophages through a thymidylate synthase/p53 axis. Ann Rheum Dis. 2016;75:2157–2165. doi: 10.1136/annrheumdis-2015-208736. [DOI] [PubMed] [Google Scholar]
- 37.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res. 2007;56:45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 38.Kaczmarek M, Nowicka A, Kozlowska M, Zurawski J, Batura-Gabryel H, Sikora J. Evaluation of the phenotype pattern of macrophages isolated from malignant and non-malignant pleural effusions. Tumour Biol. 2011;32:1123–1132. doi: 10.1007/s13277-011-0214-1. [DOI] [PubMed] [Google Scholar]
- 39.Hsu HP, Shan YS, Lai MD, Lin PW. Osteopontin-positive infiltrating tumor-associated macrophages in bulky ampullary cancer predict survival. Cancer Biol Ther. 2010;10:144–154. doi: 10.4161/cbt.10.2.12160. [DOI] [PubMed] [Google Scholar]
- 40.Zhang F, Wang HS, Wang XF, Jiang GM, Liu H, Zhang G, Wang H, Fang R, Bu XZ, Cai SH, Du J. TGF-beta induces M2-like macrophage polarization via SNAIL-mediated suppression of a pro-inflammatory phenotype. Oncotarget. 2016;7:52294–52306. doi: 10.18632/oncotarget.10561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao L, He C. Polarization of macrophages and microglia in inflammatory demyelination. Neurosci Bull. 2013;29:189–198. doi: 10.1007/s12264-013-1324-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gyorffy B, Munkacsy G, Mihaly Z, Surowiak P, Abdul-Ghani R, Schaefer R. PSMB7 is associated with anthracycline resistance and is a prognostic biomarker in breast cancer. Ejc Supplements. 2009;7:140–140. doi: 10.1038/sj.bjc.6605478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu R, Lu S, Deng Y, Yang S, He S, Cai J, Qiang F, Chen C, Zhang W, Zhao S, Qian L, Mao G, Wang Y. PSMB4 expression associates with epithelial ovarian cancer growth and poor prognosis. Arch Gynecol Obstet. 2016;293:1297–1307. doi: 10.1007/s00404-015-3904-x. [DOI] [PubMed] [Google Scholar]
- 45.Wu A, Wei J, Kong LY, Wang YT, Priebe W, Qiao W, Sawaya R, Heimberger AB. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12:1113–1125. doi: 10.1093/neuonc/noq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker KH, Sinha P, Horn LA, Clements VK, Yang H, Li J, Tracey KJ, Ostrand-Rosenberg S. HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res. 2014;74:5723–5733. doi: 10.1158/0008-5472.CAN-13-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oronsky B, Paulmurugan R, Foygel K, Scicinski J, Knox SJ, Peehl D, Zhao H, Ning S, Cabrales P, Summers TA Jr, Reid TR, Fitch WL, Kim MM, Trepel JB, Lee MJ, Kesari S, Abrouk ND, Day RM, Oronsky A, Ray CM, Carter CA. RRx-001: a systemically non-toxic M2-to-M1 macrophage stimulating and prosensitizing agent in Phase II clinical trials. Expert Opin Investig Drugs. 2017;26:109–119. doi: 10.1080/13543784.2017.1268600. [DOI] [PubMed] [Google Scholar]
- 48.Zheng PH, Guo HG, Li GC, Han SQ, Luo F, Liu Y. PSMB4 promotes multiple myeloma cell growth by activating NF-kappa B-miR-21 signaling. Biochem Biophys Res Commun. 2015;458:328–333. doi: 10.1016/j.bbrc.2015.01.110. [DOI] [PubMed] [Google Scholar]
- 49.Ri M, Iida S, Nakashima T, Miyazaki H, Mori F, Ito A, Inagaki A, Kusumoto S, Ishida T, Komatsu H, Shiotsu Y, Ueda R. Bortezomib-resistant myeloma cell lines: a role for mutated PSMB5 in preventing the accumulation of unfolded proteins and fatal ER stress. Leukemia. 2010;24:1506–1512. doi: 10.1038/leu.2010.137. [DOI] [PubMed] [Google Scholar]
