Abstract
The aim of this study was to identify specific microRNAs (miRNAs) and their regulatory roles in the process of 1, 25-dihydroxyvitamin D3-induced (VD3-induced) osteogenic differentiation of human adipose-derived Mesenchymal stem cells (hAMSCs). The differentially expressed miRNAs in VD3-induced hAMSCs was examined. The putative target genes of these miRNAs were predicted. A total of 76 conserved miRNAs, including 18 miRNAs were significantly up-regulated and 58 miRNAs were significantly downregulated, and significantly differentially expressed between the two samples. The expression of 4 upregulated miRNAs (miR-1-3p, miR-1247-5p, miR-217, and miRNA-483) and 5 downregulated miRNAs (miR-1284, miR-218, miR-582-3p, miR-187-3p, and miRNA-122-5p) were verified. The highly enriched GOs and KEGG pathway showed target genes of these miRNAs were significantly involved in multiple biological processes (signal transduction, cell differentiation, cell adhesion and cell proliferation), and several osteogenic pathways (MAPK signaling pathway, TGF-β/BMP signaling pathway, and Wnt signaling pathway). Finally, TGF-β/BMP signaling pathway was selected for target verification and function analysis. We observed that a number of osteo-genes in the TGF-β/BMP superfamily, such as BMPRI, BMPRII, TGFBRI, TGFBRII, BMP4, TGFβ, Smad2, 3, 8, were predicted to be target gene of the differentially expressed miRNAs. Among them, TGFB, BMP4, BMPRI, and Smad8, which are positive regulators in osteoblast differentiation, were confirmed to be significantly up-regulated in VD3-induced cells by qRT-PCR; while Smad6 and activinRI, which are negative regulators of the TGF-β/BMP superfamily, were shown to be significantly down-regulated. These results will help to understand the role of miRNA in the regulation of the osteogenic differentiation of hAMSCs.
Keywords: MicroRNAs; 1, 25-dihydroxyvitamin D3; osteogenic differentiation; human adipose-derived mesenchymal stem cells
Introduction
Human adipose-derived Mesenchymal stem cells (hAMSCs) are multipotent cells present in the adipose tissue, which are capable of differentiating into osteogenic, chondrogenic, and adipogenic lineages, when cultured in appropriate in vitro conditions [1-3]. Previous studies tested the ability of hAMSCs to differentiation into multiple phenotypes of adipogenesis, osteogenesis, chondrogenesis, and neurogenesis in vitro, and confirmed the hypothesis that hAMSCs were a type of multipotent adult stem cell and not solely a mixed population of unipotent progenitor cells [1,4,5]. Compared with bone-marrow-derived MSCs (BMSCs), AMSCs are easier to obtain, have relatively lower donor site morbidity, a higher yield at harvest, and can expand more rapidly in vitro than BMSCs [6-8]. In addition, the proliferation and differentiation potential of AMSCs are independent of age [8,9]. Our and others’ previous studies had successfully induced AMSCs into an osteogenic lineage in vitro and repaired bone defects using AMSCs as seed cells [10-13]. These advantages suggest that AMSCs are a promising alternative source of seed cells for bone tissue engineering and regeneration. The possibility of obtaining AMSCs from an autologous source and their ability to differentiate into bone tissue makes them ideal candidates for bone defect treatment.
Elucidating key regulatory pathways and molecules either involved in maintaining AMSCs in their undifferentiated state of during the process of osteogenic differentiation allows for a better handle on expanding and culturing AMSCs in large scale for therapeutic applications. The previous work suggested that cell signaling pathways regulated the osteogenesis of MSCs [14]. NFkB inhibits osteogenesis of mesenchymal stem cells by promoting β-catenin degradation [15]. TGF-β/BMP signaling pathway is of interest, as it has been reported to play a prominent role in promoting osteoblast differentiation and bone formation [16,17]. Our recent studies have demonstrated that the ERK and JNK cell signal pathway, which are two distinctly regulated groups of mitogen-activated protein kinases (MAPKs), play an important role in the commitment to osteogenic differentiation [18-20]. Activation of ERK and JNK is especially related to the sequential expression of osteogenesis-related genes, while blockage of them can inhibit the osteogenic differentiation of AMSCs [18-20]. In addition to cell signaling pathways, another important regulatory mechanism of cell function is the post-transcriptional modulation of gene expression by microRNAs (miRNAs).
MicroRNAs (miRNAs) are a class of endogenous, small (with a length of 18-25 nucleotides), single-stranded, noncoding RNA molecules. The primary function of miRNAs is to downregulate the expression of specific proteins at the posttranscriptional level by specifically binding to the 3’-untranslational region (3’-UTR) of target messenger RNAs (mRNAs) and subsequently preventing their translation and/or promoting their degradation [21]. An increasing body of evidence has demonstrated that miRNAs play important roles in various biological processes, including cell proliferation, differentiation, programmed apoptosis and cell death. In addition, miRNAs are involved in regulation some signal transduction pathways such as Wnt signaling pathway, NF-kb signaling pathway and MAPK signaling pathway [22-24]. The understanding of integrated gene expression and epigenetic miRNAs mechanisms should be important for the osteogenesis of hAMSCs.
Over the past several years, growing evidence has indicated that several miRNAs are involved in regulating osteogenic differentiation in hAMSCs and human mesenchymal stem cells (hMSCs). Lu zi et al. reported that miR-26a regulated the osteogenic differentiation of hADSCs via the bone morphogenetic protein (BMP) signal regulatory protein Smad1 during osteoblast differentiation [25]. Wang et al. indicated that the overpression of miR-26a promoted hAMSC osteogenesis via suppressing the expression of GSK3β protein, while GSK3β influences Wnt signalling pathway by regulating β-catenin, and subsequently altered the expression of its downstream target C/EBPα, which in turn transcriptionally regulated the expression of miR-26a by physically binding to the CTDSPL promoter region [26]. Interesting, Su et al. [27] reported that miR-26a functions oppositely in osteogenic differentiation of BMSCs and ADSCs. The distinct activation pattern and role of signaling pathways determined that miR-26a majorly targeted on GSK3β to activate Wnt signaling for promoting osteogenic differentiation of BMSCs, whereas it inhibited Smad1 to suppress BMP signaling for interfering with the osteogenic differentiation of ADSCs [27]. In addition, miR-154-5p negatively regulates ADSCs osteogenic differentiation through the Wnt/PCP pathway by directly targeting Wnt11 [28]. Overexpression of miR-218 enhanced Wnt/β-catenin signaling activity and osteogenic differentiation of hAMSCs via directly targeting SFRP2 and DKK2, which is a WNT signaling pathway antagonist [29]. The potential of adipogenesis and osteogenesis of ADSCs was also decreased by miR-34a overexpression, which was recovered by co-treatment with anti-miR-34a [30]. miR-17-5p, miR-106a, miR-22 and miR-27a are involved in the balance between osteogenic and adipogenic differentiation of AMSCs. Li et al. revealed that miR-17-5p and miR-106a could promote adipogenesis and inhibit osteogenesis of hAMSCs by directly targeting BMP2 [31]. Overexpression of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of hAMSCs by repressing HDAC6 protein expression [32]. Low-expression of miR-27a promotes the shift of MSCs from osteogenic differentiation to adipogenic differentiation in osteoporosis by targeting Mef2c [33]. miRNAs can target the tr-anscription factors which can directly regulate osteogenic differentiation of MSCs. miR204 has been shown to promote the adipogenesis and inhibit the osteogenesis of human MSCs through the direct suppression of Runx2 [34]. Many studies have pointed out the importance of miRNAs in regulation osteogenic differentiation in hAMSCs and hMSCs, however, current studies on the regulatory roles of miRNA during osteogenic differentiation have focused largely on dexamethasone-induced osteogenesis. We therefore initiated this study to investigate the osteogenic differentiation of 1, 25-dihydroxyvitamin D3-induced (VD3-induced) hAMSCs.
Apart from microarray analysis, Solexa sequencing is another common, high-throughput method used for differential miRNA expressions screening between paired samples. The most merit is that Solexa sequencing can be used to explore and identify a new miRNA that can be further validated. Microarray analysis was only used to identify the obtained target because of lack of corresponding probes [35]. Therefore, Solexa sequencing was adopted to analyze miRNA expression in osteogenic differentiation of AMSCs.
