Abstract
Objective: This study works to develop novel models that may be adopted for earlier non-invasive breathomics tests to determine pneumonia pathogens. Methods: Two types of pneumonia models were created, both in vitro and in vivo. Paraneoplasm lung tissue and specific pathogen-free (SPF) rabbits were adopted and separately challenged with sterile saline solution control or three pathogens: Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa. After inoculation, headspace air or exhaled air were absorbed by solid phase micro-extraction (SPME) fibers and subsequently analyzed with gas chromatograph Mass Spectrometer (GCMS). Results: Pneumonia and pathogen-specific discriminating VOC patterns (1H-Pyrrole-3-carbonitrile, Diethyl phthalate, Cedrol, Decanoic acid, Cyclohexane, Diisooctyl phthalate) were determined. Conclusion: Our study successfully generated nosocomial pneumonia models for pneumonia diagnosis and pathogen-discriminating breath tests. The tests may allow for earlier pneumonia and pathogen diagnoses, and may transfer empirical therapy to targeted therapy earlier, thus improving clinical outcomes.
Keywords: Breath test, VOC biomarkers, nosocomial pneumonia model, pathogen diagnosis, breathomics
Introduction
Nosocomial pneumonia is a major cause of death, morbidity, and resource utilization, notably in patients with severe underlying conditions. However, early appropriate antibiotic treatment can improve outcomes. Staphylococcus aureus, Pseudomonas aeruginosa, and Enterobacteria represent some of the most frequent fatal pathogens in nosocomial pneumonia. Early targeted therapy is essential to improving pneumonia outcomes, but at present, the pathogenic diagnosis of nosocomial pneumonia is low yielding and often requires invasive, time-consuming methods, such as bronchoscopy or lung biopsy with subsequent culturing [1,2]. Therefore, it is of utmost need to develop a non-invasive method for early diagnoses, preferably by facilitating the rapid identification of the specific pathogens.
In recent years, breath tests have been adopted in many diseases for diagnosis and monitoring. For instance, the C-13 breath test is a standard screening test for gastric HP infection; NO breath levels are also used as a biomarker of airway inflammation, although its use is limited in non-allergic patients [3,4]. Additional exhaled biomarkers, including volatile organic compounds (VOCs), have since been studied. VOCs are a group of carbon-based chemicals that are volatile at room temperature and is gaining in popularity as a rapid, non-invasive diagnosis and monitoring method in diseases such as lung cancer, asthma, chronic obstructive pulmonary disease (COPD), and cystic fibrosis [5-9]. Notably, VOCs were recently found to have the potential to replace plasma tests of lipid levels [10]. Previously, screening studies of VOCs from bacterial metabolites for detection and classification of virulent bacteria yielded both qualitative and quantitative results through direct mass spectrometric methods [11-14]. Encouraging results were reported, indicating that some bacteria have their own characteristic range of volatile metabolites detected. Thus, VOC analyses have seen increasing use in lung bacterial infections tests [15,16]. Sensor systems such as the electronic nose system (EN) have been proposed to generate VOC profiles (breathprints), but are often unspecific and rely on a complex mathematical pattern recognition technology instead of clearly identifying specific VOCs [8,17]. More convincing evidence was shown when VOCs were detected by gas chromatography mass spectrometry (GCMS) in cystic fibrosis (CF) patients, for whom 2-aminoacetophenone was measured as volatile biomarker produced by P. aeruginosa in several studies [18-22]. Biomarkers of Aspergillus spp. and Mycobacterium tuberculosis were also reported [23,24].
Schnabel et al. have detected exhaled VOCs from 100 intubated patients in the ICU; this study revealed 12 VOCs that correctly discriminated between VAP(+) and VAP(-) groups [25]. The respective sensitivity, specificity, and AUC values were 75.8% ± 13.5%, 73.0% ± 11.8%, and 0.87. However, this study did not report pathogen-specific VOCs, and the metabolic mechanism is currently unclear [25]. Another study reported breath tests of 46 intubated patients in the ICU, in which patients found to have significant pathogen loads in the lower respiratory tract presented characteristic VOC patterns [26]. In a study that enrolled 22 VAP patients, including five Staphylococcus aureus and five Candida infection patients, pathogen-specific VOCs were also found to overlap between in vitro experiments and in vivo VAP patients [27]. In addition, our previous studies have shown high discriminating efficacy of VOCs for more than 100 VAP patients with A. Baumani infection, colonization, or absence [28]. Taken together, these studies bring us promising perspectives of breath tests in pneumonia diagnosis. However, the role of VOC detection on earlier diagnoses before typical clinical presentation has yet to be determined, and the underlying metabolic pathways remain unclear [5,8].
Because GC/MS detection of VOCs is highly sensitive, the results may be influenced by interventions to the patients before sampling, such as suction, PEEP adjusting, breathing frequency, cardiac output, tubing, and drugs (especially antibiotics). Other influencing factors include gut flora status, food intake, smoking status, metabolic disorders, and liver cirrhosis [29]. It is thus difficult for such small cohort studies to abolish bias, since the metabolic pathways of VOCs are key to determining diagnostic value, and help to unveil the pathophysiology of pneumonia. Collectively, this urges the development of novel research models, especially those that can distinguish between different pathogen infection. No published research has systemically assessed lung infection models for pathogen-specific VOC detection.
