Abstract
Neuropathic pain is an important medical problem with few effective treatments. The sigma 1 receptor (σ1R) is known to be a potential target for neuropathic pain therapeutics, and antagonists for this receptor are effective in preclinical models and are currently in phase II clinical trials. Conversely, relatively little is known about σ2R, which has recently been identified as transmembrane protein 97 (Tmem97). We generated a series of σ1R and σ2R/Tmem97 agonists and antagonists and tested them for efficacy in the mouse spared nerve injury (SNI) model. In agreement with previous reports, we find that σ1R ligands given intrathecally (IT) produce relief of SNI-induced mechanical hypersensitivity. We also find that the putative σ2R/Tmem97 agonists DKR-1005, DKR-1051, and UKH-1114 (Ki ~ 46 nM) lead to relief of SNI-induced mechanical hypersensitivity, peaking at 48 h after dosing when given IT. This effect is blocked by the putative σ2R/Tmem97 antagonist SAS-0132. Systemic administration of UKH-1114 (10 mg/kg) relieves SNI-induced mechanical hypersensitivity for 48 h with a peak magnitude of effect equivalent to 100 mg/kg gabapentin and without producing any motor impairment. Finally, we find that the TMEM97 gene is expressed in mouse and human dorsal root ganglion (DRG) including populations of neurons that are involved in pain; however, the gene is also likely expressed in non-neuronal cells that may contribute to the observed behavioral effects. Our results show robust antineuropathic pain effects of σ1R and σ2R/Tmem97 ligands, demonstrate that σ2R/Tmem97 is a novel neuropathic pain target, and identify UKH-1114 as a lead molecule for further development.
Keywords: Neuropathic pain, sigma 1 receptor, sigma 2 receptor, Tmem97, dorsal root ganglion, drug discovery
Graphical Abstract
INTRODUCTION
Neuropathic pain is a common medical problem that is very poorly treated by current therapeutics. Recent meta-analyses show that the most widely prescribed drugs for neuropathic pain only achieve 50% pain relief in 1 of 4 patients for the antidepressants (e.g., monoamine oxidase inhibitors) and are effective in as few as 1 in 7 patients for the gabapentinoids.1 Better therapeutics are clearly needed for neuropathic pain. Sigma receptors (σRs) are unique transmembrane proteins expressed throughout the CNS (central nervous system) and in certain peripheral tissues. Consisting of two subtypes, the sigma 1 receptor (σ1R) and the sigma 2 receptor (σ2R), these proteins are involved in intracellular ion regulation and neuron survival.2 σ1R has been cloned and a crystal structure of the receptor obtained.3 The involvement of this receptor in pain signaling has been studied extensively, and a variety of compounds that bind to σ1R demonstrate antinociceptive effects.4 E-52862,5 a σ1R antagonist, is currently in phase II clinical trials for the treatment of neuropathic pain as both a monotherapy and in a multidrug cocktail.
Although strong evidence supports a role for σ1R in neuropathic pain, σ2R has not been explored as a pain target, and the molecular identity of this receptor has been challenging to pinpoint. Molecular cloning recently confirmed the identify of σ2R as transmembrane protein 97 (Tmem97),6 and this receptor will accordingly be referred to as σ2R/Tmem97 herein unless we are referring to the Tmem97 mouse or TMEM97 human genes. σ2R/Tmem97 is a gene product that appears to be involved in cholesterol trafficking and homeostasis and interacts with NPC-1, a protein responsible for shuttling lipids to postlysosomal locations.7–9 σ2R/Tmem97 is involved in regulating intracellular Ca2+ concentration, and certain σ2R/Tmem97 ligands can induce a transient rise in intracellular Ca2+ levels,2 while other compounds that bind to σ2R/Tmem97 suppress Ca2+ influx in the presence of an inducer.10 Some evidence suggests that compounds that bind to σ2R/Tmem97 modulate a signaling pathway involving progesterone receptor membrane component one (PGRMC111),10 a heme binding protein12 involved in cell survival2 and apoptosis.13 With respect to medical relevance, it is noteworthy that TMEM97 has been implicated in the metabolic disorder, Niemann-Pick disease,9 as well as in multiple neurodegenerative and neurological conditions.14 Indeed, small molecules that bind to σ2R/Tmem97 are emerging as potential therapeutics for a range of CNS disorders, including Alzheimer’s disease (AD)15 and schizophrenia.16
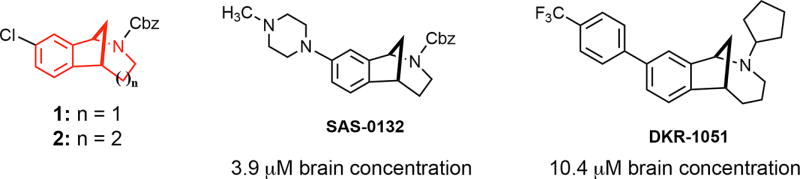
We have discovered that the chlorinated norbenzomorphan and methanobenzazocine scaffolds 1 and 2 (Figure 1) are excellent templates for preparing both selective and mixed affinity σ1R and σ2R/Tmem97 ligands.17 In the context of CNS lead development, these scaffolds are of particular interest because of their drug-like properties18,19 and the ease with which they cross the blood brain barrier, as demonstrated by representative compounds SAS-013210 and DKR-1051 (Figure 1). In vivo testing has revealed that some compounds derived from 1 and 2 have promising medicinal properties that may warrant further preclinical development. For example, certain σ2R/Tmem97 ligands are neuroprotective10 and also restore cognitive function in transgenic AD mice10,20 as well as in aged, nondiseased animals,10 the latter of which suggests σ2R/Tmem97 may be a potential target for treating mild cognitive impairment (MCI). Despite this increasing knowledge base and the emergence of tool ligands for probing σ2R/Tmem97, the involvement of σ2R/Tmem97 in neuropathic pain has not yet been reported.
Figure 1.
Norbenzomorphan and methanobenzazocine scaffolds 1 and 2 and the CNS penetrant σ2R/Tmem97 ligands SAS-0132 and DKR-1051.
Herein we describe the antineuropathic pain properties of a series of σ1R and σ2R/Tmem97 ligands in mice. We report that molecules that bind to σ2R/Tmem97 as putative agonists exert a profound effect on mechanical hypersensitivity in the spared nerve injury (SNI) model with a duration of action and potency that is superior to that of gabapentin. Small molecule modulation of σ2R/Tmem97 may thus represent a new approach for managing pain by a previously unexplored mechanism of action.
