Abstract
Transplantable murine models of ovarian high grade serous carcinoma (HGSC) remain an important research tool. We previously showed that ID8, a widely-used syngeneic model of ovarian cancer, lacked any of the frequent mutations in HGSC, and used CRISPR/Cas9 gene editing to generate derivatives with deletions in Trp53 and Brca2. Here we have used one ID8 Trp53 −/− clone to generate further mutants, with additional mutations in Brca1, Pten and Nf1, all of which are frequently mutated or deleted in HGSC. We have also generated clones with triple deletions in Trp53, Brca2 and Pten. We show that ID8 Trp53 −/−;Brca1 −/− and Trp53 −/−;Brca2 −/− cells have defective homologous recombination and increased sensitivity to both platinum and PARP inhibitor chemotherapy compared to Trp53 −/−. By contrast, loss of Pten or Nf1 increases growth rate in vivo, and reduces survival following cisplatin chemotherapy in vivo. Finally, we have also targeted Trp53 in cells isolated from a previous transgenic murine fallopian tube carcinoma model, and confirmed that loss of p53 expression in this second model accelerates intraperitoneal growth. Together, these CRISPR-generated models represent a new and simple tool to investigate the biology of HGSC, and the ID8 cell lines are freely available to researchers.
Introduction
HGSC is the commonest subtype of epithelial ovarian cancer and accounts for approximately 80% of ovarian cancer deaths. It is marked by universal TP53 mutation1 and extreme genomic instability2,3. Approximately 20% HGSC harbour mutations in BRCA1 or BRCA2 4, whilst loss of NF1 and PTEN expression is seen in around 20% cases, largely resulting from complex genomic rearrangements and structural variation5. Importantly, the complexity of HGSC means that multiple genomic abnormalities can co-exist within individual tumours5. Although robust prognostic and predictive molecular classifiers of HGSC remain elusive, patients with germline BRCA1 and BRCA2 mutations have improved overall prognosis6 with improved response to platinum7 and PARP inhibitor8 chemotherapy. Conversely, PTEN loss is a marker of poor prognosis9.
Lack of reliable immunocompetent murine models has significantly impeded HGSC research10. Recently, new genetically engineered mouse models (GEMM) of HGSC have been developed11,12, in which Trp53, Brca1, Brca2, Pten and Nf1 have been deleted in fallopian tube epithelial cells using Cre-mediated recombination. These models have great potential to expand our understanding of HGSC biology, but still require large-scale breeding programmes, and the mice can take many months to develop tumours. Thus, transplantable models remain valuable research tools.
First described in 200013, the ID8 model is a widely-used syngeneic model of ovarian cancer. However, using whole-exome sequencing, we recently showed that ID8 lacked any mutations fundamental in HGSC biology14: ID8 cells were Trp53 wild-type and retained functional p53 signalling. Brca1 and Brca2 were both wild-type and ID8 displayed competent homologous recombination DNA double strand break repair. Using CRISPR/Cas9 gene editing, we generated four Trp53 −/− and two Trp53 −/−;Brca2 −/− ID8 clones and characterised their intraperitoneal growth14.
We have now targeted three more genes critical in HGSC, Brca1, Pten and Nf1, in ID8 Trp53 −/− cells, and have also generated lines with triple deletion in Trp53, Brca2 and Pten. We have evaluated sensitivity to platinum and PARP inhibitor therapy in these cells, as well as our original Trp53 −/− and Trp53 −/−;Brca2 −/− cells. We have also targeted Trp53 in cells isolated from a previous transgenic murine fallopian tube carcinoma model15, in which Cre recombinase, under the control of the anti-Mullerian hormone type 2 receptor promoter (Amhr2-Cre), was used to delete Dicer, a key processor of microRNAs, and Pten selectively in the fallopian tube.
Our results indicate that loss of NF1 or PTEN expression increases intraperitoneal growth in ID8 cells, and is associated with a poor outcome following platinum chemotherapy. The utility of these ID8 derivatives is confirmed by increased sensitivity to both PARP inhibitor and platinum chemotherapy upon loss of BRCA1 or BRCA2 function. In the fallopian tube Dicer;Pten double knockout (DKO) cells, loss of p53 significantly increases intraperitoneal growth.
Results
CRISPR/Cas9 Brca1, Nf1 and Pten editing in ID8 cells
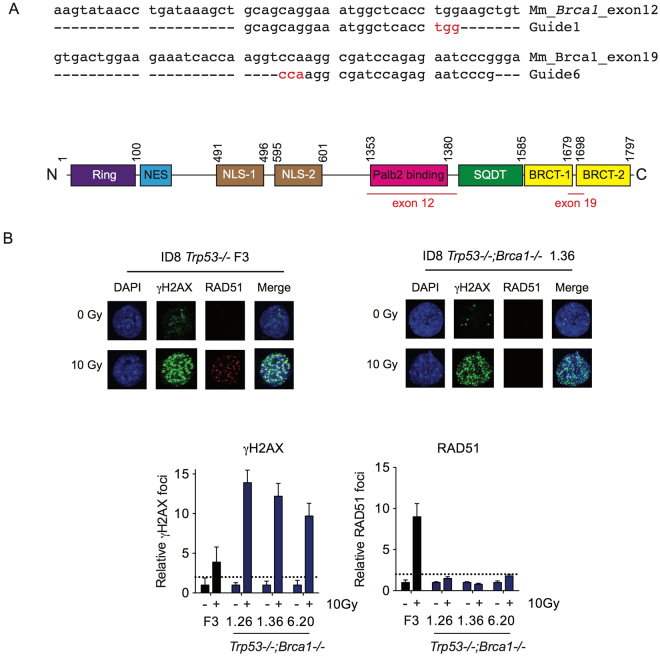
We used one ID8 Trp53 −/− clone (F3)14 to generate new sublines with deletions in Brca1, Pten and Nf1. We first targeted the Brca1 PALB2-binding domain in exon 12 (guide 1) and the BRCT-2 domain in exon 19 (guide 6) (Fig. 1A). Three single cell clones with bi-allelic deletions were isolated following transfection with the two separate guide RNA (clones 1.26, 1.36 - guide 1; clone 6.20 - guide 6; Fig. S1). All three clones failed to generate RAD51 foci in response to 10 Gy irradiation (Fig. 1B) or 10 µM rucaparib (Fig. S2), thus fulfilling criteria for defective homologous recombination (HR)16.
