Abstract
Behind every successful IVF embryo transfer, there is a great game of chance. Methods seeking to tilt the balance and increase the likelihood of implantation have been proposed and implemented with varying results, including embryo morphology, FISH-PGS, comprehensive chromosomal screening (CCS), morphokinetics, endometrial receptivity testing. It has been suggested that mitochondrial DNA (mtDNA) copy number could serve as a biomarker for embryo viability, but this concept was recently challenged. The world of IVF is left with unanswered questions: Why are there discrepancies in the reports? Should mtDNA copy number be considered to rank embryos for transfer? And in a broader sense, how well must a technique be validated before its implementation in the IVF clinic? Here, we explore these questions attempting to piece together the published data and suggest future directions to help unravel the subject matter.
Keywords: Mitochondria, Mitochondrial DNA, mtDNA, IVF embryo selection
A problem of consensus
The human genome in its entirety is composed of two elements: nuclear DNA, comprising the autosomes and sex chromosomes, and mtDNA, a 16.6 kb plasmid encoding 37 genes necessary for mitochondrial function [1]. Interestingly, mtDNA is multi-copy, and human tissues can display a wide range of mtDNA copy number per cell [2, 3].
A paper published in 2015 by Fragouli et al. and supported Reprogenetics (Livingston, NJ, USA) quantified mtDNA levels in trophectoderm (TE) biopsies of euploid blastocysts selected for transfer and reported that on average, the blastocysts that led to a clinical pregnancy had a lower mtDNA copy number than non-successful blastocysts [4]. Additionally, of the 131 euploid blastocysts analyzed, 29 had mtDNA levels above a specified maximum threshold and all failed to implant.
Shortly thereafter, a manuscript by Diez-Juan et al., supported by Igenomix (Valencia, Spain), echoed the findings of Fragouli et al. [5]. The authors analyzed 65 blastocysts and observed that the higher the mtDNA levels, the less likelyr a blastocyst was to result in clinical pregnancy. Seven samples had severely elevated mtDNA levels, and all seven failed to implant.
Both studies also looked at a series of other parameters, including the predictive power for implantation of mtDNA levels at cleavage stage embryos, with discordant results. Therefore, blastocyst stage quantitation of mtDNA emerged as a promising predictor of implantation potential.
The biological rationale for this phenomenon was postulated to tie in with the “Quiet Embryo Hypothesis,” which states that an embryo that is developing normally adopts a baseline (or quiet) metabolic activity [6]. Conversely, a stressed embryo engages in a compensatory mechanism that increases its metabolic output. By extension, elevated mtDNA numbers could, as the authors of the two original manuscripts suggest, therefore be a biomarker of a stressed embryo that is unlikely to implant. Together, these studies proposed the use of mtDNA copy number in a clinical setting. As a result, the test became commercially available at both Reprogenetics (under the name MitoGrade™) and Igenomix (under the name MitoScore™).
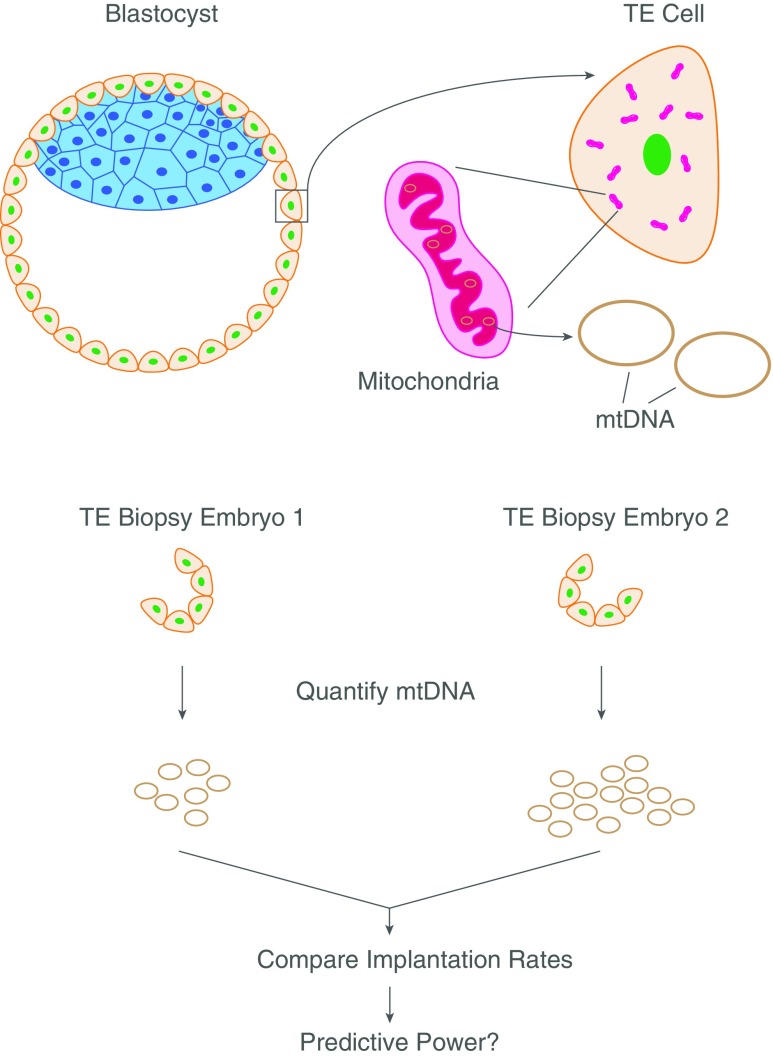
As is to be expected, laboratories across the world have become interested in the subject and are attempting to reproduce the published result in their own hands (Fig. 1). The efforts from our center (ZFC, Foster City, CA, USA) resulted in a publication in early 2017, which, as fate had it, disagreed with the previous findings [7]. In our clinic, the difference in mtDNA copy number between euploid blastocysts that upon transfer did implant and those that failed to do so was statistically insignificant, and mtDNA quantitation’s predictive value in regard to viability was negligible. Building on our initial findings, which were based on NextGen sequencing (NGS) data, we switched analysis platforms to quantitative real-time polymerase chain reaction (qPCR). After performing a cross-platform validation and testing a sample size larger than the previous two published studies (n = 241), we again came to the conclusion that mtDNA quantitation was an ineffective predictor of implantation in our clinic.
