Abstract
Accumulating evidence shows that microRNAs play important roles in cancers, including glioma. MiRNAs have been shown to participate in a variety of cellular functions including cell apoptosis, cell proliferation, neural development, and stem cell differentiation. Previous studies reported that miR-936 levels were downregulated in glioma specimens. Here, we further investigate the potential role of miR-936 in glioma. Quantitative reverse transcription-PCR was applied to detect the expression of mir-936 in glioma specimens. The direct targets of miR-936 were identified by bioinformatics analysis and were further validated by immunoblotting and luciferase reporter assay. The effects of miR-936 on glioma cell proliferation and cell cycle of glioma cells were analyzed by Cell-Counting Kit 8 assay, colony formation, 5-ethynyl-2-deoxyuridine (EDU) and flow cytometry assays. A xenograft model was used to study the effect of miR-936 on tumor growth and angiogenesis. Expression levels of miR-936 were greatly downregulated in glioma specimens, CKS1 was confirmed as a direct target of miR-936. The glioma cell cycle was blocked to G1 by negatively regulating CKS1 and its downstream signaling pathway, Akt-ERK1/2. Furthermore, overexpression of CKS1 rescued the inhibitory effects of miR-936. In vivo studies revealed that increased levels of miR-936 delayed the growth of tumors. Taken together, mir-936 may act as a glioma suppressor by targeting CKS1.
Keywords: miR-936, glioma, CKS1, proliferation
Introduction
Glioblastoma multiform (GBM) represents one of the most frequently occurring and aggressive forms of primary brain tumors in adults [1]. Despite advances in standard therapy, that surgical resection followed by radiation and chemotherapy, the prognosis still remains dismal [2]. Thus, it is essential to developing novel and effective therapeutic strategies for glioma.
MicroRNAs (miRNAs) are small, endogenous noncoding RNAs composed of 18-23 nucleotides (nt) that post-transcriptionally regulate gene expression by targeting the 3’-untranslated regions of mRNAs [3,4]. Recent studies have reported the abnormal expression of microRNAs (miRNAs) linked with various tumors, including gliomas [5]. Accumulating evidence has indicated that a subset of miRNAs are deregulated in gliomas and play important role in proliferation, invasion, migration and chemosensitivity [6-12].
The cyclin-dependent protein kinase regulatory subunit 1 (Cks1) gene which encodes a 9 KD protein Cks1 takes important roles in cell growth, proliferation and apoptosis. One of crucial functional roles of Cks1 is to mediate cell cycle progression from G1 to S phase [13]. Aberrant expression of Cks1 in multiple human cancers and is significantly associated with tumor proliferation [14-17].
In this study, we identified a novel tumor-suppressive miRNA, miR-936, and provide the first evidence for a role of the miRNA in glioma proliferation via directly targeting CKS1. Our results not only revealed that Cks1 overexpression in glioma could be the result of downregulation of miR-936, but also suggested an important role for the loss of miR-936 in proliferation and cell cycle progression in glioma, highlighting its potential as a therapeutic target.
Materials and methods
Chinese Glioma Genome Atlas (CGGA) database and human tissue samples
Thirty-nine glioma tissue specimens were obtained postoperatively from the Department of Neurosurgery, First Affiliated Hospitals of Nanjing Medical University. Of these thirty-nine samples, fourteen were low-grade glioma samples (grades I and II) and eighteen were high-grade glioma samples (grades III and IV). In addition, seven non-cancerous brain tissues (NBTs) were collected as negative controls. The Research Ethics Committee of Nanjing Medical University (Nanjing, Jiangsu, China) approved the use of GBMs and non-cancerous brain tissues, and the procedures were performed in accordance with the approved guidelines. Permissions were obtained from participants, and patients granted informed consent.
MiRNA expression profiles of 198 glioma samples of different grades were downloaded from the Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn). Among the 198 cases, 49 were diagnosed as astrocytoma (A), 14 as oligodendroglioma (O), 12 as anaplastic astrocytoma (AA), 13 as anaplastic oligodendroglioma (AO), 19 as anaplastic oligoastrocytoma (AOA), and 91 as GBM.
Cell culture
Human GBM cell lines U87, U251, U118, LN229, A172, and T98 were purchased from the Chinese Academia Sinica Cell Repository (Shanghai, China). Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Invitrogen, Carlsbad, CA, USA). Normal human astrocytes (NHAs) were obtained from Lonza (Walkersville, MD, USA) and cultured following the manufacturer’s instructions. All cells were incubated at 37°C in a fully humidified atmosphere containing 5% CO2 in filtered air.
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was isolated from normal brain tissues, human glioma specimens and cultured cells using TRIzol reagent (Invitrogen) with manufacturer’s instructions. Expression of U6 was used as an endogenous control. To detect the relative levels of miR-936 in tissues and cells, the ABI 7300 HT sequence detection system (Applied Biosystems) was used. The relative expression of each gene was calculated and normalized using the 2-ΔΔCt method. Each sample was tested in triplicate.
