Abstract
Cervical cancer is one of the most common gynecological malignancies in women worldwide. The long non-coding RNA (lncRNA) LINC00473 is increased in some human cancer tissues and it plays important roles in tumorigenesis. However, neither the expression pattern nor the biological functions of LINC00473 have been elucidated in cervical cancer so far. In the present study, gain- and loss-of-function assays showed that LINC00473 promoted cell proliferation and inhibited cell apoptosis in cervical cancer cells in vitro. Moreover, we found that LINC00473 enhanced the growth of cervical cancer cells in vivo. Mechanistic investigation showed that LINC00473 directly interacted with ILF2 and suppressed its degradation. Finally, we demonstrated that miR-34a reduced the stability of LINC00473. These findings may have important implications for developing novel therapeutic strategies for cervical cancer.
Keywords: LINC00473, ILF2, growth, miR-34a
Introduction
Cervical cancer is one of the most common gynecological malignancies in women worldwide, with a global incidence of approximately 500,000 new diagnosed cases and 260,000 cases of cancer-related deaths annually [1]. Recently, with the development and effectiveness of early screening tests for cervical cancer, the incidence and mortality rates of cervical cancer in developed countries have decreased [2,3]. Despite substantial development in understanding the molecular mechanisms and treatment for cervical cancer in recent years, the overall 5-year survival rates remain unsatisfied [4]. Therefore, it is urgently needed to identify novel prognostic markers and better understand the molecular mechanisms underlying the initiation and development of cervical cancer.
Genome-wide sequencing analyses have indicated that the vast majority of the genome is transcribed as non-coding RNAs (ncRNAs), including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) [5]. LncRNAs are generally defined as RNA transcripts with more than 200 nucleotides in length and no protein-coding potential. Increasing evidence has demonstrated that lncRNAs are implicated in a variety of pathophysiological processes, such as gene expression, cell proliferation, apoptosis, and tumorigenesis [6]. Importantly, lncRNAs were found to function as either oncogene or tumor suppressor gene to participate in the pathogenesis and development of many kinds of diseases including cancers [7]. Some functional lncRNAs have been identified in cervical cancer. For example, in epithelial ovarian carcinoma, there is a correlation between HOTAIR expression and metastatic stage. The regulation of specific matrix metalloproteinases (MMPs) and EMT-related genes is thought to be responsible for this correlation [8], suggesting that HOTAIR expression levels might be taken as a prognostic factor for ovarian carcinoma [9]. Cervical cancer tissues markedly express lower levels of MEG3. Furthermore, MEG3 down-regulation correlates positively with increased tumor size, advanced FIGO stage, metastasis of lymph nodes and HPV positivity. In addition, growth suppression and increased apoptosis of cervical cancer cells is observed after MEG3 upregulation, which demonstrates its tumor suppressive role in this cancer [10]. However, the expression and underlying mechanism of cervical cancer associated with aberrant lncRNAs remain largely unclear.
LINC00473 gene encodes an intergenic lncRNA from the chromosome 6q27 locus. LINC00473 consists of two exons and has two annotated transcript isoforms. Recently, Chen et al. showed that lncRNA LINC00473 was increased in human non-small cell lung cancer (NSCLC) and predicted poor prognosis. Moreover, they found that sustained LINC00473 expression was required for the growth and survival of LKB1-inactivated NSCLC cells by interacts with NONO, a component of the cAMP signaling pathway, thereby facilitating CRTC/CREB-mediated transcription [11]. However, neither the expression pattern nor the biological functions of LINC00473 have been elucidated in cervical cancer so far. Therefore, in the present study, we detected the expression of LINC00473 in human cervical cancer tissues and investigated the biological functions of LINC00473 in cervical cancer progression.
Materials and methods
Clinical samples
A total of 80 cervical cancer tissues and adjacent non-tumor tissues were obtained from Huai’an First People’s Hospital of Nanjing Medical University between 20010 and 2013. The clinical stage and histological diagnosis were identified on the basis of the International Federation of Gynecology and Obstetrics (FIGO) classification system. Follow-up information was collected every 3 months via telephone or by mail. This study was reviewed and approved by the Human Ethics Approval Committee of Huai’an First People’s Hospital of Nanjing Medical University. All patients signed informed consent.
Cell lines and culture conditions
Five cervical cancer cell lines, SiHa, HeLa, Caski, C4-1 and C-33a, were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and the American Type Culture Collection (ATCC; Manassas, VA, USA), respectively. All cell lines were cultured in RPMI-1640 (Gibco, Gaithersburg, MD, USA)medium supplemented with 10% fetal bovine serum (FBS) (Gibco, Gaithersburg, MD, USA). All the media contained 1% penicillin-streptomycin (100 U/ml penicillin and 100 µg/ml streptomycin).