RESULTS
Screening of σ1R and σ2R/Tmem97 Ligands for Activity in the Mouse SNI Model
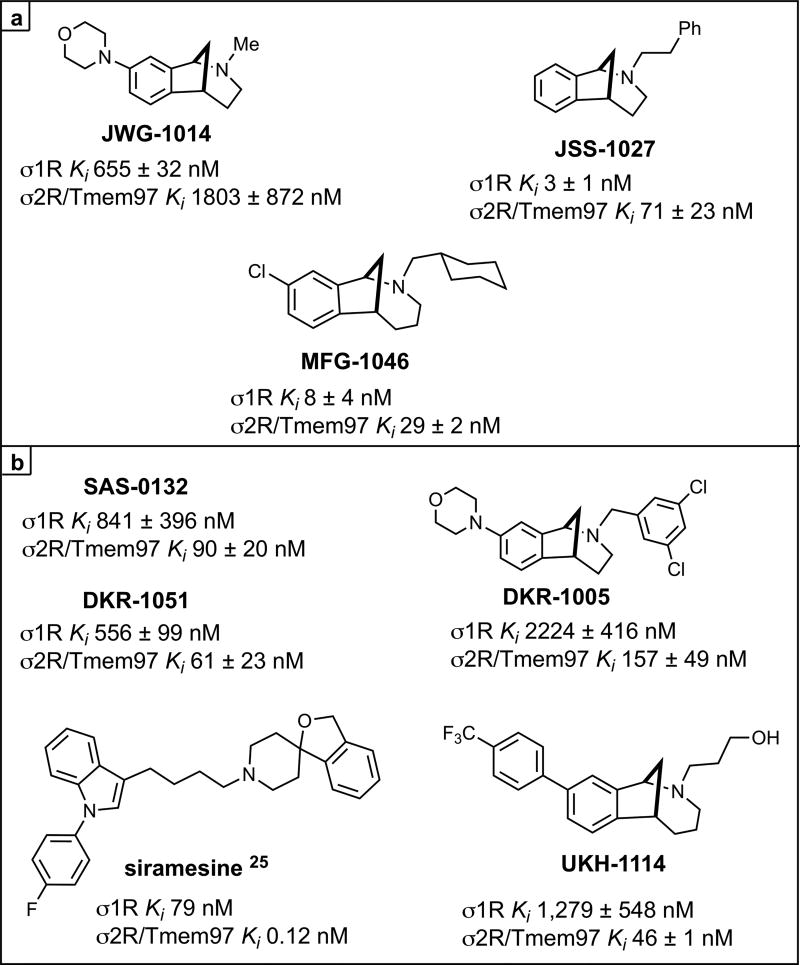
Several previous studies have demonstrated that σ1R antagonists show efficacy in rodent neuropathic pain models via a spinal mechanism of action,21–23 but the possible effect of σ2R/Tmem97 ligands in this model has not been explored. We screened a series of σ1R and σ2R/Tmem97 ligands (Figure 2) for activity in the SNI model via a single IT injection. We chose this route of administration because σ1R antagonists have been shown to be active in rodent pain models with IT injection,21,24 and because we had incomplete knowledge of compound disposition in vivo with systemic dosing.
Figure 2.
σ Receptor binding ligands.aKi are shown for each ligand at σ1R or σ2R/Tmem97 as the mean of at least two independent experiments ± standard deviation. Siramesine data is from ref 25. aFor all compounds, σ2R/Tmem97 was sourced from rat PC12 cells, with the exception of siramesine, which utilized rat brain homogentate. See Methods Section for more details.
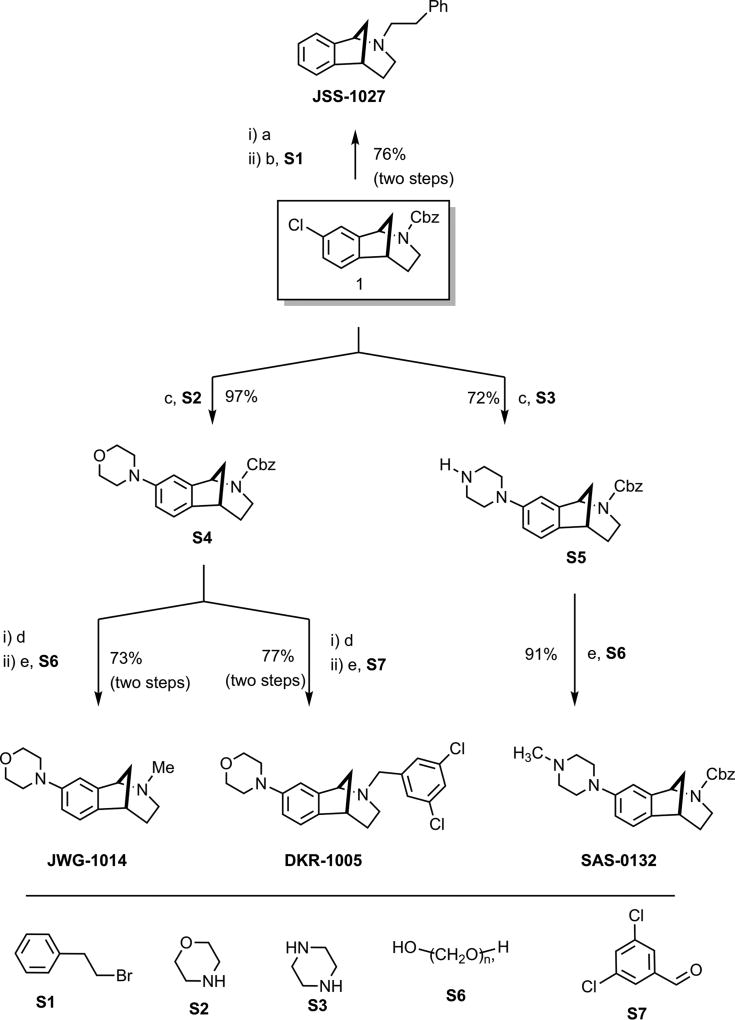
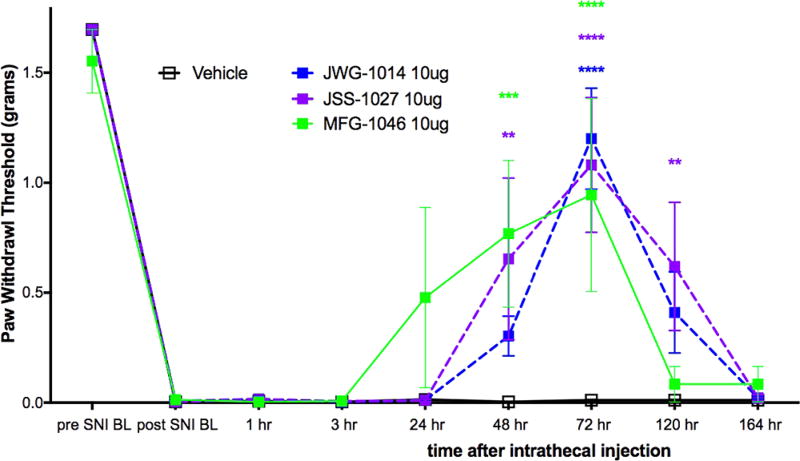
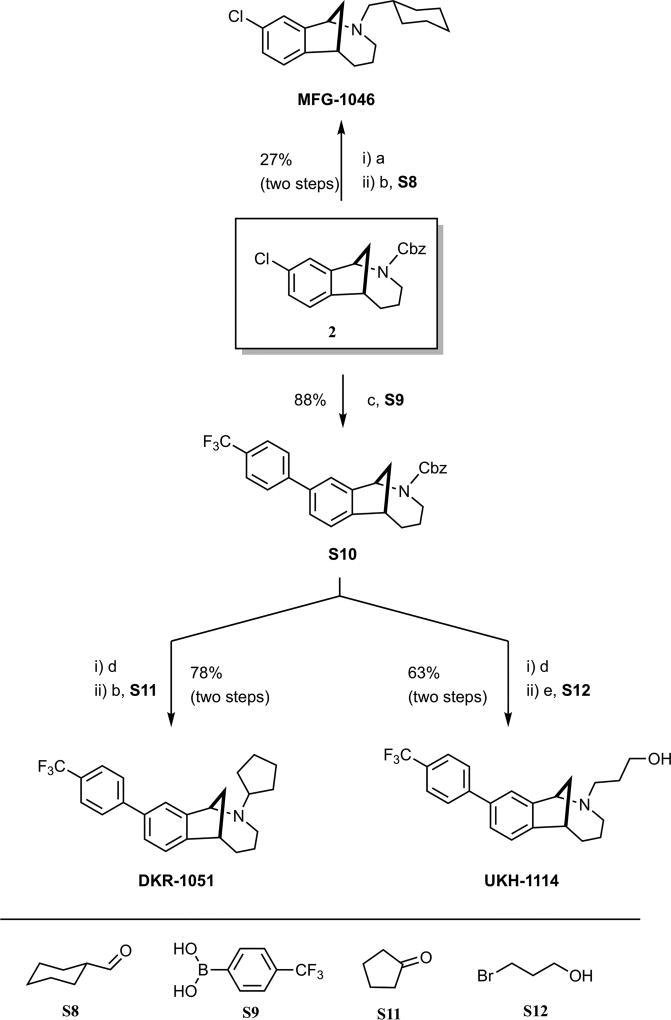