Figure 1.
Generation of Trp53 −/−;Brca1 −/− ID8 cells. (A) Design of guide RNA targeted to exons 12 and 19 of Brca1. Nucleotides in red represent PAM sequence. Schematic representation of BRCA1 protein with exons 12 and 19 highlighted in red (bottom). Numbers represent amino acid positions. (B) ID8 Trp53 −/− and Trp53 −/−;Brca1 −/− cells were irradiated (10 Gy), fixed and stained for γH2AX and RAD51, and counterstained with DAPI. RAD51 foci were counted in up to 30 untreated and irradiated cells. Bars represent mean (+/− SEM) γH2AX (left) and RAD51 (right) foci per cell; dotted lines represents two-fold increase in γH2AX and RAD51 foci/cell relative to untreated cells, suggestive of induction of DNA double strand breaks and functional homologous recombination respectively16.
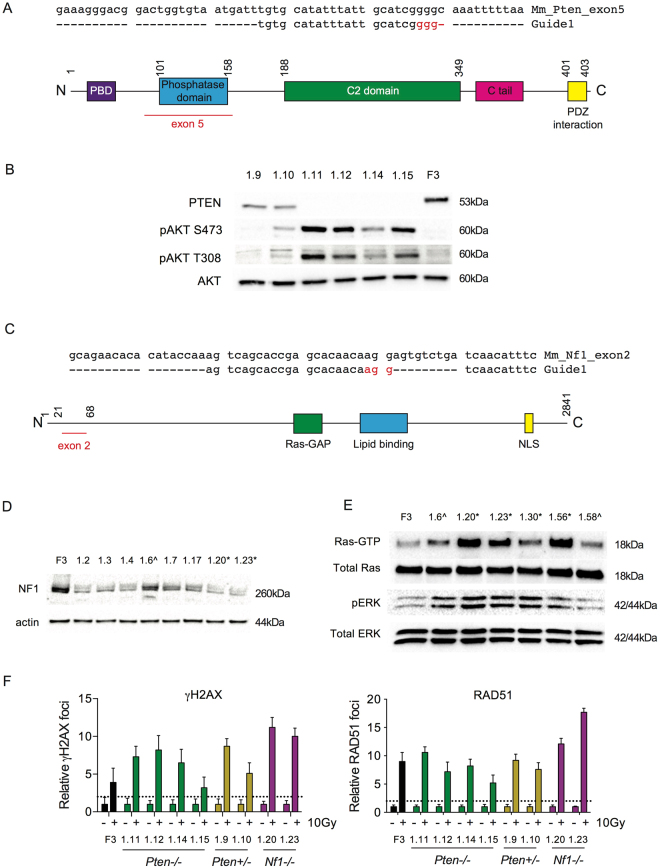
We targeted exon 5 of Pten, encoding the phosphatase domain (Fig. 2A). Four clones with bi-allelic deletions (clones 1.11, 1.12, 1.14, 1.15) and two with single allele deletions (clones 1.9, 1.10) were isolated - deletions ranged from 2 bp to >400 bp (Fig. S3). Immunoblots showed absent PTEN in Trp53 −/−;Pten −/− cells compared to F3, with reduced levels in the heterozygote clones (Fig. 2B). Bi-allelic Pten loss resulted in marked increases in AKT phosphorylation at both S473 and T308 upon serum starvation (Fig. 2B). AKT phosphorylation was less marked in one of the heterozygote clones (1.10) than in Trp53 −/−;Pten −/−, although still greater than in F3 control.
Figure 2.
Generation of Trp53 −/−;Nf1 −/− and Trp53 −/−;Pten −/− ID8 cells. (A) Design of guide RNA targeted to exon 5 of Pten. Nucleotides in red represent PAM sequence. Schematic representation of PTEN protein with exon 5 highlighted in red (bottom). Numbers represent amino acid position. (B) Immunoblot for PTEN and phospho-AKT in clones following Pten gRNA transfection. Clones 1.11, 1.12, 1.14 and 1.15 had bi-allelic Pten deletions and showed absent PTEN protein with phosphorylation of AKT at both S473 and T308 following serum starvation. Clones 1.9 and 1.10 had mono-allelic deletions. F3 = ID8 Trp53 −/−. (C) Design of guide RNA targeted to exon 2 of Nf1 as previously29. Nucleotides in red represent PAM sequence. Schematic representation of Nf1 protein with exon 2 highlighted in red (bottom). Numbers represent amino acid positions. (D) Immunoblot for Nf1 in clones following Nf1 gRNA transfection. *; clones 1.20 and 1.23 had bi-allelic indels confirmed by Sanger sequencing, ^; clone 1.6 had single allele deletion. Clones 1.2, 1.3, 1.4, 1.7 and 1.17 were not sequenced. (E) Ras-GTP co-immunoprecipitation and phospho-ERK immunoblot in ID8 clones following Nf1 gRNA transfection. *; clones 1.20, 1.23, 1.30 and 1.56 had confirmed bi-allelic indels and demonstrated increased Ras-GTP pulldown and ERK phosphorylation suggestive of activated Ras signalling. ^; clones 1.6 and 1.58 had single allele deletions. (F) ID8 Trp53 −/−;Pten −/−, Trp53 −/−;Nf1 −/− and Trp53 −/−;Pten +/− cells were irradiated (10 Gy), fixed and stained for γH2AX and RAD51, and counterstained with DAPI. RAD51 foci were counted in up to 30 untreated and irradiated cells. Bars represent foci per cell (mean +/− SEM); γH2AX (left) and RAD51 (right); dotted lines represents two-fold increase in γH2AX and RAD51 foci/cell relative to untreated cells as above.
