Abstract
There are no blood biomarkers for the diagnosis of renal cell carcinoma (RCC) in routine clinical use. We focused on the gene expression profile of peripheral blood cells obtained from RCC patients to discover novel biomarkers for RCC diagnosis. Using microarray analysis and quantitative verification, CXCL7 was shown to be significantly upregulated in the peripheral blood cells of RCC patients. Importantly, aberrant CXCL7 expression was confirmed even in peripheral blood cells obtained from early stage (pT1a) RCC patients, and the expression level of CXCL7 in peripheral blood cells was a potential independent biomarker for the diagnosis of RCC by receiver operating characteristic curve analysis (sensitivity, 70.0%; specificity, 64.0%; area under the curve = 0.722; multiple logistic regression analysis: odds ratio, 1.07; 95% confidence interval, 1.03–1.11; P = 0.0004). Moreover, CXCL7 expression in peripheral blood cells significantly decreased after resection of the primary tumor. CXCL7 is more highly expressed in PBMCs than in neutrophils from both healthy controls and RCC patients. Interestingly, CXCL7 expression in PBMCs from healthy volunteers was significantly elevated following coculture with RCC cells compared to those cocultured with normal cells as a control. These results suggest that aberrant CXCL7 expression in peripheral blood cells is induced by RCC cells and may serve as a novel biomarker in the diagnosis of RCC.
Keywords: Biomarker, blood, CXCL7, diagnosis, renal cell carcinoma
Kidney cancer is one of the most common cancers, accounting for over 140 000 deaths worldwide each year, with renal cell carcinoma (RCC) accounting for nearly 90% of all cases.1 It accounts for approximately 3% of adult malignancies and 90–95% of neoplasms arising from the kidney.1 Most RCC patients do not present with canonical symptoms, such as flank pain or macrohematuria, and it is important to detect early stage RCC patients using non‐invasive diagnostic methods along with ultrasonography or computed tomography.
Due to the delayed appearance of RCC‐related clinical signs, metastatic disease is present in 20–30% of patients at diagnosis, with treatment outcomes for such patients remaining inadequate.2 Therefore, novel methods allowing for early detection of RCC should greatly improve the prognosis of many RCC patients, and such methods may also be useful for monitoring metastatic RCC patients during systemic therapy.
Blood‐based tests, known as liquid biopsy, are non‐invasive and suitable for monitoring disease progression. Here, we focused on the gene expression profiles of peripheral blood cells obtained from RCC patients to discover novel biomarkers for RCC diagnosis using the PAXgene Blood RNA system (Becton Dickinson, Franklin Lakes, NJ, USA) to easily collect total RNA from peripheral blood cells.3 This is the first study to show that gene expression profiling of peripheral blood cells could provide biomarkers for the diagnosis of RCC.
Materials and Methods
Patients and blood samples
Whole blood (2.5 mL) was collected directly into PAXgene Blood RNA tubes (Becton Dickinson) and stored at −80°C. None of the patients had received systemic therapy, such as molecular‐targeted therapies or immunotherapies, before the collection of blood samples. Patients were staged according to the 7th AJCC TNM staging system.4 None of the healthy controls had a history of any other cancers. Post‐operation blood samples (n = 10) were collected at least 1 month after surgical resection of primary tumors to avoid the effect of surgical stress on gene expression in peripheral blood cells. This study was approved by the ethics committee of Osaka University Hospital (Suita, Japan) (#13397‐2).
RNA extraction from blood and microarray analysis
Total RNA was extracted from 204 blood samples according to the manufacturer's protocols. Total RNA (100 ng) was subjected to cDNA synthesis using the Ambion WT expression kit (Life Technologies, Carlsbad, CA, USA). cDNA was biotinylated and then fragmented with the GeneChip WT terminal labeling kit (Affymetrix, Santa Clara, CA, USA). After making the hybridization cocktail, biotinylated cDNAs were hybridized with GeneChip Human Transcriptome Array 2.0 (Affymetrix) for 16 h using 24 samples as the discovery cohort. After hybridization, the GeneChip was washed and stained using the GeneChip hybridization wash and stain kit (Affymetrix) and the GeneChip Fluidics Station 450 (Affymetrix). The GeneChip was then scanned by the GeneChip Scanner 3000 7G (Affymetrix). Partek Genomics Suite software was used for the data analysis (Partek, St. Louis, MO, USA).
