Abstract
Glioblastoma multiforme (GBM) is one of the most malignant and aggressive brain tumors with great amount of hyaluronan (HA) secretion and CD44 overexpression (HA receptor). CD44 has been suggested as a cancer stem cells (CSCs) marker. However, several clinical studies have indicated that CD44low glioma cell exhibit CSCs traits. Additionally, our previous study indicated that more CD44 expression was associated with a better prognosis in GBM patients. To determine whether CD44 is an appropriate marker of glioma stem cells (GSCs), we manipulated CD44 expression using intrinsic (CD44 knockdown, CD44kd) and extrinsic (HA supplement, HA+) methods. Our results show that CD44kd suppressed cell proliferation by retarding cell cycle progression from G0/G1 to S phase. Furthermore, it caused GSCs traits, including lower expression of differentiation marker (glial fibrillary acidic protein, GFAP), a higher level of sphere formation and higher expression of stem cell markers (CD133, nestin and Oct4). The reduction of CD44 expression induced by HA+ was accompanied by an increase in GSCs properties. Interestingly, the presence of HA+ in glioma cells with GSC traits conversely facilitated differentiation. Our data indicated that the CD44 low-expressing cells exhibit more GSCs straits, suggesting that CD44 is not an appropriate marker for GSCs. Furthermore, the preferential expression of CD44 at the invasive rim in rat glioma specimen implies that CD44 may be more important for invasion and migration instead of GSCs marker in glioma.
Keywords: CD44, glioma, hyaluronan, cancer stem cell
Introduction
Glioma is among the most aggressive human cancers. Despite recent advances in multimodal therapies, high-grade glioma remains fatal [1]. Recently, increasing evidence has indicated that the existence of a small population of stem-like cells called cancer stem cells (CSCs) within malignant tumors, which appear to have cancer-initiating capacity [2]. Examining CSCs in the tumor population may provide insight into the mechanism underlying tumorigenesis and novel therapeutic targets for optimizing therapeutic approaches to cancer.
CD44 is an important marker used to isolate a number of different CSCs lineages, such as colon, prostate, pancreatic and gastric cancer [3-6]. In glioma, CD44-enriched glioma stem cells (GSCs) exhibited significantly higher self-renewal capacity. Furthermore, increased levels of stem cell genes, including nanog, sox2, oct4, and Id1, coincided with the increased expression of the CD44 intracellular domain, suggesting a link between CD44 and a stem-like phenotype in glioma [7-9]. However, the identification of the GSCs markers using lectin microarray and LC-MS/MS revealed that CD44 was highly expressed in many glioma cell lines, including U87, U373 and T98G but not in glioblastoma (GBM)-derived stem-like cell line [10]. In addition, the GSC lines derived from primary GBM can be divided into two clusters according to their gene expression profile. The expression profiles of fetal neural stem cell-like, type I GSC lines were CD44low, CD133high and PDGFRαhigh. Conversely, type II GSC lines had adult neural stem cell characteristic, including CD44high, CD133low and PDGFRαlow [11]. The CD44low/CD133high GSCs further showed higher tumorigenicity than CD44high/CD133low GSCs [12]. Despite our previous finding that CD44 expression is correlated with GBM malignance, higher levels of CD44 expression were surprisingly correlated with higher survival in GBM patients [13]. Based on these findings, whether CD44 is an appropriate marker for GSCs remains controversial. Therefore, additional studies are needed to clarify the effect of CD44 on GSCs property. Furthermore, the CD44 signal is stimulated by the interaction with its ligand. Hyaluronan (HA), a major ligand of CD44, is the primary extracellular matrix component of brain tissue and is especially up-regulated in malignant glioma [14,15]. An increase in the accumulation of HA significantly impacts glioma cell malignancy and facilitates the development of GSCs characteristics [16,17]. However, the effect of HA-CD44 interaction on CSCs characteristics in glioma cells remains unclear.
In this study, we used shRNA to selectively reduce CD44 expression in rat (C6) and human (U87) glioma cell lines, and then cultured these cells on HA-coated plates to mimic the glioma microenvironment. Our data showed that reduced CD44 and increased GSCs traits were expressed under CD44 knockdown (CD44kd) or HA-supplemented (HA+) conditions. In contrast, HA+ combined with CD44kd or the application of the clinical drug TMZ increased CD44 expression and reduced GSCs traits. These data clearly indicate that glioma cells with lower CD44 expression exhibit more GSCs traits. Hence, we suggest that CD44 is not an appropriate marker of GSCs and that nestin and Oct4 are more suitable than CD44 as GSCs markers. Furthermore, the preferential distribution of CD44 at the invasive rim implies that CD44 may play an important role in glioma invasion.
Material and methods
Cell lines and culture
Rat glioma C6 (ATCCR CCL-107TM) and human glioma U87 (ATCCR HTB-14TM) cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 10 U/ml penicillin, 10 µg/ml streptomycin and 0.025 µg/ml amphotericin B at 37°C in a 5% CO2 incubator. Growth curves were assessed using the trypan blue dye exclusion method every 12 hours for a duration of 96 hours. The cell doubling time was calculated using the following formula: Doubling Time = [duration x log (2)]/[log (Final Concentration) - log (Initial Concentration)].
MTT assay
The dye 3-(4,5-dimethylthiazol)-2,5-diphenyltetrazilium bromide (MTT) (Sigma) was used for MTT assays as previously described [18]. A total of 3×104 cells were plated in 24-well plates and incubated overnight to allow cell attachment. Cell activity was then analyzed every 24 hours. The cells were treated with 20 μg MTT solution (500 μg/ml) for 4 hours and then solubilized with dimethysulfoxide (DMSO). Absorbance was read at 570 nm and used to determine cell activity using a multiscan reader (Dynatech, VA, USA).
RNAi transfectants for interference against CD44
The C6-sh1 and 2 cell lines were prepared by transfecting C6 rat glioma cells with the vector pBLOCK-iTTM-6-DEST, which contained a cDNA that constitutively expressed the rat CD44 shRNA, using Lipofectamine 2000. The second cell line, U87-sh, was prepared by transfecting U87 human glioma cells with the vector pLKO.1, which contained a cDNA that expressed the human CD44 shRNA, using a lentivirus. The cells were maintained in complete medium.
HA-coated culture dish
HA solution was prepared by dissolving HA powder (Mw: 1.47×106 Da; Pentapharm, Basel, Switzerland) in double-distilled water. After the HA solution had been applied to the surface of the culture dishes at 16 μg/cm2, the coated substratum was dried on a hot plate at 45°C for 30 min.
Reverse transcription PCR
Total RNA was extracted from cells using Trizol reagent (Life Technologies, Carlsbad, CA). One step RT-PCR was performed using a SuperScript™ One-Step RT-PCR with Platinum® Taq kit (Invitrogen) with 1 μg RNA, 12.5 μl 2X reaction buffer, 1 μl 10 mM primer and 1 μl superscript II platinum Taq polymerase in a final volume of 25 μl. PCR was performed at 94 °C (denaturation) for 30 s, at annealing temperature for 30 s, and at 74°C (elongation) for 1 min. All RT-PCR products were visualized using ethidium bromide staining under UV illumination after electrophoresis in a 1.2-1.5% agarose gel. The primers are listed in Table 1.
Table 1.
