Abstract
CIC/Capicua is an HMG‐box transcription factor that is well conserved during evolution. CIC recognizes the T(G/C)AATG(A/G)A sequence and represses its target genes, such as PEA3 family genes. The receptor tyrosine kinase/RAS/MAPK signals downregulate CIC and relieves CIC's target genes from the transrepressional activity; CIC thus acts as an important downstream molecule of the pathway and as a tumor suppressor. CIC loss‐of‐function mutations are frequently observed in several human neoplasms such as oligodendroglioma, and lung and gastric carcinoma. CIC is also involved in chromosomal translocation‐associated gene fusions in highly aggressive small round cell sarcoma that is biologically and clinically distinct from Ewing sarcoma. In these mutations, PEA3 family genes and other important target genes are upregulated, inducing malignant phenotypes. Downregulation of CIC abrogates the effect of MAPK inhibitors, suggesting its potential role as an important modifier of molecular target therapies for cancer. These data reveal the importance of CIC as a key molecule in signal transduction, carcinogenesis, and developing novel therapies.
Keywords: Capicua/CIC, CIC‐DUX4, oligodendroglioma, round cell sarcoma, RTK/RAS/MAPK pathway
The RTK/RAS/MAPK pathway plays a central role in development, progression, and survival of cancer cells. A number of mutations in the pathway have been identified in the broad spectrum of cancer.1 In most of the mutations, enhancement and/or prolongation of phosphorylation was found in proteins of signal mediators in the pathway. The signal is transmitted to the nuclear proteins, such as transcription factors, cofactors, and/or chromatin regulators, and the abnormal signaling disorganizes the epigenetic status.2 During malignant transformation, progression, and survival of cancer cells under therapeutic stress, the nuclear proteins and transcriptional program downstream to RTK/MAPK signaling modify cellular biological activities and their interference will be one of the critical targets of therapies.3
Multiple downstream molecules are activated in response to RTK/MAPK phosphorylation signals.4 The PEA3 family of ETS transcription factors, ETV1, ETV4, and ETV5, are known to act as such downstream nuclear proteins.5, 6, 7 The PEA3 family genes are involved in chromosomal translocation associated with prostate cancer and ES, and their overexpression promotes cell proliferation, motility, and invasion.8 As a common direct repressor of PEA3 genes, Capicua/CIC is an important RTK/MAPK downstream molecule that is contained in an ATXN1/CIC repressor complex and regulates cell proliferation.5, 9, 10
Capicua/CIC is an HMG‐box transcriptional repressor that is well conserved during evolution. There is growing evidence that CIC is involved in a variety of human cancer. These aberrations include both loss‐of‐function and gain‐of‐function mutations, indicating the pleiotropic characteristics of CIC in cancer. This review describes the functions of CIC, its mutation spectrum in human cancer and signaling pathways, and mechanistic consequences involved in these mutations.
Structure and function of Capicua/CIC
Human CIC encodes two protein isoforms, CIC‐L and CIC‐S, consisting of 2517 and 1608 amino acids, respectively (Fig. 1a).11 CIC is a mammalian homolog of Drosophila capicua that is well conserved in many organisms and there are no apparent homologs in mammals (Fig. 1b). CIC recognizes chromatin through its consensus T(G/C)AATG(A/G)A sequence (also called CIC octamer) using an HMG‐box as a DNA‐binding motif,12, 13 unlike other HMG class transcription factors most of which do not bind DNA in the sequence‐specific manner.14 The CIC HMG‐box is highly conserved among species and there are also additional conserved motifs, C1 and C2, in the C‐terminus and the central part, respectively.15, 16 The in vitro DNA binding assay using mutant CIC constructs showed that the C1 motif is required for stable DNA binding by its interaction with the HMG box.13 Thus, both the HMG‐box and C1 motif contribute to the core function of CIC, as is also suggested by the mutation spectrum in human cancer (see below).
Figure 1.