Oligonucleotides, lentiviral vectors, plasmid construction, and cell transfection
Lentiviruses carrying hsa-miR-936, hsa-miR-NC and siRNA-CKS1 or siRNA-NC were purchased from Genepharma (Shanghai, China). Stable cell lines were established by infecting U87 and U251 cells, followed by puromycin selection.
Fluorescence in situ hybridization (FISH)
The expression of miR-936 in GBM samples and NBTs was detected by FISH. The mature human miR-936 sequence is: 3’-GUCGCUUUGUCCGUUUGCCUC-5’. We used (LNA)-based probes directed against the full length mature miRNA sequence. The 5’-FAMlabelled miR-936 probe sequence is: 5’-CAGCGAAACAGGCAAACGGAG-3’, and was purchased from BioSense (Guangzhou, China). The FISH procedure followed the BioSense instructions. Briefly, frozen sections were fixed with 4% paraformaldehyde for 30 min, then washed twice with PBS. Fixed slides were then treated with proteinase K at 37°C for 10 min, followed by dehydration in 70%, 85% and 100% ethanol for 5 min. The probe was then added to the slides, which were denatured at 78°C for 5 min. Hybridization was then carried out overnight at 42°C in a humid chamber. The next day, post-hybridization washes were performed with 50% formamide with 2 × SSC at 43°C, followed by 2 × SSC washes at room temperature to remove non-specific and repetitive RNA hybridization. Finally, slides were counterstained with DAPI (Sigma) for 10 min and examined with a Zeiss LSM 700 Meta confocal microscope (Oberkochen, Germany).
Cell counting kit-8 (CCK-8) assay
U87 and U251 cells were seeded in 96-well plates at a density of 2000 cells per well. And the CCK-8 assay (Dojindo, Japan) were performing when the cells were cultured at 24, 48, 72 and 96 hs. Then, 10 μL CCK8 solution was added into each well, and cells were incubated for 1 h in a humidified incubator. Optical density was measured at 450 nm.
Colony formation assay
Colony formation assays were performed to assess the proliferation ability of transfected U87 and U251 cells, which were harvested 24 h and then seeded in a new six-well plate (200 cells/well) and cultured for approximately 2 weeks until colony formation was observed. Then, the cells were fixed in 4% paraformaldehyde and stained by Giemsa (Vetec, Shanghai, China).
5-ethynyl-2-deoxyuridine (EDU) proliferation assay
The Cell-Light EDU imaging detection kit was purchased from Life Technologies (MA, USA). Cells which had been transfected 48 h previously were incubated with 10 μM EDU for 24 h, fixed, permeabilized, and stained with both the Alexa-Fluor 594 reaction cocktail for EDU and Hoechst 33342 for cell nuclei, according to the manufacturer’s protocol. Finally, samples were imaged under a fluorescent microscope.
Cell cycle analysis
Each group of glioma cell lines was harvested by centrifugation at 1500 r/m for 5 min. Then wash the cell with phosphate buffered saline (PBS), fixed with 75% alcohol and stored at -20°C overnight. The cell were washed by PBS and incubated with a Cell Cycle Staining Kit (Multi Sciences, Hangzhou, China) for 30 minutes without light. At last the cell were analyzed by flow cytometry.
Western blot analysis
The proteins were separated on 10% SDS-PAGE and the transferred to PVDF membrane (Merck Millipore). The membranes were blocked in 5% nonfat milk at room temperature for 2 h and incubated with primary antibodies overnight at 4°C with diluted (1:1000) primary antibodies against CKS1 (ab130529, Abcam), β-actin (AF0003, Beytime), AKT (#4685, CST), P-AKT (#4060, CST), CDC2 (#9868, CST), CDK2 (#9868, CST), CDK4 (#9868, CST), Cyclin E1 (#4129, CST), ERK (#9926, CST), P-ERK (#4370, CST) followed by incubation with a horseradish peroxidase-conjugated secondary antibody (1:2000, Santa Cruz Biotechnology, CA, USA) for 2 h at room temperature. Membranes were probed using SuperSignal® Maximum Sensitivity Substrate (Thermo Fisher Scientifc).
Dual luciferase reporter assay
Mutated and wild-type (WT) putative miR-936-binding sites in the CKS1 3’-UTR region were amplified and cloned into the XbaI site of a pGL3 control vector (Invitrogen). U87 and U251 cells were seeded in a 24-well plate and con-transfected with WT or mutated 3’-UTR luciferase reporter and miR-936 mimics (RiboBio). After transfected for 24 h, the luciferase activities were analyzed by the Promega Dual Luciferase Reporter Assay System (WI, USA).