We first assessed the σ1R preferring ligands, JWG-1014, JSS-1027, and MFG-1046, all given at 10 μg doses, and observed antipain effects for all three compounds that were significantly different from vehicle. JWG-1014 and JSS-1027 (Figure 8) both demonstrated efficacy 48 h post IT injection, and this effect persisted through 120 h after ligand administration (Figure 3). Faster onset of action was observed with MFG-1046 (Figure 9), which elicited an antipain effect at 24 h that continued through 72 h after IT injection (Figure 3).
Figure 8.
Synthesis of norbenzomorphan σ2R/Tmem97 ligands.
Reagents and conditions (a) 10% Pd/C, H2, EtOH. (b) K2CO3, CH3CN, 50 °C. (c) Pd(OAc)2, JohnPhos®, NaOt-Bu, toluene, 100 °C. (d) TMSI, CH2Cl2, then HCl. (e) Na(OAc)3BH, CH3COOH, 1,2-dichloroethane.
Figure 3.
Effect of IT injection of σ1R ligands in the mouse SNI model. Compounds acting on σ1R were tested in the mouse SNI model. Mechanical sensitivity was measured at the indicated time points after IT injection of 10 μg compound. All vehicle groups, n = 6; JWG-1014, n = 5; JSS-1027, n = 5; MFG-1046, n = 4. **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Figure 9.
Synthesis of methanobenzazocine σ2R/Tmem97 ligands.
Reagents and conditions (a) TMSI, CH2Cl2, then HCl. (b) Na(OAc)3BH, 1,2-dichloroethane. (c) Pd[P(tBu)3]2, Cs2CO3, 1,4-dioxane, 98 °C. (d) 10% Pd/C, H2, EtOH. (e) K2CO3, CH3CN, 55 °C.