For Nf1, we targeted exon 2 (Fig. 2C), and used a combination of NF1 expression on immunoblot (Fig. 2D), Raf-RBD co-immunoprecipitation and ERK phosphorylation (Fig. 2E) to screen clones. Clones 1.20, 1.23, 1.30 and 1.56 all had confirmed bi-allelic deletions on Sanger sequencing, whilst 1.6 and 1.58 had single allele changes. We selected two clones (1.20 and 1.23; see Fig. S4 for sequencing data) for further evaluation as they demonstrated increased GTP-bound RAS compared to F3, indicative of activated RAS signalling, as well as increased ERK phosphorylation on immunoblot (Fig. 2E). All Trp53 −/−;Nf1 −/−, Trp53 −/−;Pten −/− and Trp53 −/−;Pten +/− cells were HR competent in 10 Gy irradiation experiments (Figs 2F and S5).
Generation of triple-deleted ID8 lines
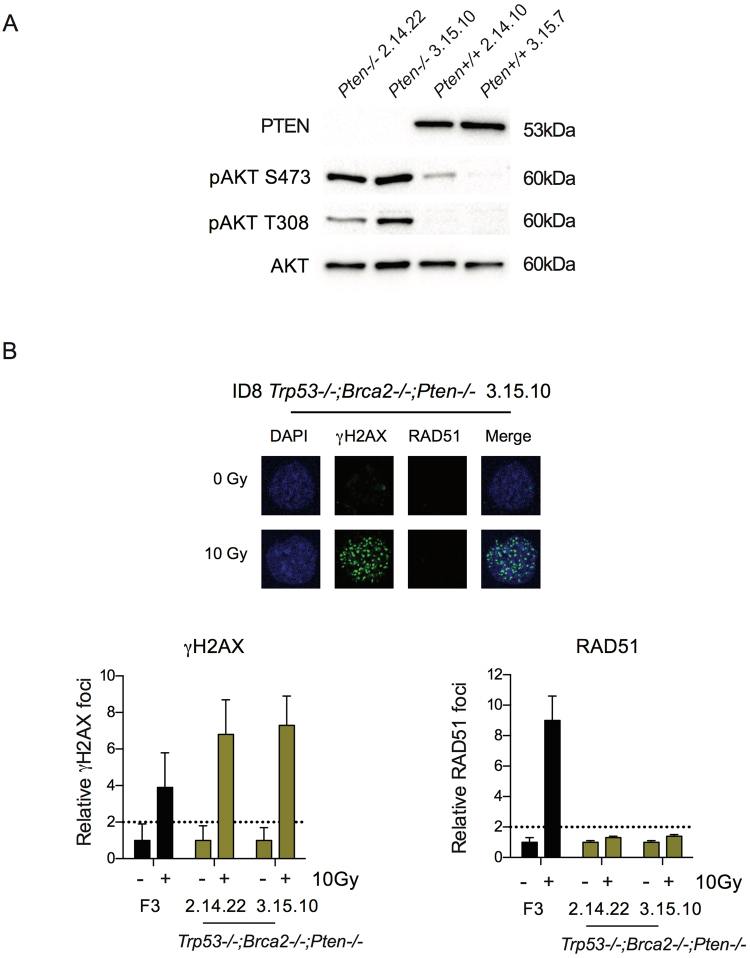
Multiple genomic abnormalities can co-exist within one HGSC tumour5,17. In addition, several of the GEMM require deletion of three genes for reliable tumorigenesis11,12. Therefore, we elected to target Pten in two of our previous Trp53 −/−;Brca2 −/− clones (2.14 and 3.1514) to generate triple deleted lines. Two clones with bi-allelic alterations (Fig. S6) were generated (2.14.22 and 3.15.10) or no alterations (2.14.10, 3.15.7). The triple deleted clones lacked PTEN expression on immunoblot (Fig. 3A), and again showed marked increases in AKT phosphorylation at both S473 and T308 upon serum starvation (Fig. 3A). Of note, the triple-deleted cells did not generate RAD51 foci formation in response to 10 Gy irradiation, suggesting that absent PTEN expression did not alter the defective HR induced by Brca2 loss (Fig. 3B).
Figure 3.
Generation of triple-deleted Trp53 −/−;Brca2 −/−;Pten −/− ID8 cells. (A) Immunoblot for PTEN and phospho-AKT following overnight serum starvation in clones isolated following Pten gRNA transfection. Clones 2.14.22 and 3.15.10, with bi-allelic Pten indels, showed absent PTEN expression and increased phosphorylation of AKT at both S473 and T308 compared to the two guide control clones, 2.14.10 and 3.15.7 that had no detectable change in Pten sequence. (B) ID8 Trp53 −/− and Trp53 −/−;Brca2 −/−;Pten −/− cells were irradiated (10 Gy), fixed and stained for γH2AX and RAD51, and counterstained with DAPI. RAD51 foci were counted in up to 30 untreated and irradiated cells. Bars represent foci per cell (mean +/− SEM); γH2AX (left) and RAD51 (right); dotted lines represent two-fold increase in γH2AX and RAD51 foci/cell relative to untreated cells as above.