Subcutaneous xenograft model
BALB/c-A nude mice at 4 weeks old were purchased from Shanghai Experimental Animal Center of the Chines Academy of Sciences and maintained under specific pathogen-free (SPF) conditions for 1 week. Animal handling and experimental procedures were processed following the Guide for the Care and Use of Laboratory Animals and agreement accessed form the Animal Experimental Ethics Committee of Nanjing Medical University. 12 nude were randomly separated into two groups. U251 cells (5 × 106 cells in 100 mL pretreated with lentivirus containing the miR-936 or negative control sequences) were subcutaneously injected into the posterior flank of the nude mice. The sizes of the tumor were measured by vernier scale every 4 days. The formula (volume = 0.5 × length × width2) was used to calculate the tumor volumes. The tumors were weighted 36 days after injection. Total proteins were extracted for specific protein levels by western blot assay.
Immunohistochemistry
The tissue was fixed in 4% paraformaldehyde, embedded in paraffin, and cut into 3.5-lm-thick sections. The tissue sections were incubated with 1:200-dilutec primary antibody overnight at 4°C, and then were incubated with biotin-labeled immunoglobulin (1:100 dilution; GeneTech) for 1 h at room temperature. After incubated with avidin-biotin-peroxidase complex followed by diaminobenzidine counterstained with hematoxylin (GeneTech), and visualized under a light microscope. Brown staining in cells was considered as positive signaling.
Statistical analysis
All experiments were performed three times and data were analyzed using GraphPad Prism 5 software. Statistical evaluation of data was performed using Student’s t-test with P<0.05 considered statistically significant.
Result
Mir-936 is down regulated in glioma tissue and cell lines
In order to investigate miRNA expression in human glioma tissues, the Chinese Glioma Genome Atlas (CGGA) data was used to analyze the expression in 198 glioma tissues (Grade II: 63; Grade III: 44 and Grade IV: 91). The result is that the expression of mir-936 was significantly higher in low grade gliomas than that in high grade gliomas (Figure 1A). Then we analyzed the prognostic value of mir-936 expression in 82 GBM cases by Kaplan-Meier survival analysis. The patients with lower expression (n = 41) of mir-936 had shorter medial overall survival times compared with who with higher expression of mir-936 (n = 41) (Figure 1C). In addition, we evaluated the expression level of miR-936 in 32 different glioma tissues and 7 non-cancerous brain tissues by qRT-PCR. The result indicated that the higher grade of the tumor the lower expression level of the mir-936 (Figure 1B). At last, we evaluated the expression level of mir-936 in U251, T98, U118, LN229, U87, A172 cell lines and as well as NHAs. Compared with NHAs, the expression level of mir-936 is obviously downregulated in the rest cell lines (Figure 1D). To further confirm the thought, we choose one representative GBM specimen and the noncancerous brain tissues for FISH analysis, and we drew a consistent result (Figure 1E).These results supported the hypothesis that mir-936 may act as a tumor suppresser in glioma.
Figure 1.
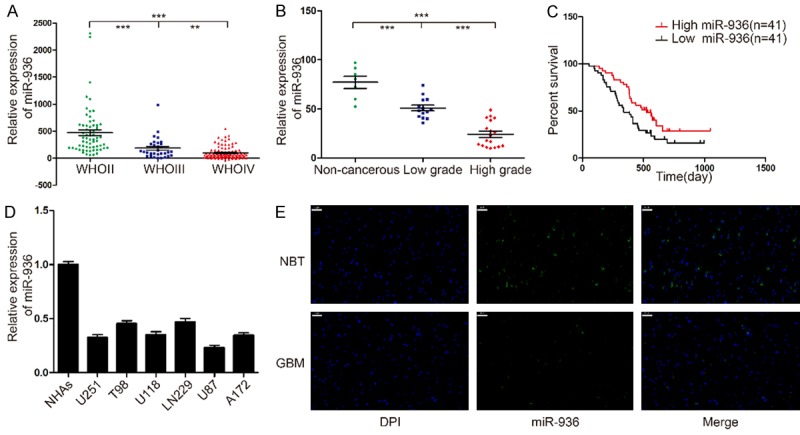
MiR-936 expression levels in human gliomas and cell lines. A. CGGA database showing miR-936 expression in different grades of glioma. B. The expression of miR-936 in 7 non-cancerous brain tissues, 14 low-grade glioma tissues and 18 high-grade glioma tissues was measured by real-time PCR, miR-936 levels in normal brain tissues were indeed higher compared with glioma specimens, and were significantly decreased with ascending pathological grade of tumor. C. Kaplan-Meier survival analysis of overall survival duration in 82 GBM patients according to miR-936 expression using CGGA. D. qRT-PCR analysis of miR-936 in NHAs and different glioma cell lines (U251, T98, U118, LN229, U87, A172). E. The different expressions of miR-936 were determined by FISH in GBM and NBT, respectively. Bar = 50 μm.
MiR-936 attenuate the proliferation of glioma cells
The downregulated expression level of miR-936 led us to investigate whether it plays a roles as suppressor in GBM development. We first evaluated the effect of mir-936 on cell proliferation by using CCK-8 and colony formation assays. Before that, in order to confirm the lentivirus for mir-936 was transfected into U87 and U251 successfully, the qRT-PCR was used and its results demonstrated that miR-936 was significantly increased compared to negative control group (Figure 2A). The CCK-8 assay showed that miR-936-transduced U251 and U87 cells exhibit significant lower growth rates than control groups (Figure 2B, 2C). The colony formation assays consistently showed that up-regulated expression of miR-936 notably reduced the number of colonies of the 2 transfected cell lines after 12 days of culture compared with the controls (Figure 2E, 2G).