Fig. 1.
Overview of the method establishing the predictive power of mtDNA levels in implantation of blastocysts during IVF
A main feature that differentiated our report from the others was that all our data stemmed from a single center. The previous studies originated from reference laboratories that had agglomerated data points from various client IVF clinics and analyzed them as a single unit. It was therefore impossible to deduce whether their findings were valid in each individual clinic or only held true when the data was analyzed in bulk. By basing our study on a single clinic, we were able to correct for numerous inter-facility variables, including culture media used, temperature, biopsy technique, and equipment. In an effort to further mitigate confounding factors in our analysis, we proceeded to investigate 24 patients that had undergone more than one transfer from the same embryo cohort and compared mtDNA levels between the embryos that had implanted and those that did not. Also in this case, mtDNA copy number was not a useful predictor of pregnancy.
Following our paper, a study originating from Reproductive Medicine Associates of New Jersey (RMA NJ, USA) performed an analysis that can be considered the gold standard in embryology for controlling for potential confounding factors: double embryo transfer (DET) [8]. The authors quantified mtDNA in male-female embryo pairs from the same cohort that were simultaneously transferred and led to a singleton pregnancy. By the gender of the born baby, it was deduced which of the embryos had successfully implanted, all the while elegantly controlling for any inter-patient variables. From the analysis of 69 such instances, the authors concluded that mtDNA quantitation did not distinguish between implanted and non-implanted embryos.
Therefore, as studies have evolved to increasing refinement of controls for confounding variables, the validity of mtDNA copy number as a predictor of implantation has come into question.
Making sense of it all
Together, the discordant results from the published studies offered a possible compromise: mtDNA copy number in embryos works as a predictor of implantation in some centers but not in others. Was it possible that the original studies had based their conclusions on a stochastic event occurring in a subset of clinics?
Indeed, follow-up reports from the Reprogenetics group in which data was stratified into individual clinics instead of pooling all numbers together showed exactly that [9, 10]. In roughly half of clinics investigated (17 out of 35), the authors were unable to identify any euploid blastocysts that contained elevated mtDNA levels, regardless of whether they had implanted or not. In the rest of clinics, the percentage of embryos with high mtDNA levels varied widely, ranging from 1 to 27% [9, 10]. From this, it becomes apparent that anything linking mtDNA levels and implantation is not a universal biological phenomenon, but rather a center-specific occurrence.
Three more single-clinic studies with large sample sizes (n > 100) were presented at the conference of Preimplantation Genetic Diagnosis International Society (PGDIS): One stemming from Repromeda (Brno, Czech Republic), one from Bahceci IVF Center (Istanbul, Turkey), and one from IVI Valencia (Valencia, Spain). All three failed to establish any predictive power of implantation by mtDNA copy number [11–13]. Importantly, both latter studies relied on the services of Igenomix to quantify mtDNA in their embryos, thereby controlling for possible technical differences regarding mtDNA quantitation.