This study aims to assess both in vitro and in vivo infection models to determine characteristic VOCs of the most common bacterial pathogens of nosocomial pneumonia (Figure 1). The development of a novel, rapid, and non-invasive diagnosis method to identify pneumonia pathogens is critical, and such a model would also help unveil the underlying metabolic pathways behind nosocomial pneumonia.
Figure 1.
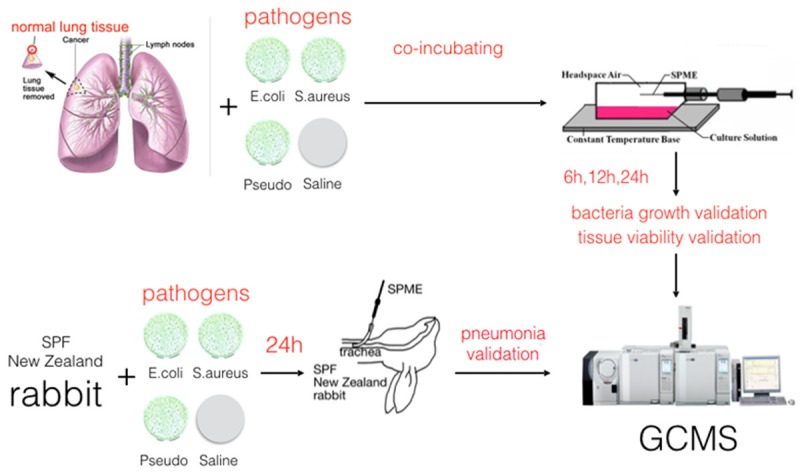
Experiment design of in vitro and in vivo pneumonia models for VOC detection. Resected paracancerous lung tissue and rabbits were challenged with three pathogens and sterile saline. Then headspace air and exhaled gas were absorbed with SPME. Those collected VOC samples underwent GCMS detection.
Materials and methods
Study subjects or animals
Specific pathogen-free New Zealand white rabbits were obtained from Animal Science College of Zhejiang University. Upon arrival, all animals were placed in quarantine for one week in cages, and fed with water and commercial feed in accordance with the most recent recommendations [30]. Study was conducted only after animal ethics approval from the appropriate ethics committee of the hospital.
Animal pneumonia model
Pneumonia was induced with a modified method; on the first day of experiment, rabbits were anaesthetized intraperitoneally with 8% chloralic hydras at 8 mg/kg. In order to avoid oral bacterial contamination, an incision was made in the neck of each rabbit and the anterior trachea exposed; subsequently, 0.5 mL pathogen solution was intratracheally injected with a 1 mL sterile syringe. Animals were then placed in the upright position for 15 seconds in order to facilitate distal alveolar migrations by gravity, sutured, and placed back into individual cages. Sterile saline was used as a control.
Paracancerous human lung tissue
Paracancerous human lung tissues were collected from resected lung cancer patients and cultured with DMEM media. Ethical approval was issued by the ethics committee of the hospital, and informed consent was obtained from each patient. Lung tissue was divided into four pieces of approximately 1 cm3 each, and incubated with either sterile saline as a control or three different bacterial pathogens (concentrations of log8 cfu.mL or log9 cfu.mL). Headspace air was detected with SPME fibers at successive intervals of lengths 6, 12, and 24 hours after incubation.
Bacterial culturing
Escherichia coli (American Type Culture Collection 25922; ATCC, Manassas, VA, USA), Pseudomonas aeruginosa (ATCC 27853), and Staphylococcus aureus (ATCC 25923) were used for preparation of the inoculum in the study. The three bacteria represent the most frequently seen pathogens in hospital-acquired pneumonia patients, and are widely used in animal pneumonia models. One day prior to inoculation, bacteria were thawed from -80°C at room temperature and pipetted onto nutrient agar plates. They were then incubated for 18 hours at 37°C in 5% CO2. On the inoculation day, the overnight bacteria were diluted in sterile saline to the concentrations of log9 cfu.mL or log8 cfu.mL.
Confirmation of pneumonia animal models
Twenty-four hours after intratracheal injection of pathogens, animals underwent VOC collection procedures and sacrificed. Positive pneumonia results were confirmed by pathologists blinded to the experiments according to the gold standard procedure for pneumonia verification.
VOCs collection and detection
In tissue models, headspace air was absorbed directly with SPME fibers at 6, 12, and 24 hours after co-incubation with the three pathogens and saline control. Afterwards, validations of bacterial growth and tissue viability were carried out; validated samples were candidates for further GCMS testing. In animal models, percutaneous tracheotomy was performed 24 hours after intratracheal injection and inoculation; a coated sterile tube was inserted and the tip tightly fixed into the trachea. Sedated animals were thus able to breath spontaneously through the tube. SPME fibers were placed inside the tube for 30 minutes to absorb the VOCs of exhaled air at room temperature, and all samples were collected in triplicate.