Figure 2.

Overexpression of miR-936 inhibits glioma cell proliferation in vitro. A. The relative expression of miR-936 in U251 and U87-MG cells was analyzed by qRT-PCR after transfection. B, C. Proliferation was determined using the CCK-8 assay following culture for 96 h. Data are presented as the means of triplicate experiments. D, F. Proliferating cells were examined using the EDU assay. Representative images are shown (original magnification, 200 ×). (**P<0.01). E, G. Long-term cell viability was evaluated using the colony formation assay. Data are presented as the means of triplicate experiments. H, I. The cell cycle phase of U251 and U87-MG cells transfected with miR-936 or negative control (miR-ctrl) lentivirus was analyzed by flow cytometry. J. Western blot analysis of CDC2, CDK2 and cyclin E1 in U251 and U87-MG cells 48 h after transfection. β-actin served as the loading control.
As the changes in proliferation often associated with cell cycle progression [18], we determined whether growth inhibition was associated with specific cell cycle changes. The effect of miR-936 on the cell cycle progression of U251 and U87 cells were characterized by flow cytometric analysis. Compared with the mir-control groups, U87 and U251 cells with miR-936 over-expression showed significant increase percentage of the cells in the G1/G0 phase (Figure 2H, 2I). To investigate the molecules involved in the cell cycle distribution, we measured the expression of CDC2, CDK2, CDK4 and Cyclin E1 in miR-936-overexpression U87 and U251 cells. All of these molecules have been previously reported to be important regulators for G1 phase [19-21]. Compared with miR-contorl group, ectopic expression of miR-936 significantly suppressed CDC2, CDk2, CDK4 and Cyclin E1 in U87 and U251 cells (Figure 2J). Collectively, our results demonstrate that miR-936 may attenuate glioma cell proliferation.
CKS1 is a direct target of miR-936 in glioma cells
It is generally accepted that miRNAs do not cause direct changes in cellular functions. The function is regulated by the expression of their downstream target genes [3,22]. We investigate the potential specific targets of miR-936 by 4 different computational methods, including TargetScan, miRanda, miRDB and miRWalk. According to these methods, CKS1 attracted our attention and Western blot analysis showed that expression of CKS1 protein was downregulated in miR-936-transfected cells (Figure 3A).The miR-936 binding sites in the 3’UTR of CKS1 were identified by miRNA algorithms (Figure 3B). To further determine the correlation between miR-936 and CKS1 levels, we measured the levels of CKS1 protein in glioma specimens. The results showed that the expression levels of CKS1 were positively correlated with the degree of the grade of glioma (Figure 3F, 3G). Then we explored the correlation between CKS1 and miR-936 in the same tissues. As show in Figure 3H, the CKS1 levels in glioma tissues were inversely correlated with miR-936 expression levels (Spearman’s correlation analysis r = -0.59). To further determine whether miR-936 could directly bind to the 3’-UTR of CKS1, we performed luciferase reporter assays. Luciferase activity in U87 cells was notably decreased after the cells were transfected with wild-type vector (Figure 3D). The activity was also significantly decreased when binding site1 was mutated, however, mutation of binding sit2 nearly rescued the decrease. We can see the similar results in U251 cells (Figure 3C, 3E). The finding above suggest that miR-936 directly regulates CKS1 expression by binding to site 2 (nt2178-2184) in the 3’-UTR of CKS1.
Figure 3.
