Abstract
The expression of microRNA (miR)‐140‐5p is known to be reduced in both pulmonary arterial hypertension (PAH) patients and monocrotaline‐induced PAH models in rat. Identification of target genes for miR‐140‐5p with bioinformatics analysis may reveal new pathways and connections in PAH. This study aimed to explore downstream target genes and relevant signaling pathways regulated by miR‐140‐5p to provide theoretical evidences for further researches on role of miR‐140‐5p in PAH. Multiple downstream target genes and upstream transcription factors (TFs) of miR‐140‐5p were predicted in the analysis. Gene ontology (GO) enrichment analysis indicated that downstream target genes of miR‐140‐5p were enriched in many biological processes, such as biological regulation, signal transduction, response to chemical stimulus, stem cell proliferation, cell surface receptor signaling pathways. Kyoto Encyclopedia of Genes and Genome (KEGG) pathway analysis found that downstream target genes were mainly located in Notch, TGF‐beta, PI3K/Akt, and Hippo signaling pathway. According to TF–miRNA–mRNA network, the important downstream target genes of miR‐140‐5p were PPI, TGF‐betaR1, smad4, JAG1, ADAM10, FGF9, PDGFRA, VEGFA, LAMC1, TLR4, and CREB. After thoroughly reviewing published literature, we found that 23 target genes and seven signaling pathways were truly inhibited by miR‐140‐5p in various tissues or cells; most of these verified targets were in accordance with our present prediction. Other predicted targets still need further verification in vivo and in vitro.
Keywords: GO, KEGG, miR‐140‐5p, target gene, transcription factor
Abbreviations
- GO
gene ontology
- KEGG
kyoto encyclopedia of genes and genome
- PAH
pulmonary arterial hypertension
- PASMC
pulmonary arterial smooth muscle cell
- TF
transcription factor
Pulmonary arterial hypertension (PAH) is a chronic progressive disease of pulmonary vasculature characterized by sustained elevation of pulmonary vascular resistance and pulmonary arterial pressure, consequently leading to right heart failure and eventual death 1. The pathogenesis of PAH is associated with genetic predisposition, inflammation, increase in vascular tone, elevation in pulmonary artery cell proliferation and resistance to apoptosis, and the presence of in situ thrombosis 2, 3, 4, 5. Effect of current treatment on PAH remains poor and available therapies to improve long‐term prognosis are limited 6, so exploring novel molecular mechanisms and generating therapeutic approaches are urgently needed.MicroRNAs (miRNAs) are small noncoding RNA molecules around 22 nucleotides long that bind the 3′‐untranslated region (UTR) of mRNA to degrade mRNA and therefore to negatively regulate relevant genes expression 7. miRNAs have the ability to target numerous genes mRNA, therefore potentially controlling a host of genes expression and the activity of multiple signaling pathways 8, 9, 10. Recent studies have shown that reduction in microRNA (miR)‐140‐5p is found in both patients with PAH and monocrotaline‐induced PAH models in rat, which is involved in the development of PAH 11, 12. Therefore, it is important to identify comprehensive downstream targets of miR‐140‐5p with bioinformatics analysis in PAH, and this might provide some critical information for the development and treatment of PAH. In this study, downstream target genes regulated by miR‐140‐5p and upstream transcription factors (TFs) regulating miR‐140‐5p expression were predicted, and the downstream target genes were analyzed for gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genome (KEGG) pathway. Next, the upstream TFs and downstream targets of miR‐140‐5p were determined according to the TF–miRNA–mRNA network. Finally, the direct downstream targets and relevant signaling pathways regulated by miR‐140‐5p were obtained in published literature and were compared with the predicted results of this study.
Materials and methods
Mature sequences of miR‐140‐5p in various species
Mature sequences of miR‐140‐5p in various species were obtained in the miRBase database (http://mirbase.org/index.shtml).
Target gene prediction of miR‐140‐5p
Identification of target genes is critical for characterizing the functions of miRNAs. In this study, miRanda (http://www.microrna.org/), TargetScan (http://www.targetscan.org/), RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/submission.html), and miRDB (http://www.mirdb.org/) databases were used to predict the target genes of miR‐140‐5p. To make our predicted target genes more convincible, only the target genes predicted by at least three databases were selected for further analyses.
Database‐based GO and KEGG pathway enrichment analysis
Target mRNA of miR‐140‐5p supported by at least three databases were used for GO analysis to predict gene functions. Integration Discovery (DAVID) software, version 6.7 (http://david.abcC.ncifcrf.gov), was used to perform GO analysis to identify biological processes, cellular components, and molecular functions of these target genes. At the same time, the probable signaling pathways in which these target genes were enriched were analyzed by KEGG database (http://www.genome.jp/kegg/). The P‐value <0.05 was considered significant.
Upstream TFs prediction of miR‐140‐5p
Human miR‐140‐5p precursor was obtained in the miRBase database and its 5000 bp upstream was defined as the miR‐140‐5p promoter. The TFs of miR‐140‐5p were predicted using MOODS‐python software (version 1.9.3) in JASPAR database (http://jaspar.binf.ku.dk/), which includes various vertebrate TFs. The P‐value <0.0001 was considered significant.
Construction of the network for TF–miR‐140‐5p–mRNA
By merging the regulatory relationships between TFs and miR‐140‐5p, miR‐140‐5p and target genes, genes and genes (TF→miRNA, miRNA→gene and gene→gene), we constructed a comprehensive TF–miR‐140‐5p–mRNA regulatory network using Gephi software (release 0.8.1‐β, http://gephi.github.io/).
Screening target genes and signaling pathways inhibited by miR‐140‐5p in published studies
To obtain downstream target genes and signaling pathways modulated by miR‐140‐5p in published studies, a comprehensive electronic search of Web of Science and PubMed databases was performed until April 20, 2017. The keyword ‘miR‐140‐5p’ in the titles or abstracts was used, and then, studies exploring the targets of miR‐140‐5p were collected.
