Abstract
SWEET proteins play an indispensable role as a sugar efflux transporter in plant development and stress responses. The SWEET genes have previously been characterized in several plants. Here, we present a comprehensive analysis of this gene family in the rubber tree, Hevea brasiliensis. There are 36 members of the SWEET gene family in this species, making it one of the largest families in plant genomes sequenced so far. Structure and phylogeny analyses of these genes in Hevea and in other species demonstrated broad evolutionary conservation. RNA‐seq analyses revealed that SWEET2, 16, and 17 might represent the main evolutionary direction of SWEET genes in plants. Our results in Hevea suggested the involvement of HbSWEET1a, 2e, 2f, and 3b in phloem loading, HbSWEET10a and 16b in laticifer sugar transport, and HbSWEET9a in nectary‐specific sugar transport. Parallel studies of RNA‐seq analyses extended to three other plant species (Manihot esculenta, Populus trichocarpa, and Arabidopsis thaliana) produced findings which implicated MeSWEET10a, 3a, and 15b in M. esculenta storage root development, and the involvement of PtSWEET16b and PtSWEET16d in P. trichocarpa xylem development. RT‐qPCR results further revealed that HbSWEET10a, 16b, and 1a play important roles in phloem sugar transport. The results from this study provide a foundation not only for further investigation into the functionality of the SWEET gene family in Hevea, especially in its sugar transport for latex production, but also for related studies of this gene family in the plant kingdom.
Keywords: gene expression, Hevea brasiliensis, structure and evolution, sugar transport, SWEET
Abbreviations
- ABA
abscisic acid
- GSDS
gene structure display server
- SRA
sequence read archive
- Suc
sucrose
SWEET (Sugars Will Eventually be Exported Transporter) proteins, which feature up to seven transmembrane TM helix domains, selectively transport different kinds of sugar substrates, including sucrose, fructose, and glucose 1. As a sugar efflux transporter, SWEET proteins play important roles in plant growth and development, stress responses, and plant–microbe interactions. Cellular sugar efflux is an essential function in many processes, such as phloem loading, nectar secretion, nourishing symbionts such as mycorrhiza, and in maternal efflux for filial tissue development 1. Sugar efflux systems can be hijacked by pathogens for access to nutrition from hosts 2, and accordingly, mutations that block recruitment of the efflux mechanism by the pathogen facilitate plant resistance to their attack 3. Previous studies on SWEETs have been focused mainly on two model plant species, namely Arabidopsis thaliana and Oryza sativa 4, 5, 6. In A. thaliana, 17 SWEET family members have been characterized, and they fall into four phylogenetic clades, in which AtSWEET1‐3 are in Clade I, AtSWEET4‐8 in Clade II, AtSWEET9‐15 in Clade III, and AtSWEET16‐17 in Clade IV. The SWEETs from most of the other plants are named following the nomenclature adopted in A. thaliana. SWEET genes, in their many isoforms, are versatile and their functions in plants are widely encompassing. For example, AtSWEET1 acts as a glucose transporter 7, with AtSWEET9 being a nectary‐specific sugar transporter which is essential for nectar production 6. AtSWEET11 and AtSWEET12 catalyze sucrose export from phloem parenchyma cells in source leaves and play a critical role in phloem loading 5. AtSWEET16 and AtSWEET17, as vacuolar SWEET proteins, function as fructose‐specific exporters, connecting the vacuolar lumen to the cytosol 8, 9. In O. sativa, OsSWEET11, located on the plasma membrane and expressed in the phloem of leaves, is presumably involved in phloem loading, as is the case with its Arabidopsis homologues, AtSWEET11 and AtSWEET12 5. OsSWEET11 and OsSWEET14 are specifically exploited by bacterial pathogens for virulence by means of direct binding of a bacterial effector to the SWEET promoter 4, 10. Recently, genome‐wide expression patterns of SWEET genes have been characterized in Brassica napus, Pyrus bretschneideri, Sorghum bicolor, and Glycine max 11, 12, 13, 14, all pointing to important roles of SWEETs in plant growth, development, and stress responses.
Natural rubber (cis‐1,4‐polyisoprene, NR) is an important industrial and strategic raw material, the sole commercial source of which is Hevea brasiliensis (the Para rubber tree, Hevea hereafter), a perennial tropical tree species 15. As sucrose is the precursor molecule for rubber biosynthesis and latex regeneration 16, understanding the mechanisms of its transport and metabolism in the rubber tree is of fundamental importance to improving rubber productivity 17. Significant progress was made in the understanding of Hevea sucrose transport and metabolism with the cloning of six sucrose transporter (SUT) genes, among which HbSUT3 (HbSUT1B) was found to be the key member responsible for sucrose loading into laticifers 18, 19. HbNIN2 has also been identified as the key gene responsible for sucrose catabolism in rubber‐producing laticifers 20. Moreover, two Hevea sucrose synthase genes, HbSUS2 and HbSUS3, were found to exert negative control over sucrose catabolism in the laticifers 21. Hevea SWEET genes have not hitherto been investigated in detail, but their characterization has recently been facilitated by the Hevea genome and transcriptome having been independently sequenced by research groups from China 22, Malaysia 23, 24, and Thailand 25, and by the availability of the proteome of Hevea latex 26.
We report here a genome‐wide analysis of the SWEET gene family in Hevea where we compare the results with those from four other plant species, viz. Manihot esculenta and Ricinus communis belonging to the same family (Euphorbiaceae) as Hevea, and two model plants, A. thaliana and Populus trichocarpa. The study encompassed a total of 127 SWEET genes, the expression patterns of which were analyzed in different tissues in response to various treatments, and at several phases of tissue development. In addition, the gene structure and phylogeny of these genes were compared to help further understanding of the roles of SWEET genes in Hevea sugar transport. As a further objective in this investigation, data on SWEET genes and their expression in four other plant species were examined, along with the results from Hevea, to compare the structure of their respective gene families and appraise the functions of their members in the plant kingdom.
Results and Discussion
Genome‐wide identification of SWEET gene families in Hevea and four other plant species
We identified all SWEET gene family members in five plant species (Hevea, A. thaliana, P. trichocarpa, M. esculenta, and R. communis) from their published genome sequences. In this exercise, the SWEET genes from the three Euphorbiaceae plants (Hevea, M. esculenta, and R. Communis) were characterized for the first time. The most recent genome and protein sequences of these species were downloaded from Phytozome v10. Local BLAST searches of the genomes were performed using the published SWEET sequences of three model plants of A. thaliana, O. sativa, and P. trichocarpa as queries 1, 5. This analysis identified a total of 127 SWEET genes in the five selected plant species, comprising 36 SWEET genes in Hevea (Table 1a) 22, 28 in M. esculenta (Table 1b), 18 in R. communis (Table 1c), 17 in A. thaliana 10, and 28 in P. trichocarpa 6. All the SWEET gene members newly identified this study were named according to the nomenclature of the A. thaliana SWEET gene family. The gene numbers of SWEET families identified here for the two model plants (A. thaliana and P. trichocarpa) matched those previously reported 5.
Table 1.
