Abstract
The 1976 outbreak of Legionnaires' disease led to the discovery of the intracellular bacterial pathogen Legionella pneumophila. Given their impact on human health, Legionella species and the mechanisms responsible for their replication within host cells are often studied in alveolar macrophages, the primary human cell type associated with disease. Despite the potential severity of individual cases of disease, Legionella are not spread from person-to-person. Thus, from the pathogen's perspective, interactions with human cells are accidents of time and space—evolutionary dead ends with no impact on Legionella's long-term survival or pathogenic trajectory. To understand Legionella as a pathogen is to understand its interaction with its natural hosts: the polyphyletic protozoa, a group of unicellular eukaryotes with a staggering amount of evolutionary diversity. While much remains to be understood about these enigmatic hosts, we summarize the current state of knowledge concerning Legionella's natural host range, the diversity of Legionella-protozoa interactions, the factors influencing these interactions, the importance of avoiding the generalization of protozoan-bacterial interactions based on a limited number of model hosts and the central role of protozoa to the biology, evolution, and persistence of Legionella in the environment.
Keywords: Legionella, amoebae, protozoa, host range, environment, Acanthamoebae, Hartmannella, Naegleria
Predator vs. prey: legionella and its natural protozoan hosts
In the environment, bacteria are targets of predation by grazing protozoa (Hahn and Höfle, 2001; Molmeret et al., 2005). In response to predation, many bacteria have developed strategies to either avoid predation or survive, and in some cases, replicate within protozoa. As bacteria are destined to encounter a large number of protozoa species in nature, their fitness will be determined by the breadth and diversity of protozoa within which they are able to grow. Though many types of bacteria are able to replicate within protozoa (Greub and Raoult, 2004), this behavior is best characterized in the bacterial pathogen Legionella, in particular Legionella pneumophila, which will be the major focus of this review.
L. pneumophila in the environment
L. pneumophila is ubiquitous in nature (Fliermans, 1996; van Heijnsbergen et al., 2015). While various species of Legionella have been isolated from soil and marine environments, freshwater systems serve as the major reservoirs of L. pneumophila (Fliermans, 1996; van Heijnsbergen et al., 2015). L. pneumophila can exist in a planktonic form however, it is more often found within mixed community biofilms (Mampel et al., 2006). L. pneumophila intercalates into existing biofilms (Lau and Ashbolt, 2009; Stewart et al., 2012) where it acquires nutrients by forming synergistic relationships with other members of the biofilm (Tison et al., 1980; Pope et al., 1982; Bohach and Snyder, 1983; Wadowsky and Yee, 1983; Stout et al., 1986; Stewart et al., 2012; Koide et al., 2014). L. pneumophila is also capable of surviving in nutrient-poor conditions by necrotrophic growth on dead cell masses (Temmerman et al., 2006). Although, its interactions with other bacteria promote L. pneumophila survival in oligotrophic environments, intracellular growth within protozoa is likely the predominant mechanism of L. pneumophila proliferation in its natural habitat (Rowbotham, 1980).
The impact of natural hosts on legionella persistence in the environment and pathogenesis
Protozoa function as natural reservoirs of L. pneumophila and promote disease in humans. The intracellular environment of the host cell protects L. pneumophila from harsh environmental conditions while providing a nutrient rich replicative niche (Greub and Raoult, 2004; Abdel-Nour et al., 2013). The ability of L. pneumophila to survive within amoebae also protects the bacteria from killing by water disinfection procedures (Plouffe et al., 1983; King et al., 1988; Kilvington and Price, 1990; Biurrun et al., 1999; Storey et al., 2004; Bouyer et al., 2007; García et al., 2008; Cervero-Aragó et al., 2014, 2015), a reciprocal relationship that also enhances survival of the host (García et al., 2007). As a consequence, L. pneumophila are commonly found in man-made potable water supply and distribution systems (Ikedo and Yabuuchi, 1986; Breiman et al., 1990; Yamamoto et al., 1992; Fields et al., 2002; Lasheras et al., 2006; Brousseau et al., 2013; Thomas et al., 2014). Although, there is one reported case of probable human-to-human transmission of Legionella (Correia et al., 2016), the vast majority of evidence suggests a non-communicable disease. Instead, human exposure predominantly occurs through the inhalation of contaminated water aerosols (Fields, 1996), which can lead to pneumonic respiratory disease. L. pneumophila passaged through amoebae are more virulent in animal models of infection compared to bacteria grown in broth culture (Cirillo et al., 1994, 1999; Barker et al., 1995; Brieland et al., 1996; Garduño et al., 2002). The earliest description of L. pneumophila's interaction with amoebae even proposed that an important route of human infection may be the inhalation of the pathogen in an amoebal-encapsulated state (Rowbotham, 1980). Thus, the interaction of L. pneumophila with protozoa is a critical determinant in both the persistence of Legionella in environmental and man-made reservoirs, and the incidence and severity of disease.
The broad host range of L. pneumophila
Many bacterial pathogens become highly specialized for growth in one or a small subset of hosts but few are able to grow in multiple hosts. Host jumping has been observed for some pathogens but often comes at a price, the inability to grow in the previous host (Ma et al., 2006). In contrast, L. pneumophila exhibits an extensive host range replicating within a diverse array of protozoan hosts that span multiple phyla, from Amoebozoa (amoebae) to Percolozoa (excavates) to Ciliophora (ciliated protozoa) (Rowbotham, 1980; Fields, 1996). The ability to maintain such a broad host range is due to the assembly of a large cohort of genes that allow L. pneumophila to adapt to variations between hosts (O'Connor et al., 2011). Moreover, the ability to continually evolve and alter the composition of its virulence gene repertoire allows L. pneumophila to adapt to shifts in protozoan populations in their natural habitats (O'Connor et al., 2011). Since the discovery that L. pneumophila can survive and replicate within free-living amoeba (Rowbotham, 1980), the relationship between L. pneumophila and its protozoa hosts has garnered significant attention, largely due to the important role of protozoa in the epidemiology of this pathogen. In this review, we expand on the early works of Rowbotham and Fields (Rowbotham, 1980, 1986; Fields, 1996) to summarize the current knowledge of the host range of L. pneumophila in environmental reservoirs and the factors that impact the outcome of Legionella-protozoa interactions.
The different fates of L. pneumophila within protozoan hosts
While L. pneumophila has an extensive host range, the fate of the bacterium once it enters the host cell can vary greatly. Several protozoa are able to efficiently deliver L. pneumophila to the lysosome for degradation, resulting in the death of the bacterium (Amaro et al., 2015). L. pneumophila predation by protozoa does not seem to be restricted to one particular group. While members of the Cercozoa phylum seem to be especially adept at digesting L. pneumophila (Amaro et al., 2015), distantly related members of the Amoebozoa phylum (Cashia limacoides, Vannella platypodia, and Vexillifera bacillipedes) are also efficient at killing L. pneumophila (Rowbotham, 1986). In contrast, many protozoa serve as hosts for L. pneumophila replication. In these cases, the Legionella-protozoa interaction is detrimental to the host: the bacteria multiply to high numbers and then kill the host as they exit the cell (Rowbotham, 1983). Alternatively, L. pneumophila can be toxic to the host in the absence of replication, a protist version of food-poisoning (Amaro et al., 2015). L. pneumophila within amoebae has been shown to inhibit both amoebae proliferation (Mengue et al., 2016) and chemotactic motility (Simon et al., 2014). The fates of the two organisms are not solely defined by this “it's you or me” relationship, as a number of intermediate outcomes have been observed. In response to extreme stress, amoebae undergo encystation, transforming into a dormant, highly resistant cyst form. While encystation restricts bacterial replication (Rowbotham, 1986; Ohno et al., 2008), L. pneumophila is able to survive the encystation process until more favorable conditions arise (Kilvington and Price, 1990; Greub and Raoult, 2003). Similarly, for some Legionella-protozoa pairs, L. pneumophila is resistant to grazing by the protozoan and thus survives within the host cell but fails to replicate (Smith-Somerville et al., 1991). Alternatively, L. pneumophila can be packaged into multi-membrane vesicles that are distinct from the replication vacuole and expelled into the extracellular environment (Rowbotham, 1983; Berk et al., 1998; Hojo et al., 2012; Amaro et al., 2015). The release of Legionella-containing pellets has been observed in both the ciliated protozoa Tetrahymena spp. (Faulkner et al., 2008; Hojo et al., 2012) and the amoebal hosts Acanthamoeba castellanii and Acanthamoeba astronyxis (Bouyer et al., 2007; Amaro et al., 2015), and does not appear to coincide with bacterial replication. Whether this process is driven by the bacterium or the host is still unclear. The pellet compartment can protect L. pneumophila from environmental stress (Bouyer et al., 2007; Koubar et al., 2011) which would be beneficial during its transition between host cells and thus a potential mechanism to ensure its survival. Consistent with this idea, a functional Type IVb secretion system, a major L. pneumophila virulence factor required for lysosome avoidance and intracellular replication, appears to be important for the release of L. pneumophila in pellets (Berk et al., 2008). Alternatively, the inability to digest the bacteria may simply trigger a host response that involves bacterial expulsion, as a similar phenomenon is observed with non-pathogenic Escherichia coli, Bacillus subtilis, and Mycobacterium luteus (Hojo et al., 2012; Denoncourt et al., 2014). Whether L. pneumophila resists predation or is expelled in pellets, the host is considered to be only partially restrictive due to the survival of L. pneumophila and its potential to transition to other host cells. Indeed, one might speculate that such intermediate host-bacterial interactions (resistance to protozoan predation in the absence of replication) might resemble the first evolutionary step toward becoming an intracellular pathogen.
Methods for defining protozoan hosts of legionella
Protozoan hosts of Legionella are defined by two main techniques: co-culture and co-isolation. When combined with microscopy, co-culture techniques allow for the direct visualization of Legionella within host cells, and by analyzing infected cells over time, bacterial replication within a particular host provides direct experimental evidence of Legionella survival and replication. When combined with plating assays to monitor bacterial numbers, co-culture methods allow bacterial growth rates, maximum growth and the impact of bacterial dose and various external conditions on the interaction to be analyzed. However, while Legionella may be able to replicate in a given host under specific laboratory conditions, the experimental system may not reflect conditions encountered in the environment and thus, biologically relevant interactions that commonly occur in nature. Co-isolation studies attempt to address this issue by examining the co-existence of protozoa and Legionella in environmental samples. In rare cases, protozoa harboring Legionella have been isolated from environmental samples providing direct evidence of their interaction in the environment (Thomas et al., 2006; Hsu et al., 2011; Kao et al., 2013). More commonly, Legionella are identified by 16S sequencing of DNA extracts from bacteria isolated by Legionella-selective culture methods on bacteriological medium (Salloum et al., 2002; Sheehan et al., 2005) or enrichment through co-culture of environmental samples with amoebae (Pagnier et al., 2008). Protozoa may be identified microscopically by fluorescence in situ hybridization (FISH) or the morphological appearance of trophozoites (Jacquier et al., 2013; Muchesa et al., 2014), or by 18S sequencing of DNA extracts following an amoebal enrichment step in which individual isolates are cultured on lawns of bacteria permissive to amoebal grazing (Greub and Raoult, 2004; Delafont et al., 2013; Muchesa et al., 2014). Thus, while most co-isolation studies do not provide direct evidence of Legionella growth within the protozoa identified, they can be used to predict environmentally relevant interactions, to substantiate experimental findings from co-culture techniques and are likely to implicate new protozoan species as potential hosts of Legionella.
