Abstract
Liquid biopsy is gaining significant attention as a tool for unveiling the molecular landscape of tumor and holds great promise for individualized medicine for cancer. Cell-free DNA serves as an extremely important component of liquid biopsy for cancer, and cell-free DNA in urine is even promising due to the remarkable advantage of urine as an ultra-noninvasive sample source over tissue and blood. Compared with the widely studied cell-free DNA in blood, less is known about the role of urinary cell-free DNA. Urinary cell-free DNA has the ability to give comprehensive and crucial information on cancer as it carries genetic messages from cells shedding directly into urine as well as transporting from circulation. As an indispensable component of liquid biopsy, urinary cell-free DNA is believed to have the potential of being a useful and ultra-noninvasive tool for cancer screening, diagnosis, prognosis, and monitoring of cancer progression and therapeutic effect. In this review, we provide the current insights into the clinical applications of urinary cell-free DNA in cancer. We also introduce the basic biological significance and some technical issues in the detection of urinary cell-free DNA.
Keywords: Liquid biopsy, urinary cell-free DNA, cancer, clinical application
Introduction
Cancer is a serious public health threat worldwide [1]. Diagnosis and treatment towards cancer mainly depend on the genomic profiles of tumors [2]. Since molecular profiling based on tissue samples is limited by tumor heterogeneity and difficulties of repeated sampling, liquid biopsy based on various circulating molecules emerges at the right moment [3-6]. Liquid biopsy is gaining significant attention as a tool for unveiling the molecular landscape of tumor and holds great promise for individualized medicine for cancer [7,8]. Cell-free DNA (cfDNA) exists as fragmented nucleic acids in various extracellular body fluids in both healthy individuals and people with diseases, serving as an extremely important tool of liquid biopsy [9]. In recent years, cfDNA in blood circulation has become a topic of interest in the field of cancer, and developed as novel biomarkers in cancer clinical management [3,10,11].
Apart from blood, cfDNA could also be detected in other body fluids, the most intriguing among which is urine. The presence of genetic materials in urine has long been established, though initially from urinary cells [12-15]. CfDNA in urine is later discovered with the ability to give comprehensive and crucial information on cancer as it carries genetic messages releasing from cells shedding directly into urine as well as transporting from circulation. Genomic aberrations detected in urinary cell-free DNA (ucfDNA) have been demonstrated to be the same as those in primary tumors. Therefore, urine could be a novel source of genetic material that reflects the genomic aberrations of cancers [16]. More importantly, urine has a remarkable advantage as an ultra-noninvasive sample source over tissue and blood [16], especially for patients who need repeated sampling to monitor cancer progression and therapeutic effect. Furthermore, unlike serum or plasma collection which requires specialized facility or equipment, the collection of urine that requires only sterile collection containers is implementable even in remote areas and is relatively time and cost efficient. As urine contains lower levels of protein than blood, the isolation of DNA fragments could therefore be technically easier [17].
Compared with the widely studied cfDNA in blood, less is known about the role of ucfDNA. UcfDNA is an important component of liquid biopsy in clinical oncology, containing a wide range of genetic information. More and more attention has been paid for the role of ucfDNA as a novel ultra-noninvasive biomarker in the clinical management of both urological and non-urological cancers, exploring the potential clinical utility of ucfDNA in screening, diagnosis, surveillance and prognosis. In this review, we put an emphasis on the clinical applications of ucfDNA in cancer. At the same time, the origin and biological characteristics of ucfDNA, as well as some important technical issues in the process of ucfDNA detection are also discussed.
Origin and characteristics of ucfDNA
The origins of ucfDNA are shown in Figure 1. UcfDNA originates either from cells shedding into urine from genitourinary tract, or from cell-free DNA in circulation passing through glomerular filtration. The latter is also known as “transrenal DNA” [18,19].
Figure 1.
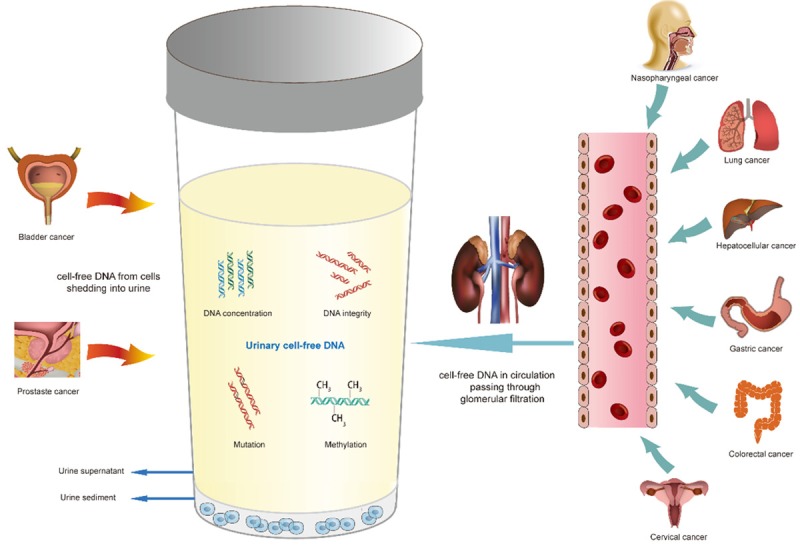
Origins of urinary cell-free DNA. Urinary cell-free DNA originates either from cells shedding into urine from genitourinary tract, or from cell-free DNA in circulation passing through glomerular filtration. Genetic alterations including DNA concentrations, integrity, mutations and methylation status are examined to evaluate the potential clinical utility of urinary cell-free DNA for both urological and non-urological cancers.
UcfDNA fragments can be classified into two categories according to their size: high-molecular-weight DNA and low-molecular-weight DNA [17,18,20]. High-molecular-weight ucfDNA fragments are usually 1 kbp or longer, deriving from the necrotic cells along the urogenital tract falling into urine or from lymphocytes that normally exist in urine [17,21]. DNA fragments up to 1.3 kbp and 19 kbp were found in male and female urine, respectively [20]. Low-molecular-weight DNA can originate either from circulation or apoptotic cells in contact with urine [16,17,22]. The size of low-molecular-weight DNA fragments in urine has been evaluated in various studies. Su et al. recovered DNA fragments between 150 and 250 bp from urine supernatant [17]. Another study drew the similar conclusion that the main part of ucfDNA ranged from 150 to 400 bp [20]. In 2008, Melkonyan et al. improved the isolation and detection methods of ucfDNA, and observed a group of shorter ucfDNA fragments of 10-150 bp, in addition to the previously discovered 150-200 bp DNA fragments [23]. Furthermore, a recent study evaluating fetal DNA in the urine of pregnant women revealed that fetal ucfDNA fragments were even shorter, with a peak of 29 to 45 bp in length [24].
UcfDNA in urological diseases has been studied more extensively, since the majority of ucfDNA originates from apoptosis or necrosis of cells exfoliated from urogenital system. It has been estimated that more than 3 × 106 epithelial cells from urogenital tract can fall into urine within one day under normal conditions [22] and may partially undergo apoptosis to release DNA fragments into urine [18]. Urological diseases, especially tumors, which can be in direct contact with urine, can also release tumor cells into urine. Genetic information in ucfDNA has therefore been widely studied in urological cancers, predominantly in bladder cancers [25] and prostate cancers [26]. Donor-derived DNA sequences were detected for the first time in 1999, in the urine of renal transplantation recipients [27]. Subsequent research uncovered that graft-derived ucfDNA in transplant recipients increased significantly during acute rejection and returned to normal after anti-rejection therapy [28,29].
Transrenal DNA, another indispensable source of ucfDNA, is gaining significant attention in recent years. Originating from blood circulation, transrenal DNA contains important genomic information from various positions all over the body. The existence of transrenal DNA has been verified in various studies. In 2000, Botezatu et al. confirmed, for the first time, from several experiments on animals and humans, the existence of ucfDNA and its ability to traverse the kidney barrier from blood [18]. Purified 32P-labeled DNA was injected subcutaneously into the peritoneal cavity of mice, and 150-160 bp fragments of this labeled DNA were found in the urine of mice within 3 days [18]. Human Raji cells were also injected into mice and urinary DNA was isolated for PCR analysis. The results showed that human-specific Alu sequences were detected in mice urine [18]. They also detected Y chromosome-specific DNA sequences in the urine of women pregnant with male fetuses and females transfused with male blood [18]. Subsequent studies also confirmed the presence of fetal cell-free DNA in maternal urine [30-32]. KRAS mutations were discovered in ucfDNA in the urine of patients with colorectal [17,18,21,33] and pancreatic cancer [18]. UcfDNA has also been evaluated in various kinds of non-urological cancers as a potential biomarker for diagnosis, treatment monitoring, and prognosis [3,16,34-36].