In vivo tumorigenesis
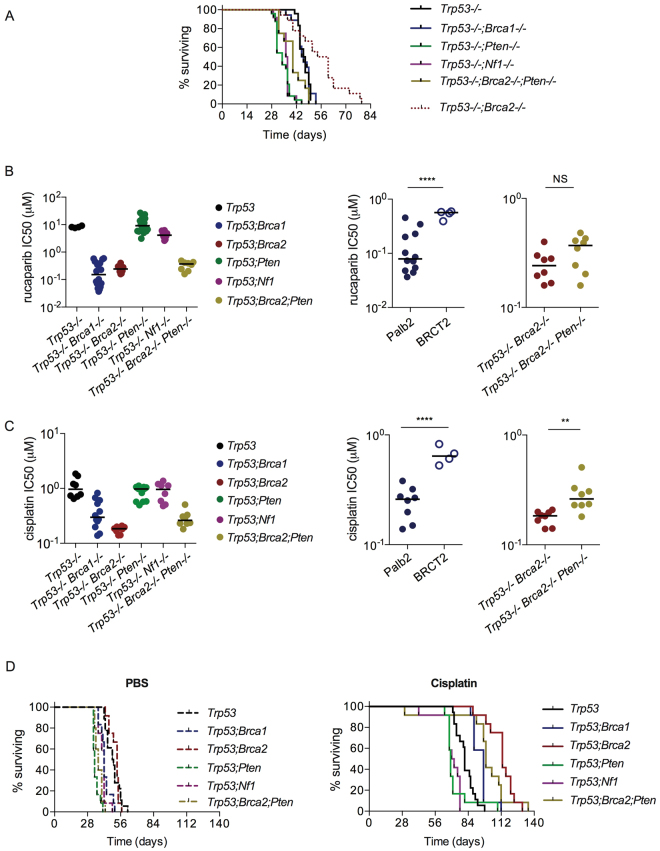
We then assessed intraperitoneal growth in female C57Bl/6 mice using at least two separate clones for each new genotype (Fig. 4A). Previously we showed that loss of p53 function significantly increased the rate of growth of intraperitoneal ID8 compared to parental controls, whilst additional loss of BRCA2 expression slowed growth relative to p53 loss alone. Results here showed that there was no difference in survival for mice bearing Trp53 −/−;Brca1 −/− tumours compared to Trp53 −/− (median time to reach humane endpoint 46 and 47 days respectively, p = NS), but deletion of either Pten or Nf1 expression significantly accelerated growth compared to p53 loss alone (Trp53 −/−;Pten −/− 34 days, p < 0.0001; Trp53 −/−;Nf1 −/− 36.5 days, p < 0.0001). Survival of mice bearing heterozygote Trp53 −/−;Pten +/− tumours (40.5 days) lay between that of Trp53 −/− and Trp53 −/−;Pten −/−, although differences were not significant (Fig. S7). The triple-deleted tumours showed accelerated growth (median time to reach humane endpoint 40 days) compared to both Trp53 −/− (p = 0.003) and Trp53 −/−;Brca2 −/− p = 0.0002), but slower than Trp53 −/−;Pten −/− (p = 0.0024).
Figure 4.
In vivo growth of Trp53 −/−;Brca1 −/−, Trp53 −/−;Nf1 −/− and Trp53 −/−;Pten1 −/− ID8 cells; platinum and PARP inhibitor sensitivity. (A) Cells (5 × 106) were injected intraperitoneally into female C57Bl/6 mice in groups of six. Two different clones were used per genotype for Trp53 −/−;Brca1 −/−, Trp53 −/−;Nf1 −/−, Trp53 −/−;Pten −/− and Trp53 −/−;Brca2 −/−;Pten −/− tumours: a single clone (F3) was used for Trp53 −/−−/− data. Mice were killed when they reached humane endpoints. The data for Trp53 −/−;Brca2 −/− have been published previously14 but are presented here for illustrative purposes. (B) Cell sensitivity to PARP inhibition. At least two clones were used per genotype, except Trp53 −/−, where a single clone (F3) was used. Each dot represents one triplicate experiment. Bars represent median. Right; results for Trp53 −/−;Brca1 −/− by site of mutation - exon 12 (Palb2 domain - clones 1.26, 1.36) and exon 19 (BRCT-2 domain - clone 6.20), and Trp53 −/−;Brca2 −/− and Trp53 −/−;Brca2 −/−;Pten −/− cells. ****p < 0.0001, **p < 0.01. (C) Cell sensitivity to cisplatin. At least two clones were used per genotype, except Trp53 −/−, where the F3 clone was used. Each dot represents one triplicate experiment. Bars represent median. Right; results for Trp53 −/−;Brca1 −/− by site of mutation - exon 12 (Palb2 domain - clones 1.26, 1.36) and exon 19 (BRCT-2 domain - clone 6.20) – and for Trp53 −/−;Brca2 −/− and Trp53 −/−;Brca2 −/−;Pten −/− cells. ****p < 0.0001. (D) Cells (5 × 106) were injected intraperitoneally into female C57Bl/6 mice in groups of six. Two different clones were used per genotype for Trp53 −/−;Brca1 −/−, Trp53 −/−;Nf1 −/− and Trp53 −/−;Pten1 −/− tumours: a single clone (F3) was used for Trp53 −/− data. Mice received cisplatin (5 mg/kg) or PBS as intraperitoneal injections (200 µl) on days 28, 35 and 42. Mice were killed when they reached humane endpoints.
Platinum and PARP inhibitor sensitivity
We then investigated the effects of specific mutations in the ID8 cells upon sensitivity to the PARP inhibitor rucaparib in vitro, and to platinum chemotherapy both in vitro and in vivo. Both Trp53 −/−;Brca1 −/− and Trp53 −/−;Brca2 −/− cells were significantly more sensitive to rucaparib than F3 Trp53 −/− (Fig. 4B, Table 1), whilst loss of Pten and Nf1 function individually had no effect on rucaparib sensitivity. There was no overall difference between the sensitivity of Trp53 −/−−/−;Brca1 −/− and Trp53 −/−;Brca2 −/− cells. However, the Brca1 BRCT-2 domain clone (6.20) was significantly less sensitive than both Brca1 Palb2-binding domain mutants (1.26, 1.36) and the Trp53 −/−;Brca2 −/− cells, which also have a deletion in the Palb2-binding domain (Fig. 4B). Loss of PTEN expression did not have a significant impact upon rucaparib sensitivity in the Trp53 −/−;Brca2 –/−;Pten −/− cells compared to Trp53 −/−;Brca2 −/− (Fig. 4B).
Table 1.
Summary of the sensitivity of ID8 clones to rucaparib and platinum.
| Genotype | Rucaparib IC50 (µM) mean +/− sd | p= | Cisplatin IC50 (µM) mean +/− sd | p= |
|---|---|---|---|---|
| Trp53 −/− | 8.23 +/− 0.62 | — | 1.10 +/− 0.47 | — |
| Trp53 −/−;Brca1 −/− | 0.24 +/− 0.21 | <0.01 | 0.38 +/− 0.22 | <0.0001 |
| Trp53 −/−;Brca2 −/− | 0.25 +/− 0.08 | <0.01 | 0.18 +/− 0.03 | <0.0001 |
| Trp53 −/−;Pten −/− | 11.2 +/− 6.94 | NS | 0.89 +/− 0.21 | NS |
| Trp53 −/−;Nf1 −/− | 4.31 +/− 1.33 | NS | 0.90 +/− 0.35 | NS |
| Trp53 −/−;Brca2 −/−;Pten −/− | 0.33 +/− 0.12 | <0.01 | 0.28 +/− 0.10 | <0.0001 |
Data represent IC50 averages from 4–8 triplicate experiments per cell line. IC50 values were compared using one-way ANOVA with Bonferroni’s test for multiple comparisons with Trp53 −/− cells as comparator.
