Abstract
Understanding the molecular origin of influenza receptor specificity is complicated by the paucity of quantitative affinity measurements, and the qualitative and variable nature of glycan array data. Further obstacles arise from the varied impact of viral glycosylation and the relatively narrow spectrum of biologically relevant receptors present on glycan arrays. A survey of receptor conformational properties is presented, leading to the conclusion that conformational entropy plays a key role in defining specificity, as does the newly reported ability of biantennary receptors that terminate in Siaα2-6Gal sequences to form bidentate interactions to two binding sites in a hemagglutinin trimer. Bidentate binding provides a functional explanation for the observation that Siaα2-6 receptors adopt an open-umbrella topology when bound to hemagglutinins from human-infective viruses, and calls for a reassessment of virus avidity and tissue tropism.
Introduction
Wild birds are the primary natural reservoir for influenza A viruses [1], and the 1918 Spanish Flu pandemic that killed an estimated 50 million people [2] is believed to have originated from spontaneous mutations in an avian influenza virus that conferred human-to-human transmissibility [3,4]. While zoonotic influenza can infect humans [5], close contact with infected animals is required [6]. Subsequent human-to-human transmission, leading to pandemics, requires that the virus undergo additional genetic alterations [5,6]. As noted by Reper-ant et al. [5], in order for a zoonotic virus to become human-infective, it must overcome three sets of barriers: animal-to-human transmission, virus-cell interaction, and human-to-human transmission. Seasonal influenza epidemics arise from human-to-human transmission of circulating strains that have undergone sufficient mutation (antigenic drift) to circumvent established immunity within the population [7].
In contrast to the Spanish Flu, the Swine Flu pandemic of 2009 was relatively mild [8]. Nevertheless it raised concerns within the World Health Organization because of the rapidity with which it spread [9]; within 6 weeks of the first case, Swine Flu had spread to over 70 countries [10] and required the development of a new vaccine. Human adaptation is of particular concern in the case of highly pathogenic avian influenza (HPAI) subtypes, such as H5N1. Although infrequent, human infection by avian H5N1 has been reported in 16 countries, resulting in approximately 60% mortality [11]. Preparedness for pandemics therefore necessitates anticipation of the virulence of emerging strains, providing motivation for developing a deeper understanding of the basis for influenza specificity. Here, we reassess the relationship between host glycan structure and influenza specificity in light of recent data that indicates critical roles for glycan substructure and dynamics.
Influenza A classification is based on the antigenic properties of the hemagglutinin (HA) and neuraminidase (NA) envelope proteins. Influenza HA is a homotrimeric glycoprotein whose protomers each comprise a globular head domain (HA1) and stalk region (HA2) [12]. Each HA1 domain contains a receptor binding site (RBS), through which the virus adheres to the host cell via binding to host glycans that contain sialic acid (Sia, neuraminic acid, Neu5Ac). There are currently 18 hemagglutinin subtypes, which are classified into two groups based on their antigenic properties: group 1 consists of H1-2, H5-6, H8-H9 H11-13, and H16; group 2 contains H3-4, H7, H10, H14, and H15. The most extensively studied HAs include H1, H3 and H5 [13•,14]. The NA protein mediates the cleaving of Sia from the host receptor glycan post cellular infection, enabling progeny virus to escape from the host cell surface [15]. Cryoelectron tomography indicates that there are approximately 300 HA proteins in the viral envelope [16], with the ratio of HA to NA varying between different strains from 4 to 6:1 [16,17]. Compound factors affect the ability of a particular strain of influenza to infect humans, including the level of exposure, the replication rate in newly infected individuals, the glycan binding preferences of the viral surface HA, and the activity of the viral surface NA [15,18–23]. Further, the enzyme activity of the NA must balance with the affinity of the HA [15,22]. If the NA is too active, relative to the affinity of the HA, it will attenuate the ability of the virus to infect the host cell. Conversely, a relatively weak NA will impair shedding of the progeny virus.
In addition to receptor specificity, zoonotic infection is also sensitive to differences in the susceptibility of the HA to pH-mediated endosomal fusion [24], and differences in the efficiency of translocation of the viral ribonucleoprotein complex to the host nucleus [25] (host adaptation). Moreover, ease of transmission and replication appears to be dependent on the distribution and composition of the receptors on host tissue. Viral attachment studies have shown that human influenza viruses adhere more strongly to human trachea and bronchi than avian viruses, and attach to different cell types [26]. Thus, the lack of a suitable receptor has been invoked as being responsible for the inefficient transmission [27] and replication of avian viruses in humans [28,29]. Much work has been done to elucidate the molecular basis for the observed tissue tropism [28,30,31,32••].
Defining receptor specificity
The canonical view of the relationship between HA receptor specificity and species infectivity is that the HA in human-infective viruses prefers to bind to glycans present on the cell surface that terminate with the Siaα2-6Gal (α2-6) sequence; in avian-infective viruses, the HA prefers to bind to glycans that terminate in Siaα2-3Gal (α2-3). Some species, such as swine, can be co-infected by viruses that prefer either α2-3 or α2-6 structures, leading to the potential for genetic reassortment (antigenic shift) in swine that results in the introduction of α2-6 binding preference (enhanced human infectivity) into a zoonotic framework [5].
The discovery of the α2-6/α2-3 infectivity relationship originated not from quantitative biophysical studies, but from more qualitative, yet robust, hemagglutination assays [33]. Rogers and Paulson’s [34••] seminal work on enzymatically modified red blood cells (RBCs) established that influenza receptor specificity depends, to an extent, on the species from which the virus was isolated. They reported that isolates of human subtype H3N2 agglutinated RBCs whose modified surface glycans terminated in the α2-6 sequence, but that these isolates did not agglutinate RBCs with α2-3 glycans. Conversely, avian isolates preferentially agglutinated RBCs containing the α2-3 linkage. While hemagglutination by influenza is a general phenomenon not limited to chicken RBCs [35], not all virus strains agglutinate all types of RBCs equally well [35,36]. Unmodified chicken RBCs contain a diversity of multiantennary glycans, roughly in an equal ratio of α2-3:α2-6 [36], but these represent only a limited subset of the glycans found on human epithelial tissue, which also include multiple lactosamine repeats in the antennae. The observation that the necessary human-type receptors are not present provides an explanation of the inability of certain human-adapted influenza strains to agglutinate chicken RBCs [36]. As noted by Ovsyannikova et al. [35], species selection of red blood cells (RBCs) is critical to determine antibody titers to influenza viruses reliably, however, further glycomics analyses are required to elucidate the origin of the differences in RBC agglutination behavior.
Affinity versus avidity
Monomeric binding affinities for HA-glycan interactions confirm the canonical view of HA specificity, but show remarkably modest differences between α2-3 and α2-6 receptors (Table 1). Avidity arising from interactions between multiple host glycans and multiple trimeric HAs on the viral surface has been invoked to explain the difference between the weak (mM) monomeric affinities for HA-glycan interactions and the sub-μM binding for whole virus [37,38•,39,40••]. Indeed, models of binding kinetics [38•,39] have shown that avidity can exponentially amplify the subtle differences in monomeric affinities, resulting in agreement with experimental virus binding kinetics.
Table 1.