Some technical issues in isolation and detection of ucfDNA
Isolation and detection of ucfDNA are of vital importance in the study of clinical value of ucfDNA in cancer. The reliability of clinical application of ucfDNA is, to a great extent, dependent on the sensitivity and reproducibility of assays in the process of isolation and detection of ucfDNA [19]. Since there are no standard protocols for isolation and detection of cfDNA in urine, multiple methods have been introduced in research papers. The concentration of cfDNA in urine is relatively low, and the fragments of ucfDNA are mostly short [23]. The development of techniques mainly focused on improving the isolation and detection sensitivity of short DNA fragments in urine [19].
CfDNA in urine could be isolated using either commercial kits or classical laboratory techniques [16,17]. Since circulating DNA fragments are usually short, the choice of commercial DNA isolation kits that are able to isolate low-molecular-weight DNA fragments should be taken into consideration. As for the quantification of ucfDNA, the most commonly used approaches include the spectrophotometric method, the fluorimetric method, as well as the amplification method [37].
All this time, methods for detection of genomic alterations in ucfDNA are mainly PCR-based assays [38,39]. To gain better detection sensitivity, short amplicons are designed and applied in both conventional and real-time PCR assays [23]. More recently, the rapid development of new molecular assays such as droplet digital PCR (ddPCR) technology and the next-generation sequencing (NGS) has largely improved the sensitivity of ucfDNA detection [40,41]. These newly developed assays have been proven to offer a deeper insight into the clinical utility of ucfDNA in cancer [42].
Clinical applications of ucfDNA in cancer
UcfDNA analysis holds promise in the clinical management of cancer. Potential applications of ucfDNA analysis include cancer detection and diagnosis, surveillance of tumor progression, monitoring of treatment response and predicting tumor prognosis (Figure 2). A summary of literature focusing on the potential clinical applications of ucfDNA in cancer is shown in Table 1.
Figure 2.
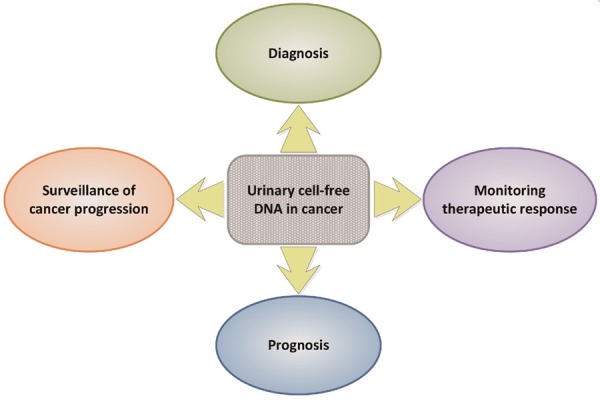
Clinical applications of urinary cell-free DNA in cancer.
Table 1.
Summary of potential clinical applications of ucfDNA in cancer
| Clinical application | Markers | Types of cancer | Detection methods | References |
|---|---|---|---|---|
| Detection/Diagnosis | UcfDNA concentration | Bladder cancer | GeneQuant Pro | Zancan et al. [54] |
| Quant-iT DNA high-sensitivity assay kit | ||||
| Real-time PCR | ||||
| NanoDrop 1000 | ||||
| UcfDNA/UCr concentration and ucfDNA integrity | Bladder cancer | PicoGreen 400-bp real-time PCR | Chang et al. [25] | |
| UcfDNA quantification | Bladder cancer | Real-time PCR | Brisuda et al. [55] | |
| TopoIIA levels | NMIBC | Real-time PCR | Kim et al. [60] | |
| UcfDNA integrity | Bladder cancer | Real-time PCR using IQ SYBR Green | Casadio et al. [38] | |
| Six microsatellite markers on chromosomes 4, 9, and 17 | Bladder cancer | PCR | Utting et al. [66] | |
| Twelve microsatellite markers on 6 chromosomes | Bladder cancer | PCR | Szarvas et al. [13] | |
| TSPAN13-to-S100A9 ratio | Prostate cancer | Real-time PCR | Yan et al. [68] | |
| UcfDNA integrity (c-Myc, BCAS1, and HER2) | Prostate cancer | Real-time PCR | Casadio et al. [39] | |
| UcfDNA integrity (c-MYC, HER2, and AR) | Prostate cancer | Real-time PCR | Salvi et al. [69] | |
| GSTP1 gene promoter hypermethylation | Prostate cancer | Methylation-specific PCR | Bryzgunova et al. [72] | |
| KRAS mutations | Advanced colorectal adenocarcinoma and advanced pancreatic cancer | PCR | Botezatu et al. [18] | |
| KRAS mutations | Colorectal cancer | Restriction-enriched PCR | Su et al. [17,33,35] | |
| mVIM | Colorectal cancer | Quantitative MethyLight PCR-based assay | Song et al. [73] | |
| TP53 mutation | Hepatocellular carcinoma | Locked nucleic acid clamp-mediated PCR assay | Lin et al. [74] | |
| HCC-associated HBV mutation | HBV-associated hepatocellular carcinoma | Real-time PCR | Lin et al. [75] | |
| HPV DNA | Cervical cancer | NGS | Guerrero-Preston et al. [80] | |
| Surveillance of cancer progression | Somatic variants | Bladder cancer | ddPCR | Birkenkamp-Demtröder et al. [81] |
| Somatic variants | UBC | OncoScan assay | Togneri et al. [82] | |
| FGFR3 and PIK3CA mutations | Bladder cancer | ddPCR | Christensen et al. [83] | |
| EGFR mutations | NSCLC | ddPCR | Li et al. [88] | |
| KRAS mutations | NSCLC | ddPCR | Wang et al. [85] | |
| Monitoring treatment response | Copy number variations | Prostate cancer | Whole genome sequencing | Xia et al. [86] |
| EGFR mutations | NSCLC | Short footprint mutation enrichment NGS | Reckamp et al. [40] | |
| EGFR mutations | NSCLC | ddPCR | Li et al. [84] | |
| Chen et al. [89] | ||||
| Husain et al. [96] | ||||
| Tchekmedyian et al. [91] | ||||
| EGFR mutations | Gastric cancer | ddPCR | Shi et al. [36] | |
| CAD-ALK gene rearrangement | Colorectal cancer | NGS | Siravegna et al. [92] | |
| BRAF V600E mutations | Colorectal neuroendocrine cancer | PCR | Klempner et al. [93] | |
| KRAS G12/G13 mutations | Advanced cancers | Mutation-enrichment NGS | Fujii et al. [41] | |
| Prognosis | EBV DNA | Nasopharyngeal carcinoma | Real-time PCR | Chan et al. [94] |
| Sengar et al. [95] | ||||
| EGFR mutations | NSCLC | ddPCR | Li et al. [84] | |
| KRAS mutations | NSCLC | ddPCR | Wang et al. [85] |
Abbreviations: ucfDNA, urinary cell-free DNA; NMIBC, non-muscle-invasive bladder cancer; GSTP1, glutathione S-transferase P1 gene; mVIM, hypermethylated vimentin gene; NGS, next-generation sequencing; ddPCR, droplet digital PCR; UBC, urothelial bladder cancer; NSCLC, non-small cell lung cancer.
Detection and diagnosis of cancer
The most studied area of the clinical applications of ucfDNA could be referred to the detection and diagnosis of cancer. A variety of DNA alterations including DNA quantification, DNA integrity, mutations and methylation status have been investigated in urine of both urological and non-urological cancer to evaluate the diagnostic utility of ucfDNA [7,16,42,43].
Urological cancer markers
Since urological cancer is in direct contact with urine flowing through the urogenital tract, urine may contain cell-free DNA sequences derived from apoptotic or necrotic tumor cells shedding into urine. Thus, urine has great potential as a desirable source of diagnostic biomarkers because of the genetic information storing in the urinary cell-free tumor DNA. Previously, genetic and epigenetic alterations in urological cancer, including renal cancer [14,44,45], bladder cancer [46-49] and prostate cancer [50-52] have been studied extensively, using DNA from urine sediment. However, ucfDNA from urine supernatant has become a more preferable target for investigating in urological cancer, and it has been considered to be superior to urine sediment because a mass of normal DNA existing in urine sediment could interfere the analytical results [13]. UcfDNA analysis in urological cancer mainly focuses on bladder cancer and prostate cancer.