The determination of VOCs was performed on a Gas Chromatograph Mass Spectrometer (GCMS-QP2010/PLUS, Shimadzu, Japan) with split-splitless injector. Desorption time of SPME was set at 3 minutes under 250°C in GC injector, while the splitless mode was maintained for 2 minutes before setting a 1:10 split ratio. The 30 m × 0.25 mm × 0.25 µm capillary column Rtx-1 (Restek) was used, and its flow velocity set at 1 mL/min; the temperature of the column oven increased from 40°C to 250°C in 40 minutes. The GCMS worked in full scan mode at the 35-400 m/z range [6].
Data analysis
The mass spectrometry library (NIST 05 and NIST 05 s) (National Institute of Standards and Technology) was used to match, identify, and search similar compounds; the highest similarity matches were presumed to be the most likely candidates. Manual checking was initiated for cautious identification if the similarity matching results were less than 80%, and the “20% rule” was applied for data selection. Briefly, a variable was adopted when nonzero data were available for at least 20% of all samples within at least one of the experimental groups. Some compounds, such as siloxanes, caryophyllene, longifolene, and cedrene were also excluded initially. All VOC values were grouped according to differing pathogens, and data subjected to Canonical Discriminant Analysis and Multivariate Discriminant Logistic Analysis on Stata MP (Version 14). Statistically significant discriminating VOCs of each group were calculated [6].
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Results
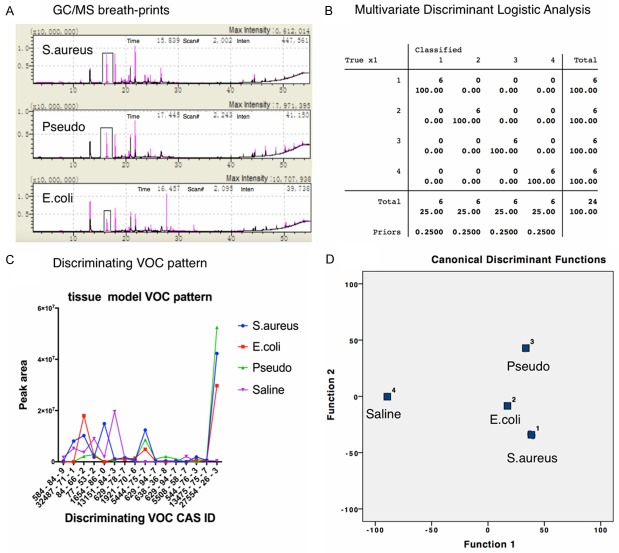
In total, six patients’ paracancerous lung tissues were collected and divided into 24 sections, followed by co-culturing separately with three pathogens (Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus) and sterile saline control. Hundreds of substances were detected in all groups; compared with control groups, infected lung tissues emitted significantly discriminating VOCs. Despite different concentrations of challenging pathogens at both log9 cfu.mL and log8 cfu.mL, VOC patterns remained the same (Figure 2). The discriminating power of pathogen-specific VOCs increased consecutively when detected at 6, 12, and 24 hours after incubation; the first timepoint was sufficient to obtain the discriminating VOCs. Compared to control, all types of infected lung tissues emitted pathogen specific VOCs, including 2, 4-diisocyanatotoluene, 1H-pyrrole-3-carbonitrile, diethyl phthalate, cedrol, decanoic acid, cyclohexane, heptadecane, pristane, benzoic acid, heneicosane, phytane, andrographolide, hexadecane, 8-hexylpentadecane, and diisooctyl phthalate (Figure 3). KEGG metabolic pathways were predicted according to the metabolic database, while heptasiloxane, octadecamethylcyclononasiloxane, and octamethylcyclotetrasiloxane were predicted to originate from artificial plastic products.
Figure 2.
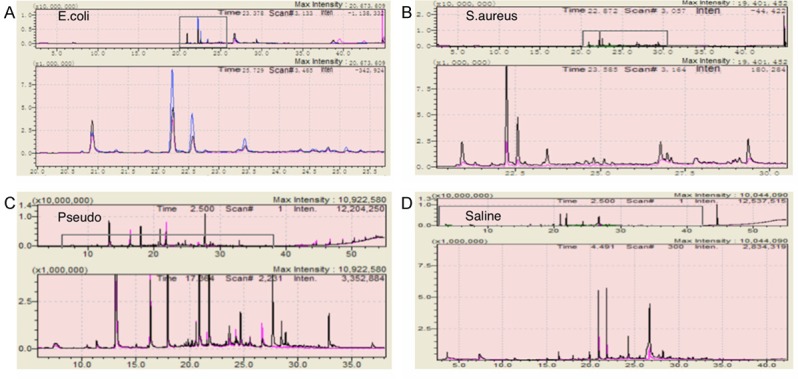
GC-MS VOC analysis of lung tissue model with different challenging pathogen concentrations (log8 CFU.mL, log9 CFU.mL). A. Lung tissue infection model incubated with E.coli. B. Lung tissue infection model incubated with S.aureus. C. Lung tissue infection model incubated with Pseudomonas. D. Lung tissue infection model incubated with sterile saline.
Figure 3.