In this study, to further clarify the regulatory effects and underlying mechanisms of miRNAs in the osteogenic differentiation of VD3-induced hAMSCs, we examined the differential expression of miRNAs in VD3 induced hAMSCs compared to non-induced hAMSCs during osteogenic differentiation via Solexa sequencing technology. The putative target genes of these miRNAs were predicted using bioinformatics analysis. The target genes of the selected differentially expressed miRNAs were also analyzed in the gene ontology (GO) and KEGG biological pathway. The results of this study will help in gaining understanding of the role of miRNA in the regulation of the osteogenic differentiation of hAMSCs.
Materials and methods
Harvest, culture and differentiation of hAMSCs
Fresh human lipoaspirates were obtained from five healthy patients (average age 38 years) who had received abdominal liposuction at the Department of Plastic and Reconstructive Surgery of Minhang Hospital. All patients gave written informed consent. All protocols for human tissue handling were approved by the Research Ethical Committee of Fudan University. Processed lipoaspirate (PLA) cell isolation and culture were performed as previously described [18]. In brief, lipoaspirates were washed intensively with an equal volume of 0.1 M phosphate buffered saline (PBS, pH 7.4) and digested with 0.075% collagenase type I (Washington Biochemical Corp, USA) at 37°C for 60 min. Enzyme activity was neutralized with low glucose Dulbecco’s modified Eagle’s medium (LG-DMEM, Gibco, USA), containing 10% fetal bovine serum (FBS, HyClone, USA), and the digested lipoaspirates were centrifuged at 1200 × g for 10 min to obtain a high-density stromal vascular fraction (SVF). The SVF was then treated with red blood cell lysing buffer (0.3 g/L ammonium chloride in 0.01 M Tris-HCl buffer, pH 7.5, Sigma) for 5 min, centrifuged at 600 × g for 10 min, and filtered through a 100-μm nylon mesh to remove undigested tissue. Cells were resuspended in LG-DMEM culture medium, containing 10% FBS, 100 mg/mL streptomycin, 100 U/mL penicillin (growth medium, GM), and plated at 4 × 104 cells/cm2 in 100 mm culture dishes (Falcon, USA), with the medium changed twice a week. When they reached 70-80% confluence, the cells were passaged and hASCs before passage three were used in the current study.
For osteogenic differentiation, hAMSCs were cultured in osteogenic medium (OM) comprising GM supplemented with 0.01 μM 1, 25-dihydroxyvitamin D3, 50 μM ascorbate-2-phosphate, and 10 mM β-glycerophosphate. Osteogenic differentiation was confirmed via alkaline phosphatase (ALP) and accumulated calcium assay. The cells cultured in GM were collected as controls.
Alkaline phosphatase (ALP) staining and alizarin red S staining
Alkaline phosphatase (ALP) staining was implicated as a marker of osteoblast differentiation which is expressed early in the process. The hAMSCs at passage three were plated into 6-well plates (Falcon) and cultured in OM. At day 10 after passage, alkaline phosphatase (ALP) histochemistry was performed using Sigma Diagnostic Kit 85.
Alizarin Red S staining was performed to detect the calcification at late stage of induction. The hAMSCs at passage three were plated onto 6-well plates (Falcon) and cultured in OM for up to 21 days. The cells were fixed with 70% ice-cold ethanol for 1 h and then incubated in 40 mM Alizarin Red S (Sigma) at pH 4.2 for 30 min at room temperature, with agitation in an orbital shaker (60 rpm). After two intensive rinses with deionized water, extracellular matric (ECM) mineral-bound staining was photographed under a Nikon TE300 phase-contrast microscope (Nikon).
Construction of small RNA libraries and solexa sequencing
Total RNA was extracted from GM and OM cell layars using Trizol reagent (Invitrogen, USA), according to the single step acid-phenol guanidinium extraction method. RNA integrity was confirmed using the 2100 Bioanalyzer (Agilent Technologies). Two sRNA libraries were constructed using homogenized and pooled total RNAs of three individuals for each group (GM and OM). For each group, 10 mg of total RNA was used for library construction with a Small RNA Sample Prep Kit (Illumina, USA) following the manufacturer’s instructions with minor modifications. Briefly, after 15% Tris-Borate-EDTA (TBE) denaturing polyacrylamide gel electrophoresis (PAGE) the 18- to 30-nt fraction of total RNA was excised, purified, and ligated to 3’ and 5’ RNA adaptors using T4 RNA ligase. The adaptor-ligated sRNAs were subjected to RT-PCR with 15 cycles of PCR amplification. The PCR products (approximately 90-bp, corresponding to sRNA + adaptors) were purified on 4% agarose gels to create the libraries. The purified libraries were used directly for cluster generation and sequencing analysis using an Illumina/Solexa G1 sequencer (Shanghai Oebiotech Co. Ltd, China).
Sequencing data analysis and identification of miRNAs
First, the low-quality reads were filtered to remove reads without the 3’ adaptor, 5’ adaptor-contaminant reads, reads without the insert fragment, reads containing poly (A) stretches, and reads of less than 18 nt. Next, the remaining sequences (clean reads) were mapped to the human genome using SOAP (http://soap. genomics.org.cn) with a tolerance of one mismatch to analyze their distribution. The sequences were aligned against known miRNA precursors and mature miRNAs deposited in the miRBase 21.0 to identify conserved miRNAs. The clean reads were compared against the sRNAs (rRNAs, tRNAs, snRNAs, snoRNA, miRNA) deposited in the GenBank and Rfam (http://www.sanger.ac.uk/resources/databases/rfam.html) databases to annotate the sRNA sequences. Because some sRNA tags might map to more than one category we used priority rules to ensure that every unique sRNA was mapped to only one annotation as follows: rRNA etc. (GenBank > Rfam) > known miRNA > repeat > exon > intron).
After identifying the conserved miRNAs, the remaining sequences of the two libraries were aligned with the integrated human transcriptome to predict novel miRNAs. Potentially novel miRNAs were analyzed in two steps, first using miRDeap 2 software and then using RNAfold software. The miRDeap 2 program was used to analyze structural features of the miRNA precursors to identify all novel miRNA candidates. The resulting structures were retained as novel miRNA candidates only if they met the criteria described by Allen et al [36] and Friedlander et al [37]. The novel human pre-miRNA sequences were checked using RNAfold to predict stem-loop structure (http://rna.tbi.univie.ac.at). The stem-loop hairpins were considered to be typical only when they fulfilled the following criteria: 1) The number of base pairs in a stem was ≥ 18 nt; 2) The number of errors in one bulge was ≤ 18; 3) The secondary structures of the hairpins were stable with a free energy of hybridization less than -20 kcal/mol; 4) The percentage of the miRNA in the stem was ≥ 80%; 5) The length of the hairpin (up and down stem plus terminal loop) was ≥ 53 nt; 6) The length of the hairpin loop was ≤ 22 nt; and 7) The percentage of A and U in the mature miRNA was 30%-70%. Any sequence that satisfied these strict criteria was considered a candidate miRNA precursor.
Differential expression analysis
We compared the expression of the known miRNAs between the two samples to identify differentially expressed miRNAs. miRNA expression in the two samples was analyzed by Log2-ratio figure and Scatter Plot. The procedure was as follows: 1) The expression of miRNA in the two samples (GM and OM) was normalized to obtain expression of the transcript per million (normalized expression (NE) = Actual miRNA count/Total count of clean reads*1,000,000). When the normalized expression of a certain miRNA was zero, we revised its expression value to 0.01. If the normalized expression of a certain miRNA was lower than 1, further differential expression analysis was conducted without this miRNA. 2) We calculated fold-change and P-value from the normalized expression and then generated the log2 ratio plot and scatter plot. Fold-change = log2 (OM-NE/GM-NE).
Validation and expression analysis of human miRNAs via qPCR
Differentially expressed miRNAs were validated using qPCR. Briefly, miRNA was isolated from the GM and OM cell layars using miRcute miRNA Isolation Kit (TIANGEN, China, DP501), and 1 microgram of total RNA from each sample was reverse-transcribed into cDNA using the NCodeTM EXPRESS SYBR® GreenERTM miRNA qPCR Kit (Invitrogen). qPCR was performed using a GeneAmp PCR system 9600 (Perkin-Elmer). Relative transcript levels were measured and normalized with β-actin levels. All primers used for qPCR are listed in Table 1. To confirm the miRNA expression profiles obtained using miRNA microarray analysis, the expressions of miRNAs were also examined using qPCR. The expression levels were normalized to that of U6, which was used as an internal control. Relative expression of a specific gene was calculated using the comparative Ct method.
Table 1.