- 50.Yang YM, Lee S, Nam CW, Ha JH, Jayaraman M, Dhanasekaran DN, Lee CH, Kwak MK, Kim SG. G alpha (12/13) inhibition enhances the anticancer effect of bortezomib through PSMB5 downregulation. Carcinogenesis. 2010;31:1230–1237. doi: 10.1093/carcin/bgq097. [DOI] [PubMed] [Google Scholar]
- 51.D’Souza AJ, Desai SD, Rudner XL, Kelly MN, Ruan S, Shellito JE. Suppression of the macrophage proteasome by ethanol impairs MHC class I antigen processing and presentation. PLoS One. 2013;8:e56890. doi: 10.1371/journal.pone.0056890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu S, Chen Z, Yang J, Chen L, Gong S, Zhou H, Guo L, Wang J. Overexpression of the PSMB5 gene contributes to bortezomib resistance in T-lymphoblastic lymphoma/leukemia cells derived from Jurkat line. Exp Hematol. 2008;36:1278–1284. doi: 10.1016/j.exphem.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 53.Balsas P, Galan-Malo P, Marzo I, Naval J. Bortezomib resistance in a myeloma cell line is associated to PSM beta 5 overexpression and polyploidy. Leuk Res. 2012;36:212–218. doi: 10.1016/j.leukres.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 54.Wang Q, Lu Y, Li R, Jiang Y, Zheng Y, Qian J, Bi E, Zhang C, Hou J, Wang S, Yi Q. Therapeutic effects of CSF1R-blocking antibodies in multiple myeloma. Leukemia. 2017 doi: 10.1038/leu.2017.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beyar-Katz O, Magidey K, Ben-Tsedek N, Alishekevitz D, Timaner M, Miller V, Lindzen M, Yarden Y, Avivi I, Shaked Y. Bortezomib-induced pro-inflammatory macrophages as a potential factor limiting anti-tumour efficacy. J Pathol. 2016;239:262–273. doi: 10.1002/path.4723. [DOI] [PubMed] [Google Scholar]
- 56.Groll M, Berkers CR, Ploegh HL, Ovaa H. Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20S proteasome. Structure. 2006;14:451–456. doi: 10.1016/j.str.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 57.Lichter DI, Danaee H, Pickard MD, Tayber O, Sintchak M, Shi H, Richardson PG, Cavenagh J, Blade J, Facon T, Niesvizky R, Alsina M, Dalton W, Sonneveld P, Lonial S, van de Velde H, Ricci D, Esseltine DL, Trepicchio WL, Mulligan G, Anderson KC. Sequence analysis of beta-subunit genes of the 20S proteasome in patients with relapsed multiple myeloma treated with bortezomib or dexamethasone. Blood. 2012;120:4513–4516. doi: 10.1182/blood-2012-05-426924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kraus M, Ruckrich T, Reich M, Gogel J, Beck A, Kammer W, Berkers CR, Burg D, Overkleeft H, Ovaa H, Driessen C. Activity patterns of proteasome subunits reflect bortezomib sensitivity of hematologic malignancies and are variable in primary human leukemia cells. Leukemia. 2007;21:84–92. doi: 10.1038/sj.leu.2404414. [DOI] [PubMed] [Google Scholar]
- 59.Coiffier B, Li W, Henitz ED, Karkera JD, Favis R, Gaffney D, Shapiro A, Theocharous P, Elsayed YA, van de Velde H, Schaffer ME, Osmanov EA, Hong X, Scheliga A, Mayer J, Offner F, Rule S, Teixeira A, Romejko-Jarosinska J, de Vos S, Crump M, Shpilberg O, Zinzani PL, Cakana A, Esseltine DL, Mulligan G, Ricci D. Prespecified candidate biomarkers identify follicular lymphoma patients who achieved longer progression-free survival with bortezomib-rituximab versus rituximab. Clin Cancer Res. 2013;19:2551–2561. doi: 10.1158/1078-0432.CCR-12-3069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.