Discriminant analysis of pathogen specific VOCs from lung tissue model. A. GC-MS analysis of VOCs from different pathogens incubated lung tissue model, blanked with sterile saline; B. Multivariate Discriminant Logistic Analysis of VOCs from different pathogen groups (1. S.aureus, 2. E.coli, 3. Pseudomonas, 4. Sterile saline); C. Discriminating VOC pattern in animal model; D. Multivariate Discriminant Analysis of VOCs from different pathogen groups (1. S.aureus, 2. E.coli, 3. Pseudomonas, 4. Sterile saline).
Following treatment as described above and pneumonia diagnosis by a pathologist blinded to pathogen and pneumonia group data, macroscopic evidence of swelling, redness, or gray congestion of animal lungs were present in all experimental groups but absent in the control group. Microscopic findings revealed evidence that polymorphonuclear leukocytes infiltrates and fibrinous exudates filled up alveoli in the lungs of the experimental groups, but not in the control cohort (Figure 4). All VOCs detected in exhaled air from the corresponding pathogen-challenged pneumonia animals were similar. Subsequently, bacterial pneumonia VOCs were compared to the control group using Multivariate Discriminant Logistic Analysis, uncovering statistically discriminating VOCs (Figure 5). Those VOCs were reported to be 1H-pyrrole-3-carbonitrile, diethyl phthalate, cedrol, decanoic acid, cyclohexane, trans-squalene, diisooctyl phthalate, and heptasiloxane.
Figure 4.
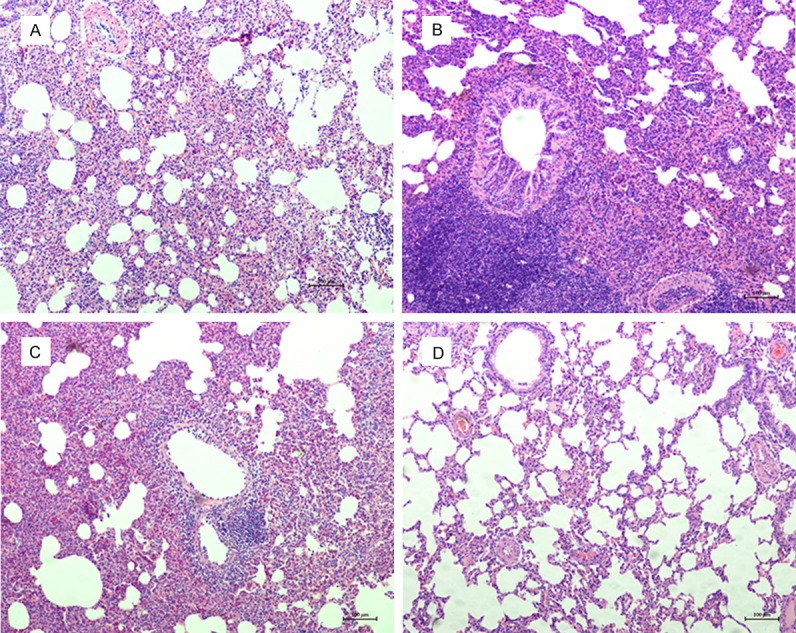
Microscopic findings revealed in vivo pneumonia evidences that polymorphonuclear leukocytes infiltrates and fibrinous exudates filled up alveoli, while the sterile saline control group was absent. A. E.coli pneumonia animal model; B. S.aureus pneumonia animal model; C. Pseudomonas pneumonia animal model; D. Sterile saline control animal model.
Figure 5.
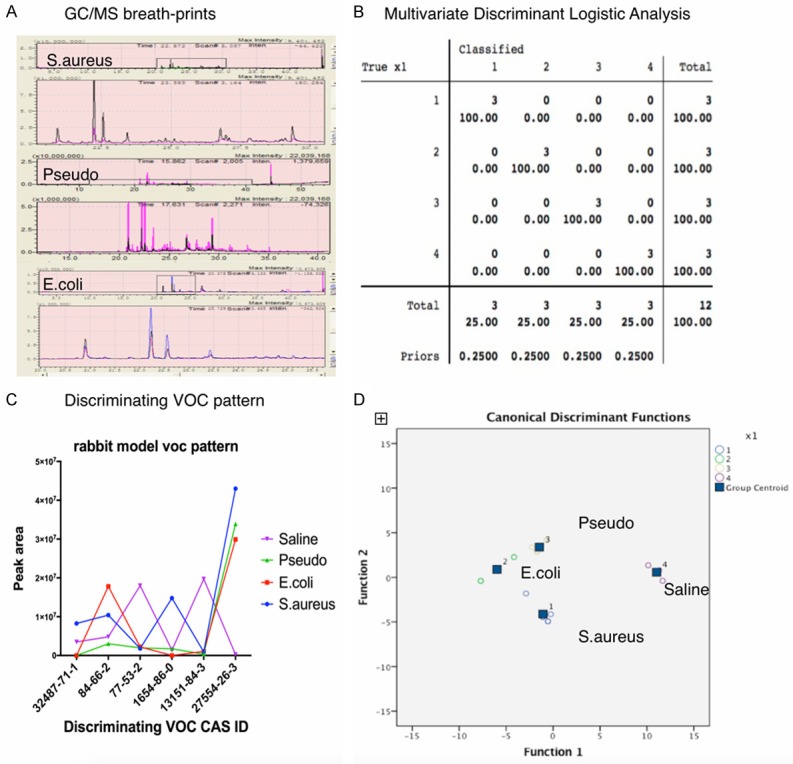
Discriminant analysis of pathogen specific VOCs from pneumonia animal model. A. GC-MS analysis of VOCs from different pathogens challenged pneumonia animal model, blanked with sterile saline; B. Multivariate Discriminant Logistic Analysis of VOCs from different pathogen groups; C. Discriminating VOC pattern in animal model; D. Multivariate Discriminant Analysis of VOCs from different pathogen groups.