Results
Mature sequences of miR‐140‐5p in various species
Mature sequences of miR‐140‐5p in various species were obtained in the miRBase database. The pre‐miR‐140‐5p was located at position 69933081 ~ 69933180 of chromosome 16, and the gene ID of human miR‐140‐5p was MIMAT0000431. As shown in Table 1, mature sequences of miR‐140‐5p were highly conserved in various species and human miR‐140‐5p was chosen for further analyses.
Table 1.
Mature sequences of miR‐140‐5p in various species
| ID | Mature name | Sequence |
|---|---|---|
| MIMAT0000151 | mmu‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0000431 | hsa‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0000573 | rno‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0001159 | gga‐miR‐140‐5p | AGUGGUUUUACCCUAUGGUAG |
| MIMAT0001836 | dre‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0002143 | ssc‐miR‐140‐5p | AGUGGUUUUACCCUAUGGUAG |
| MIMAT0006812 | oan‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGU |
| MIMAT0006197 | mml‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0012745 | mdo‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0012926 | eca‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0014557 | tgu‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0015763 | ppy‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0021765 | aca‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGU |
| MIMAT0022552 | ola‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0023767 | cgr‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0025434 | pol‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0026220 | ccr‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0032359 | ssa‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0035960 | chi‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
| MIMAT0036560 | tch‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUA |
| MIMAT0036719 | oha‐miR‐140‐5p | CAGUGGUUUUACCCUAUGGUAG |
Prediction of target genes for miR‐140‐5p
As shown in Fig. 1, the number of predicted target genes of miR‐140‐5p in miRanda, TargetScan, RNAhybrid, and miRDB databases was 2370, 428, 1017, and 262, respectively. There were 482 target genes supported by at least two databases, 123 target genes predicted by at least three databases and five target genes supported by all four databases. The target genes of miR‐140‐5p predicted by at least three databases are listed in Table 2 and were used for further analyses.
Figure 1.
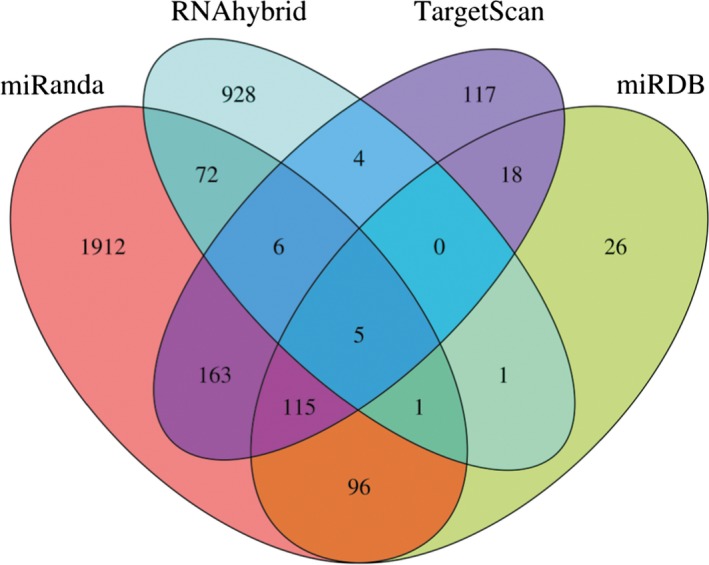
The number of predicted target genes of miR‐140‐5p.
Table 2.
The target genes of miR‐140‐5p predicted by at least three databases
| ABCA1 | ACSL6 | ADAM10 | ADAMTS5 | ADCY6 | ANKFY1 |
| ANKIB1 | AP2B1 | BACH1 | BAZ2B | BCL9 | BMP2 |
| C1R | CADM3 | CAND1 | CAPN1 | CCNYL1 | CELF1 |
| CORO2A | CREB | CTCF | CYTH2 | DNM3 | DOK4 |
| DPP10 | DPYSL2 | EGR2 | EIF4G2 | ELAVL2 | ENTPD5 |
| EPB41L2 | ERC2 | FAM175B | FBN1 | FCHO2 | FES |
| FGF9 | FLRT2 | FOXP2 | FYCO1 | GNG5 | GIT1 |
| HAND2 | HDAC4 | HDAC7 | HDGFRP3 | HNRNPH3 | HS2ST1 |
| HSPA13 | IGSF3 | IPO7 | JAG1 | KAT2B | KBTBD2 |
| KIF1B | KLF6 | KLF9 | KLK10 | LAMC1 | LHFPL2 |
| LMNB1 | LPHN2 | LRAT | LRP4 | LSM14B | LYSMD3 |
| MARK1 | MED13 | MMD | MYCBP2 | MYO10 | NAA20 |
| NAALADL2 | NCKAP1 | NCOA1 | NCSTN | NFE2L2 | NLK |
| NPL | NUCKS1 | OSBPL6 | PPPICC | PAFAH1B2 | PDGFRA |
| PPTC7 | PDE7A | PPP1R12A | PALM2‐AKAP2 | RBM39 | RFX7 |
| RNF19A | RALA | RAB10 | SEPT2 | STRADB | SYS1 |
| SLAIN1 | SAMD4 | SMOC2 | SNX2 | SRCAP | SHROOM3 |
| SIAH1 | SLC30A5 | SLC38A2 | TTYH3 ST5 | TLR4 | TTK |
| TJP1 | TSSK2 | TSPAN12 | TSC22D2 | TTYH2 | TGFBR1 |
| UBR5 | UBR5 | VEZF1 | VEGFA | WNT1 | WDFY3 |
| YOD1 | ZBTB10 | ZNF800 |
GO enrichment analysis for predicted target genes of miR‐140‐5p
GO enrichment analysis was conducted for the target genes of miR‐140‐5p predicted by at least three databases. As shown in Table 3, the target genes of miR‐140‐5p were mainly located in basement membrane (P < 0.05) and participated in the molecular functions of protein binding, activating transcription factor binding, ion binding, lipid binding, and so on (P < 0.05). In addition, the target genes of miR‐140‐5p were involved in various biological processes, including biological regulation, metabolic process, cell communication, signal transduction, response to chemical stimulus, stem cell proliferation, cell surface receptor signaling pathway (P < 0.05). Fig. 2 presents the number of target genes corresponding to each GO term.