Various types of genetic alterations were studied using ucfDNA to evaluate the possible diagnostic value of ucfDNA for bladder cancer. In 2005, Zancan et al. first investigated the ucfDNA concentrations in 35 suspected bladder cancer patients before cystoscopy [53]. UcfDNA concentrations higher than 250 ng/mL were detected in all bladder cancer patients (16/16) confirmed by cystoscopy, whereas only 36.8% patients (7/19) with negative cystoscopy had ucfDNA higher than 250 ng/mL. Thus, a higher concentration of ucfDNA in bladder cancer patients, compared to that in healthy subjects, was confirmed, indicating the promising diagnostic utility of ucfDNA concentration in bladder cancer. Subsequently, the same research team conducted a further study in 2009, on a larger cohort, including 45 bladder cancer patients and 87 healthy individuals [54]. UcfDNA concentrations were quantified using four different methods, and were found, by each of the methods, not to differ significantly between bladder cancer patients and healthy individuals. As a result, ucfDNA concentration was not considered a reliable diagnostic marker for bladder cancer. In another study performed by Chang et al., ucfDNA concentration was evaluated adjusting to urine creatinine in 46 bladder cancer patients and 98 controls [25]. The mean ucfDNA/UCr concentration in bladder cancer patients was significantly higher than that in controls, indicating that ucfDNA/UCr had the potential for diagnosing bladder cancer. Brisuda and co-workers proposed that the inconsistent results about the role of ucfDNA concentration in bladder cancer could be attributed to the use of various non-standardized methodologies [55]. They standardized the methodology and quantified the ucfDNA concentrations in 66 bladder cancer patients and 34 controls using the optimized method. According to the results shown in this study, ucfDNA levels had the ability to discriminate between the presence and absence of bladder cancer, exhibiting a potential diagnostic utility of ucfDNA concentrations in bladder cancer patients. Topoisomerase-II alpha (TopoIIA) is an isoform of DNA gyrase, and altered expression of TopoIIA exists in various cancers as well as normal tissues [56-58]. TopoIIA was reported to be associated with the progression and recurrence of primary non-muscle-invasive bladder cancer (NMIBC) [59]. The level of TopoIIA cell-free DNA in the urine of bladder cancer patients was also examined for potential diagnostic value [60]. The results showed that urinary TopoIIA cfDNA was significantly higher in bladder cancer patients compared to that in non-cancer patient controls and hematuria patients. In addition, urinary TopoIIA cfDNA level was also able to discriminate between muscle-invasive bladder cancer (MIBC) and NMIBC. This study provided the evidence for the potential diagnostic value of urinary TopoIIA cfDNA for bladder cancer [60].
It has been reported that DNA from normal cells is mainly through apoptosis, resulting in short and uniform fragments, while for tumor cells, released DNA fragments are relatively long as a result of necrosis [61,62]. Therefore, the integrity of ucfDNA in urological cancers was widely studied. DNA integrity was also examined as a diagnostic marker of bladder cancer. Chang et al. discovered that ucfDNA concentration was more reliable and accurate for diagnosis of bladder cancer when detecting longer DNA fragments using the 400-bp real-time PCR-based detection method [25]. Another study drew a similar conclusion by verifying sequences longer than 250 bp (c-Myc, BCAS1, and HER2) in ucfDNA from 51 bladder cancer patients, 46 symptomatic patients, and 32 healthy controls [38]. UcfDNA integrity analysis showed a satisfactory sensitivity and specificity in early diagnosis of bladder cancer.
Microsatellite alterations are integral and valuable markers for human cancer detection [63-65]. Six microsatellite markers on chromosomes 4, 9, and 17 were analyzed in cfDNA in urine and blood from patients with conspicuous bladder lesions [66]. It turned out that 88% of the microsatellite changes could be detected in at least one of the body fluids, thus indicating the clinical utility of ucfDNA for diagnosis and screening of bladder cancer. Similar conclusion was also obtained in a study carried out by Szarvas and co-workers [13].
UcfDNA levels, integrity, and methylation status were also extensively investigated as potential diagnostic markers of prostate cancer.
Yan et al. explored the diagnostic value of ucfDNA levels in prostate cancers using a previously described two-gene expression ratio method [67]. TSPAN13 and S100A9 were selected as candidate genes and the TSPAN13-to-S100A9 ratio was evaluated in 95 urine specimens from prostate cancer patients and benign prostatic hyperplasia controls (BPH) [68]. The TSPAN13-to-S100A9 ratio in ucfDNA was significantly higher in prostate cancer than in BPH, demonstrating the excellent potential of ucfDNA as a diagnostic biomarker for prostate cancer patients.
UcfDNA integrity was evaluated for early diagnosis of prostate cancer. In 2013, Casadio et al. examined ucfDNA integrity in 29 prostate cancer patients and 25 healthy volunteers by quantifying sequences longer than 250 bp corresponding to 3 genes (c-Myc, BCAS1, and HER2) [39]. Showing a sensitivity of 0.79 and a specificity of 0.84, ucfDNA integrity was considered to be a promising biomarker for early diagnosis of prostate cancer. After the preliminary study mentioned above, researchers subsequently performed further study of ucfDNA integrity in prostate cancer in a larger cohort, and compared the diagnostic value of ucfDNA with the traditional biomarker prostate-specific antigen (PSA) [69]. Using the same methods, the researchers evaluated DNA fragments longer than 250 bp of 3 frequently amplified genes (c-MYC, HER2, and AR) in 67 prostate cancer patients and 64 patients with benign diseases of the urogenital tract. The results showed that the sensitivity and specificity of ucfDNA analysis were lower than that of PSA, indicating that ucfDNA integrity might not be a reliable biomarker for early diagnosis of prostate cancer. The contradictory results of the two studies may be explained as follows [69]. The study cohort recruited in the pilot study was much smaller than that of the confirmatory study. Besides, patients in the confirmatory study included some with inflammation, calculi and cysts, leading to the exfoliation of inflammatory cells into urine, increasing the amount of cfDNA released and thus leading to false positive results. Despite these contradictory results of ucfDNA integrity analysis, ucfDNA could still be a potential source of other biomarkers and enable the detection of gene alterations in prostate cancer.
Promoter hypermethylation of the glutathione S-transferase P1 gene (GSTP1) is the most frequent DNA aberration observed in prostate cancer [70,71]. Methylation profile of GSTP1 gene promoter in ucfDNA was evaluated in patients with prostate cancer, patients with benign prostatic hyperplasia and healthy controls [72]. The methylation status of GSTP1 gene detected in urine was the same as that of the gene in the blood of the same patients. GSTP1 gene methylation status in ucfDNA of prostate cancer patients was significantly different from that of BPH patients and healthy controls, indicating the potential diagnostic value of ucfDNA.
Non-urological cancer markers
As early as 2000, Botezatu et al. conducted a series of experiments, which demonstrated, for the first time, the existence of transrenal cell-free DNA in animal and human models [18]. In this study, KRAS mutations were detected in the urine of four out of five advanced colorectal adenocarcinoma patients and in five out of eight patients with advanced pancreatic cancer [18]. Since then, an increasing number of ucfDNA alterations in various non-urological cancers have been discovered and evaluated as potential biomarkers for cancer diagnosis.
Since the detection of KRAS mutations in ucfDNA in colorectal cancer (CRC) [18], further studies were carried out to evaluate the potential clinical utility of KRAS mutations in the urine of CRC patients. A study carried out by Su et al. showed that mutated KRAS sequences could be identified in the urine derived from KRAS proto-oncogene mutation-positive CRC patients, and that mutated KRAS sequences were far more abundant in low molecular weight (150 to 250 bp) than in high molecular weight (greater than 1 kb) fragments. Remarkably, KRAS mutations were analyzed in paired urine and tissue samples from 20 patients with either CRC or adenomatous polyps, and a 83% concurrence was observed in urine and tissue samples from the same individuals [17]. Subsequently, Su and co-workers conducted two more studies to detect KRAS mutations in ucfDNA in the urine of CRC patients. They further examined KRAS mutations in urine, plasma and serum of CRC or adenomatous polyps patients. The results showed that the incidence of KRAS mutations in ucfDNA was significantly higher than that in serum or plasma. As a result, urine seemed to be a better source for detecting KRAS mutations in CRC, compared to serum or plasma, indicating the potential clinical practicality of ucfDNA for the detection and diagnosis of CRC or adenomatous polyps [33,35]. In addition to KRAS mutations, epigenetic DNA markers could also be detected in the urine of CRC patients. In a study conducted by Song et al., hypermethylated vimentin gene (mVIM) was evaluated in the urine from 20 CRC patients and 20 control subjects with no known neoplasia. mVIM was detected in 75% CRC patients, demonstrating significant association with CRC. In contrast, only 10% of the controls contained mVIM in their urine samples. Moreover, mVIM was mostly detected in low-molecular-weight urine DNA, confirming its origin from circulation and existence in urine in the form of cell-free DNA. Therefore, it seemed that mVIM detection in ucfDNA held promise for CRC screening and diagnosis [73].
