Abstract
Purpose
This study explores polymorphisms in the growth differentiation factor 9 (GDF9) gene (exon 1) with respect to fertility in Egyptian sheep.
Methods
Blood samples were collected, and genomic DNA was extracted from 24 Saidi and 13 Ossimi ewes. A 710 bp portion of the GDF9 gene, was amplified using specific primers, and the sequence was analyzed to clarify the phylogenetic relationship of Egyptian breed sheep. In addition, the PCR-RFLP method using Pst1 or Msp1 restriction enzymes was used to mask polymorphisms of partial exon 1 of GDF9 gene to establish molecular markers for twinning.
Results
The lambing rate percentage and litter size showed significant difference between ewes, which produce single and twin lamb for each breed individually, whereas the coefficient of variation of the Saidi breed is greater than that of the Ossimi breed. The results suggested that the GDF9 gene shared a similarity in sequence compared to six accession numbers of Ovis aries found in GenBank. Molecular phylogenetic analyses were performed based on nucleotide sequences in order to examine the position of the Egyptian breeds among many other sheep breeds. The results indicate that accession number AF078545 of O. aries is closely related with Saidi and Ossimi ewes that produce single or twin lamb using the unweighted pair group method with arithmetic mean (UPGMA) analysis. Results showed that Msp1 enzyme digestion revealed polymorphic restriction pattern consisting of one band with 710 bp for ewes producing single lamb and two bands with 710 and 600 bp for ewes producing twin lamb in Saidi sheep breed.
Conclusion
Sequence analysis and diversity of polymorphisms in the GDF9 gene (exon 1) have a novel base substitution (A–T) for detection of FecG mutations that serve as a molecular marker for twinning.
Keywords: Sheep, Litter size, Fecundity, GDF9 gene, Polymorphism, Restriction enzymes, Phylogenetic tree, PCR-RFLP
Introduction
The main objective of sheep breed in the world is one or more of the following: meat, milk, and wool production, where in Egypt, the sheep meat production is more important than fiber production and the sheep contribute 6% of the total red meet produced [1]. There are three major breeds in Egypt: Rahmani, Ossimi, and Barki. Rahmani is distributed mainly in north of the Nile delta, Ossimi in mid Egypt, and Barki in western Mediterranean coastal region. Minor breeds like Saidi and Sohagi are located in south Egypt [2].
Reproduction is a complex process, and fecundity traits such as ovulation rate and litter size can be genetically regulated by many genes with small effects and sometimes also by single genes with major effects, called fecundity (Fec) genes [3]. Various major genes have been reported to affect prolificacy in sheep, which include three related oocyte-derived components, namely, bone morphogenetic protein receptor type 1B (BMPR1B), known as FecB on chromosome 6 [4]; growth differentiation factor 9 (GDF9), known as FecG on chromosome 5 [5]; and bone morphogenetic protein 15 (BMP15), known as FecX on chromosome X [5, 6].
GDF9 is an oocyte-derived growth factor in the transforming growth factor β (TGF-β) superfamily. It is highly expressed in the oocyte and has a pivotal influence on the surrounding somatic cells, particularly granulosa, cumulus, and theca cells [7]. Paracrine interactions between the developing oocyte and its surrounding follicular cells are essential for the correct progression of both the follicle and the oocyte. GDF9 is essential for the overall process of folliculogenesis, oogenesis, and ovulation and thus plays a major role in female fertility [8].
Eight different point mutations (G1–G8) have been identified in Belclare and Cambridge prolific sheep breeds [5]. There are four mutations in GDF9 gene, identified in different sheep breeds: high fertility (FecGH) in Belclare and Cambridge breed, Thoka (FecGT) in Icelandic breed [9], Embrapa (FecGE) in Santa Ines breed [10], and the fourth not establish edits name and allele in Norwegian white sheep (Finnsheep) breed [11]. The mutation in the GDF9 gene increases the ovulation rate in heterozygous individuals, but in two out of four mutations, follicle development is disrupted in homozygous individuals resulting in infertility [12]. The FecGH mutation causes increased ovulation rate in heterozygous ewes, while homozygous ewes are sterile. But FecGE allele in homozygote state increases ovulation rate and litter size [13].
Sheep are large mammals which have many similarities to humans in terms of physiology. They are easy to handle and suffer from many diseases which affect humans. The use of animal models obtained through genetic manipulation has proven to be very useful for identifying many different diseases [14]. In Egypt, the genetic diversity of sheep in respect to these important economic genes has not been sufficiently studied. But, the sequence analysis and polymorphism of the BMP15 gene (exon 2) have been studied [15]. Results showed that FecX gene was monomorphic and disagreement with litter size; therefore, it is indispensable to survey other genes in order to establish the marker-assisted selection technique. This investigation was carried out to explore the presence of polymorphism in GDF9 gene (exon 1) using DNA sequencing and PCR-restriction fragment length polymorphism (RFLP) methods in these Egyptian sheep breeds that can act as a marker for twinning and is helpful in breeding selection for genetic improvement programs.