Quantitative RT‐PCR analysis
Five hundred nanograms of RNA were subjected to cDNA synthesis using Prime Script RT Reagent Kit (Clontech, Mountain View, CA, USA), according to the manufacturer's instructions, in a final volume of 40 μL. Quantitative RT‐PCR (qRT‐PCR) was carried out with a Thermal Cycler Dice Real Time System TP800 (TaKaRa Bio, Shiga, Japan) using SYBR premix Ex Taq II (TaKaRa Bio). The PCR conditions were 95°C for 30 s followed by 40 cycles at 95°C for 5 s and 60°C for 30 s for each gene‐specific primer. Dissociation curves were generated at the end of the amplification to confirm the absence of primer‐dimer formations. GAPDH was used as a reference gene. Quantitative RT‐PCR analysis of CXCL7 and GAPDH expression was carried out with the following oligonucleotides: CXCL7 forward primer, 5′‐CTGGCTTCCTCCACCAAAGG‐3′; CXCL7 reverse primer, 5′‐GACTTGGTTGCAATGGGTTCC‐3′; GAPDH forward primer, 5′‐GCACCGTCAAGGCTGAGAAC‐3′; GAPDH reverse primer, 5′‐TGGTGAAGACGCCAGTGGA‐3′. The PCR reactions for each sample were carried out in triplicate.
Neutrophil isolation
Neutrophils were isolated from whole‐blood samples of healthy volunteers (n = 8) and RCC patients (n = 8) using Ficoll‐Paque Plus (GE Healthcare, Uppsala, Sweden) according to the manufacturer's procedure. The purity was typically 90–95% when assessed by H&E staining. In all experiments, cell viability, as assessed by the Trypan blue exclusion method, was over approximately 95%.
Peripheral blood mononuclear cell isolation
Peripheral blood mononuclear cells from individuals were isolated from whole‐blood samples (21 mL) and collected into BD Vacutainer CPT tubes with sodium citrate (Becton Dickinson) according to the standard procedure.
Cell culture and in vitro coculture experiment
Human kidney cancer cell lines 786‐O and Caki‐2 and the non‐malignant human kidney cell line HEK293T were purchased from ATCC (Manassas, VA, USA) and maintained in basal media (RPMI for 786‐O and Caki‐2, DMEM for HEK293T) supplemented with 10% FBS and 1% gentamicin–tyrosine solution (Invitrogen, Eugene, OR, USA) in a humidified incubator set to 37°C and 5% CO2. For coculture experiments, the three cell lines were split into 35‐mm dishes (Becton Dickinson) and incubated for 2 days to reach 80–90% confluence. Transwell inserts (0.4‐μm pore size; Corning, Corning, NY, USA) were then placed into the wells and 5 × 106 PBMCs from healthy volunteers were added to each Transwell insert. After 18 h, PBMCs were collected and lysed with Buffer RLT Plus with beta‐ME (RNeasy Plus Mini Kit; Qiagen, Venlo, Netherlands) and subjected to RNA extraction and qRT‐PCR.
Statistical analysis
Statistical analysis was undertaken using JMP12 (SAS Institute, Cary, NC, USA). Results are presented as mean ± SD, and data were compared using Wilcoxon test, Wilcoxon signed‐rank test, or Dunn's multiple comparison test. Receiver operating characteristic (ROC) curve analysis was used to generate area under the curve (AUC) values for selected genes to evaluate their predictive ability for the diagnosis of RCC. Univariate and multivariate logistic regression analyses were carried out to determine independent factors for the diagnosis of RCC. Cancer‐specific survival was calculated using the Kaplan–Meier method, and differences between groups were assessed by the log–rank test. Differences were considered statistically significant when the P‐value was < 0.05.
Results
CXCL7 was upregulated in peripheral blood cells of RCC patients
To identify an immunogenic signature in peripheral blood cells, we undertook gene microarray analysis using peripheral blood cells obtained from RCC patients compared to those from healthy controls. The discovery cohort included 12 RCC patients diagnosed at Osaka University Hospital between 2013 and 2014, with 11 being diagnosed as clear cell RCC (ccRCC) and one as papillary RCC, and 12 healthy volunteers as controls. Six of the RCC patients were early stage (stage I) patients and the others were advanced (stage III–IV) (Table 1). We identified a unique gene expression signature for RCC patients, with 15 genes showing significantly higher or lower expression in RCC patients compared to healthy controls (fold‐change ≥1.5 or ≤ −1.5, P < 0.05, signal intensity ≥7.00, protein coding gene) (Table 2). These results were confirmed by qRT‐PCR (Fig. S1). To identify which of these genes could be potential biomarkers for RCC diagnosis, ROC curve analysis was carried out using the discovery cohort samples. We found that CXCL7 was the candidate gene with the highest accuracy (AUC = 0.868, P < 0.001) for the diagnosis of RCC (Fig. 1).
Table 1.