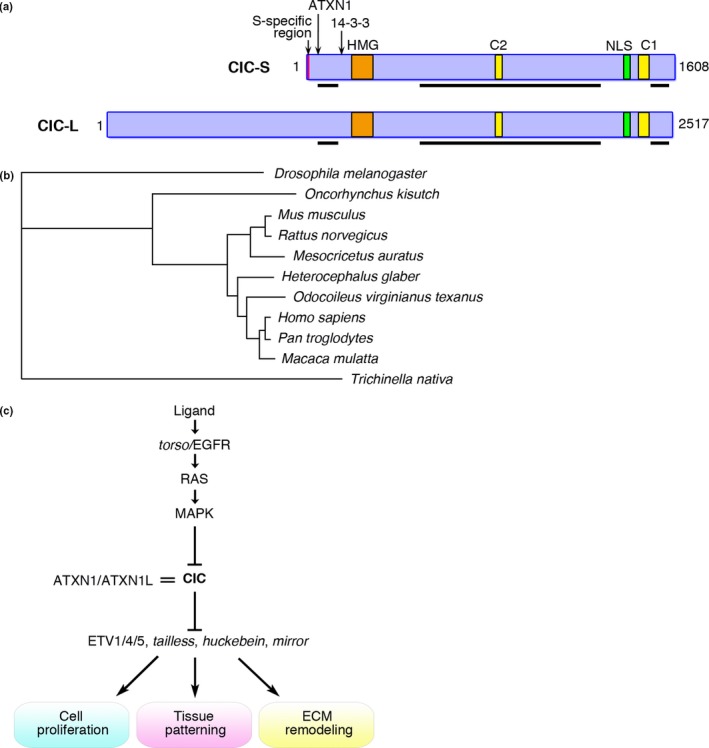
Structure, conservation, and functions of CIC. (a) Two isoforms of the CIC protein contain well‐conserved HMG‐box, C1 and C2 motifs, nuclear localization signal (NLS), and ataxin 1 (ATXN1)/ataxin 1 like (ATXN1L) and 14‐3‐3 binding sites. The numbers of amino acids for each isoform are indicated. Black bars, proline‐rich regions. Red bar, isoform S‐specific N‐terminal 22 amino acid sequence. (b) Evolutionary relationship among CIC/capicua proteins. The protein distances were calculated between two sequences and a phylogenetic tree was reconstructed by the neighbor‐joining method. (c) Molecular pathway around CIC. CIC represses its target genes such as ETV1/4/5 and RAS/MAPK signals downregulate CIC. Interaction between CIC and ATXN1 or ATXN1L is important for CIC's repression activity.
Drosophila capicua was first identified as a transcriptional repressor downstream to torso, a Drosophila RTK with partial homology to mammalian RET, PDGFR, and c‐kit (Fig. 1c).16, 17, 18 Capicua represses tailless and huckebein by interacting with groucho using the capicua C‐terminus encompassing the C1 motif during Drosophila embryogenesis. Capicua also represses mirror expression that determines the ovarian follicle cell fate.19 Moreover, ATXN1 that is mutated in human SCA1 modulates the repressional activity of capicua, interacting with the N‐terminal region of capicua.20, 21 Conversely, haploinsufficiency of Cic improved SCA1 disease phenotypes in Atxn1 mutant mice.22
The homozygous knockout mouse for Cic‐L shows the defect of alveolar organization of the lung, and the phenotype is similar with that of the compound Atxn1 and Atxn1l knockout mouse, indicating the importance of CIC/ATXN1 interaction in tissue homeostasis.23 In mutants, Etv4 repression by CIC is cancelled, resulting in upregulation of Mmp9 and aberration of ECM remodeling.23 The conditional Cic mutation lacking exons 2–6 also induced abnormal lung alveolarization with reduced alveolar surfactant protein expression.24 Moreover, hematopoietic lineage cell‐specific knockout of Cic induced remarkable autoimmune responses with increased follicular helper T cells, which was mediated by derepression of Etv5.25
Interaction between CIC and ATXN1 protein family is also important for brain development. Disruption of the ATXN1/CIC complex affects thickness of cerebral cortex, inducing multiple behavioral abnormalities in mice.26 In human, the germline heterozygous CIC truncating mutations were reported in patients of intellectual disability, attention deficit hyperactivity disorder, and autism spectrum disorder.26 The Cic‐L knockout homozygous mutant also shows downregulation of transporter genes such as Bsep and Mdr2 in hepatocytes showing bile acid accumulation.27 CIC is ubiquitously expressed in many organs except for kidney, and thus its function is important for homeostasis of multiple organs.
Functions of CIC as a downstream molecule of RTK/MAPK signaling are important for tissue patterning and cell proliferation. While CIC constitutively represses its target genes when MAPK signals are off, it is promptly downregulated by MAPK phosphorylation, inducing upregulation of PEA3 family genes that promote cellular proliferation and migration.7, 9, 28 Activation of EGFR induces MAPK‐dependent phosphorylation of CIC directly or through p90RSK, promoting CIC binding to 14‐3‐3 proteins and inhibiting the importin alpha 4 activity.9 Binding of CIC to 14‐3‐3 proteins also reduces DNA binding activity of CIC. In addition, ERK‐induced phosphorylation reduces CIC's repressor activities and eventually promotes cytoplasmic export of phosphorylated CIC from nucleus, resulting in its degradation.29, 30 CIC degradation is achieved by the ubiquitin E3 ligase complex Cullin1/SKP1/Archipelago in the ERK‐dependent manner.31 Thus, CIC expression is clearly downregulated in accordance with torso‐ and EGFR‐induced MAPK activity. Importantly, multiple cis elements to which CIC potentially binds are found as responsive elements for RTK signaling.32
There is a well‐conserved MAPK‐docking site (C2 motif, Fig. 1a), and the C2 deletion mutant is insensitive for MAPK‐induced downregulation.18, 33 The ERK‐independent downregulation of CIC by minibrain/DYRK1A is observed in Drosophila wing and eye formation.34 In addition, bicoid antagonizes downregulation of CIC in anterior patterning of Drosophila.35, 36 In this case, bicoid acts as a competitive substrate for MAPK, which renders CIC phosphorylation.