List and sequences of primers used in the RT-PCR analysis
| Annotation | Primer sequence | Cycle | T (°C) |
|---|---|---|---|
| Rat cd44 | 5’-TCCCACTATGACACATATTGC-3’ | 32 | 55 |
| 5’-ACACCTTCTCCTACTGTTGAC-3’ | |||
| Rat gfap | 5’-CAAGCCAGACCTCACAGCG-3’ | 32 | 55 |
| 5’-GGTGTCCAGGCTGGTTTCTC-3’ | |||
| Rat cd133 | 5’-AAGCAGCAAGTTGCCGGAGGAA-3’ | 35 | 61 |
| 5’-GTCATCTTCTCTGTGATGGCGTACA-3’ | |||
| Rat nestin | 5’-GCTACATACAGGACTCTGCTG-3’ | 35 | 58 |
| 5’-AAACTCTAGACTCACTGGATTCT-3’ | |||
| Rat oct4 | 5’-GAGGGATGGCATACTGTGGAC-3’ | 32 | 60 |
| 5’-GGTGTACCCCAAGGTGATCC-3’ | |||
| Rat has1 | 5’-GCTCTATGGGGCGTTCCTC-3’ | 35 | 57 |
| 5’-CACACATAAGTGGCAGGGTCC-3’ | |||
| Rat has2 | 5’-TCGGAACCACACTGTTTGGAGTG-3’ | 35 | 62 |
| 5’CCAGATGTAAGTGACTGATTTGTCCC-3’ | |||
| Rat β-actin | 5’-CCTCTGGTCGTACCAC-3’ | 25 | 47.2 |
| 5’-CAGTAACAGTCCGCCT-3’ | |||
| Human cd44 | 5’-CCAACTCCATCTGTGCAG-3’ | 32 | 50.3 |
| 5’-AACCTCCTGAAGTGCTGC-3’ | |||
| Human gfap | 5’-CGCGGCACGAACGAGTCC-3’ | 32 | 57 |
| 5’-GTGTCCAGGCTGGTTTCTCG-3’ | |||
| Human cd133 | 5’-CACTTACGGCACTCTTCACCT-3’ | 35 | 60 |
| 5’-TGCACGATGCCACTTTCTCAC-3’ | |||
| Human nestin | 5’-GGGACAGAGTTCTCCGAGCT-3’ | 35 | 67 |
| 5’-GAAGCCAGGACAGCAGGATC-3’ | |||
| Human oct4 | 5‘-GTGGAGGAAGCTGACAACAA-3’ | 35 | 60 |
| 5‘-GCCGGTTACAGAACCACACT-3’ | |||
| Human nanog | 5‘-CCTATGCCTGTGATTTGTGG-3’ | 35 | 57 |
| 5’-GTTGTTTGCCTTTGGGACT-3’ | |||
| Human β-actin | 5’-TGGAATCCTGTGGCATCCATGAAAC-3’ | 25 | 57.7 |
| 5’-TAAAACGCAGCTCAGTAACAGTCCG-3’ |
Flow cytometry
The cells were harvested and blocked in 3% BSA and then incubated with the following antibodies: rat CD44 (MA1-81319, Thermo, USA, 1:50) and human CD44 (MAB2137, Millipore, USA, 1:1000), nestin (AB5922, Chemicon, USA, 1:200), CD133 (LF-PA50121, AbFRONTIER, Korea, 1:100), Oct4 (PA1-16943, Thermo, USA, 1:100) and GFAP (G4546, Sigma, USA, 1:300). The color analyses were performed using a FACSCalibur system (Becton-Dickinson Immunocytometry Systems, San Jose, CA). The cell cycle was studied using total DNA staining. Cells were harvested and fixed with 70% ice-cold ethanol at 4°C for 1 hour. After they were centrifuged, the cells were resuspended and stained with 1 ml propidium iodide (PI) solution containing 0.2 mg RNase A, 20 μl 5% Triton X-100 and 20 μg PI (Sigma-Aldrich, USA) for 30 min. The cells were then analyzed using flow cytometry. Further analyses were performed using FLOWJo software.
Western blot analysis
The cells were collected from 10 cm culture dishes and washed with normal saline 3 times. A volume of 2 ml of lysis buffer (8 M urea, 4% CHAPS and protease inhibitor) was added to suspend the cells. The total cell lysate was centrifuged at 13,000 rpm at 4°C for 15 min to separate the soluble proteins from the cell lysate. An analytical 4-12% Bis-Tris gel (Invitrogen, USA) was used, and 30 μg of proteins were analyzed unless otherwise stated. For the immunoblotting experiments, the protein in the Bis-Tris gel was transferred to a polyvinylidene difluoride membrane using an electroblot apparatus. The membranes were then incubated for 2 h at 25°C with a the following specific primary antibody: rat nestin (MAB353, Millipore, USA, 1:1000), rat CD44 (MA1-81319, Thermo, USA, 1:10000), HAS1 (sc-23145, Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1:1000), HAS2 (sc-34068, Santa Cruz Biotechnology, Santa Cruz, CA, USA, 1:1000), or beta-actin (RB-9421-P1, Thermo, USA, 1:5000). An antibody coupled to horseradish peroxidase was used as the secondary antibody. An enhanced chemiluminescence kit (Millipore, USA) was used to detect the proteins.
Immunofluorescence
Cells were cultured on glass coverslips at 37°C overnight, washed twice with phosphate-buffered saline (PBS), and fixed with 2% paraformaldehyde on ice for 20 min. After the cells were blocked in 3% BSA for 1 hour, they were incubated overnight with the primary antibodies at 4°C. The following antibodies were used: mouse monoclonal anti-rat CD44 (Thermo, USA), rat monoclonal anti-CD44 (Millipore, USA), HAS1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), HAS2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), GFAP (Sigma, USA), Oct4 (Thermo, USA) and TGF-β (Abcam, Cambridge, UK). FITC-conjugated or R-phycoerythrin-conjugated secondary antibodies were used. After the coverslips were washed with PBS, the cell nuclei were counterstained with Hoechst dye 33342. Then, the slides were viewed and photographed using a fluorescence microscope.
Immunocytochemistry
Tissue sections were deparaffinized, treated with 3% H2O2 for 10 min at room temperature, and then microwaved in 0.01 M citrate buffer (pH 6.0) to retrieve antigenicity. The sections were blocked in 1% BSA, cultivated at 37°C for 30 min, and then incubated overnight at 4°C with a goat anti-human CD44 monoclonal antibody (1:100 dilution in PBS containing 1% BSA). The samples were washed four times with PBS, and goat anti-mouse IgG (PerkinElmer) was used as the second antibody. Immunocomplexes were visualized using a LSAB 2 HRP kit (Dako, Carpinteria, CA, USA) and 3,3’-diaminobenzidine tetrachloride as the substrate. Sections were lightly counterstained with hematoxylin, dehydrated through a graded alcohol series, cleared with xylene, and mounted with coverslips.
Giemsa staining
The cells were washed with PBS and fixed with 2 ml fixation solution (methanol (Merck, Germany): acetic acid (Merck, Germany) = 3:1) for 30 min. The cells were immersed in freshly prepared Giemsa’s buffer (6% Na2HPO4 (American Biorganics, INC) and 5% KH2PO4 (American Biorganics, INC), pH 7) for 10 min. They were then stained in Giemsa’s azure eosine methylene blue solution (Merck, Germany) for 45 min. The cells were rinsed with Giemsa’s buffer several times and then rapidly dipped in 5% acetic acid (Merck, Germany). The cells were dehydrated in 100% alcohol (Merck, Germany) and xylene (Kanto Chemical, CO). After the cells were air dried, cell morphology was observed using light microscopy.
Alcian blue stain
To analyze the distribution of hyaluronan in the rat brain, rat brain slides (10 µm) were incubated for 30 min at room temperature in 1% Alcian blue 8GX (Sigma) in 3% acetic acid, pH 2.5, and then rinsed four times.
Colony-formation assay
Cells were cultured in 12-well culture plates at 5,000 cells/well in 0.1% agar in DMEM culture medium over a 0.2% agar layer. The plates were then incubated for 14 days until the colonies were large enough to be visualized. Colonies were counted under a stereomicroscope. All experiments were performed in triplicate.
Orthotopic glioma model
All animal procedures were approved by the Institutional Animal Care and Use Committee (approval number: NCKU IACUC99075). Wistar rats (8 weeks old) were stereotaxically implanted with cells (1×105 cells/5 µl PBS) in the striatum at the following coordinates: 0.1 mm lateral and 0.3 mm anterior to bregma at a depth of 0.55 mm from the skull surface. Prior to surgery, the rats were anesthetized with sodium pentobarbital (45 mg/kg, intraperitoneally). Twenty-one days after the tumor cells were injected, all animals were sacrificed. The brains were removed following perfusion, placed in a series of sucrose gradient solutions, embedded in optimum cutting temperature compound (OCT, Tissue-Tek, Miles, Elkhart, IN), and stored at -80°C. Later, the brains were coronally sectioned, using a cryostat, into 10 µm-thick slices that were mounted on slides and then used for Giemsa staining and immunohistochemistry.
Patient samples
This study was approved and performed according to the guidelines of the Institutional Review Board of Chang Gung Memorial Hospital (approval #104-2656B). Written informed consent was obtained from all patients. The histopathological examinations were performed according to WHO criteria by a neuropathologist.
Statistical analysis
No assumptions were made regarding the data distribution, and all quantitative data are expressed as the mean ± standard deviation. Each experiment was repeated three times in duplicate. Statistical analyses of comparisons between two groups were evaluated using the one-tailed Mann-Whitney U-test. A probability value of P < 0.05 was considered statistically significant.