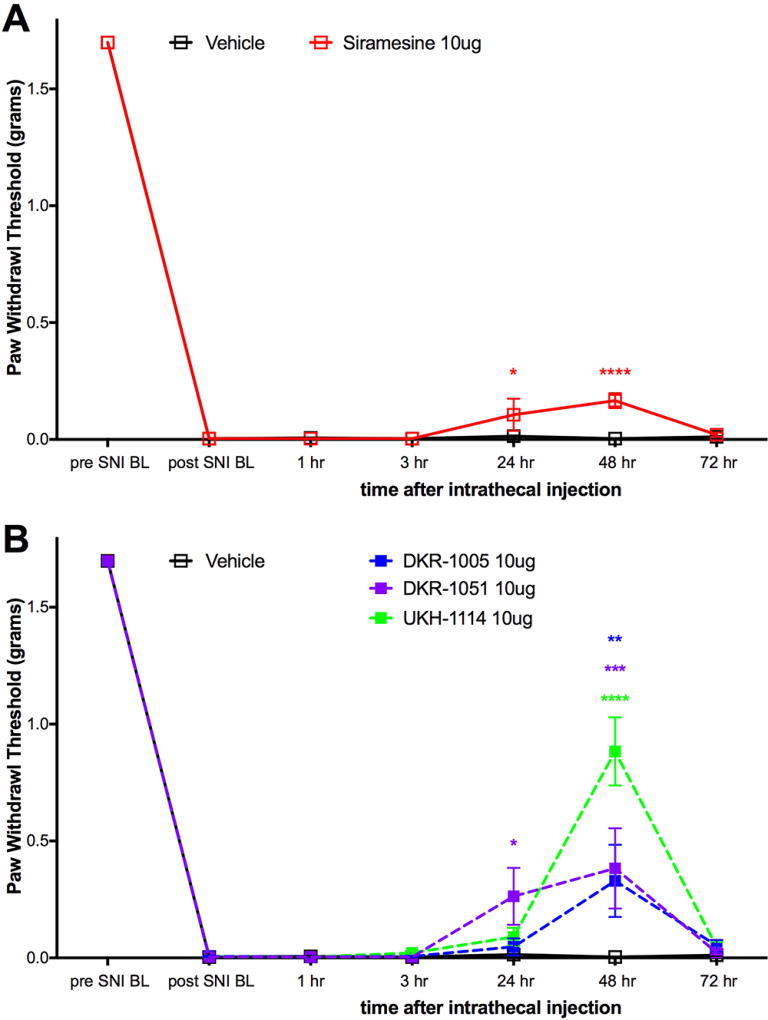
We then tested four σ2R/Tmem97 preferring ligands of the norbenzomorphan and methanobenzazocine structural class, as well as the known σ2R/Tmem97 agonist, siramesine.25–27 Siramesine induced a small inhibitory effect on mechanical hypersensitivity that was significant at 24 and 48 h (Figure 4A). On the other hand, DKR-1005, DKR-1051, and UKH-1114 all produced significant antimechanical hypersensitivity effects at either 24 or 48 h after IT injection (Figure 4B).
Figure 4.
Effect of IT injection of σ2R/Tmem97 ligands in the SNI model. Mechanical sensitivity was measured at the indicated time points after IT injection of 10 μg compound. (A) Effect of siramesine (n = 4) compared to vehicle (n = 6) is shown. (B) Effect of DKR-1005 (n = 6), DKR-1051 (n = 5) and UKH-1114 (n = 11) compared to vehicle (n = 6) is shown. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
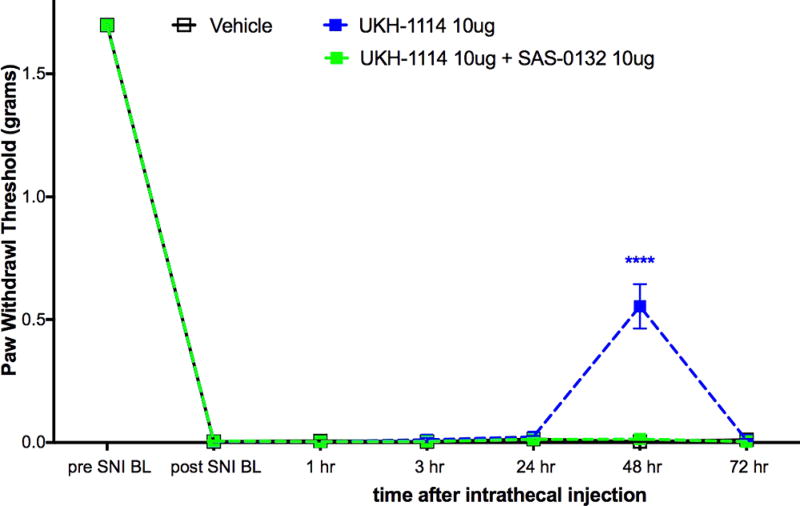
Based on the observed in vivo effects of the four σ2R/Tmem97 preferring ligands examined, tentative functional activity assignments were made. Compounds DKR-1005, DKR-1051, and UKH-1114 elicited pronounced and sustained antinociceptive effects. However, when UKH-1114 was administered with SAS-0132, the antinociceptive action of UKH-1114 was abolished (Figure 5). Because UKH-1114 elicits a significant antipain effect that is completely blocked by SAS-0132, we surmise that UKH-1114 is a σ2R/Tmem97 agonist (or partial agonist) and that σ2R/Tmem97 agonism is responsible for the antimechanical hypersensitivity effects observed with these compounds in SNI mice. On the other hand, SAS-0132 appears to function as a σ2R/Tmem97 antagonist by suppressing the action of UKH-1114. Indeed, these findings are consistent with our previous work that showed that SAS-0132 behaves as a σ2R/Tmem97 antagonist, whereas DKR-1051 acts as a σ2R agonist10 based upon their opposing effects upon Ca2+ in SK-N-SH neuroblastoma cells. Namely, treatment of SK-N-SH neuroblastoma cells with DKR-1051 induces a rapid Ca2+ transient, and this effect is attenuated when cells are pretreated with SAS-0132.
Figure 5.
Effect of UKH-1114 is blocked by the σ2R/Tmem97 antagonist SAS-0132. σ2R/Tmem antagonist SAS-0132 at 10 μg dose given at the same time as UKH-1114, also at 10 μg dose, completely blocked the effect seen with UKH-1114 given alone. n = 6 per group. ****p < 0.0001.
Systemic Administration of σ2R/Tmem97 Agonist Alleviates Neuropathic Pain
Of the putative σ2R/Tmem97 agonists we tested, UKH-1114 had the largest behavioral effect, and its chemical properties and high selectivity for σ2R/Tmem97 versus >50 other proteins (See Tables 1 and 2) make it an eligible candidate for systemic dosing.
Table 1.
Chemical Properties of UKH-1114a
| molecular weight | 375.4 |
| ClogD (7.4) | 4.3 |
| total polar surface area | 23.5 |
| hydrogen bond donors | 1 |
| hydrogen bond acceptors | 2 |
| rotatable bonds | 5 |
Calculated with ACD/I-Laboratories (https://ilab.acdlabs.com/iLab2/).
Table 2.