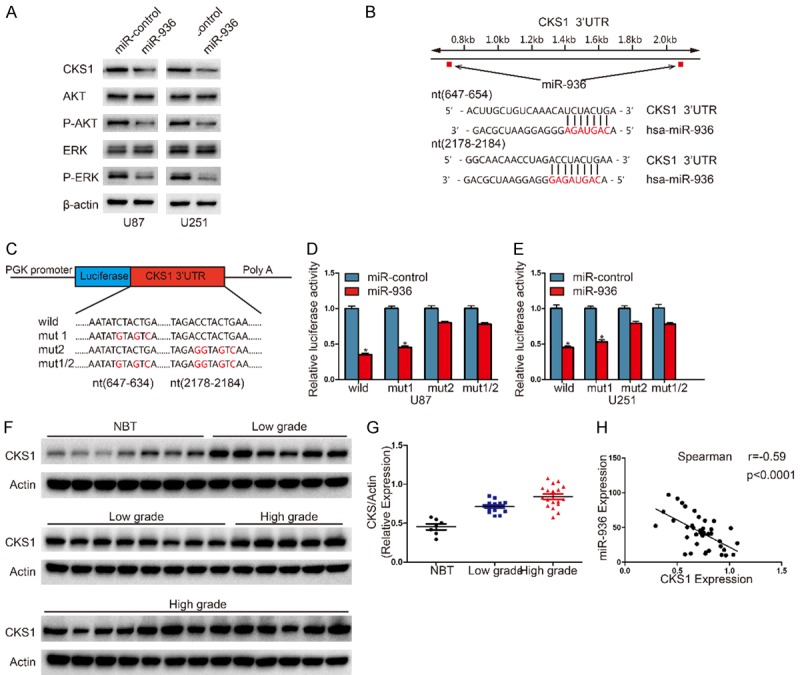
MiR-1468-5p directly and negatively regulates RRM1 in glioma cells. A. Western blot analysis of CKS1, p-AKT, AKT, p-ERK1/2 and ERK1/2 in U87 and U251 cells after transduction with miR-ctrl or miR-936. B. Predicted miR-936 binding sites in the 3’-UTR of the CKS1 gene. C. Wild-type and mutant CKS1 3’-UTR reporter constructs. D, E. Luciferase reporter assays were performed in U251 and U87 cells with co-transfection of indicated wild-type or mutant 3’-UTR constructs and miR-936 mimic. The data shown are representative of three independent experiments. Data shown are mean ± SD of three independent experiments. *P<0.05. F, G. The expression levels of CKS1 in NBTS and glioma specimens were determined by western blotting; the fold changes were normalized to β-Actin. The non-neoplastic brain tissues (n = 7) were collected from brain trauma surgery. The low-grade (n = 14) represents samples derived from grades I and II glioma tissues, whereas high-grade (n = 18) represents grades III and IV glioma tissues. Data represent the means ± SD from three independent experiments. *P<0.05, **P<0.01, ***P<0.001. H. Pearson’s correlation analysis of the relative expression levels of miR-936 and the relative protein levels of CKS1.
Down-regulation of CKS1 and miR-936 overexpression in glioma cells have the similar effect
The correlation between miR-936 and CKS1 have been verified, we wondered whether CKS1 is a downstream effecter of miR-936. In order to confirm this, we knock down the expression of CKS1 by transfected si-NC and si-CKS1 into U87 and U251 cells. The suppressed expression of CKS1 protein was indicated by western blot analysis and the activity of the proliferation pathway was inhibited (Figure 4A). Then we use CCK-8, colony formation and EDU assays to evaluate the effects of the knockdown of CKS1 on cell viability and growth. Knockdown of CKS1 expression dramatically suppressed the proliferation of GBM cells (Figure 4B-G). These results were similar with the effect of miR-936 overexpression.
Figure 4.
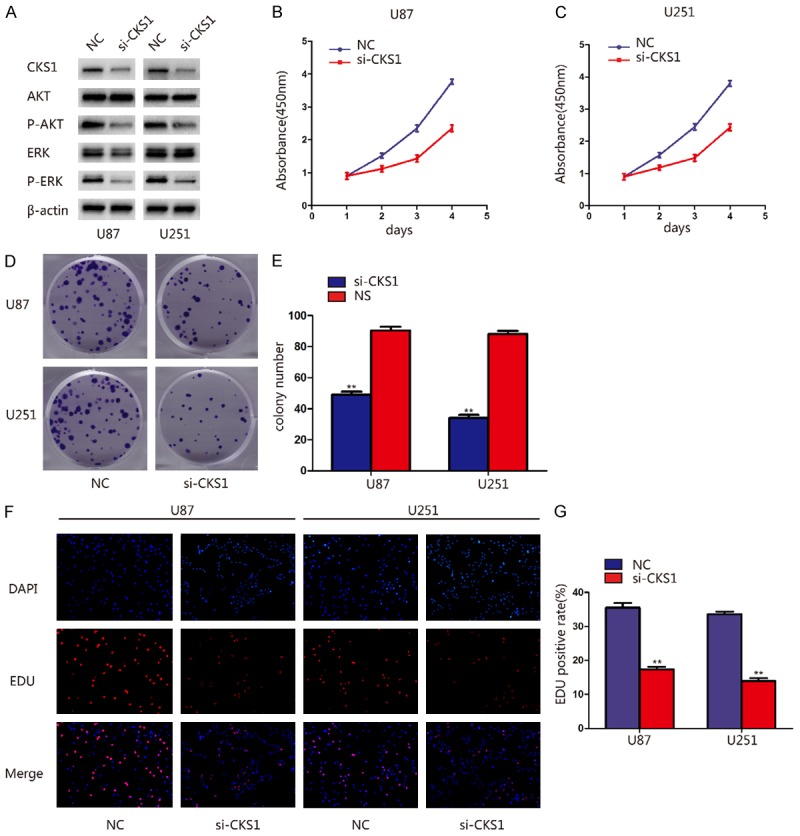
CKS1 knockdown suppressed proliferation and cell cycle progression of glioma cells. A. Western blot showing the levels of CKS1, p-AKT, AKT, p-ERK1/2 and ERK1/2 in U87 and U251 cells after transfection with siCKS1. B, C. Proliferation ability was determined using the CCK-8 assay following culture for 96 h. Data are presented as the means of triplicate experiments. D, E. Long-term cell viability was evaluated using the colony formation assay. Data are presented as the means of triplicate experiments. F, G. Proliferating cells were examined using the EDU assay. Representative images are shown (original magnification, 200 ×). (**P<0.01).
