Endocytosis – cell drinking and eating – is an essential process and thus is believed to have evolved along with the first common eukaryotic ancestors [1]. Several distinct endocytic pathways function for different classes of cargo [2]; however, after decades of study, clathrin-mediated endocytosis (CME) has become the best mechanistically understood internalization pathway [3, 4]. CME is a multi-step process initiated by the interaction of adaptor proteins (APs) with both phosphatidyl-4,5-bisphosphate (PIP2) on the plasma membrane and sorting motifs on the cytoplasmic tails of transmembrane cargo molecules. APs trigger the assembly of clathrin into curved polygonal lattices that, together with various endocytic accessory proteins, deform the membrane and concentrate cargo molecules into invaginated clathrin coated pits (CCPs). Among many players, a large GTPase, dynamin, has been suggested as a critical factor for different stages of CME.
At late stages of CME, dynamin assembles into collar-like structures around the necks of deeply invaginated CCPs and catalyzes membrane fission to release clathrin-coated vesicles (CCVs), which carry their cargo into the cell. Dynamin can also be detected in its unassembled state on nascent clathrin lattices [5, 6], where it has been suggested to play a role in regulating earlier stages of CCP maturation [7].
Primordial clathrin-mediated endocytosis without dynamin
Comparative genomic studies have identified clathrin, adaptors, and other endocytic accessory proteins, such as epsin in all eukaryotic organisms, suggesting that clathrin-mediated endocytosis (CME) existed in the earliest common eukaryotic ancestor [8]. Paradoxically, dynamin is not detected until much later in evolution [8, 9]. These findings demonstrate that although dynamin is essential for CME in higher eukaryotes, endocytosis in many unicellular organisms occurs independently of dynamin.
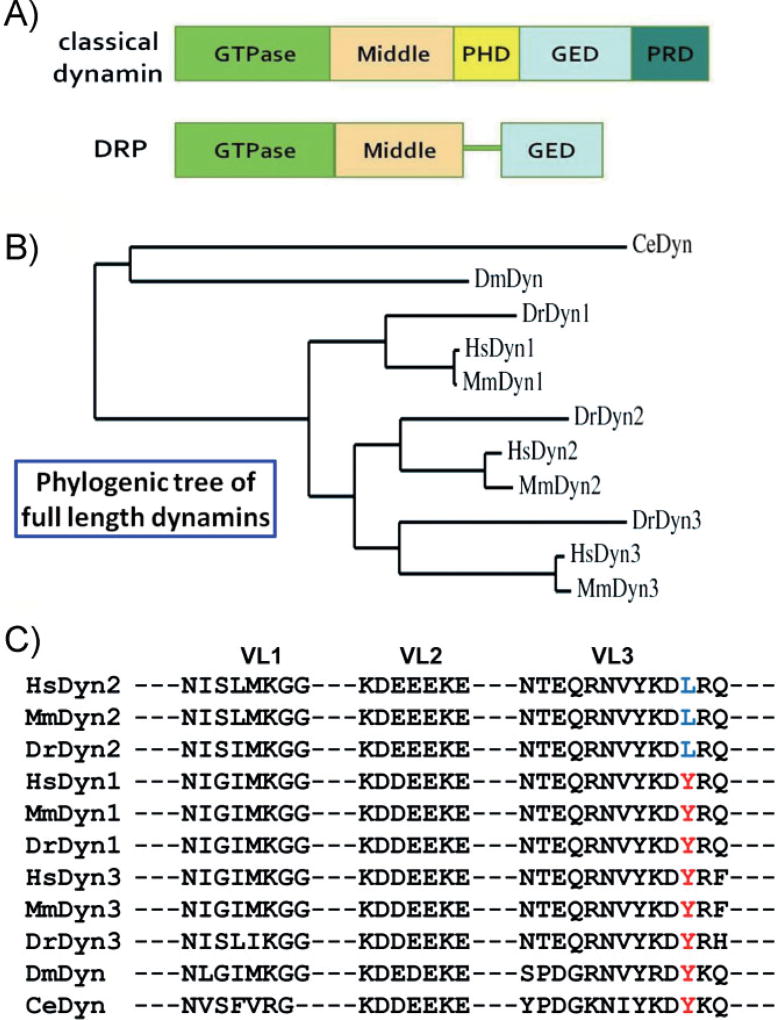
Dynamin belongs to a diverse family of large, multi-domain GTPases, named the dynamin superfamily. Within this superfamily, dynamin-related proteins (DRPs) are distinguished from “classical” dynamins because they lack both a pleckstrin homology domain (PHD) and a proline/arginine rich domain (PRD; Fig. 1A). DRPs, which appear much earlier in evolution and exist in all major eukaryote groups, are hypothesized to have originally evolved for their roles in mitochondria division and/or cytokinesis and may partially participate in endocytosis in some