Characteristics of renal cell carcinoma (RCC) patients and healthy controls
| Discovery cohort | Verification cohort | |||
|---|---|---|---|---|
| Healthy controls (n = 12) | RCC patients (n = 12) | Healthy controls (n = 50) | RCC patients (n = 130) | |
| Age, years (median) | 29–78 (64) | 36–79 (62) | 48–76 (66) | 27–83 (64) |
| Gender, male/female | 9/3 | 9/3 | 39/11 | 95/35 |
| Hb, g/dL (median) | 11.0–15.5 (14.0) | 9.2–16.2 (12.9) | 12.0–17.6 (14.2) | 4.9–17.4 (13.6) |
| CRP, mg/dL (median) | 0–0.05 (0) | 0–13.94 (0.28) | 0–0.69 (0) | 0–19.86 (0.05) |
| NLR (median) | 0.93–3.97 (1.78) | 1.15–5.87 (1.95) | 0.83–12.23 (1.96) | 0.87–9.68 (2.46) |
| TNM stage, I/II/III/IV | 6/0/4/2 | 78/9/26/17 | ||
| Histological type | ||||
| Clear cell | 11 | 111 | ||
| Papillary | 1 | 7 | ||
| Chromophobe | 0 | 6 | ||
| Unclassified | 0 | 6 | ||
Hb, hemoglobin; CRP, C‐reactive protein; NLR, neutrophil to lymphocyte ratio.
Table 2.
Fifteen genes that showed significantly higher or lower expression in renal cell carcinoma patients compared to healthy controls
| Gene | Coding protein | Fold‐change | P‐value |
|---|---|---|---|
| IFI44 | Interferon induced protein 44 like | 2.04 | 0.019 |
| MYL9 | Myosin light chain 9 | 1.78 | 0.001 |
| CMPK2 | Cytidine/uridine monophosphate kinase 2 | 1.77 | 0.032 |
| ITGB3 | Integrin α‐IIb/β‐3 | 1.72 | 0.009 |
| HERC5 | HECT and RLD domain containing e3 ubiquitin protein ligase 5 | 1.71 | 0.043 |
| ITGA2B | Integrin subunit α2b | 1.70 | 0.010 |
| TREML1 | Triggering receptor expressed on myeloid cells like 1 | 1.63 | 0.007 |
| IFIT3 | Interferon induced protein with tetratricopeptide repeats 3 | 1.61 | 0.033 |
| MX1 | MX dynamin like GTPase 1 | 1.59 | 0.039 |
| EIF2AK2 | Eukaryotic translation initiation factor 2α kinase 2 | 1.57 | 0.005 |
| SH3BGRL2 | SH3 domain binding glutamate rich protein like 2 | 1.53 | 0.011 |
| CXCL7 | C‐X‐C motif chemokine ligand 7 | 1.51 | 0.001 |
| PRKAR2B | Protein kinase CAMP‐dependent type II regulatory subunit β | 1.50 | 0.011 |
| CD79A | CD79a molecule | −1.65 | 0.022 |
| MS4A1 | Membrane spanning 4‐domains A1 | −1.67 | 0.023 |
Figure 1.
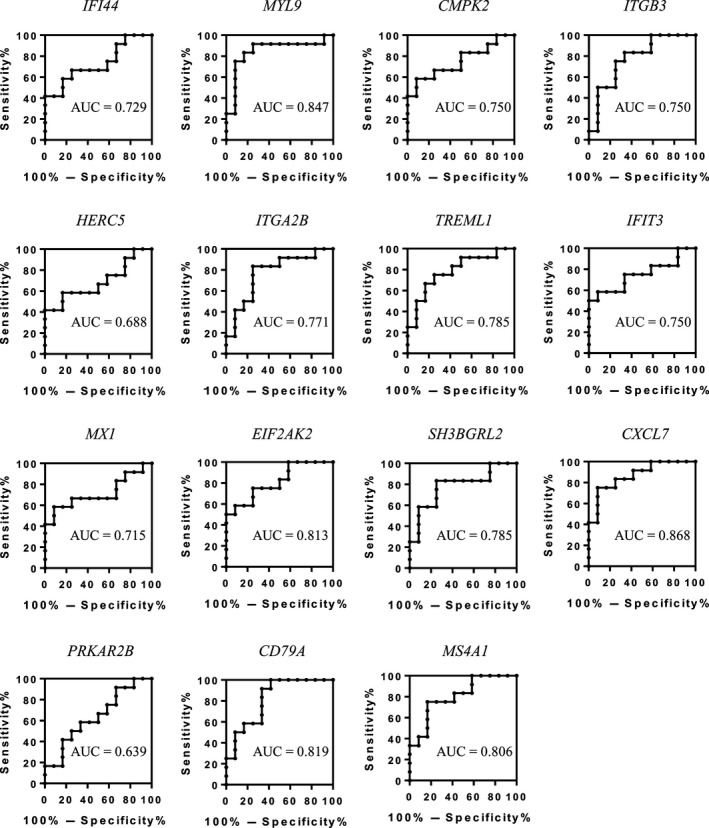
Discovery of candidate genes for the diagnosis of renal cell carcinoma. Expression levels in peripheral blood cells of 15 candidate genes identified from the discovery cohort (n = 24) were determined by quantitative RT‐PCR. These values were then subjected to receiver operating characteristic curve analysis for correlation with diagnosis of renal cell carcinoma.