Table 3.
Gene ontology (GO) analysis for predicted target genes of miR‐140‐5p
| ID | Term | P‐value | Genes annotated to the term |
| Biological processes | |||
| GO:0050794 | Regulation of cellular process | 5.39E‐06 | VEGFA| FGF9| PPP1CC|Pin1|HDAC7|PDGFRA|TGFBR1|ADAM10… |
| GO:0050789 | Regulation of biological process | 9.05E‐06 | FGF9|BMP2|LAMC1|NUMBL|PDGFRA|PPP1CC||ADAM10|TLR4|TGFBR1… |
| GO:0007154 | Cell communication | 5.69E‐05 | WNT1|PPP1CC|PDGFRA|TLR4|HDAC7|ADAM10| BMP2|TGFBR1… |
| GO:0023052 | Signaling | 6.14E‐05 | PDGFRA|PPP1CC|FGF9|WNT1|TGFBR1|BMP2|ADAM10|JAG1|TLR4… |
| GO:0044763 | Single‐organism cellular process | 8.73E‐05 | VEGFA|FGF9|LAMC1|BMP2|TLR4|WNT1|TGFBR1|PDGFRA|PPP1CC… |
| GO:0065007 | Biological regulation | 9.89E‐05 | VEGFA|BMP2|TLR4|CREB|PPP1CC|PDGFRA|ADAM10|TGFBR1… |
| GO:0007165 | Signal transduction | 0.00011 | PPP1CC|PDGFRA|WNT1|TGFBR1|FGF9|VEGFA|NCSTN|TLR4|ADAM10… |
| GO:0042221 | Response to chemical stimulus | 0.00048 | NUMBL|PPP1CC|PDGFRA|VEGFA|LAMC1|TGFBR1|FGF9|BMP2|ADAM10|TLR4… |
| GO:0072089 | Stem cell proliferation | 0.00087 | ACSL6|NUMBL|RAB10|HAND2|WNT1|BMP2… |
| GO:0007166 | Cell surface receptor signaling pathway | 0.00370 | TLR4|WNT1|BMP2|ADAM10|NCSTN|JAG1|PPP1CC|PDGFRA|FGF9… |
| GO:0050896 | Response to stimulus | 0.01555 | PPP1CC|PDGFRA|WNT1|CREB|TGFBR1|VEGFA||FGF9|BMP2|ADAM10|TLR4… |
| GO:0019538 | Protein metabolic process | 0.02054 | CREB|PPP1CC|PDGFRA|NUMBL|TLR4|ADAM10|BMP2|KAT2B|NCSTN| TGFBR1… |
| GO:0006464 | Cellular protein modification process | 0.03073 | HDAC4|CREB|ADAM10|TLR4|TGFBR1|PPP1CC|PDGFRA… |
| Molecular functions | |||
| GO:0005515 | Protein binding | 2.53E‐07 | TLR4|ADAM10|PDGFRA|WNT1|HDAC7|VEGFA|CREB|PPP1CC|TGFBR1|FGF9… |
| GO:0005488 | Binding | 0.00048 | HDAC7|JAG|LMNB1|PDGFRA|ADAM10| TLR4|FGF9|KAT2B|TGFBR1… |
| GO:0033613 | Activating transcription factor binding | 0.00320 | EGR2|NFE2L2|HDAC4|HDAC7|HAND2… |
| GO:0043167 | Ion binding | 0.00724 | VEGFA|PPP1CC|ADAM10|PDGFRA|TGFBR1|HDAC4|FGF9|HDAC7… |
| GO:0008289 | Lipid binding | 0.04471 | LAMC1|OSBPL6|FES|DNM3|MYO10|TLR4… |
| Cellular components | |||
| GO:0005604 | Basement membrane | 0.04119 | FGF9|PDGFRA|TLR4|VEGFA|SMOC2… |
Figure 2.
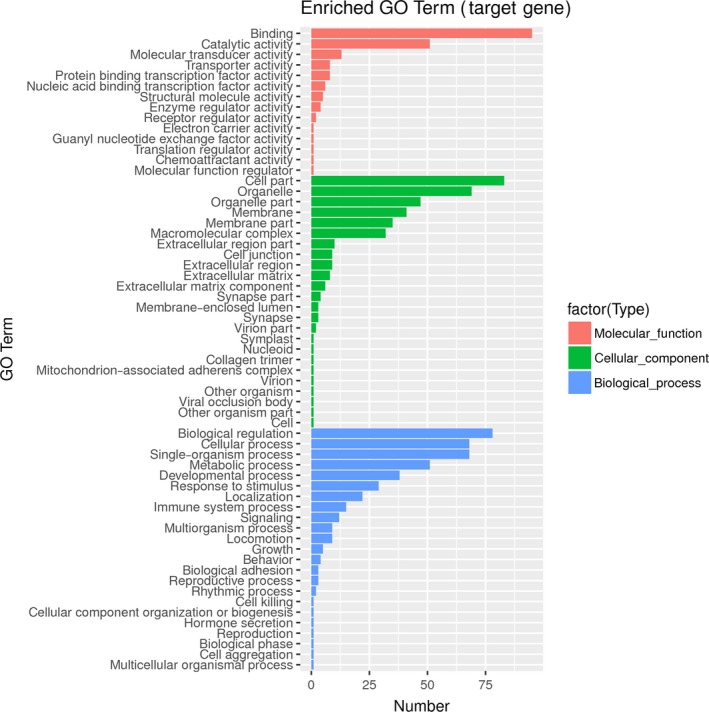
Gene ontology (GO) enrichment analysis for predicted target genes of miR‐140‐5p.