It is conceivable that all mentioned independent studies [7, 8, 11–13] randomly happened to be examples of those clinics in which mtDNA levels hold no predictive power at all. If this scenario is actually the case, future autonomous studies conducted blindly by additional clinics should reveal a clearer picture—so far, we are unaware of any clinic that having analyzed significant sample sizes, independently validated blastocyst mtDNA copy number as a biomarker of implantation.
Some guidelines on accurately quantifying mtDNA
In hopes that more independent labs with molecular biology capacity will conduct their in-house analysis of mtDNA levels, we think it appropriate to share some notes on accurate quantitation of mtDNA in human embryos. While reported cases exist of determining mtDNA copy number using fluorescent in situ hybridization (FISH) [14], in the vast majority of instances, researchers have relied on qPCR or NGS, be it in embryology or any other biomedical field.
For NGS, it is essential to use a technology that yields sufficient sequencing depth to make accurate estimates of mtDNA levels. In our hands, we have validated the Veriseq (Illumina) workflow combined with bioinformatics analysis in Geneious R9 (Biomatters), a commercially available software program that facilitates simple determination of read number by genome region [7]. Relative mtDNA copy number is calculated by dividing the number of reads mapping to the mtDNA genome by either the total number of reads or the number of reads mapping to the nuclear genome. Alternatively, one can divide the value for coverage of the mtDNA genome by that of the nuclear genome. Dividing the value obtained for mtDNA by a total or nuclear DNA value is necessary to normalize between samples for differences in cell number of the original biopsy as well as technical batch effects, such as amplification or library preparation.
Further refinement necessary for accurate quantitation is achieved by using a correction factor that normalizes for variations in nuclear DNA compositions between samples. The use of such a correction factor has been used in mtDNA quantitation in cancer [15], and we have adapted it to embryology [7]. For example, non-corrected comparisons of mtDNA levels between male and female embryos are intrinsically skewed, considering that the male genome is shorter than the female’s, thereby artificially tilting the values in the mtDNA/nuclear DNA calculations described above. We have provided a guideline of corrections for NGS analysis of mtDNA copy number that can be routinely implemented [7].
Another consideration is that the alignment algorithm used should be stringent and non-uniquely mapped reads discarded. This is particularly important for mtDNA-mapping reads due to the occurrence of nuclear mitochondrial DNA segments (NUMTs). These are sequences of mtDNA origin that have inserted as pseudogenes into the nuclear genome over evolutionary time and might skew the results if not accounted for during bioinformatic analysis.
Using qPCR for mtDNA quantitation equally relies on finding a ratio of two values (mtDNA and nuclear DNA) in order to normalize for biopsy size and technical workflow variance between samples. Several commercially available assay systems exist designed to quantify mtDNA by qPCR using mtDNA-to-nuclear DNA ratio, such as NovaQUANT™ (EMD Millipore), Human mtDNA Ratio Kit (Takara), or mtDNA Copy Number Assay (Detroit R&D). If designing the assay in-house, selecting appropriate qPCR targets for each component is crucial. Again, due to the existence of NUMTs, the locus chosen as a target for the mtDNA qPCR must be absolutely unique with as low as possible similarity to regions in the nuclear genome. The locus chosen for the nuclear assay should not be in the sex chromosomes, since this would lead to skewed results when comparing male and female embryos [7].
Fragouli et al. have suggested that their use of a nuclear multi-copy locus is superior to a single-copy locus for normalization and might explain how their results differ from ZFC’s, citing potential allele drop-out (ADO) during whole genome amplification (WGA) [4, 9]. They have tested this experimentally comparing the reproducibility of normalizing to a multi-copy sequence (Alu) to a selection of single-copy loci [9], although it is unclear whether the assays used were properly validated, such as by establishing their efficiencies by standard curves and serial dilutions (as was done in [5, 7, 8] and shown in the figures of said manuscripts). We were generally unable to confirm these reported findings, due to the insufficient information provided in describing the Alu qPCR Taqman assay used by the Reprogenetics group, since only the sequence of the probe is given and not that of the primers [4]. Unfortunately, this effectively bars any attempt to reproduce the stated observations.