Experimentally defined protozoan hosts of L. pneumophila
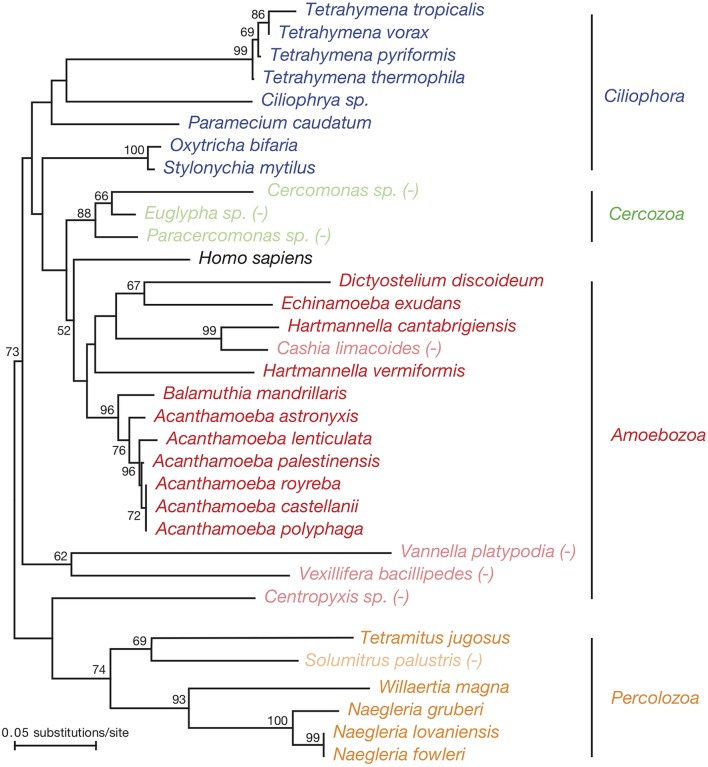
The initial discovery that L. pneumophila is capable of surviving and replicating in protozoa fostered a number of independent investigations to examine the host range of this bacterium (Table 1). Co-culture methods in combination with various microscopy techniques demonstrated growth of L. pneumophila in diverse protozoan hosts encompassing several species of Acanthamoeba (A. castellanii, Acanthamoeba polyphaga, and Acanthamoeba palestinensis), Hartmannella (Vermamoeba vermiformis, formerly Hartmannella vermiformis and Hartmannella cantabridiensis) and Naegleria (Naegleria gruberi, Naegleria lovaniensis, and Naegleria jadini) as well as Tetrahymena pyrofomis, Echinamoeba exudans, and Tetramitus jugosus (formerly Vahlkampfia jugosus) (Rowbotham, 1980, 1986; Tyndall and Domingue, 1982; Anand et al., 1983; Barbaree et al., 1986). While the list of hosts was dominated by three particular genera (Acanthamoeba, Hartmannella, and Naegleria), collectively it represented three different phyla Amoebozoa, Ciliophora, and Percolozoa and amongst them, four distantly related classes of protozoa, Discosea (Acanthamoebae), Tubulinea (Echinamoeba and Hartmannella), Heterolobosea (Naegleria and Tetramitus), and Oligohymenophorea (Tetrahymena) (Figure 1).
Table 1.
Experimentally defined protozoan hosts of L. pneumophila.
| Protozoan species | Protozoan strain | L. pneumophila serogroup (Sg): strain | Fate of L. pneumophila | Experimental evidence | References |
|---|---|---|---|---|---|
| Acanthamoeba spp. | AMI137, AMI116, AMI073, AMI191, Humidifier strain | Sg1: Lens | Intracellular multiplication | CFU counting, Phase-contrast microscopy | Rowbotham, 1980; Dupuy et al., 2016 |
| Sg2: Togus-1 | |||||
| Sg3: Bloomington-2 | |||||
| Sg5: Cambridge-2 | |||||
| Acanthamoeba sp. 155 | Sg1 | Intracellular multiplication | CFU counting, Epifluorescence microscopy | Cervero-Aragó et al., 2014, 2015 | |
| Acanthamoeba astronyxis | Isolate C37C6 | Sg1: Philadelphia-1 | Live cells are packaged in expelled pellets | Electron microscopy | Marciano-Cabral and Cabral, 2003; Amaro et al., 2015 |
| Acanthamoeba castellanii | ATCC® 30234™, CCAP 1534/2, L1501/2A, L501/2A, Neff | Sg1: JR32, Lens, Paris, Philadelphia-1, Philadelphia-2, Pontiac-1 | Intracellular multiplication | CFU counting, Electron microscopy | Rowbotham, 1980; Holden et al., 1984; Moffat and Tompkins, 1992; Hilbi et al., 2001; Bouyer et al., 2007; Tyson et al., 2013; Mengue et al., 2016 |
| Sg2: Togus-1 | |||||
| Sg3: Bloomington-2 | |||||
| Sg4: Los Angeles | |||||
| Sg6: Oxford-1 | |||||
| Neff | Sg5: Dallas 1E | Live cells are packaged in expelled pellets | Electron microscopy | Berk et al., 1998 | |
| Acanthamoeba lenticulata | PD2 | Sg1: AX71, Philadelphia-1, SC94, SC97 | Intracellular multiplication | CFU counting | Molmeret et al., 2001 |
| Sg2: AX2 | |||||
| Sg3: AX52, AX54, AX82 | |||||
| Acanthamoeba palestinensis | Sg1 | Intracellular multiplication | CFU counting, Electron microscopy, Epifluorescence microscopy, Phase contrast microscopy | Anand et al., 1983; Harf et al., 1997 | |
| Acanthamoeba polyphaga | Ap-1, L1501/3A, Puschkarew | Sg1: AA100, Corby, Nottingham-8, Leeds 1A SAP, Leeds-4, Lp02, Philadelphia-2, Pontiac-1 | Intracellular multiplication | CFU counting, Electron microscopy, Phase-contrast microscopy | Rowbotham, 1980, 1986; Kilvington and Price, 1990; Gao et al., 1997; Buse and Ashbolt, 2011 |
| Sg2: Oxford-2, Togus-1 | |||||
| Sg3: Bloomington-2 | |||||
| Sg4: Los Angeles-1 | |||||
| Sg5: Cambridge-2 | |||||
| Sg6 | |||||
| Sg7: Dallas-5, Chicago-8 | |||||
| Sg8: York-1, Concord-3 | |||||
| Puschkarew | Sg5: Dallas 1E | Intracellular Survival, Live cells are packaged in expelled pellets | CFU counting, Electron microscopy | Berk et al., 1998; Buse and Ashbolt, 2011 | |
| Acanthamoeba royreba | Sg4: Los Angeles | Intracellular multiplication | Bacteria cell count, Epifluorescence microscopy | Tyndall and Domingue, 1982 | |
| Balamuthia mandrillaris | CDC-V039 | Sg1: JR32, 130b | Intracellular multiplication | CFU counting, Phase-contrast microscopy | Shadrach et al., 2005 |
| Ciliophrya sp. | Sg1: Corby | Intracellular survival | Epifluorescence microscopy | Rasch et al., 2016 | |
| Dictyostelium discoideum | AX2, AX2-214, AX3 | Sg1: Benidorm 030E, Corby, Philadelphia-1 | Intracellular multiplication | CFU counting, Electron microscopy | Hägele et al., 2000; Solomon et al., 2000 |
| Echinamoeba exudans | SH274 | Sg1: RI-243 | Intracellular multiplication | Electron microscopy | Fields et al., 1989 |
| Hartmannella cantabrigiensis | Sg2: PR-1 | Intracellular multiplication | Electron microscopy | Rowbotham, 1986 | |
| Sg5: Leeds-10 | |||||
| Sg7: Chicago-8, Dallas-5 | |||||
| Sg8: York-1 | |||||
| Naegleria spp. | AMI242, AMI117, AMI135, AMI161 | Sg1: Lens | Intracellular multiplication | CFU counting | Dupuy et al., 2016 |
| Naegleria fowleri | Lee | Sg1: Lp02 | Intracellular multiplication | CFU counting, Electron microscopy | Newsome et al., 1985; Buse and Ashbolt, 2011 |
| Sg3: Bloomington-2 | |||||
| Sg6: Chicago-2 | |||||
| Sg5: Dallas 1E | Intracellular survival | CFU counting | Buse and Ashbolt, 2011 | ||
| Naegleria gruberi | 1518/1E | Sg2: Togus-1 | Intracellular multiplication | Phase-contrast microscopy | Rowbotham, 1980 |
| Sg3: Bloomington-2 | |||||
| Sg5: Cambridge-2 | |||||
| Naegleria jadini | B1518/2 | Sg2: Togus-1 | Intracellular multiplication | Phase-contrast microscopy | Rowbotham, 1980 |
| Sg3: Bloomington-2 | |||||
| Sg5: Cambridge-2 | |||||
| Naegleria lovaniensis | TS | Sg1: Philadelphia-1, 130b | Intracellular multiplication | Confocal microscopy, CFU counting, Bacteria cell count, Epifluorescence microscopy | Tyndall and Domingue, 1982; Declerck et al., 2005; Tyson et al., 2013, 2014 |
| Sg4: Los Angeles | |||||
| Oxytricha bifaria | Sg1: Corby | Intracellular survival | Epifluorescence microscopy | Rasch et al., 2016 | |
| Paramecium caudatum | RB-1 | Sg1: Philadelphia-1 | Intracellular multiplication | Fluorescence microscopy | Watanabe et al., 2016 |
| Stylonychia mytilus | Sg1: Corby | Intracellular survival | Epifluorescence microscopy | Rasch et al., 2016 | |
| Tetrahymena sp. | Sg1 | Intracellular multiplication | CFU counting, Epifluorescence microscopy | Barbaree et al., 1986; Berk et al., 2008 | |
| Sg1: Lp02 | Live cells are packaged in expelled pellets | Electron microscopy, Fluorescence microscopy | Berk et al., 2008 | ||
| Tetrahymena pyriformis | No. 500 | Sg1: Philadelphia-1, 130b | Intracellular multiplication | CFU counting, Electron microscopy | Fields et al., 1984, 1986; Cianciotto and Fields, 1992 |
| Sg3: SC-6-C3 | |||||
| Tetrahymena thermophila | Mating type IV | Sg1: Philadelphia-1 | Intracellular multiplication | CFU counting, Light microscopy Electron microscopy | Kikuhara et al., 1994 |
| Sg1: Philadelphia-2 | Intracellular survival | CFU counting, Light microscopy Electron microscopy | Kikuhara et al., 1994 | ||
| Inbred strain B, SB021 | Sg1: JR32 | Intracellular multiplication | Electron microscopy; Live cells are packaged in expelled pellets | Hojo et al., 2012 | |
| Tetrahymena tropicalis | Sg1: Lens, Philadelphia-1 | Live cells are packaged in expelled pellets | Electron microscopy | Faulkner et al., 2008; Koubar et al., 2011 | |
| Tetrahymena vorax | V2S | Sg1: Philadelphia-1 | Intracellular survival | Electron microscopy, Fluorescence microscopy | Smith-Somerville et al., 1991 |
| Tetramitus jugosusb (Vahlkampfia jugosa) | Sg1: Leeds 4 | Intracellular multiplication | Electron microscopy | Rowbotham, 1986 | |
| Vermamoeba vermiformisa (Hartmannella vermiformis) | ATCC® 50256™, CDC-19 | Sg1: AA100, Lens, 130b Philadelphia-1, RI-243 | Intracellular multiplication | CFU counting, Electron microscopy | Rowbotham, 1986; King et al., 1991; Wadowsky et al., 1995; Abu Kwaik, 1996; Buse and Ashbolt, 2011; Tyson et al., 2013; Dupuy et al., 2016 |
| Sg5: E-52, E-62 | |||||
| Sg6: E-66, E-67 | |||||
| Sg1: Lp02 | Intracellular survival | CFU counting | Buse and Ashbolt, 2011 | ||
| Sg3: Bloomington-2 | |||||
| Sg5: Dallas 1E | |||||
| Sg6: Chicago-2, | |||||
| Sg7: Dallas-5, PR-3 | |||||
| Willaertia magna | c2c Maky, T5[S]44, Z503 | Sg1: Lens, Paris, Philadelphia-1, 130b | Intracellular multiplication | CFU counting, Electron microscopy | Dey et al., 2009; Tyson et al., 2014 |
Figure 1.