Result
Inhibition of CD44 expression by shRNA
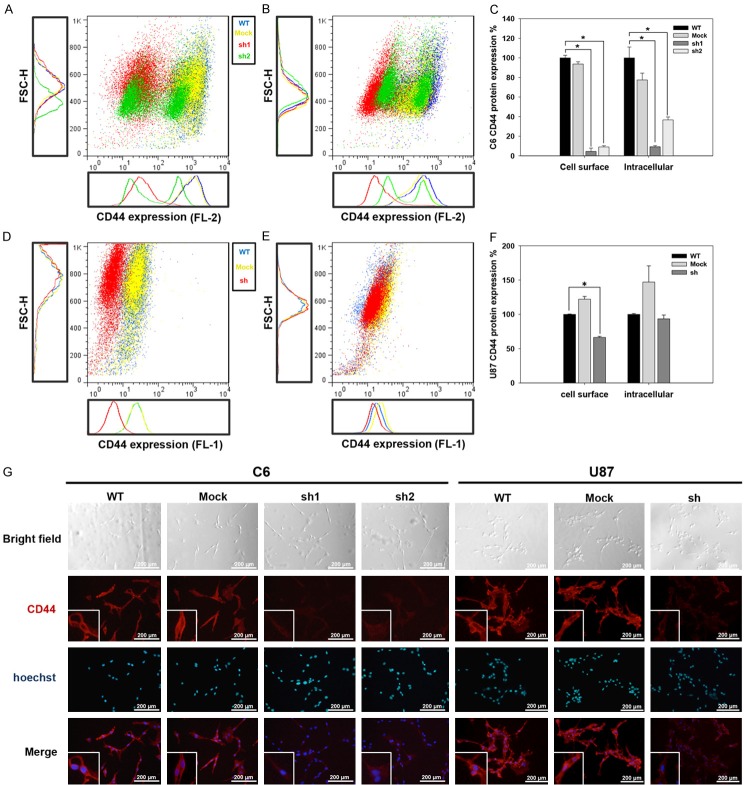
To study the effect of CD44 on GSCs traits, we used an shRNA to selectively reduce CD44 expression in rat (C6) and human (U87) glioma cell lines. Because the most dominant form of CD44 expressed in brain tumors is the standard form (CD44s), we designed shRNA fragments capable of targeting sequences specific to the standard CD44 transcript. RT-PCR analysis revealed that CD44 gene expression was reduced by 70%, 50% and 60% in C6-sh1, C6-sh2 (Supplementary Figure 1A and 1B) and U87-sh (Supplementary Figure 1C and 1D) cells, respectively. The results of protein analyses performed using flow cytometry (Figure 1A-C) and immunofluorescence assays (Figure 1G, left side) revealed a significant decrease in the expression of cell surface and intracellular CD44 protein in the C6-sh1 and C6-sh2 cells. Although the fluorescence intensity of CD44 labeling was lower in U87-sh than in U87-WT and U87-mock cells (Figure 1G, right side), only the cell surface CD44 protein was reduced in U87-sh cells according to flow cytometry (Figure 1D-F).
Figure 1.
CD44 protein expression in C6 and U87 cells. Flow cytometry and immunofluorescence analysis of CD44 protein expression in C6 and U87 cells. A-F. CD44 expression was analyzed using flow cytometry. The blue dot indicates WT, the yellow dot indicates the mock, and the red dot indicates sh. A. The expression of the cell surface CD44 in C6 cells. B. The expression of the intracellular domain of CD44 in C6 cells. C. Statistical analysis of CD44 protein expression in C6 cells. D. The expression of the cell surface CD44 in U87 cells. E. The expression of the intracellular domain of CD44 in U87 cells. F. Quantification of CD44 protein expression in U87 cells. G. Immunofluorescence detection of the CD44 protein (red in upper panel) in C6 and U87 glioma cells. Blue indicates Hoechst staining of cell nuclei (middle panel) (*p value < 0.05).
CD44 knockdown prolonged the cell cycle
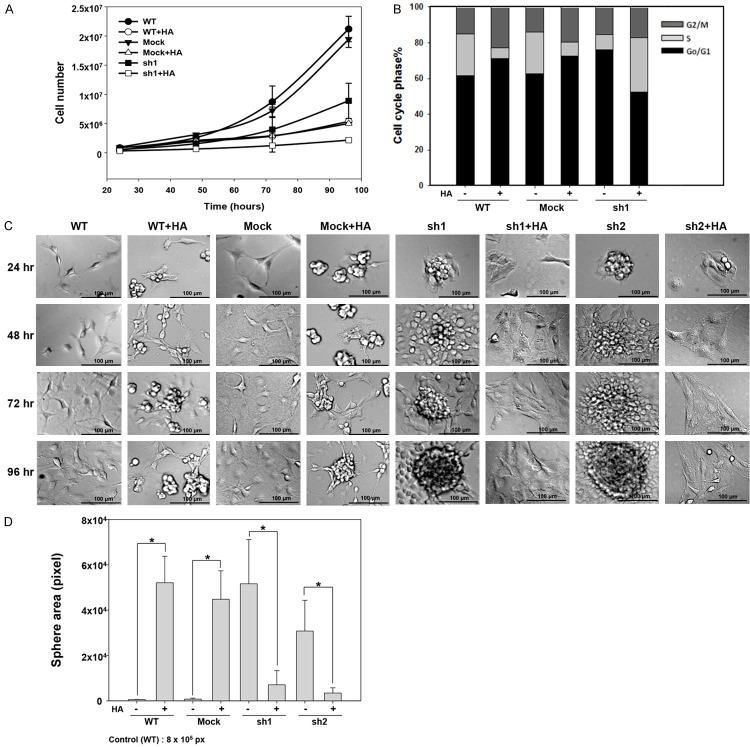
To investigate the effects of CD44 on cell growth, we counted cells every 24 hours for a period of 360 hours and then calculated growth curves using the trypan blue dye exclusion method. The results revealed that cell growth was slower in C6-sh1 and C6-sh2 cells than in C6-mock and C6-WT (control) cells for the first 7 days. However, despite the fact that the C6-mock and C6-WT cells had higher proliferation rates, these cells exhibited decreased growth on day 8. Conversely, the C6-sh1 and C6-sh2 cells continued growing until day 14 and 12, respectively, ultimately achieving a cell density similar to that in the C6-WT and C6-mock cells (Figure 2A). It worth noting that the pattern of cell growth was similar between C6-sh1 and C6-sh2 cells but that the doubling time was longer in C6-sh1 cells than in C6-sh2 cells by 6 hours (44.02 and 38.08 hours, respectively). The same result was observed in MTT assays (Supplementary Figure 2). To further confirm the effects of CD44 on cell growth, we analyzed the cell cycle in C6 cell lines. CD44 is a cell-surface glycoprotein involved in cell-cell and cell-extracellular matrix interactions. To prevent an effect of cell density, all experiments were performed at the same cell density (approximately 3×106 cells/10 cm culture dish) in this study. As shown in Table 2, in C6-sh1 cells, the decrease in the percentage of cells in S and G2/M phase and the increase in the percentage of cells in G0/G1 phase without a sub-G1 increase indicating that C6-sh1 cells were arrested in G0/G1 phase. These results demonstrated that lower CD44 expression levels prolong the cell cycle but increase long-term proliferative capacity.
Figure 2.
Cell growth, morphology and sphere formation ability in CD44kd cells. A. The growth curves of the C6 cell lines were analyzed every 24 hours over a period of 360 hours by performing cell counting using the trypan blue dye exclusion method. The total number of cells was counted at each time point. (-λ-) WT, (-○-) mock, (-τ-) sh1, and (-λ-) sh2. n = 3 (NS: non-significant difference). B. Cell density and morphology in C6 cell lines. Scale bar = 200 μm. The fourth and sixth panels are high magnification views of the sections shown in white frames in C6-sh1 and C6-sh2 cells, respectively. C. Sphere formation was observed in C6 cell lines on day 4 using Giemsa staining. D. Statistical analysis of the spherical area in C6 cell lines. E. Sphere formation was observed in U87 cell lines on day 6 using Giemsa staining. Scale bar = 500 μm. F. Statistical analysis of the spherical area in the U87 cell lines. G. The sphere-forming ability of the C6 cell lines was investigated using 3D colony-formation assays. C6 cells were seeded in 0.1% agar in DMEM culture medium over a layer of 0.2% agar, and the colonies were observed after 14 days of incubation. H. Statistical analysis of colony numbers in C6 cell lines (*p value < 0.05).
Table 2.
Cell cycle analysis of C6 cell lines. The cells were collected at the same density (approximately 3×106 cells/10 cm culture dish) and then analyzed according to PI fluorescence intensity using flow cytometry. The table shows the percentage of cells in the sub-G1, -G0/G1, -G2/M or -S phases of the cell cycle across the experimental C6 cell lines
| Cell Population | % of Gated Cells (mean ± SD) | |||
|---|---|---|---|---|
|
|
||||
| C6-WT | C6-mock | C6-sh1 | p value | |
| sub G1 | 1.74 ± 0.8 | 2.27 ± 2.28 | 2.34 ± 1.32 | 0.207315 |
| G0/G1 | 36.54 ± 1.59 | 39.88 ± 4.64 | 72.03 ± 1.16 | 0.00591¶ |
| S | 39.05 ± 2.39 | 36.52 ± 2.8 | 15.75 ± 0.72 | 0.002937¶ |
| G2/M | 22.65 ± 1.36 | 21.32 ± 4.4 | 9.87 ± 0.7 | 0.004046¶ |
The significance of the difference between C6-WT and C6-sh1 cells.