Monomeric oligosaccharide — HA binding affinities
| HA viral strain | Canonical specificity | Ligand | KD (mM) | ΔG (kcal/mol)a |
|---|---|---|---|---|
| H3N2 | ||||
| A/Hong Kong/1/1968 [38•,41•] (X-31) | α2-6 | SiaαOMe | 2.8 ± 0.3 | −3.5 |
| 3′SLN | 3.1 ± 0.4 | −3.4 | ||
| 3′SL | 3.6 ± 0.7, 3.2 ± 0.6 | −3.3, −3.4 | ||
| LSTa | 3.8 ± 0.8 | −3.3 | ||
| 6′SLN | 2.0 ± 0.2 | −3.7 | ||
| 6′SL | 1.7 ± 0.5, 2.1 ± 0.3 | −3.8, −3.6 | ||
| A/Memphis/102/72 [42•] | α2-6 | SiaαOMe | 2.0 ± 1.1 | −3.7 |
| LSTa | 8.0 | −2.9 | ||
| LSTc | 1.2 | −4.0 | ||
| H3N2 | ||||
| A/Hong Kong/1/1968 (X-31) L226Q [41•] | α2-3 | SiaαOMe | 4.7 ± 0.5 | −3.2 |
| 3′SL | 2.9 ± 0.3 | −3.4 | ||
| 6′SL | 5.9 ± 0.7 | −3.0 | ||
| H5N1 | ||||
| A/Vietnam/1194/04[38•] | α2-3 | 3′SLN | 1.1 ± 0.2 | −4.0 |
| 3′SL | 0.7 ± 0.4 | −4.3 | ||
| 6′SLN | 17 ± 3 | −2.4 | ||
| 6′SL | 21 ± 6 | −2.3 | ||
| A/Vietnam/1194/04 (ferret transmissible) N158D/N224K/Q226L/T318I [38•] |
α2-6 | 3′SLN | 32 ± 8 | −2.0 |
| 3′SL | 43 ± 12 | −1.9 | ||
| 6′SLN | 12 ± 2.5 | −2.6 | ||
| 6′SL | 17 ± 5 | −2.4 | ||
| H10N8 | ||||
| A/Jiangxi-Donghu/346/2013[43] | α2-6 | 3′SLN | 1.8 ± 0.39 | −3.7 |
| 6′SLN | 1.4 ± 0.32 | −3.9 | ||
At 25°C.
In 2012, Lin et al. [44••] reported that the avidity of H3N2 viruses for an α2-6 trisaccharide receptor decreased approximately fourfold between 1968 and 2001, then progressively decreased a further 200-fold from 2001 to 2010, to such an extent that higher virus concentrations were required to observe any binding for the 2010 strains. This decrease in binding avidity was shown to be the result of mutations (antigenic drift) that weakened specific interactions between the RBS and the glycan receptors [44••]. Recently, Peng et al. [45••] screened the HAs from a number of H3N2 viruses against a custom glycan array that included multiantennary glycans of the type found in the human respiratory tract [46••], and confirmed that binding to short, or linear, glycans had steadily decreased, consistent with the observations of Lin et al. [44••]. However, strong binding to long biantennary sialoglycans was observed that was relatively insensitive to the effects of antigenic drift.
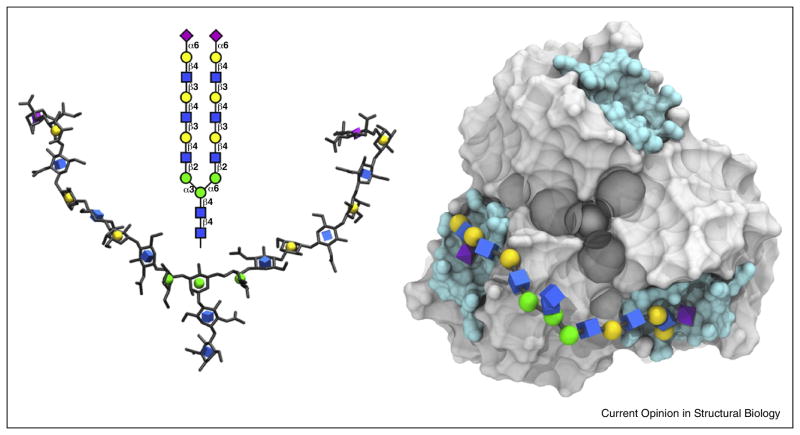
Recently, Peng et al. [45••], and de Vries et al. [47•] have raised the intriguing possibility that both branches in a biantennary glycan could bind simultaneously to two RBSs in an HA trimer, provided the branches were sufficiently long to reach two RBSs (Figure 1). Such bidentate binding would amplify the affinity of the glycan, potentially resulting in an apparent affinity of as much as the square of the monovalent KD (KD,mono2) [40••], although this would likely be reduced by entropic penalties. This binding enhancement would enable the HA to continue to retain affinity for certain biantennary glycans despite the overall negative impact of antigenic drift on receptor binding at a monovalent level. This hypothesis provides a basis for explaining the observation that, despite the general decrease in avidity displayed by H3N2 viruses [44••,48], they retain the ability to bind to biantennary glycans [45••] and, thus, to infect and transmit in the human population [48].
Figure 1.
Right: proposed [45••] bidentate binding of a biantennary α2-6 glycan (left, 3D-SNFG representation [49]) to the HA (grey surface) from a pandemic H1N1 (A/California/04/2009), residues lining the RBS are shown in cyan. The glycan is shown in the conformation required for bidentate binding.
Impact of HA glycosylation on specificity
Glycosylation of HA proteins varies both in location and composition depending on the strain of the virus [45••,50], as well as on the cell-type in which the virus was produced [51]. Over time, the number of glycosylation sites in circulating influenza strains has increased [50,52], presumably shielding the protein surface from antibody recognition and assisting the virus in evading host immune surveillance [50,53–56]. However, the more heavily glycosylated an HA1 domain, the more likely that its receptor binding ability will be impaired, either because the glycosylation directly blocks access to the RBS [57,58], or because it forms a shield through which short receptor glycans may not be able to penetrate. Increased glycosylation, thus, potentially decreases affinity and virulence [59•]. Three decades ago, it was observed that passaging of an avian infective H1N1 strain (A/WSN/1933) in mammalian (MDBK) cells led to the loss of glycosylation at N129 in the HA1 domain, leading to an increased affinity for host receptors, whereas passaging in chicken cells had no effect on glycosylation [60]. More recently, based on an analysis of 3D structures of HAs, Jayaraman et al. [57] predicted that, because of its proximity to the RBS, the loss of glycosylation at N91 in the HA from an H1N1 (A/South Carolina/1/18 and two variants, D225G and D190E/D225G) should affect receptor-binding properties. While loss of glycosylation at N91 was found to have no affect on the binding of the D190E/D225G (avian-like) variant to immobilized α2-3 oligosaccharides, it completely abrogated binding of the D225G variant to α2-3 and α2-6 oligosaccharides, and attenuated binding of wild-type HA to α2-6 oligosaccharides. The mechanism underlying the negative impact of loss of glycosylation on α2-6 binding was not identified.
In H5N1 strains, the N158 glycosylation site occupies a similar spatial position to that of N129 in H1N1 strains, and appears to produce similar effects when glycosylated (attenuation of antigenicity, reduction of affinity for α2-6 receptors [61]). H5N1 viruses lacking glycosylation at N158 transmit efficiently by direct contact among guinea-pigs [62]. In 2015, Zhang et al. [63] examined the impact of glycosylation at three sites in the HA1 of an H5N1 virus (A/Mallard/Huadong/S/2005) and reached the conclusions that loss of glycosylation at N158 was a prerequisite for binding to α2,6-modified RBCs, and viruses with a loss of glycosylation at N158 or N169 had higher lethality in mice. In 2010, Liao et al. [64••] showed that deletion of glycosylation sites in an H5 derived from a consensus-based sequence [65] led to no major change in the glycan binding profiles for α2-3 oligosaccharides.
Yang et al. [54] noted in a study of H3N2 strains that the viruses had evolved to prefer longer linear glycans, and hypothesized that this preference was related to an increase in the number of glycosylation sites in the HA1. Alymova et al. [66] also recently examined H3N2 with varying glycosylation levels, and concluded that glycosylation of the HA1 could decrease binding affinity, without reducing virulence. They further introduced the hypothesis, based on the consistent binding of the HAs to linear α2-6 sialylated polylactosamine glycans, that physiologically relevant receptor binding had not changed over the past 40 years. However, their array did not include the large biantennary glycans used by Peng et al. [45••], who concluded that H3N2 had evolved specificity for extended, branched α2-6 glycans.
While the current data regarding the impact of HA1 glycosylation show strain dependence, binding to α2-6 receptors generally appears to be markedly sensitive to variations in HA1 glycosylation. Further studies will be required to develop a clear understanding of the conditions under which HA1 glycosylation alters receptor binding and or virulence.