organisms [9–13]. The PHD and PRD are both involved in targeting dynamin to the plasma membrane. The PHD binds phosphatidyl-4,5-bisphosphate (PIP2) and is required for dynamin-membrane interactions [14], whereas the PRD interacts with several Src homology 3 domain (SH3 domain)-containing endocytic accessory factors and is required for targeting unassembled dynamin to clathrin-coated pits (CCPs) [15]. In addition, both the PHD and PRD function to regulate dynamin activity [16–18].
Figure 1. The domain structure and comparison of dynamins.
A: A comparison of the domain structures of classical dynamins and DRPs. The GTPase, middle, and GED are conserved across all dynamin superfamily members. The PHD and PRD, involved in targeting and regulating dynamin activity, are unique to classical dynamins. B: Phylogenic analysis of full-length dynamins from human (Hs), mouse (Mm), zebra fish (Dr), fly (Dm), and nematode (Ce). The rooted phylogenetic tree was generated with Phylogeny.fr [28]. C: Amino acid alignment of dynamin PHDs. Membrane-interacting VLs (VL1–3) of dynamin PHD from different organisms are shown. A residue in VL3, Y600 in Dyn 1 and 3 and L600 in Dyn2 (highlighted in red and blue, respectively), confers curvature sensitivity [23] and is diagnostic of vertebrate dynamin isoforms. PHDs were aligned with CLUSTAL 2.0.1; and the compared organisms include Homo sapiens (Hs), Mus musculus (Mm), Danio rerio (Dr), Drosophila melanogaster (Dm), and Caenorhabditis elegans (Ce).
The earliest dynamin corresponds to the neuronal isoform
The genomes of Caenorhabditis elegans and Drosophila encode a single classical dynamin, which is primarily expressed in the nervous system [19, 20]. In contrast, vertebrate genomes encode three classical dynamin genes that exhibit tissue-specific expression: dynamin-1 (Dyn1) is exclusively expressed in neurons, dynamin-2 (Dyn2) is ubiquitously expressed and dynamin-3 (Dyn3) is primarily expressed in neurons and testes. As the biochemical properties of Dyn3 have not been studied, we focus here on Dyn1 and Dyn2. Despite their high sequence identity (>75%), these isoforms have tissue-specific functions. Dyn1 (and Dyn3), but not Dyn2, can fully support the rapid, stimulus-dependent endocytosis required for synaptic vesicle recycling in neurons [19–21]; whereas Dyn2 more efficiently supports CME in nonneuronal cells [22]. This raises the question: Which is the primordial dynamin isoform, the neuronal-specific Dyn1 or the ubiquitously expressed Dyn2?