CIC functions as a tumor suppressor in human cancer
CIC's repressive function to the downstream targets in the RAS/MAPK signals suggests its role as a tumor suppressor gene in carcinogenesis. Indeed, CIC was found mutated in the majority of human oligodendroglioma, in which biallelic mutations and/or loss of CIC were frequently observed (Fig. 2a, Table 1).37, 38 In brain tumors, CIC mutations are rather specific to oligodendroglioma and are rarely observed in astrocytic tumors.39 CIC mutations in oligodendroglioma are frequently associated with IDH1 and FUBP1 mutations, suggesting a cooperative role among these three genes in tumorigenesis.39, 40, 41 Cooperative increase of intracellular 2HG, reduced clonogenicity, and slower proliferation in cell lines introduced with IDH1 and CIC double mutations was reported, however, the significance of 2HG increase in oligodendroglioma development and survival remains unclear.11
Figure 2.
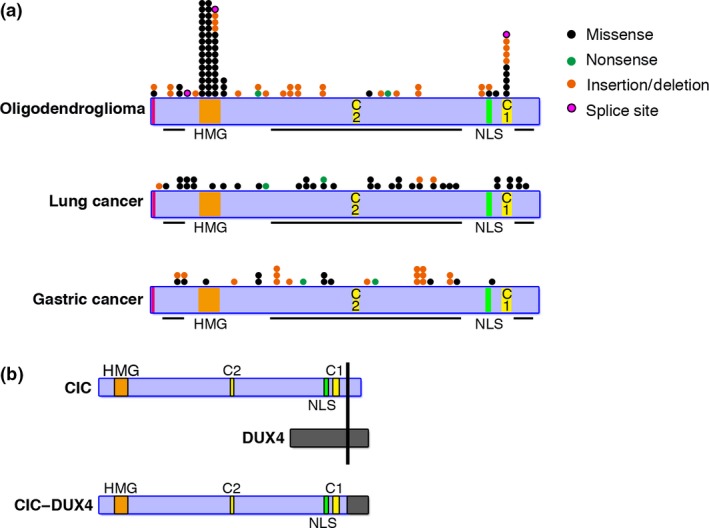
CIC mutations in human cancer. (a) Types and distributions of CIC mutations in oligodendroglioma, lung cancer, and gastric cancer. Point mutations and small indels are shown in relation to the functional domains. The types of each mutation are indicated. Isoform S‐specific mutations are observed in oligodendroglioma. (b) Structure of the CIC–DUX4 fusion protein. NLS, nuclear localization signal.
Table 1.
CIC mutations in human cancer
| Tumor type | Type of alterations | Function | Reference |
|---|---|---|---|
| Oligodendroglioma | LOH(19q and/or 1p) | LOF | 37, 38, 41 |
| Missense: 59.3% (51/86), nonsense: 4.7% (4/86), in/del: 33.7% (29/86), splice site: 3.5% (3/86) | LOF | ||
| Lung cancer | Missense: 87.5% (35/40), nonsense: 5.0% (2/40), in/del: 7.5% (3/40) | LOF | 26 |
| Gastric cancer | Missense: 37.9% (11/29), nonsense: 6.9% (2/29), in/del: 55.2% (16/29) | LOF | 26 |
| T‐ALL | Point mutation 100% (5/5) | LOF | 44 |
| CIC‐rearranged sarcoma | t(4;19)(q35;q13.1)CIC‐DUX4 fusion | GOF | 12, 52, 57 |
| t(10;19)(q26.3;q13.1) CIC‐DUX4 fusion | GOF | 57 | |
| t(X;19)(q13;q13.3) CIC‐FOXO4 fusion | GOF | 61 | |
| CNS‐PNET | t(15;19)(q14;q13.2) CIC‐NUTM1 fusion | GOF | 63 |
| in/del: 100% (1/1) | LOF | ||
| Angiosarcoma | CIC‐LEUTX fusion | GOF | 64 |
| Missense: 100% (7/7) | LOF |
GOF, gain of function; LOF, loss of function.