CXCL7 expression level in peripheral blood cells is an independent predictive biomarker for RCC diagnosis
We evaluated the predictive ability of CXCL7 expression levels in peripheral blood cells using verification cohort samples. The verification cohort included 130 RCC patients (stage I, 78; stage II, 9; stage III, 26; stage IV, 17) diagnosed at Osaka University Hospital between 2013 and 2016, and 50 healthy controls. Of the 130 RCC patients in this cohort, 111 were diagnosed with ccRCC, 7 were diagnosed with papillary RCC, 6 were diagnosed with chromophobe RCC, and 6 were diagnosed with unclassified RCC (Table 1). The expression levels of CXCL7 in peripheral blood cells from RCC patients were significantly higher than those from healthy controls (P < 0.01; Fig. 2a). Moreover, CXCL7 expression increased according to TNM stage (Fig. 2b). Elevated levels of CXCL7 mRNA were confirmed even in RCC patients with small tumors (pT1a, n = 59, P < 0.01; Fig. 2c). The ROC curve analysis showed a sensitivity of 70.0% and a specificity of 64.0% (AUC = 0.722, P < 0.001; Fig. 2d) in order to diagnose RCC. Multivariate logistic regression analysis revealed that the expression level of CXCL7 in peripheral blood cells was the significant diagnostic factor (odds ratio, 1.07; 95% confidence interval, 1.033–1.109; P < 0.001, Table 3). These results strongly suggest that CXCL7 expression level in peripheral blood cells is a potential biomarker for the diagnosis of RCC. Using Kaplan–Meyer analysis and the log–rank test, the levels of CXCL7 (low vs high, P = 0.036) showed a significant association with cancer‐specific survival rate (Fig. 3).
Figure 2.
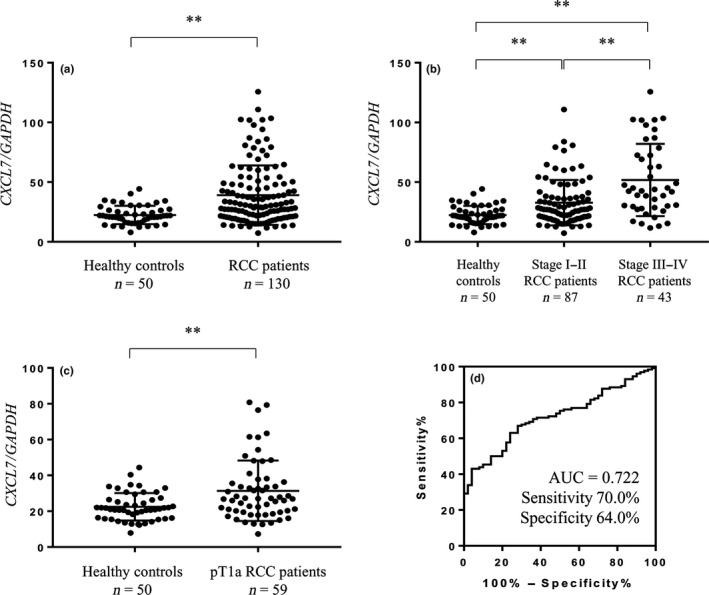
Expression level of CXCL7 in peripheral blood cells was upregulated in renal cell carcinoma (RCC) patients compared to healthy controls. (a) Expression level of CXCL7 in peripheral blood cells was quantified by quantitative RT‐PCR in the verification cohort (n = 180). **P < 0.01 (Wilcoxon test). (b) Comparison of the expression level of CXCL7 in peripheral blood cells among healthy controls, early stage RCC patients, and late stage RCC patients. **P < 0.01 (Dunn's multiple comparison test). (c) Expression level of CXCL7 in peripheral blood cells from pT1a patients. **P < 0.01 (Wilcoxon test). (d) Receiver operating characteristic curve analysis for the diagnosis of RCC using the expression level of CXCL7 in peripheral blood cells from the verification cohort (n = 180).
Table 3.
Univariate and multivariate logistic regression analyses on expression levels of CXCL7 for the diagnosis of renal cell carcinoma in the verification cohort
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P‐value | OR | 95% CI | P‐value | |
| Age | 0.969 | 0.936–1.000 | 0.055 | |||
| Gender (male vs female) | 1.358 | 0.642–3.043 | 0.437 | |||
| Hb | 0.733 | 0.591–0.885 | 0.003 | 0.820 | 0.632–1.051 | 0.124 |
| CRP | 1.441 | 1.100–2.332 | 0.046 | 1.144 | 0.853–1.851 | 0.486 |
| NLR | 1.183 | 0.947–1.543 | 0.173 | |||
| CXCL7 expression in peripheral blood cells | 1.073 | 1.040–1.115 | <0.001 | 1.066 | 1.033–1.109 | <0.001 |
CI, confidence interval; CRP, C‐reactive protein; Hb, hemoglobin; NLR, neutrophil to lymphocyte ratio; OR, odds ratio.
Figure 3.