KEGG pathway analysis for predicted target genes of miR‐140‐5p
Enriched signaling pathways for the target genes of miR‐140‐5p identified by KEGG pathway analysis were ranked according to the P‐values. As shown in Table 4, the top rankings were related to Notch, cancer‐associated pathway, TGF‐beta, PI3K/Akt, HTLV infection, Hippo, HIF‐1, alcoholism signaling pathways, and so on (P < 0.05); among them, Notch, TGF‐beta, PI3K/Akt, and Hippo signaling pathways were well known to be associated with the pathogenesis of PAH. Fig. 3 presents the rich factor, Q value, and gene number corresponding to each pathway term.
Table 4.
Kyoto Encyclopedia of Genes and Genome (KEGG) pathway analysis for predicted target genes of miR‐140‐5p
| Term | ID | Sample number | Background number | P‐value | Genes |
|---|---|---|---|---|---|
| Notch signaling pathway | hsa04330 | 4 | 52 | 0.006408 | JAG1|ADAM10|KAT2B|NCSTN |
| Pathways in cancer | hsa05200 | 9 | 337 | 0.016384 | FGF9|TGFBR1|VEGFA|SLC2A1|WNT1|BMP2|PDGFRA|LAMC1 |
| Endocrine and other factor‐regulated calcium reabsorption | hsa04961 | 3 | 48 | 0.022347 | AP2B1|ADCY6|DNM3 |
| HTLV‐I infection | hsa05166 | 7 | 268 | 0.031935 | TGFBR1|KAT2B|SLC2A1|EGR2|WNT1|PDGFRA|ADCY6 |
| Regulation of actin cytoskeleton | hsa04810 | 6 | 221 | 0.031935 | PPP1R12A|NCKAP1|FGF9|GIT1|PDGFRA|PPP1CC |
| Pancreatic cancer | hsa05212 | 3 | 66 | 0.031935 | RALA|TGFBR1|VEGFA |
| Epithelial cell signaling in Helicobacter pylori infection | hsa05120 | 3 | 66 | 0.031935 | TJP1|GIT1|ADAM10 |
| Proteoglycans in cancer | hsa05205 | 6 | 231 | 0.033735 | PPP1R12A|FGF9|VEGFA|WNT1|TLR4|PPP1CC |
| Adherence junction | hsa04520 | 3 | 74 | 0.037848 | NLK|TJP1|TGFBR1 |
| Alcoholism | hsa05034 | 5 | 183 | 0.038681 | HDAC7|HDAC4|CREB3L1|GNG5|PPP1CC |
| PI3K‐Akt signaling pathway | hsa04151 | 7 | 358 | 0.045545 | FGF9|VEGFA|PDGFRA|LAMC1|TLR4|CREB|GNG5 |
| Focal adhesion | hsa04510 | 5 | 214 | 0.045545 | PPP1R12A|VEGFA|PDGFRA|LAMC1|PPP1CC |
| Endocytosis | hsa04144 | 5 | 212 | 0.045545 | AP2B1|TGFBR1|GIT1|PDGFRA|DNM3 |
| Viral carcinogenesis | hsa05203 | 5 | 213 | 0.045545 | HDAC7|HDAC4|KAT2B|EGR2|CREB3L1 |
| Hepatitis B | hsa05161 | 4 | 151 | 0.045545 | TGFBR1|EGR2|TLR4|CREB3L1 |
| Insulin secretion | hsa04911 | 3 | 92 | 0.045545 | SLC2A1|CREB3L1|ADCY6 |
| GABAergic synapse | hsa04727 | 3 | 89 | 0.045545 | SLC38A2|GNG5|ADCY6 |
| TGF‐beta signaling pathway | hsa04350 | 3 | 83 | 0.045545 | TGFBR1|SMAD4|BMP2 |
| Gap junction | hsa04540 | 3 | 96 | 0.045545 | TJP1|PDGFRA|ADCY6 |
| Hippo signaling pathway | hsa04390 | 4 | 156 | 0.045565 | TGFBR1|WNT1|BMP2|PPP1CC |
Figure 3.
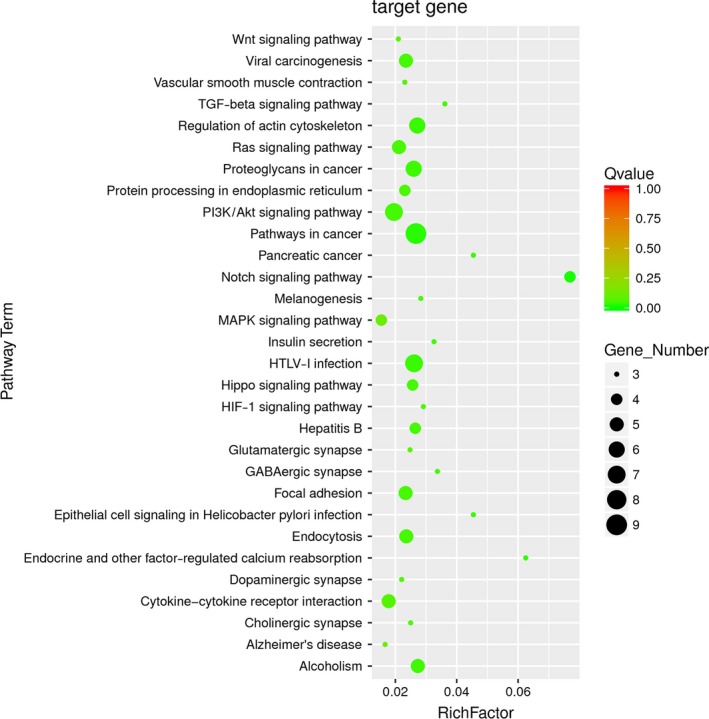
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for predicted target genes of miR‐140‐5p.