Characteristics of SWEET genes in three Euphorbiaceae members, H. brasiliensis, M. esculenta, and R. communis
| SWEETs | ID | CDS length in bp | Predicted protein | No. of introns | Group | ||
|---|---|---|---|---|---|---|---|
| Length (aa) | Isoelectric point | Mol Wt | |||||
| (A) H. brasiliensis | |||||||
| HbSWEET1a | scaffold1368_1746 | 753 | 251 | 10.09 | 27668.01 | 5 | Clade I |
| HbSWEET1b | scaffold4412_5699 | 750 | 250 | 8.82 | 27885.95 | 5 | Clade I |
| HbSWEET1c | scaffold2014_38474 | 747 | 249 | 9.81 | 27486.73 | 5 | Clade I |
| HbSWEET2a | scaffold0633_726258 | 705 | 235 | 8.81 | 26322.47 | 5 | Clade I |
| HbSWEET2b | scaffold0291_2393 | 606 | 202 | 8.58 | 22646.22 | 4 | Clade I |
| HbSWEET2c | scaffold0649_515754 | 591 | 197 | 9.54 | 21862.14 | 4 | Clade I |
| HbSWEET2d | scaffold1397_73855 | 543 | 181 | 9.98 | 20036.78 | 5 | Clade I |
| HbSWEET2e | scaffold0991_115099 | 705 | 235 | 8.53 | 25981.78 | 5 | Clade I |
| HbSWEET2f | scaffold1207_75820 | 504 | 168 | 8.02 | 18705.26 | 3 | Clade I |
| HbSWEET3a | scaffold0047_2029699 | 747 | 249 | 9.76 | 27958.14 | 5 | Clade I |
| HbSWEET3b | scaffold0802_319652 | 747 | 249 | 9.93 | 28196.37 | 5 | Clade I |
| HbSWEET4a | scaffold0250_352964 | 726 | 242 | 8.69 | 26996.28 | 5 | Clade II |
| HbSWEET4b | scaffold0371_980268 | 771 | 257 | 6.76 | 28814.09 | 5 | Clade II |
| HbSWEET4c | scaffold0371_939664 | 561 | 187 | 8.89 | 20630.72 | 3 | Clade II |
| HbSWEET5a | scaffold0121_20098 | 714 | 238 | 9.60 | 26665.18 | 5 | Clade II |
| HbSWEET5b | scaffold0190_471668 | 543 | 181 | 9.93 | 20425.77 | 3 | Clade II |
| HbSWEET6 | scaffold1545_54737 | 774 | 258 | 10.00 | 28336.02 | 4 | Clade II |
| HbSWEET7 | scaffold1143_36139 | 783 | 261 | 9.96 | 28805.48 | 4 | Clade II |
| HbSWEET9a | scaffold1512_21440 | 768 | 256 | 10.20 | 28969.04 | 5 | Clade III |
| HbSWEET9b | scaffold0030_998488 | 813 | 271 | 9.11 | 30400.98 | 5 | Clade III |
| HbSWEET10a | scaffold1273_165194 | 684 | 228 | 8.44 | 26296.42 | 5 | Clade III |
| HbSWEET10b | scaffold0491_348730 | 828 | 276 | 9.10 | 31738.97 | 5 | Clade III |
| HbSWEET10c | scaffold1273_149445 | 681 | 227 | 8.45 | 25949.84 | 3 | Clade III |
| HbSWEET10d | scaffold0491_383573 | 783 | 261 | 9.35 | 29770.06 | 5 | Clade III |
| HbSWEET10e | scaffold0462_183492 | 726 | 242 | 10.11 | 27840.30 | 4 | Clade III |
| HbSWEET10f | scaffold0491_387781 | 819 | 273 | 7.37 | 31209.06 | 5 | Clade III |
| HbSWEET11 | scaffold0807_24959 | 846 | 282 | 8.91 | 31803.87 | 5 | Clade III |
| HbSWEET12 | scaffold0807_8989 | 846 | 282 | 8.65 | 31780.94 | 5 | Clade III |
| HbSWEET15a | scaffold0177_54016 | 693 | 231 | 7.03 | 26007.72 | 3 | Clade III |
| HbSWEET15b | scaffold0868_88200 | 900 | 300 | 6.88 | 33874.84 | 5 | Clade III |
| HbSWEET16a | scaffold1307_48627 | 750 | 250 | 8.59 | 27904.85 | 5 | Clade IV |
| HbSWEET16b | scaffold0566_478727 | 732 | 244 | 7.43 | 26617.46 | 5 | Clade IV |
| HbSWEET16c | scaffold0625_502257 | 702 | 234 | 6.51 | 25723.25 | 5 | Clade IV |
| HbSWEET17a | scaffold0340_202757 | 618 | 206 | 9.54 | 22303.57 | 4 | Clade IV |
| HbSWEET17b | scaffold0340_208877 | 915 | 305 | 9.70 | 33568.20 | 5 | Clade IV |
| HbSWEET17c | scaffold0878_306703 | 678 | 226 | 8.96 | 24905.58 | 4 | Clade IV |
| (B) M. esculenta | |||||||
| MeSWEET1a | cassava4.1_014638 m | 750 | 250 | 9.63 | 27676.81 | 5 | Clade I |
| MeSWEET1b | cassava4.1_014650 m | 750 | 250 | 9.75 | 27920.23 | 5 | Clade I |
| MeSWEET2a | cassava4.1_015227 m | 702 | 234 | 8.55 | 26066.93 | 5 | Clade I |
| MeSWEET2b | cassava4.1_030719 m | 564 | 188 | 7.89 | 20898.87 | 3 | Clade I |
| MeSWEET3a | cassava4.1_026477 m | 741 | 247 | 9.63 | 27553.91 | 5 | Clade I |
| MeSWEET3b | cassava4.1_022559 m | 672 | 224 | 9.61 | 25566.19 | 3 | Clade I |
| MeSWEET4 | cassava4.1_016815 m | 582 | 194 | 9.37 | 21364.54 | 3 | Clade II |
| MeSWEET5 | cassava4.1_026390 m | 714 | 238 | 9.09 | 26997.57 | 5 | Clade II |
| MeSWEET6 | cassava4.1_014231 m | 780 | 260 | 10.12 | 28528.22 | 4 | Clade II |
| MeSWEET7 | cassava4.1_028141 m | 777 | 259 | 9.98 | 28488.95 | 4 | Clade II |
| MeSWEET9a | cassava4.1_032222 m | 678 | 226 | 9.40 | 25435.76 | 5 | Clade III |
| MeSWEET9b | cassava4.1_031208 m | 813 | 271 | 8.33 | 30284.89 | 5 | Clade III |
| MeSWEET10a | cassava4.1_013474 m | 840 | 280 | 8.18 | 31795.01 | 5 | Clade III |
| MeSWEET10b | cassava4.1_015602 m | 675 | 225 | 8.03 | 25795.72 | 3 | Clade III |
| MeSWEET10c | cassava4.1_021350 m | 843 | 281 | 9.05 | 31710.95 | 5 | Clade III |
| MeSWEET10d | cassava4.1_013519 m | 837 | 279 | 7.81 | 31755.76 | 5 | Clade III |
| MeSWEET10e | cassava4.1_032927 m | 846 | 282 | 8.44 | 31954.92 | 5 | Clade III |
| MeSWEET11 | cassava4.1_028116 m | 852 | 284 | 8.09 | 31982.66 | 5 | Clade III |
| MeSWEET12a | cassava4.1_017557 m | 522 | 174 | 6.24 | 19568.11 | 2 | Clade III |
| MeSWEET13 | cassava4.1_026944 m | 834 | 278 | 8.88 | 31415.27 | 5 | Clade III |
| MeSWEET15a | cassava4.1_026251 m | 717 | 239 | 9.63 | 27175.79 | 5 | Clade III |
| MeSWEET15b | cassava4.1_014124 m | 789 | 263 | 9.64 | 29723.69 | 6 | Clade III |
| MeSWEET16a | cassava4.1_014996 m | 723 | 241 | 8.16 | 26331.94 | 5 | Clade IV |
| MeSWEET16b | cassava4.1_015143 m | 711 | 237 | 7.24 | 25939.66 | 5 | Clade IV |
| MeSWEET17 | cassava4.1_014640 m | 750 | 250 | 9.20 | 27984.99 | 5 | Clade IV |
| MeSWEET17a | cassava4.1_032999 m | 513 | 171 | 9.59 | 18675.24 | 4 | Clade IV |
| MeSWEET17b | cassava4.1_012690 m | 906 | 302 | 9.83 | 33200.06 | 5 | Clade IV |
| MeSWEET17c | cassava4.1_014587 m | 753 | 251 | 9.76 | 27550.75 | 5 | Clade IV |
| (C) R. communis | |||||||
| RcSWEET1 | 27985.m000892 | 744 | 248 | 10.08 | 27412.66 | 5 | Clade I |
| RcSWEET2 | 30026.m001515 | 504 | 168 | 9.19 | 18787.52 | 3 | Clade I |
| RcSWEET3 | 30169.m006529 | 753 | 251 | 9.27 | 28219.28 | 5 | Clade I |
| RcSWEET4a | 29822.m003349 | 582 | 194 | 7.44 | 21739.97 | 3 | Clade II |
| RcSWEET4b | 27613.m000628 | 699 | 233 | 6.64 | 25779.37 | 0 | Clade II |
| RcSWEET4c | 29475.m000237 | 708 | 236 | 7.98 | 26033.96 | 0 | Clade II |
| RcSWEET4d | 29822.m003348 | 726 | 242 | 8.80 | 27262.76 | 5 | Clade II |
| RcSWEET5 | 30147.m013970 | 645 | 215 | 9.07 | 24416.23 | 3 | Clade II |
| RcSWEET6 | 30068.m002528 | 783 | 261 | 9.98 | 28738.3 | 4 | Clade II |
| RcSWEET9 | 29647.m002020 | 858 | 286 | 8.42 | 32111.69 | 5 | Clade III |
| RcSWEET10a | 30147.m014446 | 831 | 277 | 9.05 | 31790.91 | 5 | Clade III |
| RcSWEET10b | 30147.m014447 | 837 | 279 | 9.00 | 31743.64 | 5 | Clade III |
| RcSWEET11 | 30147.m014444 | 855 | 285 | 8.04 | 32313.45 | 5 | Clade III |
| RcSWEET12 | 30147.m014445 | 891 | 297 | 8.27 | 33206.31 | 5 | Clade III |
| RcSWEET15 | 29929.m004599 | 816 | 272 | 10.07 | 30647.06 | 6 | Clade III |
| RcSWEET16a | 29579.m000197 | 747 | 249 | 7.08 | 27723.79 | 5 | Clade IV |
| RcSWEET16b | 29726.m004066 | 732 | 244 | 8.27 | 26803.66 | 5 | Clade IV |
| RcSWEET17 | 30128.m008852 | 864 | 288 | 9.95 | 31371.19 | 5 | Clade IV |
The lengths of SWEET‐coding regions (CDS) were similar among the three Euphorbiaceae plants examined, ranging from 504 to 915 bp in Hevea, 513 to 906 bp in M. esculenta, and 504 to 891 bp in R. communis (Table 1). The molecular weights of the SWEET proteins in three Euphorbiaceae species ranged from 18.7 to 33.9 kDa, while their isoelectric points (pIs) fell between 6.24 and 10.20 (Table 1).