Subsequently, frequent and recurrent loss‐of‐function mutations and/or reduced expression of CIC have been reported in lung, stomach, and prostate cancer (Fig. 2a).7, 13, 28, 38, 42 The mutations were mostly detected around the HMG‐box and the C1 domain in oligodendroglioma (Fig. 2a).13 This characteristic distribution pattern of CIC mutations is consistent with the mechanisms that both the HMG‐box and the C1 domain are necessary for stable DNA binding of CIC.13 No mutations have been reported in the isoform L‐specific region, instead, isoform S‐specific mutations were observed in a few cases of oligodendroglioma, suggesting that the function of CIC‐S might be important in carcinogenesis. As a result of CIC loss‐of‐function mutations, repression of PEA3 family genes are cancelled, promoting cellular proliferation and migration (Fig. 3a). Interestingly, frequent mutations at the HMG‐box and C1 motif observed in oligodendroglioma were not found in lung or gastric cancer (Fig. 2a). CIC mutations might occur at the early stage of oligodendroglioma development, whereas mutations were acquired at the advanced stages in lung and gastric cancer. CIC mutations are not maintained in some cases of recurrent oligodendroglioma,43 suggesting the mutation might not be required for oligodendroglioma survival.
Figure 3.
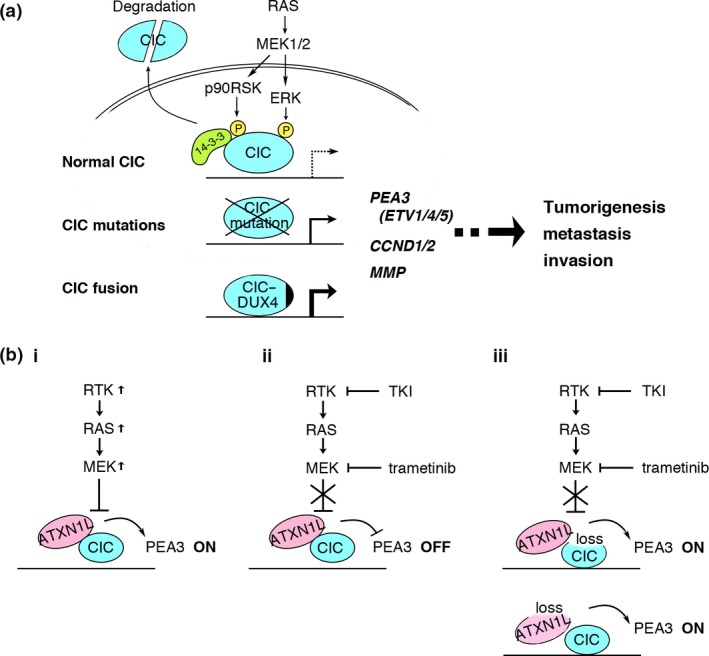
Molecular pathways around CIC in cancer. (a) Upregulation of downstream genes are achieved at the physiological level (top), CIC mutations (middle), or CIC fusion (bottom) at various levels. (b) Relationship between inhibitory drugs in the receptor tyrosine kinase (RTK)/RAS/MAPK pathway and the ataxin 1 like (ATXN1L)/CIC axis. Mutations within the pathway downregulate CIC (i), and tyrosine kinase inhibitors (TKI) and trametinib inhibit the downregulation (ii). When expression of ATXN1L or CIC is significantly reduced, the effects are cancelled (iii).
Interestingly, a glial fibrillary acidic protein‐Cre‐induced Cic mutation in mouse failed to induce oligodendroglioma.24 Instead, when the same mutation was ubiquitously induced in adult mice, T‐cell lymphoblastic leukemia developed at high penetrance. Although the result might be caused by the difference in genetic predisposition for cancer between human and mouse, it was consistent with the fact that mutations of CIC as well as the RTK/RAS/MAPK pathway genes were reported in human T‐ALL (Table 1).24, 44
CIC fusion genes in human cancer
CIC is also involved in human malignancies as gene fusions associated with chromosomal translocation involving 19q13. The CIC fusion to DUX4 in Ewing‐like small round cell sarcoma with t(4,19)(q35;q13) translocation was first identified in 2006.12 Most of the CIC coding region, except for the very C‐terminal end, is preserved in the CIC–DUX4 fusion, and both the HMG‐box and C1 domain are thus included in the fusion protein, indicating that the fusion protein possesses DNA‐binding activity (Fig. 2b). Addition of the DUX4 C‐terminal part induces conversion of CIC's transrepressional activity to transactivation, resulting in drastic upregulation of target genes such as PEA3 family genes (Fig. 3a).12 DUX4 encodes a double homeodomain protein and is located in the D4Z4 repeat that is distributed in the subtelomeric regions of the mammalian genome with predominant distribution in 4q and 10q.45, 46 Aberrant expression of DUX4 is associated with facioscapulohumeral muscular dystrophy.47, 48, 49, 50 The mechanisms of transcriptional activation of DUX4, by recruiting p300/CBP using its C‐terminal domain that is included in the CIC–DUX4 fusion, was proposed.51 As a result of DUX4 fusion, CIC acquires transcriptional activation, perhaps through recruitment of p300/CBP, and the fusion converts transrepressional activity of CIC to upregulate its target genes, thereby shows strong oncogenic activity.