Cell transfection
siRNAs that specifically target human LINC00473 or ILF2 were purchased from GenePharma (Shanghai, China). The complementary DNA (cDNA) of LINC00473 was chemically synthesized and cloned into the KpnI and BamHI sites of pcDNA expression vector (Invitrogen), namely, pcDNA-LINC00473. Cells were plated onto six-well plates and cultured for 24 h prior to transfection. Then, siRNAs or plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen). The cells were collected 48 h after transfection and applied for further functional analysis of target genes.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen). The RNA concentration and quality were determined by NanoDrop 2000 (Quawell, San Jose, CA, USA). Total RNA (1 µg) was used for first strand cDNA synthesis with a reverse transcription reaction using a reverse transcription kit (Takara, Dalian, China). The corresponding cDNA was used for quantitative real-time PCR using SYBR-Green Real-Time Master Mix (Takara). GAPDH was used as the internal control. The primers used for LINC00473 were: 5’-GGCAGCCTCAGGTTACAAAT-3’ (forward) and 5’-AGGAGCAGGTAGGGAAATGA-3’ (reverse); for GAPDH, 5’-CCCACTCCTCCACCTTTGAC-3’ (forward) and 5’-ATACCAGGAAATGAGCTTGACAA-3’ (reverse). The qRT-PCR analysis was performed on Applied Biosystems 7500 Sequence Detection System (ABI, Foster City, CA, USA). Data were analyzed using the 2-ΔΔCt method.
Western blotting
Total protein from tissues and cells were extracted using RIPA lysis buffer (Beyotime, Shanghai, China). Total protein (20 µg) was separated on SDS polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked and incubated with primary antibodies (ILF2; 1:1000; Abcam, Cambridge, MA, USA) (GAPDH; 1:2000; Abcam, Cambridge, MA, USA). Finally, the membranes were cultured with goat anti-rabbit IgG-HRP (sc2004; Santa Cruz, CA, USA) at a 1:5000 dilution. Proteins were analyzed by enhanced chemiluminescence (ECL) as described by the manufacturer’s instructions (Beyotime).
Cell proliferation assays
Transfected cervical cancer cells were seeded on a 96-well plate at a density of 2000 cells per well and incubated at 37°C. Proliferation was determined using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) kit (Keygen) at 24, 48, 72, and 96 h after transfection. The optical density (OD) was measured at 560 nm.
Cell apoptosis assay
Transfected cervical cancer cells were stained using an Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences). Then, cells were analyzed through BD FACS Canto II (BD Biosciences) and analyzed with BD FACSDiva software.
RNA immunoprecipitation
RNA immunoprecipitation (RIP) assay was conducted to determine whether LINC00473 interacts with ILF2 in cervical cancer cells using Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore). Briefly, the cells were lysed in RIP lysis buffer, and then incubated with magnetic beads-bound human anti-ILF2 antibody (Abcam) or control normal rabbit immunoglobulin G (IgG; Millipore). Afterward, the samples were incubated with proteinase K to digest protein and the immunoprecipitated RNA was isolated by QIAamp MinElut Virus Spin Kit (Qiagen, Hilden, Germany). Finally, purified RNA was analyzed by qRT-PCR.
RNA pull-down
RNA pull-down was performed as previously described [12]. In vitro biotin-labeled RNAs (LINC00473 and its antisense RNA) were transcribed with the biotin RNA labeling mix (Roche) and T7 RNA polymerase (Roche) treated with RNase-free DNase I (Promega) and purified with RNeasy Mini Kit (QIAGEN). Biotinylated RNA was incubated with nuclear extracts of breast cancer cells, and pull-down proteins were run on SDS-PAGE gels. Western blot followed.
Statistical analysis
All values were expressed as mean ± standard deviation (SD) from at least three independent experiments. GraphPad Prism V5.0 (GraphPad Software, Inc., La Jolla, CA, USA) software was used to determine statistical differences by using the Student’s t test or one-way analysis of variance (ANOVA). p<0.05 was considered to be statistically significant.
Results
Expression level of lncRNA LINC00473 in cervical cancer tissues and cell lines
To investigate the expression level of lncRNA LINC00473 in human cervical cancer tissues, qRT-PCR was performed to detect the expression of LINC00473 in 80 cervical cancer and matched adjacent para-carcinoma tissues. As shown in Figure 1A, LINC00473 was significantly upregulated in tumor tissues compared with matched adjacent para-carcinoma tissues. Furthermore, we also analyzed the expression of LINC00473 in five cervical cancer cell lines and the results revealed that SiHa cells showed the highest expression of LINC00473, while HeLa expressed the lowest level of LINC00473 expression (Figure 1B). Based on the results, the SiHa and HeLa cell lines were selected for further study.
Figure 1.
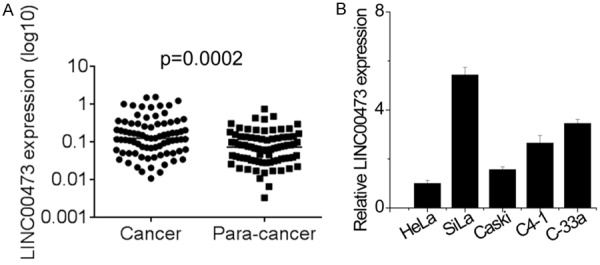
Expression level of lncRNA LINC00473 in cervical cancer tissues and cell lines. A. Expression level of lncRNA LINC00473 was detected in 80 pairs of cervical cancer tissues and matched para-cancer tissues using qRT-PCR. B. Expression level of lncRNA LINC00473 was detected in five cervical cancer cell using qRT-PCR.