In hepatocellular carcinoma (HCC), TP53 mutation and HCC-associated HBV mutation were detected in ucfDNA, indicating its potential as a diagnostic biomarker for HCC screening. Lin and co-workers investigated an HCC-associated mutation, TP53 249T hotspot mutation, in the urine of patients with HCC. It was the first report of detection of TP53 249T mutation in the urine of HCC patients. TP53 mutations were successfully detected in 9 out of 17 samples, indicating urine as a potential non-invasive sample source for HCC screening [74]. In a small pilot study on HBV-associated HCCs, HBV 1762T/1764A double mutation was detected in both circulation and urine. The results of this study showed potential clinical utility of urine for non-invasive HCC screening, although further studies with larger sample size were essential for validating the potential [75].
Molecular detection of human papillomavirus (HPV) DNA has been of considerable interest in risk assessment and screening of cervical cancer [76,77]. Urine-based HPV DNA testing has been investigated as a non-invasive approach complementary to cytology test [78]. Discovery of cell-free HPV DNA in urine could be traced back to 1999, though sensitivity was relatively low [79]. Recently, new technology such as NGS was applied to cell-free HPV DNA detection in urine, indicating that urine could be a promising material for personalized cervical cancer screening and diagnosis [80].
Surveillance of cancer progression
Cancer surveillance is important for early diagnosis of disease progression and metastasis, as well as for optimizing treatment. Various studies have explored the potential utility of ucfDNA for surveillance of cancer progression by monitoring genomic variants in urine.
A retrospective pilot study was conducted in 12 patients with recurrent or progressive/metastatic NMIBC during a 20-year follow-up from 1994 to 2015 [81]. Somatic variants in cfDNA from urine, blood and tumor were detected using NGS and ddPCR assays. Higher levels of neoplastic ucfDNA were discovered before cancer progression, in patients with progressive disease than those with recurrent disease, and tumor cfDNA could be detected in urine and blood in both early and advanced stages of bladder cancer. Therefore, disease progression in bladder cancer can be monitored by detecting genomic variants in ucfDNA. In urothelial bladder cancers (UBCs), genomic profiles were also performed in ucfDNA [82]. The data demonstrated that high sensitivity was achieved for genomic aberrations detection using ucfDNA and ucfDNA was highly representative in detecting recurrent genomic aberrations. The results were suggestive of the potential utility of ucfDNA in monitoring UBC progression. UcfDNA analysis for FGFR3 or PIK3CA mutations using ddPCR assays also demonstrated its utility in the disease surveillance for bladder cancer [83]. UcfDNA at different time points from NMIBC patient cohort were analyzed for FGFR3 or PIK3CA mutations and showed that higher levels of FGFR3 or PIK3CA mutations in ucfDNA were associated with the progression of NMIBC.
Similar utility of ucfDNA had also been demonstrated in non-small cell lung cancer (NSCLC). A total of 160 NSCLC patients at various stages of disease participated and samples were collected prospectively at 2-month intervals. A baseline sample was taken before treatment commencement. Transrenal DNA was compared with plasma DNA to ascertain the sensitivity. DdPCR was used to profile the urine and blood samples for key EGFR mutations. Serial monitoring of NSCLC patients with different disease stages showed stable molecular signatures and correlated to different treatments [84].
Another study also focused on ucfDNA alterations in NSCLC patients. DdPCR was used to detect mutant DNA in 200 NSCLC patients. Transrenal DNA was successfully detected in all the patients (100%). Overall concordance rate for mutant KRAS DNA within urine specimens and primary tissue biopsies was 95%. More importantly, longitudinal monitoring of urine specimens showed an increase in the quantity of transrenal DNA, which was highly associated with disease progression and outcome. The study indicated that urinary specimens that can be extracted non-invasively present new opportunities to track patients with KRAS mutation, who were undergoing therapy [85].
Monitoring therapeutic response
Targeted therapy for cancer is now common in the context of precision medicine. Monitoring response and resistance to targeted therapy could be indispensable during the course of treatment of cancer patients. Since tumor biopsy is constrained by disadvantages such as tumor heterogeneity and invasive repeated samplings, non-invasive specimens are gaining in importance. UcfDNA had been detected in diversified cohorts receiving certain antitumor therapies, and the dynamic tracking of genetic abnormalities in ucfDNA was therefore evaluated for monitoring the efficacy of the therapeutic process.
Whole genome sequencing was used for the first time in 2016 to evaluate copy number variations (CNV) in ucfDNA from advanced prostate cancer patients [86]. In this study, a series of tumor-associated CNVs were detected in ucfDNA before and after androgen deprivation therapy in hormone sensitive prostate cancer (HSPC) and docetaxel chemotherapy in castrate resistant prostate cancer (CRPC). Significant CNVs in 34 genomic loci were discovered during the course of therapy. In addition, a urine genomic abnormality (UGA) score algorithm was established to evaluate the ten most significant segments with CNVs. The UGA scores could reflect cancer progression status and overall survival during therapy, indicating that ucfDNA has potential clinical utility in monitoring and predicting treatment response in advanced prostate cancer.
A study carried out by Reckamp and co-workers evaluated the EGFR mutations in matched urine and plasma samples from EGFR mutant-positive advanced NSCLC patients under rociletinib treatment using a short footprint mutation enrichment NGS assay. The results showed that the sensitivity of EGFR mutation detection in urine was 72% (34 of 47 specimens) for T790M, 75% (12 of 16) for L858R and 67% (28 of 42) for exon 19 deletions, using tissue samples as a reference. Furthermore, when the specimens met the recommended volume requirement, the sensitivity increased to 93% (13 of 14 specimens) for T790M, 80% (4 of 5) for L858R, and 83% (10 of 12) for exon 19 deletions, which were comparable to plasma findings. Twelve additional T790M-positive cases with undetectable T790M mutations in tissue sample analysis were identified by testing both urine and plasma. UcfDNA in NSCLC exhibited high sensitivity and complementarity in EGFR mutation detection, and showed great potential in the diagnosis and monitoring of treatment response of NSCLC [40]. Similar results have also been obtained in other studies [87,88]. EGFR mutation status was also identified using ddPCR in ucfDNA from patients in different stages of NSCLC. It turned out that ucfDNA showed good concordance with results derived from plasma DNA in both early- and late-stage NSCLC patients, highlighting the potential clinical utility of ucfDNA in continual monitoring of NSCLC [84]. Chen et al. performed similar analysis of ucfDNA at different time points in NSCLC patients, who received EGFR tyrosine kinase inhibitor (TKI) treatment. The results showed that ucfDNA mutation status correlated closely with treatment efficacy, indicating the potential utility of urine for monitoring EGFR TKI treatment [89]. In another study carried out by Husain and co-workers, they evaluated dynamic changes in EGFR activating (exon 19del and L858R) and resistance (T790M) mutation levels in advanced NSCLC patients receiving osimertinib, and found that eight out of nine evaluable NSCLC patients had detectable T790M-mutant DNA fragments in pre-treatment baseline samples. In addition, daily monitoring of EGFR mutations in urine of NSCLC patients showed that surveillance of ucfDNA may enable early assessment of patient response and proof-of-concept studies for drug development [90]. Tchekmedyian et al. also performed ultrasensitive detection and longitudinal monitoring of EGFR mutations using non-invasive urinary circulating tumor DNA (ctDNA) liquid biopsies in five patients with NSCLC treated with EGFR TKIs. The results verified the diagnostic potential of urinary ctDNA as a non-invasive molecular diagnostic tool to assess tumor burden and response to therapy [91].
In addition to the EGFR mutations detected in the urine of lung cancer patients, EGFR mutations in ucfDNA were also studied in gastric cancer [36]. Urine EGFR mutation status was examined in 120 gastric cancer patients with EGFR mutations and 100 healthy controls. During the course of EGFR TKI treatment of gastric cancer, EGFR mutations were monitored serially for 12 months. The concordance rate of EGFR mutation status between ucfDNA and primary tissue samples was 92% at baseline and 99% at different time points in gastric cancer patients. The results suggested that ucfDNA may serve as a reliable marker for treatment monitoring in primary gastric cancer [36].
In a study performed by Siravegna et al., researchers detected a CAD-ALK gene rearrangement in a metastatic colorectal cancer patient undergoing ALK inhibitor treatment. CAD-ALK gene rearrangement in urine was tracked during the course of therapy and found to be concordant with cancer progression, which was also confirmed by radiological test. Thus, urine was believed to have the potential of monitoring tumor progression in a non-invasive way during target therapy and could be further used to monitor minimal residual disease in patients carrying gene fusions [92].