Prediction of upstream TFs for miR‐140‐5p and construction of TF–miR‐140‐5p–mRNA network
The number of predicted TFs for miR‐140‐5p with P‐value <0.0001 was 393. To reduce false‐positive results, TFs with a quality score (Q‐score) less than 10 were filtered. As shown in Table 5, the remaining TFs, including PAX5, FOXI1, IRF1, FOSL1, RUNX2, were chosen for further analyses. Finally, by merging the regulatory relationships between TFs and miR‐140‐5p, miR‐140‐5p and target genes, as well as genes and genes, we built a comprehensive TF–miR‐140‐5p–mRNA regulatory network, as shown in Fig. 4.
Table 5.
Prediction of transcription factors and binding sites of miR‐140‐5p
| Model ID | Model name | Hit position | Strand | Score | Predicted site sequence |
|---|---|---|---|---|---|
| MA0014.2 | PAX5 | 95 | − | 10.5663 | gtctcactctgttgcccat |
| MA0014.2 | PAX5 | 3874 | − | 11.6915 | gtcttgctctgttgcccag |
| MA0025.1 | NFIL3 | 722 | − | 10.0393 | TTCTTACATAA |
| MA0035.3 | Gata1 | 3391 | − | 10.0718 | acagataaaaa |
| MA0036.2 | GATA2 | 3391 | − | 10.4087 | acagataaaaattt |
| MA0041.1 | Foxd3 | 4529 | + | 10.4011 | ttttgtttgttt |
| MA0042.1 | FOXI1 | 984 | + | 11.5926 | GGATGTTTGTTT |
| MA0042.1 | FOXI1 | 4529 | + | 10.3990 | ttttgtttgttt |
| MA0046.1 | HNF1A | 4949 | + | 10.3282 | agttaataatttta |
| MA0050.2 | IRF1 | 3825 | + | 11.0065 | tttttctttttcttttctttc |
| MA0050.2 | IRF1 | 3840 | + | 12.4803 | tctttctttcttttttttttt |
| MA0050.2 | IRF1 | 3844 | + | 10.0776 | tctttcttttttttttttttt |
| MA0062.2 | GABPA | 1506 | + | 10.0387 | ccggaagtcga |
| MA0073.1 | RREB1 | 1164 | − | 10.9028 | TTTTGGTTGTTGTTTTGTTT |
| MA0073.1 | RREB1 | 3734 | + | 10.2056 | caacaaaacaaaacaaaaca |
| MA0471.1 | E2F6 | 143 | − | 10.6410 | tcttcccgcct |
| MA0477.1 | FOSL1 | 4238 | − | 11.2229 | cctgagtcacc |
| MA0478.1 | FOSL2 | 4239 | − | 10.3145 | ctgagtcacct |
| MA0481.1 | FOXP1 | 3756 | + | 10.2195 | acaaaaaaaacacaa |
| MA0481.1 | FOXP1 | 4018 | − | 10.3465 | ttttgtttttttagt |
| MA0490.1 | JUNB | 4239 | − | 10.6046 | ctgagtcacct |
| MA0491.1 | JUND | 2362 | + | 10.0256 | GAAAATGATATCACA |
| MA0493.1 | Klf1 | 4812 | + | 10.548 | caccacaccca |
| MA0511.1 | RUNX2 | 3813 | + | 11.453 | tgtgtatgtggtttt |
| MA0515.1 | Sox6 | 3772 | − | 10.2529 | gaaacaatgg |
| MA0595.1 | SREBF1 | 2000 | − | 10.1772 | gtggcgtgat |
Figure 4.
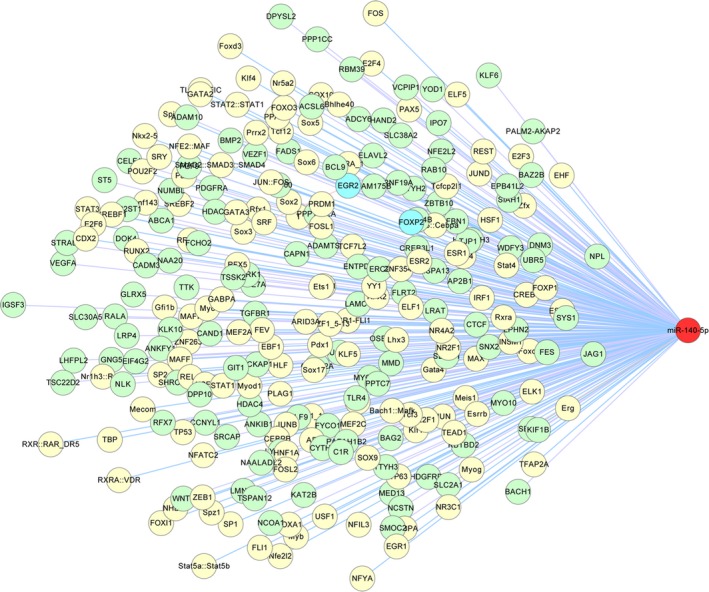
Regulatory network of TF–miR‐140‐5p–mRNA.
Screening target genes and signaling pathways modulated by miR‐140‐5p in published studies
A comprehensive electronic search of Web of Science and PubMed databases was performed until April 20, 2017, to obtain target genes and signaling pathways modulated by miR‐140‐5p in published studies. Finally, a total of 26 papers including 23 target genes and seven signaling pathways inhibited by miR‐140‐5p were obtained; most of them focus on the functions of miR‐140‐5p suppressing tumor growth, migration, and invasion in various tumor tissues and cells. Two recent studies have found that SMURF1 and Dumt1 are direct target genes of miR‐140‐5p in pulmonary arterial smooth muscle cells (PASMCs) and are involved in the pathogenesis of PAH. The details are shown in Table 6.