Correlation of lncRNA LINC00473 expression with clinicopathological factors in cervical cancer patients
To investigate the relationship between LINC00473 expression and clinicopathological features in cervical cancer, 80 patients were divided into two groups according to the median expression level of LINC00473: a low LINC00473 expression group (n=40, LINC00473 expression ratio ≤ median ratio) and a high LINC00473 expression group (n=40, LINC00473 expression ratio ≥ median ratio). The correlation between LINC00473 expression and clinicopathological factors are shown in Table 1. High LINC00473 expression was observed to be associated with tumor size (P=0.001) and FIGO stage (P=0.007). In contrast, there was no association between LINC00473 expression with age (P=0.655), lymphatic (P=0.816) and distant metastasis (P=0.653). Furthermore, we used a Kaplan-Meier survival analysis to examine the correlation between LINC00473 expression and the prognosis of patients with cervical cancer. The results showed that patients with higher LINC00473 levels had shorter overall survival time than those with lower LINC00473 levels (Figure 2). These findings suggest that elevated LINC00473 may exert as an oncogene in cervical cancer.
Table 1.
The association between LINC00473 expression and clinicopathological factors in cervical cancer patients
| Clinical parameter | LINC00473 | P value | |
|---|---|---|---|
|
| |||
| High | Low | ||
| Age (years) | |||
| ≤40 | 19 | 21 | 0.655 |
| >40 | 21 | 19 | |
| Size (cm) | |||
| ≥4 | 25 | 10 | 0.001 |
| <4 | 15 | 30 | |
| FIGO stages | |||
| I-II | 15 | 27 | 0.007 |
| III-IV | 25 | 13 | |
| Lymphatic metastasis | |||
| Yes | 25 | 26 | 0.816 |
| No | 15 | 14 | |
| Distant metastasis | |||
| Yes | 21 | 23 | 0.653 |
| No | 19 | 17 | |
P value was acquired by Pearson chi-square test. The median expression level was used as the cutoff.
Figure 2.
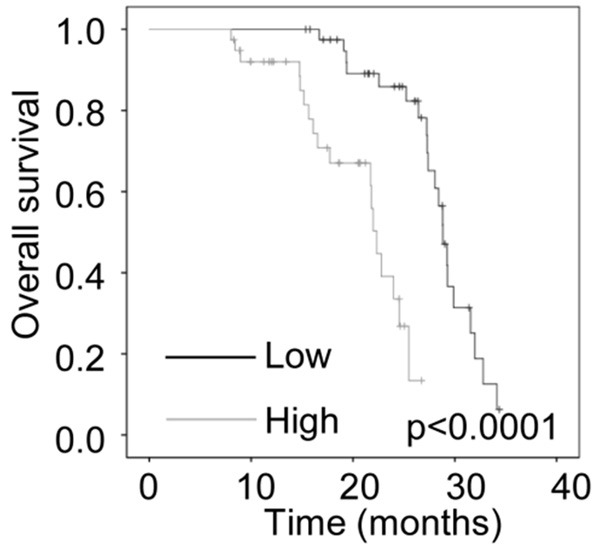
The correlation between LINC00473 and prognosis of patients with cervical cancer. Kaplan-Meier survival curve and log-rank test were used to evaluate the association of LINC00473 expression with overall survival rate. Patients were segregated into LINC00473-high group and LINC00473-low according to the median of LINC00473 expression in cervical cancer tissues.
Effects of LINC00473 on cervical cancer cell proliferation
To evaluate the role of lncRNA LINC00473 in cervical cancer cell proliferation, the expression of LINC00473 was silenced or overexpressed by siRNA or LINC00473 overexpression vector transfection, respectively. The expression of LINC00473 was decreased in the SiHa cells after transfection with LINC00473 siRNA (Figure 3A) and increased in HeLa cells after transfection with LINC00473 overexpression vectors (Figure 3B). MTT and colony formation assays revealed that depletion of LINC00473 expression suppressed the proliferation of the SiHa cells, while overexpression of LINC00473 enhanced the proliferative ability of HeLa cells (Figure 3C-F).
Figure 3.
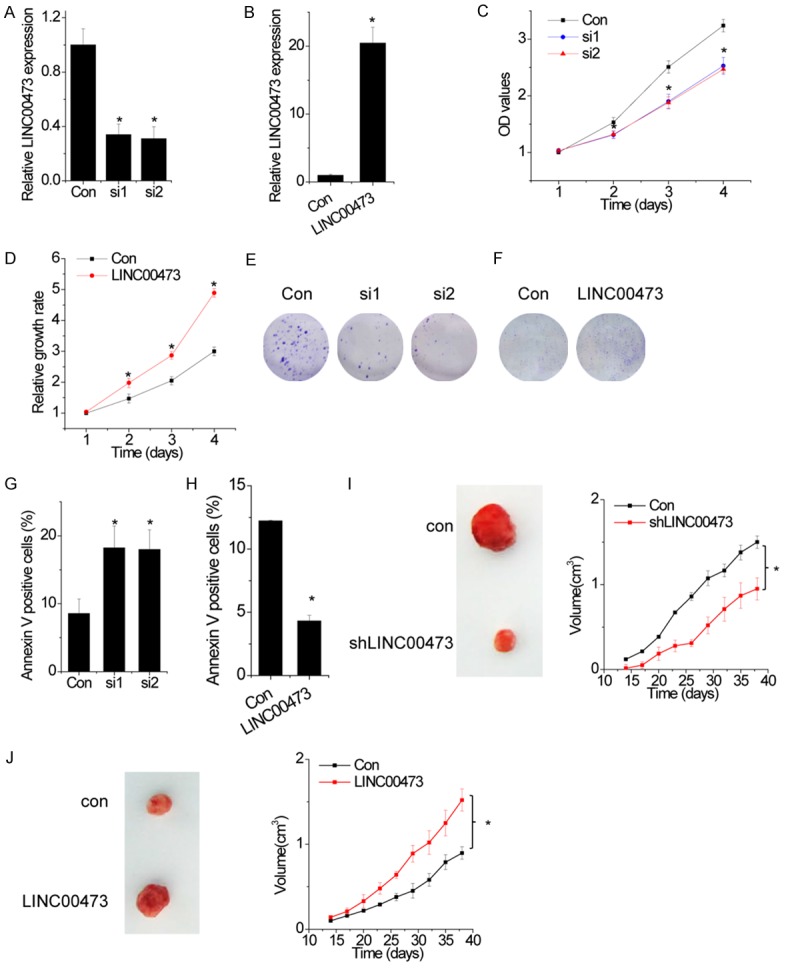