Sequences of Primers for qRT-PCR in the experiment
| Gene | Primers (F = forward; R = reverse) |
|---|---|
| Runx2 | F: 5’-GTCTTACCCCTCCTACCTGA-3’ |
| R: 5’-TGCCTGGCTCTTCTTACTGA-3’ | |
| ALP | F: 5’-ACGTGGCTAAGAATGTCATC-3’ |
| R: 5’-CTGGTAGGCGATGTCCTTA-3’ | |
| OCN | F: 5’-CAAAGGTGCAGCCTTTGTGTC-3’ |
| R: 5’-TCACAGTCCGGATTGAGCTCA-3’ | |
| PPARγ2 | F: 5’-GATACACTGTCTGCAAACATATCACAA-3 |
| R: 5’-CCACGGAGCTGATCCCAA-3’ | |
| TGFβ | F: 5’-CAGAAAACAAGGCATATAATAACAG-3’ |
| R: 5’-CAGAAAACAAGGCATATAATAACAG-3’ | |
| BMP4 | F: 5’-GGGAAGGAGTGTGGTGGTGG-3’ |
| R: 5’-CAGAAAACAAGGCATATAATAACAG-3’ | |
| BMPRI | F: 5’-TGCAAGGATTCACCGAAAGC-3’ |
| R: 5’-TGCCATCAAAGAACGGACCTAT-3’ | |
| Smad8 | F: 5’-CAGCATCTTTGTCCAGAGCC-3’ |
| R: 5’- AAAGCTCATCCGAATCGTGC-3’ | |
| Smad6 | F: 5’-TCCGAAGTCCGCTCGGTAG-3’ |
| R: 5’-TCACCGTCTCGCAGTCACT-3’ | |
| ActivinRI | F: 5’-TTCTTCCCCCTTGTTGTCCTC-3’ |
| R: 5’-ACAGGTGTAGTTGGTCTGTAGG-3’ | |
| β-actin | F: 5’-ATCATGTTTGAGACCTTCAA-3’ |
| R: 5’-CATCTCTTGCTCGAAGTCCA-3’ |
Analysis by go and the KEGG (Kyoto encyclopedia of genes and genomes) pathway
To better understand miRNA target function and classification, as well as the metabolic regulatory networks associated with human miRNAs and their targets, we used InterProScan [38] and Blast2go [39] to perform GO annotation and enrichment analysis for three ontologies, molecular function, cellular component, and biological process. The GO terms were significantly enriched in the predicted candidate target genes of the miRNAs and the genes corresponding to certain biological functions. To identify significantly enriched metabolic or signal transduction pathways among the target gene candidates compared with the whole reference gene background we used Cytoscape software V2.8.2 (http://www.cytoscape.org/) [40] and the ClueGO plug-in (http://apps.cytoscape.org/apps/cluego) [41] to decipher the KEGG (http://www.genome.jp/kegg/) [42] pathway and determine biological functions. In all the tests, the P-values were calculated using the Benjamini-corrected modified Fisher’s exact test, and P ≤ 0.05 was considered to be statistically significant.
Statistical analysis
Data are presented as mean ± SD. Student’s t test was performed to compare two groups. ANOVA was used for multiple comparisons. In both cases, differences with P < 0.05 were considered statistically significant.
Results
Osteogenic differentiation of hAMSCs
The hAMSCs at passage three were plated into 6-well plates (Falcon) and cultured in OM. Compared to the controls, there was a significant increase in the ALP staining (day 7) and Alizarin Red S staining (day 21) of the cells cultured with OM (Figure 1A).
Figure 1.

Efficiency of osteogenic differentiation of hAMSCs induced by 1, 25-dihydroxyvitamin D3 (VD3). A: Increased ALP staining and Alizarin Red S staining in VD3-induced hAMSCs. B: VD3 increased the expression of genes involved in osteogenic differentiation. Data are expressed as mean ± SE of each group of cells from three separate experiments. *P < 0.05, compared to VD3-induced hAMSCs in 1 day.
The expression of genes involved in osteogenic differentiation was determined by qRT-PCR. OM-inducing cells showed higher mRNA levels of Runx2, ALP, and OCN when compared with the controls (Figure 1B).
Characteristics and sequence analysis of the small RNAs
After deep sequencing of sRNAs (10 to 30 nt) in the two sRNA libraries, hAMSCs cultured in growth medium (GM) and osteogenic medium (OM) and removal of the low-quality sequences (reads with low sequencing quality, no 3’ adapter sequence, presence of 5’ adapter sequence, no insert fragment, less than 18 nt, or containing polyA), a total of 48,099,397 and 45,713,126 clean reads were determined for the GM and OM groups, respectively. The majority of sRNAs were 19-24 nt, and the most abundant size class in the sRNA sequence distribution was 22 nt, which accounted for 35.88% and 44.86% of the OM and GM libraries, respectively.
To assess the efficiency of high-throughput sequencing for sRNA detection, the total population of clean sRNAs were annotated and classified by alignment with GenBank and Rfam databases. The total of 46,771,188 reads in GM library (97.23%) and 44,595,886 reads in OM library (97.55%) were mapped to the human genome. The composition of the RNA classes in each library is shown in Figure 2. The classification annotation revealed that 21,275,022 and 25,570,745 reads in the GM (44.23%) and OM (55.94%) libraries, respectively, were classified as know miRNAs, while 2,483,101 and 1,720,858 reads in the GM (5.16%) and OM (3.76%) libraries, respectively, were unannotated and require further analysis for novel miRNA candidates (Figure 2).
Figure 2.
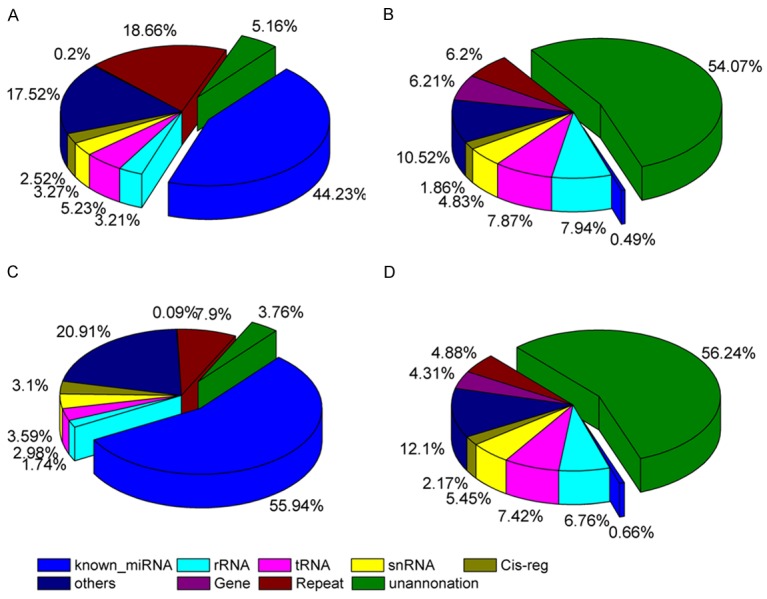
Distribution of sRNAs among different categories in the GM and OM library. A: Total number of reads in GM. B: Total number of unique sequences in GM. C: Total number of reads in OM. D: Total number of unique sequences in OM.
Identification of known conserved miRNAs
We aligned the clean reads to the precursor/mature miRNAs in the miRBase 21.0 database. The sequence and count of families were obtained (Table S1).
A total of 1,167 conserved miRNAs (1,024 from GM library and 1,034 from OM library) were identified, and 891 of these were present in both libraries. In addition, 276 miRNAs were detected in only one sRNA library. For example, miR-5680, miR-4697-3p, and miR-3657 were only identified in the GM library, whereas miR-483-5p, miR-495-5p, miR-548ax and miR-6783-3p were only identified in the OM library.
Comparison of the expression profiles of known miRNAs between the two libraries is shown in Table S2. The expression of known miRNAs was demonstrated using a Log2-ratio and scatter plot (Figure 3). A total of 76 conserved miRNAs were significantly differentially expressed (P < 0.05) between the two samples. Compared with miRNA expression in the GM library, 18 miRNAs in the OM library were significantly up-regulated with P < 0.01, whereas 58 miRNAs were significantly downregulated with P < 0.01 (Figure 3 and Table S2).
Figure 3.

Differential expression of conversed miRNAs between GM and OM library. Each point in the figure represents a miRNA. Red points represent miRNAs with fold-change > 2, blue points represent miRNAs with fold-change > 1/2 and ≤ 2, green points represent miRNAs with fold-change ≤ 1/2.