Oncogenic BRAF V600E substitutions are observed primarily in melanoma, colon cancer, and non-small cell lung cancer, and have been identified in multiple tumor types [93]. Klempner et al. reported for the first time about the recurrent BRAF V600E mutations in advanced high-grade colorectal neuroendocrine tumors and determined the BRAF alteration frequency to be 9% in 108 cases. Among these BRAF alterations, 80% were BRAF V600E. Dramatic response to BRAF-MEK combination occurred in two cases of metastatic high-grade rectal neuroendocrine carcinoma refractory to standard therapy. Following initiation of therapy, there was a rapid decrease in urinary BRAF V600E ctDNA, along with serum chromogranin A levels that paralleled clinical resolution of symptoms and preceded radiologic response. The study indicated that urinary BRAF V600E circulating tumor DNA had the promising ability to monitor paralleled disease response of colorectal neuroendocrine cancers [93].
Fujii et al. developed a quantitative, mutation-enrichment NGS test for detecting KRAS G12/G13 mutations in ucfDNA and the results were compared with those from the clinical testing of archived tumor tissue and plasma cfDNA samples from patients with advanced cancer. In 71 patients, the concordance between ucfDNA and tumor DNA was 73% (sensitivity, 63%; specificity, 96%) for all patients and 89% (sensitivity, 80%; specificity, 100%) for patients with urine samples of 90 to 110 mL. Patients had significantly fewer KRAS G12/G13 copies in ucfDNA during systemic therapy than at baseline or during disease progression. Compared with no changes or increases in ucfDNA KRAS G12/G13 copies during therapy, decreases in these measures were associated with longer median time to treatment failure [41].
Biomarkers for cancer prognosis
Cancer prognosis, including metastasis and relapse, is of vital importance to cancer patients. Non-invasive methods to determine cancer prognosis are therefore urgently required.
Circulating Epstein-Barr virus (EBV) DNA could be excreted transrenally into urine in nasopharyngeal carcinoma (NPC) patients, and was quantitatively detected in the urine of 42 out of 74 NPC patients using a 59-bp real-time PCR assay. Remarkably, the concentration of EBV DNA in plasma from patients with detectable EBV DNA in urine was also significantly higher, showing a positive correlation between plasma and urine. Therefore, urine EBV DNA analysis may be used as a non-invasive test for NPC monitoring and prognosis [94]. After the discovery of EBV DNA in urine, further study was carried out by Sengar et al. to explore the clinical utility of urine EBV DNA in NPC. It was found that EBV DNA in urine of NPC patients had high diagnostic sensitivity and correlated well with plasma EBV DNA before and after therapy, as well as predicting therapy response and survival of NPC patients. Although studies on larger cohorts are needed, urine EBV DNA is believed to be a promising prognostic biomarker in NPC [95].
In a study involving 160 patients at various stages of NSCLC, researchers detected key EGFR mutations in both urine and blood samples. Survival analysis showed good prognostic utility in late-stage patients with high ucfDNA variations and in patients that acquired T790M mutation [84]. In addition to EGFR mutations, KRAS mutations were also evaluated as a potential prognostic marker in NSCLC patients. Serial samplings were performed during different treatment cycles to gauge the predictive value. Patients with positive results at baseline had lower median overall survival than those with wildtype. Longitudinal monitoring of urine specimens showed an increase in the quantity of ucfDNA, which was closely associated with disease outcome [85].
Conclusions
Although the number of studies concerning the clinical applications of ucfDNA in cancer are still limited and mostly carried out on relatively small patient cohorts, the promising future of the potential clinical value of ucfDNA is worth expecting. As an ultra-noninvasive tool for liquid biopsy, ucfDNA has unique advantages on molecular profiling of tumor, and is believed to have a complementary and synergistic effect on serum and plasma in diagnosis, progression surveillance, treatment monitoring and prognosis for both urological and non-urological cancers. In future, we believed that the advancement of molecular assays (such as NGS and ddPCR) and the development of larger validation and prospective studies could offer a deeper insight into the clinical applications of ucfDNA in cancer.
Disclosure of conflict of interest
None.
Abbreviations
- cfDNA
cell-free DNA
- ucfDNA
urinary cell-free DNA
- ddPCR
droplet digital PCR
- NGS
next-generation sequencing
- TopoIIA
topoisomerase-II alpha
- NMIBC
non-muscle-invasive bladder cancer
- MIBC
muscle-invasive bladder cancer
- BPH
benign prostatic hyperplasia
- PSA
prostate-specific antigen
- GSTP1
glutathione S-transferase P1 gene
- CRC
colorectal cancer
- mVIM
hypermethylated vimentin gene
- HCC
hepatocellular carcinoma
- HPV
human papillomavirus
- UBC
urothelial bladder cancer
- NSCLC
non-small cell lung cancer
- CNV
copy number variation
- HSPC
hormone sensitive prostate cancer
- CRPC
castrate resistant prostate cancer
- UGA
urine genomic abnormality
- TKI
tyrosine kinase inhibitor
- ctDNA
circulating tumor DNA
- EBV
Epstein-Barr virus
- NPC
nasopharyngeal carcinoma
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Aronson SJ, Rehm HL. Building the foundation for genomics in precision medicine. Nature. 2015;526:336–342. doi: 10.1038/nature15816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 4.Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol. 2014;11:129–144. doi: 10.1038/nrclinonc.2013.253. [DOI] [PubMed] [Google Scholar]
- 5.Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mader S, Pantel K. Liquid biopsy: current status and future perspectives. Oncol Res Treat. 2017;40:404–408. doi: 10.1159/000478018. [DOI] [PubMed] [Google Scholar]
- 7.Di Meo A, Bartlett J, Cheng Y, Pasic MD, Yousef GM. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol Cancer. 2017;16:80. doi: 10.1186/s12943-017-0644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Chang S, Li G, Sun Y. Application of liquid biopsy in precision medicine: opportunities and challenges. Front Med. 2017 doi: 10.1007/s11684-017-0526-7. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 9.Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res. 2007;635:105–117. doi: 10.1016/j.mrrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Schwarzenbach H, Hoon DS, Pantel K. Cellfree nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 11.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih lM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA Jr. Detection of circulating tumor DNA in earlyand late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sardana G, Diamandis EP. Biomarkers for the diagnosis of new and recurrent prostate cancer. Biomark Med. 2012;6:587–596. doi: 10.2217/bmm.12.72. [DOI] [PubMed] [Google Scholar]
- 13.Szarvas T, Kovalszky I, Bedi K, Szendroi A, Majoros A, Riesz P, Fule T, Laszlo V, Kiss A, Romics I. Deletion analysis of tumor and urinary DNA to detect bladder cancer: urine supernatant versus urine sediment. Oncol Rep. 2007;18:405–409. [PubMed] [Google Scholar]
- 14.Cairns P. Detection of promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Ann N Y Acad Sci. 2004;1022:40–43. doi: 10.1196/annals.1318.007. [DOI] [PubMed] [Google Scholar]