Effects of LINC00473 on cervical cancer cell proliferation. A. SiHa cells were transfected with siRNAs against LINC00473. The relative expression level of LINC00473 was detected by qRT-PCR. B. HeLa cells were transfected with plasmid expressing LINC00473. The relative expression level of LINC00473 was detected by qRT-PCR. C. The cell growth rates were determined by performing MTT assay. Knockdown of LINC00473 expression in SiHa cells significantly suppressed cell proliferation, relative to control cells. D. The cell growth rates were detected by performing MTT assay. Overexpression of LINC00473 in HeLa cells significantly enhanced cell proliferation, relative to control cells. E. Representative images of colony formation induced by the LINC00473 silenced cells. F. Representative images of colony formation induced by the LINC00473 overexpressed cells. G. SiHa cells with LINC00473 downregulation were stained with a combination of annexin V and 7-AAD and analyzed by FACS. Cells positive for annexin V staining were counted as apoptotic cells, and the percentage of apoptotic cells is shown. H. HeLa cells with LINC00473 upregulation were stained with a combination of annexin V and 7-AAD and analyzed by FACS. Cells positive for annexin V staining were counted as apoptotic cells, and the percentage of apoptotic cells is shown. I. Effects of LINC00473 downregulation on tumor growth in vivo. The tumor growth curves were shown. J. Effects of LINC00473 overexpression on tumor growth in vivo. The tumor growth curves were shown. Data are shown as mean ± SD; *p<0.05.
We further analyzed cell cycle distribution and apoptosis using flow cytometry in above transfected cells. The results showed that knockdown of LINC00473 had a significantly higher percentage of Annexin V-positive cells than control cells (Figure 3G and 3H). In contrast, we observed that overexpression of LINC00473 had protective effects to apoptosis in HeLa cells. However, neither knockdown nor overexpression of LINC00473 influenced the cell cycle distribution (Data not shown). Together, these data demonstrate that LINC00473 promotes cell proliferation and inhibits cell apoptosis.
Based on the above findings that LINC00473 promoted cell proliferation in cervical cancer, we investigated the effects of LINC00473 on cancer growth in vivo. The mean volumes of xenograft tumors generated from LINC00473-knockdown SiHa cells were lower than those of tumors generated from control cells (Figure 3I). In contrast, tumors generated from LINC00473-overexpressing HeLa cells were larger than those generated from control cells (Figure 3J). These results highlight the important role of LINC00473 in cervical cancer growth.
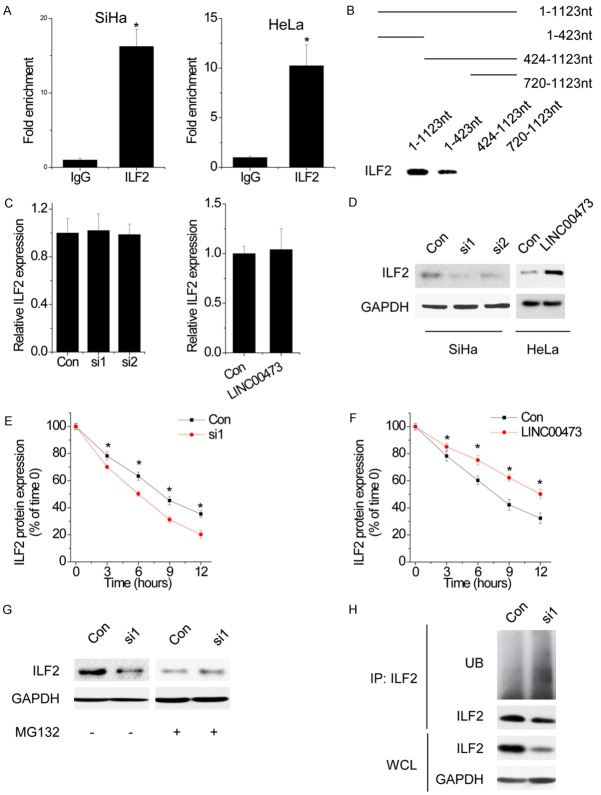
LINC00473 interacts with ILF2 and inhibits ILF2 degradation
LncRNAs might function to regulate multiple pathways via their interactions with miRNAs, mRNAs and proteins. We performed RNA pull-down and mass spectrometry analysis to search for potential LINC00473-associated proteins. Among these proteins, interleukin enhancer binding factor 2 (ILF2) was of interest because it has been reported to exert oncogenic function in cancers [13,14]. For confirmation, we detected the association between LINC00473 and ILF2 by performing RIP assay. The results showed that LINC00473 was significantly enriched by ILF2 antibody than the nonspecific IgG control antibody (Figure 4A). To further validate the association and determine the specific binding region between LINC00473 and ILF2, we performed RNA pull-down assay and deletion-mapping experiments. We found a 423 nt region at the 5’ end of LINC00473 required for the association with ILF2 (Figure 4B). Taken together, we demonstrated a specific association between LINC00473 and ILF2.