Table 6.
Target genes and signaling pathways modulated by miR‐140‐5p in published studies. NA, not available; HCC, hepatocellular carcinoma; T‐ALL, T‐cell acute lymphoblastic leukemia; Th1, T helper type 1; HSCC, hypopharyngeal squamous cell carcinoma; EPCs, endothelial progenitor cells; PH, pulmonary hypertension; HUVECs, human umbilical vein endothelial cells; BTC, biliary tract cancer; TSPCs, tendon stem/progenitor cells; LLC, Lewis lung cancer cells; MSCs, mesenchymal stem cells; TSCC, tongue squamous cell carcinoma
| Author (Year) | Target genes | Inhibited pathways | Associated functions | Cell or tissue types |
|---|---|---|---|---|
| Hu (2017) | VEGFA | NA | Inhibit cell proliferation and invasion, promote apoptosis | Glioma tissues and cells |
| Meng (2017) | HMGN5 | NA | Decrease cell resistance to chemotherapy | Osteosarcoma tissues and cells |
| Yan (2017) | Pin1 | Pin1‐dependent cancer pathway | Suppress tumor growth | HCC tissues and cells |
| Correia (2016) | TAL1 | NA | Suppress tumor growth | T‐ALL cells |
| Guan (2016) | STAT1 | NA | Suppress Th1 cell differentiation | Th1 cells |
| Jing (2016) | ADAM10 | Notch1 signaling pathway | Suppress tumor migration and invasion | HSCC tissues and cells |
| Liu (2016) | HDAC7 | NA | Protect EPCs | EPCs |
| Lv (2016) | Slug | NA | Inhibit cell migration and invasion | HCC tissues |
| Rothman (2016) | SMURF1 | BMP signaling pathway | Inhibit cell proliferation, migration, and PH development | PASMCs, rat PH models |
| Su (2016) | IGF2BP1 | NA | Decrease cell proliferation, migration, and invasion | Cervical cancer cells and tissues |
| SUN (2016) | VEGFA | NA | Decrease cell proliferation, migration, and tube formation | HUVECs |
| Wei (2016) | IP3k2 | IP3 signaling pathway | Promote chemotherapy‐induced autophagy | Human osteosarcoma cells |
| Yu (2016) | Septin 2 | NA | Suppress cell proliferation and colony formation | BTC tissues and cells |
| Zhang (2016) | Dnmt1 | NA | Inhibit cell proliferation, promote cell apoptosis | Human PH tissues, human PASMCs |
| Barter (2015) | FZD6 | Wnt signaling pathway | Promote chondrogenic differentiation | Mesenchymal stem cells |
| Chen (2015) | Pin1 | NA | Promote cell senescence | TSPCs |
| Lan (2015) | PDGFRA | NA | Inhibit cancer growth | Human ovarian cancer tissues and cells |
| Zhai (2015) | Smad2 | TGF‐β signaling pathway | Decrease cell invasion and proliferation | Colorectal cancer stem cells |
| Zhang (2015) | VEGFA | NA | Inhibit tumor progression | Colorectal cancer tissues and cells |
| Zhang (2015) | TGFBR1 | TGF‐β signaling pathway | Regulate adipocyte differentiation | Bone marrow stromal cells |
| Li (2014) | MMD | ERK signaling pathway | Inhibit cell proliferation | LLCs |
| Hwang (2014) | BMP2 | BMP signaling pathway | Suppress osteogenesis | Human MSCs |
| Karlsen (2014) | RALA | NA | Stimulate chondrogenesis | MSCs |
| Yang (2014) | ADAM10, LAMC1, HDAC7 | NA | Suppress migration and invasion | TSCC tissues and cells |
| Shi (2013) | FoxP2 | NA | Impair dendritic development and vocal learning | Zebra finch brain tissues |
| Yang (2013) | TGFBR1, FGF9 | TGF‐β and ERK signaling pathway | Suppress cell proliferation and tumor metastasis | HCC tissues and cells |
Discussion
Pulmonary arterial hypertension is a chronic life‐threatening condition requiring long‐term management 13, and its available therapies are limited 6. There is a clear and urgent need for new therapeutic options based on deeply exploring the pathogenesis of PAH. Previous studies have indicated that miR‐140‐5p is dramatically downregulated, which in turn causes the development of a variety of cancers by the loss of suppressing tumor cell migration and growth 14, 15, 16, 17. miR‐140‐5p has been recently found to be reduced in both PAH patients and MCT‐induced PAH models in rat 11, 12. However, the downstream targets regulated by miR‐140‐5p contributing to the development of PAH remain largely unknown.
In this study, we found that the target genes of miR‐140‐5p were enriched in many biological processes, such as biological regulation, metabolic process, cell communication, signal transduction, response to chemical stimulus, stem cell proliferation, cell surface receptor signaling pathway. In KEGG pathway analysis, the target genes of miR‐140‐5p were mainly located in Notch, TGF‐beta, PI3K/Akt, and Hippo signaling pathways. According to the TF–miRNA–mRNA network, the important genes potentially regulated by miR‐140‐5p included PPI, TGF‐betaR1, smad4, JAG1, ADAM10, FGF9, PDGFRA, VEGFA, TLR4, LAMC1, CREB, and the upstream TFs, which might regulate miR‐140‐5p expression including TAX5, FOXI, IRF1, GATA6, RUNX2. After thoroughly reviewing published literature, we found that 23 target genes and seven signaling pathways were truly inhibited by miR‐140‐5p in various tissues or cells; most of these downstream targets were in accordance with our present prediction.