CKS1 reintroduction attenuates the inhibitory effects of miR-936
Having determined that CKS1 is a miR-936 target, we further explored whether CKS1 was a functional miR-936 target by transfecting CKS1 plasmids into U87 and U251 cells which stably expressing miR-936 or miR-control. Overexpression of CKS1 rescued the decrease of its level caused by miR-936 overexpression. Besides, the Phosphorylation levels of AKT and ERK were changed in a similar way compared with the level of CKS1. That is, the decrease of p-AKT and p-ERK level caused by miR-936 overexpression could be rescued by up-regulation of CKS1 (Figure 5A). To further explore whether CKS1 is an important target of miR-936 in cell proliferation, the experiments on proliferation and cell cycle were conducted. Both cell cycle and proliferation experiments demonstrated that restoration of CKS1 expression antagonized the inhibitory effects of miR-936. Taken together, these results suggest that CKS1 is a functional target of miR-936 in glioma cells.
Figure 5.

CKS1 reintroduction reverses the inhibitory effect of miR-936. A, B. Western blot analysis of CKS1, p-AKT, AKT, p-ERK1/2 and ERK1/2 in the indicated cells. C, D. CCK8 assay with miR-control or miR-936-transduced U87 and U251 cells transfected with vector or CKS1; **P<0.01 and ***P<0.001. E, F. Co-transfected U87 and U251 cells were analyzed by the colony formation assay; *P<0.05 and **P<0.01. G-I. The Edu assay was performed 48 h after cotransfection; *P<0.05 and **P<0.01. J-L. Flow cytometry was conducted to determine the cell cycle distribution in each group.
MiR-936 attenuated tumor growth in nude mice and decreased CKS1 protein levels in subcutaneous tumors
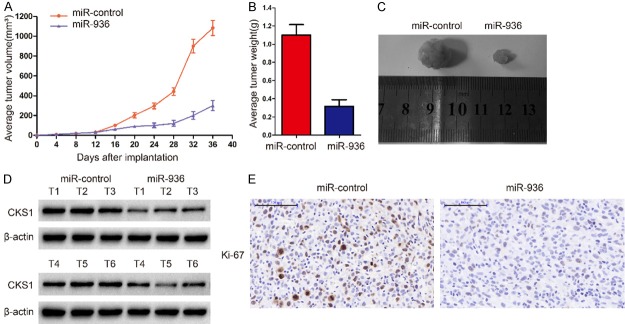
To further verify the role of miR-936 and CKS1 in glioma invasion, we extended our investigation to examine if miR-936 could retard glioblastoma growth in vivo using nude mice. MiR-936 overexpression inhibited tumor growth both in size and weight (Figure 6A-C). Consistent with our hypothesis, western blot analysis also showed that the levels of CKS1 in the miR-936-overexpressing group were lower than those of the miR-control group (Figure 6D).Further, immunohistochemistry experiments were used to measure the levels of Ki67, which are often used to reflect proliferation and angiogenesis of tumor. Consistent with the previous results, Ki67 were notably reduced in the miR-936 overexpression group (Figure 6E). Together, these findings demonstrate that miR-936 inhibits the proliferation of glioma cells in vivo.
Figure 6.
MiR-936 attenuated tumor growth in nude mice and decreased CKS1 protein level in subcutaneous tumors. A-C. Male BALB/c nude mice were subcutaneously injected with U87 cells and tumor volumes were measured every 4 days, and the tumors were harvested and photographed after 36 days. Representative images from each group are shown. The tumors from the miR-936 group were smaller and lighter than those from the miR-conrtol group (*p<0.05, **p<0.01, and ***p<0.001). D. The levels of CKS1 from the tumor tissues in the two groups were analyzed by western blot. β-actin was used as an internal control. E. Ki-67 expression was measured by immunohistochemistry in mouse tumor tissues; compared with that in the miR-control group, the level of Ki-67 in the miR-936 group was lower (magnification 200 ×).
Discussion
The proliferation ability of GBM is one of the contributing factors leading to poor prognoses, but the involved mechanisms remain unclear. Mounting evidence suggests that miRNAs play important roles in proliferation of human cancers, including GBM. In our study, the biological roles of miR-936 and its target gene CKS1 in glioma proliferation was investigated.
