Abstract
Acute aortic dissection (AAD) is a life-threatening cardiovascular disease with the high morbidity and mortality. Imaging modalities are the gold standard for the diagnosis of AAD; however, they are not always available in emergency department. Biomarker-assisted diagnosis is important for the early treatment of AAD. The aim of the present study was to identify potential microRNA (miRNA) biomarkers for AAD. Differentially expressed plasma miRNAs between AAD patients and age-matched healthy volunteers were analyzed by miRNA microarray. Quantitative RT-PCR was further performed to verify the expression of selected miRNAs (miR-4787-5p and miR-4306) with an increased number of samples. Receiver operating characteristic (ROC) analysis was used to assess the diagnostic value of miR-4787-5p and miR-4306 as biomarkers for distinguishing AAD. Using TargetScan and miRanda, miR-4787-5p and miR-4306 were selected to predict target gene related to cytokines detecting by dual luciferase assay and western blotting. Nine upregulated and twelve downregulated miRNAs were identified in the circulating plasma of AAD patients. qRT-PCR verified statistically consistent expression of two selected miRNAs with microarray analysis. ROC analyses demonstrated that miR-4787-5p and miR-4306 were specific and sensitive for the early diagnosis of AAD. Bioinformatic predictions and dual luciferase assay suggested that polycystin-1 (PKD1) and transforming growth factor-β1 (TGF-β1) were respectively direct target of miR-4787-5p and miR-4306. Furthermore, the protein expression of the downstream targets of PKD1 and TGF-β1 were significantly reduced following overexpression of miR-4787-5p and miR-4306. These results revealed that miR-4787-5p and miR-4306 could be developed as diagnostic potential biomarkers for AAD, and they could be involved in the pathogenesis of AAD.
Keywords: MiRNA-4787-5p, miRNA-4306, plasma, acute aortic dissection, polycystin 1, transforming growth factor-β1
Introduction
Acute aortic dissection (AAD) is a life-threatening disease, due to the suddenly onset, progress rapidly, high mortality. Mechanically, circulating blood flows into the media of the aorta through the rupture of the intima and forms true and false lumens. The death rate from AAD increased from 2.49 per 100,000 to 2.78 per 100,000 inhabitants between 1990 and 2010, with men more often affected than women [1-5]. Although the incidence of AAD is increasing, most cases of AAD remain unidentified or misdiagnosed owing to the silent and polytropic nature of the disease and the lack of a specific clinical phenotype [6]. Delayed diagnosis is closely linked to the prognosis; approximately 50% and 80% of patients with AAD fail to be timely diagnosed and die within 3 days and 2 weeks, respectively [2]. Therefore, early and correct diagnosis is essential for controlling the development of AD and initiating the appropriate therapy to reduce the mortality rate and improve prognosis. AD can be diagnosed correctly by multiple spiral computed tomography, transthoracic or transesophageal echocardiography, and magnetic resonance angiography; however, these tests are expensive and complicated, and they sometimes cannot be performed at the emergency room or the results are delayed. The value of biomarkers for the diagnosis of AD is increasingly being recognized because of their rapid, easy, and convenient operation at bedside. Several plasma markers such as D-dimer, calponin, elastin, CD40L, MPO, MMP-1 and TIMP-1 have been investigated as potential candidates for use as biomarkers of AAD [7,8]. However, these biomarkers have not been adopted into routine clinical practice mainly because of inadequate sensitivity and specificity. Therefore, novel biomarkers with high sensitivity and specificity are highly desirable for the diagnosis of AAD [9].
MicroRNAs (miRNAs) is a sort of short, single-stranded, noncoding RNA that plays a role in gene regulation by binding to the target miRNA directly and forming base pairing guide silencing complex [10]. miRNAs play an important role in many physiological and pathological processes, such as cell growth, apoptosis, cellular differentiation, metabolism, tumorigenesis, angiogenesis, and heart damage and protection [11-13]. In recent years, growing evidence suggests that miRNAs not only play crucial roles in the physiological processes of cardiovascular development, but also in the pathologic processes of cardiovascular diseases. miRNAs are abundantly present in plasma/serum in a remarkably stable form and can be detected using real-time PCR assays [14,15]; in addition, the expression profiles of circulating miRNAs can change in various diseases. Therefore, circulating miRNAs could be used as novel and potential biomarkers for the diagnosis and prognosis of diseases, such as cancer, heart disease, pregnancy abnormalities, diabetes, psychosis, and various infectious diseases [16-21]. Recently, circulating miRNAs have been detected in a variety of cardiovascular diseases, suggesting their value as biomarkers for improving the diagnostic accuracy of cardiovascular disease [22-24] and as predictors of cardiovascular events [25]. However, few studies have investigated circulating miRNAs in AAD and the results are inconsistent [26-28]. The present study analyzed the differential expression of circulating miRNAs between AAD patients and healthy control subjects using microarray technologies to provide evidence of the value of miRNAs as new diagnostic biomarkers for AAD, and predicted their downstream targets to suggest primarily the roles on the pathogenesis of AAD.
Materials and methods
Ethics statement
The study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University, and complied with the declaration of Helsinki. All participants gave their informed consent for the acquisition and use of patient plasma, aorta samples and anonymized clinical data prior to their inclusion in the study.
Clinical specimens
A total of 98 patients with AAD (including 47 type A and 51 type B) and 56 age- and gender-matched healthy volunteers without AAD were enrolled from the First Affiliated Hospital of Zhengzhou University between January 2015 and March 2016. Inclusion guidelines included a time interval between symptom onset and hospital admission of ≤24 h and AAD diagnosed by aortic computed tomography angiography (CTA), transoesophageal echocardiography or MRA. AAD was classified according to the Stanford classification. Exclusion criteria were patients under 18 years of age, Marfan’s syndrome, Ehlers-Danlos syndrome, Turner syndrome, familial thoracic or abdominal aortic aneurysm, aortic ulcers or aortic intramural hematoma formation, dissection caused by trauma, pregnancy, and postoperative patients. A detailed description of the clinical characteristics of the study population is presented in Table 1.
Table 1.
Clinical characters of AAD and control groups
| Characteristics | AAD (n=98) | Control (n=56) |
|---|---|---|
| Age (year) | 51.2 ± 10.9 | 52.3 ± 11.0 |
| Male | 79 (80.6%) | 43 (76.7%) |
| Smoking | 35 (35.7%) | 7 (12.5%) |
| Alcoholic | 26 (26.5%) | 4 (7.1%) |
| Hypertension | 74 (76.3%)* | 0 |
| Atherosclerosis | 42 (42.9%) | 19 (37.3%) |
| D-dimmer | 4.41 ± 6.40* | 0.14 ± 0.07 |
Data are presented as mean ± SD or n (%).