An 18S phylogenetic tree of the experimentally defined hosts of L. pneumophila. Evolutionary history was inferred using the Neighbor-Joining method based on an alignment of 18S rRNA sequences. Evolutionary analyses were performed using MEGA7 (Kumar et al., 2016). Restrictive host species that do not support L. pneumophila replication or survival are indicated by lighter shading and the annotation “(−)”. Taxonomic designations are based on the classification system outlined in Ruggiero et al. (2015).
Subsequent studies to investigate L. pneumophila pathogenesis have progressively expanded the list of protozoan hosts of this bacterium (Table 1 and Figure 1), including additional species of Acanthamoeba (Acanthamoeba lenticulata and Acanthamoeba royreba) and Naegleria (Naegleria fowleri) as well as more distantly related genera from their respective phyla such as Dictyostelium discoideum (Hägele et al., 2000; Solomon et al., 2000) and Balamuthia mandrillaris (Amoebozoa) (Shadrach et al., 2005) and Willertia magna (Percolozoa) (Dey et al., 2009; Tyson et al., 2014). Similarly, a number of additional ciliated protozoa were identified that were permissive for L. pneumophila survival, including Tetrahymena spp. (Tetrahymena tropicalis and Tetrahymena vorax), Oxytricha bifaria, Stylonychia mytilus, Paramecium caudatum and a member of the Ciliophrya genus, and in one case L. pneumophila replication (Tetrahymena thermophila), greatly expanding representation from this group (Kikuhara et al., 1994; Rasch et al., 2016; Watanabe et al., 2016). The beneficial interaction of L. pneumophila with these organisms appears to be specific as members from each of the representative phyla were also identified that were highly restrictive to L. pneumophila survival (Figure 1): T. vorax (Ciliophora), A. astronyxis, and Cashia limocoides (Amoebozoa) and Solumitrus palustris (Percolozoa) (Rowbotham, 1986; Smith-Somerville et al., 1991; Amaro et al., 2015). In addition, L. pneumophila was unable to grow in V. platypodia and V. bacillipedes (Rowbotham, 1986), which form a distantly related clade of the Amoebozoa phyla (Figure 1). Similarly, of the members of the Cercozoa phylum examined so far, Cercomonas sp., Euglypha sp., and Paracercomonas sp., all three are restrictive for L. pneumophila growth (Amaro et al., 2015; Rasch et al., 2016), suggesting that distinct orders and families within this class may be more restrictive than others. Thus, while the host range of L. pneumophila is vast, it does appear to have its limitations.
Suggested environmental hosts of L. pneumophila
Protozoa in both natural and man-made environments can alter the composition of microbial communities by eliminating bacteria through predation or augmenting populations of bacteria that are capable of replicating within these organisms (Yamamoto et al., 1992). Co-isolation techniques have been used to describe the composition of these communities within natural fresh water systems such as hot springs, thermal spas, lakes, ponds, streams, and anthropogenic reservoirs, such as cooling towers, industrial and private water networks and compost facilities. L. pneumophila is capable of surviving an array of physical conditions including temperatures ranging from 6 to 63°C (Fliermans et al., 1981). Thermal springs have been of particular interest as they boast characteristically high water temperatures, providing optimal conditions for L. pneumophila growth (Hsu et al., 2011; Ji et al., 2014; Rasch et al., 2016). Artificial aquatic reservoirs are of considerable epidemiological significance and typically support higher numbers of bacteria compared to natural water systems (Yamamoto et al., 1992), likely due to higher average water temperatures (Ikedo and Yabuuchi, 1986; Fields et al., 2002; Lasheras et al., 2006). The results of these population level analyses have validated many of the co-culture defined hosts of L. pneumophila while identifying several additional potential hosts (Table 2).
Table 2.
Suggested protozoan hosts of L. pneumophila.
| Protozoa | Environment source | Identification method used | References |
|---|---|---|---|
| Acanthamoebidae | Cooling towers | Identified morphologically via microscopy | Yamamoto et al., 1992 |
| Acanthamoeba spp. | Compost facilities | Sequence analysis | Conza et al., 2013, 2014 |
| Cooling towers | Identified morphologically via microscopy | Kurtz et al., 1982 | |
| Sequence analysis | Declerck et al., 2007 | ||
| Drinking water systems | Sequence analysis | Marciano-Cabral et al., 2010; Valster et al., 2011; Ji et al., 2014 | |
| Hospital water networks | Identified morphologically via microscopy | Rohr et al., 1998; Steinert et al., 1998 | |
| Industrial water networks | Identified morphologically via microscopy; Sequence analysis | Scheikl et al., 2014 | |
| Natural water systems | Sequence analysis | Declerck et al., 2007; Hsu et al., 2011; Ji et al., 2014 | |
| Acanthamoeba castellanii | Compost facilities | Sequence analysis | Conza et al., 2013 |
| Acanthamoeba hatchetti | Compost facilities | Sequence analysis | Conza et al., 2013, 2014 |
| Hospital water network | Identified morphologically via microscopy | Breiman et al., 1990 | |
| Natural water systems | Sequence analysis | Hsu et al., 2015 | |
| Acanthamoeba jacobsi | Natural water systems | Sequence analysis | Hsu et al., 2011 |
| Acanthamoeba lenticulata | Compost facilities | Sequence analysis | Conza et al., 2013 |
| Acanthamoeba palestinensis | Natural water systems | Sequence analysis | Kao et al., 2013 |
| Acanthamoeba polyphaga | Compost facilities | Sequence analysis | Conza et al., 2013, 2014 |
| Cooling towers | Not specified | Rowbotham, 1986 | |
| Natural water systems | Sequence analysis | Hsu et al., 2009 | |
| Amoebidae | Cooling towers | Identified morphologically via microscopy | Yamamoto et al., 1992 |
| Aspidiscidae | Cooling towers | Identified morphologically via microscopy | Yamamoto et al., 1992 |
| Bodonidae | Cooling towers | Identified morphologically via microscopy | Yamamoto et al., 1992 |
| Cashia limacoides | Cooling towers | Not specified | Rowbotham, 1986 |
| Centropyxis sp. | Natural water systems | Identified morphologically via microscopy | Rasch et al., 2016 |
| Ciliophrya sp. | Natural water systems | Identified morphologically via microscopy | Rasch et al., 2016 |
| Colpodidae | Cooling towers | Identified morphologically via microscopy | Yamamoto et al., 1992 |
| Comandonia operculata | Hospital water network | Identified morphologically via microscopy | Breiman et al., 1990 |
| Cyclidium spp. | Cooling towers | Identified morphologically via microscopy | Barbaree et al., 1986 |
| Diphylleia rotans | Sewage treatment systems | Sequence analysis | Valster et al., 2010 |
| Echinamoeba spp. | Hospital water networks | Identified morphologically via microscopy | Rohr et al., 1998 |
| Echinamoeba exudans | Drinking water systems | Sequence analysis | Valster et al., 2011 |
| Hospital water networks | Identified morphologically via microscopy | Fields et al., 1989 | |
| Echinamoeba thermarum | Drinking water systems | Sequence analysis | Valster et al., 2011 |
| Cooling towers | Sequence analysis | Valster et al., 2010 | |
| Euglypha sp. | Natural water systems | Identified morphologically via microscopy | Rasch et al., 2016 |
| Filamoeba nolandi | Hospital water networks | Identified morphologically via microscopy | Breiman et al., 1990 |
| Flamella balnearia | Compost facilities | Sequence analysis | Conza et al., 2013 |
| Hartmannellidae | Cooling towers | Identified morphologically via microscopy | Yamamoto et al., 1992 |
| Hartmannella spp. | Cooling towers | Sequence analysis | Declerck et al., 2007 |
| Identified morphologically via microscopy | Kurtz et al., 1982 | ||
| Hospital water networks | Identified morphologically via microscopy | Fields et al., 1989; Breiman et al., 1990; Nahapetian et al., 1991 | |
| Natural water systems | FISH; Identified morphologically via microscopy | Zbikowska et al., 2014 | |
| Sequence analysis | Declerck et al., 2007 | ||
| Hartmannella cantabrigiensis | Hospital water networks | Identified morphologically via microscopy | Rowbotham, 1986; Fields et al., 1989 |
| Learamoeba waccamawenis | Compost facilities | Sequence analysis | Conza et al., 2013, 2014 |
| Mayorella spp. | Hospital water networks | Identified morphologically via microscopy | Steinert et al., 1998 |
| Naegleria spp. | Cooling towers | Identified morphologically via microscopy | Barbaree et al., 1986 |
| Sequence analysis | Declerck et al., 2007 | ||
| Compost facilities | Sequence analysis | Conza et al., 2013, 2014 | |
| Drinking water systems | Sequence analysis | Marciano-Cabral et al., 2010; Ji et al., 2014 | |
| Hospital water networks | Identified morphologically via microscopy | Nahapetian et al., 1991; Rohr et al., 1998 | |
| Industrial water networks | Identified morphologically via microscopy | Scheikl et al., 2014 | |
| Natural water systems | Sequence analysis | Declerck et al., 2007; Hsu et al., 2011; Ji et al., 2014 | |
| FISH; Identified morphologically via microscopy | Zbikowska et al., 2014 | ||
| Naegleria australiensis | Compost facilities | Sequence analysis | Conza et al., 2013 |
| Natural water systems | Sequence analysis | Huang and Hsu, 2010 | |
| Naegleria fowleri | Thermal saline bath | FISH; Identified morphologically via microscopy | Zbikowska et al., 2013 |
| Natural water systems | FISH; Identified morphologically via microscopy | Zbikowska et al., 2014 | |
| Naegleria gruberi | Compost facilities | Sequence analysis | Conza et al., 2013 |
| Natural water systems | Sequence analysis | Hsu et al., 2015 | |
| Naegleria lovaniensis | Natural water systems | Sequence analysis | Huang and Hsu, 2010; Kao et al., 2013 |
| Naegleria pagei | Natural water systems | Sequence analysis | Huang and Hsu, 2010 |
| Neoparamoeba spp. | Drinking water systems | Sequence analysis | Valster et al., 2011 |
| Natural water systems | Sequence analysis | Valster et al., 2010 | |