Figure 4.
LINC00473 interacts with ILF2 and inhibits ILF2 degradation. A. The interaction of LINC00473 with ILF2 was confirmed by an RNA immunoprecipitation (RIP) assay. B. Deletion mapping analysis of ILF2-binding domains of LINC00473. Shown are the following: schematic diagram of LINC00473 full-length and truncated fragments (top panel) and western blot of ILF2 in RNA pull-down samples by different LINC00473 fragments (bottom panel). C. The ILF2 mRNA expression level in control and LINC00473 silencing or overexpressing cells. D. The ILF2 protein expression level in control and LINC00473-silencing or-overexpressing cells. E. The stability of ILF2 protein over time was measured by western blot relative to time 0 after blocking new protein synthesis with CHX in control and LINC00473-silencing SiHa cells. F. The stability of ILF2 protein over time was measured by western blot relative to time 0 after blocking new protein synthesis with CHX in control and LINC00473-overexpressing HeLa cells. G. ILF2 protein expression in control and LINC00473-silencing SiHa cells treated with vehicle control or MG132. H. SiHa cells transfected with control siRNA or LINC00473 siRNA were cultured for 48 h. Cell lysates were immunoprecipitated with anti-ILF2 antibody, and the immunocomplexes were immunoblotted with antibodies against UB and ILF2. Data are shown as mean ± SD; *p<0.05.
Next, we determined the functional relationship between of LINC00473 and ILF2. Overexpression or knockdown of LINC00473 have no effect on ILF2 mRNA (Figure 4C). However, silence of LINC00473 significantly increased the protein level of ILF2 in SiHa, and overexpression of LINC00473 showed the opposite effect in HeLa cells (Figure 4D). These results suggested that the association of LINC00473 and ILF2 may influence the stability of ILF2 protein. To further confirm the LINC00473-mediated ILF2 regulation, we treated control and LINC00473-silencing SiHa cells with cycloheximide (CHX) and analyzed the stability of ILF2. We found that the half-life of ILF2 was much shorter in LINC00473-silencing cells than in control cells, whereas LINC00473 overexpression elongated the half-life of ILF2 (Figure 4E and 4F). In agreement with this observation, when MG132 was added into the culture medium to inhibit proteasome degradation, the endogenous ILF2 protein expression in LINC00473 knockdown cells was significantly increased and reached a level that was comparable to that in control cells (Figure 4G), and higher ILF2 ubiquitination levels were also observed in LINC00473 knockdown cells treated with MG132 (Figure 4H). Taken together, these data suggested that LINC00473 is important for the ILF2 protein stability.
ILF2 is critical for the function of LINC00473 in cervical cancer
Next, we investigated the role of ILF2 in cervical cancer cells. We found that knockdown of ILF2 significantly suppressed cell proliferation and induced apoptosis in SiHa cells (Figure 5A-C). On the contrary, upregulation of ILF2 enhanced the proliferative rate and inhibited apoptosis in HeLa cells (Figure 5D-F). These data suggest that ILF2-mediated cell phenotypes was similar to LINC00473.
Figure 5.
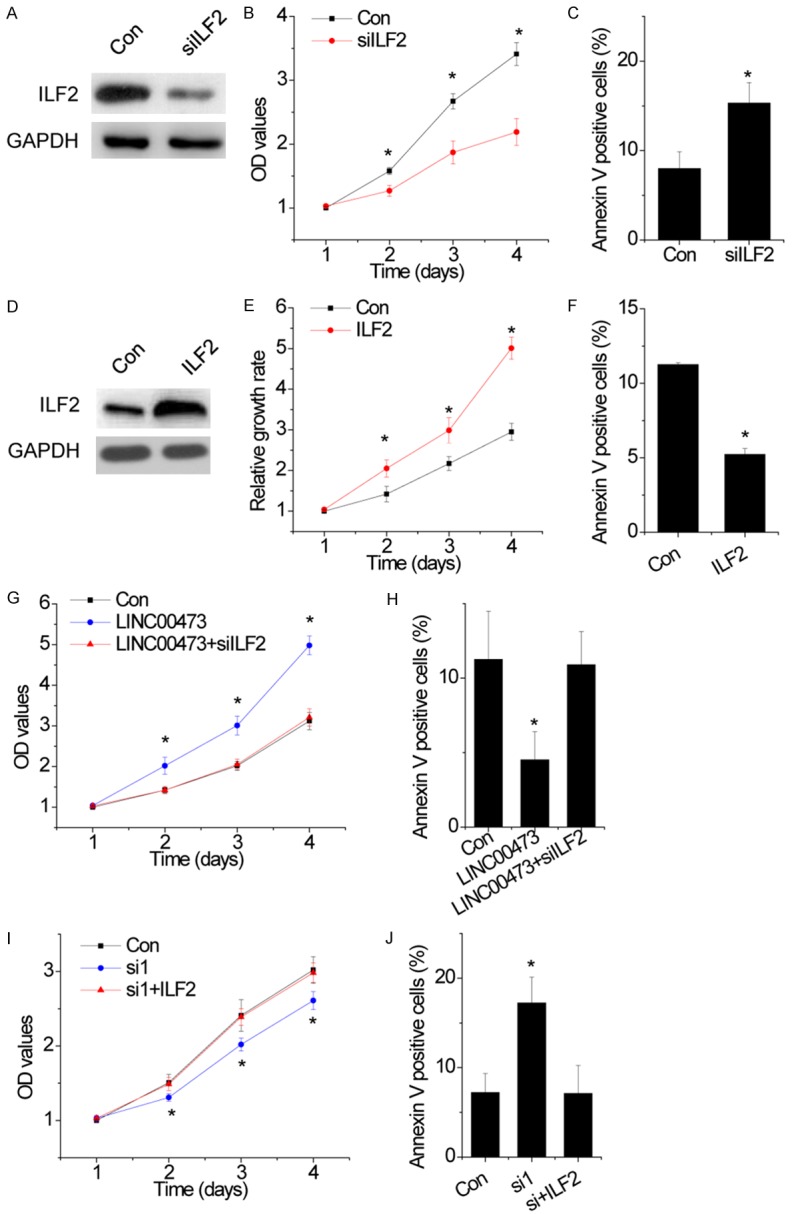