Phylogenetic analysis of the SWEET gene families
In order to establish the phylogenetic relationships in the SWEET gene families among Hevea and the four other plant species, we aligned the 127 SWEET protein sequences in plants and constructed a phylogenetic tree as shown in Fig. 1 (Table S1). The plant SWEET proteins were clustered into four major groups with high bootstrap values, designated Clades I to IV. The 36 Hevea SWEET genes were dispersed among the four groups with 11, 7, 12, and 6 isoforms, respectively, in Clades I, II, III, and IV. Similarly, the SWEET family of genes in the other four species were also clustered into the above four groups, with 3, 5, 7, and 2 isoforms, respectively, in A. thaliana, 11, 3, 8, and 6 in P. trichocarpa, 6, 4, 12, and 6 in M. esculenta, and 3, 6, 6, and 3 in R. communis (Table 1, Fig. 1). Phylogenetic analysis as well as amino acid sequence comparison revealed universal existence of paralogous SWEET gene pairs and clusters in the five species. In Hevea, nine such SWEET gene pairs (HbSWEET2a/2b in Clade I, HbSWEET2c/2d in Clade I, HbSWEET2e/2f in Clade I, HbSWEET4a/4b in Clade II, HbSWEET5a/5b in Clade II, HbSWEET10e/10f in Clade III, HbSWEET15a/15b in Clade III, HbSWEET16b/16c in Clade IV, and HbSWEET17a/17c in Clade IV) and one gene cluster (HbSWEET1a, 1b, and 1c in Clade I) were identified. Except for the pairs of HbSWEET2c/2d, HbSWEET2e/2f, and HbSWEET10e/10f, the Ka/Ks values of the other paralogous gene pairs were less than 0.5, showing that these genes had undergone a purifying selection (Table 2). The different expression patterns exhibited by the two genes in most of the gene pairs suggested that a functional divergence had occurred after gene duplication (Fig. 4). In A. thaliana, there were two SWEET gene pairs (AtSWEET6/7 and AtSWEET16/17) and one paralogous gene cluster (AtSWEET11, 12, 13, 14). In P. trichocarpa, there were two SWEET gene pairs (PtSWEET15a/15b and PtSWEET17a/17b) and four paralogous gene clusters (PtSWEET1a, 1b, 1c, 1d; PtSWEET2a, 2b, 2c; PtSWEET3a, 3b, 3c; and PtSWEET10a, 10b, 10c, 10d). In M. esculenta and R. communis, there was only one SWEET gene pair (MeSWEET10d/10e, RcSWEET4b/4c) in each species.
Figure 1.
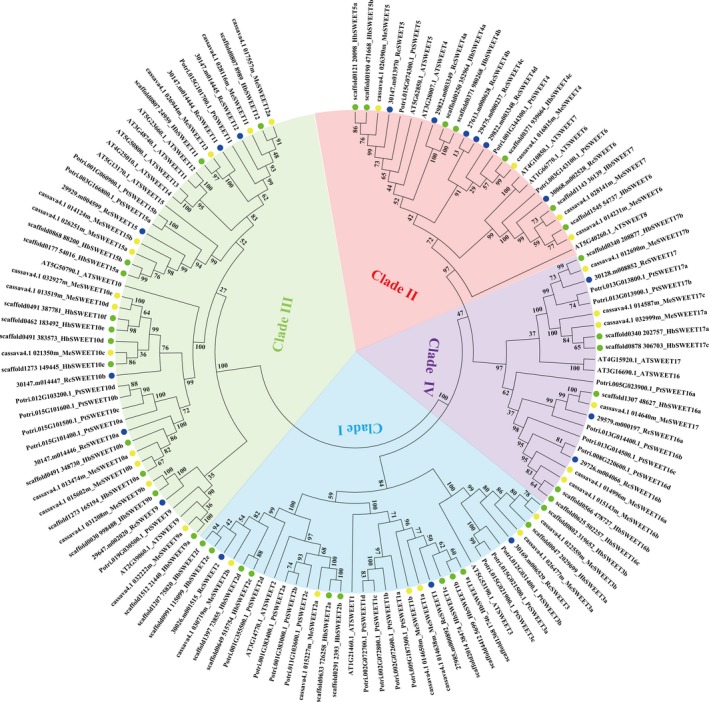
Phylogenetic analysis of the SWEET genes in H. brasiliensis and four other plant species. An unrooted phylogenetic tree of plant SWEET proteins was constructed using the neighbor‐joining method with the MEGA 6.0 program. Plant species and their SWEET proteins are as follows: H. brasiliensis, HbSWEETs (36), marked with green dots; A. thaliana, AtSWEETs (17); P. trichocarpa, PtSWEETs (28); M. esculenta, MeSWEETs (28), marked with yellow dots; R. communis, RcSWEETs (18), marked with blue dots.
Table 2.
Divergence between paralogous HbSWEET gene pairs in H. brasiliensis. The synonymous (Ks) and nonsynonymous (Ka) substitution rates between gene duplicate pairs were calculated by KaKs‐Calculator. MA, a model that averages parameters across 14 candidate models
| Gene pairs | Method | Ka | Ks | Ka/Ks | P‐Value (Fisher) | Length | S‐Sites | N‐Sites |
|---|---|---|---|---|---|---|---|---|
| HbSWEET1b/1c | MA | 0.6391 | 1.2846 | 0.4975 | 4.17E‐17 | 669 | 191.017 | 477.983 |
| HbSWEET2a/2b | MA | 0.6391 | 1.2846 | 0.4975 | 4.17E‐17 | 669 | 191.017 | 477.983 |
| HbSWEET2c/2d | MA | 0.1047 | 0.1065 | 0.9835 | 0.8669 | 543 | 137.643 | 405.357 |
| HbSWEET2e/2f | MA | 0.0080 | 0.0085 | 0.9479 | 0.5795 | 504 | 98.1862 | 405.814 |
| HbSWEET4a/4b | MA | 0.1419 | 0.4445 | 0.3192 | 4.48E‐12 | 699 | 193.225 | 505.775 |
| HbSWEET5a/5b | MA | 0.1044 | 0.2728 | 0.3828 | 2.03E‐05 | 543 | 142.305 | 400.695 |
| HbSWEET10e/10f | MA | 0.1524 | 0.1124 | 1.3556 | 0.2410 | 696 | 185.281 | 510.719 |
| HbSWEET15a/15b | MA | 0.0750 | 0.1856 | 0.4042 | 0.0001 | 690 | 179.627 | 510.373 |
| HbSWEET16b/16c | MA | 0.0584 | 0.3780 | 0.1544 | 6.24E‐18 | 684 | 186.783 | 497.217 |
| HbSWEET17a/17c | MA | 0.0806 | 0.2715 | 0.2970 | 7.67E‐08 | 615 | 157.587 | 457.413 |
Figure 4.