UKH-1114 Binding Profile at Non-Sigma Receptor Sites
| target | Ki (nM) | target | Ki (nM) |
|---|---|---|---|
| 5HT1A | a | Beta2 | a |
| 5HT1B | a | Beta3 | a |
| 5HT1D | a | BZP rat brain | a |
| 5HT1e | a | calcium channel | >10 000 |
| 5HT2A | a | D1 | a |
| 5HT2B | a | D2 | a |
| 5HT2C | a | D3 | a |
| 5HT3 | a | D4 | a |
| 5HT5a | a | D5 | a |
| 5HT6 | a | DOR | a |
| 5HT7 | a | GabaA | a |
| A2B2 | a | H1 | a |
| A2B4 | a | H3 | a |
| A3B2 | a | KOR | 1383 |
| A3B4 | a | M1 | a |
| A4B2 | a | M2 | a |
| A4B2b | a | M3 | a |
| A4B4 | a | M4 | a |
| A7 | a | M5 | a |
| A7b | a | MOR | a |
| Alpha1a | a | NET | 1046 |
| Alpha1b | a | NMDA | 6724 |
| Alpha1d | a | PBR | a |
| Alpha2a | a | SERT | a |
| Alpha2b | a | V1A | >10 000 |
| Alpha1c | a | V1B | >10 000 |
| AMPA | >10 000 | V2 | >10 000 |
| Beta1 | a |
<50% inhibition of radioligand binding at 10 μM.
Sourced from rodent brain.
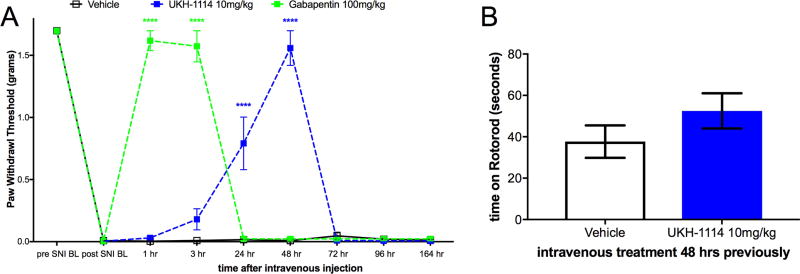
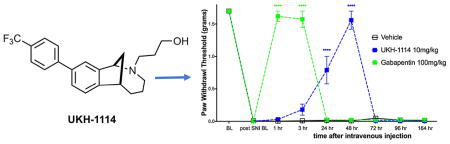
Therefore, we injected UKH-1114 IV in mice at 10 mg/kg and compared the effect of the compound to the gold-standard antineuropathic pain treatment, gabapentin (100 mg/kg). Gabapentin completely reversed mechanical hypersensitivity at 1 and 3 h after injection, but animals were fully mechanically hypersensitive again 24 h after IV injection. UKH-1114 also produced a complete reversal of mechanical hypersensitivity but with a different time course (Figure 6A). A significant effect was observed at both 24 and 48 h after injection indicating that pain relief from this mechanism lasts longer than that produced with a 10-fold larger dose of gabapentin.
Figure 6.
Systemic dosing with UKH-1114 leads to alleviation of neuropathic pain without motor effects. (A) Vehicle (n = 6), gabapentin (100 mg/kg, n = 6), or UKH-1114 (10 mg/kg, n = 6) were given IV and mechanical testing was done at the indicated time points. (B) Rotorod testing was done 48 h after IV injection on an accelerating rotorod reaching a maximum of 40 rotations per minute over 200 s. Latency to fall is shown on the third trial (n = 6 per group). ****p < 0.0001.
Because the mechanism of action of σ2R/Tmem97 ligands for neuropathic pain relief is not known, we were concerned that this effect could be produced by motor impairment. To test this possibility we used the rotorod test. SNI mice were given IV vehicle or UKH-1114 and tested at the peak time point for alleviation of neuropathic pain, 48 h after injection. There was no effect of UKH-1114 on motor performance (Figure 6B) ruling out the possibility of motor impairment.
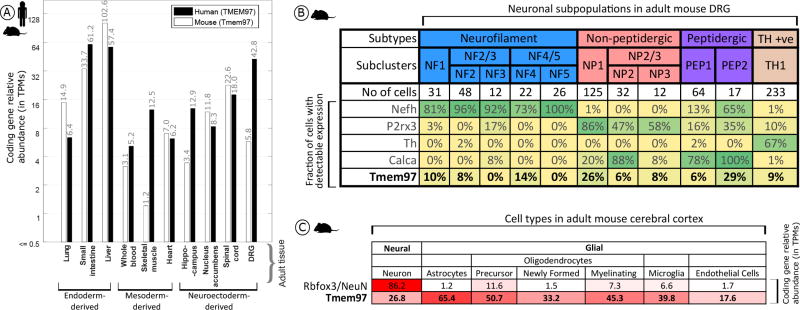
Tmem97 Gene Expression Analysis in Mouse and Human Tissues
To gain insight into where σ2R/Tmem97 is expressed, we quantified Tmem97 mRNA relative abundance in mouse DRG as well as additional tissues, and their orthologous tissues in humans, based on publicly available sequencing data (human data sets: GTex project,28 ENCODE project,29 Uhlén et al.,30 Duff et al.,31 and the UTD DRG project;32 mouse data sets: mouse ENCODE project,33 Rakic et al.,34 Gerhold et al.,35 Eipper-Mains et al.,36 and Huan et al.37). We find that Tmem97 gene expression and its human orthologue (TMEM97) are ubiquitously expressed in a wide range of tissues, with higher expression in the human and mouse gastrointestinal (GI) tract commensurate with its role in cholesterol trafficking7 (Figure 7A). While expression levels in the human DRG is high, both single cell38 and bulk35 RNA-seq for mouse DRGs identify gene expression, but at lower levels than in human (Figure 7A). TMEM97/Tmem97 is also relatively highly expressed in human and mouse spinal cord. While the detection rate for Tmem97 across mouse DRG neuronal subpopulations is relatively low, it is clearly expressed in subpopulations of peptidergic and nonpeptidergic nociceptors (Figure 7B). Given the high TPM levels in mouse and human DRG, Tmem97 may also be expressed in non-neuronal cells in DRG (e.g., satellite glial cells or Schwann cells). Unfortunately, mouse DRG glial transcriptomes have not been characterized, so we turned to a CNS tissue where these cell populations have available transcriptomes. We find that in adult cerebral cortex,39 Tmem97 expression in cortical glial cells can be enriched 2-fold or more over neuronal expression levels (Figure 7C), lending credence to the hypothesis of glial expression of Tmem97 in the DRG and/or spinal cord.