Conceptually, at the blastocyst stage, the effect of ADO during WGA is equal between the two methods, considering that for TE biopsies, the initial copy number of unique loci is 10–20 (stemming from five to ten cells). This effectively converts a single-copy locus into a multi-copy sequence for the test. A WGA system such as Sureplex (Rubicon) has an estimated 10% allele drop-out rate according to one study [16]. Whether one out of ten copies is lost, or a thousand copies out of a million are lost during WGA, the result will be the same. It is worthwhile noting that many genomic multi-copy sequences (such as Alu) comprise self-transposable elements, translating into variability in number amongst the human population [17–19]. By their very nature, such sequences are therefore not ideal tools for standardization across samples. Regardless of whether a single- or multi-copy test used, any error associated with ADO would spread evenly across embryo groups and would not lead to preferentially skewed numbers in either implanted or non-implanted group of transferred euploid blastocysts. Importantly, the Treff et al. study also relied on a multi-copy gene for normalization, and their findings, show that mtDNA copy number is not predictive of implantation [7, 8].At ZFC, analysis of 60 embryos using a multi-copy sequence as a normalizer in fresh DNA material showed even distribution of mtDNA content in implanted and non-implanted groups (unpublished results).
From a general standpoint, a cross-platform validation is appropriate to substantiate mtDNA quantitation methods. Simply put, an embryo should show similar mtDNA levels regardless of quantitation method used. In ZFC’s case, we performed such a validation between NGS by Veriseq (Illumina) and qPCR with Taqman assays (Applied Biosystems) targeting ND6 and CYTB for mtDNA sequences and RNaseP single-copy nuclear locus normalization [7]. The experiment showed statistically significant linearity, in other words, embryos with mtDNA levels ranging from very low to very high showed near identical results with either technique used.
Perspective and challenges
It is reasonable to demand rigorous evaluation of a test before implementation in a clinical setting—something that is true in IVF as in all fields of biomedicine. This has not been the case for mtDNA quantitation in blastocyst embryos. It has become obvious that the initial conclusions were over-interpretations that must be carefully qualified and re-assessed. We share the view presented by others that a potentially interesting observation has been pushed into the clinic too hastily [20].
We hope that laboratories with means to conduct molecular biology experiments will test mtDNA levels in their embryos. Also, we encourage clinics that rely on reference laboratories for genetic screening that are currently receiving information on mtDNA copy number to conduct their own internal, independent analysis of the predictive power of mtDNA levels. Regardless of outcome, positive or negative, we urge the findings to be shared with the IVF community. We still await a study originating from an independent laboratory not connected to entities with commercial interests in the subject confirming any connection between mtDNA levels and implantation prediction.
If such studies were to appear, and indeed it was confirmed that mtDNA copy number has predictive power of implantation in some clinics but not in others, a whole slew of new questions would emerge. How could this be explained from a biological standpoint? Would we be dealing with an iatrogenic effect? What is different about those clinics with high mtDNA copy number embryos?
From a mechanistic point of view, the link between mtDNA copy number and the Quiet Embryo Hypothesis is, at this stage, speculative at best. It is well possible that a stressed embryo engages in elevating its metabolic output as a compensatory mechanism, but a relationship between perceived cellular stress and increased numbers of DNA molecules in mitochondria remains undocumented. A confounding aspect of the narrative is that a mitochondrial organelle can contain a broad range of mtDNA copies. High mtDNA content in a cell does not equate large quantities of mitochondria. Also, elevated numbers of mitochondria do not necessarily mean high mitochondrial function or metabolic activity. More research is needed to explore and support such a model.
Needless to say, the paradigm to validate a biomarker for implantation is a well-designed clinical trial (CT). Reprogenetics and Igenomix have each started a CT to assess the effectiveness of mtDNA quantitation in embryo selection. The status of both CTs is currently “recruiting” according to www.clinicaltrials.gov, which means results will not be available for some time, but we are hopeful the findings will shed light on the subject matter.
It is important to note that the Reprogenetics’ CT (accessible online under NCT02673125) is specifically testing patients from clinics that show “> 20% MitoGrade elevated embryos.” From their own data, it is apparent that this only applies to two out of 35 clinics that have participated with them [9]. It will be crucial to keep their findings in perspective and avoid conclusions that generalize potentially sporadic observations. Otherwise, the preferential selection of data in this CT would constitute a blatant case of confirmation bias.
If IVF continues to strive for ever higher implantation rates (and most importantly live birth rates), it also needs to navigate through murky waters of evidence-based medicine. Fortunately, science is self-correcting such that future well-conducted studies will reveal the real role of mtDNA quantitation in IVF… if any.