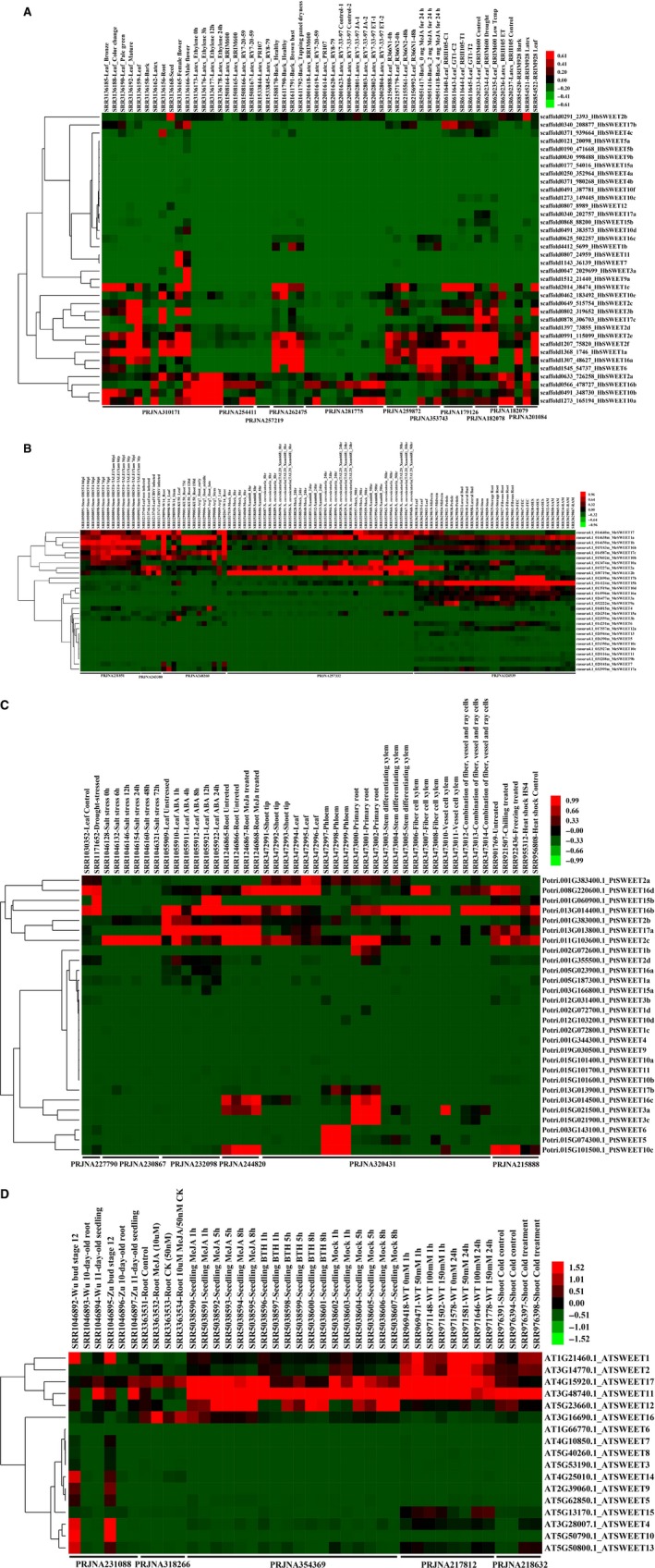
Expression analyses of the SWEET genes based on Solexa sequencing. (A), Hierarchical clustering and differential expression analysis of the HbSWEET genes in seven tissues (leaf, bark, latex, root, seed, female flower, and male flower), at four developmental stages of leaves (bronze, color change, pale‐green and mature), during ethephon treatment (0 h, 3 h, 12 h, and 24 h, PRJNA310171), latex (RRIM600 and RY7‐20‐59, PRJNA254411; clones PR107 and RY879, PRJNA257219), brown bast and tapping panel dryness (PRJNA262475), ET and JA treatment (PRJNA281775), Clone FX 3864 response to GCL012 (PRJNA259872), MeJA (PRJNA353743), Corynespora cassiicola tolerance (PRJNA179126), abiotic stress (drought, low temperature, PRJNA182078 and ethephon treatment, PRJNA182079), and tissues (leaf, bark, and latex, PRJNA201084); (B), Hierarchical clustering and differential expression analysis of the MeSWEET genes in different tissues (root, leaf, stem, PRJNA248260), infected by pathogenic Xanthomonas (PRJNA231851), CBSV virus (PRJNA243380), tissues (PRJNA324539), and bacterial blight pathogen (PRJNA257332); (C), Hierarchical clustering and differential expression analysis of the PtSWEET genes under ABA stimulation (0 h, 1 h, 4 h, 8 h, 12 h, and 24 h, PRJNA232098), methyl jasmonate stimulation (PRJNA244820), chilling, freezing, and heat shock (PRJNA207974, PRJNA215888), salinity stress (0 h, 6 h, 12 h, 24 h, and 72 h, PRJNA230867), tissues (PRJNA320431), drought stress (PRJNA227790); (D), Hierarchical clustering and differential expression analysis of the AtSWEET genes in different tissues (floral bud, root, seeding, PRJNA231088), MeJA or BTH (PRJNA354369), MeJA and CK (PRJNA318266), cold stress (PRJNA218632), salt stress (0 mm, 50 mm, 100 mm, 150 mm, PRJNA217812).
Upon further examining the genomic locations, we found that some SWEET genes in the same clade were located adjacent to each other. For example, in Hevea, HbSWEET4b and HbSWEET4c were located on scaffold0371, HbSWEET10a and HbSWEET10c on scaffold1273, HbSWEET10b, HbSWEET10d, and HbSWEET10f on scaffold0491, HbSWEET11 and HbSWEET12 on scaffold0807, and HbSWEET17a and HbSWEET17b on scaffold0340. In P. trichocarpa, PtSWEET1b, PtSWEET1c, and PtSWEET1d were located adjacent to each other on chromosome 2, PtSWEET3a and PtSWEET3c on chromosome 15, PtSWEET10a, PtSWEET10b, PtSWEET10c, and PtSWEET11 on chromosome 15, PtSWEET16b, PtSWEET16c, PtSWEET17a, and PtSWEET17b on chromosome 13. In R. communis, RcSWEET4a and RcSWEET4d were located on scaffold39822, and RcSWEET10a, RcSWEET10b, RcSWEET11, and RcSWEET12 on scaffold30147. These adjacent gene pairs and clusters had apparently been derived from tandem duplication events.
Structural organization of SWEET genes
The exon–intron structures of the 127 SWEET genes in five plant species were determined based on the predicted sequences. As shown in Fig. 2A, most Hevea SWEET members within the same groups share similar gene structures in terms of intron number, domain localization, and exon length. Although the lengths vary, introns are inserted into nearly the same locations of the gene ORFs. Most SWEET members contain 3–5 introns. Of the 36 members in Hevea, for example, 24 have 5 introns, 7 have 4 introns, and 5 have 3 introns (Fig. 2A, Table 1a). In the total of 127 SWEET genes among the five plant species, there were only three SWEET members with no introns, namely RcSWEET4b, RcSWEET4c, and AtSWEET6, all of which were clustered in Clade II (Fig. 2A‐E). Some SWEET members lacked exons at the 5′ end, such as HbSEET2f, 4c, 5b, 15a, 10e, and 10c, MeSWEET4, 3b, 2b, 12a, and 10b, RcSWEET2, 5, and 4a, and PtSWEET1c, 16c, and 15a (Fig. 2A‐E). Most SWEET members contain 4–7 TM helix domains, and 25 of the 127 members lost one to three of the seven TM helix typical of plant SWEETs (Fig. 3). In addition, the lengths of most AtSWEET and RcSWEET genes are shorter than those of the other plant SWEET genes, perhaps reflecting a relationship between gene length and genome size of a given species.