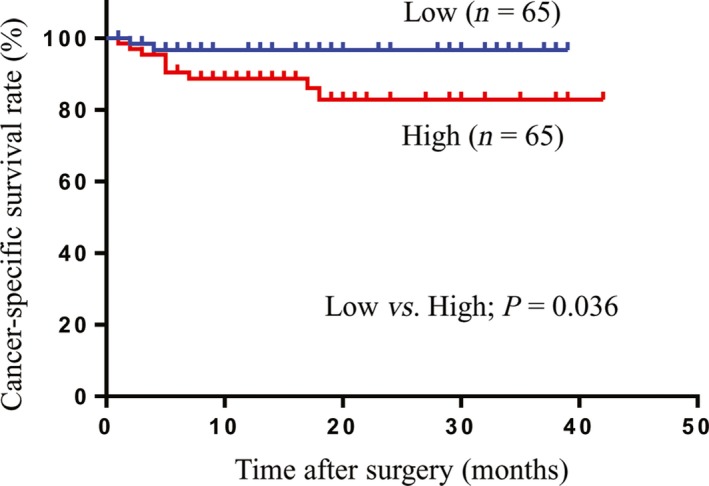
Association between the expression level of CXCL7 in peripheral blood cells with cancer‐specific survival in patients with renal cell carcinoma. Cut‐off value of CXCL7 expression is 30.66 (median) (Kaplan–Meier method, log–rank test).
We also evaluated the usefulness of CXCL7 using only ccRCC patients, the major histological type of RCC.5 The expression levels of CXCL7 in peripheral blood cells from ccRCC patients (n = 111) were significantly higher than those from healthy controls (P < 0.01; Fig. S2a) and increased according to TNM stage (Fig. S2b). Even in pT1a patients, CXCL7 mRNA was elevated compared to healthy controls (P < 0.01; Fig. S2c). The ROC curve analysis showed a sensitivity of 70.3% and a specificity of 62.0% (AUC = 0.712, P < 0.001; Fig. S2d) for the diagnosis of ccRCC. Multivariate logistic regression analysis revealed that the expression level of CXCL7 in peripheral blood cells was the significant diagnostic factor (odds ratio, 1.065; 95% confidence interval, 1.032–1.108; P < 0.001; Table S1). These results strongly suggest that the CXCL7 expression level in peripheral blood cells is a potential biomarker for the diagnosis of ccRCC.
Next, we evaluated the expression levels of CXCL7 in peripheral blood cells obtained from RCC patients (n = 10: stage I, 3; stage II, 2; stage III, 3; stage IV, 2) before and after surgical removal of the primary tumor. Interestingly, expression levels of CXCL7 in peripheral blood cells were significantly lower in postoperative states than in preoperative states (P < 0.01; Fig. 4).
Figure 4.
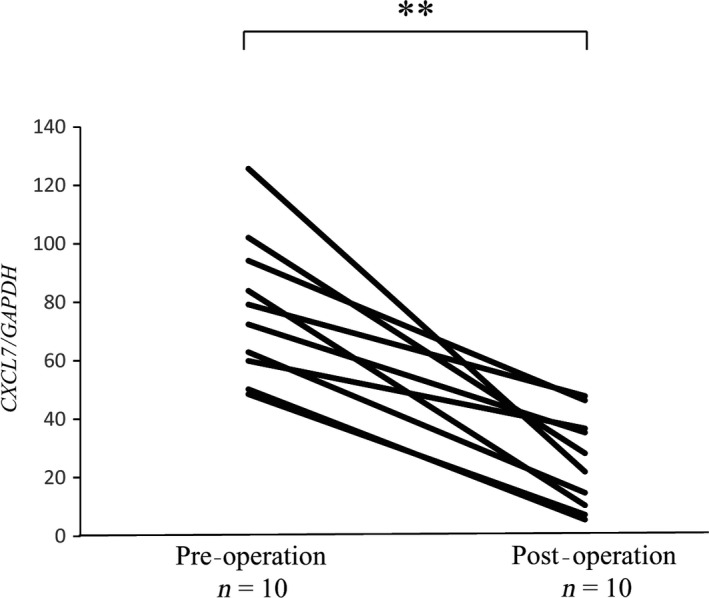
Expression levels of CXCL7 in peripheral blood cells decreased after removal of primary tumors in patients with renal cell carcinoma. Changes in the expression levels of CXCL7 in peripheral blood cells before (pre‐operation) and after (post‐operation) surgical removal of the primary tumor were quantified by quantitative RT‐PCR (n = 10). **P < 0.01 (Wilcoxon signed‐rank test).