P < 0.05;
AAD: acute aortic dissection.
Plasma and aorta samples of collection
Peripheral venous blood samples (5 ml) were drawn into tubes containing EDTA. The AAD blood samples were collected within 24 h after symptom onset before surgery or interventional therapy. Blood samples from volunteers were collected in the morning on an empty stomach. The blood was then centrifuged at 1200 g for 10 min at 4°C. The supernatant (plasma) was separated from the cellular layer by pipetting, centrifuged at 12,000 g for 10 min at 4°C and transferred into RNase/DNase-free tubes. Furthermore, ascending aorta specimen were obtained from AAD patients (type A, n=13) undergoing surgical replacement and from age- and gender-matched multi organ donors (n=5) without aortic diseases. The plasma and aorta samples were prepared as described previously and stored at -80°C in liquid nitrogen until RNA extraction. A maximum of 2 h between the blood draw and separation into plasma was the standardized protocol in this study.
RNA extraction and miRNA microarray analysis
Total RNA was extracted and purified from plasma samples using the mirVanaTM miRNA Isolation Kit (Ambion, Austin, TX, USA), following the manufacturer’s instructions and checked for a RIN number to inspect RNA integration by an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, US). miRNA in total RNA was labeled with miRNA Complete Labeling and Hyb Kit (Agilent Technologies) following the manufacturer’s instructions, labeling section. Each slide was hybridized with 100 ng Cy3-labeled RNA using miRNA Complete Labeling and Hyb Kit (Agilent Technologies) in a hybridization oven (Agilent Technologies) at 55°C with agitation at 20 rpm for 20 h according to the manufacturer’s instructions, hybridization section. After hybridization, slides were washed in staining dishes (Thermo Shandon, Waltham, MA, US) with Gene Expression Wash Buffer Kit (Agilent Technologies,). Slides were then scanned with Agilent Microarray Scanner (Agilent Technologies,) and Feature Extraction software 10.7 (Agilent technologies) with default settings. Raw data were normalized by Quantile algorithm, Gene Spring Software 12.6 (Agilent Technologies). The mean normalized signal from biological replicates was used for differential expression analysis. miRNAs with a change in expression levels of at least 2-fold (P < 0.05) between groups were selected for further analysis. The experiment was repeated three times.
miRNA analysis by quantitative reverse transcription polymerase chain reaction
In order to get a more reliable result, we increased the number of samples. The expression levels of individual miRNAs were measured using quantitative reverse transcription polymerase chain reaction (qRT-PCR) using Maxima SYBR Green qPCR Master Mix kit (Thermo Fisher, Pittsburgh, PA, US). After selecting, we got the two up-regulated miRNAs (miR-4787-5P and miR-4306) in accordance with the result of miRNA microarray analysis. The differentially expressed miRNAs between AAD and Control groups were selected for further verification in plasma and aorta samples. The relative expression was calculated using the ΔCt method. Briefly, total RNA was reverse transcribed to cDNA using a cDNA Synthesis Kit (Thermo Fisher). The Bulge-LoopTM miRNA qRT-PCR Primer Set (Shanghai Genepharma Co. Ltd, Shanghai, China) was used to detect and quantify the expression of miR-4787-5P and miR-4306 according to the manufacturer’s instructions; miR-16 was used as the internal control for the plasma samples. The cDNA was amplified in duplicates on the ABI PRISM 7900HT Real-time PCR System with primers (miRNA-4787-5P: F5’-GGCGGGGGTGGCG-3’; R5’-TATGGTTGTTCTCTGCTCTGTCTC-3’; miRNA-4306: F5’-AAAGCGCCGCTGGAGAGA-3’; R5’-TATGGTTGTTCACGACTCCTTCAC-3’; miRNA-16 F5’-AAGCACCTAGCAGCACGTAAATA-3’; R5’-TATGGTTTTGACGACTGTGTGAT-3’) and probes according to the manufacturer’s instructions. The expression levels of target miRNAs in plasma and aorta samples were normalized to the endogenous control. Data were analyzed by the ΔCt method as follows: ΔCt = Ct value(miR-16) - Ct value(miRNA of interest). All data were represented by 2-ΔCt value.
Target gene prediction
Prediction of miRNA target genes was performed by computational algorithms according to the base-pairing rules between miRNA and mRNA target sites, location of binding sequences within the 3’ untranslated region (3’-UTR) of the target, and conservation of target binding sequences within genomes. TargetScan and miRanda were used to predict miR-4787-5p and miR-4306 targets related to proteins detected in this study. The target genes of differentially expressed miRNAs were predicted by online tools including TargetScan v5.1 (http://www.targetscan. org/), Sanger (http://www.sanger.ac.uk/), Pictar (http://pictar.bio.nyu.edu/), and Miranda v5 (http://miRNA.sanger.ac.uk/).
Dual luciferase assay
Human polycystin 1 (PKD1) fragments containing putative binding sites for miR-4787-5p were amplified by PCR from human genomic DNA with the following primers: forward 5’-AACGAGCTCGGCTCCCAGGGTGGAGGAAGGTGA-3’, reverse 5’-CTGCTCGAGGCAGTCAGACAGCTCTTTTATTGA-3’. Mutant PKD1 3’-UTRs were constructed with the ACCCCCG to TGGGGGC mutation and obtained by overlap extension PCR. Fragments (215 bp) were cloned into a pmirGLO reporter vector (Promega, Madison, WI, US) with SacI and XhoI sites, downstream of the luciferase gene, to generate the recombinant vectors pmir-GLO-PKD1-wt and pmir-GLO-PKD1-mut. Human transforming growth factor-β1 (TGF-β1) fragments containing putative binding sites for miR-4306 were amplified by PCR from human genomic DNA with the following primers: forward 5’-ATCGAGCTCACATGATCGTGCGCTCCTGCAAGTG-3’, reverse 5’-ACTCTCGAGGCATCTCAGAGTGTTGCTATGGTGA-3’. Mutant TGF-β1 3’-UTRs were constructed with the TCTCTCC to AGAGAGG mutation and obtained by overlap extension PCR. Fragments (420 bp) were cloned into a pmirGLO reporter vector (Promega) with SacI and XhoI sites, downstream of the luciferase gene, to generate the recombinant vectors pmir-GLO-TGF-β1-wt and pmir-GLO-TGF-β1-mut. Gels were run under the same experimental conditions. For the luciferase reporter assay, 293T cells were transiently co-transfected with miRNAs (miR-4787-5p/miR-4306 mimic or scrambled miR-4787-5p/miR-4306 negative control) and reporter vectors (wild-type or mutant-type reporter vectors) using Lipofectamine 2000. Luciferase activities were measured using a Dual Luciferase Assay Kit (Promega) according to the manufacturer’s instructions at 48 h post-transfection.