| Oxytricha bifaria | Natural water systems | Identified morphologically via microscopy | Rasch et al., 2016 |
| Paravahlkampfia ustianaa (Vahlkampfia ustiana) | Hospital water networks | Identified morphologically via microscopy | Breiman et al., 1990 |
| Pleuronematidae | Cooling towers | Identified morphologically via microscopy | Yamamoto et al., 1992 |
| Rhinosporidium sp. | Tap water system | Sequence analysis | Valster et al., 2010 |
| Saccamoeba spp. | Hospital water networks | Identified morphologically via microscopy | Rohr et al., 1998 |
| Singhamoeba horticola | Compost facilities | Sequence analysis | Conza et al., 2013, 2014 |
| Stenamoeba spp. | Compost facilities | Sequence analysis | Conza et al., 2013, 2014 |
| Stenamoeba limacina | Compost facilities | Sequence analysis | Conza et al., 2014 |
| Stylonychia mytilus | Natural water systems | Identified morphologically via microscopy | Rasch et al., 2016 |
| Tetrahymenidae | Cooling towers | Identified morphologically via microscopy | Yamamoto et al., 1992 |
| Tetrahymena spp. | Cooling towers | Identified morphologically via microscopy | Barbaree et al., 1986 |
| Tetramitus spp. | Compost facilities | Sequence analysis | Conza et al., 2013 |
| Tetramitus entericab (Vahlkampfia enterica) | Compost facilities | Sequence analysis | Conza et al., 2013 |
| Vahlkampfia spp. | Compost facilities | Sequence analysis | Conza et al., 2014 |
| Cooling towers | Sequence analysis | Declerck et al., 2007 | |
| Drinking water systems | Sequence analysis | Marciano-Cabral et al., 2010 | |
| Hospital water networks | Identified morphologically via microscopy | Breiman et al., 1990; Rohr et al., 1998; Steinert et al., 1998 | |
| Natural water systems | Sequence analysis | Declerck et al., 2007; Hsu et al., 2011 | |
| Vahlkampfia avara | Compost facilities | Sequence analysis | Conza et al., 2013, 2014 |
| Vannella spp. | Hospital water networks | Identified morphologically via microscopy | Rohr et al., 1998 |
| Vannella platypodia | Cooling towers | Not specified | Rowbotham, 1986 |
| Vermamoeba vermiformisc (Hartmannella vermiformis) | Compost facilities | Sequence analysis | Conza et al., 2013, 2014 |
| Drinking water systems | Sequence analysis | Valster et al., 2011; Ji et al., 2014 | |
| Hospital water networks | Identified morphologically via microscopy | Rowbotham, 1986; Fields et al., 1989; Breiman et al., 1990; Rohr et al., 1998 | |
| Sequence analysis | Thomas et al., 2006 | ||
| Industrial water networks | Identified morphologically via microscopy | Scheikl et al., 2014 | |
| Natural water systems | Sequence analysis | Hsu et al., 2011, 2015; Ji et al., 2014 | |
| Sequence analysis | Kao et al., 2013 | ||
| Sequence analysis | Valster et al., 2010 | ||
| Tap water systems | Sequence analysis | Valster et al., 2010 | |
| Vexillifera bacillipedes | Cooling towers | Not specified | Rowbotham, 1986 |
| Vorticellidae | Cooling towers | Identified morphologically via microscopy | Yamamoto et al., 1992 |
| Willaertia spp. | Cooling towers | Sequence analysis | Declerck et al., 2007 |
| Natural water systems | Sequence analysis | Declerck et al., 2007 | |
| Willaertia magna | Compost facilities | Sequence analysis | Conza et al., 2013 |
Vahlkampfia ustiana has been renamed Paravahlkampfia ustiana.
Vahlkampfia enterica has been renamed Tetramitus enterica.
Hartmannella vermiformis has been renamed Vermamoeba vermiformis (Smirnov et al., 2011).
There is tremendous concordance between co-culture-confirmed Legionella-protozoa interactions and the results of co-isolation studies (Tables 1, 2). With the exception of Balamuthia and Dictyostelium, all protozoan genera shown to support intracellular growth in laboratory co-culture studies reside with L. pneumophila in the environment (Table 2). While this is not surprising for Acanthamoeba, Hartmannella, and Naegleria, as these are some of the most abundant protozoa in nature, in many cases co-isolation studies identified the same species of these genera. In particular, three of the protozoa identified, A. palestinensis, N. lovaniensis, and V. vermiformis that had been shown to support L. pneumophila replication in co-culture experiments (Anand et al., 1983; Rowbotham, 1986; Declerck et al., 2005; Thomas et al., 2006) were isolated from water samples harboring L. pneumophila (Kao et al., 2013). Similarly, amoebal enrichment assays resulted in the isolation of Acanthamoeba jacobsi harboring L. pneumophila directly from a thermal spring water sample (Hsu et al., 2011). These results identify A. jacobsi as a new host of L. pneumophila and provide direct evidence of an interaction between L. pneumophila and these four protozoan hosts in the environment. The lack of co-isolation of L. pneumophila with either Balamuthia or Dictyostelium species is likely because these protozoa are typically found in soil and the majority of samples analyzed were isolated from aquatic environments (Dunnebacke et al., 2004; Vadell and Cavender, 2007). The high degree of correlation between the co-culture and co-isolation studies supports the role of these organisms as natural hosts of L. pneumophila in environmental reservoirs.
Co-isolation studies predict a number of additional phyla and classes of protozoa may support L. pneumophila survival or growth (Table 2). In addition to the Amoebozoa, Ciliophora, and Percolozoa phyla, protozoa from Apusozoa (Diphylleia rotans), Cercozoa (Euglypha sp.), Euglenozoa (Bodonidae sp.), and Opsithokonta (Rhinosporidium sp.) were identified. Two additional classes of protozoa from previously identified phyla are also represented, Variosea (Flamella balnearia) and Oligohymenophorea with representatives encompassing four different families spanning three orders within this group. For those classes of protozoa already identified as hosts by co-culture experiments, three additional orders, Thecamoebida (Stenamoeba limacina), Arcellinida (Centropyxis sp.), and Sporadotricina (Aspidiscidae family) and five genera (Comandonia operculata, C. limacoides, Paravahlkampfia ustiana, Learamoeba waccamawenis, and Singhamoeba horticola) were identified. Finally, of the known hosts of L. pneumophila from co-culture experiments, additional species of Acanthamoeba (A. jacobsi), Naegleria (Naegleria pagei and Naegleria australiensis), Tetramitus (Tetramius enterica), and Vahlkampfia (Valkampfia avara) were also isolated. Combined, co-isolation and co-culture experiments represent 7 of the 8 phyla of the protozoa kingdom, 12 of the 41 classes within these phyla and 21 of the 82 defined orders, demonstrating the tremendous diversity amongst L. pneumophila hosts.
Protozoa more commonly found associated with L. pneumophila in environmental reservoirs may indicate that they are more likely to be true hosts of the bacterium. While the Acanthamoeba spp., Naegleria spp., Vahlkampfia spp., and Hartmannella spp. (including Vermamoeba vermifomis) are commonly found in multiple sources (Table 2), particular protozoa appear to co-reside with L. pneumophila in more than one environmental sample (Table 2). A. hatchetti, A. polyphaga, H. cantabrigensis, N. fowleri, N. lovaniensis, Neoparamoeabe sp., and Willertia sp. have been isolated from both natural and man-made water sources (Table 2), suggesting that these protozoa may function as hosts of L. pneumophila in both natural reservoirs and potable water. Both E. exudans and Echinamoeba thermarum have been identified in more than one potable water sample (Table 2), suggesting these amoebae may play more prominent roles in the epidemiology of L. pneumophila. A higher incidence of specific protozoa with L. pneumophila may indicate a stronger likelihood that these protozoa are responsible for the persistence of L. pneumophila in environmental reservoirs.
Not all protozoa species isolated from the same environmental source are hosts of L. pneumophila. Of several species of free-living amoeba collected from a cooling tower, only A. polyphaga supported intracellular growth of L. pneumophila whereas L. pneumophila failed to replicate within C. limacoides, V. platypodia, and V. bacillipedes (Rowbotham, 1986). Similarly, of several ciliated protozoa species in biofilm samples isolated from a thermal spa, L. pneumophila was able to infect Ciliophrya sp., O. bifaria, and S. mytilus, but no intracellular bacteria were detected within Euglypha sp. or Centropyxis sp. (Rasch et al., 2016). Thus, L. pneumophila is able to persist in environments comprised of both L. pneumophila-restrictive and permissive protozoan hosts. The relative abundancy of L. pneumophila in different environmental niches may reflect mixed populations of these two types of protozoa. Alternatively, in some circumstances L. pneumophila may deplete entire populations of permissive hosts, enriching for resistant species of protozoa that remain. Thus, the absence of certain types of protozoa may not necessarily rule them out as contributors to L. pneumophila growth and persistence in the environment.
The distribution of protozoa between the types of water sources examined (natural water reservoirs, cooling towers, potable water distribution system, and compost sites; Table 2) was relatively uniform with a few notable exceptions. Amoebozoa and Percolozoa, making up the majority of the protozoa identified, were found in all water sources. Amoebozoa were more predominant in cooling towers and potable water systems. The lower abundance of Percolozoa in cooling towers coincided with a higher abundance of Ciliophora (ciliated protozoa) whereas in potable water, an enrichment in organisms from the Tubulinea class of Amoebozoa, in particular Echinamoeba was observed. In contrast, fewer members of the Discosea class were reported and in particular, no members of the Centramoebida order despite their presence in all other sites. The perseverance of L. pneumophila within various water environments despite variation in the protozoa composition demonstrates the highly adaptive nature of this bacterium to fluctuations in host population dynamics.