Materials and methods
Experimental animals
The Saidi and Ossimi sheep breeds used in this study were selected based on their single/twin production in three repetitive production cycles. The data of 24 Saidi and 13 Ossimi ewe sheep were collected from different farms belonging to the Ministry of Agriculture, Egypt. These data were used to study the reproduction traits, i.e., lambing rate (%) and litter size. Twenty-four Saidi individuals (12 ewes which produce twins and 12 which produce single) and 13 Ossimi individuals (7 ewes which produce twins and 6 which produce single) were used to study the polymorphism.
Blood sampling
Whole blood samples (5 ml) were collected from the jugular vein of each 24 Saidi and 13 Ossimi ewes in vacationer glass tubes containing EDTA (1 mg/ml). Blood samples were transferred to the laboratory in an ice box kept at 4 °C until used. The experimental procedures were performed according to protocols approved by the Biological Studies Animal Care and Use Committee of Egypt. All efforts were made to minimize any discomfort during blood collection.
DNA extraction
Genomic DNA was extracted from 150 μl blood samples using xanthogenate protocol described by Tillett and Neilan [16]. The quantified DNA was stored at −20 °C until further processing.
PCR amplification of GDF9 gene
Specific primers 5′-GAATTGAACCTAGCCCACCCAC-3′ and 5′-AGCCTACATCAACCCATGAGGC-3′ were used to amplify GDF9 gene (exon 1), which corresponded to the GenBank accession number AF078545, according to Hanrahan et al. [5]. The primer was synthesized by Invitrogen, Biotechnology Co. Ltd. (USA). The PCR amplification was performed in 25 μl total volume, each PCR reaction mixture containing 12.5 μl Master Mix (OnePCR™), 1 μl of each primer, 2 μl of genomic DNA (50 ng/μl), and 8.5 μl of sterile deionized water. PCR conditions were as follows: an initial denaturation step at 94 °C for 5 min, 35 cycles of 94 °C for 1 min, 62 °C for 1 min and 72 °C for 2 min, and a final extension step at 72 °C for 10 min using thermal cycler 2720 (Applied Biosystems, USA). PCR products were checked by electrophoresis using 1.8% agarose gel in 1× TAE buffer. The products were then purified using the QIAquick Gel Extraction kit no. 28706 (QIAGEN) following the manufacturer’s instructions and sequenced by automated DNA sequencing reactions, which were performed using a Sequencing Ready Reaction Kit (Life Technologies) in conjunction with ABI-PRISM and ABI-PRISM Big Dye Terminator Cycler.
DNA sequence and phylogenetic analysis
A consensus sequence of GDF9 fragments from both Saidi and Ossimi ewes which produce twins and single was constructed by using the SeqMan™ II 4.05 package for Windows 32. These sequences were subjected to alignment with GDF9 sequences of the GenBank, EMBL, DDBJ, and PDB from breeds of Ovis aries using the BLASTN 2.2.18 and BLASTP 2.2.18 (Basic Local Alignment Search Tool) algorithm at http://www.ncbi.npm. The MEGA version 5.2 program was used to generate a phylogenetic tree using the unweighted pair group method with arithmetic mean (UPGMA) method according to Sneath and Sokal [17]. The evolutionary distances were computed using the maximum composite likelihood method [18].
Polymorphism detection, genotyping, and association analysis
Thirty-seven PCR products of partial GDF9 gene (exon 1) were digested using Pst1 and Msp1 restriction enzymes (Fermentas, Germany, #ER0611 and ER0541, respectively) according to the manufacturer’s instructions. A final reaction volume of 32 μl contained 10 μl PCR product, 18 μl H2O free of nuclease, 2 μl of 10× buffer, and 2 μl (5 units) of each restriction enzyme. The final volume of the mixture was mixed gently and spun down for a few seconds and then incubated for 18 h at 37 °C in water bath and stopped at 65 °C for 10 min. Restriction digestion products were checked by electrophoresis using 3% agarose gel in 1× TAE buffer and staining with ethidium bromide. The 100-bp ladder was used as a molecular size marker. The digestion PCR products were analyzed, and moreover, the allele and genotype frequencies of GDF9 gene were determined.
Statistical analysis
Data were statistically analyzed using the SPSS program, version 16.0 [19]. Means of lambing rate percentage and litter size of each breed were compared for main effects and their interaction by Duncan’s multiple range test [20], when significant F values were obtained (P < 0.05). The coefficient of variance (CV) percentage was calculated for each breed individually according to the formula .
Results
Reproduction traits
Fertility traits have a major impact on efficiency and profitability in lamb meat production. In this respect, the lambing rate and litter size of ewes which produce single and twin lamb from Saidi and Ossimi sheep breeds, respectively, reflected ovulation rate, which is an important economic value. The lambing rate percentage and litter size showed a significant difference between ewes which produce single and twin lamb in each Saidi and Ossimi sheep breed (Table 1), while the CVs of the Saidi and Ossimi breeds for lambing rate percentage are 13.29 and 2.42 and also for litter size are 11.36 and 1.68, respectively (Table 1).
Table 1.