A similar pattern was seen for cisplatin in vitro. Both Trp53 −/−;Brca1 −/− and Trp53 −/−;Brca2 −/− cells were significantly more sensitive than F3 Trp53 −/− cells, whilst loss of PTEN and NF1 expression again had no effect (Fig. 3C, Table 1). As with rucaparib, the Palb2-binding domain mutants in both Brca1 and Brca2 were more sensitive than the Brca1 BRCT-2 mutant (Fig. 4C). Interestingly, there was a significant reduction in cisplatin sensitivity in the triple deleted Trp53 −/−;Brca2 −/−;Pten −/− cells compared to Trp53 −/−;Brca2 −/− (Fig. 4C).
In vivo, there was a wide variation in survival following three doses of intraperitoneal cisplatin (5 mg/kg on days 28, 35 and 42 only). Mice bearing control Trp53 −/− tumours took a median of 81 days to reach humane endpoints (Fig. 4D, Table 2). Trp53 −/−;Pten −/− and Trp53 −/−;Nf1 −/− tumours produced the worst survival (median 69 and 71 days respectively; p < 0.0001 and p = 0.0001 respectively compared to Trp53 −/−). For Trp53 −/−;Brca1 −/−, survival was extended to 97 days (p = 0.0003 compared to Trp53 −/−), whilst the longest survival was seen with Trp53 −/−;Brca2 −/− tumours (median 113 days), which was significantly longer than both Trp53 −/− and Trp53 −/−;Brca1 −/− (p < 0.0001 for both comparisons). Mice bearing the triple-deleted Trp53 −/−;Brca2 −/−;Pten −/− tumours survived 99 days, which was significantly more than Trp53 −/− (p < 0.0001; Table 2) but less than Trp53 −/−;Brca2 −/−, although this latter comparison did not reach statistical significance (p = 0.078). In the Trp53 −/−;Brca1 −/− experiments, median survival for the PALB2 mutant (clone 1.36) was longer (97 days) than for the BRCT-2 mutant (clone 6.20; 89 days), although again this did not reach statistical significance (p = 0.066) (Fig. S8).
Table 2.
Summary of survival following cisplatin treatment in vivo.
| Genotype | Median survival cisplatin (days) | Hazard ratio (log-rank) | p= |
|---|---|---|---|
| Trp53 −/− | 81 | — | — |
| Trp53 −/−;Brca1 −/− | 97 | 0.34 | 0.0003 |
| Trp53 −/−;Brca2 −/− | 113 | 0.23 | <0.0001 |
| Trp53 −/−;Pten −/− | 69 | 4.07 | <0.0001 |
| Trp53 −/−;Nf1 −/− | 71 | 3.19 | 0.0001 |
| Trp53 −/−;Brca2 −/−;Pten −/− | 99 | 0.29 | <0.0001 |
Data represent median survival (time to reach humane endpoint) for mice bearing ID8 clones treated with cisplatin (5 mg/kg) on days 28, 35 and 42. Data were compared using log-rank test.
Trp53 knockout in Dicer−/−;Pten−/− DKO cells
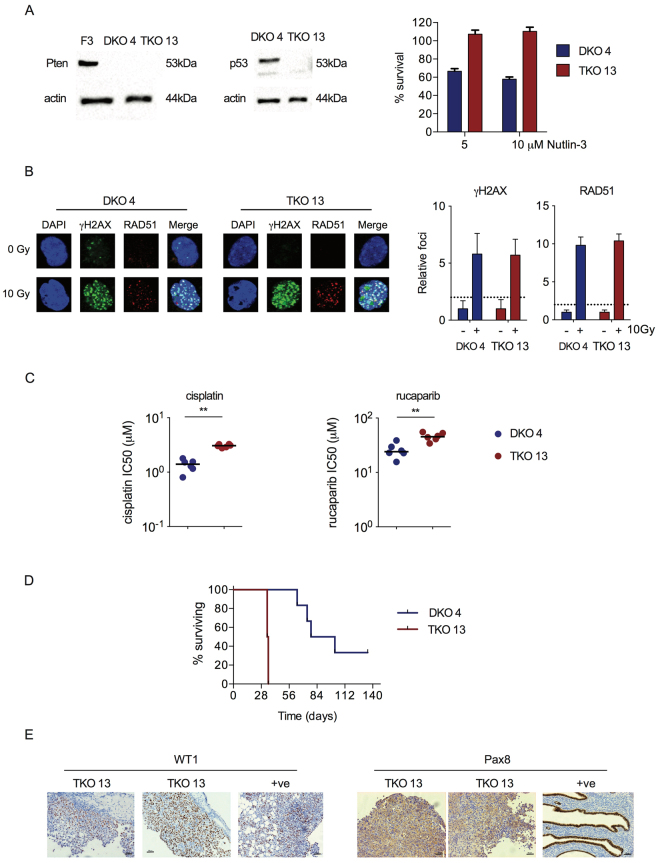
Finally, we targeted Trp53 in OvidT 497 Dicer −/−;Pten −/− DKO fallopian tube carcinoma cells using the same guide RNA used to generate ID8 Trp53 −/− clone F3. Triple knockout (TKO; Dicer −/−;Pten −/−;Trp53 −/−) clone 13 had a biallelic deletion in exon 5 (Fig. S8), with absent p53 on immunoblot, and increased resistance to Nutlin-3 compared to cells from a DKO control clone (clone 4) that had been transfected with the Trp53 gRNA but contained no Trp53 mutation (Fig. 5A). Both DKO and TKO cells retained functional HR (Fig. 5B). However, interestingly, Trp53 loss induced small but statistically significant changes in sensitivity to both cisplatin and rucaparib in TKO cells in vitro (Fig. 5C). Loss of p53 function produced a highly significant reduction in time to reach humane endpoints - median survival (Fig. 5D; median 34.5 days vs 90 days; p < 0.0001). In vivo, TKO tumours demonstrated nuclear WT1 staining (Fig. 5E). However, there was only weak cytoplasmic PAX8 staining, in contrast to strong nuclear staining seen in fallopian tube epithelial cells (Fig. 5E).
