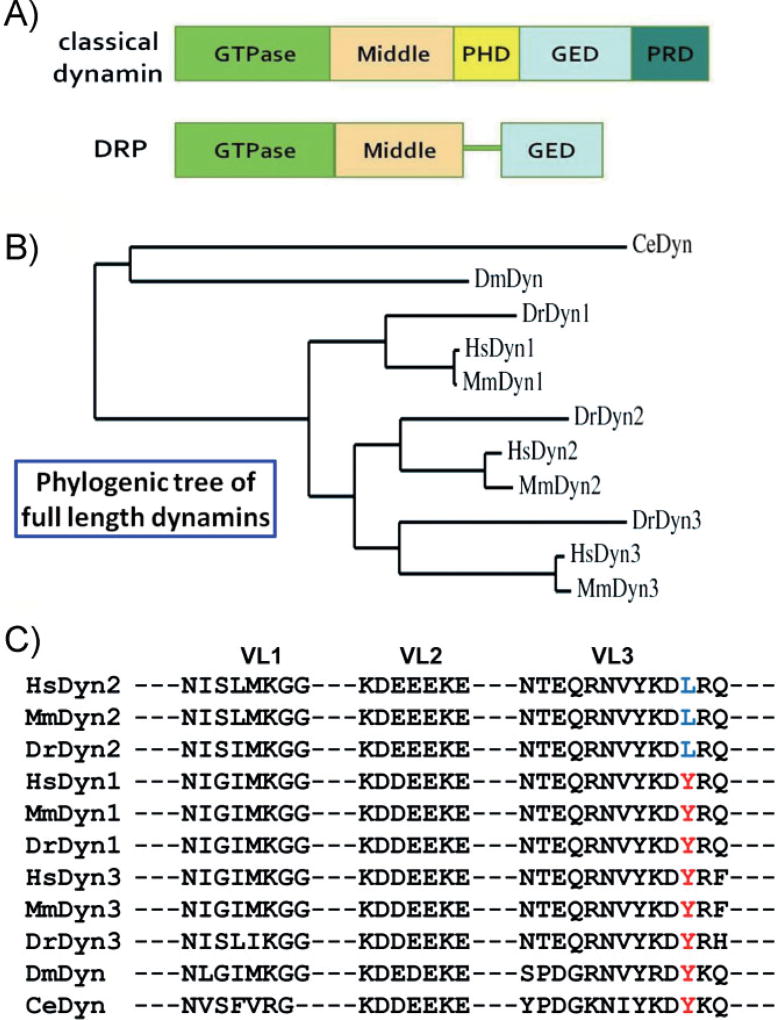
Figure 1. The domain structure and comparison of dynamins.
A: A comparison of the domain structures of classical dynamins and DRPs. The GTPase, middle, and GED are conserved across all dynamin superfamily members. The PHD and PRD, involved in targeting and regulating dynamin activity, are unique to classical dynamins. B: Phylogenic analysis of full-length dynamins from human (Hs), mouse (Mm), zebra fish (Dr), fly (Dm), and nematode (Ce). The rooted phylogenetic tree was generated with Phylogeny.fr [28]. C: Amino acid alignment of dynamin PHDs. Membrane-interacting VLs (VL1–3) of dynamin PHD from different organisms are shown. A residue in VL3, Y600 in Dyn 1 and 3 and L600 in Dyn2 (highlighted in red and blue, respectively), confers curvature sensitivity [23] and is diagnostic of vertebrate dynamin isoforms. PHDs were aligned with CLUSTAL 2.0.1; and the compared organisms include Homo sapiens (Hs), Mus musculus (Mm), Danio rerio (Dr), Drosophila melanogaster (Dm), and Caenorhabditis elegans (Ce).