CD44kd induced changes in cell morphology and enhanced the ability of cells to form spheres
To investigate cell morphology, cells were seeded on culture dishes at a density of 1×106 cells/10 cm dish. The morphologies of C6-WT, C6-mock, C6-sh1 and C6-sh2 cells were observed every 24 hours for a period of 96 hours. We found that the C6-WT and C6-mock cells had a normal fibroblast morphology, while C6-sh1 and C6-sh2 cells had irregular morphologies and began to aggregate after 24 hours. After 72 hours, these cells had formed a tight colony (patch), and the cells were spherical in shape by 96 hours (Figure 2B). We used Giemsa staining to analyze sphere formation in the C6 and U87 cell lines on day 4 and day 6, respectively. The C6-sh1, C6-sh2 and U87-sh cells showed greater patch formation than the WT and mock cells (Figure 2C and 2E), and the number of spheres in these cells was also higher than that observed in WT and mock cells (Figure 2D and 2F). In addition to the traditional 2D monolayer culture, sphere-forming ability was also investigated using 3D colony-formation assay. The C6 cells were seeded in 12-well culture plates at 5,000 cells/well in 0.1% agar in DMEM culture medium over a 0.2% agar layer, and the colonies were counted after 14 days of incubation. As shown in Figure 2G and 2H, the C6-sh1 cells had 5.2-fold higher colony-formation ability than the C6-WT and C6-mock cells. Sphere-forming ability rapidly emerged as a potential indicator that is applied to isolate embryonic stem cells (ESCs) and adult stem cells (ASCs), and for studying the biology of each of these cell types. Our data show that glioma cells expressing low levels of CD44 were better able to form spheres, suggesting that cells with low CD44 expression may possess stem cell-like properties.
Stem cell marker expression was increased by CD44kd
In many cell types, cell proliferation is mediated by the coupling of growth arrest and cellular differentiation. Our results show that the shRNA-mediated depletion of CD44s decreased glioma cell growth and led to cell cycle arrest at the G0/G1 phase, implying a differentiated phenotype [19]. However, our results also revealed that glioma cells expressing low levels of CD44 have greater sphere-forming capacity. To further clarify the effects of CD44 on cell differentiation, the expression of the astrocyte and glial differentiation marker GFAP was analyzed [20]. Our data showed that GFAP gene expression was 37% lower in C6-sh1 cells than in C6-WT cells (Figure 3A and 3B). Similarly, GFAP gene expression was 50% lower in U87-sh cells than in U87-WT cells (Figure 3C and 3D). Furthermore, the expression of the GFAP protein was 79% lower in C6-sh1 cells than in C6-WT cells (Figure 3E) and 43% lower in U87-sh cells than in U87-WT cells (Figure 3F). These results indicate that a reduction in CD44 expression produced dedifferentiated glioma cells. It was previously reported that stem cells are relatively quiescent compared to their differentiated offspring [21]. Our results show that the proliferation and differentiation rates of CD44kd cells are similar to those of stem cells. To investigate whether the stem cell-like properties of glioma cells are affected by CD44, we analyzed the expression of stem cell markers, including CD133, nestin, Nanog and Oct4, in C6 and U87 cells. The results of RT-PCR and flow cytometry indicated that in C6-sh1 cells, CD133 mRNA expression was 1.7-fold higher (Figure 3G) and CD133 protein expression was 3.6-fold higher than in C6-WT cells (Figure 3H). The expression pattern of nestin was similar to that of CD133. Specifically, the expression levels of both the nestin mRNA and protein were 2.1-fold higher in C6-sh1 cells (Figure 3G and 3H). Furthermore, Oct4, which is involved in self-renewal in undifferentiated ESCs, was also expressed at higher levels in CD44kd cells. The gene and protein expression of Oct4 were 2-fold and 2.2-fold higher in C6-sh1 cells expression was 2-fold higher in C6-sh1 cells (Figure 3G and 3H). Similarly, the expression levels of stem cell markers were higher in U87-sh cells. The gene expression levels of CD133, nestin, Oct4, and Nanog were 1.7-, 2.3-, 1.6-, and 2-fold higher in U87-sh cells (Figure 3I). And the protein expression levels of the CD133, nestin and Oct4 were increased in U87-sh cells by 3.8-, 2.4-, and 1.8-fold, respectively (Figure 3J). Furthermore, the distribution of these markers was also investigated in a rat model of glioma. As shown in Figure 4, both CD44 and GFAP were highly expressed at the margin; however, Oct4 was highly expressed in the core region of gliomas. These results indicate that a lower level of CD44 resulted in dedifferentiation characteristics and caused glioma cells to present properties similar to those observed in stem cells in both rat and human glioma cells.
Figure 3.
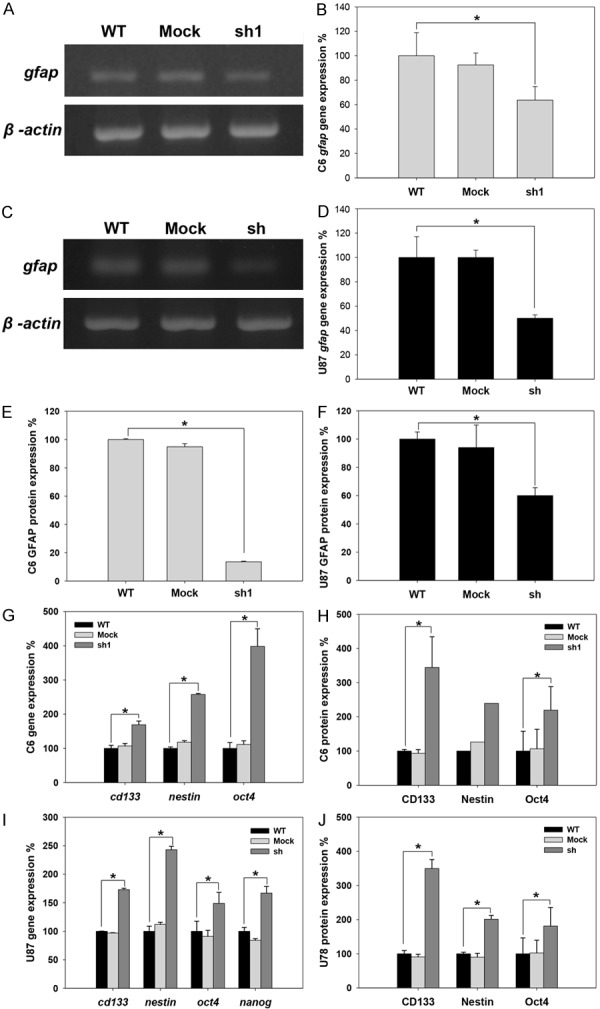
Expression level of the glial cell differentiation marker (GFAP) and stem cell markers (CD133, nestin, oct4 and nanog) in C6 and U87 cell lines. The expression levels of stem cell markers were analyzed at the same cell density (approximately 3×106 cells/10 cm culture dish) at both the transcriptional and translational levels in C6 and U87 cells. A. The expression of the GFAP mRNA in C6 cell lines. B. Quantification of GFAP mRNA expression in C6 cell lines. C. The expression level of GFAP mRNA in U87 cell lines. D. Quantification of GFAP mRNA expression in U87 cell lines. E. Statistical analysis of GFAP protein expression in C6 cell lines detected using flow cytometry. F. Statistical analysis of GFAP protein expression in U87 cell lines detected using flow cytometry. G. The expression levels of the CD133, nestin and Oct4 mRNAs were detected in C6 cell lines using RT-PCR. H. The protein expression levels of CD133, nestin and Oct4 were detected in C6 cell lines. CD133 and Oct4 expression levels were detected using flow cytometry, and nestin expression was detected using Western blot analysis. I. The gene expression levels of CD133, nestin, Oct4 and nanog were detected in U87 cells using RT-PCR. J. The expression levels of the CD133, nestin and Oct4 proteins were detected in U87 cell lines using flow cytometry (*p value < 0.05).
Figure 4.
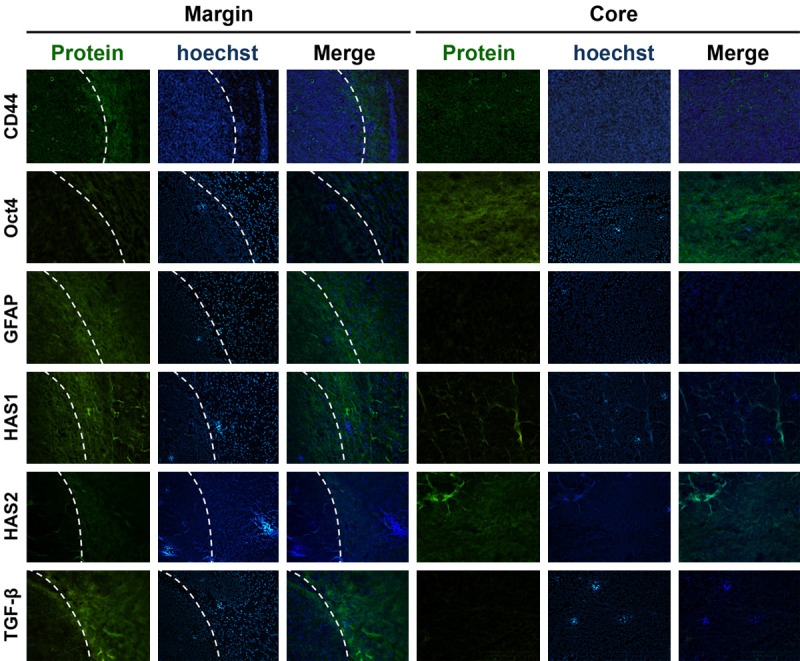
The distribution of the CD44, Oct4, GFAP, TGF-B, HAS1 and HAS2 proteins in rat glioma. Photomicrograph of a C6 rat glioma section stained using immunofluorescence for CD44, Oct4, GFAP, TGF-B, HAS1 and HAS2 (green) at its margin (left) and core (right). The white dotted line in the left panel indicates the edge that separates the tumor core from the invasive rim. Blue indicates Hoechst-stained cell nuclei.