Relating HA structure to receptor specificity
Examination of pandemic HA sequences permits the identification of mutations in the RBS that appear to play a role in switching the virus specificity. A pair of mutations identified as E190D and G225D in H1N1 viruses has been shown to be critical for switching the binding preference from α2-3 to α2-6 glycans [3,4,56], and appears to have been responsible for the Spanish Flu pandemic [67]. Mutation at only one of these sites within an H1 typically leads to dual α2-3 and α2-6 receptor binding [3,56,68]. A different pair of mutations (Q226L and G228S) enabled the H2N2 and H3N2 pandemic viruses to gain specificity for α2-6 glycans [69]. However, these observations should not be considered to be specificity ‘rules’ — as part of a study to engineer α2-6 specificity into an H5N1 (A/Vietnam/1203/04), introduction of the E190D and G225D double mutations remarkably eliminated binding to all α2-3 and α2-6 glycans examined [70]. Additional host-adaptation is required in order to achieve this specificity switch in H5N1 viruses [71–74]. Very recently de Vries et al. [47•] have shown that three mutations (V186K/G, K193T, and G228S) switch H7N9 influenza to human-type receptor specificity, with a binding profile practically identical to pandemic H1N1 A/California/04/2009.
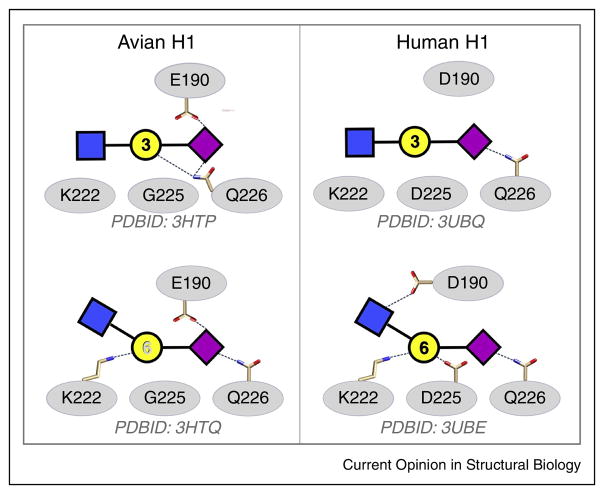
The 3D structures of HA-oligosaccharide complexes are essential for understanding, and potentially predicting, the effect of mutations in HA on receptor specificity, and the structural features of influenza HA-glycan co-complexes have been well described [13•,15,75,76•]. However, despite the large and growing number of co-crystal structures of HA-oligosaccharide complexes, rationalization of the observed specificity preferences in terms of 3D structural properties is far from straightforward [77•]. As a case in point, consider the complexes of HAs from avian-infective (A/Wild Duck/JX/12416/2005) and human-infective (A/California/04/2009) H1N1 viruses (Figure 2). These HAs have been co-crystallized with both α2-3 and α2-6 glycans, and therefore provide an opportunity to illustrate the differences in hydrogen-bonding patterns in human-adapted or avian-adapted HAs. It is clear from an examination of the hydrogen bond patterns between α2-3 and α2-6 oligosaccharides with the human-adapted HA (Figure 2, lower panels) that the α2-6 receptor makes several additional interactions (involving D190, D225 and K222) relative to the α2-3. These interactions are consistent with the observed α2-6 preference for human-adapted HAs. Why avian-adapted HAs generally bind more weakly, if at all, to α2-6 oligosaccharides is far less clear from these structures (Figure 2 upper panels). Indeed, as noted by Lin et al. [78••], the mode of binding observed for the avian-adapted HA is not consistent with the view that the avian HA favors α2-3 receptors over α2-6.
Figure 2.
Schematic representations of the binding modes for representative avian-adapted (left) and human-adapted (right) HAs from H1N1 viruses binding to α2-3 (upper) or α2-6 (lower) receptor analogs.
The answer to these structural riddles must lie in properties that are not as readily apparent as hydrogen bond networks. These include van der Waals contacts, as well as contributions from conformational entropy, which may be significantly different for the two types of ligand.
Conformational entropy — the missing link
Given the relatively plastic nature of glycans, binding to a protein incurs an entropic penalty proportional to the degree of conformational constriction, and this has been proposed as an unfavorable contribution in HA-glycan binding [79]. Notably, computational analyses, based on molecular dynamics simulations of crystallographic HA co-complexes, fail to reproduce the observed binding specificities unless entropic contributions are explicitly included [80,81••]. The magnitude of the entropy penalty S for each rotatable bond W that becomes constrained upon binding can be estimated from Boltzmann’s expression (S = R ln W) [82], or calculated from observed conformational populations [80,81••,83]. In α2-3 linkages the φ-angle (C1′–C2′–O3–C3) typically populates two rotamers in solution (anti and –gauche with respect to C1′) [84], but only one when bound to an HA, resulting in an estimated entropic penalty of approximately 0.4 kcal/mol (at 25°C). The α2-6 linkage has an additional rotatable bond that leads to multiple conformations, giving rise to an estimated entropic penalty of at least 1.5 kcal/mol [80,81••,83,85]. Furthermore, in the case of α2-6 glycans, a curled or open-umbrella topology places more of the glycan substructure in contact with the HA surface than in the case of α2-3 glycans that adopt linear or cone-like topologies. These additional glycan-HA interactions can result in entropic penalties for α2-6 glycans that are larger than those for α2-3 glycans by as much as 5 kcal/mol [81••].
Bidentate binding would also be expected to lead to a heightened entropic penalty, due to the overall restriction of motion for such large, flexible glycans, and in particular for the 1–6 linkage in the glycan core. Additionally, interactions between the amino acid side chains and the receptor in the RBS [82,86] may be entropically disfavored. For example, for K222 to form its hydrogen bond with the receptor, the long flexible side chain pays an entropic penalty of up to 2 kcal/mol (using S = R ln W) [82]. The more constrained a flexible ligand is by enthalpically favorable interactions, such as hydrogen bonds and van der Waals contacts, the higher the entropic penalty paid by the system [87], leading to the key concept of enthalpy–entropy compensation [88].
In order to prefer binding to α2-6 glycans over α2-3, the HA must evolve to form proportionally more or stronger interactions with the α2-6 receptor. Thus, although crystallography demonstrates that an avian-adapted HA can form as many (or more) interactions with an α2-6 glycan [78••], the resultant entropically disfavored stiffening of the α2-6 receptor results in a net free energy preference for the α2-3 glycan. For this reason, the number of receptor-HA interactions (Figure 2) is a poor metric for assessing subtle differences in affinity/specificity.
Relating glycan structure to specificity
Glycan array screening has been extensively applied to help define the specificity of influenza hemagglutinins. Overall, the data support the view that HAs from avian-adapted strains prefer α2-3 glycans, while human-infective strains generally prefer α2-6 [44••,45••,46••,48,70,89–97]. Nevertheless, glycan array screening has also brought to light many exceptions to the accepted view of specificity, and raised new and unanswered questions, particularly related to variations in response as a function of monosaccharide modifications (sulfation, acetylation, etc.) and glycan substructure [48,97]. Common modifications to the Sia residue include acetylation of the glyceryl side chain (typically at the 9-position), or 5-N-glycolylation (Neu5Gc), which generally attenuate binding to HA from human-infective virus [98•,99]. Remarkably, in contrast to the effect of acetylation, a 9-O-lactoyl group appears to restore affinity (H1N1 and H3N2) to levels comparable to the non-derivatized sialoside [98•]. Neu5Gc is not produced in humans [100], but can be abundant in non-human species; for example, Neu5Gc-containing glycans are the dominant moieties on epithelial cells from equine trachea [101]. Not surprisingly therefore, HAs from some (but not all) equine-infective influenza strains bind preferentially to glycans containing this modification [102], whereas HA from human-infective strains generally do not [98•], explaining the equine/ human zoonotic transmission barrier [101].