Figure 2.
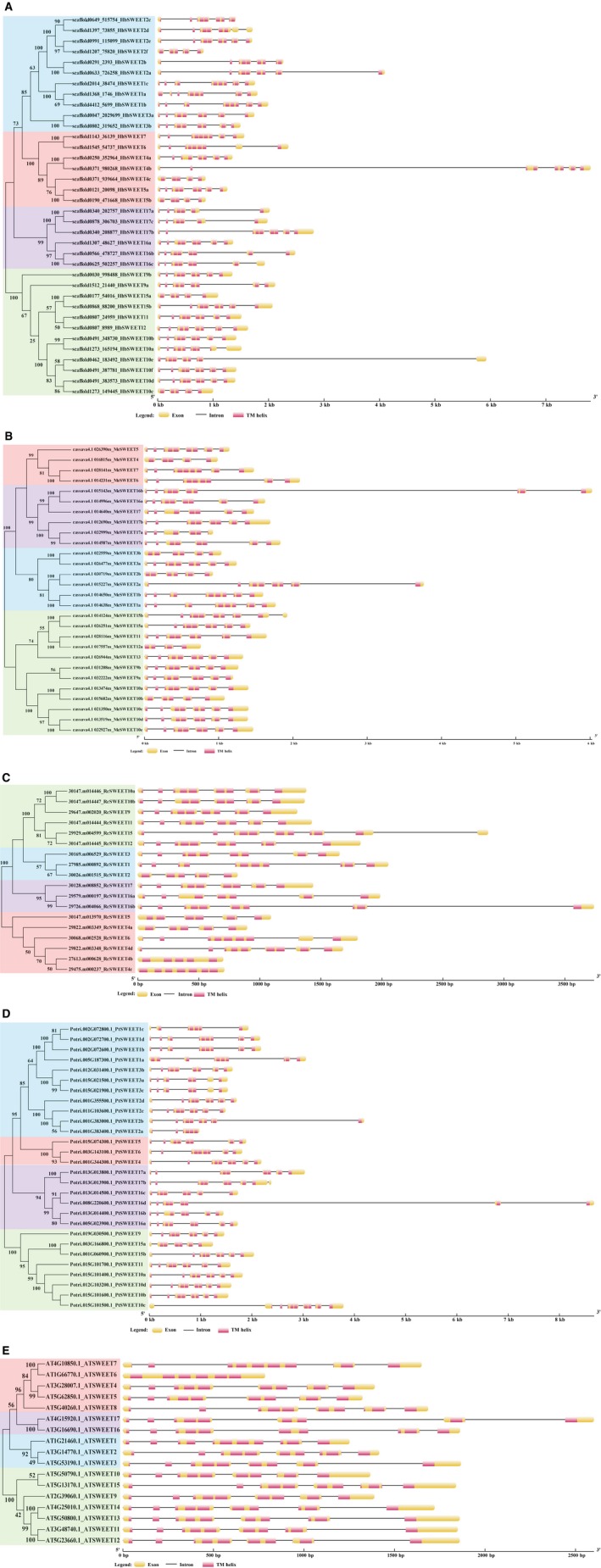
Structural organization of SWEET genes from H. brasiliensis and four other plant species. (A) to (E), structural organization of SWEET genes in H. brasiliensis, M. esculenta, R. communis, P. trichocarpa, and A. thaliana, respectively. Exons and introns are represented by boxes and black lines, respectively. The TM helix domain is represented by pink boxes. The sizes of exons and introns are proportional to their sequence lengths. Background shading: Clade 1, blue; Clade II, red; Clade III, Green; and Clade IV, Purple.
Figure 3.

Multiple sequence alignment for the predicted amino acid sequences of the SWEET genes from H. brasiliensis and four other plant species. Sequence alignment was performed using DNAMAN 6.0 software (http://www.lynnon.com/). Identical amino acids are shaded, and gaps are indicated by dots.
Tissue expression of SWEET genes
To investigate the functions of SWEET genes, gene expression profiles in different tissues were analyzed by using Solexa sequencing data in Hevea, M. esculenta, P. trichocarpa, and A. thaliana (Tables S2, S3). Analysis of gene expression from the Sequence Read Archive (SRA), adopted in the present study, has limitations as the data were compiled from different sources where genetic differences in the tested tissues and dissimilarities in the experimental conditions can make comparisons difficult. Nonetheless, such an analysis provides a broad overview of the functionalities of the various Hevea SWEETs relative to their counterparts in other plant species. The results provide useful indicators as to which SWEET genes are most commonly expressed from among the numerous isoforms. These results would serve as a guide for future follow‐up research where more exacting methodologies can be employed.
As shown in Fig. 4A, the expression levels of four SWEET genes (HbSWEET1a, 2e, 2f, and 3b) in Hevea source leaves were significantly higher than those of the other members, while three SWEET genes (HbSWEET1c, 10a, and 10b) were mainly expressed in sink leaves. In the other three plant species, MeSWEET17, MeSWEET2a, MeSWEET15b, PtSWEET2a, and PtSWEET16d were highly expressed in leaves and AtSWEET11 was mainly expressed in seedling plants (11 days old) (Fig. 4B‐D). Some of the above‐mentioned SWEET members might be involved in phloem loading and leaf development. In Hevea bark where rubber‐producing laticifers reside, HbSWEET1a and HbSWEET16a showed a predominance expression, while in latex, the cytoplasm of laticifers, HbSWEET2a, HbSWEET10a, HbSWEET10b and HbSWEET16b were the predominant isoforms. These SWEET genes might play an important role in sugar transport between the laticifers and their neighboring bark tissues, and contribute to the regulation of sucrose concentrations in laticifers together with the sucrose transporter responsible for apoplasmic sucrose uptake of laticifers, HbSUT3 18, 19. In A. thaliana, AtSWEET9 has been identified as a nectary‐specific sugar transporter 6. In Hevea, HbSWEET9a exhibited a male flower‐specific abundant expression and might have a similar function in nectary production as its A. thaliana orthologue, AtSWEET9. In addition, 14 other Hevea SWEET genes, viz. HbSWEET1a, 1c, 2a, 2e, 2f, 3a, 3b, 7, 10a, 10b, 10e, 11, 16b, and 17b, were also expressed at high levels in flowers (Fig. 4A). No SRA expression data in flowers were found in the other three plant species. In Hevea roots, seven SWEET genes (HbSWEET1a, 2a, 2f, 3b, 4c, 10e, and 17c) were abundantly expressed. In the other plant species, MeSWEET10a, MeSWEET3a, and MeSWEET15b were expressed at high levels in the storage roots of M. esculenta; their activities may be related to starch formation. Six P. trichocarpa SWEET genes (PtSWEET2a, 2c, 3a, 3c, 16b, and 16c) were expressed at high levels in the roots. On the other hand, most of A. thaliana SWEET genes showed low or no expression in the roots. PtSWEET16b and PtSWEET16d exhibited high expression in xylem fiber cells that may be related to xylem formation. There were many SWEET genes showing universal expressions in most tissues examined. These included HbSWEET1c, 10e, 2c, 3b, 17c, 2d, 2e, 2f, 1a, 16a, 6, 2a, 16b, 10b, and 10a, MeSWEET17, 1a, 1b, 16b, 17c, 10b, 10a, 2a, 2b, 17b, 15b, 10d, 16a, 3a, and 9a, PtSWEET2a, 16d, 15b, 16b, 2b, 17a, 2c, 16c, 3a, and 10c, and AtSWEET1, 2, 17, 11, 12, and 16. Interestingly, isoforms of SWEET2, 16, and 17 were observed among the universally expressed SWEET genes in all plant species examined, which might represent the main evolutionary direction of SWEET genes in plants. As shown in Fig. 4A, transcripts of 11 HbSWEET genes (HbSWEET4a, 4b, 5a, 5b, 9b, 10c, 10f, 12, 15a, 15b, and 17a) were barely detectable in almost all the tissues and all the treatments examined. Such genes comprise a large portion (~ 1/3) of the total HbSWEET gene family. This character seems to be shared by the SWEET gene families in other plant species. For example, similar expression patterns were observed for seven of 28 SWEET genes in M. esculenta (Fig. 4B), 12 of 28 in P. trichocarpa (Fig. 4C), and 4 of 17 in A. thaliana (Fig. 4D). This result suggests that the SWEET gene families in higher plants might have experienced an event of gene expansion followed by nonfunctionalization in the course of evolution. A similar phenomenon has been reported in our studies for the CDPK and CDPK‐related kinase gene families in Hevea 27. In addition, we found that most genes in Clade II have low or no expansion in all tissues examined in the four plant species.