Opposing distribution patterns of HA and CD44 in human and rat glioma
CD44 stimulates intracellular signal transduction to regulate cellular processes by interacting with its ligand. It has been consistently observed that the concentration of HA, the major ligand of CD44, is higher in glioma regions than in normal brain tissue. Therefore, we investigated the distribution of HA and CD44 in human and rat gliomas. As shown in Figure 5A, CD44 expression was higher in human GBM than in lower-malignancy gliomas (Figure 5Aa-f). However, a higher level of CD44 expression region was associated with a lower HA concentration in human specimens (Figure 5Ag-l). This antagonistic phenomenon between HA and CD44 was also observed in a rat glioma model. As shown in Figure 5B, both CD44 and HA were highly distributed at glioma regions in rat brains, but the HA concentration was higher in the core region (Figure 5Bb), while CD44 expression was higher at margin of the glioma region (Figure 5Bc). These data indicate that a higher concentration of HA is associated with a lower level of CD44 in both rat and human gliomas. To further investigate the relationship between CD44 and HA in glioma, culture dishes were coated with high molecular weight HA (HMW-HA) (1.47×106 Da) to mimic the glioma microenvironment because an abundance of HMW-HA has been observed in brain tumors [22,23]. C6 cells were seeded on culture dishes with or without a coating of HA (16 μg/cm2) at a cell density 1×106/10 cm dish, and the expression of CD44 was analyzed at both the transcriptional and translational levels. The in vivo results showed that both the mRNA and protein expression level of CD44 were decreased by HA+ in C6-WT and C6-mock cells. Interestingly, CD44 expression was increased in C6-sh1 and C6-sh2 cells grown by HA+ (Figure 5C-E).
Figure 5.
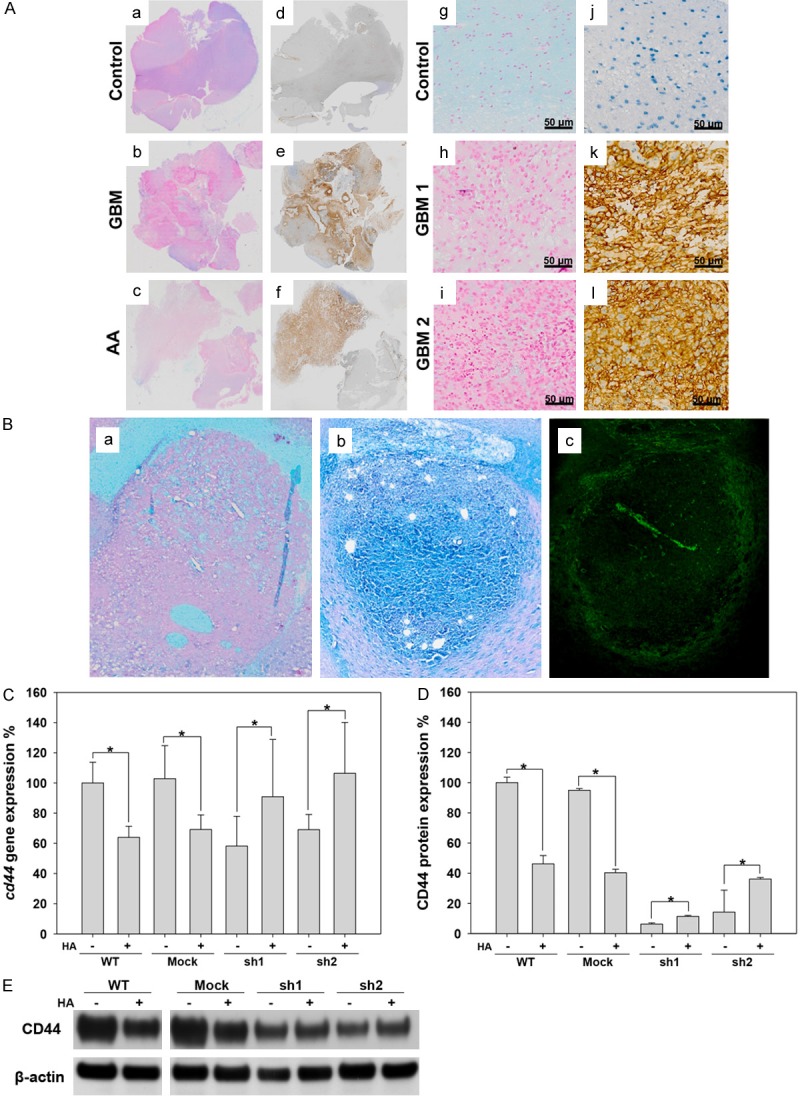
The distribution of CD44 and HA in glioma. A. a-f: The distribution of HA and CD44 in control (trauma), astrocytoma (AA) and GBM patients. g-l: Enlargement of sections from human brains with trauma and GBM that were stained for HA and CD44. Right panel: CD44. Left panel: HA. B. The distribution of HA and CD44 in a C6 rat glioma. a: Alcian blue staining for HA in a rat brain injected with only PBS (control). b: Alcian blue staining for HA in a C6 rat glioma. c: Immunofluorescence staining for CD44 (green) in a C6 rat glioma. C. The expression of cd44 mRNA detected using RT-PCR in C6 cell lines. D. Flow cytometry analysis of CD44 expression in C6 cell lines. E. Western blot analysis of CD44 in C6 cell lines.
Effects of HA on cell growth and morphology in C6 glioma cell lines
To investigate the effect of HA on cell growth, we performed cell counting every 24 hours for a duration of 96 hours using the trypan blue dye exclusion method. Cell morphology was also observed during this period (Figure 6). The data showed that HA+ decreased cell growth in both C6-WT, C6-mock and C6-sh1 cells (Figure 6A). In addition, as shown in Figure 6B, an analysis of the cell cycle revealed that the percentage of cells in G0/G1 phase was higher and the percentage of cells in S phase was lower in C6-WT and C6-mock cells treated with HA+. In contrast, HA+ decreased the percentage of G0/G1 phase cells and increased the percentage of S phase cells in C6-sh1 cells.
Figure 6.
Cell proliferation and sphere formation affected by HA. A. Growth curve in C6 cell lines treated with HA+. n = 3. B. Cell cycle analysis of C6 cell lines treated with HA+. C. Morphology of C6 cell lines treated with HA+. Scale bar = 100 μm. D. Statistical analysis of sphere formation ratios in C6 cell lines (*p value < 0.05).
Our analysis of cell morphology showed that the C6-WT and C6-mock cells had a spindle-like shape, but the morphology of the C6-WT and C6-mock cells that were grown with HA+ was irregular, and the cells began to aggregate after 24 hours and to form tight colonies (patches) after 96 hours. As our previous data showed, C6-sh1 and C6-sh2 cells expressed a spherical morphology; however, treatment with HA+ eliminated the sphere-forming capability of C6-sh1 and C6-sh2 cells and caused their cellular morphology to become flat and irregular (Figure 6C). The quantity of sphere area also showed the similar result (Figure 6D).
Effects of HA on stem cell markers in C6 glioma cell lines
As previously described, CD44 expression, which was affected by HA+, can itself affect GSCs properties in C6 cells. To further clarify the effect of HA on GSCs characteristics, we investigated the expression of GSCs markers in C6 cell lines treated with HA+. As shown in Figure 7, an analysis of expression at both the transcriptional and translational levels showed that GFAP expression was decreased by HA+ in C6-WT and C6-mock cells but increased by HA+ in C6-sh1 and C6-sh2 cells (Figure 7A, 7B). In contrast, the expression levels of both the mRNAs and the proteins of the stem cell markers CD133, Oct4 and nestin were higher in C6-WT and C6-mock cells but lower in C6-sh1 and C6-sh2 cells treated with HA+ (Figure 7C-H). These data revealed that the HA+ caused CD44 high-expressing cells (C6-WT and C6-mock) to exhibit stem cell-like characteristics but caused CD44 low-expressing cells (C6-sh1 and C6-sh2) to exhibit more differentiated characteristics.
Figure 7.