- 15.Yokota M, Tatsumi N, Tsuda I, Takubo T, Hiyoshi M. DNA extraction from human urinary sediment. J Clin Lab Anal. 1998;12:88–91. doi: 10.1002/(SICI)1098-2825(1998)12:2<88::AID-JCLA3>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salvi S, Martignano F, Molinari C, Gurioli G, Calistri D, De Giorgi U, Conteduca V, Casadio V. The potential use of urine cell free DNA as a marker for cancer. Expert Rev Mol Diagn. 2016;16:1283–1290. doi: 10.1080/14737159.2016.1254551. [DOI] [PubMed] [Google Scholar]
- 17.Su YH, Wang M, Brenner DE, Ng A, Melkonyan H, Umansky S, Syngal S, Block TM. Human urine contains small, 150 to 250 nucleotidesized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J Mol Diagn. 2004;6:101–107. doi: 10.1016/S1525-1578(10)60497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Botezatu I, Serdyuk O, Potapova G, Shelepov V, Alechina R, Molyaka Y, Ananev V, Bazin I, Garin A, Narimanov M, Knysh V, Melkonyan H, Umansky S, Lichtenstein A. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin Chem. 2000;46:1078–1084. [PubMed] [Google Scholar]
- 19.Lichtenstein AV, Melkonyan HS, Tomei LD, Umansky SR. Circulating nucleic acids and apoptosis. Ann N Y Acad Sci. 2001;945:239–249. doi: 10.1111/j.1749-6632.2001.tb03892.x. [DOI] [PubMed] [Google Scholar]
- 20.Bryzgunova OE, Skvortsova TE, Kolesnikova EV, Starikov AV, Rykova EY, Vlassov VV, Laktionov PP. Isolation and comparative study of cell-free nucleic acids from human urine. Ann N Y Acad Sci. 2006;1075:334–340. doi: 10.1196/annals.1368.045. [DOI] [PubMed] [Google Scholar]
- 21.Su YH, Wang M, Block TM, Landt O, Botezatu I, Serdyuk O, Lichtenstein A, Melkonyan H, Tomei LD, Umansky S. Transrenal DNA as a diagnostic tool: important technical notes. Ann N Y Acad Sci. 2004;1022:81–89. doi: 10.1196/annals.1318.014. [DOI] [PubMed] [Google Scholar]
- 22.Bryzgunova OE, Laktionov PP. Extracellular nucleic acids in urine: sources, structure, diagnostic potential. Acta Naturae. 2015;7:48–54. [PMC free article] [PubMed] [Google Scholar]
- 23.Melkonyan HS, Feaver WJ, Meyer E, Scheinker V, Shekhtman EM, Xin Z, Umansky SR. Transrenal nucleic acids: from proof of principle to clinical tests. Ann N Y Acad Sci. 2008;1137:73–81. doi: 10.1196/annals.1448.015. [DOI] [PubMed] [Google Scholar]
- 24.Tsui NB, Jiang P, Chow KC, Su X, Leung TY, Sun H, Chan KC, Chiu RW, Lo YM. High resolution size analysis of fetal DNA in the urine of pregnant women by paired-end massively parallel sequencing. PLoS One. 2012;7:e48319. doi: 10.1371/journal.pone.0048319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang HW, Tsui KH, Shen LC, Huang HW, Wang SN, Chang PL. Urinary cell-free DNA as a potential tumor marker for bladder cancer. Int J Biol Markers. 2007;22:287–294. doi: 10.1177/172460080702200408. [DOI] [PubMed] [Google Scholar]
- 26.He WS, Bishop KS. The potential use of cell-free-circulating-tumor DNA as a biomarker for prostate cancer. Expert Rev Mol Diagn. 2016;16:839–852. doi: 10.1080/14737159.2016.1197121. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Tong KL, Li PK, Chan AY, Yeung CK, Pang CC, Wong TY, Lee KC, Lo YM. Presence of donor- and recipient-derived DNA in cell-free urine samples of renal transplantation recipients: urinary DNA chimerism. Clin Chem. 1999;45:1741–1746. [PubMed] [Google Scholar]
- 28.Zhong XY, Hahn D, Troeger C, Klemm A, Stein G, Thomson P, Holzgreve W, Hahn S. Cellfree DNA in urine: a marker for kidney graft rejection, but not for prenatal diagnosis? Ann N Y Acad Sci. 2001;945:250–257. [PubMed] [Google Scholar]
- 29.Zhang Z, Ohkohchi N, Sakurada M, Mizuno Y, Miyagi T, Satomi S, Okazaki H. Diagnosis of acute rejection by analysis of urinary DNA of donor origin in renal transplant recipients. Transplant Proc. 2001;33:380–381. doi: 10.1016/s0041-1345(00)02057-1. [DOI] [PubMed] [Google Scholar]
- 30.Al-Yatama MK, Mustafa AS, Ali S, Abraham S, Khan Z, Khaja N. Detection of Y chromosome-specific DNA in the plasma and urine of pregnant women using nested polymerase chain reaction. Prenat Diagn. 2001;21:399–402. doi: 10.1002/pd.69. [DOI] [PubMed] [Google Scholar]
- 31.Koide K, Sekizawa A, Iwasaki M, Matsuoka R, Honma S, Farina A, Saito H, Okai T. Fragmentation of cell-free fetal DNA in plasma and urine of pregnant women. Prenat Diagn. 2005;25:604–607. doi: 10.1002/pd.1213. [DOI] [PubMed] [Google Scholar]
- 32.Majer S, Bauer M, Magnet E, Strele A, Giegerl E, Eder M, Lang U, Pertl B. Maternal urine for prenatal diagnosis--an analysis of cell-free fetal DNA in maternal urine and plasma in the third trimester. Prenat Diagn. 2007;27:1219–1223. doi: 10.1002/pd.1875. [DOI] [PubMed] [Google Scholar]
- 33.Su YH, Wang M, Brenner DE, Norton PA, Block TM. Detection of mutated K-ras DNA in urine, plasma, and serum of patients with colorectal carcinoma or adenomatous polyps. Ann N Y Acad Sci. 2008;1137:197–206. doi: 10.1196/annals.1448.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su YH, Lin SY, Song W, Jain S. DNA markers in molecular diagnostics for hepatocellular carcinoma. Expert Rev Mol Diagn. 2014;14:803–817. doi: 10.1586/14737159.2014.946908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su YH, Wang M, Aiamkitsumrit B, Brenner DE, Block TM. Detection of a K-ras mutation in urine of patients with colorectal cancer. Cancer Biomark. 2005;1:177–182. doi: 10.3233/cbm-2005-12-305. [DOI] [PubMed] [Google Scholar]
- 36.Shi XQ, Xue WH, Zhao SF, Zhang XJ, Sun W. Dynamic tracing for epidermal growth factor receptor mutations in urinary circulating DNA in gastric cancer patients. Tumour Biol. 2017;39:1010428317691681. doi: 10.1177/1010428317691681. [DOI] [PubMed] [Google Scholar]
- 37.Ralla B, Stephan C, Meller S, Dietrich D, Kristiansen G, Jung K. Nucleic acid-based biomarkers in body fluids of patients with urologic malignancies. Crit Rev Clin Lab Sci. 2014;51:200–231. doi: 10.3109/10408363.2014.914888. [DOI] [PubMed] [Google Scholar]
- 38.Casadio V, Calistri D, Tebaldi M, Bravaccini S, Gunelli R, Martorana G, Bertaccini A, Serra L, Scarpi E, Amadori D, Silvestrini R, Zoli W. Urine cell-free DNA integrity as a marker for early bladder cancer diagnosis: preliminary data. Urol Oncol. 2013;31:1744–1750. doi: 10.1016/j.urolonc.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 39.Casadio V, Calistri D, Salvi S, Gunelli R, Carretta E, Amadori D, Silvestrini R, Zoli W. Urine cell-free DNA integrity as a marker for early prostate cancer diagnosis: a pilot study. Biomed Res Int. 2013;2013:270457. doi: 10.1155/2013/270457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reckamp KL, Melnikova VO, Karlovich C, Sequist LV, Camidge DR, Wakelee H, Perol M, Oxnard GR, Kosco K, Croucher P, Samuelsz E, Vibat CR, Guerrero S, Geis J, Berz D, Mann E, Matheny S, Rolfe L, Raponi M, Erlander MG, Gadgeel S. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol. 2016;11:1690–1700. doi: 10.1016/j.jtho.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 41.Fujii T, Barzi A, Sartore-Bianchi A, Cassingena A, Siravegna G, Karp DD, Piha-Paul SA, Subbiah V, Tsimberidou AM, Huang HJ, Veronese S, Di Nicolantonio F, Pingle S, Vibat CR, Hancock S, Berz D, Melnikova VO, Erlander MG, Luthra R, Kopetz ES, Meric-Bernstam F, Siena S, Lenz HJ, Bardelli A, Janku F. Mutation-Enrichment next-generation sequencing for quantitative detection of KRAS mutations in urine cell-free DNA from patients with advanced cancers. Clin Cancer Res. 2017;23:3657–3666. doi: 10.1158/1078-0432.CCR-16-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salvi S, Gurioli G, De Giorgi U, Conteduca V, Tedaldi G, Calistri D, Casadio V. Cell-free DNA as a diagnostic marker for cancer: current insights. Onco Targets Ther. 2016;9:6549–6559. doi: 10.2147/OTT.S100901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan AK, Chiu RW, Lo YM. Cell-free nucleic acids in plasma, serum and urine: a new tool in molecular diagnosis. Ann Clin Biochem. 2003;40:122–130. doi: 10.1258/000456303763046030. [DOI] [PubMed] [Google Scholar]