Expression profile of SWEET genes in response to hormone and stress treatments
Expression levels of SWEET genes in Hevea were also examined under various kinds of hormone and stress treatments. Ethephon, an ethylene generator, is widely used to stimulate latex production of the rubber tree, but the yield‐stimulating mechanisms are still poorly understood 15, 22. As shown in Fig. 4A, expressions of HbSWEET10a were obviously upregulated by ethephon treatment in latex. In addition, HbSWEET10a was the predominant SWEET isoform in latex, the expression of which was higher than any of the other SWEET members, suggesting its important role in sugar transport of laticifers. Expressions of HbSWEET10a and HbSWEET2a appeared to be regulated by methyl jasmonate (MeJA) although in differing manners. Expressions of HbSWEET2c, HbSWEET2d, and HbSWEET3 were downregulated under drought treatment. Under low temperature treatment, expressions of HbSWEET1c were upregulated, whereas HbSWEET2c, 2d, 16a, and 17c were downregulated. Expressions of Hevea SWEET genes were also regulated by other kinds of stress treatments. For example, the expressions of HbSWEET1b, HbSWEET1c, and HbSWEET10e were affected by tapping panel dryness, a complex physiological disorder affecting latex production severely 28; HbSWEET17b expressions were downregulated under the infection of Corynespora cassiicola, a fungal pathogen causing a leaf fall disease in Hevea 29.
The expression levels of SWEET genes in M. esculenta, P. trichocarpa, and A. thaliana were also examined when the plants were subjected to treatments of hormones and different stresses, including fungus infection, drought, and cold (Fig. 4B‐D). The expressions of six MeSWEETs (MeSWEET1a, 10a, 10b, 15b, 17, and 17c) were affected by fungus infection. Expressions of PtSWEET2b and PtSWEET16d were induced by MeJA in roots. Expressions of PtSWEET15b were induced by drought and ABA (abscisic acid) treatments. In the model plant A. thaliana (Fig. 4D), the expressions of ATSWEET16 and ATSWEET17 were upregulated by MeJA in roots. In seedlings, expressions of ATSWEET12 were upregulated by MeJA, while those of ATSWEET16 were downregulated. Under cold treatment, expressions of ATSWEET1 and ATSWEET2 were upregulated, while those of ATSWEET16 and ATSWEET17 were downregulated.
Expression analyses of HbSWEET10a, HbSWEET16b, and HbSWEET1a based on qPCR
Rubber is synthesized and stored in the cytoplasm (latex) of highly specialized cells called laticifers that are differentiated from the cambium and arranged in rings. To further examine the expression of HbSWEET genes in latex and bark, quantitative RT‐PCR (qPCR) analyses of HbSWEET10a, HbSWEET16b, and HbSWEET1a were performed. As shown in Fig. 5, the results from qPCR were in broad agreement with the sequencing‐based expression analyses. HbSWEET10a and HbSWEET16b were mainly expressed in latex; HbSWEET1a was mainly expressed in bark and flower (Fig. 5A). HbSWEET10a was obviously upregulated by ethephon treatment in latex, while HbSWEET16b was obviously downregulated after 24 hours of ethephon treatment in latex, which agrees well with the results based on RNA‐seq (Fig. 4A, 5B, Table S3‐1). We also further examined the expression of HbSWEET1a under ethephon treatment in bark, while HbSWEET1a was obviously upregulated (Fig. 5B).
Figure 5.
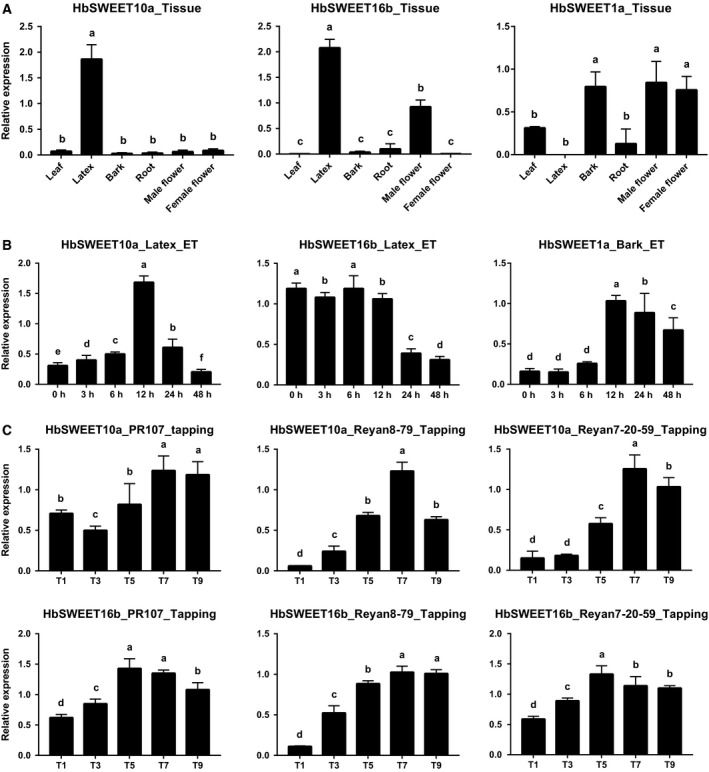
Expression analyses of HbSWEET10a, HbSWEET16b, and HbSWEET1a based on qPCR. (A), Expression of HbSWEET10a, HbSWEET16b, and HbSWEET1a transcripts in six tissues (leaf, latex, bark, root, male flower, and female flower). (B), Effect of Ethrel (2‐chloroethylphosphonic acid) treatment on HbSWEET10a and HbSWEET16b expression in latex, and HbSWEET1a expression in bark. (C), Transcript abundance of HbSWEET10a and HbSWEET16b in the first, third, fifth, seventh, and ninth tappings (T1, T3, T5, T7, and T9) on virgin Hevea trees of clones PR107, Reyan8‐79, and Reyan7‐20‐59. Values are means ± stdev of three biological replicates. Different letters indicate significant differences (Student's t‐test, P < 0.05).
The process of rubber harvesting, namely tapping, produces a conspicuous stimulating effect on latex production in virgin Hevea trees, and it has been partially ascribed to an enhanced sucrose uptake and sucrose catabolism in the laticifers 18, 20. As shown in Fig. 5C, HbSWEET10a and HbSWEET16b were obviously upregulated by tapping in three different clones PR107, Reyan7‐33‐97, and Reyan8‐79. All above results revealed that HbSWEET10a, 16b, and 1a might play an important role in sugar transport in laticifer and bark.
Conclusion
In this study, a genome‐wide analysis of SWEET gene families was undertaken for the first time in Hevea, M. esculenta, and R. communis. In silico analysis of the Hevea genome database facilitated the identification of 36 SWEET genes. The phylogenetic analysis of 127 SWEETs from Hevea and four other plant species (A. thaliana, P. trichocarpa, M. esculenta, and R. communis) classified all these SWEETs into four major groups. Members within each group might have had common evolutionary origins as seen from the sharing of similar protein motifs, exon–intron structures, and basic molecular functions. Solexa sequencing analyses revealed that SWEET2, 16, and 17 were universally expressed in different tissues of all the plant species examined, possibly representing the main evolutionary direction of plant SWEET gene families. Extensive expressional analyses in different tissues and in response to various experimental treatments, including hormones, and biotic and abiotic stresses, identified multiple tissue‐specific SWEET isoforms and isoforms showing striking responses to some of the treatments in Hevea and three other plant species (A. thaliana, P. trichocarpa, and M. esculenta). These results indicate versatile roles of SWEETs in plant growth, development, and stress responses and provide a foundation for further functional investigation of the SWEET gene families in the plant kingdom.