CIC–DUX4‐positive sarcomas are composed of small‐ to medium‐sized, round to ovoid cells without any line of differentiation. CIC–DUX4‐positive sarcoma shows a poor outcome; it was reported that overall survival of CDS patients was worse than that of ES patients, and phenotypes of CDS are distinct from those of ES.52, 53, 54 We have generated an ex vivo mouse model for human CDS by introducing CIC–DUX4 into embryonic mesenchymal cells.55 CIC–DUX4 expression induced small round cell sarcoma of aggressive growth with significantly shorter latency than that of the ES model.56 The model faithfully recapitulates the phenotype of human CDS with upregulation of CIC–DUX4 target genes such as PEA3 family genes. ETV4 is a good marker of CDS,52, 57, 58 and analysis of the CDS mouse model identified CCND2 and mucin 5AC as additional biomarkers.55
The DUX4 sequences are originated from both 4q and 10q.46, 53, 57, 59 DUX4 is also involved in translocation associated with human B‐cell lymphoblastic leukemia, and the C‐terminal region of DUX4 is deleted in these cases,60 suggesting the functional role of the C‐terminal region might be different depends on cancer types. A CIC fusion with a non‐DUX4 gene, FOXO4, was observed in a rare cases of small round cell sarcoma.61 Another cluster of CIC–NUTM1 fusion‐positive tumor was found in primitive neuroectodermal tumors of the central nervous system showing a small cell phenotype.62 Moreover, CIC mutations, including CIC–LEUTX fusion, were reported in 9 of 120 cases of angiosarcoma, and PEA3 family genes were also upregulated in CIC mutated cases.63 Although it remains to be clarified whether these non‐DUX4 fusions also convert CIC's repressor function, the HMG‐box was retained in both CIC–FOXO4 and CIC–NUTM1, suggesting similar functional modulation in non‐DUX4 fusion proteins. Reported CIC fusion genes are summarized in Table 1.
Molecular targeted therapy using CIC and future directions
The unique mutations of CIC in human cancer are characterized as a mixture of loss‐of‐function and gain‐of‐function mutations, both of which upregulate downstream target genes such as ETV4 (Fig. 3a). Many CIC target genes upregulated in CDS are also found upregulated following CRISPR/Cas9‐mediated KO in isogenic cell lines.64 The RTK/RAS/MAPK pathway is a common target of molecular targeted therapy, and acquired resistance for these therapies has been frequently observed.1 Therefore, downstream modifiers such as CIC are good alternative targets for the therapy.
As CIC suppresses MAPK downstream signals, downregulation of CIC may be one of the resistance mechanisms for targeted therapies. Indeed, reduced expression of ATXN1L that abrogates the CIC function are found to promote resistance to MAPK pathway inhibition in KRAS mutated pancreatic cancer cells.65 In this study, Wang et al. identified CIC as a gene that modulates the sensitivity for MEK1/2 inhibitor trametinib by CRISPR/Cas9‐mediated screening. The exact mechanism to explain how ATXN1L is downmodulated to reduce CIC protein and sensitivity to trametinib remains to be investigated, however, the result suggests importance of the ATXN1L–CIC axis for targeted therapy against the genetic mutations in the RTK/RAS/MAPK pathway (Fig. 3b).
To improve RTK/RAS/MAPK targeting it may be useful to assess the ATXN1L and CIC expression levels to predict the effect of inhibitory drugs, thus CIC can be used as a biological indicator of therapeutic effect. Furthermore, inhibition of CIC phosphorylation is a good alternative therapeutic approach. To this end, the reagent that mimics bicoid that blocks the CIC C2 motif from p90RSK binding might be a useful tool. The COP9 signalosome subunit 1b is another guardian of CIC that acts in an MAPK‐independent manner.31 Targeting CIC mutations in carcinoma and sarcoma is more challenging, however, epigenetic therapies that modulate transcription of CIC target genes should be considered. These therapies are effective and ideal for various cancers in which CIC plays a key role in cancer cell survival as downstream of the RTK/RAS/MAPK pathway and as a causative oncogene/tumor suppressor. In addition, it may be useful to evaluate the expression of CIC and ATXN1L to predict the effects of tyrosine kinase inhibitors and MEK inhibitors.