Least squares means and coefficient of variance percentage for lambing rate and litter size of Saidi and Ossimi sheep breeds
| Traits | CV (%) | ||
|---|---|---|---|
| Producing single lamb | Producing twin lamb | ||
| Saidi ewes | |||
| Lambing rate (%) | 79.43 b | 88.33 a | 13.29 |
| Litter size | 1.21 b | 1.42 a | 11.36 |
| Ossimi ewes | |||
| Lambing rate (%) | 81.50 b | 84.00 a | 2.42 |
| Litter size | 1.12 b | 1.15 a | 1.68 |
Values in the same row with different letters differ significantly (P < 0.05). Lambing rate (%) = (Number of ewes lambing / Number of ewes mated) × 100. Litter size = Total number of lambs birth / Number of ewes lambing. Coefficient of variance percentage = (S/X) × 100
Sequencing of GDF9 gene (exon 1) and phylogenetic analysis
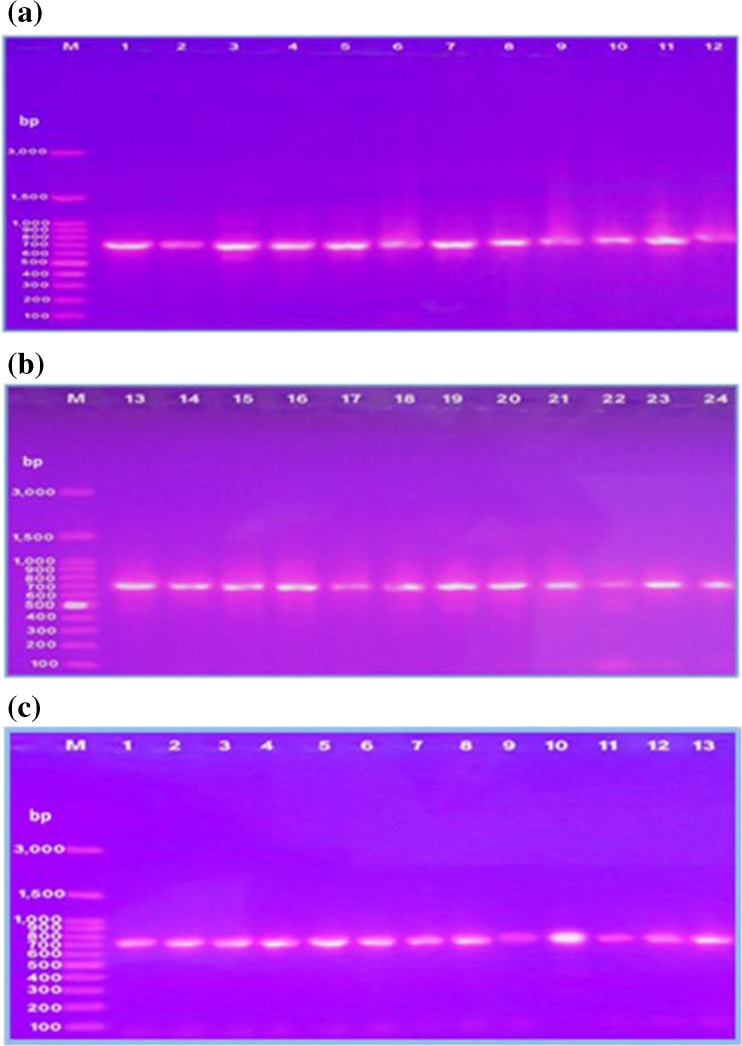
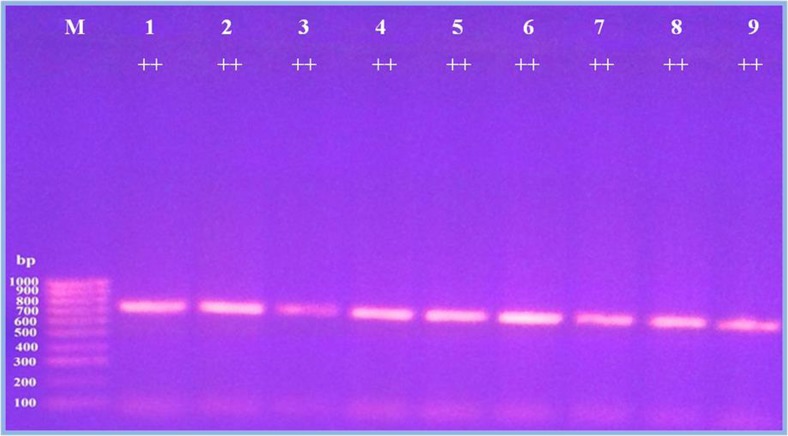
A single fragment of approximately 710-bp nucleotide sequences was amplified from each ewe individual (24 Saidi and 13 Ossimi) sheep breads (Fig. 1). Alignments of two sequences from Saidi ewes that produce single or twin lamb revealed 99.86% similarity between them and base substitution A to T comparing Saidi ewes that produce single lamb with Saidi ewes that produce twin lamb, whereas alignments of two sequences from Ossimi ewes that produce single or twin lamb revealed 100% similarity between them.
Fig. 1.