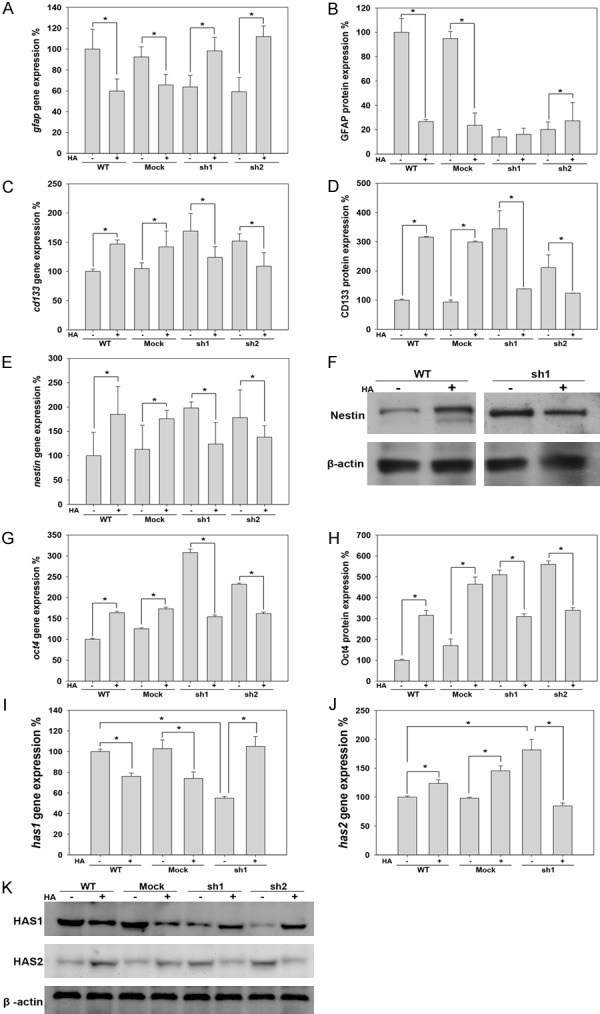
Expression of the differentiation marker (GFAP), stem cell markers (CD133, nestin and oct4) and HAS in response to HA in C6 cell lines. A. The expression of the gfap mRNA was detected using RT-PCR. B. GFAP protein expression in C6 cell lines was analyzed using flow cytometry. C. The expression of the cd133 mRNA was detected using RT-PCR. D. CD133 protein expression was analyzed in C6 cell lines using flow cytometry. E. The expression of the nestin mRNA was detected using RT-PCR. F. Western blot analyses of Nestin protein expression in C6 cell lines. G. The expression of the oct4 mRNA was detected in C6 cell lines using RT-PCR. H. Western blot analyses of Oct4 protein expression in C6 cell lines. I. The expression of the has1 mRNA was detected using RT-PCR. J. The expression of the has2 mRNA was detected using RT-PCR. K. Western blot analyses of HAS1 and HAS2 protein expression levels in C6 cell lines (*p value < 0.05).
Based on these results, we suggest that HA+ caused the diverse cell differential direction in glioma cells that express different levels of CD44. These results indicate that the effect of HA on GSCs characteristics and cell differentiation is probably mediated by CD44.
Effects of HA on HA metabolism-associated gene expression in C6 glioma cell lines
The direction of cell differentiation could be affected by HA with different molecular weight [18]. In mammalian cells, HA is synthesized in different molecular weight forms depending on tissue-specific HA synthases (HAS). HAS2 synthesizes HMW-HA, while the comparatively low molecular weight HA (LMW-HA) is synthesized by HAS1 and HAS3 [24]. Because our data showed that the exogenous HMW-HA causes distinct direction of differentiation in glioma cells, depending on the level of CD44 expression, we sought to investigate the expression of endogenous HAS expression in this study. As shown in Figure 7, HAS1 expression was decreased by CD44kd or HA+ in C6-WT and C6-mock cells, and the decrease in HAS1 expression observed in CD44kd cells was recovered by HA+ (Figure 7I and 7K). The pattern of HAS2 expression was opposite that of HAS1 (Figure 7J and 7K). The expression of HAS3 was not detected in C6 gliomas. The in vitro results showed that HAS1 distribution was similar to CD44 that was high at the margin, while HAS2 was highly expressed in the core region in a rat glioma model (Figure 4). The TGF-β has been shown to decrease HAS2 but increase HAS1 expression and the TGF-β receptor is co-expressed with CD44 [25,26]. Hence, to further clarify the relationship between CD44 and HAS, the expression of TGF-β was investigated in this study. As shown in Figure 4, the distribution of TGF-β is consistent with CD44 and HAS1 that is high at the margin in rat glioma tissues.
Treatment with TMZ elevated the expression of CD44 and GSCs markers in C6 and U87 glioma cell lines
TMZ is the current standard clinical treatment in glioma patients [27]. In this study, we treated C6 and U87 cells with 25 μg/ml TMZ and then analyzed the expression of CD44 and GSCs markers, including CD133, Oct4, nanog and nestin, after 96 hours to investigate the effect of TMZ on GSCs traits. As shown in Figure 8, the expression of CD44 was significantly higher after treatment with TMZ. Conversely, all of the GSCs markers were diminished by TMZ treatment in both C6 and U87 cells (Figure 8). This result demonstrates that TMZ significantly increased CD44 expression and that this effect was accompanied by a reduction in GSCs characteristics.
Figure 8.
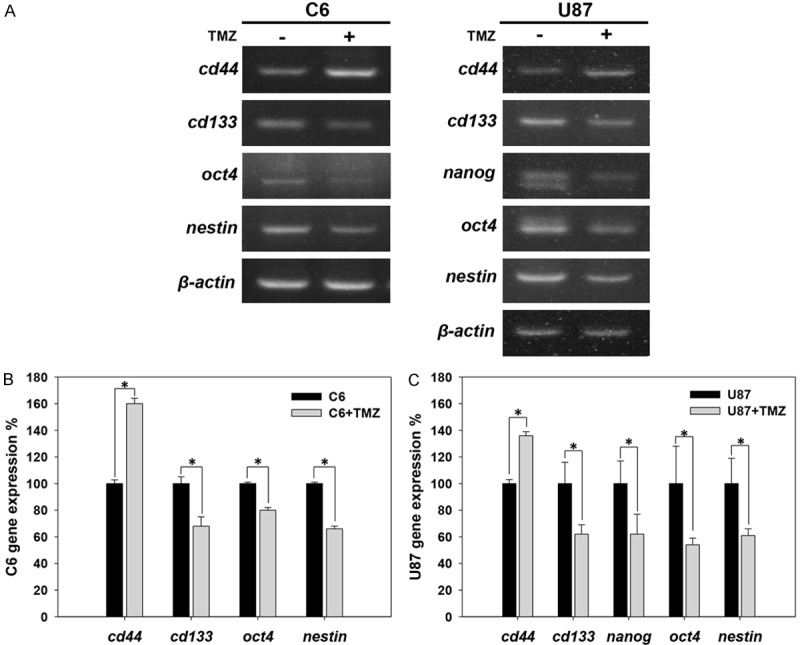
Expression of CD44 and GSCs markers in response to TMZ in C6 and U87 glioma cell lines. The expression levels of cd44, cd133, oct4, nanog and nestin in C6 and U87 cells treated with TMZ were analyzed using RT-PCR. (A) Analysis of cd44, cd133, oct4, nanog and nestin gene expression at 96 hours after treatment with 25 μg/ml TMZ. The results were analyzed using RT-PCR. The bar graph shows the quantification of the expression of these genes in (B) C6 and (C) U87 cells treated with or without TMZ (*p value < 0.05).
Discussion
The reason for expanding research into CSCs identification and targeting is that cancers arise from CSCs, and cancer might therefore be defeated if CSCs can be specifically eliminated. CD44 has been recognized as a CSCs marker in a variety of cancer types [28]. However, in this study, we provide evidence showing that reducing CD44 using different methods (CD44kd, HA+) induced the expression of GSCs markers and characteristics, including CD133, nestin, and Oct4 expression, sphere formation capacity and long-term proliferation. In contrast, applying HA+ in combination with CD44kd or treatment with TMZ increased CD44 expression and reduced GSCs characteristics. Therefore, we deduced that CD44 is not an appropriate marker for GSCs.
Over the past decade, various surface markers have been used to isolate potential CSCs in different tumors. One marker of considerable interest in GSCs is CD44 [7]. The decrease in brain tumor growth observed in CD44-/- mice suggests that only CD44high cells exhibit GSCs properties [9]. However, GBM has been known to have a histologically complex and heterogeneous cellular composition [29,30]. These data, combined with the presence of a variety of CSCs markers, persuaded us that GSCs might also be heterogeneous and not represented by one particular phenotype. This assumption is supported by previous studies, which have explored the complexity of GSCs marker expression that CD133- cells can possess GSCs properties as well as CD133+ cells derived from GBM [31-33]. Recently, several studies have indicated that GSCs lines derived from primary GBM can be divided into distinct types according to their gene expression profiles and that both CD44low/CD133high and CD44high/CD133low GSCs have sphere-forming ability and high tumorigenicity [11,12]. Furthermore, CD44 is highly expressed in several glioma cell lines, including U87, U373, and T98G cells, but was undetectable in a GBM-derived stem-like cell line [10]. In combination with our data that CD44 low-expressing glioma cells exhibit GSCs traits, these data indicating that GSCs are heterogeneous.