Materials and methods
Database search for SWEET genes in H. brasiliensis and four other plant species
Sequences of A. thaliana and P. trichocarpa SWEET genes were downloaded from the A. thaliana Information Resource (http://www.Arabidopsis.org/) and GenBank (http://www.ncbi.nlm.nih.gov/genbank). The genome and protein sequences of A. thaliana 30, P. trichocarpa 31, M. esculenta 32, and R. communis 33 were downloaded from Phytozome v10 (http://www.phytozome.net/). The H. brasiliensis genome and transcriptome data were obtained from GenBank (http://www.ncbi.nlm.nih.gov/nuccore/448814761) 22. Local BLAST alignment was performed using published SWEET sequences from A. thaliana and P. trichocarpa as queries to search against the deduced proteome of each species for the candidate SWEETs from H. brasiliensis, A. thaliana, P. trichocarpa, M. esculenta, and R. communis. All putative candidates were manually verified with the InterProScan server (http://www.ebi.ac.uk/Tools/pfa/iprscan/) to confirm the presence of protein kinase and TM helix domains.
Phylogenetic and gene structure analyses
Multiple alignments of the amino acid sequences of SWEETs from five species were performed using the Clustal X (version 1.83) program. The phylogenetic tree was constructed with MEGA6.0 34 by employing the neighbor‐joining (NJ) method with a bootstrap test for 1000 replicates. Exon–intron structures of the six species SWEET genes were analyzed by comparing the cDNA and their genomic DNA sequences through the web server GSDS 2.0 (http://gsds.cbi.pku.edu.cn/). The KaKs‐Calculator program (https://sourceforge.net/projects/kakscalculator2/) was used to calculate the nonsynonymous (Ka) and synonymous (Ks) substitutions in coding regions.
Expression analysis based on Solexa sequencing
For Solexa sequencing‐based expression analyses, Sequence Read Archive (SRA) data were downloaded from the NCBI database (Table S2) 27. The sequences included those for H. brasiliensis (C. cassiicola tolerance, PRJNA179126; abiotic stress, PRJNA182078 and PRJNA182079; tissues, PRJNA201084 35; tissues, leaf development and ethephon treatment, PRJNA310171 22, 27; Clone FX 3864 response to GCL012, PRJNA259872; ET and JA treatment, PRJNA281775 36; brown bast and tapping panel dryness, PRJNA262475 28; Hevea clones PR107 and RY879, PRJNA257219 37; RRIM600 and RY7‐20‐59, PRJNA254411; MeJA, PRJNA353743); M. esculenta (Xanthomonas tolerance, PRJNA231851; CBSV virus infected, PRJNA243380; tissue, PRJNA248260; bacterial blight pathogen infected, PRJNA257332; tissue, PRJNA324539); A. thaliana (salt stress, PRJNA217812; tissues, PRJNA231088; cold stress, PRJNA218632; MeJA or BTH, PRJNA354369; MeJA and CK, PRJNA318266); P. trichocarpa (ABA stimulation, PRJNA232098; methyl jasmonate treatment, PRJNA244820; chilling, freezing and heat shock, PRJNA207974 and PRJNA215888; salinity stress, PRJNA230867; tissue, PRJNA320431; drought stress, PRJNA227790). Raw RNA‐seq reads were processed to trim terminal low‐quality bases and adapter sequences via an in‐house custom pipeline. The clean reads were then mapped to the genome using Bowtie2, and RSEM software was used for quantifying transcript abundance with default parameters 38.
Quantitative reverse transcriptase PCR (qPCR)
To examine the expression of HbSWEET10a, HbSWEET16b, and HbSWEET1a in latex and bark, quantitative RT‐PCR (qPCR) was performed. Unless otherwise noted, Reyan7–33–97 (synonym for CATAS7–33–97 or RY3–33–97), Reyan8–79 (synonym for CATAS8–79 or RY8–79), and PR107 rubber trees (H. brasiliensis) selected for QPCR in this study were cultivated at the experimental plantation of the Rubber Research Institute of the Chinese Academy of Tropical Agricultural Sciences (Danzhou, Hainan, China). These trees were regularly tapped for latex collection in a half spiral pattern, every 3 days, without Ethrel stimulation (S/2, d/3). To study the tissue‐specific expression of HbSWEET genes, different tissues were collected from 10‐year‐old mature trees of clone Reyan7‐33‐97 that had been tapped for the last 2 years. The same type of tree was also used to examine the effect of Ethrel on expression. To analyze the effect of tapping on HbSWEET genes expression, 8‐year‐old mature virgin (never tapped) trees of clones PR107, Reyan8‐79, and Reyan7‐20‐59 were selected. The reaction was performed using the Light Cycler 2.0 system (Roche Diagnostics, Penzberg, Germany) using SYBR Green premix kit (TaKaRa) according to the manufacturer's instructions. The primer pairs used for the HbSWEET genes were 5′‐CTGCACATGC AACTCACTCACA‐3′ (F) and 5′‐CATCGGGTGGTGTAATGCTCT‐3′(R) (HbSWEET10a), 5′‐GT TCGCCTCTTGCTGCCA‐3′ (F) and 5′‐AAGTCCAAATCCCTCCGTTCA‐3′ (R) (HbSWEET16b), 5′‐TCTCCTTTCCGCCTGGTATG‐3′ (F) and 5′‐GCTCTTCTCCTTCCTTGGTGC‐3′ (R) (HbSWEET1a). For genes as internal control, YLS8 was used in gene expression analyses in the latex responding to tapping and Ethrel treatment, RH2b was used for tissue expression as recommended by Li et al. (2011) 39. The details for experimental manipulations and data analysis were as described by Tang et al. (2010) 18.
Authors contributions
CRT conceived and designed the experiments. JLS, XHX, and JYQ performed the experiments. XHX and YJF analyzed the data. JLS, XHX, and CRT wrote the manuscript. All authors read and approved the final manuscript.
Supporting information
Table S1. SWEET Accessions.
Table S2. Basic information for Solexa sequencing data of Hevea brasiliensis and three other plant species.
Table S3. RNA‐seq analysis of the expressions of SWEET genes in Hevea brasiliensis and three other plant species.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31400565, 31570672) and the Natural Science Foundation of Hainan Province (317279).