Coculture with RCC cells upregulates CXCL7 expression in PBMCs
To determine whether CXCL7 was expressed in neutrophils or PBMCs, we extracted neutrophils and PBMCs from whole blood from healthy controls (n = 8) and cancer patients (n = 8). CXCL7 was more highly expressed in PBMCs than in neutrophils, both for healthy controls and RCC patients (both were approximately 20‐fold, P < 0.01; Fig. S3). Next, to examine the effect of RCC cells on CXCL7 expression in PBMCs, we cocultured PBMCs from healthy controls with RCC cell lines (786‐O or Caki‐2 cells) or HEK293T cells as a control. Coculture with RCC cells led to significant upregulation of CXCL7 in PBMCs compared to the HEK293T cells, suggesting that RCC cells upregulated CXCL7 expression in PBMCs (P < 0.05; Fig. 5).
Figure 5.
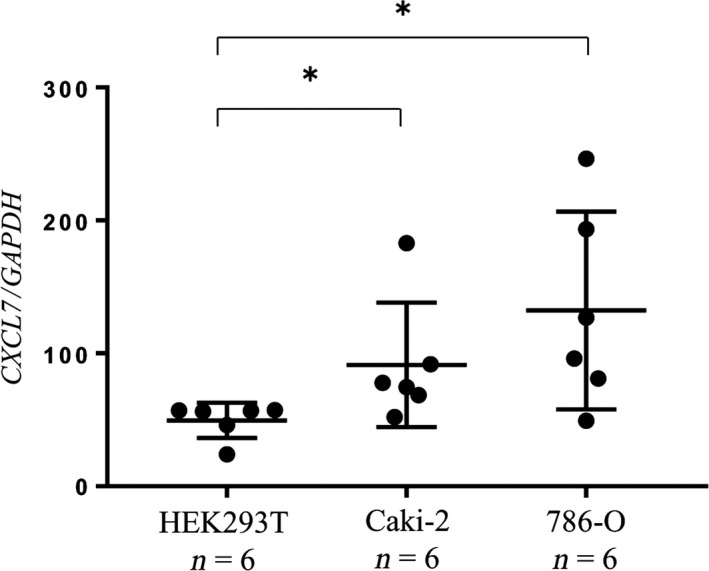
Renal cancer cells upregulated CXCL7 expression in PBMCs. Human kidney cancer cells were cocultured with PBMCs from healthy controls. The expression levels of CXCL7 in PBMCs exposed to HEK293T cells, 786‐O cells, or Caki‐2 cells were quantified by quantitative RT‐PCR. *P < 0.05 (Wilcoxon signed‐rank test).
Discussion
There are few reported blood biomarkers for the diagnosis of RCC patients. A number of miRNAs and proteins in serum or plasma were investigated as potential biomarkers for RCC, but they were not clinically useful for the diagnosis of RCC.6, 7, 8 Renal cell carcinoma is considered to be an immunogenic tumor,9 and immunotherapeutic approaches, such as interleukin‐2, α‐interferon, and immune checkpoint inhibitors have been used.10, 11, 12 Previously, expression levels of several genes in PBMCs have been reported to correlate with the prognosis of advanced RCC patients using microarray analysis and a hierarchical clustering method. However, it remains inconvenient to extract PBMCs from whole peripheral blood and difficult to ensure RNA stability in this context, hence these methods are likely not suitable for RCC diagnosis in clinical use.13 We focused on gene expression profiling of whole peripheral blood cells using the PAXgene Blood RNA system (Becton Dickinson) to collect blood samples. This system was developed to overcome those disadvantages on sample collection, transport, and storage of blood, stabilization of intracellular RNA in a closed tube, and subsequent isolation and purification of intracellular RNA from whole blood.3 As detailed in a previous report, this system has been used for gene expression profiling of peripheral blood cells from patients with cancer of the digestive system compared to healthy volunteers. The researchers discovered a gene signature profile in peripheral blood cells that differentiated patients with gastrointestinal tract cancers from healthy controls, and produced a custom‐made microarray chip for commercial use in medical checkups.14 In our approach, we aimed to develop a simpler, faster, and more cost‐effective diagnostic method for RCC based on qRT‐PCR analysis of a handful of differentially expressed genes. From our initial microarray study, three genes (CXCL7, CD79A, and EIF2AK2) were selected using Bayesian analysis.15 However, of these three targets, only the upregulation of CXCL7 in peripheral blood cells from RCC patients was shown to be statistically significant in our verification cohort (data not shown). Importantly, the expression level of CXCL7 in peripheral blood cells was significantly upregulated not only in late stage but also early stage RCC patients (Figs 1,2). Moreover, we found that high expression levels of CXCL7 in the peripheral blood of RCC patients correlated with significantly worse prognosis (Fig. 3).