Prediction of novel miRNAs
After identifying the known conserved miRNAs described above, the remaining sequences of the two libraries were aligned with the human integrated transcriptome to predict potential novel miRNA candidates. We used miRDeep2 software to predict novel miRNA candidates. RNAfold software was also used to predict the typical secondary structures of the miRNA precursors and remove pseudo-pre-miRNAs. In total, 311 hairpin structures were predicted and 424 potential novel miRNA candidates with lengths ranging from 18 to 27 nt (Table S3).
Validation of miRNAs by qRT-PCR
The Solexa results from GM and OM group were further validated individually by qPCR. We chose four miRNAs with upregulated expression in OM with endometriosis: miR-1-3p, miR-1247-5p, miR-217, miRNA-483 and five miRNAs with downregulated expression in OM with endometriosis: miR-1284, miR-218, miR-582-3p, miR-187-3p, miRNA-122-5p for qPCR validation. As shown in Figure 4, qPCR results showed that the expression of miR-1-3p, miR-1247-5p, miR-217, and miRNA-483 was significantly upregulated, while the expression of miR-1284, miR-218, miR-582-3p, miR-187-3p, and miRNA-122-5p was significantly downregulated in the OM group compared to the GM group. Thus, the qPCR results fully supported the reliability of miRNA expression profiles of OM by Solexa sequencing analysis.
Figure 4.
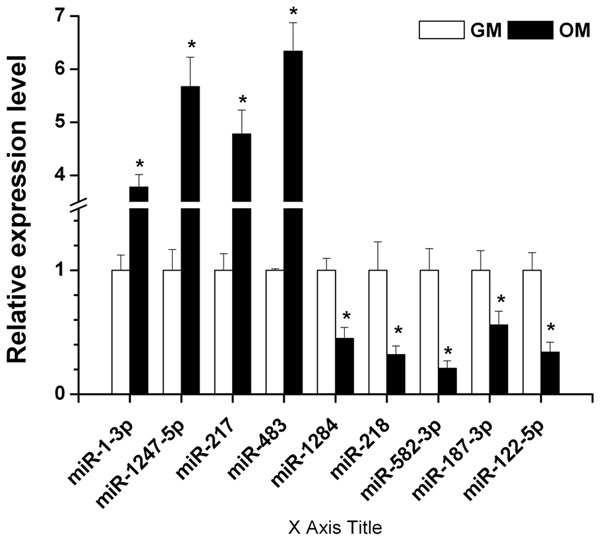
Validation of miRNAs with significantly differential expression using qPCR. Expression levels of five miRNAs were measured by qPCR. Four miRNAs (miR-1-3p, miR-1247-5p, miR-217, and miRNA-483) were validated to be significantly up-regulated, while five miRNAs (miR-1284, miR-218, miR-582-3p, miR-187-3p, miRNA-122-5p) were validated to be significantly down-regulated in VD3-induced hAMSCs. Data are expressed as mean ± SE of each group of cells from three separate experiments. *P < 0.05, compared to uninduced hAMSCs.
Differentially expressed miRNAs target the transcription factors
The transcription factors, such as Runx2, PPARγ, and Osterix, are believed to play key roles in osteogenic differentiation. miRNAs can control the osteogenic activity of transcription factors and effect the osteogenic differentiation of MSCs. To assess whether miRNAs control the osteogenic activity of transcription factors, we applied three miRNA target prediction tools (i.e., TargetScan, PicTar, and RNA22) to search for Runx2, PPARγ, and Osterix-targeting miRNAs. We identified several miRNAs that are predicted to target the Runx2, PPARγ, and Osterix (Table 2).
Table 2.
Predicted miRNAs targeted different transcription factors involved osteogenesis of stem cells
| Transcription factors | Function | miRNA |
|---|---|---|
| Runx2 | Osteogenesis, adipogenesis and chondrogenesis | miR-122-5p, miR-1271-3p, miR-1284, miR-20b-5p, miR-217, miR-218-2-3p, miR-3134, miR-338-3p, miR-3362, miR-449c-5p, miR-486-5p, miR-548, miR-5680, miR-5706, miR-582-3p, miR-605-5p, miR-671-5p, miR-6734-5p, miR-6735-3p, miR-7-1-3p |
| PPARγ | Adipogenesis, osteogenesis | miR-494-5p, miR-548, miR-5706, miR-4504, miR-3662, miR-335-3p |
| Osterix | Osteogenesis | miR-4689, miR-486-3p, miR-5680, miR-5706, miR-671-5p, miR-6780a-5p, miR-4665-5p, miR-33b-3p, miR-196a-3p, miR-1910-5p, miR-18a-3p, miR-1284 |
Gene ontology (GO) enrichment and KEGG pathway analyses of target genes
To further understand the role of these miRNAs in physiological functions and biologic processes during osteogenic differentiation of hAMSCs, miRNA target gene prediction was performed based on miRNA/mRNA interactions to provide some molecular insight into their function. A total of 34,707 annotated mRNA transcripts were predicted as putative target genes for 76 differentially expressed miRNAs (Table S4). A total of 5718 significantly enriched GO terms (P < 0.05) were indentified. The top 20 most enriched GO terms in biological process, cellular component and molecular function were show in Figure 5. The GO analysis showed a broad range of processes related to positive regulation of transcription from RNA polymerase II promoter (GO: 0045944), signal transduction (GO: 0007165), cell differentiation (GO: 0030154), cell adhesion (GO: 0007155) and cell proliferation (GO: 0008283) in biological process, cytoplasm (GO: 0005737), nucleus (GO: 0005634) in cellular component, protein binding (GO: 0005515) in molecular function.
Figure 5.
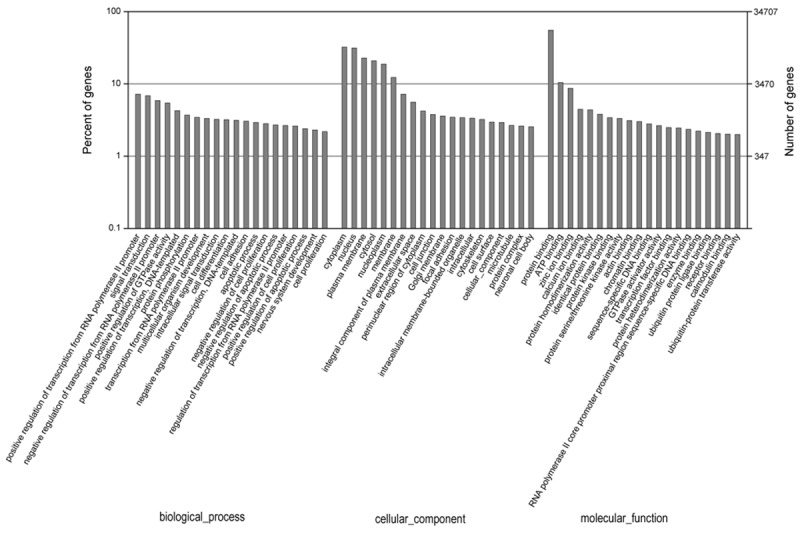
Gene ontology classification annotated by gene 2 go for target genes of differentially expressed miRNAs. The figure shows partial GO enrichment for the predicted target genes in ontologies of biological processes, cellular component, and molecular function.
The predicted target genes were classified according to KEGG function annotations to identify the pathways that were actively regulated by miRNAs (Table S5). As indicated by KEGG analysis, the signaling pathways were found to be involved in MAPK signaling pathway, TGF-β/BMP signaling pathway, calcium signaling pathway, Wnt signaling pathway and Ras signaling pathway, which involved in osteogenic differentiation of stem cells. However, as most GO and KEGG assignments and distributions are related to proliferation, development, adhesion and metabolism, our results indicated that the target genes are involved in a wide range of regulatory function in cell differentiation.
TGFβ/BMP signaling pathway were differentially expressed during VD3-induced osteogenic differentiation
TGF-β/BMP pathways are major signaling cascades responsible for osteogenesis. Our data showed that TGF-β/BMP signaling pathway appeared to be significantly enriched in bioinformatics analyse. Furthermore, we observed that a number of osteo-genes in the TGF-β/BMP superfamily, such as BMPRI, BMPRII, TGFBRI, TGFBRII, BMP4, TGFβ, Smad2, 3, 8, were predicted to be target gene of the differentially expressed miRNAs (Table 3). Among them, TGFβ, BMP4, BMPRI, and Smad8, which are positive regulators in osteoblast differentiation, were confirmed to be significantly up-regulated in VD3-induced cells by qRT-PCR; while Smad6 and activinRI, which are negative regulators of the TGF-β/BMP superfamily, were shown to be significantly down-regulated (Figure 6).