References
- 1.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Seli E. Mitochondrial DNA as a biomarker for in-vitro fertilization outcome. Curr Opin Obstet Gynecol. 2016;28(3):158–163. doi: 10.1097/GCO.0000000000000274. [DOI] [PubMed] [Google Scholar]
- 3.Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod. 2010;83(1):52–62. doi: 10.1095/biolreprod.109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel CE, et al. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet. 2015;11(6):e1005241. doi: 10.1371/journal.pgen.1005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diez-Juan A, Rubio C, Marin C, Martinez S, Al-Asmar N, Riboldi M, et al. Mitochondrial DNA content as a viability score in human euploid embryos: less is better. Fertil Steril. 2015;104(3):534–41.e1. doi: 10.1016/j.fertnstert.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 6.Leese HJ. Quiet please, do not disturb: a hypothesis of embryo metabolism and viability. BioEssays. 2002;24(9):845–849. doi: 10.1002/bies.10137. [DOI] [PubMed] [Google Scholar]
- 7.Victor AR, Brake AJ, Tyndall JC, Griffin DK, Zouves CG, Barnes FL, et al. Accurate quantitation of mitochondrial DNA reveals uniform levels in human blastocysts irrespective of ploidy, age, or implantation potential. Fertil Steril. 2017;107(1):34–42.e3. doi: 10.1016/j.fertnstert.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 8.Treff NR, Zhan Y, Tao X, Olcha M, Han M, Rajchel J, et al. Levels of trophectoderm mitochondrial DNA do not predict the reproductive potential of sibling embryos. Hum Reprod. 2017;32(4):954–962. doi: 10.1093/humrep/dex034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravichandran K, McCaffrey C, Grifo J, Morales A, Perloe M, Munne S et al. Mitochondrial DNA quantification as a tool for embryo viability assessment: retrospective analysis of data from single euploid blastocyst transfers. Hum Reprod. 2017:1–11. doi:10.1093/humrep/dex070. [DOI] [PubMed]
- 10.Fragouli E, McCaffrey C, Ravichandran K, Spath K, Grifo JA, Munné S, Wells D. Clinical implications of mitochondrial DNA quantification on pregnancy outcomes: a blinded prospective non-selection study. Hum Reprod. 2017;1-8. 10.1093/humrep/dex292. [DOI] [PubMed]
- 11.Hornak M, Horak J, Kubicek D, Travnik P, Vesely J, Vesela K. Mitochondria quantification in human IVF embryos using next-generation sequencing-based protocol. Bologna: Preimplantation Genetic Diagnosis International Society (PGDIS); 2016. [Google Scholar]
- 12.Ogur C, Gultomruk M, Caferler J, Capar B, Findikli N, Bahceci M. Maternal age has no influence on mitochondrial DNA (mtDNA) content in chromosomally normal embryos. Valencia: Preimplantation Genetic Diagnosis International Society (PGDIS); 2017. [Google Scholar]
- 13.De Los Santos MJ, Mercader A, Delgado A, Escrich L, Buendia P, Rubio C, et al. Mitochondrial DNA copy number measured by mitoscore is associated to trophectoderm quality. Valencia: Preimplantation Genetic Diagnosis International Society (PGDIS); 2017. [Google Scholar]
- 14.Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, et al. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci. 2001;21(9):3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reznik E, Miller ML, Senbabaoglu Y, Riaz N, Sarungbam J, Tickoo SK et al. Mitochondrial DNA copy number variation across human cancers. Elife. 2016;5. doi:10.7554/eLife.10769. [DOI] [PMC free article] [PubMed]
- 16.Deleye L, De Coninck D, Christodoulou C, Sante T, Dheedene A, Heindryckx B, et al. Whole genome amplification with SurePlex results in better copy number alteration detection using sequencing data compared to the MALBAC method. Sci Rep. 2015;5:11711. doi: 10.1038/srep11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hormozdiari F, Alkan C, Ventura M, Hajirasouliha I, Malig M, Hach F, et al. Alu repeat discovery and characterization within human genomes. Genome Res. 2011;21(6):840–849. doi: 10.1101/gr.115956.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23(4):183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Wildschutte JH, Baron A, Diroff NM, Kidd JM. Discovery and characterization of Alu repeat sequences via precise local read assembly. Nucleic Acids Res. 2015;43(21):10292–10307. doi: 10.1093/nar/gkv1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott RT., Jr Enhanced techniques to “power” embryonic mitochondria research. Fertil Steril. 2017;107(1):59–60. doi: 10.1016/j.fertnstert.2016.11.024. [DOI] [PubMed] [Google Scholar]