Metagenomics
Although co-isolation studies provide valuable insights into the microbial communities that support L. pneumophila, these methods cannot adequately define the full diversity of these communities (Kunin et al., 2008). While enrichment steps are often necessary to identify low abundance organisms, they create experimental bottlenecks and biases by selecting against protozoa that cannot be cultured using standard protocols (Hugenholtz and Tyson, 2008; Gomez-Alvarez et al., 2012) and Legionella isolates with host specificities that do not overlap with amoebal species commonly used in these techniques (Evstigneeva et al., 2009). Metagenome-based analyses may circumvent the limitations inherent to culture-based approaches and provide a more comprehensive, unbiased profile of these communities (Hugenholtz and Tyson, 2008; Gomez-Alvarez et al., 2009). For example, metagenomic studies of samples from three separate watersheds showed both a high level of diversity in the population of Legionella (encompassing 15 different species) and a correlation between the levels of Amoebozoa present in the water and the abundance of Legionella isolates (Peabody et al., 2017). Monitoring the abundance of Legionella, Hartmannella, and Naegleria from two environmental water sources over the course of a standard water purification procedure suggested a correlation between the abundance of Legionella and Naegleria, but not Hartmannella (Lin et al., 2014). In general however, metagenomics studies have been somewhat difficult to interpret. Often individual sites are dominated by one or a few amoebal species and the relative abundance of L. pneumophila is extremely low compared to other bacteria (Liu et al., 2012; Delafont et al., 2013): these features make it difficult to correlate the presence of L. pneumophila with specific protozoa. As the sensitivity and depth of metagenomics analysis improves, metagenomics will most certainly be a source of tremendous insight into the full repertoire of protozoan hosts of L. pneumophila.
Factors affecting the outcome of legionella-protozoa interactions
The outcome of the interaction between L. pneumophila and protozoa can be influenced by a number of factors; the identity of the host cell, variations in the predatory behavior or feeding preferences of the host, the strain or species of the bacterium, the relative abundance of the two organisms, the external environment, and other microorganisms.
The identity of the host cell can greatly impact the outcome of the infection. While some hosts are permissive for L. pneumophila replication, others are restrictive, either impeding bacterial growth or in extreme cases, survival (Amaro et al., 2015). The maximum amount and rate of L. pneumophila growth between hosts can vary significantly (Declerck et al., 2005). For example, L. pneumophila can achieve up to 10,000-fold growth in A. castellanii but only 10-fold growth in N. lovaniensis over the same time period (Declerck et al., 2005). Similarly, L. pneumophila strain Paris grows robustly in A. castellanii and V. vermiformis but is defective for growth in W. magna (Dey et al., 2009). Moreover, the differential growth of L. pneumophila Paris varies between different strains of W. magna, with robust growth in strain T5[S]44 (Tyson et al., 2014) but failure to grow in strains c2c Maky or Z502 (Dey et al., 2009). Thus, some hosts are more optimal than others for L. pneumophila survival and replication.
The predatory behavior and feeding preferences of the host can also influence Legionella-protozoa interactions. For example, the L. pneumophila auto-inducer LAI-1 disrupts chemotactic migration of D. discoideum (Simon et al., 2015) and promotes L. pneumophila uptake in both D. discoideum and A. castellanii (Tiaden et al., 2010). By restricting amoebal movement, L. pneumophila may localize feeding to the site of the bacteria—such modulation may also enrich for specific types of amoebae that support L. pneumophila replication. The LAI-1 biosynthesis genes are not conserved in all Legionella species (Burstein et al., 2016) suggesting that individual species may differentially promote their interaction with amoebae or do so via different mechanisms. Consistent with this idea, the host cell receptors that mediate L. pneumophila adhesion to V. vermiformis, A. castellanii, A. polyphaga, and N. lovaniensis and the underlying mechanisms governing bacterial uptake vary between these amoebal hosts (Venkataraman et al., 1997; Harb et al., 1998; Declerck et al., 2005, 2007). As a consequence, bacterial uptake can vary between protozoa. Indeed, A. castellanii has been shown to ingest L. pneumophila with much greater efficiency than N. lovaniensis (Declerck et al., 2005). Variations in sensing, targeting, adhesion and phagocytosis of bacteria can influence the affinity, specificity, frequency and duration with which L. pneumophila interacts with specific protozoa and thus, the impact of their cohabitation on the persistence of L. pneumophila in environmental reservoirs.
The genetic composition of the bacterium can greatly impact its fate within the host cell, as the survival and replication of different strains and species of Legionella can vary dramatically. Despite the growth defect of L. pneumophila Paris in Willertia magna, both the L. pneumophila Philadelphia-1, Lens and 130b strains are able to replicate in this amoebal host (Dey et al., 2009; Tyson et al., 2014). Similarly, comparisons between clinical and environmental isolates of L. pneumophila showed that while one clinical isolate was highly adept at growing in A. lenticulata another was severely defective and the relative amounts of replication of the environmental isolates in this host were somewhere in between (Molmeret et al., 2001). Similar differences are observed between species of Legionella. While L. pneumophila, Legionella steelei, Legionella dumoffii, and Legionella norrlandica are able to grow within A. castellanii, several other species including Legionella longbeachae, Legionella jordanis, and Legionella anisa are unable to do so (Neumeister et al., 1997; Edelstein et al., 2012; Rizzardi et al., 2014). Thus, the fate of both the bacterium and the host cell is greatly determined by the inherent properties of each organism.
The outcome of a Legionella-protozoa interaction is not only influenced by their respective identities but the relative abundance of each organism. For instance, when L. pneumophila is present at low levels they are digested for nutrients by Tetrahymena sp. but when the bacteria reach a threshold concentration, they are packaged into vesicles and secreted in pellets (Berk et al., 2008; Hojo et al., 2012). The greater the number of bacteria present, the greater the production and secretion of these bacterial pellets. Similar packaging and secretion of other types of bacteria (Denoncourt et al., 2014) suggests this may be a mechanism by which protozoa compensate for over-eating, or stock-pile food (Hojo et al., 2012).
The external environment can have a profound effect on Legionella-protozoa interactions. For example, temperature can greatly impact the intracellular fate of L. pneumophila. Although, intracellular replication of L. pneumophila in A. castellanii occurs at a range of temperatures (Rowbotham, 1981), intracellular growth is significantly reduced at lower temperatures (Ohno et al., 2008). Within more restrictive hosts, such as A. polyphaga, intracellular replication only occurs at higher temperatures whereas below 25°C, L. pneumophila is readily consumed (Nagington and Smith, 1980). In contrast, in Tetrahymena spp. L. pneumophila exhibits robust intracellular growth at 35°C (Fields et al., 1984; Barbaree et al., 1986; Kikuhara et al., 1994) but at lower temperatures, L. pneumophila is packaged into vesicles and secreted into the environment (Faulkner et al., 2008; Koubar et al., 2011). The factors affecting intracellular growth of L. pneumophila are not mutually exclusive, as different combinations of the strain of L. pneumophila, the host cell type and temperature can significantly alter intracellular growth of the bacterium (Buse and Ashbolt, 2011).
Much of the research examining Legionella-protozoa interactions has focused on specific bacterial-host pairings, which cannot address the impact of other organisms on these interactions. L. pneumophila naturally inhabits complex microbial communities, which could have both positive and negative impacts on L. pneumophila survival and population dynamics. For example, A. castellanii harboring the endosymbiont Neochlamydia S13 are unable to support L. pneumophila replication despite efficient uptake and lack of degradation in the lysosome (Ishida et al., 2014). The impact of Neochlamydia S13 on L. pneumophila replication is specific because L. pneumophila is able to replicate in A. castellanii infected with the endosymbiont Protochlamydia R18. Moreover, curing A. castellanii of Neochlamyida S13 restores intracellular growth of L. pneumophila, suggesting that the presence of the endosymbiont renders A. castellani resistant to L. pneumophila pathogenesis. In contrast, L. pneumophila has been shown to promote the intracellular growth of Brucella neotomae when the two pathogens share the same vacuole (Kang and Kirby, 2017). While sharing resources does not appear to affect L. pneumophila, it is conceivable that L. pneumophila may similarly benefit from the activities of other bacteria when it finds itself in more restrictive protozoan hosts.
Future directions
A critical challenge in understanding the molecular mechanisms of L. pneumophila pathogenesis, evolution and environmental persistence is the staggering diversity of the protozoan hosts that support L. pneumophila replication. Indeed, such diversity is thought to be responsible for shaping L. pneumophila into a generalist pathogen with a broad host range—a feature clearly important for pathogenesis in humans. Rather than having a single, defined “natural host,” L. pneumophila wanders from host to host and is constantly shaped by these disparate interactions. Such a lifestyle is a challenge for researchers studying these bacteria: (1) many protozoa remain poorly characterized, difficult to culture, and/or unsequenced; (2) the shear diversity of protozoa and complexity of natural interactions makes experimental analysis of phenotypes under “physiologically relevant” conditions extremely daunting (which hosts should be used and under what chemical and physical conditions should the interaction be studied?); and (3) how can non-binary interactions with mixed bacterial and host populations be examined in a reproducible and informative fashion? Given the importance of protozoa to L. pneumophila biology (and pathogen evolution in general), we strongly advocate efforts for the sequencing and detailed study of these organisms. While it is enticing to retreat to the comfort of studying Legionella-host interactions in mammalian macrophages and perhaps one or two model protozoa, an exciting, informative, frustrating, and messy reality remains largely unexplored. Perhaps once the diversity of bacterial-protozoan behaviors is better understood, a panel of model hosts could be chosen not based on ease of culture, but instead to capture the greatest breadth of this diversity.
Author contributions
TO, DB, AE, and GZ wrote the manuscript. GZ and AE generated the phylogenetic tree.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jason Park, Sara Rego, Soma Ghosh, and Mohammad Hossain for thoughtful review of the manuscript. This work was supported by the National Institutes of Health, Grant 1R21AI119580-01 (TO) and the Natural Sciences and Engineering Research Council of Canada, Grant RGPIN-2014-03641 (AE).