Figure 5.
Generation and evaluation of Dicer −/−;Pten −/−;Trp53 −/− TKO cells. (A) OvidT 497 Dicer −/−;Pten −/− (DKO) cells were transfected with PX459 encoding Trp53 gRNA. Clone 4 contained no Trp53 mutation; clone 13 (TKO) contained bi-allelic Trp53 exon 5 mutations. Expression of PTEN and p53 was assessed by immunoblot (left). F3 = ID8 Trp53 −/−. Sensitivity to Nutlin-3 was assessed by MTT assay (right). (B) Homologous recombination was assessed in DKO 4 and TKO 13 cells as previously. (C) Sensitivity of DKO 4 and TKO 13 cells to cisplatin. Each dot represents one triplicate experiment. Bars represent median. *p < 0.01. (D) Cells (5 × 106) were injected intraperitoneally into female C57Bl/6 mice in groups of six. Mice were killed when they reached humane endpoints. Excised tumours were fixed in formalin and stained for WT1 and PAX8. Each TKO 13 section comes from a separate mouse. Positive controls (+ve) are ID8 tumour (WT1) and normal mouse fallopian tube (PAX8), both from14. Bars represent 50 µm.
Discussion
Here we have extended our previous results, and generated further derivatives of the ID8 murine ovarian carcinoma model using CRISPR/Cas9 gene editing. Using one of our previous Trp53 −/− clones, we have generated further double mutants, with deletions in Brca1, Pten and Nf1 in addition to loss Trp53, as well as triple mutants lacking Trp53, Brca2 and Pten. We have also generated a Trp53 mutant derivative of a transplantable murine fallopian tube carcinoma cell line. Collectively, our results indicate that loss of genes known to be mutated or deleted in HGSC can alter intraperitoneal tumour growth and response to therapy. Moreover, the responses to therapy mirror those seen in patients, reinforcing the utility of these models.
As expected, we show that loss of BRCA1 or BRCA2 function induces defective homologous recombination and sensitises cells to both platinum and PARP inhibitor therapy. There was no overall difference in rucaparib sensitivity between Brca1 −/− or Brca2 −/− cells, which accords with results from part 1 of the ARIEL2 trial of rucaparib, in which radiological response rates were almost identical in BRCA1 (79%) and BRCA2-mutated (82%) tumours8. However, our data show that mice bearing Trp53 −/−;Brca2 −/− tumours have the longest survival following platinum treatment. It is now clear that patients with germline BRCA2 mutations have significantly greater long term survival than patients without mutations and even those with germline BRCA1 mutations6, which may reflect extreme sensitivity to platinum-based chemotherapy.
Our results suggest that the location of the mutation within Brca1 might influence sensitivity - the two clones with mutations in the PALB2-binding domain were significantly more sensitive to both cisplatin and rucaparib in vitro than the BRCT-2 domain mutant, although the difference in survival following in vivo platinum treatment was not significant (p = 0.066). In patients with HGSC, it remains unclear whether the type and location of mutations within BRCA1 and BRCA2 influence response to treatment or overall survival. The Australian Ovarian Cancer Study found no overall effect of location within the gene, nor of mutation type, on relapse or survival18. However, the sample size (n = 134 mutation carriers) was possibly too small to allow enrichment of mutations in one specific region. A retrospective analysis of 445 women with germline BRCA1/2 mutations (79% of whom had had breast and/or ovarian cancer) found a non-significant (p = 0.06) reduction in overall survival in those with BRCA1 exon 20 mutations compared to those with mutations in BRCA1 exons 2 or 1119. However, again the number of cases with mutation in a specific exon was low. By contrast, in a transgenic murine breast cancer model, nonsynonymous missense mutation in the Brca1 RING domain (C61G) was associated with poorer response to cisplatin and PARP inhibitor therapy than a complete Brca1 knockout, despite similar tumour morphology and copy number alterations20. Large consortia and meta-analyses will help to address genotype/phenotype relationships, especially relating to survival, but our cells may help to answer specific questions relating to protein function and chemotherapy sensitivity, and CRISPR/Cas9 technology allows for further exon-specific deletions to be created.
One striking feature of our results was the increased rate of intra-tumoural growth and reduced survival following platinum treatment of tumours with double Trp53;Pten or Trp53;Nf1 mutations compared to those with Trp53 mutations alone. Moreover, additional loss of PTEN expression significantly accelerated the growth of Trp53 −/−;Brca2 −/− tumours, and significantly reduced platinum sensitivity in vitro. There are few clear prognostic data on the influence of NF1 loss on HGSC patient survival, but our results correlate with data showing that loss of PTEN expression is associated with poor outcome9. We also show clearly that loss of PTEN expression alone does not induce defective homologous recombination in ID8 cells, although the additional loss of PTEN does not abrogate defective homologous recombination or PARP inhibitor sensitivity seen in Trp53 −/−;Brca2 −/− cells. We found OvidT 497 Dicer −/−;Pten −/− DKO cells to be HR competent, as defined by radiation-induced RAD51 foci formation, and that loss of p53 function in the Dicer −/−;Pten −/−;Trp53 −/− TKO cells induced a degree of both platinum and PARP inhibitor resistance. This contrasts with data from HCT116 cells21 and from endometrioid endometrial carcinoma22, whereby loss of PTEN was associated with partial loss of HR and sensitivity to PARP inhibition. In PTEN-deficient prostate carcinoma cells, treatment with a PARP inhibitor induced apoptosis but only in the absence of p5323. Overall, these data imply that the relationship between PTEN, p53 and DNA double strand break repair is complex and varies by tumour type; however, our results do suggest that classification of PTEN-mutant HGSC as HR defective in the absence of mutations in BRCA1 or BRCA2 4 may be erroneous.