PCR amplification of partial exon 1 of GDF9 gene from Egyptian sheep breed. a, b From Saidi sheep breed. c From Ossimi sheep breed. M 100-bp DNA ladder
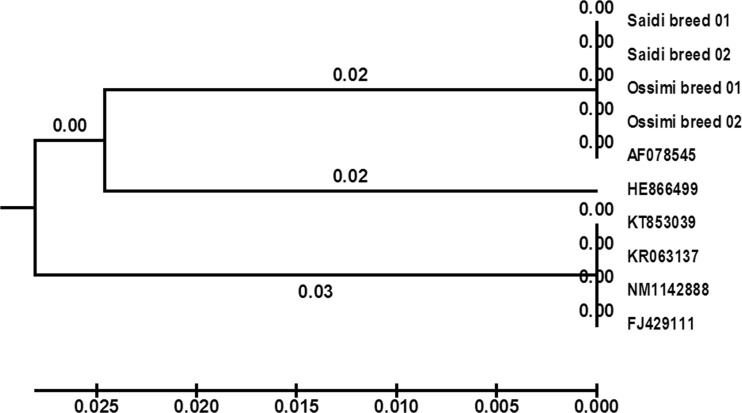
The topology of UPGMA tree of Saidi and Ossimi sheep breads with six accession numbers of O. aries in the GenBank database was represented in a monophyletic group (Fig. 2). The DNA sequences of GDF9 gene successfully grouped Egyptian breeds and O. aries sheep into two main clusters. The first cluster is extremely diverse and consisted of four accession numbers (KT853039, KR063137, NM1142888, and FJ429111). The second cluster had Egyptian sheep breeds and closely related with one accession number (AF078545), whereas accession number (HE866499) was the most distant. Multiple sequence alignment between Egyptian sheep breeds and accession number AF078545 showed that the nucleotide no. 1727 (T) was similar in all Ossimi sheep breeds and Saidi ewes that produce twin lamb and changed to (A) in Saidi ewes that produce single lamb. This base substitution (A–T) maybe in the 5′-regulatory region of sheep GDF9 gene.
Fig. 2.
UPGMA dendrogram of ten Ovis aries sheep generated based on Sneath and Sokal distances. Branch lengths are shown above the branches of clades. Saidi breed 01: ewes which produce twin lamb, Saidi breed 02: ewes which produce single lamb, Ossimi breed 01: ewes which produce twin lamb, and Ossimi breed 02: ewes which produce single lamb
RFLP analysis and genotyping
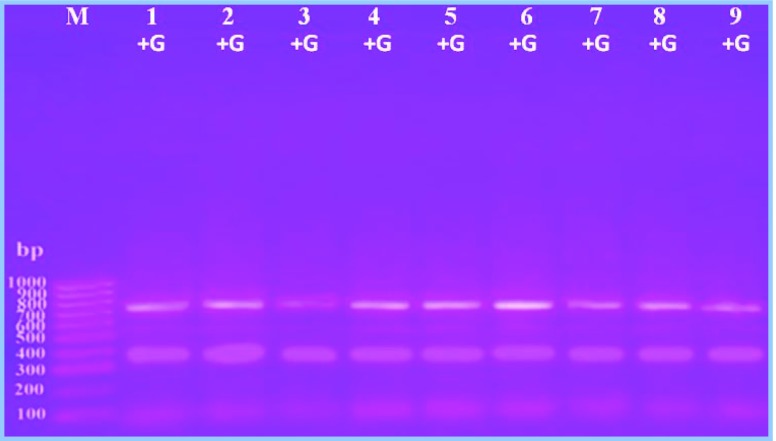
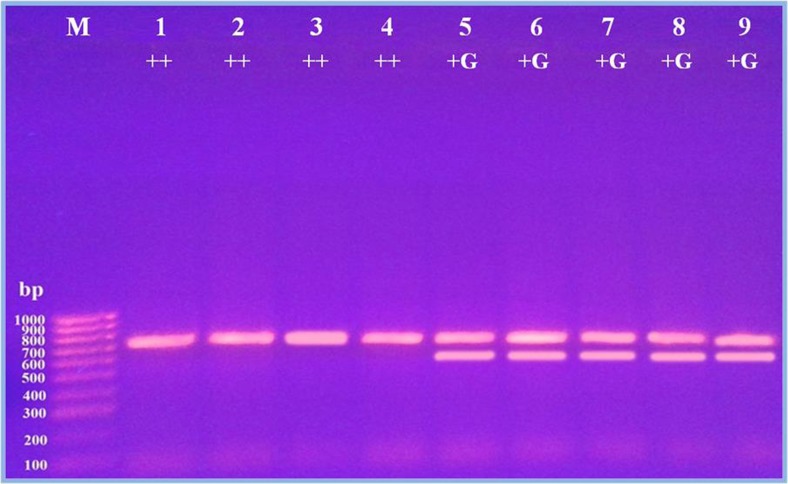
The PCR-RFLP approach has been used previously to genotype prolific sheep [21]. The PCR products of GDF9 gene (exon 1) digested by Pst1 or Msp1 restriction enzymes were used to survey molecular marker for twinning. In the present study, Saidi and Ossimi sheep breeds were screened for Pst1 enzyme digestion (Fig. 3); the results revealed a monomorphic type of restriction pattern consisting of two bands with 710 and 400 bp in Saidi or Ossimi sheep breeds. On the other hand, Msp1 enzyme digestion revealed two types of restriction pattern (Figs. 4 and 5). In Saidi sheep breed, polymorphic restriction pattern consisted of one band with 710 bp for ewes producing single lamb and two bands with 710 and 600 bp for ewes producing twin lamb (Fig. 4). In Ossimi sheep breed, monomorphic restriction pattern consisting of one band with 710 bp was observed in ewe producing single or twin lamb (Fig. 5).