Western blotting
In order to further confirm the results of target gene prediction, we chose the PKD1 (PC1) and TGF-β1 for Western blotting verification in aortic sample. Aortic tissue were dissolved in sample buffer at 37°C for 10 minutes and electrophoresed in SDS gels for transfer onto nitrocellulose membranes. Membranes were probed with primary antibodies against PC1 (1:1000, Millipore, Billerica, MA, USA) and TGF-β1 (1:1000; Santa Cruz, CA, USA) in phosphate-buffered saline (PBS). After washing, membranes were incubated with horseradish peroxidase-labeled secondary antibody (Santa Cruz Biotechnology, 1:1000 dilution in PBS). Protein-antibody reactions were detected by chemiluminescence using Kodak X-Omat films. The relative amount of proteins on the blots was determined by densitometric analysis using a HP 3c laser scanner and the program SigmaGel. Protein standards were included in each blot to normalize the densitometric data to a known amount of protein loaded.
Statistical analysis
The Statistical Package for Social Sciences, version 19.0 (SPSS, Chicago, IL, US) was used for the statistical analysis. Continuous variables were expressed as the mean ± standard deviation (SD) and categorical data were expressed as numbers and proportions. The statistically significance of the microarray results were analyzed by fold change and the Student’s t test. The threshold value used to screen differentially expressed miRNAs was a fold change ≥2.0 or ≤0.5 (P < 0.01). The diagnostic value for differentiating between AAD patients and the control was assessed by calculating the area under the receiver-operator characteristic (ROC) curve (AUC). Validation of ROC results was performed by the leave-one-out cross-validation method. In the validation, first, by using the subset of all but one sample, a ROC model was built, and the cut-off value was defined to maximize the sum of sensitivity and specificity. Then, using the cut-off value, the model was used to predict the left-out recorded samples. A P-value of < 0.05 denoted the presence of a statistically significant difference.
Results
Patient characteristics
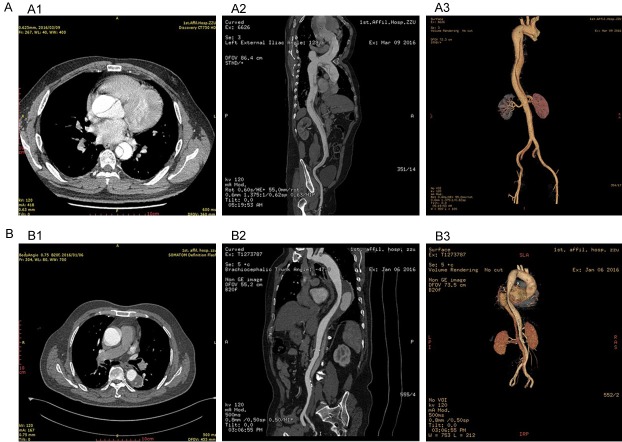
In the present study, 98 patients with AAD (including 47 type A and 51 type B) and 56 controls were enrolled. The clinical characteristics of each group are listed in Table 1. Age and gender were comparable between the two groups. There were no statistically significant differences in clinical characteristics between the AAD and control groups except for D-dimer and hypertension. Most AAD patients (76.3%) had concomitant hypertension and almost half of patients (42.9%) had atherosclerosis. Typical images of AAD patients are shown in Figure 1, which shows the vessel sections involved and type.
Figure 1.
The typical images in AAD. A: Image A1-3 are typical pictures in type A aortic dissection with aortic computed tomography angiography; B: Image B1-3 are typical pictures in type B aortic dissection with aortic computed tomography angiography.
Differentially expressed miRNAs between two groups in miRNA microarray analysis
Four patients with AAD and four normal controls were selected for miRNA microarray analysis. The miRNA expression profiles in the AAD and control groups were first analyzed. After normalization and logarithmic transformation of the miRNA microarray data, a Box Plot graph was constructed to observe the overall characteristics and to evaluate the distribution and dispersion of the data. The expression profiles of 21 miRNAs differentially regulated between the AAD and control groups were used to separate samples into biologically interpretable groups (Figure 2). Among these, nine miRNAs were upregulated by more than two-fold in the AAD group compared with the control group, while 12 miRNAs were downregulated by more than two-fold. Table 2 shows the differentially expressed miRNAs and fold changes in expression in the plasma of AAD patients.
Figure 2.
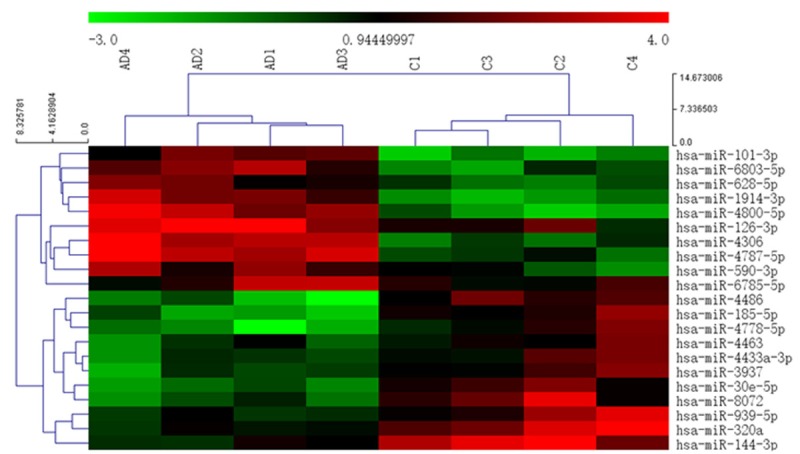
The miRNAs profiles differentiate the AAD group from the control group. Hierarchical clustering of 9 miRNAs whose expression was significantly altered (fold change > 2, P < 0.01, FDR < 0.05) in the AAD and control groups. The color stands for the intensity of the signal.
Table 2.