Several studies have shown that activation of Notch3 pathway is involved in the pathogenesis of PAH 18, 19. We have previously shown that activation of Notch3 promotes PASMC proliferation and inhibition of Notch3 pathway prevents monocrotaline‐induced development of PAH in rat 20, 21. JAG1 and ADAM10 are indispensable components of Notch signaling pathway, which were predicted as downstream targets of miR‐140‐5p in our analysis, suggesting that lack of miR‐140‐5p might promote the development of PAH by upregulation of JAG1 and ADAM10 genes and therefore activation of Notch3 cascade. In addition, activation of TGF‐beta1/Smad4 signaling promotes a proliferative PASMC phenotype and induces PAH in rat 22, 23. We found that TGF‐betaR1 and smad4 were possible downstream targets of miR‐140‐5p, reduction in miR‐140‐5p in PAH might stimulate TGF‐beta1/Smad4 pathway by upregulating TGF‐betaR1 and smad4. Previous studies have demonstrated that PDGF, TLR4, VEGFA, and FGF contribute to the pathogenesis of PAH via activating various signaling pathways, especially PI3K/Akt cascade 24, 25, 26, 27, 28. CREB, an important transcription factor lying downstream of PI3K/Akt pathway, mediates the partial functions of PI3K/Akt 29. In our analysis, PDGF, TLR4, VEGFA, FGF, and CREB were positively predicted as downstream targets of miR‐140‐5p, implying that miR‐140‐5p negatively regulates the functions of PI3K/Akt cascade by targeting FGF9, PDGFRA, VEGFA, TLR4, or CREB gene. Recent studies have also shown that Hippo signaling is associated with the development of PAH, which can be activated by PPI 30, 31. Our present results suggested that PPI was a direct target gene of miR‐140‐5p and might mediate miR‐140‐5p regulation of Hippo signaling.
Our predicted network provided potential target genes and relevant signaling pathways that might be modulated by miR‐140‐5p contribution to the development of PAH. Several targets and pathways predicted in our analysis, such as TGF‐betaR1, ADAM10, FGF9, PDGFRA, VEGFA and Notch, PI3K/Akt, TGF‐beta cascades, have been demonstrated to mediate the effects of miR‐140‐5p on antiproliferation and prodifferentiation in several cell types in published studies 16, 17, 32, 33. While the other targets predicted in our study, including PPI, smad4, JAG1, LAMC1, TLR4, and CREB as well as Hippo signaling pathway, have not been confirmed in the published literature, they still need further verification in vivo and in vitro.
Author contributions
ML and FL designed the study; WS, YW, LC, and QW analyzed and interpreted the data; WF, XY, QZ, and JW organized the results; FL wrote the manuscript.
Acknowledgement
This work was supported by Chinese National Science Foundation (No. 81670051 and No. 81330002).
References
- 1. Grant JS, White K, MacLean MR and Baker AH (2013) MicroRNAs in pulmonary arterial remodeling. Cell Mol Life Sci 70, 4479–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Higasa K, Ogawa A, Terao C, Shimizu M, Kosugi S, Yamada R, Date H, Matsubara H and Matsuda F (2017) A burden of rare variants in BMPR2 and KCNK3 contributes to a risk of familial pulmonary arterial hypertension. BMC Pulm Med 17, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Malenfant S, Neyron AS, Paulin R, Potus F, Meloche J, Provencher S and Bonnet S (2013) Signal transduction in the development of pulmonary arterial hypertension. Pulm Circ 3, 278–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rabinovitch M (2012) Molecular pathogenesis of pulmonary arterial hypertension. J Clin Invest 122, 4306–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vaillancourt M, Ruffenach G, Meloche J and Bonnet S (2015) Adaptation and remodelling of the pulmonary circulation in pulmonary hypertension. Can J Cardiol 31, 407–415. [DOI] [PubMed] [Google Scholar]
- 6. McLaughlin VV (2011) Looking to the future: a new decade of pulmonary arterial hypertension therapy. Eur Respir Rev 20, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Negi V and Chan SY (2017) Discerning functional hierarchies of microRNAs in pulmonary hypertension. JCI Insight 2, e91327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bienertova‐Vasku J, Novak J and Vasku A (2015) MicroRNAs in pulmonary arterial hypertension: pathogenesis, diagnosis and treatment. J Am Soc Hypertens 9, 221–234. [DOI] [PubMed] [Google Scholar]
- 9. Bockmeyer CL, Maegel L, Janciauskiene S, Rische J, Lehmann U, Maus UA, Nickel N, Haverich A, Hoeper MM, Golpon HA et al (2012) Plexiform vasculopathy of severe pulmonary arterial hypertension and microRNA expression. J Heart Lung Transplant 31, 764–772. [DOI] [PubMed] [Google Scholar]
- 10. Caruso P, MacLean MR, Khanin R, McClure J, Soon E, Southgate M, MacDonald RA, Greig JA, Robertson KE, Masson R et al (2010) Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler Thromb Vasc Biol 30, 716–723. [DOI] [PubMed] [Google Scholar]
- 11. Rothman AM, Arnold ND, Pickworth JA, Iremonger J, Ciuclan L, Allen RM, Guth‐Gundel S, Southwood M, Morrell NW, Thomas M et al (2016) MicroRNA‐140‐5p and SMURF1 regulate pulmonary arterial hypertension. J Clin Invest 126, 2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y and Xu J (2016) MiR‐140‐5p regulates hypoxia‐mediated human pulmonary artery smooth muscle cell proliferation, apoptosis and differentiation by targeting Dnmt1 and promoting SOD2 expression. Biochem Biophys Res Commun 473, 342–348. [DOI] [PubMed] [Google Scholar]