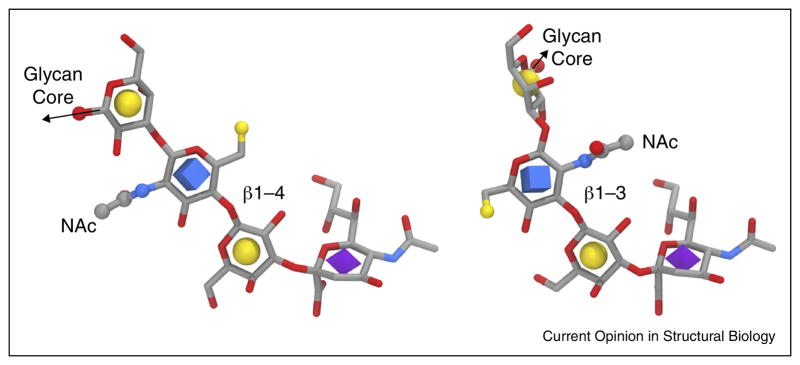
The sensitivity of binding to glycan substructure is an essential component when defining influenza specificity, but its assessment is complicated by the diversity of possible glycan structures, the influence of glycan substructure on the 3D structure of the sialylated terminus (Figure 3), and the differential impact of mutations in the RBS on interactions with glycan modifications [103]. It is impossible to separate the impact of modifications in the glycan from the overall context of the glycan 3D shape, just as it is impossible to discuss the significance of mutations in the HA independently from the context of the particular subtype. For example, the Gal-2 residue may be linked to GlcNAc-3 at either the 3-position or 4-position (Galβ1-3GlcNAc or Galβ1-4GlcNAc). This chemically subtle difference is often left undefined in glycomic analyses [36], and yet has a dramatic impact on the orientation of the GlcNAc-3 residue relative to Gal-2, flipping the positions of the NAc and O6 moieties in the GlcNAc by approximately 180° degrees in the RBS (Figure 3). This difference in glycan substructure would be expected to have a noticeable influence on binding when the HA has evolved to prefer a receptor in which the GlcNAc is modified by sulfation at O6. For example, the HA from an equine H3N8 binds preferentially to 6-sulfated sialosides, but only when the Gal-2-GlcNAc-3 linkage is present in the β1-4 form [102]. For H5 subtypes, 6-O-sulfation of the receptor enhances binding [104] and was predicted to lead to the formation of a salt bridge between the sulfate moiety and K193 [105], which was recently confirmed by crystallography [106]. Similar favorable electrostatic interactions were observed between the same sulfated receptor and K158A in an avian H10 [43].
Figure 3.
HA receptor structures indicating the influence of the Gal-2 — GlcNAc3 linkage type (left: β1-4, right: β1-3) on conformation and presentation. The structures were retrieved from PDB IDs 4YYA and 4NRL, respectively, and aligned relative to the Sia residues. Note the reversal of the N-acetyl moieties relative to the Sia residues. The GlcNAc 6-position, which may be sulfated, is shown as a small yellow sphere.
Another common modification of α2-3 sialosides is α-fucosylation at the 3-position or 4-position of GlcNAc-3. The site of fucosylation depends on the nature of the Gal-2-GlcNAc-3 linkage (β1-4 or β1-3), generating the well-known sialyl LeX (SLex) and SLea motifs, respectively. Whether or not fucosylation attenuates affinity has been suggested to depend on the presence or absence of steric collisions with bulky side chains at positions 222 and/or 227 [102,107–109]. Given the prevalence of SLex in mucins, they have been proposed as providing a barrier to infection [110].
In contrast to the 3D properties of the RBS, less attention has been given to a systematic analysis of the conformations of the receptors in the complexes, although it has frequently been observed that the α2-3 linkage adopts a ‘trans’ orientation, resulting in a cone-like topology of the glycan relative to the HA surface [111•]. The ‘cis’ orientation of the α2-6 linkage [112,113] has been further noted to lead such ligands to form a compact, curled, or folded conformation [114] that results in the receptor spanning a larger region of the HA surface, referred to as an open-umbrella topology [111•]. The use of the ‘cis-’ descriptor for the Siaα2-6Gal φ-angle has become widespread, however it is not useful when comparing the conformation of such linkages in HA complexes, as to date all such linkages adopt this conformation when co-complexed with HAs (Table 2). The conformation of the ψ angle in Siaα2-6Gal linkages does however vary, populating only two states, herein denoted ‘anti-ψ’ or ‘eclipsed-ψ’. Moreover, the terms ‘cis’ and ‘trans’ imply that the orientation of the bond is fixed, as in a double bond. As this is not the case for α2-6 or α2-3 linkages, we will refer to the so-called ‘cis’ orientation as ‘gauche’, and the ‘trans’ as ‘anti’. The receptor conformational properties extracted from well-resolved HA-oligosaccharide co-complexes are presented in Table 2.
Table 2.
Ligand conformation and HAs co-complexed to α2-6 and α2-3 oligosaccharides (resolution ≤ 2.5 Å)
| PDB ID, ligand | α2-6 | PDB ID, ligand | α2-3 | HA virus strain | Amino acids at 190,193,222, 225, 226, 227, 228 |
|---|---|---|---|---|---|
| ψa | φb | ||||
| H1N1 | |||||
| 1RVT, LSTc | anti | 1RV0, LSTa | –c | A/Swine/Iowa/15/1930 | D,S,K,G,Q,A,G |
| 1RVZ, LSTc | anti | 1RVX, LSTa | anti | A/Puerto Rico/8/1934 | E,D,K,D,Q,A,G |
| 3UBE, LSTc | anti | 3UBJ, LSTa | anti | A/California/04/2009 | D,S,K,D,Q,E,G |
| 3UBN, 6′SLN | anti | 3UBQ, 3′SLN | anti | A/California/04/2009 | D,S,K,D,Q,E,G |
| 3HTQd, LSTc | anti | 3HTPd, LSTa | anti | A/Wild Duck/JX/12416/2005 | E,T,K,G,Q,A,G |
| H2N2 | |||||
| 2WR1, LSTc | anti | 2WR2, LSTa | anti | A/Chicken/New York/29878/91 | E,T,K,G,Q,G,G |
| 2WR4, LSTc | anti | 2WR3, LSTa | anti | A/Duck/Ontario/1977 | E,T,K,G,Q,G,G |
| 2WR7, LSTc | anti | – | A/Singapore/1/57 | E,T,K,G,L,G,S | |
| H3N2 | |||||
| 2YP3, 6′SLN | anti | 2YP5, 3′SLN | –c | A/Finland/486/2004 | D,S,R,D,I,P,S |
| 2YP4, LSTc | anti | A/Finland/486/2004 | D,S,R,D,I,P,S | ||
| 2YP8, 6′SLN | –c | 2YP9, 3′SLN | –c | A/Hong Kong/4443/2005 | D,F,R,N,I,P,S |
| 2YPGd, LSTc | anti | A/Aichi/2/1968-X31 | E,S,W,G,L,S,S | ||
| H3N8 | |||||
| 4WA2, 3′SLN | anti | A/harbor seal/Massachusetts/1/2011 | E,N,L,G,Q,S,G | ||
| H5N1 | |||||
| 1JSO, LSTc | –c | 1JSN, LSTa | anti | A/Duck/Singapore/3/1997 | E,K,K,G,Q,S,G |
| 4BGX, 6′SLN | eclipsedb | 4BGYd, 3′SLN | anti | A/Vietnam/1194/2004 | E,K,K,G,Q,S,G |
| 4BH0, 6′SLN | eclipsed | 4BH1, 3′SLN | anti | A/Turkey/Turkey/1/2005 | E,R,K,G,Q,S,G |
| 4BH3, 6′SLN | anti | 4BH4, 3′SLN | –gaucheb | A/Vietnam/1203/2004 (N158D,N224Q,Q226L,T318I) | E,K,K,G,L,S,G |
| 4KDO, LSTc | anti | 4KDN, LSTa | –c | A/Vietnam/1203/2004 (N158D/N224K/Q226L/T318I) | E,K,K,G,L,S,G |
| 4CQR, 6′SLN | eclipsed | 4CQQd, 3′SLN | –gauche | A/Vietnam/1194/2004 (S227N,Q196R) | E,K,K,G,Q,R,G |
| 4CQU, 6′SLN | eclipsed | 5AJM, 3′SLN | –gauche | A/Vietnam/1194/2004 (N186K) | E,K,K,G,Q,S,G |
| 4CQY, LSTa | anti | A/turkey/Turkey/1/2005 (Δ133/I155T) | E,R,K,G,Q,S,G | ||
| 4CQX, 6′SLN | eclipsed | 4CQW, 3′SLN | anti | A/turkey/Turkey/1/2005 (Δ133/I155T) | E,R,K,G,Q,S,G |
| H6N1 | |||||
| 5BR6a, LSTc | anti | 5BR3d, LSTa | –gauche | A/Taiwan/2/2013 | V,N,A,G,Q,R,S |
| 4XKF, LSTa | –gauche | A/Taiwan/2/2013 | V,N,A,G,Q,R,S | ||
| 4XKG, 6′SLN | eclipsed | 4XKE, 3′SLN | –gauche | A/Taiwan/2/2013 | V,N,A,G,Q,R,S |
| H7N3 | |||||
| 4BSH, 6′SLN | eclipsed | 4BSI, 3′SLNd | anti | A/Turkey/Italy/214845/2002 | E,K,Q,G,Q,S,G |
| H7N9 | |||||
| 4BSB, LSTc | anti | A/Anhui/1/2013 (L20,T135) | E,K,Q,G,L,S,G | ||
| 4BSCd, 6′SLN | anti | 4BSD, 3′SLN | –gauche | A/Anhui/1/2013 (L20,T135) | E,K,Q,G,L,S,G |
| 4BSEd, LSTc | anti | A/Anhui/1/2013 (V20,A135) | E,K,Q,G,L,S,G | ||
| 4LKKa, 6′SLNLN | anti | 4LKJd, 3′SLNLN | anti | A/Anhui/1/2013 (L226Q) | E,K,Q,G,Q,S,G |