CXCL7 is a platelet‐derived growth factor and belongs to the CXC chemokine family. It is also a potent chemoattractant and activator of neutrophils.16 Overexpression of CXCL7 in cancer cells is thought to promote cell proliferation in vivo and in vitro.17 As a ligand, CXCL7 binds to C‐X‐C motif chemokine receptor 1/2 and induces tumor angiogenesis and tumor cell migration.18
As mentioned above, CXCL7 was more highly expressed in PBMCs than in neutrophils, both for healthy controls and RCC patients (Fig. S3). The mechanism by which the expression level of CXCL7 in PBMCs is elevated remains unclear. It has been reported that canine dendritic cells treated with interleukin (IL)‐1β, IL‐6, tumor necrosis factor‐α, or transforming growth factor‐β expressed significantly higher levels of CXCL7 mRNA and protein than when treated with γ‐interferon or lipopolysaccharide.19 Other reports showed that expression of CXCL7 was induced by the central pro‐inflammatory cytokine IL‐1β.17 Kidney cancer cells produce both pro‐inflammatory and T‐cell inhibitory cytokines such as IL‐6, tumor necrosis factor‐α, and transforming growth factor‐β that potentially could influence the immune response of the host.20 These cytokines have also been reported to have a positive correlation with tumor size in RCC.21 The reports indicated that kidney cancer cells induce these cytokines and upregulate the expression level of CXCL7 in peripheral blood cells of RCC patients. Here, we investigated whether RCC cells truly upregulated CXCL7 expression in PBMCs independently from other factors, for instance, as a general consequence of cancer‐related inflammation. Our in vitro study suggests that RCC cells regulated the CXCL7 expression in PBMCs. Further study is needed to clarify the mechanism by which PBMCs are influenced by cancer cells, a phenomenon that has previously been reported in lung cancer.22
In the present study, only the expression level of CXCL7 was identified as an independent biomarker for the diagnosis of RCC in multivariate analysis (Table 3). Although hemoglobin, C‐reactive protein, and neutrophil to leukocyte ratio are well known as prognostic factors for advanced RCC patients,23, 24, 25, 26 our results suggested that CXCL7 was more sensitive and useful to detect the earlier‐stage RCC patients than these three parameters, which are mainly intended for the diagnosis of advanced stage RCC.
Next, we confirmed that the expression level of CXCL7 in peripheral blood cells is significantly decreased after resection of the primary tumor, even when metastatic tumor remains. These results suggest that CXCL7 expression in peripheral blood cells may reflect the total tumor burden in each individual, potentially making it a useful biomarker for monitoring patients during systemic therapy (Fig. 4).
Finally, we also examined peripheral blood from patients with other urological cancers, such as prostate cancer and urothelial carcinoma. Although most were advanced cancers, the expression level of CXCL7 was not significantly upregulated in those patients (data not shown).
In conclusion, our results indicate that RCC cells upregulate the expression level of CXCL7 in PBMCs, making CXCL7 a potential biomarker for detecting RCC, even in early stage disease. Furthermore, we hope that in the future this biomarker will be useful for monitoring the state of RCC during the observation period after surgical resection of local RCC, as well as during systemic therapies such as molecular targeted drugs and checkpoint inhibitors.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Confirmation of microarray results by quantitative RT‐PCR.
Fig. S2. Expression levels of CXCL7 in peripheral blood cells were upregulated in patients with clear cell renal cell carcinoma compared to healthy controls.
Fig. S3. CXCL7 was predominantly expressed in PBMCs.
Table S1. Univariate and multivariate logistic regression analyses on the expression levels of CXCL7 for the diagnosis of clear cell renal cell carcinoma in the verification cohort.
Acknowledgments
We thank Kengo Kamatani (Osaka University Graduate School of Engineering Science) for his help with the statistical analysis. This work was supported by the Japan Society for the Promotion of Science (KAKENHI grant no. 16K20140).
Cancer Sci 108 (2017) 2495–2502
Funding Information
Japan Society for the Promotion of Science (16K20140).