The full-length sequence of the single dynamin from invertebrates is equally similar to each of the three dynamins from vertebrates, and thus it is uncertain as to which dynamin isoform corresponds to the progenitor (Fig. 1B). However, we recently discovered that Dyn1 and Dyn2 have quantitatively distinct curvature-generating or curvature-sensing properties that are important for their tissue-specific functions [23]. Dyn1 is a potent curvature-generator; whereas Dyn2 is primarily a curvature-sensor. Strikingly, we identified a single amino acid in the membrane-interacting variable loop (VL) 3 of the PHD (Y600 in Dyn1 and L600 in Dyn2) that is necessary and sufficient to account for these different properties and thus serves to distinguish Dyn1 from Dyn2 [23]. An examination of the sequences of classical dynamins from C. elegans, Drosophila, or Dyn1 and Dyn3 from vertebrates revealed a conserved tyrosine at this residue (Fig. 1C). The mutation to leucine occurs only in Dyn2, and only in vertebrates. These results suggest a counter-intuitive evolutionary trend, namely that the neuronal dynamins, Dyn1 and Dyn3, are the primordial forms, and the ubiquitous Dyn2 isoform evolved later. This speculation is consistent with the high expression of classical dynamin in the nervous system of C. elegans and Drosophila [19, 20].
Dynamin homologues detected in pre-metazoan cells
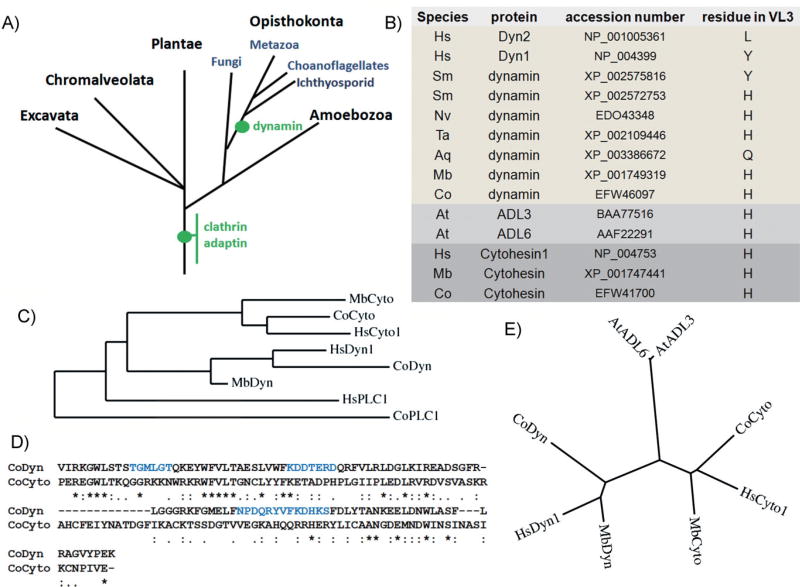
To gain further insight into the evolution of classical dynamin, we carried out a phylogenetic analysis focusing, first, on the dynamin-specific PHD. We used the dynamin PHD as the signature to look for classical dynamins. Briefly, the PHD sequences from known dynamins were aligned, and HMMER3 was used to construct a Hidden Markov model [24]. This model was then used to search the UniproKB/TrEMBL protein database, and the hits were then examined for GTPase, middle, GTPase effector domain (GED), and PRD sequences to identify homologues that shared the same domain structure as classical dynamins. While previously thought to have appeared only in metazoa [8, 9], we were able to identify dynamin homologues in the choanoflagellate Monosiga brevicollis and the Ichthyosporid Capsaspora owczarzaki (E-values for their PHD are 2.1e−43 and 2.9e−41, respectively). These protists are the closest relatives of metazoans [24]. We also identified dynamins in the earliest metazoans, including Porifera (sponges), Placozoa, and Cnidaria; animals that lack a central nervous system. It is important to note that the dynamins we have identified in early metazoans ranging from C. owczarzaki to Schistosoma mansoni represent putative/predicted proteins and whether these proteins function in CME-related or other roles remains to be determined. Nonetheless, these observations demonstrate that dynamin evolved prior to the evolution of a central nervous system (Fig. 2A).
Figure 2. Evolution of the dynamin PHD.