Fig. 3.
Digestion product of partial exon 1 of GDF9 gene from Saidi or Ossimi sheep breeds with Pst1 restriction enzyme. Lanes 1–4: single-producing ewe, lanes 5–9: twin-producing ewe, M: 100-bp DNA ladder
Fig. 4.
Digestion product of partial exon 1 of GDF9 gene from Saidi breed with Msp1 restriction enzyme. Lanes 1–4: single-producing ewe, lanes 5–9: twin-producing ewe, M: 100-bp DNA ladder
Fig. 5.
Digestion product of partial exon 1 of GDF9 gene from Ossimi breed with Msp1 restriction enzyme. Lanes 1–3: single-producing ewe, lanes 4–9: twin-producing ewe, M: 100-bp DNA ladder
The allele and genotype frequencies of GDF9 gene based on Msp1 restriction enzyme digestion are presented in Table 2. The allele frequencies are 0.73 and 0.27 in Saidi sheep breed and 1.0 and 0.0 in Ossimi sheep breed for Fec + and Fec G, respectively. The Saidi and Ossimi sheep genotype frequencies Fec ++, Fec +G, and Fec GG were 0.533, 0.394, and 0.073 and 1.0, 0.0, and 0.0 according to the results of Msp1 enzyme, respectively (Table 2).
Table 2.
Allele and expected genotype frequencies of GDF9 in Saidi and Ossimi sheep breeds
| Restriction enzymes | Sheep breed | No. of ewes | Expected genotype frequencies | Allele frequencies | |||
|---|---|---|---|---|---|---|---|
| ++ | +G | GG | + | G | |||
| Msp1 | Saidi | 24 | 0.533 | 0.394 | 0.073 | 0.73 | 0.27 |
| Ossimi | 13 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | |
Discussion
Many genes and their functions are highly conserved throughout the animal kingdom. It’ is very similar between species, including cell proliferation, metabolism, and growth regulation. This homology across species is a key to considering the possibility of studying diseases and their underlying molecular mechanisms in animal models of genetic disease. Indeed, animal models have been crucial in understanding both genetic diseases [22] and non-genetic diseases [23]. Sheep are relatively outbred and much more human-like than mice, due to their closer genetic and physiological composition [24].
GDF9 is expressed in oocytes and is an attractive candidate gene for primary ovarian insufficiency (POI) because it is, like BMP15, a member of the TGF gene family. Increased frequencies of certain novel variants have been detected in European, Caucasian, and Asian patients [25–27], but not in Japanese and New Zealand populations [28–30].
Ovulation is a complex mechanism that differs among species and depends both on genetic and environmental factors. Mammals can be either mono- or polyovulatory based on how many oocytes mature and are released during ovulation. Ruminants typically release a single oocyte per ovulation compared to pigs and rodents which have high ovulation rates [31]. The ovulation rate even differs between breeds. In sheep, it ranges from one egg per ovulation in Texel and Suffolk to ten eggs per ovulation in the prolific Booroola Merino breed [4, 32]. Litter size differs between and within sheep breeds, and it depends on ovulation rate and is affected by the number of fertilized oocytes. Our results showed that litter size differs between and within sheep breeds and in accordance with those obtained [9]. The genetics of sheep litter size has been investigated by many researchers [5, 33–35]. The GDF9 gene, also called FecG, is located on chromosome 5 and codes for oocyte-derived GDF9 and is essential for normal folliculogenesis. The growth factor is present in oocytes from the primary stage of follicular development until ovulation [5]. Multiple genes were identified having substantial effects on reproduction traits, and some of them are most important being affecting prolificacy in animals. High litter size is an economically important trait that enhances sheep productivity in terms of producing a higher number of lambs, meat, and wool [33].
Bodensteiner et al. [36] reported the nucleotide sequence of the ovine GDF9 gene (GenBank accession number AF078545). Ovine GDF9 spans approximately 2.5 kb and contains two exons and one intron. Exon 1 extends from 1780 to 2178 bp, while exon 2 extends from 3302 to 4266 bp. Multiple sequence alignment between Egyptian sheep breeds and accession number AF078545 showed that the nucleotide No. 1727 (T) was similar in all Ossimi sheep breeds and Saidi ewes that produce twin lamb and changed to (A) in Saidi ewes that produce single lamb. This base substitution (A–T) maybe in the 5′-regulatory region of sheep GDF9 gene. In sheep, polymorphic sequence variations have been identified in GDF9 gene. Nucleotide changes (G-A) in G1 allele [5], (C-T) in FecGH allele [5], (A-C) in FecTT allele [12], (T-G) in FecGE allele [13], (C-T) in FecGV allele [37], and (G-A) [11] were identified. One nucleotide transition mutation (A125G) and one 5-bp deletion/insertion (TTCTT) mutation at the 163–167 locus in 5′-regulatory region of sheep GPR54 gene were identified in Small Tail Han and Corriedale sheep [38]. The transition mutation did not have a significant effect on prolificacy, but deletion mutation (particularly C nucleotide) seemed to have a significant effect on prolificacy.