References
- 1. Ferlay J, Soerjomataram I, Dikshit R et al Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–86. [DOI] [PubMed] [Google Scholar]
- 2. Pal SK, Ghate SR, Li N et al Real‐world survival outcomes and prognostic factors among patients receiving first targeted therapy for advanced renal cell carcinoma: a SEER‐medicare database analysis. Clin Genitourin Cancer 2017; 15: e573–82. [DOI] [PubMed] [Google Scholar]
- 3. Chai V, Vassilakos A, Lee Y, Wright JA, Young AH. Optimization of the PAXgene blood RNA extraction system for gene expression analysis of clinical samples. J Clin Lab Anal 2005; 19: 182–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Edge SB. AJCC Cancer Staging Manual, 7th edn New York; London: Springer, 2010. [Google Scholar]
- 5. Choueiri TK, Motzer RJ. Systemic therapy for metastatic renal‐cell carcinoma. N Engl J Med 2017; 376: 354–66. [DOI] [PubMed] [Google Scholar]
- 6. Wang C, Hu J, Lu M et al A panel of five serum miRNAs as a potential diagnostic tool for early‐stage renal cell carcinoma. Sci Rep 2015; 5: 7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fedorko M, Stanik M, Iliev R et al Combination of MiR‐378 and MiR‐210 serum levels enables sensitive detection of renal cell carcinoma. Int J Mol Sci 2015; 16: 23382–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Twine NC, Stover JA, Marshall B et al Disease‐associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. Cancer Res 2003; 63: 6069–75. [PubMed] [Google Scholar]
- 9. Santoni M, Berardi R, Amantini C et al Role of natural and adaptive immunity in renal cell carcinoma response to VEGFR‐TKIs and mTOR inhibitor. Int J Cancer 2014; 134: 2772–7. [DOI] [PubMed] [Google Scholar]
- 10. Motzer Robert J, Hutson Thomas E, Tomczak P et al Sunitinib versus interferon alfa in metastatic renal‐cell carcinoma. N Engl J Med 2007; 356: 115–24. [DOI] [PubMed] [Google Scholar]
- 11. Rini BI, Escudier B, Tomczak P et al Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): a randomised phase 3 trial. Lancet 2011; 378: 1931–9. [DOI] [PubMed] [Google Scholar]
- 12. Motzer RJ, Escudier B, Tomczak P et al Axitinib versus sorafenib as second‐line treatment for advanced renal cell carcinoma: overall survival analysis and updated results from a randomised phase 3 trial. Lancet Oncol 2013; 14: 552–62. [DOI] [PubMed] [Google Scholar]
- 13. Burczynski ME, Twine NC, Dukart G et al Transcriptional profiles in peripheral blood mononuclear cells prognostic of clinical outcomes in patients with advanced renal cell carcinoma. Clin Cancer Res 2005; 11: 1181–9. [PubMed] [Google Scholar]
- 14. Honda M, Sakai Y, Yamashita T et al Differential gene expression profiling in blood from patients with digestive system cancers. Biochem Biophys Res Commun 2010; 400: 7–15. [DOI] [PubMed] [Google Scholar]
- 15. Robert C. The Bayesian Choice: From Decision‐Theoretic Foundations to Computational Implementation, 2nd edn New York: Springer Science & Business Media, 2007; 343–89. [Google Scholar]
- 16. Yan Z, Zhang J, Holt JC et al Structural requirements of platelet chemokines for neutrophil activation. Blood 1994; 84: 2329–39. [PubMed] [Google Scholar]
- 17. Grépin R, Guyot M, Giuliano S et al The CXCL7/CXCR1/2 axis is a key driver in the growth of clear cell renal cell carcinoma. Cancer Res 2014; 74: 873–83. [DOI] [PubMed] [Google Scholar]
- 18. Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett 2008; 267: 226–44. [DOI] [PubMed] [Google Scholar]
- 19. Wang YS, Liao KW, Chen MF, Huang YC, Chu RM, Chi KH. Canine CXCL7 and its functional expression in dendritic cells undergoing maturation. Vet Immunol Immunopathol 2010; 135: 128–36. [DOI] [PubMed] [Google Scholar]
- 20. Lahn M, Fisch P, Köhler G et al Pro‐inflammatory and T cell inhibitory cytokines are secreted at high levels in tumor cell cultures of human renal cell carcinoma. Eur Urol 1999; 35: 70–80. [DOI] [PubMed] [Google Scholar]
- 21. Yoshida N, Ikemoto S, Narita K et al Interleukin‐6, tumour necrosis factor alpha and interleukin‐1beta in patients with renal cell carcinoma. Br J Cancer 2002; 86: 1396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang DH, Rutledge JR, Patel AA, Heerdt BG, Augenlicht LH, Korst RJ. The effect of lung cancer on cytokine expression in peripheral blood mononuclear cells. PLoS ONE 2013; 8: e64456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Martino M, Klatte T, Seemann C et al Validation of serum C‐reactive protein (CRP) as an independent prognostic factor for disease‐free survival in patients with localised renal cell carcinoma (RCC). BJU Int 2013; 111: 348–53. [DOI] [PubMed] [Google Scholar]
- 24. Fujita T, Iwamura M, Ishii D et al C‐reactive protein as a prognostic marker for advanced renal cell carcinoma treated with sunitinib. Int J Urol 2012; 19: 908–13. [DOI] [PubMed] [Google Scholar]
- 25. Pichler M, Hutterer GC, Stoeckigt C et al Validation of the pre‐treatment neutrophil‐lymphocyte ratio as a prognostic factor in a large European cohort of renal cell carcinoma patients. Br J Cancer 2013; 108: 901–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hutterer GC, Krieger D, Mrsic E et al Preoperative leucocytosis, thrombocytosis and anemia as potential prognostic factors in non‐metastatic renal cell carcinoma. Anticancer Res 2015; 35: 3463–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Confirmation of microarray results by quantitative RT‐PCR.
Fig. S2. Expression levels of CXCL7 in peripheral blood cells were upregulated in patients with clear cell renal cell carcinoma compared to healthy controls.
Fig. S3. CXCL7 was predominantly expressed in PBMCs.
Table S1. Univariate and multivariate logistic regression analyses on the expression levels of CXCL7 for the diagnosis of clear cell renal cell carcinoma in the verification cohort.