As a new discovered miRNA, miR-936 has not been reported before. Through CGGA database analysis, we found that the expression level of miR-936 was negatively correlated with glioma grade. We than analyzed expression of miR-936 in clinical tissue samples, independent qRT-PCR, several glioma cell lines (especially U87 and U251), and NHAs. Our findings showed that miR-936 was frequently downregulated in both glioma tissues and cell lines and low miR-936 levels were linked to higher incidence of high-grade gliomas. Subsequent functional assays revealed that restoration of miR-936 induced a significant reduction in cell proliferation, which suggested that miR-936 may acts as a tumor suppressor and involved in glioma proliferation. To explore the effect of miR-936 on the proliferation of glioma cells, we transfected miR-936 lentivirus into the cells. The performance of the stably transfected glioma cells in CCK-8, colony formation and EDU assays showed that the cells proliferation capacity was notably decreased in both short and long term. Consistent with the flow cytometry assay result that miR-936 could arrest cells at G1/S phase, the western blot show that CDC2, CDk2, Cyclin E1 were significantly decreased after overexpression of miR-936.
In this study, we have demonstrated that the CKS1 oncogene is a target of miR-936 in vitro and in vivo. First, luciferase reporter assays confirmed that miR-936 directly recognizes the 3’-UTR of CKS1 transcripts. Second, CKS1 expression was significantly decreased in glioma cells stably expressing miR-936. Third, an inverse correlation between CKS1 protein and miR-936 levels in clinical samples was found. Finally, the inhibition of CKS1 protein by miR-936 inhibited subcutaneous tumor growth. As a direct target of miR-936, CKS1 displayed an important functional role affecting the proliferation of U251 and U87 glioma cell lines.
The expression of CKS1 is strongly associated with aggressive breast tumors, prostate carcinomas, non-small cell lung cancers, and several other malignancies [14,15,23,24]. The underlying mechanism of CKS1 in glioma proliferation remains unclear. It has been reported that CKS1 activates the p27/cdk2 signaling pathway [25-29]. Whether CKS1 promotes glioma proliferation by activating the p27/cdk2 pathway requires further investigation. Additionly, CKS1 overexpression can also promotes cell drug resistance by activates STAT3 and MEK/ERK through SKP2 and p27K [15,30,31]. The interaction of CKS1 with STAT3 and MEK/ERK and their role in glioma drug resistance should be explored in the future.
In summary, we found that miR-936 is significantly downregulated in glioma tissues and cell lines, and we identified a link between miR-936 and CKS1, which is a novel factor in glioma tumorigenesis. Moreover, we demonstrated, for the first time, that the role of the miR-936/CKS1 axis was to regulate glioma proliferation and cell cycle progression. This novel miR-936/CKS1 axis may further our understanding of the molecular mechanisms involved in glioma proliferation, and targeting miR-936/CKS1 may serve as a promising therapeutic strategy for glioma treatment.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (81300998 and 81471269), National Natural Science Foundation of Jiangsu Province (BK20131022 and BK20160047), Jiangsu Province’s Key Discipline of Medicine (XK201117), Jiangsu Province and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Specially Appointed Professor Foundation of Jiangsu Province (ky216r201307).
Disclosure of conflict of interest
None.
References
- 1.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro Oncol. 2012;14(Suppl 5):v1–49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Shi L, Zhang J, Pan T, Zhou J, Gong W, Liu N, Fu Z, You Y. MiR-125b is critical for the suppression of human U251 glioma stem cell proliferation. Brain Res. 2010;1312:120–126. doi: 10.1016/j.brainres.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 7.Sun L, Yan W, Wang Y, Sun G, Luo H, Zhang J, Wang X, You Y, Yang Z, Liu N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011;1389:9–18. doi: 10.1016/j.brainres.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Wang YY, Sun G, Luo H, Wang XF, Lan FM, Yue X, Fu LS, Pu PY, Kang CS, Liu N, You YP. MiR-21 modulates hTERT through a STAT3-dependent manner on glioblastoma cell growth. CNS Neurosci Ther. 2012;18:722–728. doi: 10.1111/j.1755-5949.2012.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi ZM, Wang XF, Qian X, Tao T, Wang L, Chen QD, Wang XR, Cao L, Wang YY, Zhang JX, Jiang T, Kang CS, Jiang BH, Liu N, You YP. MiRNA-181b suppresses IGF-1R and functions as a tumor suppressor gene in gliomas. RNA. 2013;19:552–560. doi: 10.1261/rna.035972.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang L, Shi ZM, Jiang CF, Liu X, Chen QD, Qian X, Li DM, Ge X, Wang XF, Liu LZ, You YP, Liu N, Jiang BH. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget. 2014;5:5416–5427. doi: 10.18632/oncotarget.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang R, Luo H, Wang S, Chen Z, Hua L, Wang HW, Chen W, Yuan Y, Zhou X, Li D, Shen S, Jiang T, You Y, Liu N, Wang H. MiR-622 suppresses proliferation, invasion and migration by directly targeting activating transcription factor 2 in glioma cells. J Neurooncol. 2015;121:63–72. doi: 10.1007/s11060-014-1607-y. [DOI] [PubMed] [Google Scholar]