- 44.Eisenberger CF, Schoenberg M, Enger C, Hortopan S, Shah S, Chow NH, Marshall FF, Sidransky D. Diagnosis of renal cancer by molecular urinalysis. J Natl Cancer Inst. 1999;91:2028–2032. doi: 10.1093/jnci/91.23.2028. [DOI] [PubMed] [Google Scholar]
- 45.Hoque MO, Begum S, Topaloglu O, Jeronimo C, Mambo E, Westra WH, Califano JA, Sidransky D. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64:5511–5517. doi: 10.1158/0008-5472.CAN-04-0799. [DOI] [PubMed] [Google Scholar]
- 46.Chung W, Bondaruk J, Jelinek J, Lotan Y, Liang S, Czerniak B, Issa JP. Detection of bladder cancer using novel DNA methylation biomarkers in urine sediments. Cancer Epidemiol Biomarkers Prev. 2011;20:1483–1491. doi: 10.1158/1055-9965.EPI-11-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Yu Y, Ye R, Zhang D, Li Q, An D, Fang L, Lin Y, Hou Y, Xu A, Fu Y, Lu W, Chen X, Chen M, Zhang M, Jiang H, Zhang C, Dong P, Li C, Chen J, Yang G, Liu C, Cai Z, Zhou F, Wu S. An epigenetic biomarker combination of PCDH17 and POU4F2 detects bladder cancer accurately by methylation analyses of urine sediment DNA in Han Chinese. Oncotarget. 2016;7:2754–2764. doi: 10.18632/oncotarget.6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Renard I, Joniau S, van Cleynenbreugel B, Collette C, Naome C, Vlassenbroeck I, Nicolas H, de Leval J, Straub J, Van Criekinge W, Hamida W, Hellel M, Thomas A, de Leval L, Bierau K, Waltregny D. Identification and validation of the methylated TWIST1 and NID2 genes through real-time methylation-specific polymerase chain reaction assays for the noninvasive detection of primary bladder cancer in urine samples. Eur Urol. 2010;58:96–104. doi: 10.1016/j.eururo.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 49.Kinde I, Munari E, Faraj SF, Hruban RH, Schoenberg M, Bivalacqua T, Allaf M, Springer S, Wang Y, Diaz LA Jr, Kinzler KW, Vogelstein B, Papadopoulos N, Netto GJ. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013;73:7162–7167. doi: 10.1158/0008-5472.CAN-13-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeronimo C, Usadel H, Henrique R, Silva C, Oliveira J, Lopes C, Sidransky D. Quantitative GSTP1 hypermethylation in bodily fluids of patients with prostate cancer. Urology. 2002;60:1131–1135. doi: 10.1016/s0090-4295(02)01949-0. [DOI] [PubMed] [Google Scholar]
- 51.Goessl C, Krause H, Muller M, Heicappell R, Schrader M, Sachsinger J, Miller K. Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 2000;60:5941–5945. [PubMed] [Google Scholar]
- 52.Goessl C, Muller M, Heicappell R, Krause H, Miller K. DNA-based detection of prostate cancer in blood, urine, and ejaculates. Ann N Y Acad Sci. 2001;945:51–58. doi: 10.1111/j.1749-6632.2001.tb03863.x. [DOI] [PubMed] [Google Scholar]
- 53.Zancan M, Franceschini R, Mimmo C, Vianello M, Di Tonno F, Mazzariol C, Malossini G, Gion M. Free DNA in urine: a new marker for bladder cancer? Preliminary data. Int J Biol Markers. 2005;20:134–136. doi: 10.1177/172460080502000209. [DOI] [PubMed] [Google Scholar]
- 54.Zancan M, Galdi F, Di Tonno F, Mazzariol C, Orlando C, Malentacchi F, Agostini M, Maran M, Del Bianco P, Fabricio AS, Murer B, Pianon C, Gion M. Evaluation of cell-free DNA in urine as a marker for bladder cancer diagnosis. Int J Biol Markers. 2009;24:147–155. doi: 10.1177/172460080902400304. [DOI] [PubMed] [Google Scholar]
- 55.Brisuda A, Pazourkova E, Soukup V, Horinek A, Hrbacek J, Capoun O, Svobodova I, Pospisilova S, Korabecna M, Mares J, Hanus T, Babjuk M. Urinary cell-free DNA quantification as non-invasive biomarker in patients with bladder cancer. Urol Int. 2016;96:25–31. doi: 10.1159/000438828. [DOI] [PubMed] [Google Scholar]
- 56.Nakopoulou L, Zervas A, Lazaris AC, Constantinides C, Stravodimos C, Davaris P, Dimopoulos C. Predictive value of topoisomerase II alpha immunostaining in urothelial bladder carcinoma. J Clin Pathol. 2001;54:309–313. doi: 10.1136/jcp.54.4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohashi Y, Sasano H, Yamaki H, Shizawa S, Kikuchi A, Shineha R, Akaishi T, Satomi S, Nagura H. Topoisomerase II alpha expression in esophageal squamous cell carcinoma. Anticancer Res. 1999;19:1873–1880. [PubMed] [Google Scholar]
- 58.Dingemans AM, Witlox MA, Stallaert RA, van der Valk P, Postmus PE, Giaccone G. Expression of DNA topoisomerase IIalpha and topoisomerase IIbeta genes predicts survival and response to chemotherapy in patients with small cell lung cancer. Clin Cancer Res. 1999;5:2048–2058. [PubMed] [Google Scholar]
- 59.Kim EJ, Lee YS, Kim YJ, Kim MJ, Ha YS, Jeong P, Lee OJ, Kim WJ. Clinical implications and prognostic values of topoisomerase-II alpha expression in primary non-muscle-invasive bladder cancer. Urology. 2010;75:1516, e1519–1513. doi: 10.1016/j.urology.2009.08.055. [DOI] [PubMed] [Google Scholar]
- 60.Kim YH, Yan C, Lee IS, Piao XM, Byun YJ, Jeong P, Kim WT, Yun SJ, Kim WJ. Value of urinary topoisomerase-IIA cell-free DNA for diagnosis of bladder cancer. Investig Clin Urol. 2016;57:106–112. doi: 10.4111/icu.2016.57.2.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 62.Sozzi G, Conte D, Leon M, Ciricione R, Roz L, Ratcliffe C, Roz E, Cirenei N, Bellomi M, Pelosi G, Pierotti MA, Pastorino U. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J. Clin. Oncol. 2003;21:3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 63.Qureshi KN, Lunec J, Neal DE. Molecular biological changes in bladder cancer. Cancer Surv. 1998;31:77–97. [PubMed] [Google Scholar]
- 64.Mao L, Lee DJ, Tockman MS, Erozan YS, Askin F, Sidransky D. Microsatellite alterations as clonal markers for the detection of human cancer. Proc Natl Acad Sci U S A. 1994;91:9871–9875. doi: 10.1073/pnas.91.21.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, Murphree AL, Strong LC, White RL. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 66.Utting M, Werner W, Dahse R, Schubert J, Junker K. Microsatellite analysis of free tumor DNA in urine, serum, and plasma of patients: a minimally invasive method for the detection of bladder cancer. Clin Cancer Res. 2002;8:35–40. [PubMed] [Google Scholar]
- 67.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A, Muir B, Mohapatra G, Salunga R, Tuggle JT, Tran Y, Tran D, Tassin A, Amon P, Wang W, Wang W, Enright E, Stecker K, Estepa-Sabal E, Smith B, Younger J, Balis U, Michaelson J, Bhan A, Habin K, Baer TM, Brugge J, Haber DA, Erlander MG, Sgroi DC. A twogene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 68.Yan C, Kim YH, Kang HW, Seo SP, Jeong P, Lee IS, Kim D, Kim JM, Choi YH, Moon SK, Yun SJ, Kim WJ. Urinary Nucleic Acid TSPAN13-to-S100A9 Ratio as a Diagnostic Marker in Prostate Cancer. J Korean Med Sci. 2015;30:1784–1792. doi: 10.3346/jkms.2015.30.12.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salvi S, Gurioli G, Martignano F, Foca F, Gunelli R, Cicchetti G, De Giorgi U, Zoli W, Calistri D, Casadio V. Urine cell-free DNA integrity analysis for early detection of prostate cancer patients. Dis Markers. 2015;2015:574120. doi: 10.1155/2015/574120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee WH, Isaacs WB, Bova GS, Nelson WG. CG island methylation changes near the GSTP1 gene in prostatic carcinoma cells detected using the polymerase chain reaction: a new prostate cancer biomarker. Cancer Epidemiol Biomarkers Prev. 1997;6:443–450. [PubMed] [Google Scholar]
- 71.Eng C, Herman JG, Baylin SB. A bird’s eye view of global methylation. Nat Genet. 2000;24:101–102. doi: 10.1038/72730. [DOI] [PubMed] [Google Scholar]
- 72.Bryzgunova OE, Morozkin ES, Yarmoschuk SV, Vlassov VV, Laktionov PP. Methylation-specific sequencing of GSTP1 gene promoter in circulating/extracellular DNA from blood and urine of healthy donors and prostate cancer patients. Ann N Y Acad Sci. 2008;1137:222–225. doi: 10.1196/annals.1448.039. [DOI] [PubMed] [Google Scholar]