| 4N62, 3′SL(6S)N | +gauchee | A/Shanghai/2/2013 (L226) | E,K,Q,G,L,S,G | ||
| H9N2 | |||||
| 1JSI, LSTc | anti | 1JSH, LSTa | –gauche | A/Swine/Hong Kong/9/1998 | V,N,L,G,L,H,G |
| H10N2 | |||||
| 4CYZ, LSTa | anti | A/mallard/Sweden/51/2002 | E,D,Q,G,Q,S,G | ||
| H10N8 | |||||
| 4D00, 6′SLN | anti and eclipsedf | A/Jiangxi-Donghu/346/2013 | E,D,Q,G,Q,S,G | ||
For α2-6 linkages, ψ (C2′–O6–C6–C5) adopts either an anti (188° ± 23) or eclipsed (113° ± 7) conformation. The remaining glycosidic angles adopt a single conformation characterized by average φ (C1′–C2′–O6–C6) = −55° ± 11 (–gauche, a.k.a. ‘cis’) and average ω (O6–C6–C5–O5) = 63° ± 17, with the exceptions of 5BR6, where φ = 201°, and ω = −41°, 4LKK, where ω = 157°.
For α2-3 linkages, φ (C1′–C2′–O3–C3) adopts either an anti (185° ± 10, a.k.a. ‘trans’) or –gauche (−57° ± 8, a.k.a. ‘cis’) conformation. The ψ (C2–O3–C3–C4) glycosidic angle adopt a single conformation average ψ = 100° ± 11.
Only the Sia residue is resolved.
Resolution > 2.5 Å.
Distorted Sia-1 and GlcNAc-3 rings, high B-factors.
Eclipsed in chain E.
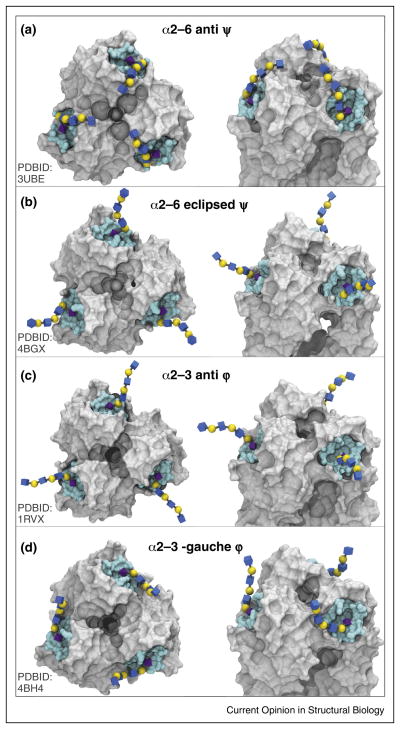
An examination of the data in Table 2 indicates that α2-6 linkages adopt two conformations when bound to HAs, which can be defined by the value of the ψ (C2′–O6–C6–C5) angle. Two shapes are also adopted by bound α2-3 linkages, which vary in the φ (C1′C2′O3-C3) angle. The significance of these shapes, with respect to the presentation of the receptor in the RBS is illustrated in Figure 4. Figure 4 illustrates that the open-umbrella topology is associated with the ‘curled’ anti-ψ conformation of an α2-6 linkage (panel A), while the cone-like topology results from the ‘extended’ anti-φ conformation of the α2-3 linkage (panel C). Presented in panels B and D are the alternative conformations of α2-6 (eclipsed-ψ) and α2-3 (–gauche-φ) linkages. The trisaccharides in the crystal structures presented in Figure 4 have each been extended to contain three lactosamine repeats to clearly illustrate the impact of the Sia-Gal linkage conformation on the orientation of the glycans. This analysis is consistent with the observations by Xu et al. [81••] that the division of the glycan topologies into only cone-like or open-umbrella is insufficient to capture the diversity of glycan conformations in HA complexes.
Figure 4.
Top (left) and side (right) views of the HA1 domains of four HA co-complexes that illustrate the four common ligand conformations seen in HA-oligosaccharide co-complexes.
The recent proposal by Peng et al. [45••] that multi-antennary α2-6 glycans can form bidentate interactions with trimeric HAs casts new light on the origin of glycan substructure differences. There are several constraints on the ability of a glycan to exhibit bidentate binding. One such constraint is the ability of the antennae to span the distance between two RBSs without steric blocking by HA surface residues, or by HA glycosylation. Another constraint arises from the topologies of the termini of individual glycan branches, which must facilitate orientations conducive to bidentate binding. As shown in Figure 4, only α2-6 receptors in a curled anti-ψ conformation satisfy this latter requirement; no known conformations of the α2-3 receptors promote bidentate binding. Although the α2-3 oligosaccharides in the –gauche φ-conformation (panel D) reach upward from the RBS rather than away (as in panels B and C), their spatial divergence from each other precludes their origination as branches of a single bian-tennary glycan. Biantennary binding requires that the bound oligosaccharides converge toward a common point in the glycan core (as in panel A). Ultimately, the inability of biantennary α2-3 receptors to form bidentate complexes arises from the linear shape of the α2-3 linkage, which controls the relative orientation of the Siaα2-3Gal disaccharide.
The observation that the α2-3 linkage precludes bidentate binding provides further insight into the functional significance of the cone like (α2-3) versus open-umbrella (α2-6) topologies [111•]. The curled anti-ψ conformation of the α2-6 glycans promotes the formation of a bidentate complex, which may also be stabilized by glycan-protein interactions associated with the larger contact area of the open-umbrella topology. Thus, while both α2-3 and α2-6 glycans may in principle form multimeric interactions with an HA, only the α2-6 receptors appear to be able to form bidentate interactions. When glycan density is sufficiently high that two or more glycans can bind simultaneously to the same HA, bidentate binding may offer little enhancement to affinity [95]. However, the ability to form bidentate interactions provides a unique opportunity for the virus to achieve avidity-enhanced binding to α2-6 receptors on a single glycan. This unique capability explains why, despite the overall loss of avidity [44••], human-adapted H3N2 viruses retain affinity for a subset of long biantennary α2-6 glycans [45••]. Tissue tropism therefore needs to be interpreted not only in terms of composition and spatial distribution of the glycans, but also in the relative density of α2-3 and α2-6 glycans.
Conclusions
Although glycan array screening is a convenient method for examining specificity, developing structure–activity relationships solely on the basis of such data is perilous. Glycan array data should generally be treated qualitatively given that the data are sensitive to numerous factors, including glycan density, glycan linker chemistry, analyte concentration, and detection method [115]. While it is possible to determine surface binding constants (KD,surf) using glycan arrays [64••,116], offering an important advantage by quantifying the binding properties of each of the glycans in an array, these protocols are not yet in widespread use. A further factor that significantly complicates the interpretation of array data is the extremely limited diversity of even the largest arrays [46••]. This limitation has obvious consequences for ligand discovery [45••], and for the elucidation of structure-specificity relationships. Although at present, data from glycan array screening need generally to be treated qualitatively, community-wide standards are being developed [117], which together with more quantitative approaches to data processing [64••] and computational analysis [118•,119•], will enhance the interpretability of such data. A powerful example of the generation and use of quantitative surface KD values from glycan array screening was reported by Wong et al. [120••]. They were able to dissect the energetic contributions made by each monosaccharide, including the sulfate moieties, in an array of sialosides binding to HAs, showing that the sulfate could enhance binding by nearly 100-fold. Further, by comparing the relative binding energies for each receptor, they were able to conclude that there is likely a competition between favorable binding interactions in the RBS, which the sulfate group maximizes and the fucose sterically blocks.