Table 3.
Predicted target genes of differentially expressed miRNAs in the “TGF-β/BMP signaling pathway” gene ontology group
| Targe genes | Function | miRNAs |
|---|---|---|
| BMPRI | BMP receptor, osteogenesis | miR-541-3p |
| BMPRII | BMP receptor, osteogenesis | miR-4689 |
| TGFβRI | Receptor for growth and differentiation factor | miR-1247-3p |
| TGFβRII | Receptor for growth and differentiation factor | miR-1247-3p |
| BMP 4 | Ostegenesis, signal transduction | miR-18a-3p |
| TGFβ | Osteogenesis, proliferation, signal transduction | miR-4467, miR-210-3p, miR-4665-5p, miR-4697-3p, miR-4780a-5p |
| Smad2/3 | Osteogenesis | miR-3180-3p, miR-486-5p, miR-671-5p |
| Smad6 | Osteogenesis, signal transduction | miR-3940-3p |
| Smad8 | Osteogenesis | miR-4523 |
Figure 6.
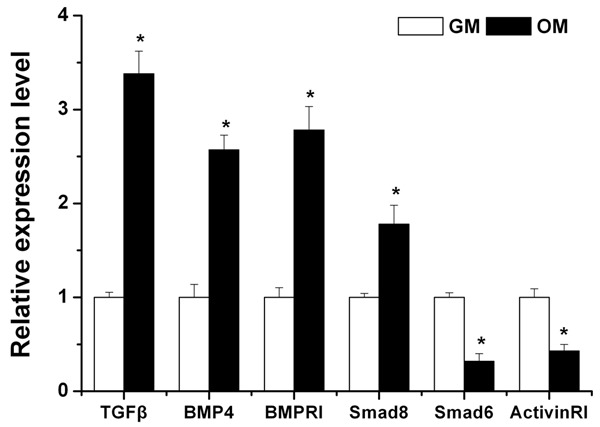
Six osteogenic genes in the TGF-β/BMP superfamily which were predicted to be target genes of the differentially expressed miRNAs were confirmed to be significantly regulated in VD3-induced hAMSCs by qPCR. Data are expressed as mean ± SE of each group of cells from three separate experiments. *P < 0.05, compared to uninduced hAMSCs.
Discussion
Human adipose-derived Mesenchymal stem cells (hAMSCs) were first isolated from human adipose tissue by Zuk et al in 2001 [3]. hAMSCs exhibited reproducible growth and differentiation potential in culture, when driven with osteogenic, adipogenic, myogenic, or chondrogenic lineage-specific culture media. Our and other several previous studies suggested a high osteogenic potential for hAMSCs, but the molecular mechanisms that under lie hAMSCs osteogenic differentiation are still unknown [11,13,18,19]. There are complex pathway regulations involved in cell differentiation at both transcriptional and post-transcriptional levels. Recently, it has been shown that miRNAs, small noncoding RNAs, regulated osteoblast differentiation and bone formation positively by negative regulation of target genes expression at the post-transcriptional level [21,24]. In this study, we first used Solexa sequencing technology to profile miRNA expression and screen miRNAs with differential significant wxpression (p < 0.05) before and after osteogenic differentiation of hAMSCs induced by VD3.
Previous screening studies on osteogenic differentiation of hAMSCs and MSCs suggested that altered expression of a number of miRNAs involved in osteogenesis, but most of these studies were conducted by microarray assay. Zhang et al. found that eight miRNAs differently expressed pre- and post-osteogenic differentiation of hAMSCs induced by DEX, among which four miRNAs (miR-17, miR-20a, miR-20b, and miR-106a) were up-regulated and four miRNAs (miR-31, miR-125a-5p, miR-125b, and miR-193a) were down-regulated [26]. Li et al. reported that sixteen differentially expressed miRNAs were identified in osteogenic differentiation of BMSCs induced by DEX, of which nine miRNAs were up-regulated (hsa-miR-155, hsa-miR-196a, hsa-miR-199a-5p, hsa-miR-130a, hsa-miR-26a, hsa-miR-221, hsa-miR-23a, hsa-miR-22, and hsa-miR-27a) and 7 miRNAs were down-regulated (hsa-miR-21, hsa-miR-140-3p, hsa-miR-214, hsa-miR-744, has-miR-320a, hsa-miR-320b, and hsa-miR-320c) [43]. Gong et al. studied the differentially expressed miRNAs by Satb2 overexpression in murine bone arrow stromal cells using miRNA microarray and found that ten down-regulated miRNAs including miR-27a, miR-125a-5p, and miR-466f-3p, and 18 up-regulated miRNAs including miR-17, miR-20a and miR-210 were differentially expressed [44]. However, only four miRNAs (miR-17, miR-20a, miR-27a, and miR 125a-5p) were observed in tow studies of these. In this study, we investigated the differentially expressed miRNAs in osteogenic differentiation of hAMSCs induced by VD3 and found that the expression of 58 miRNAs was significantly decreased, wheres the expression of 18 miRNAs was significantly increased. Among the 76 miRNAs, there are only 11 miRNAs observed during osteogenic and chondrocytic differentiation of hAMSC and/or stem cells. The expression of miR-1-3q was most repressed upon hypertrophic differentiation. Transfection of human chondrocytic HCS-2/8 cells and chicken normal chondrocytes with miR-1-3q led to repressed expression of aggrecan, the major cartilaginous proteoglycan gene [45]. The expression of miR-7 was repressed during osteogenic differentiation of PDGFRα + cells [46]. miR-20b-5p was up-regulated during the osteogenic differentiation of AMSCs induced by DEX [26]. miR-144-3p is down-regulated during osteoblast differentiation of C3H10 T1/2 cells. Overexpression of miR-144-3p inhibited osteogenic differentiation, whereas inhibition of miR-144-3p reversed this process [47]. miR-210-5p could promote osteoblast differentiation but inhibit adipocyte differentiation in BMSCs [48], while miR-210-3p showed a significantly downregulated expression during osteogenesis of BMSCs [49]. miR-217 is a modest inhibitor of Runx2 in MC3T3 cells but more roust in ATDC5 cells and targets RUNX2 and TRPS1 to control the osteogenic and chondrogenic differentiation, respectively [49,50]. Over-expression of miR-338-3p can inhibit the osteogenic differentiation of BMSCs [51]. Overexpression of miR-429 promoted osteogenic differentiation of in MC3T3-E1 cells [52]. Overexpression of miR-486-5p inhibits proliferation, adipogenic and osteogenic differentiation of hAMSCs [53]. Kockdown of human OstemiR miR-541-3p increased Osteopontin/SPP1 expression and calcification in hMSC osteoblastic differentiation, indicating that miR-541-3p is a negative regulator of osteoblastic differentiation [54]. In addition, the same family of miR-25, miR-29, miR-218, miR-335, miR-196, and miR-18a were reported to regulate the osteogenic differentiation of differentiation cell types [29,50,55-61]. However, the remaining 59 miRNAs were identified for the first time in the osteogenic differentiation of hAMSCs. In the face of great difference between this study and previous studies, the main reasons may stem from the osteogenic inducer and analytical methods.
Solexa sequencing, as one of high-throughput sequencing technologies, can produce highly accurate, reproducible, and quantitative readouts of small RNAs, including those expressed at low levels. The efficacy of solexa sequencing in miRNA profile analysis was validated in many studies. Importantly, compared to microarray analysis, solexa sequencing is capable of identifying new unreported miRNAs. Thus, solexa sequencing adopted in our research should be another key reason for the new identified miRNAs. In this study, 311 hairpin structures were predicted and 424 potential novel miRNA candidates with lengths ranging from 18 to 27 nt, which will greatly enrich the human miRBase.