Differentially expressed miRNAs in AAD patients by plasma microarray analysis
| Up-regulated miRNAs | Fold change | Down-regulated miRNAs | Fold change |
|---|---|---|---|
| hsa-miR-126-3p | 7.421 | hsa-miR-4486 | 0.453 |
| hsa-miR-4787-5p | 6.210 | hsa-miR-4463 | 0.441 |
| hsa-miR-4800-5p | 5.477 | hsa-miR-4778-5p | 0.426 |
| hsa-miR-4306 | 4.251 | hsa-miR-8072 | 0.413 |
| hsa-miR-6803-5p | 4.474 | hsa-miR-30e-5p | 0.403 |
| hsa-miR-628-5p | 3.457 | hsa-miR-6785-5p | 0.395 |
| hsa-miR-590-3p | 3.307 | hsa-miR-4433a-3p | 0.394 |
| hsa-miR-1914-3p | 2.425 | hsa-miR-144-3p | 0.388 |
| hsa-miR-101-3p | 2.378 | hsa-miR-939-5p | 0.386 |
| hsa-miR-320a | 0.236 | ||
| hsa-miR-3937 | 0.231 | ||
| hsa-miR-185-5p | 0.175 |
AAD, acute aortic dissection.
qRT-PCR analysis of differentially expressed miRNAs between the two groups
To further determine the expression levels of selected miRNAs, we respectively examined the levels of miR-4787-5p and miR-4306 in both plasma (AAD=98, controls=56) and aortic samples (AAD=13, controls=5) with qRT-PCR. As shown in Figure 3, circulating miR-4787-5p expression was higher in the AAD group (1.15 ± 0.87) than in the control group (0.26 ± 0.22); and circulating miR-4306 expression was higher in AAD patients (1.27 ± 0.96) than in the control group (0.39 ± 0.24). The differences in miR-4787-5p and miR-4306 expression between the two groups were statistically significant (P < 0.01). In addition, there were higher tissue expression of miR-4787-5p in the AAD group (6.67 ± 1.76) than in the control group (1.85 ± 0.45); and higher tissue expression of miR-4306 in AAD patients (4.90 ± 1.61) than in the control group (1.92 ± 0.62). The differences between the two groups were statistically significant (P < 0.05).
Figure 3.
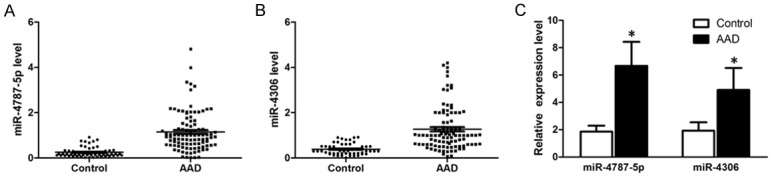
A. Differential expression of the circulating miR-4787-5p between the AAD patients and controls. The plasma levels of miR-4787-5p were significantly increased in AAD patients (n=98) compared to controls (n=56). B. Differential expression of the circulating miR-4306 between the AAD patients and controls. The plasma levels of miR-4306 were significantly increased in AAD patients (n=98) compared to controls (n=56). C. Differential expression of the tissue miR-4787-5p and miR-4306 between the AAD patients and controls. The tissue levels of miR-4787-5p and miR-4306 were significantly increased in AAD patients (n=13) compared to controls (n=5).
Diagnostic performance of miR-4787-5p and miR-4306 in AAD
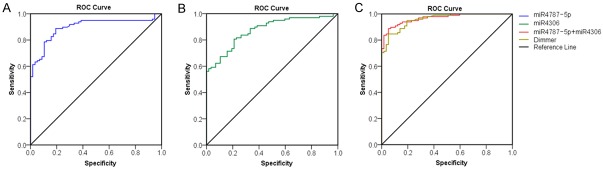
To investigate the possibility that these miRNAs may serve as new and potential biomarkers for AAD, ROC analysis was performed in the two recruited cohorts. The diagnostic performances of miR-4787-5p and miR-4306 for AAD were assessed. As shown in Figure 4A and 4B, the AUC values for miR-4787-5p and miR-4306 were 0.898 (95% confidence interval [CI], 0.847-0.948) and 0.874 (95% CI, 0.820-0.927), respectively. This result demonstrated that miR-4787-5p and miR-4306 had marked sensitivity and specificity for AAD. In Figure 4C, the AUC for the combination value of miR-4787-5p and miR-4306 was 0.961 (95% CI, 0.820-0.927), whereas the AUC measured for D-dimmer was 0.958. This result demonstrated that the combination value of miR-4787-5p and miR-4306 was similar or superior to that of D-dimmer for the early diagnosis of AAD.
Figure 4.
Comparisons of the sensitivity and specificity of the diagnosis by plasma miR-4787-5P and miR-4306 in the AAD patients. ROC curves were constructed to evaluate the diagnostic values of has-miRNA-4787-5P and has-miRNA-4306 for the AAD patients. AUC: area under the ROC curve. A. The ROC Curve of miR-4787-5P; B. The ROC Curve of miR-4306; C. The ROC Curve of the combination value of miR-4787-5p and miR-4306 compared with D-dimmer.
PKD1 is a direct target of miR-4787-5p and TGF-β1 is a direct target of miR-4306
Bioinformatics analysis using the TargetScan and miRanda algorithms predicted that the 3’-UTR of PKD1 contains binding sites for miR-4787-5p (Figure 5A). To determine whether PKD1 is a direct target of miR-4787-5p, recombinant vectors were digested using SacI and XhoI and a dual luciferase reporter system containing either wild-type or mutant 3’-UTR of PKD1 was used for analysis. Co-transfection with miR-4787-5p significantly suppressed the luciferase activity of the reporter containing the wild-type 3’-UTR, whereas it had no effect on the activity of the reporter containing the mutant 3’-UTR (P < 0.01, Figure 5B). This result indicated that PKD1 is a direct target of miR-4787-5p.
Figure 5.