The Dicer −/−;Pten −/−;Trp53 −/− TKO cells also demonstrate clearly that loss of p53 function significantly enhances intra-peritoneal tumour growth. This is in keeping with our original ID8 work14 and data from the Dicer −/−;Pten −/−;Trp53 LSL-R172H/+ TKO mice, in which expression of mutant p53 resulted in a more aggressive phenotype than Dicer −/−;Pten −/− alone24. Unfortunately, no transplantable cell lines have been described from the Dicer −/−;Pten −/−;Trp53 LSL-R172H/+ TKO mice with which to allow comparison of tumours lacking p53 expression with those harbouring a potential gain-of-function point mutation. One potential criticism of the ID8 model is that it is likely to be of ovarian origin, whereas the majority of HGSC cases arise from precursor lesions within the distal fallopian tube25,26. The Dicer −/−;Pten −/− DKO represents a novel model of tubal carcinogenesis, although the exact cell of origin remains unclear15. Our immunohistochemistry show the DKO and TKO tumours to express WT1 but not PAX8, suggesting that they have either lost PAX8 expression during tumorigenesis, or may not arise in the secretory cells of the tubal fimbria– interestingly, in transgenic Pten −/−;Trp53 LSL-R172H/+ mice, tumours were still able to form following removal of the fallopian tumours, suggesting that HGSC can arise in the ovaries24.
In conclusion, we have generated new transplantable murine models that recreate further key mutations of HGSC. These cell lines will be powerful tools, alongside transgenic models and primary patient material, to elucidate HGSC biology and chemotherapy resistance, and all of the ID8 lines are freely available to other researchers upon request.
Materials and Methods
Cells
The production of ID8 Trp53 −/− cells was described previously14. OvidT 479 cells were isolated from tumours arising in Dicer-Pten double knockout (DKO) mice (Amhr2 cre/+ Dicer flox/flox Pten flox/flox) as previously described15,27.
CRISPR/Cas9 and selection
Guide RNAs targeting the PALB2-binding domain in exon 12 and the BRCT-2 domain in exon 19 of Brca1 were designed using two open-access software programs, CHOPCHOP (https://chopchop.rc.fas.harvard.edu/) and CRISPR design (http://crispr.mit.edu/), and ligated into BbsI-linearized pSpCas9(BB)-2A-Puro [PX45928, a gift from Feng Zhang via Addgene]. The gRNA for Pten was a gift from Douglas Strathdee (CRUK Beatson Institute, Glasgow, UK) and targeted exon 5, encoding the phosphatase domain. The gRNA for Nf1 was designed to target exon 2 as previously described29. gRNAs for Pten and Nf1 were ligated into PX33030 (a gift from Walter Jackson via Addgene, ref 78621).
Cells were transfected as previously14, omitting puromycin selection for Pten and Nf1 targeting. PCR primers spanning target sites of deletion are listed in Table S1. PCR products were cloned using InFusion kit (ClonTech) and clones with large PCR deletions were selected for subsequent analysis. Remaining clones were screened using the Surveyor Nuclease Assay (Integrated DNA Technology). Mutations were confirmed by Sanger sequencing. All sequence alignment was performed using MAFFT version 7 (http://mafft.cbrc.jp/).
γH2AX/Rad51 assay
24 h treatment following treatment with 10 µM rucaparib (Clovis Oncology, Boulder, CO), or following 10 Gy irradiation, cells were fixed and stained for γH2AX and Rad51 foci as previously14 using a Zeiss 710 confocal microscope and Zen software (Zeiss). Microscope settings were as follows: 20x objective, 1024 × 1024 frame, 6.25 s scan time. Lasers used were 561 nm (red), 488 nm (green) and 405 nm (DAPI). Fluorophores were ALEXA-568 (for RAD51) and ALEXA-488 (for γH2AX). Foci were counted in at least 30 nuclei per condition without any image manipulation. Images presented in figures were captured from raw Zen files using Photoshop CS5 v12.1 software (Abode), and processed in their entirety using the ‘AutoContrast’ tool, and then cropped to show single representative cells. Classification of homologous recombination competence was performed as previously16.
Immunoblot and co-immunoprecipitation
For detection of phospho-AKT, cells were serum-starved for 16 hours prior to lysis. Antibodies used were: phospho-AKT (T308) (Cell Signalling 13038), phospho-AKT (S473) (Cell Signaling 40600), AKT (Cell Signalling 4685) and PTEN (Cell Signalling 9552). Co-immunoprecipitation of Ras-GTP was performed using Ras pulldown activation assay kit (BK008-S; Cytoskeleton, Denver CO, USA). 5 × 105 cells were plated on 10 cm dishes for 72 hours, then serum-starved for 16 hours prior to lysis. All subsequent steps were performed according to manufacturer’s instructions. The same lysates were probed for phospho-ERK (Cell Signalling 4695) and total ERK.
All immunoblot images were acquired using a BioRad ChemiDoc MP imaging system and Image Lab 5.0 software (both BioRad, Watford UK). Images were visualised using Photoshop CS5 v12.1 software (Abode), and processed in their entirety using the ‘AutoContrast’ tool, and then cropped to include relevant lanes only. All immunoblot images presented derive from single blots and no Photoshop touch-up tools were used in any image. All loading controls come from blots stripped and re-probed.
In vitro cytotoxicity
ID8 cells (3 × 104 cells/well of a 24 well plate) were treated with 3 nM–30 μM rucaparib (Clovis Oncology, Boulder CO, USA) or 10 nM–1000 μM cisplatin (Accord Healthcare, Harrow, UK via Beatson West of Scotland Cancer Centre chemotherapy pharmacy) 4 hours after initial plating. 68 hours thereafter, cell survival was assessed using sulphorhodamine B (rucaparib) or MTT31 (cisplatin) assay.
In vivo experiments and immunohistochemistry
All experiments complied with the UK welfare guidelines32 and were conducted under specific personal and project license authority (70/8645) with ethical approval (University of Glasgow) in dedicated animal facilities. 5 × 106 cells were inoculated intraperitoneally (IP) in 6–8 week old female C57Bl/6 mice (Charles River Laboratories, UK) in groups of 6. A minimum of two clones per genotype were used in all experiments. Mice were monitored regularly and killed upon reaching UK Home Office limits. All decisions about animal welfare and experiment endpoints were made by D.A. or S.M. independently of main study investigators to prevent bias. Cisplatin (5 mg/kg in 200 µl PBS) or PBS was administered intraperitoneally on days 28, 35 and 42 only. Ascites was collected and all visible tumour deposits dissected out, and either snap frozen or fixed in neutral-buffered 4% paraformaldehyde. 5 µm sections from formalin-fixed paraffin-embedded tumours were stained (Dako Autostainer, Dako, UK). The following antibodies were used for immunohistochemistry - WT1 (Can (R9), Abcam, 1:250); PAX8 (ZR-1, Zeta, 1:30). Images were captured on a Zeiss Observer Z1 microscope (20x objective) using Zen software (Zeiss), visualised using Photoshop CS5 v12.1 software (Abode), and processed in their entirety using the ‘AutoContrast’ tool.