Understanding the regulatory mechanisms of osteogenic differentiation of stem cells is helpful for treatment of various bone diseases. Recent development in molecular biology and gene technology has enabled us to partially unravel the molecular mechanisms that regulate osteogenic differentiation. To our current knowledge, the transcription factors, such as Runx2, PPARγ, and Osterix, are believed to play key roles in osteogenic differentiation [62-69]. In this study, a part of significantly different expressed miRNAs target the transcription factors, such as Runx2, PPARγ, and Osterix. Runx2 was considered to be essential for osteogenic differentiation, and studies have demonstrated that Runx2 binds to important downstream target genes, such as major osteogenesis-related genes, including ALP and OCN, to regulate the osteogenic differentiation of stem cells. Zhang et al. reported that a panel of 11 Runx2-targeting miRNAs (miR-23a, miR-30c, miR-34c, miR-133a, miR-135a, miR-137, miR-204, miR-205, miR-217, miR-218, and miR-338) is expressed in a lineage-related pattern in mesenchymal cell types and regulation the osteogenic and chondrogenic differentiation [70]. Osterix, which acts downstream to Runx2, is a zinc-finger-containing transcription factor essential for embryonic osteoblast differentiation and bone formation [66,67,71,72]. PPARγ, is one of the most important factors in balancing adipogenesis and osteogenesis in MSCs. Overexpression of PPARγ, positively regulates adipogenesis but negatively regulates osteogenesis [62,69]. miR-548d-5p promoted osteogenic differentiation of MSCs by suppressing the expression of PPARγ [73]. Runx2, Osterix and PPARγ are predicted targets of differently expressed miRNAs during osteogenic differentiation in hAMSCs, and they define the cell differentiation via miRNA regulation.
In this study, in order to further investigate the regulatory role of these differentially miRNAs, bioinformatics analysis will be used to explain the annotation of their functions. Abundant high-enrichment GOs of target genes regulated by differential miRNAs might involve several biological processes such as signal transduction (GO: 0007165), cell differentiation (GO: 0030154), cell adhesion (GO: 0007155) and cell proliferation (GO: 0008283). In addition, specific KEGG pathways targeted by differential miRNAs were found involved in MAPK signaling pathway, TGF-β/BMP signaling pathway, calcium signaling pathway, Wnt signaling pathway and Ras signaling pathway. Among those, the TGF-β/BMP signaling pathways have been reported to play a prominent role in promoting osteoblast differentiation and bone formation. Using online software, putative targets of differentially expressed miRNAs involving in the TGF-β/BMP signaling pathways were partly classified according to their contribution in osteogenic differentiation. Our results are therefore supportive of a critical role for these pathways in osteogenic differentiation of hAMSCs.
TGF-β/BMP pathways are major signaling cascades responsible for osteogenesis [14,16,17]. Our data showed that some key regulators of osteogenesis such as BMPRI, BMPRII, TGFBRI, TGFBRII, BMP4, TGF-β, Smad2, 3, 6, 8 and ActivinR1 which function as positive/negative regulator of osteogenesis and signal transduction mediators are included. BMP4 and TGF-β are members of the TGF-β/BMP superfamily and play a key regulatory role during bone formation and repair. TGF-β and BMP receptors including TGF-β receptors (TGFBRI, TGFBRII) and BMP receptors (BMPRI, BMPRII) are involved in bone formation [74,75]. TGF-β bind to TGFB receptor complex to regulate cellular functions via the phosphorylation of Smad2/3, while BMPs bind to BMP receptor complex to regulate cellular functions including cell differentiation and growth via the phosphorylation of Smad1/5/8 [14]. In this study, our data suggested that BMPRI, BMPRII, TGFBRI, TGFBRII, BMP4, TGF-β, Smad2, 3, 8 were predicted to be target gene of the differentially expressed miRNAs and were partly confirmed to be significantly up-regulated in osteogenic differentiation of hAMSCs induced by VD3.
Smad6 is one of the inhibitory Smad family or I-Smads [14,76]. Chen et al. reported that Smad6 is involved in a negative feedback loop regulating BMP signaling and is required to limit BMP signaling during bone formation [16]. ActivinRI, as a type I receptor, transmits signals from activin together with the type II receptor ActivinRII. Yosuke Mizuno et al. reported that miR-210 acts as a positive regulator of osteoblastic differentiation by inhibiting the TGF-β/activin signaling pathway through inhibition of AcvR1b [75]. Kamiya et al. showed that loss-of-function of ActivinRI in osteoblasts increases bone mass and activates canonical Wnt signaling through suppression of Wnt inhibitors SOST and DKK1 [77]. Our data showed that Smad6 and ActivinRI which are negative regulators of the TGF-β/BMP pathway, were significantly down-regulated in VD3-induced osteogenic differentiation of hAMSCs. It can be supposed that the differentially expressed miRNAs might regulate VD3-induced osteogenic differentiation by inhibiting these negative regulators of TGF-β/BMP pathway.
Conclusion
In this study, we investigated the differentially expressed miRNAs in osteogenic differentiation of hAMSCs induced by VD3 and found that the expression of 58 miRNAs was significantly decreased, wheres the expression of 18 miRNAs was significantly increased. Furthermore, our findings suggest that a number of the miRNAs, of which target genes are involved in the transcription factors, such as Runx2, PPARγ, Osterix, and β-catenin and TGF-β/BMP signaling pathway, play an important role in VD3-induced osteogenic differentiation. We hope our results will facilitate our understanding of the mechanism of VD3-induced osteogenic differentiation of hAMSCs, and subsequently provide high performance seed cells for tissue-engineered bone regeneration.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant nos. 81301335 and 81772433), and Shanghai Municipal Commission of Health and Family Planning Foundation (Grant no. 201440337).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Dai R, Wang Z, Samanipour R, Koo KI, Kim K. Adipose-derived stem cells for tissue engineering and regenerative medicine applications. Stem Cells Int. 2016;2016:6737345. doi: 10.1155/2016/6737345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma T, Sun J, Zhao Z, Lei W, Chen Y, Wang X, Yang J, Shen Z. A brief review: adipose-derived stem cells and their therapeutic potential in cardiovascular diseases. Stem Cell Res Ther. 2017;8:124. doi: 10.1186/s13287-017-0585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, Benhaim P, Lorenz HP, Hedrick MH. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 4.Bunnell BA, Flaat M, Gagliardi C, Patel B, Ripoll C. Adipose-derived stem cells: isolation, expansion and differentiation. Methods. 2008;45:115–120. doi: 10.1016/j.ymeth.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P, Hedrick MH. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrow KL, Hoyland JA, Richardson SM. Human adipose-derived stem cells exhibit enhanced proliferative capacity and retain multipotency longer than donor-matched bone marrow mesenchymal stem cells during expansion in vitro. Stem Cells Int. 2017;2017:2541275. doi: 10.1155/2017/2541275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowan CM, Shi YY, Aalami OO, Chou YF, Mari C, Thomas R, Quarto N, Contag CH, Wu B, Longaker MT. Adipose-derived adult stromal cells heal critical-size mouse calvarial defects. Nat Biotechnol. 2004;22:560–567. doi: 10.1038/nbt958. [DOI] [PubMed] [Google Scholar]
- 8.De Ugarte DA, Morizono K, Elbarbary A, Alfonso Z, Zuk PA, Zhu M, Dragoo JL, Ashjian P, Thomas B, Benhaim P, Chen I, Fraser J, Hedrick MH. Comparison of multi-lineage cells from human adipose tissue and bone marrow. Cells Tissues Organs. 2003;174:101–109. doi: 10.1159/000071150. [DOI] [PubMed] [Google Scholar]