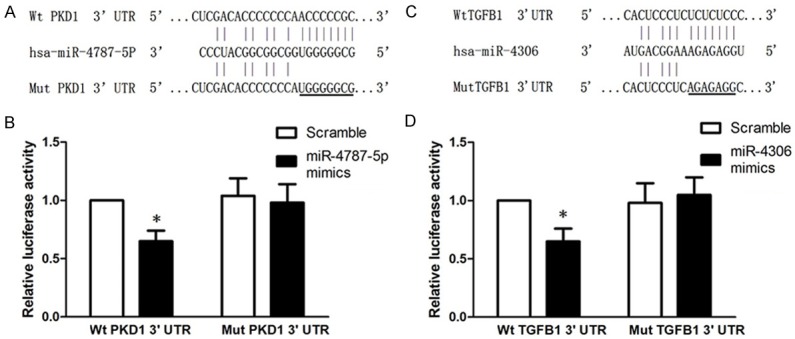
PKD1 and TGF-β1 are respectively direct target of miR-4787-5p and miR-4306. PKD1 was identified as a target of miR-4787-5p in 293T. A. Putative miR-4787-5p binding sequences in the PKD1. B. Dual luciferase reporter co-transfection with miR-4787-5p and wild-type or mutant 3’-UTRs of PKD1. C. Putative miR-4306 binding sequences in the TGF-β1. D. Dual luciferase reporter co-transfection with miR-4306 and wild-type or mutant 3’-UTRs of TGF-β1. The reporter containing the wild-type 3’-UTR significantly suppressed luciferase activity. *P < 0.01, compared with Scramble/Wt PKD1 3’-UTR group.
Similarly, the 3’-UTR of TGF-β1 contains binding sites for miR-4306, as predicted by TargetScan and miRanda (Figure 5C). Co-transfection with miR-4306 significantly suppressed the luciferase activity of the reporter containing the wild-type 3’-UTR but not that of the mutant reporter (P < 0.01, Figure 5D). This indicated that TGF-β1 is a direct target of miR-4306.
Western blotting analysis of predicted target proteins between the two groups
To further confirm the relationship between miRNAs and predicted target proteins, we examined the protein levels of PKD1 and TGF-β1 by western blotting. As shown in Figure 6, protein expression level of PKD1 was lower in the AAD group (1.03 ± 0.23) than in the control group (0.30 ± 0.14); and protein expression level of TGF-β1 was lower in AAD patients (1.09 ± 0.24) than in the control group (0.25 ± 0.12). The differences in PKD1 and TGF-β1 protein expression between the two groups were statistically significant (P < 0.05). The PKD1 and TGF-B1 original western blot image for Figure 6A and 6B could be found in Supplementary Data at the end of this article.
Figure 6.
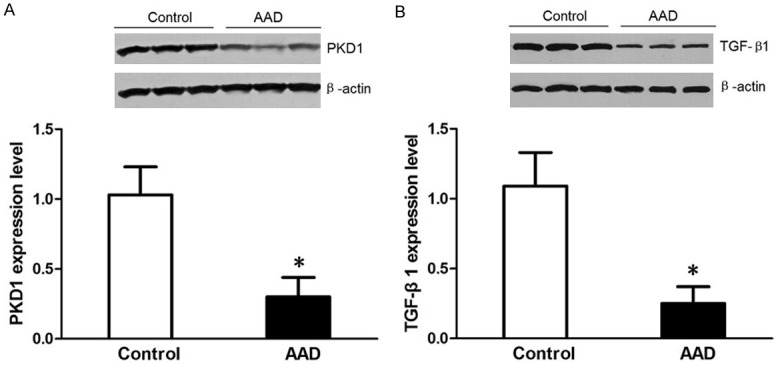
Validation for the expression of PKD1 (A) and TGF-β1 (B) in the aortic tissue by Western blotting. In the AAD group compared with control, PKD1 and TGF-β1 are underexpressed.
Discussion
AAD is a common condition that results from the weakening of the arterial wall and is characterized by the separation of the aortic media caused by the flow of extraluminal blood, which can cause death from aortic rupture. The pathogenesis and gene regulatory networks involved in this disease remain unclear. In the early stages of AAD, pathological changes such as the rupture of the aortic intima result in the flow of circulating blood into the media of the aorta; this is followed by the formation of true and false lumens and the release of gene regulatory products, such as smooth muscle myosin heavy chain [29,30], BB-isozyme of creatine kinase [31], calponin [32], elastin, and D-dimer into the bloodstream. These potential biomarkers have not met the requirements of a ‘gold standard’ because of inadequate sensitivity and specificity. Therefore, it is important to identify new biomarkers with high sensitivity and specificity for the early diagnosis of AAD.
Based on their tissue specificity, rapid release kinetics, and stability in plasma, circulating miRNAs have been proposed as potentially useful and novel biomarkers for detecting various cardiovascular diseases [33], including heart failure [23], myocardial infarction [34], coronary artery disease, hypertension [19], and pulmonary arterial hypertension [35]. Increasing evidence has confirmed that miRNAs are important factors controlling gene expression. Circulating miRNAs are not only novel biomarkers for the diagnosis of cardiovascular diseases, but also play an important role in the pathogenesis, development, and prognosis of diseases.
In recent years, studies evaluated the miRNA expression profile of aortic tissues or plasma in AAD patients. However, only partial AAD types (type A AAD or thoracic aortic dissection) were assessed, which is not representative of the general status of AAD patients. Therefore, in the present study, we evaluated the miRNA expression profile in the plasma of all AAD patients (including type A and type B) and healthy volunteers for the first time to reveal the potential diagnostic value of miRNAs in AAD. Microarray techniques revealed a set of differentially expressed miRNAs, with 9 upregulated and 12 downregulated miRNAs in the plasma of AAD patients compared with that of controls. qRT-PCR of miR-4787-5p and miR-4306 further validated the reliability of the microarray result. In our study, we found that the levels of miR-4787-5p and miR-4306 were significantly higher in patients with AAD than in the control group. The findings of the present study have not been reported in previous studies. Prediction of miRNA targets indicated that the two dysregulated miRNAs mainly target genes associated with cell-cell adhesion, extracellular matrix metabolism, inflammatory factors, ion channels, cytoskeleton organization, cancer, and multiple signaling pathways related to cellular processes [36-38]. These target genes are associated with cellular proliferation, differentiation, and apoptosis, which are involved in the pathogenesis of AAD. The present results indicate that the two miRNAs identified may not only participate in the pathological processes of AAD, but could also be potential biomarkers for AAD.
In the present study, we analyzed the concentration of D-dimer in the AAD and control groups. Although D-dimer is a sensitive marker for patients with AAD and is widely used in clinical practice, the pooled specificity is 0.56 (95% CI: 0.51-0.60) by meta-analysis result from abnormal increasing in other diseases such as pulmonary embolism (PE) [39]. We performed ROC curve analyses to determine the diagnostic value of miR-4787-5p and miR-4306 in comparison with D-dimer. We found that although the diagnostic performance of D-dimer was superior to that of either of the two miRNAs, the combination value of miR-4787-5p and miR-4306 was similar to that of D-dimer for the early diagnosis of AAD. The results of these analyses indicated that circulating miR-4787-5p and miR-4306 are specific and sensitive for AAD and may be promising novel biomarkers for the early diagnosis of AAD.