RFLP is a technique in molecular biology to differentiate minor nucleotide sequence variations in homologous fragments of DNA. The technique relies on the specificity of restriction endonucleases, which are highly sequence-specific and cut the double-stranded DNA only at their recognition sites. The action of restriction enzymes produces variable lengths of homologous DNA molecules containing minor sequence variations or polymorphisms, and the cleaved fragments in the digested DNA can be separated by electrophoretic techniques. RFLP has a good repeatability and stability. The polymorphisms of three candidate genes in five Egyptian and Saudi sheep breeds (Barki, Ossimi, Rahmani, Najdi, and Harri) were detected by PCR-RFLP. The results showed that the polymorphism frequencies of FecB gene with Avall digestion are significantly imbalanced in five breeds [39]. The PCR-RFLP approach has been used previously to genotype prolific sheep [21]. The presence of one copy of GDF9 gene increases the fecundity rate in Saidi sheep and the same result reported by Hanrahan et al. [5], Juengel et al. [40], Liao et al. [41], and Ala Noshahr and Rafat [42]. This indicates that the presence of one copy of mutant GDF9 gene increases fecundity rate in sheep; the present study showed the same result reported by Hanrahan et al. [5], Davis et al. [9], Juengel et al. [40], and Liao et al. [41]. Ewes heterozygous for GDF9 mutations have increased ovulation rates, whereas homozygous ewes are sterile due to a failure of normal ovarian follicular development [5, 6]. Generally, many different loci effect reproduction and ovulation rate between different breeds of sheep, more than genetic background, are under control of age, season, and nutrition. According to these and the high prolificacy in these breeds, it is concluded that high prolificacy may be under control of other factors such as age, season, and nutrition or maybe there is another major gene in Egyptian sheep. Ewes heterozygous for GDF9 have increased lambing rate and litter size, whereas wild-type ewes had reduced ovulation rate. On the other hand, analysis of polymorphism for GDF9 (FecGH) loci in Shal sheep indicates that the genetic factor responsible for twinning or multiple lambing rates is not related to the reported mutated alleles at the GDF9 major gene in this breed [43].
In conclusion, there are fecundity genes with major effect on ovulation rate and litter size in different sheep breeds. The FecG gene was polymorphic and in agreement with the litter size to establish marker-assisted selection method. The knowledge of fecundity genes is important to understand the process of fertility and infertility in mammals and thereby be able to treat genetic disorders associated to reproduction.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study has been carried out in accordance with EU directive 2010/63/EU for animal experiments (http://ec.europa.eu/environment/chemicals//lab_animals/legislation_en.htm ).
References
- 1.Abulyazid I, Abdalla MS, Sharada HM, Hassanin WF. Prolificacy detection in Egyptian sheep using RFLP-specific PCR. Egypt Acad J Biolog Sci. 2011;3:1–4. [Google Scholar]
- 2.ICARDA. ICARDA Annual Report 2006. International Center for Agricultural Research in the Dry Areas. Aleppo, Syria; 2007. p.130.