References
- Abdel-Nour M., Duncan C., Low D. E., Guyard C. (2013). Biofilms: the Stronghold of Legionella pneumophila. Int. J. Mol. Sci. 14, 21660–21675. 10.3390/ijms141121660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Kwaik Y. (1996). The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62, 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro F., Wang W., Gilbert J. A., Anderson O. R., Shuman H. A. (2015). Diverse protist grazers select for virulence-related traits in Legionella. ISME J. 9, 1607–1618. 10.1038/ismej.2014.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand C. M., Skinner A. R., Malic A., Kurtz J. B. (1983). Interaction of L. pneumophilia and a free living amoeba (Acanthamoeba palestinensis). J. Hyg. 91, 167–178. 10.1017/S0022172400060174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbaree J. M., Fields B. S., Feeley J. C., Gorman G. W., Martin W. T. (1986). Isolation of protozoa from water associated with a legionellosis outbreak and demonstration of intracellular multiplication of Legionella pneumophila. Appl. Environ. Microbiol. 51, 422–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J., Scaife H., Brown M. R. (1995). Intraphagocytic growth induces an antibiotic-resistant phenotype of Legionella pneumophila. Antimicrob. Agents Chemother. 39, 2684–2688. 10.1128/AAC.39.12.2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk S. G., Faulkner G., Garduño E., Joy M. C., Ortiz-Jimenez M. A., Garduño R. A. (2008). Packaging of live Legionella pneumophila into pellets expelled by Tetrahymena spp. does not require bacterial replication and depends on a Dot/Icm-mediated survival mechanism. Appl. Environ. Microbiol. 74, 2187–2199. 10.1128/AEM.01214-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk S. G., Ting R. S., Turner G. W., Ashburn R. J. (1998). Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 64, 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biurrun A., Caballero L., Pelaz C., León E., Gago A. (1999). Treatment of a Legionella pneumophila-colonized water distribution system using copper-silver ionization and continuous chlorination. Infect. Control. Hosp. Epidemiol. 20, 426–428. 10.1086/501645 [DOI] [PubMed] [Google Scholar]
- Bohach G. A., Snyder I. S. (1983). Characterization of surfaces involved in adherence of Legionella pneumophila to Fischerella species. Infect. Immun. 42, 318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyer S., Imbert C., Rodier M. H., Héchard Y. (2007). Long-term survival of Legionella pneumophila associated with Acanthamoeba castellanii vesicles. Environ. Microbiol. 9, 1341–1344 10.1111/j.1462-2920.2006.01229.x [DOI] [PubMed] [Google Scholar]
- Breiman R. F., Fields B. S., Sanden G. N., Volmer L., Meier A., Spika J. S. (1990). Association of shower use with Legionnaires' disease. Possible role of amoebae. JAMA 263, 2924–2926. 10.1001/jama.1990.03440210074036 [DOI] [PubMed] [Google Scholar]
- Brieland J., McClain M., Heath L., Chrisp C., Huffnagle G., LeGendre M., et al. (1996). Coinoculation with Hartmannella vermiformis enhances replicative Legionella pneumophila lung infection in a murine model of Legionnaires' disease. Infect. Immun. 64, 2449–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brousseau N., Lévesque B., Guillemet T. A., Cantin P., Gauvin D., Giroux J.-P., et al. (2013). Contamination of public whirlpool spas: factors associated with the presence of Legionella spp., Pseudomonas aeruginosa and Escherichia coli. Int. J. Environ. Health Res. 23, 1–15. 10.1080/09603123.2012.678001 [DOI] [PubMed] [Google Scholar]
- Burstein D., Amaro F., Zusman T., Lifshitz Z., Cohen O., Gilbert J. A., et al. (2016). Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nat. Genet. 48, 167–175. 10.1038/ng.3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buse H. Y., Ashbolt N. J. (2011). Differential growth of Legionella pneumophila strains within a range of amoebae at various temperatures associated with in-premise plumbing. Lett. Appl. Microbiol. 53, 217–224 10.1111/j.1472-765X.2011.03094.x [DOI] [PubMed] [Google Scholar]
- Cervero-Aragó S., Rodríguez-Martínez S., Puertas-Bennasar A., Araujo R. M. (2015). Effect of common drinking water disinfectants, chlorine and heat, on free Legionella and amoebae-associated Legionella. PLoS ONE 10:e0134726. 10.1371/journal.pone.0134726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervero-Aragó S., Sommer R., Araujo R. M. (2014). Effect of UV irradiation (253.7 nm) on free Legionella and Legionella associated with its amoebae hosts. Water Res. 67, 299–309. 10.1016/j.watres.2014.09.023 [DOI] [PubMed] [Google Scholar]
- Cianciotto N. P., Fields B. S. (1992). Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. U.S.A. 89, 5188–5191. 10.1073/pnas.89.11.5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo J. D., Cirillo S. L., Yan L., Bermudez L. E., Falkow S., Tompkins L. S. (1999). Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67, 4427–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo J. D., Falkow S., Tompkins L. S. (1994). Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62, 3254–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conza L., Casati Pagani S., Gaia V. (2014). Influence of climate and geography on the occurrence of Legionella and amoebae in composting facilities. BMC Res. Notes 7:831. 10.1186/1756-0500-7-831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conza L., Pagani S. C., Gaia V. (2013). Presence of Legionella and free-living amoebae in composts and bioaerosols from composting facilities. PLoS ONE 8:e68244. 10.1371/journal.pone.0068244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia A. M., Ferreira J. S., Borges V., Nunes A., Gomes B., Capucho R., et al. (2016). Probable person-to-person transmission of Legionnaires' disease. New. Eng. J. Med. 374, 497–498. 10.1056/NEJMc1505356 [DOI] [PubMed] [Google Scholar]
- De Jonckheere J. F., Brown S. (2005). The identification of vahlkampfiid amoebae by ITS sequencing. Protist 1, 89–96. 10.1016/j.protis.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Declerck P., Behets J., van Hoef V., Ollevier F. (2007). Detection of Legionella spp. and some of their amoeba hosts in floating biofilms from anthropogenic and natural aquatic environments. Water Res. 41, 3159–3167. 10.1016/j.watres.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Declerck P., Behets J., Delaedt Y., Margineanu A., Lammertyn E., Ollevier F. (2005). Impact of non-Legionella bacteria on the uptake and intracellular replication of Legionella pneumophila in Acanthamoeba castellanii and Naegleria lovaniensis. Microb. Ecol. 50, 536–549. 10.1007/s00248-005-0258-0 [DOI] [PubMed] [Google Scholar]
- Delafont V., Brouke A., Bouchon D., Moulin L., Héchard Y. (2013). Microbiome of free-living amoebae isolated from drinking water. Water Res. 47, 6958–6965. 10.1016/j.watres.2013.07.047 [DOI] [PubMed] [Google Scholar]
- Denoncourt A. M., Paquet V. E., Charette S. J. (2014). Potential role of bacteria packaging by protozoa in the persistence and transmission of pathogenic bacteria. Front. Microbiol. 5:240. 10.3389/fmicb.2014.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey R., Bodennec J., Mameri M. O., Pernin P. (2009). Free-living freshwater amoebae differ in their susceptibility to the pathogenic bacterium Legionella pneumophila. FEMS Microbiol. Lett. 290, 10–17. 10.1111/j.1574-6968.2008.01387.x [DOI] [PubMed] [Google Scholar]
- Dunnebacke T. H., Schuster F. L., Yagi S., Booton G. C. (2004). Balamuthia mandrillaris from soil samples. Microbiology 150, 2837–2842. 10.1099/mic.0.27218-0 [DOI] [PubMed] [Google Scholar]
- Dupuy M., Binet M., Bouteleux C., Herbelin P., Soreau S., Héchard Y. (2016). Permissiveness of freshly isolated environmental strains of amoebae for growth of Legionella pneumophila. FEMS Microbiol. Lett. 363:fnw022. 10.1093/femsle/fnw022 [DOI] [PubMed] [Google Scholar]
- Edelstein P. H., Edelstein M. A., Shephard L. J., Ward K. W., Ratcliff R. M. (2012). Legionella steelei sp. nov., isolated from human respiratory specimens in California, USA, and South Australia. Int. J. Syst. Evol. Microbiol. 62, 1766–1771. 10.1099/ijs.0.035709-0 [DOI] [PubMed] [Google Scholar]
- Evstigneeva A., Raoult D., Karpachevskiy L., La Scola B. (2009). Amoeba co-culture of soil specimens recovered 33 different bacteria, including four new species and Streptococcus pneumoniae. Microbiology 155, 657–664. 10.1099/mic.0.022970-0 [DOI] [PubMed] [Google Scholar]
- Faulkner G., Berk S. G., Garduño E., Ortiz-Jiménez M. A., Garduño R. A. (2008). Passage through Tetrahymena tropicalis triggers a rapid morphological differentiation in Legionella pneumophila. J. Bacteriol. 190, 7728–7738. 10.1128/JB.00751-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. S. (1996). The molecular ecology of Legionellae. Trends Microbiol. 4, 286–290. 10.1016/0966-842X(96)10041-X [DOI] [PubMed] [Google Scholar]
- Fields B. S., Barbaree J. M., Shotts E. B., Jr., Feeley J. C., Morrill W. E., Sanden G. N., et al. (1986). Comparison of guinea pig and protozoan models for determining virulence of Legionella species. Infect. Immun. 53, 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. S., Benson R. F., Besser R. E. (2002). Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15, 506–526. 10.1128/CMR.15.3.506-526.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields B. S., Sanden G. N., Barbaree J. M., Morrill W. E., Wadowsky R. M., White E. H., et al. (1989). Intracellular multiplication of Legionella pneumophila in amoebae isolated from hospital hot water tanks. Curr. Microbiol. 18, 131–137. 10.1007/BF01570838 [DOI] [Google Scholar]
- Fields B. S., Shotts E. B., Feeley J. C., Gorman G. W., Martin W. T. (1984). Proliferation of Legionella pneumophila as an intracellular parasite of the ciliated protozoan Tetrahymena pyriformis. Appl. Environ. Microbiol. 47, 467–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliermans C. B. (1996). Ecology of Legionella: from data to knowledge with a little wisdom. Microb. Ecol. 32, 203–228. 10.1007/BF00185888 [DOI] [PubMed] [Google Scholar]
- Fliermans C. B., Cherry W. B., Orrison L. H., Smith S. J., Tison D. L., Pope D. H. (1981). Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L. Y., Harb O. S., Abu Kwaik Y. (1997). Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect. Immun. 65, 4738–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M. T., Baladrón B., Gil V., Tarancon M. L., Vilasau A., Ibañez A., et al. (2008). Persistence of chlorine-sensitive Legionella pneumophila in hyperchlorinated installations. J. Appl. Microbiol. 105, 837–847. 10.1111/j.1365-2672.2008.03804.x [DOI] [PubMed] [Google Scholar]
- García M. T., Jones S., Pelaz C., Millar R. D., Abu Kwaik Y. (2007). Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environ. Microbiol. 9, 1267–1277. 10.1111/j.1462-2920.2007.01245.x [DOI] [PubMed] [Google Scholar]
- Garduño R. A., Garduño E., Hiltz M., Hoffman P. S. (2002). Intracellular growth of Legionella pneumophila gives rise to a differentiated form dissimilar to stationary-phase forms. Infect. Immun. 70, 6273–6283. 10.1128/IAI.70.11.6273-6283.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alvarez V., Revetta R. P., Santo Domingo J. W. (2012). Metagenomic analyses of drinking water receiving different disinfection treatments. Appl. Environ. Microbiol. 78, 6095–6102. 10.1128/AEM.01018-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alvarez V., Teal T. K., Schmidt T. M. (2009). Systematic artifacts in metagenomes from complex microbial communities. ISME J. 3, 1314–1317. 10.1038/ismej.2009.72 [DOI] [PubMed] [Google Scholar]