Cancer cells use multiple signaling pathways that regulate biological processes such as proliferation, immortalization, self‐renewal, migration, and invasion. The Cic‐L homozygous KO mice showed abnormal remodeling of ECM in the lung.23 This phenotype is closely recapitulated as upregulation of the ECM gene set in the CDS mouse model.55 In malignancies, mutant CIC could orchestrate biological activities of cancer cells in both cell autonomous and non‐autonomous manners.
In conclusion, CIC acts as a modulator in the pathway and both loss‐of‐function and gain‐of‐function mutations of CIC dysregulate the targets, such as the PEA3 family transcription factors, CCND1/D2, and MMPs, resulting in abnormal cellular growth, invasion, and metastasis. Preservation of CIC's tumor suppressor functions are thus of great importance for prevention and therapies against malignant disorders.
Disclosure Statement
Takuro Nakamura has received a commercial research grant from Otsuka Pharmaceutical Co. Ltd. The other authors have no conflict of interest.
Abbreviations
- 2HG
2‐hydroxyglutarate
- ATXN1
ataxin 1
- ATXN1L
ataxin 1 like
- CCND1/D2
cyclin D1/D2
- CDS
CIC–DUX4‐positive sarcoma
- CIC
Capicua transcriptional repressor
- CNS‐PNET
primitive neuroectodermal tumors of the central nervous system
- DUX4
double homeobox 4
- ECM
extracellular matrix
- EGFR
epidermal growth factor receptor
- ES
Ewing sarcoma
- RTK
receptor tyrosine kinase
- SCA1
spinocerebellar ataxia type 1
- TKI
tyrosine kinase inhibitor
Acknowledgements
This work was supported by a Grant‐in‐Aid for Scientific Research from the Japan Society for the Promotion of Science (26250029 to TN and 16K07131 to MT).
Cancer Sci 108 (2017) 2319–2325
Funding information
Japan Society for the Promotion of Science.
References
- 1. Schmitt MW, Loeb LA, Salk JJ. The influence of subclonal resistance mutations on targeted cancer therapy. Nat Rev Clin Oncol 2016; 13: 335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nabet B, Broin PO, Reyes JM, et al Deregulation of the Ras‐Erk signaling axis modulates the enhancer landscape. Cell Rep 2015; 12: 1300–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zawistowski JS, Bevill SM, Goulet DR, et al Enhancer remodeling during adaptive bypass to MEK inhibition is attenuated by pharmacologic targeting of the P‐TEFb complex. Cancer Discov 2017; 7: 303–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell 2017; 170: 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jimenez G, Shvartsman SY, Paroush Z. The Capicua repressor – a general sensor of RTK signaling in development and disease. J Cell Sci 2012; 125: 1383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin Y, Ha N, Fores M, et al EGFR/Ras signaling controls Drosophila intestinal stem cell proliferation via Capicua‐regulated genes. PLoS Genet 2015; 11: e1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liao S, Davoli T, Leng Y, Li MZ, Xu Q, Elledge SJ. A genetic interaction analysis identifies cancer drivers that modify EGFR dependency. Genes Dev 2017; 31: 184–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oh S, Shin S, Janknecht R. ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta 2012; 1826: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dissanayake K, Toth R, Blakey J, et al ERK/p90(RSK)/14‐3‐3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicua. Biochem J 2011; 433: 515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tseng ASK, Tapon N, Kanda H, et al Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/Ras signaling pathway. Curr Biol 2007; 17: 728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chittaranjan S, Chan S, Yang C, et al Mutations in CIC and IDH1 cooperatively regulate 2‐hydroxyglutarate levels and cell clonogenicity. Oncotarget 2014; 5: 7960–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawamura‐Saito M, Yamazaki Y, Kaneko K, et al Fusion between CIC and DUX4 up‐regulates PEA3 family genes in Ewing‐like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet 2006; 15: 2125–37. [DOI] [PubMed] [Google Scholar]
- 13. Fores M, Simon‐Carrasco L, Ajuria L, et al A new mode of DNA binding distinguishes Capicua from other HMG‐box factors and explains its mutation patterns in cancer. PLoS Genet 2017; 13: e1006622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development 2013; 140: 4129–44. [DOI] [PubMed] [Google Scholar]
- 15. Fores M, Ajuria L, Samper N, et al Origins of Context‐dependent gene repression by Capicua. PLoS Genet 2015; 11: e1004902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jimenez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by Torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev 2000; 14: 224–31. [PMC free article] [PubMed] [Google Scholar]