A: Model for major acquisition of selected proteins in eukaryotes. The evolutionary relationships of various eukaryotic taxa are shown [8], and the positions of acquisitions of gene families are indicated (green dots). B: Table showing the identity of the diagnostic residue in VL3 in dynamins from various organisms. In lower organisms the residue is a histidine, which is conserved in the VL3 of the related PHD from cytohesin (ArfGEF). C: A common ancestor for PHDs of cytohesins and dynamins. PHDs from dynamins, cytohesins, and phospholipase Cγ 1 (PLC1) are aligned and plotted as the rooted phylogenetic tree. D: Alignment of PHDs from an early premetazoan organism C. owczarzaki (Co). Shown is the PHD from dynamin and cytohesin. PHDs are aligned with CLUSTAL 2.0.1, and the VLs are colored in blue. Asterisk indicates identical amino acid, and period or colon represent similar amino acids. E: Unrooted phylogenetic tree of PHDs from dynamins, cytohesins, and Arabidopsis dynamin-like proteins, ADL3 and ADL6, is plotted with Phylogeny.fr. The abbreviation of organisms include Homo sapiens (Hs), S. mansoni (Sm), Nematostella vectensis (Nv), Trichoplax adhaerens (Ta), Amphimedon queenslandica (Aq), M. brevicollis (Mb), C. owczarzaki (Co), and A. thaliana (At).
Efficient curvature generation: A necessity for synaptic transmission
What does the functionally diagnostic residue (tyrosine) within the VL3 tell us about dynamin evolution? Interestingly, the early dynamin homologues encode neither a tyrosine nor a leucine at position 600, but rather a histidine (Fig. 2B). We detect the first appearance of the tyrosine residue in Platyhelminthes (flatworms, e.g. S. mansoni), the most primitive extant animals that possess a central nervous system (Fig. 2B). Thus, mutation to the curvature generating tyrosine residue appears coincident with the development of synaptic transmission.
Based on these observations we hypothesize that dynamin-Y600 arose from the ancient dynamin-H600 in conjunction with the specific role of CME in synaptic transmission. CME at the synapse must be rapid and tightly regulated in coordination with excitation-dependent synaptic vesicle release. Moreover, the products of CME, i.e. recycled synaptic vesicles, must be uniform in size to deliver their quantal signals. The evolution of neuronal dynamins may reflect a situation where selection favored these requirements for regulated CME. In this scenario the PHD and PRD may have evolved as regulatory domains controlling the spatial and temporal activation of dynamin for clathrin-mediated synaptic vesicle recycling.
We further speculate that the later evolution of Dyn2 also reflects its regulatory role in controlling the fidelity and timing of CME in higher organisms that must cope with complex and diverse cargos and the regulation of trans-membrane signaling receptors in non-neuronal cells. The high sensitivity to membrane curvature conferred by the tyrosine to leucine mutation in the PHD of Dyn2 suggests that Dyn2 can only self-assemble on the narrow neck of mature CCPs. In this way, Dyn2 could function as an assembly switch that monitors the progression of CCP maturation and determines the timing of clathrin-coated vesicle (CCV) completion and release [23].
An evolutionary glimpse into this transition may be seen in the parasitic flatworm, S. mansoni. Unlike most invertebrates, which express only a single dynamin isoform, we could detect two classical dynamins encoded in its genome: one with histidine in VL3 and the other with tyrosine (Fig. 2B). With the same domain structure as classical dynamin, including a dynamin-specific GTPase domain and PRD, it is likely that both of these isoforms function to support CME. Thus, it will be interesting to determine whether they exhibit tissue-specific expression patterns. Furthermore, it will be intriguing to compare the biochemical properties of S. mansoni dynamin-H600 with dynamin-Y600. The positively charged, but pH-sensitive histidine residue in VL3 may function to regulate its interactions with negatively charged membranes.
The orgin of dynamin’s pleckstrin homology domain
The PHD is the 11th most common domain in the human genome [25]. Although all PHDs have similar three-dimensional structures, their primary sequences are not conserved. From where did the dynamin progenitor coopt its PHD? To address this question we searched for the PHD most similar to that of dynamin and identified the PHD from the guanine-nucleotide exchange factors for Arf (ArfGEF) cytohesin. The phylogenetic tree of PHDs shows a common ancestor of PHD from dynamins and cytohesins (Fig. 2C). A comparison of the PHD sequences of dynamin and cytohesin from the pre-metazoan organism, C. owczarzaki showed a striking 22% similarity (Fig. 2D), as compared to virtually no identity with the PIP2-binding phospholipase C (PLC)γ1 PH domain from the same organism.