- 12.Xu X, Bao Z, Liu Y, Ji J, Liu N. MicroRNA-98 Attenuates cell migration and invasion in glioma by directly targeting Pre-B cell leukemia homeobox 3. Cell Mol Neurobiol. 2017 doi: 10.1007/s10571-017-0466-4. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynard GJ, Reynolds W, Verma R, Deshaies RJ. Cks1 is required for G(1) cyclin-cyclindependent kinase activity in budding yeast. Mol Cell Biol. 2000;20:5858–5864. doi: 10.1128/mcb.20.16.5858-5864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan Y, Zhang Y, Wang J, Lin C, Ittmann MM, Wang F. Aberrant expression of Cks1 and Cks2 contributes to prostate tumorigenesis by promoting proliferation and inhibiting programmed cell death. Int J Cancer. 2008;123:543–551. doi: 10.1002/ijc.23548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang XC, Tian J, Tian LL, Wu HL, Meng AM, Ma TH, Xiao J, Xiao XL, Li CH. Role of Cks1 amplification and overexpression in breast cancer. Biochem Biophys Res Commun. 2009;379:1107–1113. doi: 10.1016/j.bbrc.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 16.Inui N, Kitagawa K, Miwa S, Hattori T, Chida K, Nakamura H, Kitagawa M. High expression of Cks1 in human non-small cell lung carcinomas. Biochem Biophys Res Commun. 2003;303:978–984. doi: 10.1016/s0006-291x(03)00469-8. [DOI] [PubMed] [Google Scholar]
- 17.Lee SW, Kang SB, Lee DS, Lee JU. Akt and Cks1 are related with lymph node metastasis in gastric adenocarcinoma. Hepatogastroenterology. 2013;60:932–937. doi: 10.5754/hge121214. [DOI] [PubMed] [Google Scholar]
- 18.Hardwick LJ, Ali FR, Azzarelli R, Philpott A. Cell cycle regulation of proliferation versus differentiation in the central nervous system. Cell Tissue Res. 2015;359:187–200. doi: 10.1007/s00441-014-1895-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miele A, Braastad CD, Holmes WF, Mitra P, Medina R, Xie R, Zaidi SK, Ye X, Wei Y, Harper JW, van Wijnen AJ, Stein JL, Stein GS. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol Cell Biol. 2005;25:6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandhu C, Donovan J, Bhattacharya N, Stampfer M, Worland P, Slingerland J. Reduction of Cdc25A contributes to cyclin E1-Cdk2 inhibition at senescence in human mammary epithelial cells. Oncogene. 2000;19:5314–5323. doi: 10.1038/sj.onc.1203908. [DOI] [PubMed] [Google Scholar]
- 21.Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 22.Romero-Cordoba SL, Salido-Guadarrama I, Rodriguez-Dorantes M, Hidalgo-Miranda A. miRNA biogenesis: biological impact in the development of cancer. Cancer Biol Ther. 2014;15:1444–1455. doi: 10.4161/15384047.2014.955442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapira M, Ben-Izhak O, Linn S, Futerman B, Minkov I, Hershko DD. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer. 2005;103:1336–1346. doi: 10.1002/cncr.20917. [DOI] [PubMed] [Google Scholar]
- 24.Slotky M, Shapira M, Ben-Izhak O, Linn S, Futerman B, Tsalic M, Hershko DD. The expression of the ubiquitin ligase subunit Cks1 in human breast cancer. Breast Cancer Res. 2005;7:R737–744. doi: 10.1186/bcr1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 26.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 27.Carrano AC, Pagano M. Role of the F-box protein Skp2 in adhesion-dependent cell cycle progression. J Cell Biol. 2001;153:1381–1390. doi: 10.1083/jcb.153.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat Cell Biol. 2001;3:321–324. doi: 10.1038/35060126. [DOI] [PubMed] [Google Scholar]
- 29.Spruck C, Strohmaier H, Watson M, Smith AP, Ryan A, Krek TW, Reed SI. A CDK-independent function of mammalian Cks1: targeting of SCF(Skp2) to the CDK inhibitor p27Kip1. Mol Cell. 2001;7:639–650. doi: 10.1016/s1097-2765(01)00210-6. [DOI] [PubMed] [Google Scholar]
- 30.Shi L, Wang S, Zangari M, Xu H, Cao TM, Xu C, Wu Y, Xiao F, Liu Y, Yang Y, Salama M, Li G, Tricot G, Zhan F. Over-expression of CKS1B activates both MEK/ERK and JAK/STAT3 signaling pathways and promotes myeloma cell drug-resistance. Oncotarget. 2010;1:22–33. doi: 10.18632/oncotarget.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S, Tricot G, Shi L, Xiong W, Zeng Z, Xu H, Zangari M, Barlogie B, Shaughnessy JD Jr, Zhan F. RARalpha2 expression is associated with disease progression and plays a crucial role in efficacy of ATRA treatment in myeloma. Blood. 2009;114:600–607. doi: 10.1182/blood-2008-12-194126. [DOI] [PMC free article] [PubMed] [Google Scholar]