In addition to the genetic traits inherent in cancer cells, the tumor microenvironment is thought to significantly impact tumor onset and progression [34]. In the central nervous system, HA has been shown to have an effect on the differentiation of glial cells [35,36]. Here, we show that in wild type glioma cells, HMW-HA maintained the cells in a quiescent GSCs state and down-regulated CD44. Furthermore, cells treated with TMZ efficiently increased the expression of CD44 and reduced the levels of other GSCs markers. These results further confirm that CD44 and GSCs traits are negatively correlated, indicating that CD44 is not a suitable marker for GSCs.
Although HA+-induced GSCs properties were observed in wild type glioma cells, it is interesting that combining HA+ with CD44kd conversely triggered cell differentiation by increasing CD44 expression (Figure 9). HA induces different biological signals depending on its molecular weight. An accumulation of HMW-HA maintains progenitor cells in a quiescent state, whereas LMW-HA promotes cell differentiation [35,36]. Here, we show that CD44 low-expressing glioma cells, which express high levels of HAS2 and low levels of HAS1, exhibit GSCs-like properties. The overexpression of HAS2 has been reported to promote CSCs characteristics, including anchorage-independent growth and tumorigenicity [37]. Thus, we suggest that the quiescent GSCs traits observed in CD44 low-expressing glioma cells might be maintained by the accumulation of HMW-HA. In contrast, CD44 high-expressing glioma cells express low levels of HAS2 and high levels of HAS1, which may result in the accumulation of LMW-HA and facilitate cell differentiation (Figure 9). TGF-β has been reported to down-regulate HAS2 and up-regulate HAS1 expression [25,26]. Here, we show that the distribution of TGF-β was contrary to HAS2 but consistent with HAS1 and CD44. Together, these results indicate that the diverse directions of differentiation observed in glioma cells expressing different levels of CD44 may be manipulated through the CD44/TGF-β/HAS pathway.
Figure 9.
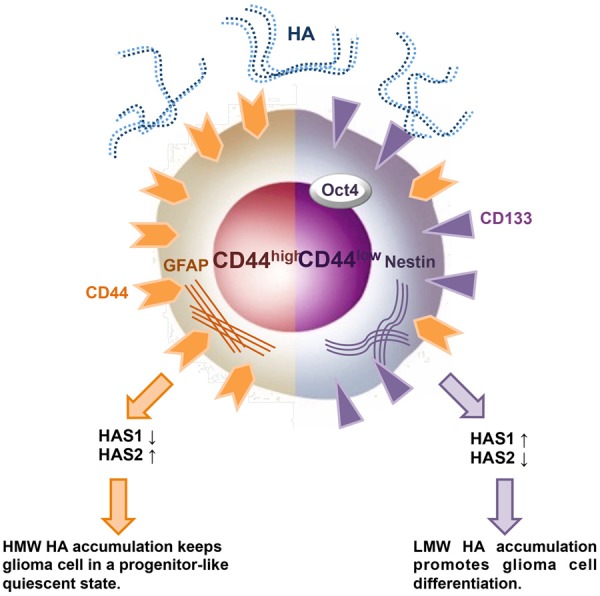
A model of cell differentiation in glioma cells expressing different levels of CD44 in an HA-rich environment. The model summarizes the results of this study, which shows that in an HA-rich environment, the different directions of differentiation occurs in cell lines expressing different levels of CD44. The effects of the HA-CD44 interaction on the levels of HAS1 and HAS2 are also shown. Orange and purple are used to indicate the effect of HA on cell dedifferentiation in mature glioma cells (CD44high) and stem cell-like glioma cells (CD44low).
In addition to being a CSCs marker, CD44 has also been implicated in many important biological processes in cancer. Similar to our results, the absence of CD44 decreases the initial proliferation rate; nevertheless, CD44kd cells exhibit higher proliferative ability in endothelial and GBM cells [38,39], suggesting that CD44 maintains mitotic activity for an extended period of time. CD44 expression is also associated with a metastatic phenotype [40-42]. The CD44+ GSCs had enriched invasion and migration pathways [43], and consistent with our result that CD44 was preferentially expressed at the invasive rim, implying that CD44 is important for invasion and migration [44].
In conclusion, in this study, we provided evidence showing that CD44 is not an appropriate marker for GSCs. Instead, the major roles of CD44 in glioma may be proliferation, migration and invasion. The GSCs traits observed in CD44 low-expressing cells were probably induced by elevated levels of Oct4 and nestin, which are more appropriate GSCs markers and generally consistent across all CSCs. This result likely explains the longer median survival time observed in GBM patients with higher CD44 expression in our previous clinical study [13].
Acknowledgements
This research was supported by the National Health Research Institutes [grant numbers NHRI-EX93-9223NI and NHRI-EX106-10502NI], Taiwan; and through the National Cheng Kung University [Research Grant 1700-B10] and Chang Gung Memorial Hospital [Research Grant CMRPG3D1063 and CMRPG3D1113].
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Kalokhe G, Grimm SA, Chandler JP, Helenowski I, Rademaker A, Raizer JJ. Metastatic glioblastoma: case presentations and a review of the literature. J Neurooncol. 2012;107:21–27. doi: 10.1007/s11060-011-0731-1. [DOI] [PubMed] [Google Scholar]
- 2.Pollak J, Rai KG, Paddison PJ, Rostomily RC, Ramirez JM. Transcriptional profiling of glioblastoma stem-like cells reveals enrichment of ion channels with functional implications for malignancy. AACR. 2016;76:2523. [Google Scholar]
- 3.Bucay N, Sekhon K, Yang T, Majid S, Shahryari V, Hsieh C, Mitsui Y, Deng G, Tabatabai ZL, Yamamura S, Calin GA, Dahiya R, Tanaka Y, Saini S. MicroRNA-383 located in frequently deleted chromosomal locus 8p22 regulates CD44 in prostate cancer. Oncogene. 2017;36:2667–2679. doi: 10.1038/onc.2016.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponnurangam S, Dandawate PR, Dhar A, Tawfik OW, Parab RR, Mishra PD, Ranadive P, Sharma R, Mahajan G, Umar S, Weir SJ, Sugumar A, Jensen RA, Padhye SB, Balakrishnan A, Anant S, Subramaniam D. Quinomycin A targets Notch signaling pathway in pancreatic cancer stem cells. Oncotarget. 2016;7:3217–3232. doi: 10.18632/oncotarget.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon C, Park DJ, Schmidt B, Thomas NJ, Lee HJ, Kim TS, Janjigian YY, Cohen DJ, Yoon SS. CD44 expression denotes a subpopulation of gastric cancer cells in which Hedgehog signaling promotes chemotherapy resistance. Clin Cancer Res. 2014;20:3974–3988. doi: 10.1158/1078-0432.CCR-14-0011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Su YJ, Lai HM, Chang YW, Chen GY, Lee JL. Direct reprogramming of stem cell properties in colon cancer cells by CD44. EMBO J. 2011;30:3186–3199. doi: 10.1038/emboj.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, Prieto-Sanchez RM, Barba I, Martinez-Saez E, Prudkin L, Cuartas I, Raventos C, Martinez-Ricarte F, Poca MA, Garcia-Dorado D, Lahn MM, Yingling JM, Rodon J, Sahuquillo J, Baselga J, Seoane J. TGF-beta receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Stamenkovic I, Yu Q. CD44 attenuates activation of the hippo signaling pathway and is a prime therapeutic target for glioblastoma. Cancer Res. 2010;70:2455–2464. doi: 10.1158/0008-5472.CAN-09-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pietras A, Katz AM, Ekstrom EJ, Wee B, Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT, Holland EC. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14:357–369. doi: 10.1016/j.stem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He J, Liu Y, Xie X, Zhu T, Soules M, DiMeco F, Vescovi AL, Fan X, Lubman DM. Identification of cell surface glycoprotein markers for glioblastoma-derived stem-like cells using a lectin microarray and LC-MS/MS approach. J Proteome Res. 2010;9:2565–2572. doi: 10.1021/pr100012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lottaz C, Beier D, Meyer K, Kumar P, Hermann A, Schwarz J, Junker M, Oefner PJ, Bogdahn U, Wischhusen J, Spang R, Storch A, Beier CP. Transcriptional profiles of CD133+ and CD133- glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res. 2010;70:2030–2040. doi: 10.1158/0008-5472.CAN-09-1707. [DOI] [PubMed] [Google Scholar]