ILF2 is critical for the function of LINC00473 in cervical cancer. A. The protein level of ILF2 in control and ILF2 silenced SiHa cells. B. The cell growth rates were determined by performing MTT assay. Knockdown of ILF2 expression in SiHa cells significantly suppressed cell proliferation, relative to control cells. C. SiHa cells with ILF2 downregulation were stained with a combination of annexin V and 7-AAD and analyzed by FACS. Cells positive for annexin V staining were counted as apoptotic cells, and the percentage of apoptotic cells is shown. D. The protein level of ILF2 in control and ILF2 overexpressed HeLa cells. E. The cell growth rates were detected by performing MTT assay. Overexpression of ILF2 in HeLa cells significantly enhanced cell proliferation, relative to control cells. F. HeLa cells with ILF2 upregulation were stained with a combination of annexin V and 7-AAD and analyzed by FACS. Cells positive for annexin V staining were counted as apoptotic cells, and the percentage of apoptotic cells is shown. G. Knockdown of IFL2 abolished the proliferation increased by LINC00473 overexpression in HeLa cells. H. Knockdown of IFL2 abolished the apoptosis reduced by LINC00473 overexpression in HeLa cells. I. Upregulation of ILF2 resecued the proliferation decreased by LINC00473 knockdown in SiHa cells. J. Upregulation of ILF2 resecued the apoptosis increased by LINC00473 knockdown in SiHa cells. Data are shown as mean ± SD; *p<0.05.
Finally, we determined whether IIF2 was essential for LINC00473-induced cell proliferation. Downregulation of ILF2 almost abolished the enhancement of proliferation and inhibition of apoptosis mediated by LNC00473 overexpression in HeLa cells (Figure 5G and 5H). In contrast, restoring expression of ILF2 rescued the suppression of proliferation and promotion of cell apoptosis induced by LNC00473 knockdown (Figure 5I and 5J). Our data demonstrate that ILF2 is critical for the effect of LINC00473 on cervical cancer cells.
LINC00473 is a direct target of miR-34a
Interactions between lncRNAs and microRNAs (miRNAs), which are important classes of non-coding RNAs in eukaryotes, provide an additional layer of control in gene regulation. Using Microinspector software, we found a set of miRNAs that putatively bind to LINC00473. Among these miRNA candidates, we found that miR-34a directly binds to LINC00473. Dual-luciferase assays showed a significant decrease in luciferase activities following cotransfection of miR-34a and the wild type (WT) LINC00473 expression vector, but not a mutant LINC00473 (MUT, mutant in miR-34a binding site) (Figure 6A). We further clarified the regulatory relationship between LINC00473 and miR-34a. Overexpression of miR-34a significantly inhibited LINC00473 expression (Figure 6B), whereas overexpression of LINC00473 did not affect miR-34a expression, suggesting that LINC00473 is targeted by miR-34a (Figure 6C). Overexpression of miR-34a also decreased the LINC00473 half-life (Figure 6D). In addition, miR-34a overexpression significantly suppressed cellular proliferation in LINC00473-overexpressing cells (Figure 6D). Collectively, these data demonstrated that miR-34a directly binds to LINC00473 and serves as a negative upstream regulator of LINC00473-mediated proliferation.
Figure 6.