Plants encode multiple dynamins and DRPs. The PHDs of the classical Arabidopsis thaliana dynamins, ADL3 and ADL6, also share a high degree of similarity to cytohesin PHDs (~20% similarity). The unrooted phylogenetic tree shows that these PHDs belong to distinct groups, yet separated by similar distances (Fig. 2E). Thus, although the dynamins in plants emerged as a result of convergent evolution and do not share a common ancestor with animal dynamins, they may have independently derived their PHDs from the ancestor of cytohesins. Notably, all the PHDs in cytohesins and plant dynamins have histidine at the residue corresponding to H600 in primitive animal dynamins (Fig. 2B). Based on these observations, we hypothesize that early dynamin progenitors may have co-opted their PHD from an ancestral cytohesin, which functions through the small GTPase, ADP-ribosylation factor (Arf), to regulate coated vesicle formation at the plasma membrane. This observation is interesting in light of the suggestion that Arf and dynamin may play mechanistically similar roles as fidelity monitors and fission machinery components to mediate vesicle release [26].
While studies have shown that mammalian Dyn1 and Dyn2 are necessary and sufficient to catalyze membrane fission in vitro [27], it will be fascinating to test whether dynamins from more primitive organisms share this capacity or whether this property of dynamin emerged during subsequent evolution.
Conclusions and outlook
Based on our understanding of dynamin isoform specificity and our in silico search for primitive dynamin homologues, we hypothesize that dynamin first evolved for vesicular trafficking in premetazoans, having co-opted a PHD from the ancestor of the ArfGEF, cytohesin. Subsequent evolution of a neuron-specific isoform coincided with the development of neurotransmission in metazoans and the need for rapid and tightly regulated endocytosis for synaptic vesicle recycling. The ubiquitously expressed Dyn2 isoform, which is first detected in vertebrates, may regulate more diverse tissue- and cargo-specific roles for CME. Further analyses of the biochemical properties of these dynamin sequence variants and their cellular functions will be needed to test these speculations. However, the evolutionary path of dynamin, especially its late appearance relative to other major CME components, suggests a more specialized role in regulating this essential cellular process.
Acknowledgments
We thank Joel Dacks (University of Alberta) for critically reading an early draft of this manuscript and for invaluable advice on comprehensive sequence analysis. We also thank Xin Zhang (The Scripps Research Institute) for inspiring discussion. This is TSRI manuscript number 21711. Y.-W. L. was supported by Muscular Dystrophy Association Grant MDA-114824 and this work was supported by GM42455 and MH61345 to SLS.
Abbreviations
- ADL
Arabidopsis dynamin-like protein
- Arf
ADP-ribosylation factor
- ArfGEF
guanine-nucleotide exchange factors for Arf
- CCP
clathrin-coated pit
- CCV
clathrin-coated vesicle
- CME
clathrin-mediated endocytosis
- DRP
dynamin-related protein
- GED
GTPase effector domain
- PHD
pleckstrin homology domain
- PIP2
phosphatidyl-4,5-bisphosphate
- PRD
proline/arginine rich domain
- PLC
phospholipase C
- SH3 domain
Src homology 3 domain
- VL
variable loop.