Crystallographic studies provide unique and crucial atomic-level insight into HA-receptor interactions, but in the absence of entropic considerations, do not necessarily enable a clear rationalization of specificity. Such interpretations would greatly benefit from the generation of additional quantitative monomeric affinity measurements, as well as from modeling, which may guide the choice of targets for crystallography and array screening. Lastly, variations in HA glycosylation [45••,50,51] can impact affinity and virulence [59•], and should be considered in any analysis of specificity.
On the basis of agglutination data, glycan array screening, and (albeit limited) biophysical affinity measurements, avian-infective HAs have a clear preference for α2-3 glycans, consistent with the inability of these HAs to compensate for the entropic penalty associated with binding α2-6 glycans. The specificity of human-adapted HAs for α2-6 glycans is more complex, in part because the virus may retain residual affinity for α2-3-receptors, while evolving the ability to bind to α2-6-receptors. For preferential binding of α2-6 linked glycans, mutations must occur in the RBS that overcome the entropic penalty associated with binding to the more flexible α2-6 receptor, and/or which favor the formation of bidentate interactions with multiantennary glycans. The preference for bound α2-6 glycans to adopt an anti-ψ angle (required for bidentate binding) is seen in all well-resolved crystal structures of HAs from human transmissible viruses. This suggests that bidentate binding may be a general mechanism adopted by influenza A to boost affinity for α2-6 receptors, enabling human-to-human transmission.
This review has hopefully illustrated that, despite the challenges in reconciling all of the data relating to influenza A specificity, a molecular interpretation is emerging. The implications of glycan linkage α2-3 or α2-6 on specificity extend beyond the direct interactions between the terminal Sia-Gal sequence and the HA to more macroscopic features, such as the ability to form multi-dentate complexes and the need to overcome the inherent entropic penalty associated with binding to α2-6 glycans. A complete understanding of specificity requires a continuous reevaluation of the paradigms with a view to integrating all available data into a holistic analysis.
Footnotes
Conflicts of interest
The authors have declared no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Morens DM, Taubenberger JK. Historical thoughts on influenza viral ecosystems, or behold a pale horse, dead dogs, failing fowl, and sick swine. Influenza Other Respir Viruses. 2010;4:327–337. doi: 10.1111/j.1750-2659.2010.00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson NPAS, Mueller J. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull Hist Med. 2002;76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 3.Glaser L, et al. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol. 2005;79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tumpey TM, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 5.Reperant LA, Kuiken T, Osterhaus AD. Adaptive pathways of zoonotic influenza viruses: from exposure to establishment in humans. Vaccine. 2012;30:4419–4434. doi: 10.1016/j.vaccine.2012.04.049. [DOI] [PubMed] [Google Scholar]
- 6.Wong SS, Yuen KY. Avian influenza virus infections in humans. Chest. 2006;129:156–168. doi: 10.1378/chest.129.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith GJ, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 8.Al Hajjar S, McIntosh K. The first influenza pandemic of the 21st century. Ann Saudi Med. 2010;30:1–10. doi: 10.4103/0256-4947.59365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fineberg HV. Pandemic preparedness and response —lessons from the H1N1 influenza of 2009. N Engl J Med. 2014;370:1335–1342. doi: 10.1056/NEJMra1208802. [DOI] [PubMed] [Google Scholar]
- 10.Swerdlow DL, Finelli L, Bridges CB. 2009 H1N1 influenza pandemic: field and epidemiologic investigations in the United States at the start of the first pandemic of the 21st century. Clin Infect Dis. 2011;52(Suppl 1):S1–S3. doi: 10.1093/cid/ciq005. [DOI] [PubMed] [Google Scholar]
- 11.Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20:243–267. doi: 10.1128/CMR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 13•.Sriwilaijaroen N, Suzuki Y. Molecular basis of the structure and function of H1 hemagglutinin of influenza virus. Proc Jpn Acad Ser B Phys Biol Sci. 2012;88:226–249. doi: 10.2183/pjab.88.226. A particularly detailed and thorough review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medina RA, Garcia-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. 2010;285:28403–28409. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris A, et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci U S A. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subbarao K, Joseph T. Scientific barriers to developing vaccines against avian influenza viruses. Nat Rev Immunol. 2007;7:267–278. doi: 10.1038/nri2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gustin KM, et al. Comparison of the levels of infectious virus in respirable aerosols exhaled by ferrets infected with influenza viruses exhibiting diverse transmissibility phenotypes. J Virol. 2013;87:7864–7873. doi: 10.1128/JVI.00719-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thangavel RR, Bouvier NM. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods. 2014;410:60–79. doi: 10.1016/j.jim.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heldt FS, Frensing T, Reichl U. Modeling the intracellular dynamics of influenza virus replication to understand the control of viral RNA synthesis. J Virol. 2012;86:7806–7817. doi: 10.1128/JVI.00080-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frensing T, et al. Influenza virus intracellular replication dynamics, release kinetics, and particle morphology during propagation in MDCK cells. Appl Microbiol Biotechnol. 2016;100:7181–7192. doi: 10.1007/s00253-016-7542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H. Tissue and host tropism of influenza viruses: importance of quantitative analysis. Sci China C Life Sci. 2009;52:1101–1110. doi: 10.1007/s11427-009-0161-x. [DOI] [PubMed] [Google Scholar]
- 24.Schrauwen EJ, Fouchier RA. Host adaptation and transmission of influenza A viruses in mammals. Emerg Microbes Infect. 2014;3:e9. doi: 10.1038/emi.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabriel G, et al. Differential use of importin-alpha isoforms governs cell tropism and host adaptation of influenza virus. Nat Commun. 2011;2:156. doi: 10.1038/ncomms1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Riel D, et al. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng H, et al. Tropism and infectivity of influenza virus, including highly pathogenic avian H5N1 virus, in ferret tracheal differentiated primary epithelial cell cultures. J Virol. 2013;87:2597–2607. doi: 10.1128/JVI.02885-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito T, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakdawala SS, et al. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature. 2015;526:122–125. doi: 10.1038/nature15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couceiro JN, Paulson JC, Baum LG. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 31.Nicholls JM, et al. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res. 2007;8:73. doi: 10.1186/1465-9921-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.de Graaf M, Fouchier RA. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33:823–841. doi: 10.1002/embj.201387442. A thorough review of influenza HA-glycan interactions and their impact on transmission and pathogenesis from the perspective of tissue tropism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller GL, Stanley WM. Quantitative aspects of the red blood cell agglutination test for influenza virus. J Exp Med. 1944;79:185–195. doi: 10.1084/jem.79.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Rogers GN, Paulson J. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. doi: 10.1016/0042-6822(83)90150-2. The authors established HA binding preferences using RBCs with enzymatically modified glycan sialylation. This work forms the basis for the α2,3/α2,6 specificity paradigm. [DOI] [PubMed] [Google Scholar]
- 35.Ovsyannikova IG, et al. Turkey versus guinea pig red blood cells: hemagglutination differences alter hemagglutination inhibition responses against influenza A/H1N1. Viral Immunol. 2014;27:174–178. doi: 10.1089/vim.2013.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aich U, et al. Glycomics-based analysis of chicken red blood cells provides insight into the selectivity of the viral agglutination assay. FEBS J. 2011;278:1699–1712. doi: 10.1111/j.1742-4658.2011.08096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glick GD, et al. Ligand recognition by influenza virus. The binding of bivalent sialosides. J Biol Chem. 1991;266:23660–23669. [PubMed] [Google Scholar]
- 38•.Xiong X, et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature. 2013;497:392–396. doi: 10.1038/nature12144. Monomeric binding affinities determined for H3 and H5 HAs by microscale thermophoresis (MST), confirming that avidity is essential for explaining virus specificity. [DOI] [PubMed] [Google Scholar]