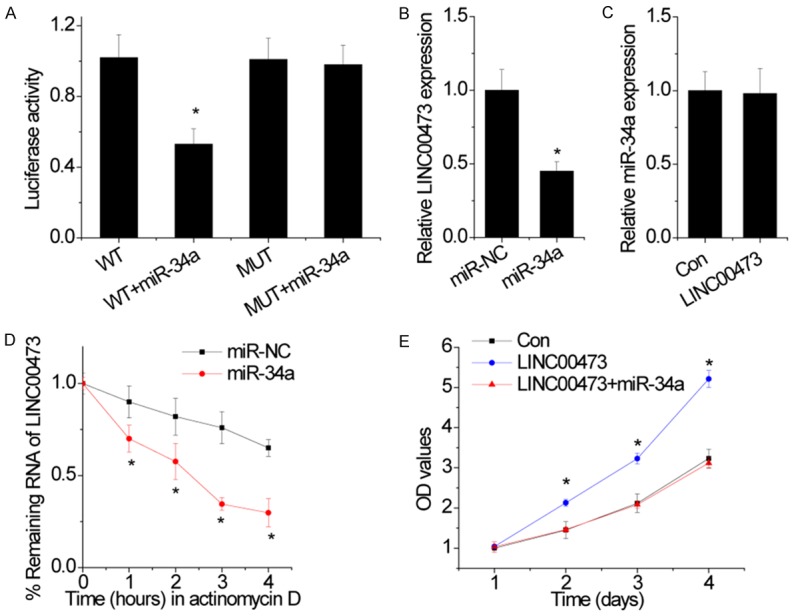
LINC00473 is a direct target of miR-34a. A. Dual-luciferase assays showed a decrease in reporter activity following cotransfection of wild-type LINC00473 (WT) and miR-34a in cells, whereas the cotransfection of mutant LINC00473 (MUT) and miR-34a had no effect on reporter activity. B. The effect of miR-34a on LINC00473 expression was detected by qRT-PCR. C. The effect of LINC00473 on miR-34a expression was detected by qRT-PCR. D. The overexpression of miR-34a shortened the half-life of LINC00473. E. miR-34a abolished the proliferation increased by LINC00473 overexpression, as confirmed in MTT assays. Data are shown as mean ± SD; *p<0.05.
Discussion
In the present study, for the first time, we demonstrated that LINC00473 was increased in cervical cancer. Upreguation of LINC00473 was associated with bigger tumor size, higher FIGO stage and poorer prognosis of patients with cervical cancer. Functional assays showed that LINC00473 promoted cell proliferation and inhibited cell apoptosis. These findings suggest that LINC00473 functions as an oncogene in cervical cancer. An increasing number of studies have demonstrated that the study of post-transcriptional regulation of lncRNAs is emerging as a research field. Consequently, it was discovered that lncRNAs might function as possible scaffold transcripts that enhance the E3-mediated ubiquitination of substrate proteins [15]. Here, we demonstrated that the lncRNA LINC00473 could promote the proliferative ability of cervical cancer cells via the regulation of ILF2 protein degradation. These findings revealed an important role of LINC00473 in the proliferation of cervical caner cells and uncovered a novel function of LINC00473 as a mediator that inhibits the ubiquitination of ILF2 in cervical caner.
ILF2, which also known as nuclear factor 45 (NF-45), a subunit of nuclear factor of activated T cells (NF-AT), is encoded by a gene located on human chromosome 1 (1q21.3) [16]. It can be expressed in normal tissues such as testis, brain, and kidney and is primarily distributed in the nucleus [17]. ILF2 is a transcription factor that interacts with ILF3 (NF90) to regulate the expression of IL-2 gene and HS4-dependent IL-13 gene at the antigen receptor response element (ARRE)/nuclear factor of activated T-cells (NFAT) DNA target sequence [18]. ILF2 regulates gene expression at multiple levels including RNA transcription, processing, and translation [19,20]. Recently, a great deal of studies have indicated that high expression of ILF2, which was observed in cervical cancer, non-small-cell lung cancer, hepatocellular carcinoma, and esophageal squamous cell cancer, was significantly related to the poor prognosis of these malignant tumors [14]. Cells deficient in ILF2 exhibit reduced internal ribosome entry site (IRES)-mediated translation of X-linked inhibitor of apoptosis protein (XIAP) and cellular inhibitor of apoptosis protein 1 (cIAP1) [21-24]. Although the regulation and function of ILF2 have been extensively investigated, the biological functions of ILF2 and regulatory mechanisms of ILF2 expression in cervical cancer have not been fully understood. Our study demonstrated that ILF2 also functioned as oncogene, which promoted cell proliferation in cervical cancer. It has been reported that miR-7 negatively regulated ILF2 expression in a post-transcriptional manner [25]. Our findings revealed a novel regulatory mechanism mediated by a lncRNA. RIP and RNA pull-down assays showed that LINC00473 directly interacted with ILF2. LINC00473 did not influence the mRNA level of ILF2, while could increase ILF2 protein level, suggesting that LINC00473 regulated ILF2 expression in a post-translational manner. Furthermore, our results showed that LINC00473 significantly elongated the half-life of ILF2 protein.
Both lncRNAs and miRNAs play critical roles in transcriptional regulation, and are involved in many human cancers. Interactions between lncRNAs and miRNAs have been reported recently. For example, CHRF serves as an endogenous ‘sponge’ of miR-489 to regulate Myd88 expression and hypertrophy [26], and miR-200 family targets lncRNA-ATB [27]. Here, we also present strong evidence that LINC00473 is silenced by miR-34a in cervical cancer cells.
In conclusion. we found that the lncRNA LINC00473, a target of miR-34a, increased the proliferation of cervical cancer cells by regulating the stability of ILF2. These findings further define the importance of lncRNAs in tumor progression and suggest that LINC00473 may be a potential therapeutic target for cervical cancer.