- Greub G., Raoult D. (2003). Morphology of Legionella pneumophila according to their location within Hartmanella vermiformis. Res. Microbiol. 154, 619–621. 10.1016/j.resmic.2003.08.003 [DOI] [PubMed] [Google Scholar]
- Greub G., Raoult D. (2004). Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17, 413–433. 10.1128/CMR.17.2.413-433.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hägele S., Köhler R., Merkert H., Schleicher M., Hacker J., Steinert M. (2000). Dictyostelium discoideum: a new host model system for intracellular pathogens of the genus Legionella. Cell. Microbiol. 2, 165–171. 10.1046/j.1462-5822.2000.00044.x [DOI] [PubMed] [Google Scholar]
- Hahn M. W., Höfle M. G. (2001). Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35, 113–121. 10.1111/j.1574-6941.2001.tb00794.x [DOI] [PubMed] [Google Scholar]
- Harb O. S., Venkataraman C., Haack B. J., Gao L. Y., Kwaik Y. A. (1998). Heterogeneity in the attachment and uptake mechanisms of the Legionnaires' disease bacterium, Legionella pneumophila, by protozoan hosts. Appl. Environ. Microbiol. 64, 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harf C., Goffinet S., Meunier O., Monteil H., Colin D. A. (1997). Flow cytometric determination of endocytosis of viable labelled Legionella pneumophila by Acanthamoeba palestinensis. Cytometry 27, 269–274. [DOI] [PubMed] [Google Scholar]
- Hilbi H., Segal G., Shuman H. A. (2001). Icm/dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol. Microbiol. 42, 603–617. 10.1046/j.1365-2958.2001.02645.x [DOI] [PubMed] [Google Scholar]
- Hojo F., Sato D., Matsuo J., Miyake M., Nakamura S., Kunichika M., et al. (2012). Ciliates expel environmental Legionella-laden pellets to stockpile food. Appl. Environ. Microbiol. 78, 5247–5257. 10.1128/AEM.00421-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden E. P., Winkler H. H., Wood D. O., Leinbach E. D. (1984). Intracellular growth of Legionella pneumophila within Acanthamoeba castellanii Neff. Infect. Immun. 45, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu B. M., Huang C. C., Chen J. S., Chen N. H., Huang J. T. (2011). Comparison of potentially pathogenic free-living amoeba hosts by Legionella spp. in substrate-associated biofilms and floating biofilms from spring environments. Water Res. 45, 5171–5183. 10.1016/j.watres.2011.07.019 [DOI] [PubMed] [Google Scholar]
- Hsu B. M., Lin C. L., Shih F. C. (2009). Survey of pathogenic free-living amoebae and Legionella spp. in mud spring recreation area. Water Res. 43, 2817–2828. 10.1016/j.watres.2009.04.002 [DOI] [PubMed] [Google Scholar]
- Hsu T. K., Wu S. F., Hsu B. M., Kao P. M., Tao C. W., Shen S. M., et al. (2015). Surveillance of parasitic Legionella in surface waters by using immunomagnetic separation and amoebae enrichment. Pathog. Glob. Health 109, 328–335. 10.1179/2047773215Y.0000000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. W., Hsu B. M. (2010). Survey of Naegleria and its resisting bacteria-Legionella in hot spring water of Taiwan using molecular method. Parasitol. Res. 106, 1395–1402. 10.1007/s00436-010-1815-0 [DOI] [PubMed] [Google Scholar]
- Hugenholtz P., Tyson G. W. (2008). Microbiology: metagenomics. Nature 455, 481–483. 10.1038/455481a [DOI] [PubMed] [Google Scholar]
- Ikedo M., Yabuuchi E. (1986). Ecological studies of Legionella species. I. Viable counts of Legionella pneumophila in cooling tower water. Microbiol. Immunol. 30, 413–423. 10.1111/j.1348-0421.1986.tb02967.x [DOI] [PubMed] [Google Scholar]
- Ishida K., Sekizuka T., Hayashida K., Matsuo J., Takeuchi F., Kuroda M., et al. (2014). Amoebal endosymbiont Neochlamydia genome sequence illuminates the bacterial role in the defense of the host amoebae against Legionella pneumophila. PLoS ONE 9:e95166. 10.1371/journal.pone.0095166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N., Aeby S., Lienard J., Greub G. (2013). Discovery of new intracellular pathogens by amoebal coculture and amoebal enrichment approaches. J. Vis. Exp. 80:51055 10.3791/51055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W. T., Hsu B. M., Chang T. Y., Hsu T. K., Kao P. M., Huang K. H., et al. (2014). Surveillance and evaluation of the infection risk of free-living amoebae and Legionella in different aquatic environments. Sci. Total Environ. 499, 212–219. 10.1016/j.scitotenv.2014.07.116 [DOI] [PubMed] [Google Scholar]
- Kang Y. S., Kirby J. E. (2017). Promotion and rescue of intracellular Brucella neotomae replication during coinfection with Legionella pneumophila. Infect. Immun. 85, e00991–e00916. 10.1128/IAI.00991-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao P. M., Tung M. C., Hsu B. M., Hsu S. Y., Huang J. T., Liu J. H., et al. (2013). Differential Legionella spp. survival between intracellular and extracellular forms in thermal spring environments. Environ. Sci. Pollut. Res. Int. 20, 3098–3106. 10.1007/s11356-012-1159-7 [DOI] [PubMed] [Google Scholar]
- Kikuhara H., Ogawa M., Miyamoto H., Nikaido Y., Yoshida S. (1994). Intracellular multiplication of Legionella pneumophila in Tetrahymena thermophila. J. UOEH 16, 263–275. 10.7888/juoeh.16.263 [DOI] [PubMed] [Google Scholar]
- Kilvington S., Price J. (1990). Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J. Appl. Bacteriol. 68, 519–525. 10.1111/j.1365-2672.1990.tb02904.x [DOI] [PubMed] [Google Scholar]
- King C. H., Fields B. S., Shotts E. B., White E. H. (1991). Effects of cytochalasin D and methylamine on intracellular growth of Legionella pneumophila in amoebae and human monocyte-like cells. Infect. Immun. 59, 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C. H., Shotts E. B., Jr., Wooley R. E., Porter K. G. (1988). Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54, 3023–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide M., Higa F., Tateyama M., Cash H. L., Hokama A., Fujita J. (2014). Role of Brevundimonas vesicularis in supporting the growth of Legionella in nutrient-poor environments. New Microbiol. 37, 33–39. [PubMed] [Google Scholar]
- Koubar M., Rodier M. H., Garduño R. A., Frere J. (2011). Passage through Tetrahymena tropicalis enhances the resistance to stress and the infectivity of Legionella pneumophila. FEMS Microbiol. Lett. 325, 10–15. 10.1111/j.1574-6968.2011.02402.x [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V., Copeland A., Lapidus A., Mavromatis K., Hugenholtz P. (2008). A bioinformatician's guide to metagenomics. Microbiol. Mol. Biol. Rev. 72, 557–578. 10.1128/MMBR.00009-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz J. B., Bartlett C. L., Newton U. A., White R. A., Jones N. L. (1982). Legionella pneumophila in cooling water systems. Report of a survey of cooling towers in London and a pilot trial of selected biocides. J. Hyg. 88, 369–381. 10.1017/S0022172400070248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasheras A., Boulestreau H., Rogues A. M., Ohayon-Courtes C., Labadie J. C., Gachie J. P. (2006). Influence of amoebae and physical and chemical characteristics of water on presence and proliferation of Legionella species in hospital water systems. Am. J. Infect. Control 34, 520–525. 10.1016/j.ajic.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Lau H. Y., Ashbolt N. J. (2009). The role of biofilms and protozoa in Legionella pathogenesis: implications for drinking water. J. Appl. Microbiol. 107, 368–378. 10.1111/j.1365-2672.2009.04208.x [DOI] [PubMed] [Google Scholar]
- Lin W., Yu Z., Zhang H., Thompson I. P. (2014). Diversity and dynamics of microbial communities at each step of treatment plant for potable water generation. Water Res. 52, 218–230. 10.1016/j.watres.2013.10.071 [DOI] [PubMed] [Google Scholar]
- Liu R., Yu Z., Guo H., Liu M., Zhang H., Yang M. (2012). Pyrosequencing analysis of eukaryotic and bacterial communities in faucet biofilms. Sci. Total Environ. 435–436, 124–131. 10.1016/j.scitotenv.2012.07.022 [DOI] [PubMed] [Google Scholar]
- Ma W., Dong F. F., Stavrinides J., Guttman D. S. (2006). Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2:e209. 10.1371/journal.pgen.0020209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mampel J., Spirig T., Weber S. S., Haagensen J. A., Molin S., Hilbi H. (2006). Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl. Environ. Microbiol. 72, 2885–2895. 10.1128/AEM.72.4.2885-2895.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano-Cabral F., Cabral G. (2003). Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 16, 273–307. 10.1128/CMR.16.2.273-307.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano-Cabral F., Jamerson M., Kaneshiro E. S. (2010). Free-living amoebae, Legionella and Mycobacterium in tap water supplied by a municipal drinking water utility in the USA. J. Water Health 8, 71–82. 10.2166/wh.2009.129 [DOI] [PubMed] [Google Scholar]
- Mengue L., Régnacq M., Aucher W., Portier E., Héchard Y., Samba-Louaka A. (2016). Legionella pneumophila prevents proliferation of its natural host Acanthamoeba castellanii. Sci. Rep. 6:36448. 10.1038/srep36448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J. F., Tompkins L. S. (1992). A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M., Horn M., Wagner M., Santic M., Abu Kwaik Y. (2005). Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71, 20–28. 10.1128/AEM.71.1.20-28.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M., Jarraud S., Mori J. P., Pernin P., Forey F., Reyrolle M., et al. (2001). Different growth rates in amoeba of genotypically related environmental and clinical Legionella pneumophila strains isolated from a thermal spa. Epidemiol. Infect. 126, 231–239. 10.1017/S0950268801005258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchesa P., Mwamba O., Barnard T. G., Bartie C. (2014). Detection of free-living amoebae using amoebal enrichment in a wastewater treatment plant of Gauteng Province, South Africa. Biomed. Res. Int. 2014:575297. 10.1155/2014/575297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagington J., Smith D. J. (1980). Pontiac fever and amoebae. Lancet 316, 1241 10.1016/S0140-6736(80)92494-0 [DOI] [PubMed] [Google Scholar]
- Nahapetian K., Challemel O., Beurtin D., Dubrou S., Gounon P., Squinazi F. (1991). The intracellular multiplication of Legionella pneumophila in protozoa from hospital plumbing systems. Res. Microbiol. 142, 677–685. 10.1016/0923-2508(91)90081-K [DOI] [PubMed] [Google Scholar]
- Neumeister B., Schöniger S., Faigle M., Eichner M., Dietz K. (1997). Multiplication of different Legionella species in Mono Mac 6 cells and in Acanthamoeba castellanii. Appl. Environ. Microbiol. 63, 1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome A. L., Baker R. L., Miller R. D., Arnold R. R. (1985). Interactions between Naegleria fowleri and Legionella pneumophila. Infect. Immun. 50, 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor T. J., Adepoju Y., Boyd D., Isberg R. R. (2011). Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc. Nat. Acad. Sci. U.S.A. 108, 14733–14740. 10.1073/pnas.1111678108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno A., Kato N., Sakamoto R., Kimura S., Yamaguchi K. (2008). Temperature-dependent parasitic relationship between Legionella pneumophila and a free-living amoeba (Acanthamoeba castellanii). Appl. Environ. Microbiol. 74, 4585–4588. 10.1128/AEM.00083-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnier I., Raoult D., La Scola B. (2008). Isolation and identification of amoeba-resisting bacteria from water in human environment by using an Acanthamoeba polyphaga co-culture procedure. Environ. Microbiol. 10, 1135–1144. 10.1111/j.1462-2920.2007.01530.x [DOI] [PubMed] [Google Scholar]