- 73.Song BP, Jain S, Lin SY, Chen Q, Block TM, Song W, Brenner DE, Su YH. Detection of hypermethylated vimentin in urine of patients with colorectal cancer. J Mol Diagn. 2012;14:112–119. doi: 10.1016/j.jmoldx.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin SY, Dhillon V, Jain S, Chang TT, Hu CT, Lin YJ, Chen SH, Chang KC, Song W, Yu L, Block TM, Su YH. A locked nucleic acid clamp-mediated PCR assay for detection of a p53 codon 249 hotspot mutation in urine. J Mol Diagn. 2011;13:474–484. doi: 10.1016/j.jmoldx.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin SY, Jain S, Song W, Hu CT, Su YH. Strategic assay developments for detection of HBV 1762T/1764A double mutation in urine of patients with HBV-associated hepatocellular carcinomas. Hepatocellular Carcinoma-Clinical Research. 2012:139–154. [Google Scholar]
- 76.Sehgal A, Gupta S, Parashari A, Sodhani P, Singh V. Urine HPV-DNA detection for cervical cancer screening: prospects and prejudices. J Obstet Gynaecol. 2009;29:583–589. doi: 10.1080/01443610903061736. [DOI] [PubMed] [Google Scholar]
- 77.Daponte A, Pournaras S, Mademtzis I, Hadjichristodoulou C, Kostopoulou E, Maniatis AN, Messinis IE. Evaluation of high-risk human papillomavirus types PCR detection in paired urine and cervical samples of women with abnormal cytology. J Clin Virol. 2006;36:189–193. doi: 10.1016/j.jcv.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 78.Vorsters A, Micalessi I, Bilcke J, Ieven M, Bogers J, Van Damme P. Detection of human papillomavirus DNA in urine. A review of the literature. Eur J Clin Microbiol Infect Dis. 2012;31:627–640. doi: 10.1007/s10096-011-1358-z. [DOI] [PubMed] [Google Scholar]
- 79.Strauss S, Jordens JZ, McBride D, Sonnex C, Edwards S, Desselberger U, Watt P, Gray JJ. Detection and typing of human papillomavirus DNA in paired urine and cervical scrapes. Eur J Epidemiol. 1999;15:537–543. doi: 10.1023/a:1007574231879. [DOI] [PubMed] [Google Scholar]
- 80.Guerrero-Preston R, Valle BL, Jedlicka A, Turaga N, Folawiyo O, Pirini F, Lawson F, Vergura A, Noordhuis M, Dziedzic A, Perez G, Renehan M, Guerrero-Diaz C, De Jesus Rodriguez E, Diaz-Montes T, Rodriguez Orengo J, Mendez K, Romaguera J, Trock BJ, Florea L, Sidransky D. Molecular triage of premalignant lesions in liquid-based cervical cytology and circulating cell-free DNA from urine, using a panel of methylated human papilloma virus and host genes. Cancer Prev Res (Phila) 2016;9:915–924. doi: 10.1158/1940-6207.CAPR-16-0138. [DOI] [PubMed] [Google Scholar]
- 81.Birkenkamp-Demtroder K, Nordentoft I, Christensen E, Hoyer S, Reinert T, Vang S, Borre M, Agerbaek M, Jensen JB, Orntoft TF, Dyrskjot L. Genomic alterations in liquid biopsies from patients with bladder cancer. Eur Urol. 2016;70:75–82. doi: 10.1016/j.eururo.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 82.Togneri FS, Ward DG, Foster JM, Devall AJ, Wojtowicz P, Alyas S, Vasques FR, Oumie A, James ND, Cheng KK, Zeegers MP, Deshmukh N, O’Sullivan B, Taniere P, Spink KG, McMullan DJ, Griffiths M, Bryan RT. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur J Hum Genet. 2016;24:1167–1174. doi: 10.1038/ejhg.2015.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christensen E, Birkenkamp-Demtroder K, Nordentoft I, Hoyer S, van der Keur K, van Kessel K, Zwarthoff E, Agerbaek M, Orntoft TF, Jensen JB, Dyrskjot L. Liquid biopsy analysis of FGFR3 and PIK3CA hotspot mutations for disease surveillance in bladder cancer. Eur Urol. 2017;71:961–969. doi: 10.1016/j.eururo.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 84.Li F, Huang J, Ji D, Meng Q, Wang C, Chen S, Wang X, Zhu Z, Jiang C, Shi Y, Liu S, Li C. Utility of urinary circulating tumor DNA for EGFR mutation detection in different stages of non-small cell lung cancer patients. Clin Transl Oncol. 2017 doi: 10.1007/s12094-017-1669-3. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 85.Wang X, Meng Q, Wang C, Li F, Zhu Z, Liu S, Shi Y, Huang J, Chen S, Li C. Investigation of transrenal KRAS mutation in late stage NSCLC patients correlates to disease progression. Biomarkers. 2017;22:654–660. doi: 10.1080/1354750X.2016.1269202. [DOI] [PubMed] [Google Scholar]
- 86.Xia Y, Huang CC, Dittmar R, Du M, Wang Y, Liu H, Shenoy N, Wang L, Kohli M. Copy number variations in urine cell free DNA as biomarkers in advanced prostate cancer. Oncotarget. 2016;7:35818–35831. doi: 10.18632/oncotarget.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wakelee HA, Gadgeel SM, Goldman JW, Reckamp KL, Karlovich CA, Melnikova V, Soria JC, Yu HA, Solomon BJ, Perol M, Neal JW, Liu SV, Raponi M, Despain D, Erlander MG, Matheny SL, Yurasov S, Camidge DR, Sequist LV. Epidermal growth factor receptor (EGFR) genotyping of matched urine, plasma and tumor tissue from non-small cell lung cancer (NSCLC) patients (pts) treated with rociletinib. J Clin Oncol. 2016;34(Suppl):9001–9001. [Google Scholar]
- 88.Lin CC, Huang WL, Wei F, Su WC, Wong DT. Emerging platforms using liquid biopsy to detect EGFR mutations in lung cancer. Expert Rev Mol Diagn. 2015;15:1427–1440. doi: 10.1586/14737159.2015.1094379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen S, Zhao J, Cui L, Liu Y. Urinary circulating DNA detection for dynamic tracking of EGFR mutations for NSCLC patients treated with EGFR-TKIs. Clin Transl Oncol. 2017;19:332–340. doi: 10.1007/s12094-016-1534-9. [DOI] [PubMed] [Google Scholar]
- 90.Husain H, Melnikova VO, Kosco K, Woodward B, More S, Pingle SC, Weihe E, Park BH, Tewari M, Erlander MG, Cohen E, Lippman SM, Kurzrock R. Monitoring daily dynamics of early tumor response to targeted therapy by detecting circulating tumor DNA in urine. Clin Cancer Res. 2017;23:4716–4723. doi: 10.1158/1078-0432.CCR-17-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tchekmedyian N, Mudad R, Blanco FF, Raymond VM, Garst J, Erlander MG, Haura E, Berz D. Longitudinal monitoring of ctDNA EGFR mutation burden from urine correlates with patient response to EGFR TKIs: a case series. Lung Cancer. 2017;108:22–28. doi: 10.1016/j.lungcan.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 92.Siravegna G, Sartore-Bianchi A, Mussolin B, Cassingena A, Amatu A, Novara L, Buscarino M, Corti G, Crisafulli G, Bartolini A, Tosi F, Erlander M, Di Nicolantonio F, Siena S, Bardelli A. Tracking a CAD-ALK gene rearrangement in urine and blood of a colorectal cancer patient treated with an ALK inhibitor. Ann Oncol. 2017;28:1302–1308. doi: 10.1093/annonc/mdx095. [DOI] [PubMed] [Google Scholar]
- 93.Klempner SJ, Gershenhorn B, Tran P, Lee TK, Erlander MG, Gowen K, Schrock AB, Morosini D, Ross JS, Miller VA, Stephens PJ, Ou SH, Ali SM. BRAFV600E mutations in high-grade colorectal neuroendocrine tumors may predict responsiveness to BRAF-MEK combination therapy. Cancer Discov. 2016;6:594–600. doi: 10.1158/2159-8290.CD-15-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chan KC, Leung SF, Yeung SW, Chan AT, Lo YM. Quantitative analysis of the transrenal excretion of circulating EBV DNA in nasopharyngeal carcinoma patients. Clin Cancer Res. 2008;14:4809–4813. doi: 10.1158/1078-0432.CCR-08-1112. [DOI] [PubMed] [Google Scholar]
- 95.Sengar M, Chorghe S, Jadhav K, Singh S, Laskar SG, Pai P, Aggarwal JP, D’Cruz A, Chaturvedi P, Deshpande M, Chaukar D, Budrukkar A, Gupta T, Murthy V, Kane S, Thakur M, Rangarajan V, Kannan S, Shet T, Kode J. Cell-free Epstein-Barr virus-DNA in patients with nasopharyngeal carcinoma: plasma versus urine. Head Neck. 2016;38(Suppl 1):E1666–1673. doi: 10.1002/hed.24297. [DOI] [PubMed] [Google Scholar]
- 96.Husain H, Kosco K, Guerrero S, Lu T, Vibat C, Erlander M, Melnikova V. Detection of EGFR T790M mutation in urinary circulating tumor DNA from metastatic non-small cell lung cancer patients. Annals of Oncology. 2015;26:i10–i10. [Google Scholar]