- 3.Drouilhet L, Lecerf F, Bodin L, Fabre S, Mulsant P. Fine mapping of the FecL locus influencing prolificacy in Lacaune sheep. Anim Genet. 2009;40:804–812. doi: 10.1111/j.1365-2052.2009.01919.x. [DOI] [PubMed] [Google Scholar]
- 4.Souza CJ, Macdougall C, Campbell BK, Mcneilly AS, Baird DT. The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1B (BMPR1B) gene. J Endocrinol. 2001;169:1–6. doi: 10.1677/joe.0.169R001. [DOI] [PubMed] [Google Scholar]
- 5.Hanrahan JP, Gregan SM, Mulsant P, Mullen M, Davis GH, Powell R, Galloway SM. Mutations in the genes for oocyte-derived growth factors GDF-9 and BMP-15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries) Biol Reprod. 2004;70:900–909. doi: 10.1095/biolreprod.103.023093. [DOI] [PubMed] [Google Scholar]
- 6.Galloway SM, McNatty KP, Cambridge LM, Laitinen MP, Juengel JL, Jokiranta TS, et al. Mutations in an oocyte-derived growth factor gene (BMP-15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nature Genet. 2000;3:279–283. doi: 10.1038/77033. [DOI] [PubMed] [Google Scholar]
- 7.Otsuka F, McTavish K, Shimasaki S. Integral role of GDF-9 and BMP-15 in ovarian function. Mol Reprod Dev. 2011;78:9–21. doi: 10.1002/mrd.21265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro F, Cruz M, Leal C. Role of growth differentiation factor 9 and bone morphogenetic protein 15 in ovarian function and their importance in mammalian female fertility. Asian Australas J Anim Sci. 2015;29:1065–1074. doi: 10.5713/ajas.15.0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis GH. Major genes affecting ovulation rate in sheep. Genet Sel Evol. 2005;37:11–23. doi: 10.1186/1297-9686-37-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demars J, Fabre S, Sarry J, Rossetti R, Gilbert H, Persani L, et al. Genome-wide association studies identify two novel BMP-15 mutations responsible for an atypical hyper prolificacy phenotype in sheep. PLoS Genet. 2013;9:e1003482. doi: 10.1371/journal.pgen.1003482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vage DI, Husdal M, Matthew PK, Klemetsdal G, Boman IA. A missense mutation in growth differentiation factor-9 (GDF-9) is strongly associated with litter size in sheep. BMC Genet. 2013;14:1. doi: 10.1186/1471-2156-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicol L, Bishop SC, Pong-Wong R, Bendixen C, Holm EL, Rhind SM, et al. Homozygosity for a single base-pair mutation in the oocytespecificGDF-9 gene results in sterility in Thoka sheep. Reproduction. 2009;138:921–933. doi: 10.1530/REP-09-0193. [DOI] [PubMed] [Google Scholar]
- 13.Silva BD, Castro EA, Souza CJ, Paiva SR, Sartori R, Franco MM, et al. A new polymorphism in the growth and differentiation factor-9 (GDF-9) gene is associated with increased ovulation rate and prolificacy in homozygous sheep. Anim Genet. 2011;42:89–92. doi: 10.1111/j.1365-2052.2010.02078.x. [DOI] [PubMed] [Google Scholar]
- 14.Yamamoto N, Christenson LK, McAllister JM, Strauss JF. Growth differentiation factor-9 inhibits 3'5'-adenosine monophosphate-stimulated steroidogenesis in human granulosa and theca cells. J Clin Endocrinol Metab. 2002;87:2849–2856. doi: 10.1210/jcem.87.6.8551. [DOI] [PubMed] [Google Scholar]
- 15.El Fiky ZA, Hassan GM, Nassar MI. Genetic polymorphism detection in bone morphogenetic protein15 (BMP15) gene related to fecundity in two Egyptian sheep breeds. Biotech Anim Husbandry. 2017;33(1):37–53. doi: 10.2298/BAH1701037E. [DOI] [Google Scholar]
- 16.Tillett D, Neilan BA. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J Phycol. 2000;36:251–258. doi: 10.1046/j.1529-8817.2000.99079.x. [DOI] [Google Scholar]
- 17.Sneath PH, Sokal RR. Numerical Taxonomy: The Principles and Practice of Numerical Classification. 1st ed. San Francisco: Freeman; 1973.
- 18.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Academy Science. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SPSS. Statistical software Package for the Social Sciences, SPSS Inc., version 16, Chicago; 2007.
- 20.Duncan DB. The multiple range and multiple F test. Biometrics. 1955;11:1–42. doi: 10.2307/3001478. [DOI] [Google Scholar]
- 21.Wilson T, Xi-Yang WU, Juengel JL, Ross IK, Lumsden JM, Lord EA, et al. Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol Reprod. 2001;64:1225–1235. doi: 10.1095/biolreprod64.4.1225. [DOI] [PubMed] [Google Scholar]