To further study the role of miRNAs in the pathogenic mechanism of AAD, we performed bioinformatics analysis using Target Scan and miRanda, which predicted that the 3’-UTRs of PKD1 and TGF-β1 contain binding sites for miR-4787-5p and miR-4306. We thus hypothesized that the direct target of miR-4787-5p is the cell-cell adhesion protein PKD1, while the direct target of miR-4306 is TGF-β1. This hypothesis was confirmed using a dual luciferase reporter system. Furthermore, we confirmed the negative regulation relationship by western blotting between PKD1 and miR-4787-5p, TGF-β1 and miR-4306. The PKD1 gene product, PC1, is a very large protein consisting of 4302 amino acids with a 3074 amino acid extracellular amino-terminus, 197 amino acid cytosolic carboxy-terminus and eleven transmembrane domains. PC1 is expressed in a variety of cell types and tissues including renal epithelium, hepatic bile ducts, pancreatic ducts, vascular smooth muscle cells (SMCs), and endothelial cells [40,41]. At the subcellular level, PC1 is localized to the primary cilium on the apical surface of epithelial cells, in addition to other reported locations in the cell such as the lateral membrane at sites of intercellular adhesion and at desmosomes [42]. Mutation of the PKD1 gene is a major cause of autosomal dominant polycystic kidney disease (ADPKD), which is associated with cardiovascular complications such as hypertension and dissecting aortic aneurysms [43]. The previous studies showed that PKD1 is downregulated in the dissected aorta [44,45], leading to dysfunction of cell-cell adhesion, which could be caused by the upregulation of miR-4787-5p. The recent work by Feng et al demonstrated that PKD1 downregulation might induce AAD formation by modulating the phenotype of VSMCs. These study suggest that the pathology of aortic dissection results from a loss of gene function. Therefore, miR-4787-5p may be involved in the pathogenesis of AAD. Similarly, studies showed that the dysregulation or mutation of TGF-β1 and activation of the TGF-β signaling pathway are involved in aortic aneurysms and dissections [46-49]. Therefore, we inferred that miR-4306 may be an important factor in the pathogenesis of aortic aneurysm and aortic dissection. However, further research is necessary to clarify the role of miRNAs in the pathogenesis of AAD.
In summary, our study analyzed differentially expressed miRNAs between patients with AAD and healthy control subjects, and detected two miRNAs that were upregulated in the circulating plasma of AAD patients. Currently, we are verifying the validity and sensitivity of the two miRNAs in a large sample of AAD patients and controls by qRT-PCR. Furthermore, we are also testing the sensitivity and specificity of the two miRNAs in AAD by ROC analysis. We identified the direct targets of miR-4787-5p and miR-4306 by dual luciferase reporter assay and western blotting. These results indicated miR-4787-5p and miR-4306 not only might be the potential novel biomarkers for the early diagnosis of AAD, but also might be involved in the pathogenesis of AAD. The present study had some limitations, including the small sample size and the lack of other disease controls. Therefore, additional studies with larger cohorts of healthy volunteers and other patients are needed to demonstrate the diagnostic value of miR-4787-5p and miR-4306 as biomarkers for AAD.
Acknowledgements
This work was supported by the grants from the Key University Research Program of Henan province (No. 17B320018), the Education Department of Henan province science and technology research projects (14A320026) and the Scientific and Technological Project of Henan Province (No. 201503029).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Sampson UK, Norman PE, Fowkes FG, Aboyans V, Song Y, Harrell FE Jr, Forouzanfar MH, Naghavi M, Denenberg JO, McDermott MM, Criqui MH, Mensah GA, Ezzati M, Murray C. Estimation of global and regional incidence and prevalence of abdominal aortic aneurysms 1990 to 2010. Glob Heart. 2014;9:159–170. doi: 10.1016/j.gheart.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 2.Hiratzka LF, Bakris GL, Beckman JA, Bersin RM, Carr VF, Casey DE Jr, Eagle KA, Hermann LK, Isselbacher EM, Kazerooni EA, Kouchoukos NT, Lytle BW, Milewicz DM, Reich DL, Sen S, Shinn JA, Svensson LG, Williams DM American College of Cardiology Foundation; American Heart Association Task Force on Practice Guidelines; American Association for Thoracic Surgery; American College of Radiology; American Stroke Association; Society of Cardiovascular Anesthesiologists; Society for Cardiovascular Angiography and Interventions; Society of Interventional Radiology; Society of Thoracic Surgeons; Society for Vascular Medicine. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with thoracic aortic disease: executive summary. A report of the American college of cardiology foundation/American heart association task force on practice guidelines, American association for thoracic surgery, American college of radiology, American stroke association, society of cardiovascular anesthesiologists, society for cardiovascular angiography and interventions, society of interventional radiology, society of thoracic surgeons, and society for vascular medicine. Catheter Cardiovasc Interv. 2010;76:E43–86. doi: 10.1002/ccd.22537. [DOI] [PubMed] [Google Scholar]
- 3.Siddiqi HK, Eagle KA. Acute aortic dissection in women: challenges and opportunities. Expert Rev Cardiovasc Ther. 2013;11:1527–1539. doi: 10.1586/14779072.2013.845085. [DOI] [PubMed] [Google Scholar]
- 4.Divchev D, Najjar T, Tillwich F, Rehders T, Palisch H, Nienaber CA. Predicting long-term outcomes of acute aortic dissection: a focus on gender. Expert Rev Cardiovasc Ther. 2015;13:325–331. doi: 10.1586/14779072.2015.1004313. [DOI] [PubMed] [Google Scholar]