Statistics
All analyses were performed using Prism v6.0 (Graphpad, CA). One-way ANOVA with Bonferroni’s test for multiple comparisons and unpaired t-test were used to compare IC50 values. Mouse survival was compared using the log-rank test. p < 0.05 was considered significant.
Electronic supplementary material
Acknowledgements
This work was funded by the University of Glasgow, Cancer Research UK [grants C16420/A12995 (I.A. McNeish), C608/A15973 (I.A. McNeish)] and Marsha Rivkin Cancer Research Center. All animal work was performed in the Biological Services Unit facilities at the Cancer Research UK Beatson Institute (Cancer Research UK grant C596/A17196).
Author Contributions
Study design: J.B.W., I.A. McN. Experimental conduct: J.B.W., M.F., S.M., J.P., B.K., S.D., D.S., K.B. Supervision: D.M., M.M., J.K., S.C., K.B., I.A. McN. Writing: J.B.W., I.A. McN. Acquisition of funding: I.A. McN. All authors read and approved the manuscript prior to submission.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
A correction to this article is available online at https://doi.org/10.1038/s41598-018-24099-3.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17119-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ahmed AA, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macintyre, G. et al. Copy-number signatures and mutational processes in ovarian carcinoma. bioRxiv, 10.1101/174201 (2017). [DOI] [PMC free article] [PubMed]
- 4.TCGA Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patch A-M, et al. Whole–genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 6.Candido Dos Reis FJ, et al. Germline mutation in BRCA1 or BRCA2 and ten-year survival for women diagnosed with epithelial ovarian cancer. Clin Cancer Res. 2015;21:652–657. doi: 10.1158/1078-0432.CCR-14-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pennington KP, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swisher EM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 9.Martins FC, et al. Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome biology. 2014;15:526. doi: 10.1186/s13059-014-0526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaughan, S. et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat. Rev. Cancer11, 719–725, nrc3144 [pii]10.1038/nrc3144 [doi] (2011). [DOI] [PMC free article] [PubMed]
- 11.Perets R, et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in brca;tp53; pten models. Cancer Cell. 2013;24:751–765. doi: 10.1016/j.ccr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhai, Y. et al. High-grade serous carcinomas arise in the mouse oviduct via defects linked to the human disease. J Pathol, 10.1002/path.4927 (2017). [DOI] [PMC free article] [PubMed]
- 13.Roby KF, et al. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis. 2000;21:585–591. doi: 10.1093/carcin/21.4.585. [DOI] [PubMed] [Google Scholar]
- 14.Walton J, et al. CRISPR/Cas9-mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high grade serous carcinoma. Cancer Res. 2016;76:6118–6129. doi: 10.1158/0008-5472.CAN-16-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, et al. High-grade serous ovarian cancer arises from fallopian tube in a mouse model. Proc Natl Acad Sci USA. 2012;109:3921–3926. doi: 10.1073/pnas.1117135109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukhopadhyay A, et al. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin. Cancer Res. 2010;16:2344–2351. doi: 10.1158/1078-0432.CCR-09-2758. [DOI] [PubMed] [Google Scholar]
- 17.Bashashati A, et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol. 2013;231:21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alsop, K. et al. BRCA Mutation Frequency and Patterns of Treatment Response in BRCA Mutation-Positive Women With Ovarian Cancer: A Report From the Australian Ovarian Cancer Study Group. J Clin Oncol30, 2654-2663, doi:JCO.2011.39.8545 [pii]10.1200/JCO.2011.39.8545 [doi] (2012). [DOI] [PMC free article] [PubMed]
- 19.Bayraktar S, et al. Genotype-Phenotype Correlations by Ethnicity and Mutation Location in BRCA Mutation Carriers. The breast journal. 2015;21:260–267. doi: 10.1111/tbj.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drost, R. et al. BRCA1 RING Function Is Essential for Tumor Suppression but Dispensable for Therapy Resistance. Cancer Cell20, 797–809, doi:S1535-6108(11)00438-7 [pii] 10.1016/j.ccr.2011.11.014 [doi] (2011). [DOI] [PubMed]
- 21.Mendes-Pereira AM, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol. Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dedes KJ, et al. PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci Transl Med. 2010;2:53ra75. doi: 10.1126/scitranslmed.3001538. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Billalabeitia E, et al. Vulnerabilities of PTEN-TP53-deficient prostate cancers to compound PARP-PI3K inhibition. Cancer Discov. 2014;4:896–904. doi: 10.1158/2159-8290.CD-13-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Coffey DM, Ma L, Matzuk MM. The ovary is an alternative site of origin for high-grade serous ovarian cancer in mice. Endocrinology. 2015;156:1975–1981. doi: 10.1210/en.2014-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piek JM, et al. Dysplastic changes in prophylactically removed Fallopian tubes of women predisposed to developing ovarian cancer. J Pathol. 2001;195:451–456. doi: 10.1002/path.1000. [DOI] [PubMed] [Google Scholar]
- 26.Lee Y, et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol. 2007;211:26–35. doi: 10.1002/path.2091. [DOI] [PubMed] [Google Scholar]
- 27.Hua Y, et al. Epithelialization of mouse ovarian tumor cells originating in the fallopian tube stroma. Oncotarget. 2016;7:66077–66086. doi: 10.18632/oncotarget.11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuckermann M, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaczmarczyk L, Mende Y, Zevnik B, Jackson WS. Manipulating the Prion Protein Gene Sequence and Expression Levels with CRISPR/Cas9. PLoS One. 2016;11:e0154604. doi: 10.1371/journal.pone.0154604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 32.Workman, P. et al. Guidelines for the welfare and use of animals in cancer research. Br. J. Cancer102, 1555-1577, doi:6605642 [pii] 10.1038/sj.bjc.6605642 [doi] (2010). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.