- 17. Li WX. Functions and mechanisms of receptor tyrosine kinase torso signaling: Lessons from Drosophila embryonic terminal development. Dev Dyn 2005; 232: 545–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Astigarraga S, Grossman R, Diaz‐Delfin J, Caelles C, Paroush Z, Jimenez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J 2007; 26: 668–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Atkey MR, Lachance JFB, Walczak M, Rebello T, Nilson LA. Capicua regulates follicle cell fate in the Drosophila ovary through repression of mirror . Development 2006; 133: 2115–23. [DOI] [PubMed] [Google Scholar]
- 20. Lam YC, Bowman AB, Jafar‐Nejad P, et al ATAXIN‐1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 2006; 127: 1335–47. [DOI] [PubMed] [Google Scholar]
- 21. Kim E, Lu HC, Zoghbi HY, Song JJ. Structural basis of protein complex formation and reconfiguration by polyglutamine disease protein Ataxin‐1 and Capicua. Genes Dev 2013; 27: 590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fryer JD, Yu P, Kang H, et al Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science 2011; 334: 690–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee Y, Fryer JD, Kang H, et al ATXN1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev Cell 2011; 21: 746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simon‐Carrasco L, Grana O, Salmon M, et al Inactivation of Capicua in adult mice causes T‐cell lymphoblastic lymphoma. Genes Dev 2017; 31: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park S, Lee S, Lee CG, et al Capicua deficiency induces autoimmunity and promotes follicular helper T cell differentiation via derepression of ETV5. Nat Commun 2017; 8: 16037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu HC, Tan Q, Rousseaux MWC, et al Disruption of the ATXN1‐CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans. Nat Genet 2017; 49: 527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim E, Park S, Choi N, et al Deficiency of Capicua disrupts bile acid homeostasis. Sci Rep 2015; 5: 8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Okimoto RA, Breitenbuecher F, Olivas VR, et al Inactivation of Capicua drives cancer metastasis. Nat Genet 2017; 49: 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lim B, Samper N, Lu H, Rushlow C, Jimenez G, Shvartsman SY. Kinetics of gene derepression by ERK signaling. Proc Natl Acad Sci USA 2013; 110: 10330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grimm O, Zini VS, Kim Y, Casanova J, Shvartsman SY, Wieschaus E. Torso RTK controls Capicua degradation by changing its subcellular localization. Development 2012; 139: 3962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suisse A, He DQ, Legent K, Treisman JE. COP9 signalosome subunits protect Capicua from MAPK‐dependent and –independent mechanisms of degradation. Development 2017; 144: 2673–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ajuria L, Nieva C, Winkler C, et al Capicua DNA‐binding sites are general response elements for RTK signaling in Drosophila . Development 2011; 138: 915–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim Y, Paroush Z, Nairz K, Hafen E, Jimenez G, Shvartsman SY. Substrate‐dependent control of MAPK phosphorylation in vivo . Mol Syst Biol 2011; 7: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang L, Paul S, Trieu KG, et al Minibrain and Wings apart control organ growth and tissue patterning through down‐regulation of Capicua. Proc Natl Acad Sci USA 2016; 113: 10583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim Y, Coppey M, Grossman R, et al MAPK substrate competition integrates patterning signals in the Drosophila embryo. Curr Biol 2010; 20: 446–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim Y, Andreu MJ, Lim B, et al Gene regulation by MAPK substrate competition. Dev Cell 2011; 20: 880–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bettegowda C, Agrawal N, Jiao Y, et al Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 2011; 333: 1453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yip S, Butterfield YS, Morozova O, et al Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol 2012; 226: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jiao Y, Killela PJ, Reitman ZJ, et al Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 2012; 3: 709–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eisenreich S, Abou‐El‐Ardat K, Szafranski K, et al Novel CIC point mutations and an exon‐spanning, homozygous deletion identified in oligodendroglial tumors by a comprehensive genomic approach including transcriptome sequencing. PLoS One 2013; 8: e76623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sahm F, Koelsche C, Meyer J, et al CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta Neuropathol 2012; 123: 853–60. [DOI] [PubMed] [Google Scholar]