Figure 7.
Expression analysis for Tmem97 (A), TMEM97 (human), and Tmem97 (mouse) gene expression across orthologous tissues, with greater expression in the mouse and human GI tract and the human DRG. (B) Analysis of mouse single cell data reveals a maximum detection rate of 29% for Tmem97 across all sensory neuron subpopulations as contrasted with 67% or more for known subpopulation marker genes. (C) Cortical expression of Tmem97 as contrasted with the neuronal marker NeuN. Tmem97 expression spans both neuronal and non-neuronal cells, with ~1.5–2.5-fold higher expression in non-neuronal cells.
DISCUSSION
Several primary conclusions may be reached based upon the work described herein. First, our results using distinct σ1R binding ligands are consistent with previous demonstrations that σ1R antagonists reduce nerve injury-induced mechanical hypersensitivity.21,40 This observation suggests that the σ1R binding ligands described herein might be antagonists. Second, we find that σ2R/Tmem97 ligands DKR-1005, DKR-1051, and UKH-1114 bind σ2R/Tmem97 with high affinity and produce antinociceptive effects when administered IT to SNI mice. UKH-1114 is the most efficacious of these compounds in vivo, producing a strong antinociceptive effect when administered IV that was longer lasting and equally efficacious at 1/10 the dose of gabapentin. UKH-1114 is highly selective for σ2R/Tmem97 binding with an affinity 28-fold greater at σ2R/Tmem97 than at σ1R, and with negligible affinity for 55 other targets (Table 2). While TMEM97/Tmem97 is expressed in DRG and spinal cord of humans and mice, the gene is likely expressed in a mix of neuronal and non-neuronal cells that may include key glial and/or immune cells that are thought to play an important role in the pathogenesis of neuropathic pain.41 Therefore, we conclude the σ2R/Tmem97 is a promising target for the generation of neuropathic pain drugs.
An interesting aspect of our behavioral findings is the relatively long onset of action of the compounds that we tested compared to rapid onset actions in previous studies with distinct ligands.21,40,42,43 There could be several reasons for this observation. One is that most previous studies have looked at short time points after administration of σ1R antagonists (30 or 45 min) or at repeated dosing effects21,40,42,43 so it is possible that effects with a slower onset are missed by these acute dosing schedules but are part of the sustained efficacy observed with repeated dosing. Another possibility is that while the mechanism of action of σ1R antagonists in pain was first thought to involve a spinal mechanism of action,21–23 more recent studies suggest an action of DRG neurons where σ1R is also expressed.44 It is therefore probable that the time to onset of effects is due to the time that is needed for the drug to diffuse to sites of action that are more distant from the injection site in the intrathecal space. Because σ1Rs are found on the nuclear envelop of DRG neurons,44 yet another possibility is that the effect of these compounds is transcriptional. If this were the case, this would also potentially take longer to manifest as a behavioral change. These considerations aside, our results are consistent with previous studies where σ1R antagonists produced long lasting relief of neuropathic mechanical hypersensitivity when given IT.21 Another important consideration here is that all of the compounds we examined as σ1R binding ligands also have affinity at σ2R/Tmem97 that could alter their behavioral effects, and some of them have affinity in the low nM range (JSS-1027 and MFG-1046).
To our knowledge, σ2R/Tmem97 ligands have not been previously tested for antinociceptive effects. The σ2R/Tmem97 ligands described herein have high affinity for σ2R/Tmem97, have good specificity relative to σ1R binding, and have little affinity for other targets in a small specificity screen. Indeed, UKH-1114 displays exceptional selectivity for σ2R/Tmem97 over >50 other receptors and channels. Although the previously described σ2R/Tmem97 agonist, siramesine, produced only a small effect in SNI mice, several other putative σ2R/Tmem97 agonists produced strong effects when given IT in the SNI model. This effect was blocked in the case of UKH-1114 by the known σ2R/Tmem97 antagonist, SAS-0132, strongly implicating σ2R/Tmem97 agonism as the mode of action responsible for antinociception. UKH-1114 was chosen to test via systemic administration in the SNI model. This compound produced a strong antimechanical hypersensitivity effect that was long lasting and devoid of motor impairment. The peak magnitude of effect was equivalent to the standard of care neuropathic pain drug, gabapentin, but it was much longer lasting at 10-fold lower dose. We observed a relatively long onset to antinociceptive action with putative σ2R/Tmem97 agonists, like σ1R ligands, although this onset to action was shorter when UKH-1114 was given IV. This may be explained by the expression of the Tmem97 gene, which is clearly expressed in structures outside of the intrathecal space and has high expression in the DRG and in non-neuronal cells in the DRG and CNS, suggesting the possibility of immune cell expression. We mined publicly available data sets and found that Tmem97 mRNA is expressed in many mouse and human tissues, but in the mouse CNS, expression levels are apparently higher in many non-neuronal cells types, including astrocytes and microglia. This is consistent with the known expression of TMEM97 in human glioma cells.45 While beyond the scope of the present experiments, the discovery of tool ligands to manipulate σ2R/Tmem97 function will allow for testing the role of this receptor in a variety of cell types in the pain pathway.
More work is clearly needed to understand the role of σ2R/Tmem97 in pain, but these unprecedented results provide compelling support for developing σ2R/Tmem97 agonists as a novel strategy for the pharmacological management of neuropathic pain, bringing forth the possibility for patients to experience enhanced pain relief and reduced side-effects by modulating a receptor not targeted by currently approved FDA drugs. We are in the process of identifying other σ2R/Tmem97 agonists to further explore their utility as potential therapeutics for neuropathic pain.