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Jia HL, Huang JM, Liang YC, Tan H, Geng HZ, Guo LY, Yao SZ. Identification of biomarkers for lymph node metastasis in earlystage cervical cancer by tissue-based proteomics. Br J Cancer. 2014;110:1748–1758. doi: 10.1038/bjc.2014.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedroza-Torres A, Fernández-Retana J, Peralta-Zaragoza O, Jacobo-Herrera N, Cantú de Leon D, Cerna-Cortés JF, Lopez-Camarillo C, Pérez-Plasencia C. A microRNA expression signature for clinical response in locally advanced cervical cancer. Gynecol Oncol. 2016;142:557–565. doi: 10.1016/j.ygyno.2016.07.093. [DOI] [PubMed] [Google Scholar]
- 5.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127:761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu JJ, Wang Y, Ding JX, Jin HY, Yang G, Hua KQ. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res. 2015;333:238–248. doi: 10.1016/j.yexcr.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol Ther. 2016;17:104–113. doi: 10.1080/15384047.2015.1108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Li JL, Lin S, Cao C, Gimbrone NT, Yang R, Fu DA, Carper MB, Haura EB, Schabath MB, Lu J, Amelio AL, Cress WD, Kaye FJ, Wu L. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest. 2016;126:2267–2279. doi: 10.1172/JCI85250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, Jean S, Li C, Huang Q, Katsaros D, Montone KT, Tanyi JL, Lu Y, Boyd J, Nathanson KL, Li H, Mills GB, Zhang L. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin ZH, Jiang XW, Shi WB, Gui QL, Yu DF. Expression and Clinical Significance of ILF2 in Gastric Cancer. Dis Markers. 2017;2017:4387081. doi: 10.1155/2017/4387081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni T, Mao G, Xue Q, Liu Y, Chen B, Cui X, Lv L, Jia L, Wang Y, Ji L. Upregulated expression of ILF2 in non-small cell lung cancer is associated with tumor cell proliferation and poor prognosis. J Mol Histol. 2015;46:325–335. doi: 10.1007/s10735-015-9624-5. [DOI] [PubMed] [Google Scholar]
- 15.Dong Y, Yoshitomi T, Hu JF, Cui J. Long noncoding RNAs coordinate functions between mitochondria and the nucleus. Epigenetics Chromatin. 2017;10:41. doi: 10.1186/s13072-017-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcoulatos P, Avgerinos E, Tsantzalos DV, Vamvakopoulos NC. Mapping interleukin enhancer binding factor 3 gene (ILF3) to human chromosome 19 (19q11-qter and 19p11-p13.1) by polymerase chain reaction amplification of human-rodent somatic cell hybrid DNA templates. J Interferon Cytokine Res. 1998;18:351–355. doi: 10.1089/jir.1998.18.351. [DOI] [PubMed] [Google Scholar]
- 17.Zhao G, Shi L, Qiu D, Hu H, Kao PN. NF45/ILF2 tissue expression, promoter analysis, and interleukin-2 transactivating function. Exp Cell Res. 2005;305:312–323. doi: 10.1016/j.yexcr.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 18.Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ, Corthesy B. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem. 1994;269:20691–20699. [PubMed] [Google Scholar]
- 19.Guan D, Altan-Bonnet N, Parrott AM, Arrigo CJ, Li Q, Khaleduzzaman M, Li H, Lee CG, Pe’ery T, Mathews MB. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol Cell Biol. 2008;28:4629–4641. doi: 10.1128/MCB.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiesler P, Haynes PA, Shi L, Kao PN, Wysocki VH, Vercelli D. NF45 and NF90 regulate HS4-dependent interleukin-13 transcription in T cells. J Biol Chem. 2010;285:8256–8267. doi: 10.1074/jbc.M109.041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faye MD, Graber TE, Liu P, Thakor N, Baird SD, Durie D, Holcik M. Nucleotide composition of cellular internal ribosome entry sites defines dependence on NF45 and predicts a posttranscriptional mitotic regulon. Mol Cell Biol. 2013;33:307–318. doi: 10.1128/MCB.00546-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graber TE, Baird SD, Kao PN, Mathews MB, Holcik M. NF45 functions as an IRES transacting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death Differ. 2010;17:719–729. doi: 10.1038/cdd.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni S, Zhu J, Zhang J, Zhang S, Li M, Ni R, Liu J, Qiu H, Chen W, Wang H, Guo W. Expression and clinical role of NF45 as a novel cell cycle protein in esophageal squamous cell carcinoma (ESCC) Tumour Biol. 2015;36:747–756. doi: 10.1007/s13277-014-2683-5. [DOI] [PubMed] [Google Scholar]
- 24.Cheng S, Jiang X, Ding C, Du C, Owusu-Ansah KG, Weng X, Hu W, Peng C, Lv Z, Tong R, Xiao H, Xie H, Zhou L, Wu J, Zheng S. Expression and critical role of interleukin enhancer binding factor 2 in hepatocellular carcinoma. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17081373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bi Y, Shen W, Min M, Liu Y. MicroRNA-7 functions as a tumor-suppressor gene by regulating ILF2 in pancreatic carcinoma. Int J Mol Med. 2017;39:900–906. doi: 10.3892/ijmm.2017.2894. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Wu Q, Han L, Yan W, Ji X, Han R, Yang J, Yuan J, Ni C. miR-489 inhibits silica-induced pulmonary fibrosis by targeting MyD88 and Smad3 and is negatively regulated by lncRNA CHRF. Sci Rep. 2016;6:30921. doi: 10.1038/srep30921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]