- 9.Cartwright MJ, Tchkonia T, Kirkland JL. Aging in adipocytes: potential impact of inherent, depot-specific mechanisms. Exp Gerontol. 2007;42:463–471. doi: 10.1016/j.exger.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvira-Gonzalez J, Sanchez-Garces MA, Cairo JR, Del Pozo MR, Sanchez CM, Gay-Escoda C. Assessment of bone regeneration using adipose-derived stem cells in critical-size alveolar ridge defects: an experimental study in a dog model. Int J Oral Maxillofac Implants. 2016;31:196–203. doi: 10.11607/jomi.4190. [DOI] [PubMed] [Google Scholar]
- 11.Gu H, Xiong Z, Yin X, Li B, Mei N, Li G, Wang C. Bone regeneration in a rabbit ulna defect model: use of allogeneic adipose-derivedstem cells with low immunogenicity. Cell Tissue Res. 2014;358:453–464. doi: 10.1007/s00441-014-1952-3. [DOI] [PubMed] [Google Scholar]
- 12.Liu G, Zhang Y, Liu B, Sun J, Li W, Cui L. Bone regeneration in a canine cranial model using allogeneic adipose derived stem cells and coral scaffold. Biomaterials. 2013;34:2655–2664. doi: 10.1016/j.biomaterials.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Garces MA, Alvira-Gonzalez J, Sanchez CM, Barbany Cairo JR, Del Pozo MR, Gay-Escoda C. Bone regeneration using adipose-derived stem cells with fibronectin in dehiscence-type defects associated with dental implants: an experimental study in a dog model. Int J Oral Maxillofac Implants. 2017;32:e97–e106. doi: 10.11607/jomi.5169. [DOI] [PubMed] [Google Scholar]
- 14.Hayrapetyan A, Jansen JA, van den Beucken JJ. Signaling pathways involved in osteogenesis and their application for bone regenerative medicine. Tissue Eng Part B Rev. 2015;21:75–87. doi: 10.1089/ten.TEB.2014.0119. [DOI] [PubMed] [Google Scholar]
- 15.Chang J, Liu F, Lee M, Wu B, Ting K, Zara JN, Soo C, Al Hezaimi K, Zou W, Chen X, Mooney DJ, Wang CY. NF-kappaB inhibits osteogenic differentiation of mesenchymal stem cells by promoting beta-catenin degradation. Proc Natl Acad Sci U S A. 2013;110:9469–9474. doi: 10.1073/pnas.1300532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen G, Deng C, Li YP. TGF-beta and BMP signaling in osteoblast differentiation and bone formation. Int J Biol Sci. 2012;8:272–288. doi: 10.7150/ijbs.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu M, Chen G, Li YP. TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu H, Guo F, Zhou X, Gong L, Zhang Y, Zhai W, Chen L, Cen L, Yin S, Chang J, Cui L. The stimulation of osteogenic differentiation of human adipose-derived stem cells by ionic products from akermanite dissolution via activation of the ERK pathway. Biomaterials. 2011;32:7023–7033. doi: 10.1016/j.biomaterials.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Gu H, Huang Z, Yin X, Zhang J, Gong L, Chen J, Rong K, Xu J, Lu L, Cui L. Role of c-Jun Nterminal kinase in the osteogenic and adipogenic differentiation of human adipose-derived mesenchymal stem cells. Exp Cell Res. 2015;339:112–121. doi: 10.1016/j.yexcr.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Liu Q, Cen L, Zhou H, Yin S, Liu G, Liu W, Cao Y, Cui L. The role of the extracellular signalrelated kinase signaling pathway in osteogenic differentiation of human adipose-derived stem cells and in adipogenic transition initiated by dexamethasone. Tissue Eng Part A. 2009;15:3487–3497. doi: 10.1089/ten.TEA.2009.0175. [DOI] [PubMed] [Google Scholar]
- 21.Huang C, Geng J, Jiang S. MicroRNAs in regulation of osteogenic differentiation of mesenchymal stem cells. Cell Tissue Res. 2017;368:229–238. doi: 10.1007/s00441-016-2462-2. [DOI] [PubMed] [Google Scholar]
- 22.Kang H, Hata A. The role of microRNAs in cell fate determination of mesenchymal stem cells: balancing adipogenesis and osteogenesis. BMB Rep. 2015;48:319–323. doi: 10.5483/BMBRep.2015.48.6.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi Y, Uehara S, Udagawa N, Takahashi N. Regulation of bone metabolism by Wnt signals. J Biochem. 2016;159:387–392. doi: 10.1093/jb/mvv124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng S, Gao D, Gao C, Wei P, Niu M, Shuai C. MicroRNAs regulate signaling pathways in osteogenic differentiation of mesenchymal stem cells (Review) Mol Med Rep. 2016;14:623–629. doi: 10.3892/mmr.2016.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luzi E, Marini F, Sala SC, Tognarini I, Galli G, Brandi ML. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J Bone Miner Res. 2008;23:287–295. doi: 10.1359/jbmr.071011. [DOI] [PubMed] [Google Scholar]
- 26.Zhang ZJ, Zhang H, Kang Y, Sheng PY, Ma YC, Yang ZB, Zhang ZQ, Fu M, He AS, Liao WM. miRNA expression profile during osteogenic differentiation of human adipose-derived stem cells. J Cell Biochem. 2012;113:888–898. doi: 10.1002/jcb.23418. [DOI] [PubMed] [Google Scholar]
- 27.Su X, Liao L, Shuai Y, Jing H, Liu S, Zhou H, Liu Y, Jin Y. MiR-26a functions oppositely in osteogenic differentiation of BMSCs and ADSCs depending on distinct activation and roles of Wnt and BMP signaling pathway. Cell Death Dis. 2015;6:e1851. doi: 10.1038/cddis.2015.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Hu C, Han L, Liu L, Jing W, Tang W, Tian W, Long J. MiR-154-5p regulates osteogenic differentiation of adipose-derived mesenchymal stem cells under tensile stress through the Wnt/PCP pathway by targeting Wnt11. Bone. 2015;78:130–141. doi: 10.1016/j.bone.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Zhang WB, Zhong WJ, Wang L. A signalamplification circuit between miR-218 and Wnt/beta-catenin signal promotes human adipose tissue-derived stem cells osteogenic differentiation. Bone. 2014;58:59–66. doi: 10.1016/j.bone.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 30.Park H, Park H, Pak HJ, Yang DY, Kim YH, Choi WJ, Park SJ, Cho JA, Lee KW. miR-34a inhibits differentiation of human adipose tissuederived stem cells by regulating cell cycle and senescence induction. Differentiation. 2015;90:91–100. doi: 10.1016/j.diff.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Li T, Wang S, Wei J, Fan J, Li J, Han Q, Liao L, Shao C, Zhao RC. miR-17-5p and miR-106a are involved in the balance between osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Stem Cell Res. 2013;10:313–324. doi: 10.1016/j.scr.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Huang S, Wang S, Bian C, Yang Z, Zhou H, Zeng Y, Li H, Han Q, Zhao RC. Upregulation of miR-22 promotes osteogenic differentiation and inhibits adipogenic differentiation of human adipose tissue-derived mesenchymal stem cells by repressing HDAC6 protein expression. Stem Cells Dev. 2012;21:2531–2540. doi: 10.1089/scd.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.You L, Pan L, Chen L, Gu W, Chen J. MiR-27a is essential for the shift from osteogenic differentiation to adipogenic differentiation of mesenchymal stem cells in postmenopausal osteoporosis. Cell Physiol Biochem. 2016;39:253–265. doi: 10.1159/000445621. [DOI] [PubMed] [Google Scholar]
- 34.He H, Chen K, Wang F, Zhao L, Wan X, Wang L, Mo Z. miR-204-5p promotes the adipogenic differentiation of human adipose-derived mesenchymal stem cells by modulating DVL3 expression and suppressing Wnt/beta-catenin signaling. Int J Mol Med. 2015;35:1587–1595. doi: 10.3892/ijmm.2015.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Huang W, Ren C, Zhao M, Jiang X, Fang X, Xia X. Analysis of serum microRNA profile by solexa sequencing in women with Endometriosis. Reprod Sci. 2016;23:1359–1370. doi: 10.1177/1933719116641761. [DOI] [PubMed] [Google Scholar]
- 36.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 38.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 40.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M. Molecular network analysis of diseases and drugs in KEGG. Methods Mol Biol. 2013;939:263–275. doi: 10.1007/978-1-62703-107-3_17. [DOI] [PubMed] [Google Scholar]
- 43.Li T, Li H, Li T, Fan J, Zhao RC, Weng X. MicroRNA expression profile of dexamethasoneinduced human bone marrow-derived mesenchymal stem cells during osteogenic differentiation. J Cell Biochem. 2014;115:1683–1691. doi: 10.1002/jcb.24831. [DOI] [PubMed] [Google Scholar]
- 44.Gong Y, Xu F, Zhang L, Qian Y, Chen J, Huang H, Yu Y. MicroRNA expression signature for Satb2-induced osteogenic differentiation in bone marrow stromal cells. Mol Cell Biochem. 2014;387:227–239. doi: 10.1007/s11010-013-1888-z. [DOI] [PubMed] [Google Scholar]