After analyzing pooled data from both the lung tissue and animal models, we consistently found common pneumonia and pathogen-specific VOC patterns (Figures 6, 7 and Table 1). These pathogen-discriminating VOCs are 1H-pyrrole-3-carbonitrile, diethyl phthalate, cedrol, decanoic acid, cyclohexane, and diisooctyl phthalate, while possible KEGG metabolic pathways were predicted according to the metabolic database.
Figure 6.
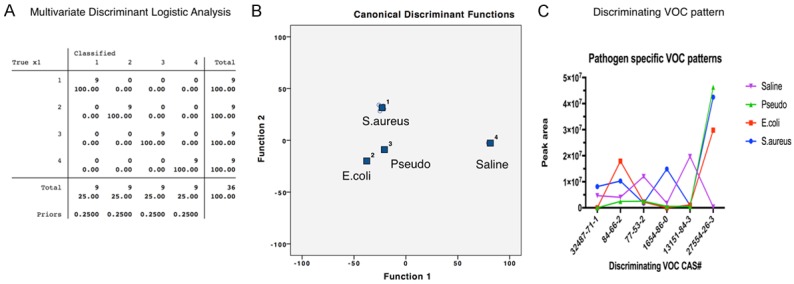
Discriminant analysis of pathogen specific VOCs for both pneumonia models. A. Multivariate Discriminant Logistic Analysis of VOCs from different pathogen chanllenged lung tissue and animal model groups with sterile saline control (1. S.aureus, 2. E.coli, 3. Pseudomonas, 4. Sterile saline); B. Multivariate Discriminant Analysis of VOCs from different pathogen groups and sterile saline control. C. Patterns of discriminating VOCs from different pathogen groups and sterile saline control.
Figure 7.
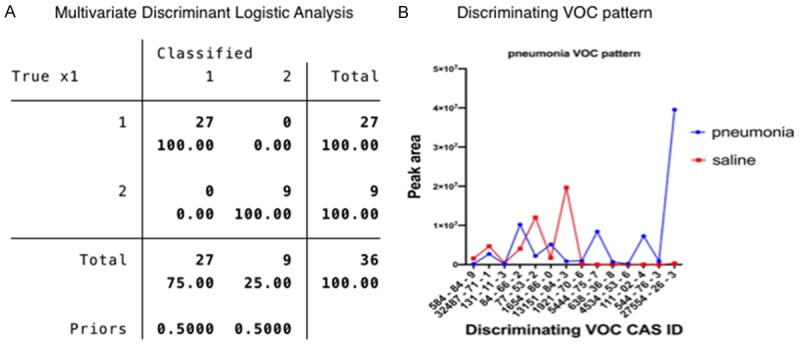
Discriminant analysis of pathogen specific VOCs from both pneumonia models and saline control.
Table 1.
Pathogen discriminating VOCs from in vitro and in vivo pneumonia models with predicted metabolic pathways
| Peak | Predicted metabolic pathway | ||||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| R.T. (min) | CAS# | Compounds | S.aureus | E.coli | Pseudo | Saline | |
|
|
|||||||
| vitro|vivo | vitro|vivo | vitro|vivo | vitro|vivo | ||||
| 22.216 | 32487-71-1 | 1H-Pyrrole-3-carbonitrile | ++|++ | -|- | -|- | ++|++ | Unkown |
| 24.334 | 84-66-2 | Diethyl phthalate | +++|+++ | +++|+++ | ++|++ | ++|++ | Aminobenzoate degradation |
| 27.118 | 77-53-2 | Cedrol | ++|++ | ++|++ | ++|++ | +++|+++ | Sesquiterpenoid biosynthesis |
| 27.809 | 1654-86-0 | Decanoic acid | +++|+++ | -|- | ++|- | ++|++ | Fatty acid biosynthesis |
| 28.061 | 13151-84-3 | Cyclohexane | +|+ | ++|++ | +|+ | +++|+++ | Benzoate degradation |
| 44.528 | 27554-26-3 | Diisooctyl phthalate | +++|+++ | +++|+++ | +++|+++ | +|+ | Unknown |
“-” denotes absence, “±” denotes less than 105, “+” denotes 105, “++” denotes 106, “+++” denotes 107.