METHODS
Laboratory Animals
All animal procedures were approved by The University of Texas at Dallas Institutional animal care and use committee (IACUC) and were in accordance with National Institutes of Health Guidelines. All of the experiments were performed on male C57Bl6/J mice obtained from Envigo at 4 weeks of age. Mice were housed in the University of Texas at Dallas Animal Care Facility for at least 1 week prior to the start of behavior testing and surgery. Animals had ad libitum access to food and water and were on a 12 h noninverted light/dark cycle. Experimenters were blinded to treatment groups in behavioral experiments. Sample size was estimated by performing a power calculation using G*Power (version 3.1.9.2). With 80% power, an expectation of d = 2.2 effect size in behavioral experiments, and alpha set to 0.05, the sample size required was calculated as n = 5 per group. Standard deviation (set at 0.3) for the power calculation was based on previously published mechanical threshold data.46–48
Behavioral Testing
Mechanical sensitivity was assessed using stimulation of the hindpaw of the mouse with calibrated von Frey filaments from Stoelting. We used the modified up–down method described previously.49 Following baseline testing, neuropathic pain was induced in mice using the SNI surgery model. This surgery consists of exposing and cutting the peroneal and tibial branches of the sciatic nerve leaving the sural nerve intact.50 Two-weeks postsurgery, mechanical sensitivity testing was repeated to ensure that mechanical hypersensitivity had indeed been produced. Following this test, groups of SNI mice were treated with intrathecal (IT) injections51 of test compounds given in a volume of 5 μL made up in sterile saline and mechanical sensitivity was assessed at time points indicated in figures. Screening for the effect of IT injection of test compounds on SNI-induced mechanical hypersensitivity was done between 21 and 60 days after SNI. Some groups of mice were tested with multiple compounds to reduce the number of animals that were needed for this study. If mice were given multiple injections they were always spaced by at least 14 days between injections. Some animals received a maximum of 3 IT injections but most received 2. In some experiments drugs were given by intravenous (IV) injection through the tail vein in a volume of 100 μL in sterile saline and mechanical sensitivity was assessed at time points indicated in figures. RotoRod testing was done on an automated rotorod device set to accelerate to a final speed of 40 rotations per minute over a 200 s time course. Mice were trained on the rotorod twice and latency to fall was recorded on the third trial.
Chemical Synthesis and Characterization
Toluene was dried by filtration through one column of activated, neutral alumina followed by one column of Q5 reactant52 and was determined to have less than 50 ppm of H2O by Karl Fischer coulometric moisture analysis. Acetonitrile (CH3CN), diethyl ether (Et2O), ethyl acetate (EtOAc), methanol (MeOH), methylene chloride (CH2Cl2), 1,2-dichloroethane (DCE), triethylamine (Et3N), and diisopropylethylamine (i-Pr2NEt) were used without further purification. All reagents were reagent grade and used without purification unless otherwise noted. All reactions involving air or moisture sensitive reagents or intermediates were performed under an inert atmosphere of nitrogen or argon in glassware that was flame or oven-dried. Reaction temperatures refer to the temperature of the cooling/heating bath. Volatile solvents were removed under reduced pressure using a Buchi rotary evaporator at 20–30 °C (bath temperature). Thin layer chromatography was run on precoated plates of silica gel with a 0.25 mm thickness containing 60F-254 indicator (EMD Millipore). Chromatography was performed using forced flow (flash chromatography) and the indicated solvent system on 230–400 mesh silica gel (Silicycle flash F60) according to the method of Still and colleagues53 unless otherwise noted. Radial Preparative Liquid Chromatography (radial plc) was performed on a Chromatotron using glass plates coated with Merck, TLC grade 7749 silica gel with gypsum binder and fluorescent indicator.
Proton nuclear magnetic resonance (1H NMR) and carbon nuclear magnetic resonance (13C NMR) spectra were obtained at the indicated field as solutions in CDCl3 unless otherwise indicated. Chemical shifts are referenced to the deuterated solvent (e.g., for CDCl3, δ = 7.26 ppm and 77.0 ppm for 1H and 13C NMR, respectively) and are reported in parts per million (ppm, δ) relative to tetramethylsilane (TMS, δ = 0.00 ppm). Coupling constants (J) are reported in Hz and the splitting abbreviations used are s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet; comp, overlapping multiplets of magnetically nonequivalent protons; br, broad; app, apparent. Molecular mass was determined using an LCMS system comprised of an Agilent 1200 Series HPLC and an Agilent 6130 single quadrupole mass spectrometer. Samples were injected onto a Phenomenex Gemini C18 column (5 μm, 2.1 × 50 mm) and eluted at 0.7 mL/min using a gradient of 10–90% acetonitrile, 0.1% formic acid (11 min linear ramp). Positive mode electrospray ionization ESI was used to verify the identity of the major component. All compounds submitted for in vivo testing were >95% purity as determined by LC via AUC at 214- and 254 nm. Experimental and characterization data for 1,54 S4, S5, SAS-0132,55 and DKR-1005 have been reported previously, and the preparation of the nor-chloro analogue of 2 has been described.56
(±)-2-Phenethyl-2,3,4,5-tetrahydro-1H-1,5-methanobenzo[c]-azepine (JSS-1027)
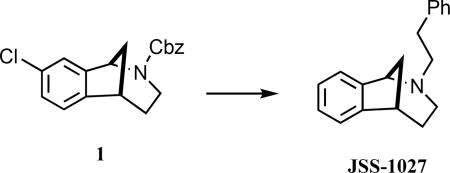
A mixture of 1 (150 mg, 0.45 mmol) and 10% Pd/C (45 wt %, 60 mg) in ethanol (5 mL) was stirred under an atmosphere of H2 for 6.5 h. The catalyst was removed by filtration through a cotton plug, and the combined filtrates were concentrated under reduced pressure to provide 71 mg of a light brown oil, which was dissolved in CH3CN (5 mL). (2-Phenyl)ethyl bromide (S1, 0.13 mL, 0.90 mmol) and K2CO3 (248 mg, 1.8 mmol) were added, and the mixture was heated at 50 °C for 20 h. The reaction was cooled to room temperature and the solids removed by filtration. The combined filtrates were concentrated to give a yellow oil that was purified using radial plc, eluting with 100% hexanes → 20% EtOAc/hexanes, to give 90 mg (76%) of JSS-1027 as a light-yellow oil: 1H NMR (500 MHz, CDCl3) δ 7.36–7.30 (m, 3 H), 7.29–7.16 (comp, 6 H), 4.08 (d, J = 5.0 Hz, 1 H), 3.19 (m, 1 H), 2.94 (td, J = 12.5, 5.9 Hz, 1 H), 2.84 (td, J = 10.0, 5.0 Hz, 1 H), 2.77 (dd, J = 15.0, 5.0 Hz, 1 H), 2.65 (td, J = 10.0, 5.0 Hz, 1 H), 2.37 (td, J = 12.5, 5.9 Hz, 1 H), 2.28 (m, 1 H), 2.07 (td, J = 10.0, 5.0 Hz, 1 H), 2.02 (d, J = 10.0 Hz, 1 H), 1.59 (m, 1 H), 1.48 (J = 10.0, 5.0 Hz, 1 H); 13C NMR (125 MHz, CDCl3) δ 146.4, 140.5, 138.7, 128.8, 128.4, 127.7, 126.2, 126.1, 124.1, 122.5, 63.7, 58.3, 46.9, 44.5, 39.9, 34.4, 30.2; IR (thin film) 2942, 2811, 1472, 1121 cm−1; HRMS (ESI) m/z calcd for C19H22N (M+H)+, 264.1747; found, 264.1748.