References
- 1.de Duve C. The origin of eukaryotes: a reappraisal. Nat Rev Genet. 2007;8:395–403. doi: 10.1038/nrg2071. [DOI] [PubMed] [Google Scholar]
- 2.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 3.Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin’s role in clathrin-mediated endocytosis. Biochem Soc Trans. 2009;37:1022–6. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid SL, Frolov VA. Dynamin: functional design of a membrane fission catalyst. Annu Rev Cell Dev Biol. 2011;27:79–105. doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- 5.Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–34. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evergren E, Tomilin N, Vasylieva E, Sergeeva V, et al. A pre-embedding immunogold approach for detection of synaptic endocytic proteins in situ. J Neurosci Methods. 2004;135:169–74. doi: 10.1016/j.jneumeth.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Sever S, Damke H, Schmid SL. Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J Cell Biol. 2000;150:1137–48. doi: 10.1083/jcb.150.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Field MC, Gabernet-Castello C, Dacks JB. Reconstructing the evolution of the endocytic system: insights from genomics and molecular cell biology. Adv Exp Med Biol. 2007;607:84–96. doi: 10.1007/978-0-387-74021-8_7. [DOI] [PubMed] [Google Scholar]
- 9.Elde NC, Morgan G, Winey M, Sperling L, et al. Elucidation of clathrin-mediated endocytosis in Tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLoS Genet. 2005;1:e52. doi: 10.1371/journal.pgen.0010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osteryoung KW, Nunnari J. The division of endosymbiotic organelles. Science. 2003;302:1698–704. doi: 10.1126/science.1082192. [DOI] [PubMed] [Google Scholar]
- 11.Miyagishima SY, Kuwayama H, Urushihara H, Nakanishi H. Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc Natl Acad Sci USA. 2008;105:15202–7. doi: 10.1073/pnas.0802412105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Bliek AM. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- 13.Smaczynska-de R, II, Allwood EG, Mishra R, Booth WI, et al. Yeast dynamin vps1 and amphiphysin rvs167 function together during endocytosis. Traffic. 2012;13:317–28. doi: 10.1111/j.1600-0854.2011.01311.x. [DOI] [PubMed] [Google Scholar]
- 14.Lee A, Frank DW, Marks MS, Lemmon MA. Dominant-negative inhibition of receptor- mediated endocytosis by a dynamin-1 mutant with a defective pleckstrin homology domain. Curr Biol. 1999;9:261–4. doi: 10.1016/s0960-9822(99)80115-8. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto PM, Herskovits JS, Vallee RB. Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J Biol Chem. 1997;272:11629–35. doi: 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- 16.Muhlberg AB, Warnock DE, Schmid SL. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 1997;16:6676–83. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenniston JA, Lemmon MA. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29:3054–67. doi: 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barylko B, Wang L, Binns DD, Ross JA, et al. The proline/arginine-rich domain is a major determinant of dynamin self-activation. Biochemistry. 2010;49:10592–4. doi: 10.1021/bi101343p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen MS, Burgess CC, Vallee RB, Wadsworth SC. Developmental stage-and tissue-specific expression of shibire, a Drosophila gene involved in endocytosis. J Cell Sci. 1992;103:619–28. doi: 10.1242/jcs.103.3.619. [DOI] [PubMed] [Google Scholar]
- 20.Labrousse AM, Shurland DL, van der Bliek AM. Contribution of the GTPase domain to the subcellular localization of dynamin in the nematode Caenorhabditis elegans. Mol Biol Cell. 1998;9:3227–39. doi: 10.1091/mbc.9.11.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson SM, Brasnjo G, Hayashi M, Wolfel M, et al. A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science. 2007;316:570–4. doi: 10.1126/science.1140621. [DOI] [PubMed] [Google Scholar]
- 22.Liu YW, Surka MC, Schroeter T, Lukiyanchuk V, et al. Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knockout cells. Mol Biol Cell. 2008;19:5347–59. doi: 10.1091/mbc.E08-08-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu YW, Neumann S, Ramachandran R, Ferguson SM, et al. Differential curvature sensing and generating activities of dynamin isoforms provide opportunities for tissue-specific regulation. Proc Natl Acad Sci USA. 2011;108:E234–42. doi: 10.1073/pnas.1102710108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, et al. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25:664–72. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- 25.Lemmon MA. Pleckstrin homology domains: not just for phosphoinositides. Biochem Soc Trans. 2004;32:707–11. doi: 10.1042/BST0320707. [DOI] [PubMed] [Google Scholar]
- 26.Pucadyil TJ, Schmid SL. Conserved functions of membrane active GTPases in coated vesicle formation. Science. 2009;325:1217–20. doi: 10.1126/science.1171004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–75. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dereeper A, Guignon V, Blanc G, Audic S, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]