- 45.Sumiyoshi K, Kubota S, Ohgawara T, Kawata K, Nishida T, Shimo T, Yamashiro T, Takigawa M. Identification of miR-1 as a micro RNA that supports late-stage differentiation of growth cartilage cells. Biochem Biophys Res Commun. 2010;402:286–290. doi: 10.1016/j.bbrc.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 46.Oishi T, Uezumi A, Kanaji A, Yamamoto N, Yamaguchi A, Yamada H, Tsuchida K. Osteogenic differentiation capacity of human skeletal muscle-derived progenitor cells. PLoS One. 2013;8:e56641. doi: 10.1371/journal.pone.0056641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao M, Huang J, Gui K, Xiong M, Cai G, Xu J, Wang K, Liu D, Zhang X, Yin W. The downregulation of miR-144 is associated with the growth and invasion of osteosarcoma cells through the regulation of TAGLN expression. Int J Mol Med. 2014;34:1565–1572. doi: 10.3892/ijmm.2014.1963. [DOI] [PubMed] [Google Scholar]
- 48.Liu XD, Cai F, Liu L, Zhang Y, Yang AL. MicroRNA-210 is involved in the regulation of postmenopausal osteoporosis through promotion of VEGF expression and osteoblast differentiation. Biol Chem. 2015;396:339–347. doi: 10.1515/hsz-2014-0268. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Xie RL, Gordon J, LeBlanc K, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Control of mesenchymal lineage progression by microRNAs targeting skeletal gene regulators Trps1 and Runx2. J Biol Chem. 2012;287:21926–21935. doi: 10.1074/jbc.M112.340398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tornero-Esteban P, Rodriguez-Rodriguez L, Abasolo L, Tome M, Lopez-Romero P, Herranz E, Gonzalez MA, Marco F, Moro E, Fernandez-Gutierrez B, Lamas JR. Signature of microRNA expression during osteogenic differentiation of bone marrow MSCs reveals a putative role of miR-335-5p in osteoarthritis. BMC Musculoskelet Disord. 2015;16:182. doi: 10.1186/s12891-015-0652-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, Sun Q, Wan C, Li L, Zhang L, Chen Z. MicroRNA-338-3p regulates osteogenic differentiation of mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2. J Cell Physiol. 2014;229:1494–1502. doi: 10.1002/jcp.24591. [DOI] [PubMed] [Google Scholar]
- 52.Huang J, Peng J, Cao G, Lu S, Liu L, Li Z, Zhou M, Feng M, Shen H. Hypoxia-induced microRNA-429 promotes differentiation of MC3T3-E1 osteoblastic cells by mediating ZFPM2 expression. Cell Physiol Biochem. 2016;39:1177–1186. doi: 10.1159/000447824. [DOI] [PubMed] [Google Scholar]
- 53.Kim YJ, Hwang SH, Lee SY, Shin KK, Cho HH, Bae YC, Jung JS. miR-486-5p induces replicative senescence of human adipose tissuederived mesenchymal stem cells and its expression is controlled by high glucose. Stem Cells Dev. 2012;21:1749–1760. doi: 10.1089/scd.2011.0429. [DOI] [PubMed] [Google Scholar]
- 54.Eguchi T, Watanabe K, Hara ES, Ono M, Kuboki T, Calderwood SK. OstemiR: a novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS One. 2013;8:e58796. doi: 10.1371/journal.pone.0058796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Candini O, Spano C, Murgia A, Grisendi G, Veronesi E, Piccinno MS, Ferracin M, Negrini M, Giacobbi F, Bambi F, Horwitz EM, Conte P, Paolucci P, Dominici M. Mesenchymal progenitors aging highlights a miR-196 switch targeting HOXB7 as master regulator of proliferation and osteogenesis. Stem Cells. 2015;33:939–950. doi: 10.1002/stem.1897. [DOI] [PubMed] [Google Scholar]
- 56.Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang W, Jafferji M, Gordon JA, Li Z, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem. 2012;287:42084–42092. doi: 10.1074/jbc.M112.377515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim YJ, Bae SW, Yu SS, Bae YC, Jung JS. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J Bone Miner Res. 2009;24:816–825. doi: 10.1359/jbmr.081230. [DOI] [PubMed] [Google Scholar]
- 58.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmieri A, Pezzetti F, Spinelli G, Arlotti M, Avantaggiato A, Scarano A, Scapoli L, Zollino I, Carinci F. PerioGlas regulates osteoblast RNA interfering. J Prosthodont. 2008;17:522–526. doi: 10.1111/j.1532-849X.2008.00331.x. [DOI] [PubMed] [Google Scholar]
- 60.Roberto VP, Tiago DM, Silva IA, Cancela ML. MiR-29a is an enhancer of mineral deposition in bone-derived systems. Arch Biochem Biophys. 2014;564:173–183. doi: 10.1016/j.abb.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Q, Zhao ZN, Cheng JT, Zhang B, Xu J, Huang F, Zhao RN, Chen YJ. Ibandronate promotes osteogenic differentiation of periodontal ligament stem cells by regulating the expression of microRNAs. Biochem Biophys Res Commun. 2011;404:127–132. doi: 10.1016/j.bbrc.2010.11.079. [DOI] [PubMed] [Google Scholar]
- 62.Graneli C, Karlsson C, Brisby H, Lindahl A, Thomsen P. The effects of PPAR-gamma inhibition on gene expression and the progression of induced osteogenic differentiation of human mesenchymal stem cells. Connect Tissue Res. 2014;55:262–274. doi: 10.3109/03008207.2014.910198. [DOI] [PubMed] [Google Scholar]
- 63.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 64.Komori T. Regulation of osteoblast differentiation by transcription factors. J Cell Biochem. 2006;99:1233–1239. doi: 10.1002/jcb.20958. [DOI] [PubMed] [Google Scholar]
- 65.Laxman N, Rubin CJ, Mallmin H, Nilsson O, Pastinen T, Grundberg E, Kindmark A. Global miRNA expression and correlation with mRNA levels in primary human bone cells. RNA. 2015;21:1433–1443. doi: 10.1261/rna.049148.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu J, Qu S, Yao B, Xu Y, Jin Y, Shi K, Shui Y, Pan S, Chen L, Ma C. Osterix acetylation at K307 and K312 enhances its transcriptional activity and is required for osteoblast differentiation. Oncotarget. 2016;7:37471–37486. doi: 10.18632/oncotarget.9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 68.Yu S, Geng Q, Pan Q, Liu Z, Ding S, Xiang Q, Sun F, Wang C, Huang Y, Hong A. MiR-690, a Runx2-targeted miRNA, regulates osteogenic differentiation of C2C12 myogenic progenitor cells by targeting NF-kappaB p65. Cell Biosci. 2016;6:10. doi: 10.1186/s13578-016-0073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhuang H, Zhang X, Zhu C, Tang X, Yu F, Shang GW, Cai X. Molecular mechanisms of PPARgamma governing MSC osteogenic and adipogenic differentiation. Curr Stem Cell Res Ther. 2016;11:255–264. doi: 10.2174/1574888x10666150531173309. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, Stein GS. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci U S A. 2011;108:9863–9868. doi: 10.1073/pnas.1018493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maehata Y, Takamizawa S, Ozawa S, Kato Y, Sato S, Kubota E, Hata R. Both direct and collagen-mediated signals are required for active vitamin D3-elicited differentiation of human osteoblastic cells: roles of osterix, an osteoblast-related transcription factor. Matrix Biol. 2006;25:47–58. doi: 10.1016/j.matbio.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 72.Shi K, Ma C. Reply to “On microRNA-214 suppressing osteogenic differentiation of C2C12 myoblast cells by targeting Osterix”. Bone. 2013;57:328–334. doi: 10.1016/j.bone.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 73.Zhang W, Zhang L, Zhou Y, Ji X, Liu J, Liu D, Yin P, Peng Y, Hao M, Zhang L, Tang P. Synergistic effects of BMP9 and miR-548d-5p on promoting osteogenic differentiation of mesenchymal stem cells. Biomed Res Int. 2015;2015:309747. doi: 10.1155/2015/309747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ehrlich M, Gutman O, Knaus P, Henis YI. Oligomeric interactions of TGF-beta and BMP receptors. FEBS Lett. 2012;586:1885–1896. doi: 10.1016/j.febslet.2012.01.040. [DOI] [PubMed] [Google Scholar]
- 75.Mizuno Y, Tokuzawa Y, Ninomiya Y, Yagi K, Yatsuka-Kanesaki Y, Suda T, Fukuda T, Katagiri T, Kondoh Y, Amemiya T, Tashiro H, Okazaki Y. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009;583:2263–2268. doi: 10.1016/j.febslet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 76.Hill CS. Transcriptional control by the SMADs. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a022079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kamiya N, Kaartinen VM, Mishina Y. Lossof-function of ACVR1 in osteoblasts increases bone mass and activates canonical Wnt signaling through suppression of Wnt inhibitors SOST and DKK1. Biochem Biophys Res Commun. 2011;414:326–330. doi: 10.1016/j.bbrc.2011.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