- Peabody M. A., Caravas J. A., Morrison S. S., Mercante J. W., Prystajecky N. A., Raphael B. H., et al. (2017). Characterization of Legionella species from watersheds in British Columbia, Canada. mSphere 2, e00246–e00217. 10.1128/mSphere.00246-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe J. F., Webster L. R., Hackman B. (1983). Relationship between colonization of hospital building with Legionella pneumophila and hot water temperatures. Appl. Environ. Microbiol. 46, 769–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope D. H., Soracco R. J., Gill H. K., Fliermans C. B. (1982). Growth of Legionella pneumophila in two-membered cultures with green algae and cyanobacteria. Curr. Microbiol. 7, 319–321. 10.1007/BF01566871 [DOI] [Google Scholar]
- Rasch J., Krüger S., Fontvieille S., Ünal C. M., Michel R., Labrosse A., et al. (2016). Legionella-protozoa-nematode interactions in aquatic biofilms and influence of Mip on Caenorhabditis elegans colonization. Int. J. Med. Microbiol. 306, 443–451. 10.1016/j.ijmm.2016.05.012 [DOI] [PubMed] [Google Scholar]
- Rizzardi K., Winiecka-Krusnell J., Ramliden M., Alm E., Andersson S., Byfors S. (2014). Legionella norrlandica sp. nov., isolated from the biopurification system of a wood processing plant in northern Sweden. Int. J. Syst. Evol. Microbiol. 65, 598–603. 10.1099/ijs.0.068940-0 [DOI] [PubMed] [Google Scholar]
- Rohr U., Weber S., Michel R., Selenka F., Wilhelm M. (1998). Comparison of free-living amoebae in hot water systems of hospitals with isolates from moist sanitary areas by identifying genera and determining temperature tolerance. Appl. Environ. Microbiol. 64, 1822–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham T. J. (1980). Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33, 1179–1183. 10.1136/jcp.33.12.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham T. J. (1981). Pontiac fever, amoebae and Legionellae. Lancet 317, 40–41. 10.1016/S0140-6736(81)90141-0 [DOI] [PubMed] [Google Scholar]
- Rowbotham T. J. (1983). Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J. Clin. Pathol. 36, 978–986. 10.1136/jcp.36.9.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbotham T. J. (1986). Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22, 678–689. [PubMed] [Google Scholar]
- Ruggiero M. A., Gordon D. P., Orrell T. M., Bailly N., Bourgoin T., Brusca R. C., et al. (2015). A higher level classification of all living organisms. PLoS ONE 10:e0119248 10.1371/journal.pone.0119248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salloum G., Meugnier H., Reyrolle M., Grimont F., Grimont P. A., Etienne J., et al. (2002). Identification of Legionella species by ribotyping and other molecular methods. Res. Microbiol. 153, 679–686. 10.1016/S0923-2508(02)01381-5 [DOI] [PubMed] [Google Scholar]
- Scheikl U., Sommer R., Kirschner A., Rameder A., Schrammel B., Zweimüller I., et al. (2014). Free-living amoebae (FLA) co-occurring with legionellae in industrial waters. Eur. J. Protistol. 50, 422–429. 10.1016/j.ejop.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadrach W. S., Rydzewski K., Laube U., Holland G., Ozel M., Kiderlen A. F., et al. (2005). Balamuthia mandrillaris, free-living amoeba and opportunistic agent of encephalitis, is a potential host for Legionella pneumophila bacteria. Appl. Environ. Microbiol. 71, 2244–2249. 10.1128/AEM.71.5.2244-2249.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan K. B., Henson J. M., Ferris M. J. (2005). Legionella species diversity in an acidic biofilm community in Yellowstone National Park. Appl. Environ. Microbiol. 71, 507–511. 10.1128/AEM.71.1.507-511.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S., Schell U., Heuer N., Hager D., Albers M. F., Matthias J., et al. (2015). Inter-kingdom signaling by the Legionella quorum sensing molecule LAI-1 modulates cell migration through an IQGAP1-Cdc42-ARHGEF9-dependent pathway. PLoS Pathog. 11:e1005307. 10.1371/journal.ppat.1005307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S., Wagner M. A., Rothmeier E., Müller-Taubenberger A., Hilbi H. (2014). Icm/Dot-dependent inhibition of phagocyte migration by Legionella is antagonized by a translocated Ran GTPase activator. Cell. Microbiol. 16, 977–992. 10.1111/cmi.12258 [DOI] [PubMed] [Google Scholar]
- Smirnov A. V., Chao E., Nassonova E. S., Cavalier-Smith T. (2011). A revised classification of naked lobose Amoebae (Amoebozoa: Lobosa). Protist 162, 545–570. 10.1016/j.protis.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Smith-Somerville H. E., Huryn V. B., Walker C., Winters A. L. (1991). Survival of Legionella pneumophila in the cold-water ciliate Tetrahymena vorax. Appl. Environ. Microbiol. 57, 2742–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon J. M., Rupper A., Cardelli J. A., Isberg R. R. (2000). Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immun. 68, 2939–2947. 10.1128/IAI.68.5.2939-2947.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M., Ockert G., Lück C., Hacker J. (1998). Regrowth of Legionella pneumophila in a heat-disinfected plumbing system. Zentralbl. Bakteriol. 288, 331–342. 10.1016/S0934-8840(98)80005-4 [DOI] [PubMed] [Google Scholar]
- Stewart C. R., Muthye V., Cianciotto N. P. (2012). Legionella pneumophila persists within biofilms formed by Klebsiella pneumoniae, Flavobacterium sp., and Pseudomonas fluorescens under dynamic flow conditions. PLoS ONE 7:e50560. 10.1371/journal.pone.0050560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey M. V., Winiecka-Krusnell J., Ashbolt N. J., Stenström T. A. (2004). The efficacy of heat and chlorine treatment against thermotolerant Acanthamoebae and Legionellae. Scand. J. Infect. Dis. 36, 656–662. 10.1080/00365540410020785 [DOI] [PubMed] [Google Scholar]
- Stout J. E., Best M. G., Yu V. L., Rihs J. D. (1986). A note on symbiosis of Legionella pneumophila and Tatlockia micdadei with human respiratory flora. J. Appl. Bacteriol. 60, 297–299. 10.1111/j.1365-2672.1986.tb01736.x [DOI] [PubMed] [Google Scholar]
- Temmerman R., Vervaeren H., Noseda B., Boon N., Verstraete W. (2006). Necrotrophic growth of Legionella pneumophila. Appl. Environ. Microbiol. 72, 4323–4328. 10.1128/AEM.00070-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. M., Thomas T., Stuetz R. M., Ashbolt N. J. (2014). Your garden hose: a potential health risk due to Legionella spp. growth facilitated by free-living amoebae. Environ. Sci. Technol. 48, 10456–10464 10.1021/es502652n [DOI] [PubMed] [Google Scholar]
- Thomas V., Herrera-Rimann K., Blanc D. S., Greub G. (2006). Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl. Environ. Microbiol. 72, 2428–2438. 10.1128/AEM.72.4.2428-2438.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiaden A., Spirig T., Sahr T., Wälti M. A., Boucke K., Buchrieser C., et al. (2010). The autoinducer synthase LqsA and putative sensor kinase LqsS regulate phagocyte interactions, extracellular filaments and a genomic island of Legionella pneumophila. Environ. Microbiol. 12, 1243–1259. 10.1111/j.1462-2920.2010.02167.x [DOI] [PubMed] [Google Scholar]
- Tison D. L., Pope D. H., Cherry W. B., Fliermans C. B. (1980). Growth of Legionella pneumophila in association with blue-green algae (cyanobacteria). Appl. Environ. Microbiol. 39, 456–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndall R. L., Domingue E. L. (1982). Cocultivation of Legionella pneumophila and free-living amoebae. Appl. Environ. Microbiol. 44, 954–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J. Y., Pearce M. M., Vargas P., Bagchi S., Mulhern B. J., Cianciotto N. P. (2013). Multiple Legionella pneumophila Type II secretion substrates, including a novel protein, contribute to differential infection of the amoebae Acanthamoeba castellanii, Hartmannella vermiformis, and Naegleria lovaniensis. Infect. Immun. 81, 1399–1410. 10.1128/IAI.00045-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J. Y., Vargas P., Cianciotto N. P. (2014). The novel Legionella pneumophila type II secretion substrate NttC contributes to infection of amoebae Hartmannella vermiformis and Willaertia magna. Microbiology 160, 2732–2744. 10.1099/mic.0.082750-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadell E. M., Cavender J. C. (2007). Dictyostelids living in the soils of the Atlantic Forest, Iguazú Region, Misiones, Argentina: description of new species. Mycologia 99, 112–124. 10.1080/15572536.2007.11832606 [DOI] [PubMed] [Google Scholar]
- Valster R. M., Wullings B. A., van den Berg R., van der Kooij D. (2011). Relationships between free-living protozoa, cultivable Legionella spp., and water quality characteristics in three drinking water supplies in the Caribbean. Appl. Environ. Microbiol. 77, 7321–7328. 10.1128/AEM.05575-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valster R. M., Wullings B. A., van der Kooij D. (2010). Detection of protozoan hosts for Legionella pneumophila in engineered water systems by using a biofilm batch test. Appl. Environ. Microbiol. 76, 7144–7153. 10.1128/AEM.00926-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heijnsbergen E., Schalk J. A., Euser S. M., Brandsema P. S., den Boer J. W., de Roda Husman A. M. (2015). Confirmed and potential sources of Legionella reviewed. Environ. Sci. Technol. 49, 4797–4815. 10.1021/acs.est.5b00142 [DOI] [PubMed] [Google Scholar]
- Venkataraman C., Haack B. J., Bondada S., Abu Kwaik Y. (1997). Identification of a Gal/GalNAc lectin in the protozoan Hartmannella vermiformis as a potential receptor for attachment and invasion by the Legionnaires' disease bacterium. J. Exp. Med. 186, 537–547. 10.1084/jem.186.4.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadowsky R. M., Wang L., Laus S., Dowling J. N., Kuchta J. M., States S. J., et al. (1995). Gentamicin-containing peptone-yeast extract medium for cocultivation of Hartmannella vermiformis ATCC 50256 and virulent strains of Legionella pneumophila. Appl. Environ. Microbiol. 61, 4464–4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadowsky R. M., Yee R. B. (1983). Satellite growth of Legionella pneumophila with an environmental isolate of Flavobacterium breve. Appl. Environ. Microbiol. 46, 1447–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Nakao R., Fujishima M., Tachibana M., Shimizu T., Watarai M. (2016). Ciliate Paramecium is a natural reservoir of Legionella pneumophila. Sci. Rep. 6:24322. 10.1038/srep24322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Sugiura M., Kusunoki S., Ezaki T., Ikedo M., Yabuuchi E. (1992). Factors stimulating propagation of Legionellae in cooling tower water. Appl. Environ. Microbiol. 58, 1394–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbikowska E., Kletkiewicz H., Walczak M., Burkowska A. (2014). Coexistence of Legionella pneumophila bacteria and free-living amoebae in lakes serving as a cooling system of a power plant. Water Air Soil Pollut. 225:2066. 10.1007/s11270-014-2066-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbikowska E., Walczak M., Krawiec A. (2013). Distribution of Legionella pneumophila bacteria and Naegleria and Hartmannella amoebae in thermal saline baths used in balneotherapy. Parasitol. Res. 112, 77–83. 10.1007/s00436-012-3106-4 [DOI] [PMC free article] [PubMed] [Google Scholar]