Discussion
This study suggests that it may be possible to determine pathogens of nosocomial pneumonia via a rapid, direct, and non-invasive breath test. VOCs are a group of chemicals that are volatile at room temperature, and the source of exhaled VOCs can be endogenous or exogenous. Endogenous VOCs are volatile metabolites from conducting airways, alveoli, or systemic VOCs generated elsewhere in the body and transported to the lungs via blood circulation; some endogenous VOCs can be absorbed in lungs before detection [5,8]. GCMS coupled with solid phase micro-extraction is adopted as a standard VOCs detection method. Using this method, we have successfully detected VOCs in lung cancer patients and established characteristic diagnostic patterns for lung cancer; excitingly, this study shows that pathogen-specific VOCs were found in both in vitro and in vivo models [6].
Here, we developed two nosocomial pneumonia models: the paracancerous lung tissue infection model, and the rabbit pneumonia model. The former was employed to explore different pathogen concentrations and detection times, while the latter is an ideal model for in vivo validation. Modified animal models of bacterial pneumonia were developed, and intratracheal injection was introduced to avoid possible oral bacteria contaminations. This animal model can decrease the potential influences caused by oral or gastrointestinal bacteria contamination, and mimics ventilator-associated pneumonia (VAP). All three common pathogens (Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus) were used in both pneumonia models, and common VOCs were emitted from both. These results further suggest that VOCs detection would be a promising method for rapid pneumonia pathogen determination. Results revealed that the number of discriminating VOCs found from the in vitro model are higher than that of the in vivo model; one possible reason could be the reabsorptive ability of the bloodstream in animal lungs, and further study is needed to explore this metabolic mechanism. However, because GCMS detection is very sensitive, the results in this study may have been affected by different lung tissue viability, or bacteria growth status during experiments. Large cohorts of research on characteristic pathogen breathprints of pneumonia are required for future studies.
In this study, before co-incubation of bacteria with lung tissue or animals, the headspace air of each cultured pathogen had been detected in our lab, and characteristic VOCs of each were found. Interestingly, in both infected lung tissues and pneumonia animal models, characteristic VOCs were different from those of cultured pathogens. These results may be caused by decreased abundance of pathogens in pneumonia models at early stages of infection, and VOCs may originate from the host response to such pathogens. Thus, pathogen-discriminating VOCs may be detectable even with small amounts of pathogens at early stages, when bacteria are less abundant. Because host responses of infection and colonization are different, these VOCs might also be employed to discriminate bacteria airway colonization from infection. For instance, previous breath test research has shown that VOCs from cystic fibrosis patients were different when comparing the colonization and infection of pseudomonas [31]. Electronic nose technology may also be adopted to detect A. fumigatus colonization in cystic fibrosis patients [17]. Our previous studies also showed high discriminating efficacy of VOCs for more than 100 VAP patients with A. Baumani infection, colonization, and absence [28].
Other pathogen detection research has also been published, including those studying PCR and MALDI-TOF MS [32,33]. Unfortunately, these procedures require expensive equipment, skilled technicians, and hours of sample processing. Furthermore, these methods are too sensitive and cannot abolish the influence of contamination. Prior research has found that genotypically different strains of Pseudomonas emitted variant VOCs [34]. Thus, combining breath testing with existing methods could increase the precision of pathogen detection; VOC detection has intriguing potential for the rapid classification of drug resistant pathogens and may serve as a drug sensitivity predictor.
Conclusion
In conclusion, this study generated two models for a rapid, non-invasive breath test that can discriminate between three most common pathogens in nosocomial pneumonia. It may potentiate a promising point-of-care test to determine pneumonia pathogens, thus guiding earlier antibiotic treatment. Our models are just the initial step for further research on the underlying metabolic pathways of nosocomial pneumonia. It is anticipated that bedside VOC profiling will eventually enable rapid pathogen diagnosis of pneumonia, and serve as a useful alternative to medical practitioners in the future.
Acknowledgements
This work was supported as a key project of the Natural Science Foundation of China [Grant number 31627801], a key project of the Natural Science Foundation of Zhejiang [Grant number 2013C03044-2], and the Health and Family Planning Commission of Zhejiang Province [Grant number 2016150952].
Disclosure of conflict of interest
None.