- 39.Xu HF, Shaw DE. A simple model of multivalent adhesion and its application to influenza infection. Biophys J. 2016;110:218–233. doi: 10.1016/j.bpj.2015.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Mammen M, Choi S-K, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. The paper provides a detailed assessment of the impact of multivalent interactions on binding kinetics. [DOI] [PubMed] [Google Scholar]
- 41•.Sauter NK, et al. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry. 1989;28:8388–8396. doi: 10.1021/bi00447a018. Monomeric binding affinities were determined by NMR for H3-glycan interactions, showing that specificity arises from very small differences in binding free energies. [DOI] [PubMed] [Google Scholar]
- 42•.Pritchett TJ, et al. Recognition of monovalent sialosides by influenza virus H3 hemagglutinin. Virology. 1987;160:502–506. doi: 10.1016/0042-6822(87)90026-2. The authors report IC50 data for small sialosides inhibiting virus–RBC interactions. The paper is a classic pre-glycan-array illustration of determining the α2,3/α2,6 specificity preferences of HAs. [DOI] [PubMed] [Google Scholar]
- 43.Vachieri SG, et al. Receptor binding by H10 influenza viruses. Nature. 2014;511:475–477. doi: 10.1038/nature13443. [DOI] [PubMed] [Google Scholar]
- 44••.Lin YP, et al. Evolution of the receptor binding properties of the influenza A (H3N2) hemagglutinin. Proc Natl Acad Sci U S A. 2012;109:21474–21479. doi: 10.1073/pnas.1218841110. The authors illustrate that H3 avidity loss has resulted from key mutations in the RBS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Peng W, et al. Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe. 2017;21:23–34. doi: 10.1016/j.chom.2016.11.004. The authors screen a glycan array that contains a larger diversity of glycans of the type present on human epithelial tissue. They propose that avidity loss from antigenic drift in HAs can be overcome by binding to long branched glycans that terminate in α2,6 linkages that can form bidentate interactions with the HA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Walther T, et al. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013;9:e1003223. doi: 10.1371/journal.ppat.1003223. An extensive glycomics analysis of glycan structure and distribution on human tissue, in which the authors note that none of the current glycan arrays contain all of the key glycans present in human airways, and thus glycan array data may not be predictive of replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.de Vries RP, et al. Three mutations switch H7N9 influenza to human-type receptor specificity. PLoS Pathog. 2017;13:e1006390. doi: 10.1371/journal.ppat.1006390. The authors predict and confirm specificity-switching mutations with potential pandemic significance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gulati S, et al. Human H3N2 influenza viruses isolated from 1968 to 2012 show varying preference for receptor substructures with no apparent consequences for disease or spread. PLOS ONE. 2013;8:e66325. doi: 10.1371/journal.pone.0066325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thieker DF, et al. 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology. 2016;26:786–787. doi: 10.1093/glycob/cww076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun S, et al. Glycosylation site alteration in the evolution of influenza A (H1N1) viruses. PLoS ONE. 2011;6:e22844. doi: 10.1371/journal.pone.0022844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.An Y, et al. Comparative glycomics analysis of influenza hemagglutinin (H5N1) produced in vaccine relevant cell platforms. J Proteome Res. 2013;12:3707–3720. doi: 10.1021/pr400329k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An Y, et al. Glycosylation analysis of engineered H3N2 influenza A virus hemagglutinins with sequentially added historically relevant glycosylation sites. J Proteome Res. 2015;14:3957–3969. doi: 10.1021/acs.jproteome.5b00416. [DOI] [PubMed] [Google Scholar]
- 53.Sun X, et al. N-linked glycosylation of the hemagglutinin protein influences virulence and antigenicity of the 1918 pandemic and seasonal H1N1 influenza A viruses. J Virol. 2013;87:8756–8766. doi: 10.1128/JVI.00593-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang H, et al. Structure and receptor binding preferences of recombinant human A (H3N2) virus hemagglutinins. Virology. 2015;477:18–31. doi: 10.1016/j.virol.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, et al. Glycosylation on hemagglutinin affects the virulence and pathogenicity of pandemic H1N1/2009 influenza A virus in mice. PLOS ONE. 2013;8:e61397. doi: 10.1371/journal.pone.0061397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevens J, et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 57.Jayaraman A, et al. Glycosylation at Asn91 of H1N1 haemagglutinin affects binding to glycan receptors. Biochem J. 2012;444:429–435. doi: 10.1042/BJ20112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mir-Shekari SY, et al. The glycosylation of the influenza A virus hemagglutinin by mammalian cells. A site-specific study. J Biol Chem. 1997;272:4027–4036. doi: 10.1074/jbc.272.7.4027. [DOI] [PubMed] [Google Scholar]
- 59•.Tate MD, et al. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses. 2014;6:1294–1316. doi: 10.3390/v6031294. The authors illustrate the importance of HA glycosylation on viral fitness and antigenicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deom CM, Caton AJ, Schulze IT. Host cell-mediated selection of a mutant influenza A virus that has lost a complex oligosaccharide from the tip of the hemagglutinin. Proc Natl Acad Sci U S A. 1986;83:3771–3775. doi: 10.1073/pnas.83.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W, et al. Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J Virol. 2010;84:6570–6577. doi: 10.1128/JVI.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao Y, et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 2009;5:e1000709. doi: 10.1371/journal.ppat.1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X, et al. Hemagglutinin glycosylation modulates the pathogenicity and antigenicity of the H5N1 avian influenza virus. Vet Microbiol. 2015;175:244–256. doi: 10.1016/j.vetmic.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 64••.Liao H-Y, et al. Differential receptor binding affinities of influenza hemagglutinins on glycan arrays. J Am Chem Soc. 2010;132:14849–14856. doi: 10.1021/ja104657b. This paper introduces an analytical method for obtaining surface KD values from glycan array data. Such approaches facilitate quantitative comparisons of ligand binding. [DOI] [PubMed] [Google Scholar]
- 65.Chen MW, et al. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc Natl Acad Sci U S A. 2008;105:13538–13543. doi: 10.1073/pnas.0806901105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alymova IV, et al. Glycosylation changes in the globular head of H3N2 influenza hemagglutinin modulate receptor binding without affecting virus virulence. Sci Rep. 2016;6:36216. doi: 10.1038/srep36216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reid AH, et al. 1918 influenza pandemic caused by highly conserved viruses with two receptor-binding variants. Emerg Infect Dis. 2003;9:1249–1253. doi: 10.3201/eid0910.020789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang W, et al. Molecular basis of the receptor binding specificity switch of the hemagglutinins from both the 1918 and 2009 pandemic influenza A viruses by a D225G substitution. J Virol. 2013;87:5949–5958. doi: 10.1128/JVI.00545-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Connor RJ, et al. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 70.Stevens J, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 71.Stevens J, et al. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J Mol Biol. 2008;381:1382–1394. doi: 10.1016/j.jmb.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen L-M, et al. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology. 2012;422:105–113. doi: 10.1016/j.virol.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imai M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Herfst S, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 76•.Russell RJ, et al. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology. 2004;325:287–296. doi: 10.1016/j.virol.2004.04.040. The authors illustrate the challenges associated with rationalizing HA specificity based only on HA-oligosaccharide 3D structures. [DOI] [PubMed] [Google Scholar]
- 77•.Raman R, et al. Glycan-receptor specificity as a useful tool for characterization and surveillance of influenza A virus. Trends Microbiol. 2014;22:632–641. doi: 10.1016/j.tim.2014.07.002. The authors illustrate the challenges associated with rationalizing HA specificity based only on HA-oligosaccharide 3D structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78••.Lin T, et al. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology. 2009;392:73–81. doi: 10.1016/j.virol.2009.06.028. The paper presents a rare structure of an avian H1, and the authors note the inability of the HA-oligosaccharide co-complexes to explain specificity. [DOI] [PubMed] [Google Scholar]
- 79.vonderLieth CW, Kozar T. Towards a better semiquantitative estimation of binding constants: molecular dynamics exploration of the conformational behavior of isolated sialyllactose and sialyllactose complexed with influenza A hemagglutinin. THEOCHEM. 1996;368:213–222. [Google Scholar]