- 12.Fu J, Yang QY, Sai K, Chen FR, Pang JC, Ng HK, Kwan AL, Chen ZP. TGM2 inhibition attenuates ID1 expression in CD44-high glioma-initiating cells. Neuro Oncol. 2013;15:1353–1365. doi: 10.1093/neuonc/not079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei KC, Huang CY, Chen PY, Feng LY, Wu TW, Chen SM, Tsai HC, Lu YJ, Tsang NM, Tseng CK, Pai PC, Shin JW. Evaluation of the prognostic value of CD44 in glioblastoma multiforme. Anticancer Res. 2010;30:253–259. [PubMed] [Google Scholar]
- 14.Laurent TC, Fraser JR. Hyaluronan. FASEB J. 1992;6:2397–2404. [PubMed] [Google Scholar]
- 15.Poukka MJ, Haapasalo H, Rilla KJ, Tyynelä-Korhonen K, Soini Y, Pasonen-Seppänen SM. Elevated expression of hyaluronan synthase 2 associates with poor prognosis in diffusely infiltrating astrocytomas. AACR. 2016;76:3111. doi: 10.1186/s12885-018-4569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedron S, Becka E, Harley BA. Regulation of glioma cell phenotype in 3D matrices by hyaluronic acid. Biomaterials. 2013;34:7408–7417. doi: 10.1016/j.biomaterials.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 17.Florczyk SJ, Wang K, Jana S, Wood DL, Sytsma SK, Sham JG, Kievit FM, Zhang M. Porous chitosan-hyaluronic acid scaffolds as a mimic of glioblastoma microenvironment ECM. Biomaterials. 2013;34:10143–10150. doi: 10.1016/j.biomaterials.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghatak S, Maytin EV, Mack JA, Hascall VC, Atanelishvili I, Moreno Rodriguez R, Markwald RR, Misra S. Roles of proteoglycans and glycosaminoglycans in wound healing and fibrosis. Int J Cell Biol. 2015;2015:834893. doi: 10.1155/2015/834893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott RE, Florine DL, Wille JJ, Yun K. Coupling of growth arrest and differentiation at a distinct state in the G1 phase of the cell cycle: GD. Proc Natl Acad Sci U S A. 1982;79:845–849. doi: 10.1073/pnas.79.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santos GC, Carvalho KC, Falzoni R, Simoes AC, Rocha RM, Lopes A, Vassallo J, Reis LF, Soares FA, da Cunha IW. Glial fibrillary acidic protein in tumor types with cartilaginous differentiation. Mod Pathol. 2009;22:1321–1327. doi: 10.1038/modpathol.2009.99. [DOI] [PubMed] [Google Scholar]
- 21.Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 22.Laurent UB, Laurent TC, Hellsing LK, Persson L, Hartman M, Lilja K. Hyaluronan in human cerebrospinal fluid. Acta Neurol Scand. 1996;94:194–206. doi: 10.1111/j.1600-0404.1996.tb07052.x. [DOI] [PubMed] [Google Scholar]
- 23.Liu CM, Chang CH, Yu CH, Hsu CC, Huang LL. Hyaluronan substratum induces multidrug resistance in human mesenchymal stem cells via CD44 signaling. Cell Tissue Res. 2009;336:465–475. doi: 10.1007/s00441-009-0780-3. [DOI] [PubMed] [Google Scholar]
- 24.Stern R. Devising a pathway for hyaluronan catabolism: are we there yet? Glycobiology. 2003;13:105R–115R. doi: 10.1093/glycob/cwg112. [DOI] [PubMed] [Google Scholar]
- 25.Pasonen-Seppanen S, Karvinen S, Torronen K, Hyttinen JM, Jokela T, Lammi MJ, Tammi MI, Tammi R. EGF upregulates, whereas TGF-beta downregulates, the hyaluronan synthases Has2 and Has3 in organotypic keratinocyte cultures: correlations with epidermal proliferation and differentiation. J Invest Dermatol. 2003;120:1038–1044. doi: 10.1046/j.1523-1747.2003.12249.x. [DOI] [PubMed] [Google Scholar]
- 26.Stuhlmeier KM, Pollaschek C. Differential effect of transforming growth factor beta (TGF-beta) on the genes encoding hyaluronan synthases and utilization of the p38 MAPK pathway in TGF-beta-induced hyaluronan synthase 1 activation. J Biol Chem. 2004;279:8753–8760. doi: 10.1074/jbc.M303945200. [DOI] [PubMed] [Google Scholar]
- 27.Wick W, Platten M, Meisner C, Felsberg J, Tabatabai G, Simon M, Nikkhah G, Papsdorf K, Steinbach JP, Sabel M, Combs SE, Vesper J, Braun C, Meixensberger J, Ketter R, Mayer-Steinacker R, Reifenberger G, Weller M NOA-08 Study Group of Neuro-oncology Working Group (NOA) of German Cancer Society. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012;13:707–715. doi: 10.1016/S1470-2045(12)70164-X. [DOI] [PubMed] [Google Scholar]
- 28.Jang E, Kim E, Son HY, Lim EK, Lee H, Choi Y, Park K, Han S, Suh JS, Huh YM, Haam S. Nanovesicle-mediated systemic delivery of microRNA-34a for CD44 overexpressing gastric cancer stem cell therapy. Biomaterials. 2016;105:12–24. doi: 10.1016/j.biomaterials.2016.07.036. [DOI] [PubMed] [Google Scholar]
- 29.Wiltshire RN, Rasheed BK, Friedman HS, Friedman AH, Bigner SH. Comparative genetic patterns of glioblastoma multiforme: potential diagnostic tool for tumor classification. Neuro Oncol. 2000;2:164–173. doi: 10.1093/neuonc/2.3.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joo KM, Kim SY, Jin X, Song SY, Kong DS, Lee JI, Jeon JW, Kim MH, Kang BG, Jung Y, Jin J, Hong SC, Park WY, Lee DS, Kim H, Nam DH. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 2008;88:808–815. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- 32.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 33.Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. CD133+ and CD133- glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 34.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Back SA, Tuohy TM, Chen H, Wallingford N, Craig A, Struve J, Luo NL, Banine F, Liu Y, Chang A, Trapp BD, Bebo BF Jr, Rao MS, Sherman LS. Hyaluronan accumulates in demyelinated lesions and inhibits oligodendrocyte progenitor maturation. Nat Med. 2005;11:966–972. doi: 10.1038/nm1279. [DOI] [PubMed] [Google Scholar]
- 36.Moshayedi P, Carmichael ST. Hyaluronan, neural stem cells and tissue reconstruction after acute ischemic stroke. Biomatter. 2013;3:e23863. doi: 10.4161/biom.23863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kosaki R, Watanabe K, Yamaguchi Y. Overproduction of hyaluronan by expression of the hyaluronan synthase Has2 enhances anchorage-independent growth and tumorigenicity. Cancer Res. 1999;59:1141–1145. [PubMed] [Google Scholar]
- 38.Tsuneki M, Madri JA. CD44 regulation of endothelial cell proliferation and apoptosis via modulation of CD31 and VE-cadherin expression. J Biol Chem. 2014;289:5357–5370. doi: 10.1074/jbc.M113.529313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maherally Z, Smith JR, An Q, Pilkington GJ. Receptors for hyaluronic acid and poliovirus: a combinatorial role in glioma invasion? PLoS One. 2012;7:e30691. doi: 10.1371/journal.pone.0030691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Y, Ruan B, Liu W, Wang J, Yang X, Zhang Z, Li X, Duan J, Zhang F, Ding R. Knockdown of CD44 inhibits the invasion and metastasis of hepatocellular carcinoma both in vitro and in vivo by reversing epithelial-mesenchymal transition. Oncotarget. 2015;6:7828. doi: 10.18632/oncotarget.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh SK, Fiorelli R, Kupp R, Rajan S, Szeto E, Cascio CL, Maire CL, Sun Y, Alberta JA, Eschbacher JM. Post-translational modifications of OLIG2 regulate glioma invasion through the TGF-β pathway. Cell Rep. 2016;16:950–966. doi: 10.1016/j.celrep.2016.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei F, Wang Q, Su Q, Huang H, Luan J, Xu X, Wang J. miR-373 Inhibits Glioma Cell U251 Migration and Invasion by Down-Regulating CD44 and TGFBR2. Cell Mol Neurobiol. 2016;36:1389–1397. doi: 10.1007/s10571-016-0338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown DV, Daniel PM, D’Abaco GM, Gogos A, Ng W, Morokoff AP, Mantamadiotis T. Coexpression analysis of CD133 and CD44 identifies proneural and mesenchymal subtypes of glioblastoma multiforme. Oncotarget. 2015;6:6267–6280. doi: 10.18632/oncotarget.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khoshyomn S, Penar PL, Wadsworth MP, Taatjes DJ. Localization of CD44 at the invasive margin of glioblastomas by immunoelectron microscopy. Ultrastruct Pathol. 1997;21:517–525. doi: 10.3109/01913129709016368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.