- 22.Frugier T, Mitchell NL, Tammen I, Houweling PJ, Arthur KG, Kay GW. A new large animal model of CLN5 neuronal ceroid lipofuscinosis in Borderdale sheep is caused by a nucleotide substitution at a consensus splice site (c.571 + 1G > A) leading to excision of exon 3. Neurobiol Dis. 2008;29:306–315. doi: 10.1016/j.nbd.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zarrinkalam MR, Beard H, Schultz CG, Moore RJ. Validation of the sheep as a large animal model for the study of vertebral osteoporosis. Eur Spine J. 2009;18:244–253. doi: 10.1007/s00586-008-0813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer DN, Neverman NJ, Chen JZ, Chang CT, Houweling PJ, Barry LA. Recent studies of ovine neuronal ceroid lipofuscinoses from BARN, the Batten Animal Research Network. Biochim Biophys Acta. 1852;2015:2279–2286. doi: 10.1016/j.bbadis.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Dixit H, Rao LK, Padmalatha V, Kanakavalli M, Deenadayal M, Gupta N, et al. Mutational screening of the coding region of growth differentiation factor 9 gene in Indian women with ovarian failure. Menopause. 2005;6:749–754. doi: 10.1097/01.gme.0000184424.96437.7a. [DOI] [PubMed] [Google Scholar]
- 26.Laissue P, Christin-Maitre S, Touraine P, Kuttenn F, Ritvos O, Aittomaki K, et al. Mutations and sequence variants in GDF9 and BMP15 in patients with premature ovarian failure. Eur J Endocrinol. 2006;5:739–744. doi: 10.1530/eje.1.02135. [DOI] [PubMed] [Google Scholar]
- 27.Kovanci E, Rohozinski J, Simpson JL, Heard MJ, Bishop CE, Carson SA. Growth differentiating factor-9 mutations may be associated with premature ovarian failure. Fertil Steril. 2007;1:143–146. doi: 10.1016/j.fertnstert.2006.05.079. [DOI] [PubMed] [Google Scholar]
- 28.Zhang P, Shi YH, Wang LC, Chen ZJ. Sequence variants in exons of the BMP-15 gene in Chinese patients with premature ovarian failure. Acta Obstet Gynecol Scand. 2007;5:585–589. doi: 10.1080/00016340701269492. [DOI] [PubMed] [Google Scholar]
- 29.Takebayashi K, Takakura K, Wang HQ, Kimura F, Kasahara K, Noda Y. Mutation analysis of the growth differentiation factor-9 and-9B genes in patients with premature ovarian failure and polycystic ovary syndrome. Fertil Steril. 2000;5:976–979. doi: 10.1016/S0015-0282(00)01539-9. [DOI] [PubMed] [Google Scholar]
- 30.Chand AL, Ponnampalam AP, Harris SE, Winship IM. ShellingAN. Mutational analysis of BMP15 and GDF9 as candidate genes for premature ovarian failure. Fertil Steril. 2006;4:1009–1012. doi: 10.1016/j.fertnstert.2006.02.107. [DOI] [PubMed] [Google Scholar]
- 31.Montgomery GW, Galloway SM, Davis GH, McNatty KP. Genes controlling ovulation rate in sheep. Reproduction. 2001;121:843–852. doi: 10.1530/rep.0.1210843. [DOI] [PubMed] [Google Scholar]
- 32.Hanrahan JP. Ovulation rate of Suffolk and Texel ewes. Animal Production Report from Dunsinea, Moorepark and Western Research Centers; 1984.
- 33.Mishra C. Genetic basis of prolificacy in sheep. Int J Livest Res. 2014;4:46–57. doi: 10.5455/ijlr.20131227083421. [DOI] [Google Scholar]
- 34.Wan Somarny WM, Roziatul Erin AR, Suhaimi AH, Nurulhuda MO, MohdHifzan R. A study of major prolificacy genes in Malin and Dorper sheep in Malaysia. J Trop Agric and Fd Sci. 2013;41:265–272. [Google Scholar]
- 35.Zamani P, Nadri S, Saffaripour R, Ahmadi A, Dashti F, Abdoli R. A new mutation in exon 2 of the bone morphogenetic protein 15 gene is associated with increase in prolificacy of Mehraban and Lori sheep. Trop Anim Health Prod. 2015;47:855–860. doi: 10.1007/s11250-015-0799-2. [DOI] [PubMed] [Google Scholar]
- 36.Bodensteiner KJ, Clay CM, Moeller CL, Sawyer HR. Molecular cloning of the ovine growth differentiation factor 9 gene and expression of growth differentiation factor 9 in ovine and bovine ovaries. Biol Reprod. 1999;60:381–386. doi: 10.1095/biolreprod60.2.381. [DOI] [PubMed] [Google Scholar]
- 37.Souza CJ, McNeilly AS, Benavides MV, Melo EO, Moraes JC. Novel GDF9 polymorphism determining higher ovulation rate and litter size in sheep. 2012. https://ainfo.cnptia.embrapa.br/digital/bitstream/item/72796/1/Souza-SRF-2012O005.pdf.
- 38.Tang QQ, Chu MX, Cao GL, Fang L, Di T, Feng T, et al. Association between polymorphism of GPR54 gene and litter size in Small Tail Han sheep. Livest Sci. 2012;143:97–103. doi: 10.1016/j.livsci.2011.09.005. [DOI] [Google Scholar]
- 39.Elkorshy N, Mahrous FK, Salem ML. Genetic polymorphism detection in four genes in Egyptian and Saudi sheep breeds. World Appl Sci J. 2013;27:33–43. [Google Scholar]
- 40.Juengel JL, Hudson NL, Whiting L, McNatty KP. Effects of immunization against bone morphogenetic protein 15 and growth differentiation factor 9 on ovulation rate, fertilization, and pregnancy in ewes. Biol Reprod. 2004;70:557–561. doi: 10.1095/biolreprod.103.023333. [DOI] [PubMed] [Google Scholar]
- 41.Liao WX, Moore RK, Shimasaki S. Functional and molecular characterization of naturally occurring mutations in the oocyte secreted factors bone morphogenetic protein-15 and growth and differentiation factor-9. J Biol Chem. 2004;17:17391–17396. doi: 10.1074/jbc.M401050200. [DOI] [PubMed] [Google Scholar]
- 42.AlaNoshahr F, Rafat SA. Genetic polymorphism of GDF9 gene in Iranian Moghani sheep breed. Iran J Appl Anim Sci. 2014;4:887–890. [Google Scholar]
- 43.Ghaffari M, Nejati-Javaremi A, Rahimi-Mianji G. Lack of polymorphism in the oocyte derived growth factor (GDF9) gene in the Shal breed of sheep. S Afr J of Anim Sci. 2009;39:355–360. [Google Scholar]