- 5.Mehta RH, Manfredini R, Hassan F, Sechtem U, Bossone E, Oh JK, Cooper JV, Smith DE, Portaluppi F, Penn M, Hutchison S, Nienaber CA, Isselbacher EM, Eagle KA International Registry of Acute Aortic Dissection (IRAD) Investigators. Chronobiological patterns of acute aortic dissection. Circulation. 2002;106:1110–1115. doi: 10.1161/01.cir.0000027568.39540.4b. [DOI] [PubMed] [Google Scholar]
- 6.Yiu RS, Cheng SW. Natural history and risk factors for rupture of thoracic aortic arch aneurysms. J Vasc Surg. 2016;63:1189–1194. doi: 10.1016/j.jvs.2015.12.043. [DOI] [PubMed] [Google Scholar]
- 7.Wen D, Du X, Dong JZ, Zhou XL, Ma CS. Value of D-dimer and C reactive protein in predicting inhospital death in acute aortic dissection. Heart. 2013;99:1192–1197. doi: 10.1136/heartjnl-2013-304158. [DOI] [PubMed] [Google Scholar]
- 8.Vianello E, Dozio E, Rigolini R, Marrocco-Trischitta MM, Tacchini L, Trimarchi S, Corsi Romanelli MM. Acute phase of aortic dissection: a pilot study on CD40L, MPO, and MMP-1, -2, 9 and TIMP-1 circulating levels in elderly patients. Immun Ageing. 2016;13:9. doi: 10.1186/s12979-016-0063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor RA, Iyer NS. A decision analysis to determine a testing threshold for computed tomographic angiography and D-dimer in the evaluation of aortic dissection. Am J Emerg Med. 2013;31:1047–1055. doi: 10.1016/j.ajem.2013.03.039. [DOI] [PubMed] [Google Scholar]
- 10.Chitwood DH, Timmermans MC. Target mimics modulate miRNAs. Nat Genet. 2007;39:935–936. doi: 10.1038/ng0807-935. [DOI] [PubMed] [Google Scholar]
- 11.Sen CK, Ghatak S. miRNA control of tissue repair and regeneration. Am J Pathol. 2015;185:2629–2640. doi: 10.1016/j.ajpath.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Choi E, Cha MJ, Hwang KC. Looking into a conceptual framework of ROS-miRNA-atrial fibrillation. Int J Mol Sci. 2014;15:21754–21776. doi: 10.3390/ijms151221754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng L, Chun-guang Q, Bei-fang L, Xue-zhi D, Zi-hao W, Yun-fu L, Yan-ping D, Yang-gui L, Weiguo L, Tian-yong H, Zhen-wen H. Clinical impact of circulating miR-133, miR-1291 and miR-663b in plasma of patients with acute myocardial infarction. Diagn Pathol. 2014;9:89. doi: 10.1186/1746-1596-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, Long G, Zhao C, Li H, Chaugai S, Wang Y, Chen C, Wang DW. Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS One. 2014;9:e105734. doi: 10.1371/journal.pone.0105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 17.Mouillet JF, Chu T, Hubel CA, Nelson DM, Parks WT, Sadovsky Y. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta. 2010;31:781–784. doi: 10.1016/j.placenta.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jickling GC, Sharp FR. Blood biomarkers of ischemic stroke. Neurotherapeutics. 2011;8:349–360. doi: 10.1007/s13311-011-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, Ma X, Lau WB, Rong R, Yu X, Wang B, Li Y, Xiao C, Zhang M, Wang S, Yu L, Chen AF, Yang X, Cai J. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124:175–184. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- 20.Shi W, Du J, Qi Y, Liang G, Wang T, Li S, Xie S, Zeshan B, Xiao Z. Aberrant expression of serum miRNAs in schizophrenia. J Psychiatr Res. 2012;46:198–204. doi: 10.1016/j.jpsychires.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 21.Cui L, Qi Y, Li H, Ge Y, Zhao K, Qi X, Guo X, Shi Z, Zhou M, Zhu B, Guo Y, Li J, Stratton CW, Tang YW, Wang H. Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-footand-mouth disease. PLoS One. 2011;6:e27071. doi: 10.1371/journal.pone.0027071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oerlemans MI, Mosterd A, Dekker MS, de Vrey EA, van Mil A, Pasterkamp G, Doevendans PA, Hoes AW, Sluijter JP. Early assessment of acute coronary syndromes in the emergency department: the potential diagnostic value of circulating microRNAs. EMBO Mol Med. 2012;4:1176–1185. doi: 10.1002/emmm.201201749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, Pinto YM. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 24.D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC, Spazzafumo L, De Simone M, Micheli B, Biglioli P, Achilli F, Martelli F, Maggiolini S, Marenzi G, Pompilio G, Capogrossi MC. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–2773. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, Mayr A, Weger S, Schett G, Shah A, Boulanger CM, Willeit J, Chowienczyk PJ, Kiechl S, Mayr M. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol. 2012;60:290–299. doi: 10.1016/j.jacc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 26.Liao M, Zou S, Weng J, Hou L, Yang L, Zhao Z, Bao J, Jing Z. A microRNA profile comparison between thoracic aortic dissection and normal thoracic aorta indicates the potential role of microRNAs in contributing to thoracic aortic dissection pathogenesis. J Vasc Surg. 2011;53:1341–1349. e3. doi: 10.1016/j.jvs.2010.11.113. [DOI] [PubMed] [Google Scholar]
- 27.Hu ZY, Luo JF, Zhong SL, Xue L, Chen YF, Fan RX. [MicroRNAs expression in normal and dissected aortic tissue] . Zhonghua Xin Xue Guan Bing Za Zhi. 2012;40:406–410. [PubMed] [Google Scholar]
- 28.Wang XJ, Huang B, Yang YM, Zhang L, Su WJ, Tian L, Lu TY, Zhang S, Fan XH, Hui RT. Differential expression of microRNAs in aortic tissue and plasma in patients with acute aortic dissection. J Geriatr Cardiol. 2015;12:655–661. doi: 10.11909/j.issn.1671-5411.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh H, Suzuki T, Hiroi Y, Ohtaki E, Suzuki S, Yazaki Y, Nagai R. Diagnosis of aortic dissection by immunoassay for circulating smooth muscle myosin. Lancet. 1995;345:191–192. doi: 10.1016/s0140-6736(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki T, Katoh H, Watanabe M, Kurabayashi M, Hiramori K, Hori S, Nobuyoshi M, Tanaka H, Kodama K, Sato H, Suzuki S, Tsuchio Y, Yazaki Y, Nagai R. Novel biochemical diagnostic method for aortic dissection. Results of a prospective study using an immunoassay of smooth muscle myosin heavy chain. Circulation. 1996;93:1244–1249. doi: 10.1161/01.cir.93.6.1244. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T, Katoh H, Kurabayashi M, Yazaki Y, Nagai R. Biochemical diagnosis of aortic dissection by raised concentrations of creatine kinase BB-isozyme. Lancet. 1997;350:784–785. doi: 10.1016/S0140-6736(05)62569-X. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki T, Distante A, Zizza A, Trimarchi S, Villani M, Salerno Uriarte JA, de Luca Tupputi Schinosa L, Renzulli A, Sabino F, Nowak R, Birkhahn R, Hollander JE, Counselman F, Bossone E, Eagle K International Registry of Acute Aortic Dissection Substudy on Biomarkers (IRAD-Bio) Investigators. Preliminary experience with the smooth muscle troponin-like protein, calponin, as a novel biomarker for diagnosing acute aortic dissection. Eur Heart J. 2008;29:1439–1445. doi: 10.1093/eurheartj/ehn162. [DOI] [PubMed] [Google Scholar]