- 80.Newhouse EI, et al. Mechanism of glycan receptor recognition and specificity switch for avian, swine, and human adapted influenza virus hemagglutinins: a molecular dynamics perspective. J Am Chem Soc. 2009;131:17422–17430. doi: 10.1021/ja904052q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81••.Xu D, et al. Distinct glycan topology for avian and human sialopentasaccharide receptor analogues upon binding different hemagglutinins: a molecular dynamics perspective. J Mol Biol. 2009;387:465–491. doi: 10.1016/j.jmb.2009.01.040. A good example of the strengths and limitations of computational simulations of HA-glycan complexes, which also indicated the importance of conformational entropy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Doig AJ, Sternberg MJE. Side-chain conformational entropy in protein folding. Protein Sci. 1995;4:2247–2251. doi: 10.1002/pro.5560041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kadirvelraj R, et al. Structure and binding analysis of polyporus squamosus lectin in complex with the Neu5Acα2-6Galβ1-4GlcNAc human-type influenza receptor. Glycobiology. 2011;21:973–984. doi: 10.1093/glycob/cwr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Veluraja K, Rao VSR. Theoretical studies on the conformation of monosialogangliosides and disialogangliosides. Carbohydr Polym. 1983;3:175–192. [Google Scholar]
- 85.Sawada T, Fedorov DG, Kitaura K. Role of the key mutation in the selective binding of avian and human influenza hemagglutinin to sialosides revealed by quantum-mechanical calculations. J Am Chem Soc. 2010;132:16862–16872. doi: 10.1021/ja105051e. [DOI] [PubMed] [Google Scholar]
- 86.Kadirvelraj R, et al. Involvement of water in carbohydrate–protein binding: concanavalin a revisited. J Am Chem Soc. 2008;130:16933–16942. doi: 10.1021/ja8039663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Komath SS, Kenoth R, Swamy MJ. Thermodynamic analysis of saccharide binding to snake gourd (Trichosanthes anguina) seed lectin. Fluorescence and absorption spectroscopic studies. Eur J Biochem. 2001;268:111–119. doi: 10.1046/j.1432-1327.2001.01852.x. [DOI] [PubMed] [Google Scholar]
- 88.Sharp K. Entropy–enthalpy compensation: fact or artifact? Protein Sci. 2001;10:661–667. doi: 10.1110/ps.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stevens D, et al. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nature. 2006;4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blixt O, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci U S A. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Childs RA, et al. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol. 2009;27:797–799. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Srinivasan K, et al. Quantitative description of glycan-receptor binding of influenza A virus H7 hemagglutinin. PLOS ONE. 2013;8:e49597. doi: 10.1371/journal.pone.0049597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McBride R, Paulson JC, de Vries RP. A miniaturized glycan microarray assay for assessing avidity and specificity of influenza A virus hemagglutinins. J Vis Exp. 2016;111:1–11. doi: 10.3791/53847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang ML, et al. Determination of receptor specificities for whole influenza viruses using multivalent glycan arrays. Chem Commun (Camb) 2015;51:5326–5329. doi: 10.1039/c4cc08613a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nycholat CM, et al. Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew Chem Int Ed Engl. 2012;51:4860–4863. doi: 10.1002/anie.201200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kumari K, et al. Receptor binding specificity of recent human H3N2 influenza viruses. Virol J. 2007;4:42. doi: 10.1186/1743-422X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao N, et al. Association analyses of large-scale glycan microarray data reveal novel host-specific substructures in influenza A virus binding glycans. Sci Rep. 2015;5:15778. doi: 10.1038/srep15778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98•.Song X, et al. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J Biol Chem. 2011;286:31610–31622. doi: 10.1074/jbc.M111.274217. The paper demonstrates the critical impact of Sia modifications on affinity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bradley KC, et al. Analysis of influenza virus hemagglutinin receptor binding mutants with limited receptor recognition properties and conditional replication characteristics. J Virol. 2011;85:12387–12398. doi: 10.1128/JVI.05570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Varki A. Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am J Phys Anthropol. 2001;(Suppl 33):54–69. doi: 10.1002/ajpa.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suzuki Y, et al. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol. 2000;74:11825–11831. doi: 10.1128/jvi.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gambaryan AS, et al. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol. 2012;86:4370–4379. doi: 10.1128/JVI.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stevens J, et al. Receptor specificity of influenza A H3N2 viruses isolated in mammalian cells and embryonated chicken eggs. J Virol. 2010;84:8287–8299. doi: 10.1128/JVI.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gambaryan AS, et al. H5N1 chicken influenza viruses display a high binding affinity for Neu5Acalpha2-3Galbeta1-4(6-HSO3)GlcNAc-containing receptors. Virology. 2004;326:310–316. doi: 10.1016/j.virol.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 105.Gambaryan A, et al. Evolution of the receptor binding phenotype of influenza A (H5) viruses. Virology. 2006;344:432–438. doi: 10.1016/j.virol.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 106.Xiong X, et al. Recognition of sulphated and fucosylated receptor sialosides by A/Vietnam/1194/2004 (H5N1) influenza virus. Virus Res. 2013;178:12–14. doi: 10.1016/j.virusres.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 107.Gambaryan AS, et al. 6-Sulfo sialyl Lewis X is the common receptor determinant recognized by H5, H6, H7 and H9 influenza viruses of terrestrial poultry. Virol J. 2008;5:1–10. doi: 10.1186/1743-422X-5-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Collins PJ, et al. Recent evolution of equine influenza and the origin of canine influenza. Proc Natl Acad Sci U S A. 2014;111:11175–11180. doi: 10.1073/pnas.1406606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hiono T, et al. Amino acid residues at positions 222 and 227 of the hemagglutinin together with the neuraminidase determine binding of H5 avian influenza viruses to sialyl Lewis X. Arch Virol. 2016;161:307–316. doi: 10.1007/s00705-015-2660-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Linden SK, et al. Mucins in the mucosal barrier to infection. Mucosal Immunol. 2008;1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111•.Chandrasekaran A, et al. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. The paper introduces the cone-like and open-umbrella topology descriptions for α2-3 and α2-6 glycan-HA co-complexes. [DOI] [PubMed] [Google Scholar]
- 112.Zhang W, et al. An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level. Science. 2013;340:1463–1467. doi: 10.1126/science.1236787. [DOI] [PubMed] [Google Scholar]
- 113.Viswanathan K, et al. Glycans as receptors for influenza pathogenesis. Glycoconj J. 2010;27:561–570. doi: 10.1007/s10719-010-9303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu R, et al. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol. 2012;86:982–990. doi: 10.1128/JVI.06322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Oyelaran O, Gildersleeve JC. Glycan arrays: recent advances and future challenges. Curr Opin Chem Biol. 2009;13:406–413. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Raman R, et al. Glycan receptor specificity as a useful tool for characterization and surveillance of influenza A virus. Trends Microbiol. 2014;22:632–641. doi: 10.1016/j.tim.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y, et al. The minimum information required for a glycomics experiment (MIRAGE) project: improving the standards for reporting glycan microarray-based data. Glycobiology. 2017;27:280–284. doi: 10.1093/glycob/cww118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118•.Grant OC, et al. Combining 3D structure with glycan array data provides insight into the origin of glycan specificity. Glycobiology. 2016;26:772–783. doi: 10.1093/glycob/cww020. The paper illustrates an approach to interpreting glycan array data in terms of 3D binding motifs, providing a mechanism for rationalizing glycan-substructure effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119•.Grant OC, et al. Presentation, presentation, presentation! Molecular-level insight into linker effects on glycan array screening data. Glycobiology. 2014;24:17–25. doi: 10.1093/glycob/cwt083. The paper illustrates the use of structure modeling to detect artifacts in glycan array data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120••.Wang C-C, et al. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc Natl Acad Sci U S A. 2009;106:18137–18142. doi: 10.1073/pnas.0909696106. The paper shows the insights gained from quantifying binding data from glycan array screening. [DOI] [PMC free article] [PubMed] [Google Scholar]