References
- 1. Chen LQ (2014) SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol 201, 1150–1155. [DOI] [PubMed] [Google Scholar]
- 2. Antony G, Zhou J, Huang S, Li T, Liu B, White F and Yang B (2010) Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene os‐11n3. Plant Cell 22, 3864–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Q, Yuan M, Zhou Y, Xianghua LI, Xiao J and Wang S (2011) A paralog of the mtn3/saliva family recessively confers race‐specific resistance to xanthomonas oryzae, in rice. Plant, Cell Environ 34, 1958–1969. [DOI] [PubMed] [Google Scholar]
- 4. Streubel J, Pesce C, Hutin M, Koebnik R, Boch J and Szurek B (2013) Five phylogenetically close rice SWEET genes confer TAL effector‐mediated susceptibility to xanthomonas oryzae pv. oryzae . New Phytol 200, 808–819. [DOI] [PubMed] [Google Scholar]
- 5. Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR and Frommer WB (2012) Sucrose efflux mediated by sweet proteins as a key step for phloem transport. Science 335, 207–211. [DOI] [PubMed] [Google Scholar]
- 6. Lin IW, Sosso D, Chen LQ, Gase K, Kim SG, Kessler D, Klinkenberg PM, Gorder MK, Hou BH, Qu XQ et al (2014) Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 508, 546–549. [DOI] [PubMed] [Google Scholar]
- 7. Xuan YH, Hu YB, Chen LQ, Sosso D, Ducat DC, Hou BH and Frommer WB (2013) Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc Natl Acad Sci USA 110, E3685–E3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klemens PA, Patzke K, Deitmer J, Spinner L, Le HR, Bellini C, Bedu M, Chardon F, Krapp A and Neuhaus HE (2013) Overexpression of the vacuolar sugar carrier ATSWEET16 modifies germination, growth, and stress tolerance in arabidopsis. Plant Physiol 163, 1338–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo WJ, Nagy R, Chen HY, Pfrunder S, Yu YC, Santelia D, Frommer WB and Martinoia E (2014) SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of arabidopsis roots and leaves. Plant Physiol 164, 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B et al (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jian H, Lu K, Yang B, Wang T, Zhang L, Zhang A, Wang J, Liu L, Qu C and Li J (2016) Genome‐wide analysis and expression profiling of the SUC and SWEET gene families of sucrose transporters in oilseed rape (brassica napusl.). Front Plant Sci 7, 1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J, Qin M, Qiao X, Cheng Y, Li X, Zhang H and Wu J (2017) A new insight into the evolution and functional divergence of sweet transporters in chinese white pear (pyrus bretschneideri). Plant Cell Physiol 58, 839–850. [DOI] [PubMed] [Google Scholar]
- 13. Mizuno H, Kasuga S and Kawahigashi H (2016) The sorghum sweet, gene family: stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol Biofuels 9, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patil G, Valliyodan B, Deshmukh R, Prince S, Nicander B, Zhao M, Sonah H, Song L, Lin L, Chaudhary J et al (2015) Soybean (glycine max) sweet gene family: insights through comparative genomics, transcriptome profiling and whole genome re‐sequence analysis. BMC Genom 16, 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. D'Auzac J, Jacob JL, Prévôt JC, Clément A, Gallois R, Crestin H, Lacote R, Pujade‐Renaud V and Gohet E (1997) The regulation of cis‐polyisoprene production (natural rubber) from Hevea brasiliensis In Recent Research Developments in Plant Physiology (Pandalai SG, ed), pp. 273–332. Trivandrum, India, Research Singpost. [Google Scholar]
- 16. Sando T, Takaoka C, Mukai Y, Yamashita A, Hattori M, Ogasawara N, Fukusaki E and Kobayashi A (2008) Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, hevea brasiliensis . Biosci Biotechnol Biochem 72, 2049–2060. [DOI] [PubMed] [Google Scholar]
- 17. Tupy J (1989) Sucrose supply and utilization for latex production In Physiology of Rubber Tree Latex (d'Auzac J, Jacob JL. and Chrestin H, eds), pp. 179–199. C.R.C. Press, Boca Raton, FL. [Google Scholar]
- 18. Tang CR, Huang DB, Yang JH, Liu SJ, Sakr S, Li HP, Zhou Y and Qin Y (2010) The sucrose transporter HbSUT3 plays an active role in sucrose loading to laticifer and rubber productivity in exploited trees of hevea brasiliensis (para rubber tree). Plant, Cell Environ 33, 1708–1720. [DOI] [PubMed] [Google Scholar]
- 19. Dusotoitcoucaud A, Kongsawadworakul P, Maurousset L, Viboonjun U, Brunel N, Pujaderenaud V, Chrestin H and Sakr S (2010) Ethylene stimulation of latex yield depends on the expression of a sucrose transporter (HbSUT1b) in rubber tree (hevea brasiliensis). Tree Physiol 30, 1586–1598. [DOI] [PubMed] [Google Scholar]
- 20. Liu S, Lan J, Zhou B, Qin Y, Zhou Y, Xiao X, Yang J, Gou J, Qi J, Huang Y et al (2015) HbNIN2, a cytosolic alkaline/neutral‐invertase, is responsible for sucrose catabolism in rubber‐producing laticifers of hevea brasiliensis (para rubber tree). New Phytol 206, 709–725. [DOI] [PubMed] [Google Scholar]
- 21. Xiao X, Tang C, Fang Y, Yang M, Zhou B, Qi J and Zhang Y (2014) Structure and expression profile of the sucrose synthase gene family in the rubber tree: indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J 281, 291–305. [DOI] [PubMed] [Google Scholar]
- 22. Tang C, Meng Y, Fang Y, Luo Y, Gao S, Xiao X, An Z, Zhou B, Zhang B, Tan X et al (2016) The rubber tree genome reveals new insights into rubber production and species adaptation. Nat Plants 2, 16073. [DOI] [PubMed] [Google Scholar]
- 23. Rahman AY, Usharraj AO, Misra BB, Thottathil GP, Jayasekaran K, Feng Y, Hou S, Ong SY, Ng FL, Lee LS et al (2013) Draft genome sequence of the rubber tree Hevea brasiliensis . BMC Genom 14, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lau NS, Makita Y, Kawashima M, Taylor TD, Kondo S, Othman AS, Shu‐Chien AC and Matsui M (2016) The rubber tree genome shows expansion of gene family associated with rubber biosynthesis. Sci Rep 6, 28594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pootakham W, Sonthirod C, Naktang C, Ruangareerate P, Yoocha T, Sangsrakru D, Theerawattanasuk K, Rattanawong R, Lekawipat N and Tangphatsornruang S (2017) De novo hybrid assembly of the rubber tree genome reveals evidence of paleotetraploidy in hevea species. Sci Rep 7, 41457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang X, Shi M, Wang D, Chen Y, Cai F, Zhang S, Wang L, Tong Z and Tian WM (2013) Comparative proteomics of primary and secondary lutoids reveals that chitinase and glucanase play a crucial combined role in rubber particle aggregation in hevea brasiliensis . J Proteome Res 12, 5146–5159. [DOI] [PubMed] [Google Scholar]
- 27. Xiao XH, Yang M, Sui JL, Qi JY, Fang YJ, Hu SN and Tang CR (2017) The calcium‐dependent protein kinase (CDPK) and CDPK‐related kinase gene families in Hevea brasiliensis‐comparison with five other plant species in structure, evolution, and expression. FEBS Open Bio 7, 4–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu JP, Xia ZQ, Tian XY and Li YJ (2015) Transcriptome sequencing and analysis of rubber tree (Hevea brasiliensis, muell.) to discover putative genes associated with tapping panel dryness (TPD). BMC Genom 16, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Breton F, Sanier C and D'Auzac J (2000) Role of cassiicolin, a host‐selective toxin, in pathogenicity of Corynespora cassiicola, causal agent of a leaf fall disease of Hevea. J Rubber Res 3, 115–128. [Google Scholar]
- 30. Initiative AG (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 31. Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A et al (2006) The genome of black cottonwood, Populus trichocarpa (torr. & gray). Science 313, 1596–1604. [DOI] [PubMed] [Google Scholar]
- 32. Wang W, Feng B, Xiao J, Xia Z, Zhou X, Li P, Zhang W, Wang Y, Møller BL, Zhang P et al (2014) Cassava genome from a wild ancestor to cultivated varieties. Nat Commun 5, 5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J and Puiu D (2010) Draft genome sequence of the oilseed species Ricinus communis. nat biotechnol. Nat Biotechnol 28, 951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamura K, Stecher G, Peterson D, Filipski A and Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chow KS, Ghazali AK, Hoh CC and Mohd‐Zainuddin Z (2014) Rna sequencing read depth requirement for optimal transcriptome coverage in hevea brasiliensis . BMC Res Notes 7, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li D, Zeng R, Li Y, Zhao M, Chao J, Li Y, Wang K, Zhu L, Tian WM and Liang C (2016) Gene expression analysis and SNP/InDEL discovery to investigate yield heterosis of two rubber tree F1 hybrids. Sci Rep 6, 24984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chao J, Chen Y, Wu S and Tian WM (2015) Comparative transcriptome analysis of latex from rubber tree clone CATAS8‐79 and PR107 reveals new cues for the regulation of latex regeneration and duration of latex flow. BMC Plant Biol 5, 120–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li B and Dewey CN (2011) RSEM: accurate transcript quantification from rna‐seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H, Qin Y, Xiao X and Tang C (2011) Screening of valid reference genes for real‐time RT‐PCR data normalization in Hevea brasiliensis and expression validation of a sucrose transporter gene HbSUT3. Plant Sci 181, 132–139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. SWEET Accessions.
Table S2. Basic information for Solexa sequencing data of Hevea brasiliensis and three other plant species.
Table S3. RNA‐seq analysis of the expressions of SWEET genes in Hevea brasiliensis and three other plant species.