References
- 1.Barbier F, Andremont A, Wolff M, Bouadma L. Hospital-acquired pneumonia and ventilator-associated pneumonia: recent advances in epidemiology and management. Curr Opin Pulm Med. 2013;19:216–228. doi: 10.1097/MCP.0b013e32835f27be. [DOI] [PubMed] [Google Scholar]
- 2.American Thoracic Society; Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005;171:388–416. doi: 10.1164/rccm.200405-644ST. [DOI] [PubMed] [Google Scholar]
- 3.Montuschi P, Mores N, Trove A, Mondino C, Barnes PJ. The electronic nose in respiratory medicine. Respiration. 2013;85:72–84. doi: 10.1159/000340044. [DOI] [PubMed] [Google Scholar]
- 4.Malerba M, Montuschi P. Non-invasive biomarkers of lung inflammation in smoking subjects. Curr Med Chem. 2012;19:187–196. doi: 10.2174/092986712803414204. [DOI] [PubMed] [Google Scholar]
- 5.van de Kant KD, van der Sande LJ, Jobsis Q, van Schayck OC, Dompeling E. Clinical use of exhaled volatile organic compounds in pulmonary diseases: a systematic review. Respir Res. 2012;13:117. doi: 10.1186/1465-9921-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng J, Wang X, Qian F, Vogel S, Xiao L, Ranjan R, Park H, Karpurapu M, Ye RD, Park GY, Christman JW. Protective role of reactive oxygen species in endotoxin-induced lung inflammation through modulation of IL-10 expression. J Immunol. 2012;188:5734–5740. doi: 10.4049/jimmunol.1101323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Kant KD, van Berkel JJ, Jobsis Q, Lima Passos V, Klaassen EM, van der Sande L, van Schayck OC, de Jongste JC, van Schooten FJ, Derks E, Dompeling E, Dallinga JW. Exhaled breath profiling in diagnosing wheezy preschool children. Eur Respir J. 2013;41:183–188. doi: 10.1183/09031936.00122411. [DOI] [PubMed] [Google Scholar]
- 8.van der Schee MP, Paff T, Brinkman P, van Aalderen WM, Haarman EG, Sterk PJ. Breathomics in lung disease. Chest. 2015;147:224–231. doi: 10.1378/chest.14-0781. [DOI] [PubMed] [Google Scholar]
- 9.Cavaleiro Rufo J, Madureira J, Oliveira Fernandes E, Moreira A. Volatile organic compounds in asthma diagnosis: a systematic review and meta-analysis. Allergy. 2016;71:175–188. doi: 10.1111/all.12793. [DOI] [PubMed] [Google Scholar]
- 10.Minh Tdo C, Oliver SR, Flores RL, Ngo J, Meinardi S, Carlson MK, Midyett J, Rowland FS, Blake DR, Galassetti PR. Noninvasive measurement of plasma triglycerides and free fatty acids from exhaled breath. J Diabetes Sci Technol. 2012;6:86–101. doi: 10.1177/193229681200600112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zechman JM, Aldinger S, Labows JN Jr. Characterization of pathogenic bacteria by automated headspace concentration-gas chromatography. J Chromatogr. 1986;377:49–57. doi: 10.1016/s0378-4347(00)80760-4. [DOI] [PubMed] [Google Scholar]
- 12.Buszewski B, Ulanowska A, Ligor T, Jackowski M, Klodzinska E, Szeliga J. Identification of volatile organic compounds secreted from cancer tissues and bacterial cultures. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;868:88–94. doi: 10.1016/j.jchromb.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 13.Scotter JM, Allardyce RA, Langford VS, Hill A, Murdoch DR. The rapid evaluation of bacterial growth in blood cultures by selected ion flow tube-mass spectrometry (SIFT-MS) and comparison with the BacT/ALERT automated blood culture system. J Microbiol Methods. 2006;65:628–631. doi: 10.1016/j.mimet.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 14.O’Hara M, Mayhew CA. A preliminary comparison of volatile organic compounds in the headspace of cultures of staphylococcus aureus grown in nutrient, dextrose and brain heart bovine broths measured using a proton transfer reaction mass spectrometer. J Breath Res. 2009;3:027001. doi: 10.1088/1752-7155/3/2/027001. [DOI] [PubMed] [Google Scholar]
- 15.Turner AP, Magan N. Electronic noses and disease diagnostics. Nat Rev Microbiol. 2004;2:161–166. doi: 10.1038/nrmicro823. [DOI] [PubMed] [Google Scholar]
- 16.Eisenmann A, Amann A, Said M, Datta B, Ledochowski M. Implementation and interpretation of hydrogen breath tests. J Breath Res. 2008;2:046002. doi: 10.1088/1752-7155/2/4/046002. [DOI] [PubMed] [Google Scholar]
- 17.de Heer K, Kok MG, Fens N, Weersink EJ, Zwinderman AH, van der Schee MP, Visser CE, van Oers MH, Sterk PJ. Detection of airway colonization by aspergillus fumigatus by use of electronic nose technology in patients with cystic fibrosis. J Clin Microbiol. 2016;54:569–575. doi: 10.1128/JCM.02214-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorn RM, Reynolds DM, Greenman J. Multivariate analysis of bacterial volatile compound profiles for discrimination between selected species and strains in vitro. J Microbiol Methods. 2011;84:258–264. doi: 10.1016/j.mimet.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhu J, Bean HD, Kuo YM, Hill JE. Fast detection of volatile organic compounds from bacterial cultures by secondary electrospray ionization-mass spectrometry. J Clin Microbiol. 2010;48:4426–4431. doi: 10.1128/JCM.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox CD, Parker J. Use of 2-aminoacetophenone production in identification of Pseudomonas aeruginosa. J Clin Microbiol. 