- 42. Choi N, Park J, Lee JS, et al miR‐93/miR‐106b/miR‐375‐CIC‐CRABP1: a novel regulatory axis in prostate cancer progression. Oncotarget 2015; 6: 23533–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aihara K, Mukasa A, Nagae G, et al Genetic and epigenetic stability of oligodendrogliomas at recurrence. Acta Neuropathol Commun 2017; 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Atak ZK, Gianfelici V, Hulselmans G, et al Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T‐cell acute lymphoblastic leukemia. PLoS Genet 2013; 9: e1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ding H, Beckers MC, Plaisance S, Marynen P, Collen D, Belayew A. Characterization of a double homeodomain protein (DUX1) encoded by a cDNA homologous to 3.3 kb dispersed repeated elements. Hum Mol Genet 1998; 7: 1681–94. [DOI] [PubMed] [Google Scholar]
- 46. van Overveld PGM, Lemmers RJFL, Deidda G, et al Interchromosomal repeat array interactions between chromosomes 4 and 10: a model for subtelomeric plasticity. Hum Mol Genet 2000; 9: 2879–84. [DOI] [PubMed] [Google Scholar]
- 47. Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 2002; 110: 339–48. [DOI] [PubMed] [Google Scholar]
- 48. Zeng W, de Gree JC, Chen YY, et al Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet 2009; 5: e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Overveld PG, Lemmers RJ, Sandkuijl LA, et al Hypomethylation of D4Z4 in 4q‐linked facioscapulohumeral muscular dystrophy. Nat Genet 2003; 35: 315–7. [DOI] [PubMed] [Google Scholar]
- 50. Dixit M, Ansseau E, Tassin A, et al DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA 2007; 104: 18157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Choi SH, Gearhart MD, Cui Z, et al DUX4 recruits p300/CBP through its C‐terminus and induces global H3K27 acetylation changes. Nucl Acids Res 2016; 44: 5161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoshida A, Goto K, Kodaira M, et al Cic‐rearranged sarcomas: A study of 20 cases and comparisons with Ewing sarcomas. Am J Surg Pathol 2016; 51: 313–23. [DOI] [PubMed] [Google Scholar]
- 53. Specht K, Sung YS, Zhang L, Richter GH, Fletcher CD, Antonescu CR. Distinct transcriptional signature and immunoprofile of CIC‐DUX4 fusion‐positive round cell tumors compared to EWSR1‐reaerranged Ewing sarcomas: further evidence toward distinct pathologic entities. Genes Chromosom Cancer 2014; 53: 622–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Antonescu CR, Owosho AA, Zhang L, et al Sarcomas with CIC‐rearrangements are a distinct pathologic entity with aggressive outcome. Am J Surg Pathol 2017; 41: 941–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoshimoto T, Tanaka M, Homme Y, et al CIC‐DUX4 induces small round cell sarcomas distinct from Ewing sarcoma. Cancer Res 2017; 77: 2927–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tanaka M, Yamazaki Y, Kanno Y, et al Ewing's sarcoma precursors are highly enriched in embryonic osteochondrogenic progenitors. J Clin Invest 2014; 121: 3061–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Italiano A, Sung YS, Zhang L, et al High prevalence of CIC fusion with double homeobox (DUX4) transcription factors in EWSR1‐negative undifferentiated small blue round cell sarcomas. Genes Chromosom Cancer 2012; 51: 207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Le Guellec S, Velasco V, Perot G, et al ETV4 is a useful marker for the diagnosis of CIC‐rearranged undifferentiated round‐cell sarcomas: a study of 127 cases including mimicking lesions. Mod Pathol 2016; 29: 1523–31. [DOI] [PubMed] [Google Scholar]
- 59. Gambarotti M, Benini S, Gamberi G, et al CIC‐DUX4 fusion‐positive round‐cell sarcomas of soft tissue and bone: a single‐institution morphological and molecular analysis of seven cases. Histopathol 2016; 69: 624–34. [DOI] [PubMed] [Google Scholar]
- 60. Yasuda T, Tsuzuki S, Kawazu M, et al Recurrent DUX4 fusions in B‐cell lymphoblastic leukemia of adolescents and young adults. Nat Genet 2016; 48: 569–74. [DOI] [PubMed] [Google Scholar]
- 61. Sugita S, Arai Y, Tonooka A, et al A novel CIC‐FOXO4 gene fusion in undifferentiated small round cell sarcoma: a genetically distinct variant of Ewing‐like sarcoma. Am J Surg Pathol 2014; 38: 1571–6. [DOI] [PubMed] [Google Scholar]
- 62. Sturm D, Orr BA, Toprak UH, et al New brain tumor entities emerge from molecular classification of CNS‐PNETs. Cell 2016; 164: 1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Huang SC, Zhang L, Sung YS, et al Recurrent CIC gene abnormalities in angiosarcomas. A molecular study of 120 cases with concurrent investigation of PLCG1, KDR, MYC, and FLT4 gene alterations. Am J Surg Pathol 2016; 40: 645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. LeBlanc VG, Firme M, Song J, et al Comparative transcriptome analysis of isogenic cell line models and primary cancers links capicua (CIC) loss to activation of the MAPK signalling cascade. J Pathol 2017; 242: 206–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang B, Krall EB, Aguirre AJ, et al ATXN1L, CIC, and ETS transcription factors modulate sensitivity to MAPK pathway inhibition. Cell Rep 2017; 18: 1543–57. [DOI] [PMC free article] [PubMed] [Google Scholar]