- 13. Hoeper MM, Bogaard HJ, Condliffe R, Frantz R, Khanna D, Kurzyna M, Langleben D, Manes A, Satoh T, Torres F et al (2014) Definitions and diagnosis of pulmonary hypertension. Turk Kardiyol Dern Ars 42(Suppl 1), 55–66. [PubMed] [Google Scholar]
- 14. Kai Y, Peng W, Ling W, Jiebing H and Zhuan B (2014) Reciprocal effects between microRNA‐140‐5p and ADAM10 suppress migration and invasion of human tongue cancer cells. Biochem Biophys Res Commun 448, 308–314. [DOI] [PubMed] [Google Scholar]
- 15. Li W and He F (2014) Monocyte to macrophage differentiation‐associated (MMD) targeted by miR‐140‐5p regulates tumor growth in non‐small cell lung cancer. Biochem Biophys Res Commun 450, 844–850. [DOI] [PubMed] [Google Scholar]
- 16. Yang H, Fang F, Chang R and Yang L (2013) MicroRNA‐140‐5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology 58, 205–217. [DOI] [PubMed] [Google Scholar]
- 17. Jing P, Sa N, Liu X, Liu X and Xu W (2016) MicroR‐140‐5p suppresses tumor cell migration and invasion by targeting ADAM10‐mediated Notch1 signaling pathway in hypopharyngeal squamous cell carcinoma. Exp Mol Pathol 100, 132–138. [DOI] [PubMed] [Google Scholar]
- 18. Yu YR, Mao L, Piantadosi CA and Gunn MD (2013) CCR2 deficiency, dysregulation of Notch signaling, and spontaneous pulmonary arterial hypertension. Am J Respir Cell Mol Biol 48, 647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xia Y, Bhattacharyya A, Roszell EE, Sandig M and Mequanint K (2012) The role of endothelial cell‐bound Jagged1 in Notch3‐induced human coronary artery smooth muscle cell differentiation. Biomaterials 33, 2462–2472. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y, Xie X, Zhu Y, Liu L, Feng W, Pan Y, Zhai C, Ke R, Li S, Song Y et al (2015) Inhibition of Notch3 prevents monocrotaline‐induced pulmonary arterial hypertension. Exp Lung Res 41, 435–443. [DOI] [PubMed] [Google Scholar]
- 21. Song Y, Zhang Y, Jiang H, Zhu Y, Liu L, Feng W, Yang L, Wang Y and Li M (2015) Activation of Notch3 promotes pulmonary arterial smooth muscle cells proliferation via Hes1/p27Kip1 signaling pathway. FEBS Open Bio 5, 656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aschner Y and Downey GP (2016) Transforming growth factor‐beta: master regulator of the respiratory system in health and disease. Am J Respir Cell Mol Biol 54, 647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chai SD, Liu T, Dong MF, Li ZK, Tang PZ, Wang JT and Ma SJ (2016) Inactivated Pseudomonas aeruginosa inhibits hypoxia‐induced pulmonary hypertension by preventing TGF‐beta1/Smad signaling. Braz J Med Biol Res 49, e5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bauer EM, Chanthaphavong RS, Sodhi CP, Hackam DJ, Billiar TR and Bauer PM (2014) Genetic deletion of toll‐like receptor 4 on platelets attenuates experimental pulmonary hypertension. Circ Res 114, 1596–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun Z (2014) Platelet TLR4: a critical link in pulmonary arterial hypertension. Circ Res 114, 1551–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Song Y, Wu Y, Su X, Zhu Y, Liu L, Pan Y, Zhu B, Yang L, Gao L and Li M (2016) Activation of AMPK inhibits PDGF‐induced pulmonary arterial smooth muscle cells proliferation and its potential mechanisms. Pharmacol Res 107, 117–124. [DOI] [PubMed] [Google Scholar]
- 27. Chuang JI, Huang JY, Tsai SJ, Sun HS, Yang SH, Chuang PC, Huang BM and Ching CH (2015) FGF9‐induced changes in cellular redox status and HO‐1 upregulation are FGFR‐dependent and proceed through both ERK and AKT to induce CREB and Nrf2 activation. Free Radic Biol Med 89, 274–286. [DOI] [PubMed] [Google Scholar]
- 28. Zheng Y, Ma H, Hu E, Huang Z, Cheng X and Xiong C (2015) Inhibition of FGFR signaling with PD173074 ameliorates monocrotaline‐induced pulmonary arterial hypertension and rescues BMPR‐II expression. J Cardiovasc Pharmacol 66, 504–514. [DOI] [PubMed] [Google Scholar]
- 29. Garat CV, Crossno JT Jr, Sullivan TM, Reusch JE and Klemm DJ (2013) Inhibition of phosphatidylinositol 3‐kinase/Akt signaling attenuates hypoxia‐induced pulmonary artery remodeling and suppresses CREB depletion in arterial smooth muscle cells. J Cardiovasc Pharmacol 62, 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lv XB, Liu CY, Wang Z, Sun YP, Xiong Y, Lei QY and Guan KL (2015) PARD3 induces TAZ activation and cell growth by promoting LATS1 and PP1 interaction. EMBO Rep 16, 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boucherat O, Bonnet S and Paulin R (2016) The HIPPO‐thesis of pulmonary hypertension. Am J Respir Crit Care Med 194, 787–789. [DOI] [PubMed] [Google Scholar]
- 32. Lan H, Chen W, He G and Yang S (2015) miR‐140‐5p inhibits ovarian cancer growth partially by repression of PDGFRA. Biomed Pharmacother 75, 117–122. [DOI] [PubMed] [Google Scholar]
- 33. Zhang W, Zou C, Pan L, Xu Y, Qi W, Ma G, Hou Y and Jiang P (2015) MicroRNA‐140‐5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem 37, 1123–1133. [DOI] [PubMed] [Google Scholar]