- 33.Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet. 2010;3:484–488. doi: 10.1161/CIRCGENETICS.110.958363. [DOI] [PubMed] [Google Scholar]
- 34.Li LM, Cai WB, Ye Q, Liu JM, Li X, Liao XX. Comparison of plasma microRNA-1 and cardiac troponin T in early diagnosis of patients with acute myocardial infarction. World J Emerg Med. 2014;5:182–186. doi: 10.5847/wjem.j.issn.1920-8642.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhodes CJ, Wharton J, Boon RA, Roexe T, Tsang H, Wojciak-Stothard B, Chakrabarti A, Howard LS, Gibbs JS, Lawrie A, Condliffe R, Elliot CA, Kiely DG, Huson L, Ghofrani HA, Tiede H, Schermuly R, Zeiher AM, Dimmeler S, Wilkins MR. Reduced microRNA-150 is associated with poor survival in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2013;187:294–302. doi: 10.1164/rccm.201205-0839OC. [DOI] [PubMed] [Google Scholar]
- 36.Mody HR, Hung SW, AlSaggar M, Griffin J, Govindarajan R. Inhibition of S-adenosylmethionine-dependent methyltransferase attenuates TGFβ1-induced EMT and metastasis in pancreatic cancer: putative roles of miR-663a and miR-4787-5p. Mol Cancer Res. 2016;14:1124–1135. doi: 10.1158/1541-7786.MCR-16-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pu Q, Huang Y, Lu Y, Peng Y, Zhang J, Feng G, Wang C, Liu L, Dai Y. Tissue-specific and plasma microRNA profiles could be promising biomarkers of histological classification and TNM stage in non-small cell lung cancer. Thorac Cancer. 2016;7:348–354. doi: 10.1111/1759-7714.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lirun K, Sewe M, Yong W. A pilot study: the effect of Roux-en-Y gastric bypass on the serum microRNAs of the type 2 diabetes patient. Obes Surg. 2015;25:2386–2392. doi: 10.1007/s11695-015-1711-x. [DOI] [PubMed] [Google Scholar]
- 39.Shimony A, Filion KB, Mottillo S, Dourian T, Eisenberg MJ. Meta-analysis of usefulness of d-dimer to diagnose acute aortic dissection. Am J Cardiol. 2011;107:1227–1234. doi: 10.1016/j.amjcard.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 40.Hassane S, Claij N, Jodar M, Dedman A, Lauritzen I, Duprat F, Koenderman JS, van der Wal A, Breuning MH, de Heer E, Honore E, DeRuiter MC, Peters DJ. Pkd1-inactivation in vascular smooth muscle cells and adaptation to hypertension. Lab Invest. 2011;91:24–32. doi: 10.1038/labinvest.2010.159. [DOI] [PubMed] [Google Scholar]
- 41.Du J, Fu J, Xia XM, Shen B. The functions of TRPP2 in the vascular system. Acta Pharmacol Sin. 2016;37:13–18. doi: 10.1038/aps.2015.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fedeles SV, Gallagher AR, Somlo S. Polycystin-1: a master regulator of intersecting cystic pathways. Trends Mol Med. 2014;20:251–260. doi: 10.1016/j.molmed.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim DY, Park JH. Genetic Mechanisms of ADPKD. Adv Exp Med Biol. 2016;933:13–22. doi: 10.1007/978-981-10-2041-4_2. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Yu C, Chang Q, Luo X, Qiu J, Liu S. Comparison of gene expression profiles in aortic dissection and normal human aortic tissues. Biomed Rep. 2016;5:421–427. doi: 10.3892/br.2016.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao M, Zou S, Weng J, Hou L, Yang L, Zhao Z, Bao J, Jing Z. A microRNA profile comparison between thoracic aortic dissection and normal thoracic aorta indicates the potential role of microRNAs in contributing to thoracic aortic dissection pathogenesis. J Vasc Surg. 2011;53:1341–1349. doi: 10.1016/j.jvs.2010.11.113. [DOI] [PubMed] [Google Scholar]
- 46.Huang CK, Luo J, Lai KP, Wang R, Pang H, Chang E, Yan C, Sparks J, Lee SO, Cho J, Chang C. Androgen receptor promotes abdominal aortic aneurysm development via modulating inflammatory interleukin-1alpha and transforming growth factor-beta1 expression. Hypertension. 2015;66:881–891. doi: 10.1161/HYPERTENSIONAHA.115.05654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertoli-Avella AM, Gillis E, Morisaki H, Verhagen JM, de Graaf BM, van de Beek G, Gallo E, Kruithof BP, Venselaar H, Myers LA, Laga S, Doyle AJ, Oswald G, van Cappellen GW, Yamanaka I, van der Helm RM, Beverloo B, de Klein A, Pardo L, Lammens M, Evers C, Devriendt K, Dumoulein M, Timmermans J, Bruggenwirth HT, Verheijen F, Rodrigus I, Baynam G, Kempers M, Saenen J, Van Craenenbroeck EM, Minatoya K, Matsukawa R, Tsukube T, Kubo N, Hofstra R, Goumans MJ, Bekkers JA, Roos-Hesselink JW, van de Laar IM, Dietz HC, Van Laer L, Morisaki T, Wessels MW, Loeys BL. Mutations in a TGF-beta ligand, TGFB3, cause syndromic aortic aneurysms and dissections. J Am Coll Cardiol. 2015;65:1324–1336. doi: 10.1016/j.jacc.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Wang ZW, Wu HB, Li Z, Li LC. Hactorbeta1 induces matrix metalloproteinase-9 expression in rat vascular smooth muscle cells via ROS-dependent ERK-NF-kappaB pathways. Mol Cell Biochem. 2013;375:11–21. doi: 10.1007/s11010-012-1512-7. [DOI] [PubMed] [Google Scholar]
- 49.Humphrey JD. Possible mechanical roles of glycosaminoglycans in thoracic aortic dissection and associations with dysregulated transforming growth factor-beta. J Vasc Res. 2013;50:1–10. doi: 10.1159/000342436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.