1979;9:479–484. doi: 10.1128/jcm.9.4.479-484.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labows JN, McGinley KJ, Webster GF, Leyden JJ. Headspace analysis of volatile metabolites of Pseudomonas aeruginosa and related species by gas chromatography-mass spectrometry. J Clin Microbiol. 1980;12:521–526. doi: 10.1128/jcm.12.4.521-526.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preti G, Thaler E, Hanson CW, Troy M, Eades J, Gelperin A. Volatile compounds characteristic of sinus-related bacteria and infected sinus mucus: analysis by solid-phase microextraction and gas chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2011–2018. doi: 10.1016/j.jchromb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 23.Chambers ST, Bhandari S, Scott-Thomas A, Syhre M. Novel diagnostics: progress toward a breath test for invasive aspergillus fumigatus. Med Mycol. 2011;49(Suppl 1):S54–61. doi: 10.3109/13693786.2010.508187. [DOI] [PubMed] [Google Scholar]
- 24.Phillips M, Basa-Dalay V, Blais J, Bothamley G, Chaturvedi A, Modi KD, Pandya M, Natividad MP, Patel U, Ramraje NN, Schmitt P, Udwadia ZF. Point-of-care breath test for biomarkers of active pulmonary tuberculosis. Tuberculosis (Edinb) 2012;92:314–320. doi: 10.1016/j.tube.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Schnabel R, Fijten R, Smolinska A, Dallinga J, Boumans ML, Stobberingh E, Boots A, Roekaerts P, Bergmans D, van Schooten FJ. Analysis of volatile organic compounds in exhaled breath to diagnose ventilator-associated pneumonia. Sci Rep. 2015;5:17179. doi: 10.1038/srep17179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fowler SJ, Basanta-Sanchez M, Xu Y, Goodacre R, Dark PM. Surveillance for lower airway pathogens in mechanically ventilated patients by metabolomic analysis of exhaled breath: a case-control study. Thorax. 2015;70:320–325. doi: 10.1136/thoraxjnl-2014-206273. [DOI] [PubMed] [Google Scholar]
- 27.Filipiak W, Sponring A, Baur MM, Filipiak A, Ager C, Wiesenhofer H, Nagl M, Troppmair J, Amann A. Molecular analysis of volatile metabolites released specifically by staphylococcus aureus and pseudomonas aeruginosa. BMC Microbiol. 2012;12:113. doi: 10.1186/1471-2180-12-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Zou Y, Wang Y, Wang F, Lang L, Wang P, Zhou Y, Ying K. Breath analysis for noninvasively differentiating acinetobacter baumannii ventilator-associated pneumonia from its respiratory tract colonization of ventilated patients. J Breath Res. 2016;10:027102. doi: 10.1088/1752-7155/10/2/027102. [DOI] [PubMed] [Google Scholar]
- 29.Filipiak W, Beer R, Sponring A, Filipiak A, Ager C, Schiefecker A, Lanthaler S, Helbok R, Nagl M, Troppmair J, Amann A. Breath analysis for in vivo detection of pathogens related to ventilator-associated pneumonia in intensive care patients: a prospective pilot study. J Breath Res. 2015;9:016004. doi: 10.1088/1752-7155/9/1/016004. [DOI] [PubMed] [Google Scholar]
- 30.National research council (U.S.). Committee for the update of the guide for the care and use of laboratory animals., Institute for laboratory animal research (U.S.) and national academies press (U.S.) Guide for the care and use of laboratory animals. Washington, D.C.: National Academies Press; 2011. [Google Scholar]
- 31.Robroeks CM, van Berkel JJ, Dallinga JW, Jobsis Q, Zimmermann LJ, Hendriks HJ, Wouters MF, van der Grinten CP, van de Kant KD, van Schooten FJ, Dompeling E. Metabolomics of volatile organic compounds in cystic fibrosis patients and controls. Pediatr Res. 2010;68:75–80. doi: 10.1203/PDR.0b013e3181df4ea0. [DOI] [PubMed] [Google Scholar]
- 32.Charretier Y, Dauwalder O, Franceschi C, Degout-Charmette E, Zambardi G, Cecchini T, Bardet C, Lacoux X, Dufour P, Veron L, Rostaing H, Lanet V, Fortin T, Beaulieu C, Perrot N, Dechaume D, Pons S, Girard V, Salvador A, Durand G, Mallard F, Theretz A, Broyer P, Chatellier S, Gervasi G, Van Nuenen M, Ann Roitsch C, Van Belkum A, Lemoine J, Vandenesch F, Charrier JP. Rapid bacterial identification, resistance, virulence and type profiling using selected reaction monitoring mass spectrometry. Sci Rep. 2015;5:13944. doi: 10.1038/srep13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent JL, Brealey D, Libert N, Abidi NE, O’Dwyer M, Zacharowski K, Mikaszewska-Sokolewicz M, Schrenzel J, Simon F, Wilks M, Picard-Maureau M, Chalfin DB, Ecker DJ, Sampath R, Singer M Rapid Diagnosis of Infections in the Critically Ill Team. Rapid diagnosis of infection in the critically Ill, a multicenter study of molecular detection in bloodstream infections, pneumonia, and sterile site infections. Crit Care Med. 2015;43:2283–2291. doi: 10.1097/CCM.0000000000001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shestivska V, Spanel P, Dryahina K, Sovova K, Smith D, Musilek M, Nemec A. Variability in the concentrations of volatile metabolites emitted by genotypically different strains of Pseudomonas aeruginosa. J Appl Microbiol. 2012;113:701–713. doi: 10.1111/j.1365-2672.2012.05370.x. [DOI] [PubMed] [Google Scholar]