(±)-2-Methyl-2,3,4,5-tetrahydro-1H-1,5-methanobenzo[c]-azepin-8-yl)morpholine (JWG-1014)
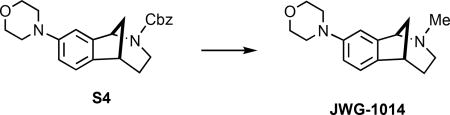
Iodotrimethylsilane (318 mg, 0.226 mL 1.59 mmol) was added in one portion to a solution of carbamate S4 (300 mg, 0.797 mmol) in CH2Cl2 (12 mL) at 0 °C in a flask wrapped in foil. The cooling bath was removed after 1 h, and the reaction was stirred for an additional 4 h, whereupon the mixture was cooled to 0 °C and methanolic HCl (2.0 M, 5 mL) added. After stirring for 5 min, the volatiles were removed under reduced pressure, 2 N HCl(aq) (20 mL) was added, and the aqueous mixture was extracted with Et2O (2 × 15 mL). The pH of the aqueous layer was adjusted to ~9 by the slow addition of 2.2 M NaOH(aq) and then extracted with CH2Cl2 (3 × 20 mL). The combined organic extracts were dried (Na2SO4) and concentrated under reduced pressure to afford 155 mg (79%) of the secondary amine intermediate, which was of sufficient purity (>95% LC and 1H NMR) to be used in the next step without purification. Paraformaldehyde (S6, 66 mg, 2.2 mmol) was added to a solution of secondary amine (28 mg, 0.11 mmol) in DCE (1 mL) followed by the addition of sodium triacetoxyborohydride (117 mg, 0.55 mmol) and acetic acid (10.5 μL, 0.18 mmol). The mixture was stirred for 24 h and then quenched by the addition of 2.2 M NaOH(aq) (~1 mL) and H2O (~2 mL). After stirring for 5 more min, the layers were separated, H2O was added to the aqueous mixture (~2 mL) followed by extraction with CH2Cl2 (3 × 20 mL). Brine was added to the aqueous mixture, which was then extracted with CH2Cl2 (2 × 20 mL). The combined organic extracts were dried (MgSO4), filtered, and concentrated under reduced pressure to give the N-methylamine, which was purified via flash column chromatography (SiO2), eluting with 100% hexanes → 10% EtOAc/hexanes → 100% EtOAc → 10% MeOH/EtOAc → 20% MeOH/EtOAc to give 26 mg (92%; 73% over two steps) of JWG-1014 as a light yellow oil: 1H NMR (500 MHz, CDCl3) δ 7.12 (d, J = 10.0 Hz, 1 H), 6.81 (s, 1 H), 6.80 (d, J = 10.0 Hz, 1 H), 3.88 (t, J = 5.0 Hz, 4 H), 3.83 (d, J = 5.0 Hz, 1 H), 3.15 (t, J = 5.0 Hz, 4 H), 3.11–3.07 (m, 1 H), 2.61 (dd, J = 7.5 Hz, 1 H), 2.22 (s, 3 H), 2.04 (d, J = 10.0 Hz, 1 H), 1.98–1.85 (comp, 3 H), 1.54–1.47 (m, 1 H); 13C NMR (125 MHz, CDCl3) δ 150.3, 138.9, 138.1, 123.0, 115.3, 113.0, 67.0, 66.1, 50.2, 48.4, 44.4, 43.2, 38.4, 29.9; IR (thin film) 2935, 2846, 1492, 1245, 1121 cm−1; HRMS (ESI) m/z calcd for C16H23N2O (M+H)+, 259.1805; found, 259.1803.
(±)-Benzyl-9-chloro-3,4,5,6-tetrahydro-1,6-methanobenzo[c]-azocine-2(1H)-carboxylate (2).56
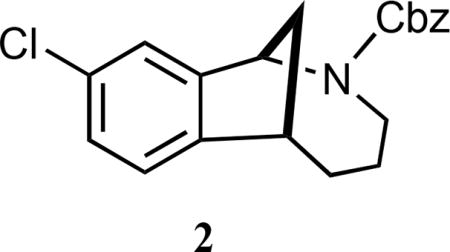
White solid (crystals from CH3CN); mp 116–119 °C; 1H NMR (500 MHz, CDCl3, as a mixture of rotamers) δ 7.53 (br s, 0.4 H), 7.40 (d, J = 10.0 Hz, 1 H), 7.34 (t, J = 7.5 Hz, 1 H), 7.30–7.15 (comp, 3.6 H), 7.14–7.07 (m, 1 H), 6.99 (t, J = 7.5 Hz, 1 H), 5.46 (d, J = 7.5 Hz, 0.4 H), 5.29 (d, J = 7.5 Hz, 0.6 H), 5.18 (d, J = 15.0 Hz, 0.6 H), 5.15–5.08 (m, 1 H), 5.01 (d, J = 15.0 Hz, 0.4 H), 4.08 (dd, J = 15.0, 8.8 Hz, 0.6 H), 3.83 (dd, J = 15.0, 8.8 Hz, 0.4 H), 3.33–3.26 (m, 1 H), 3.20–3.07 (m, 1 H), 2.31–2.17 (m, 1 H), 2.08 (t, J = 15.0 Hz, 1 H), 1.78–1.65 (comp, 2 H), 1.53–1.39 (m, 1 H), 0.98–0.85 (m, 1 H); 13C NMR (125 MHz, CDCl3, as a mixture of rotamers) δ 155.6, 155.4, 147.5, 147.3, 145.1, 145.0, 136.8, 136.5, 132.6, 132.4, 128.7, 128.5 (2 C), 128.4, 128.2, 127.9, 127.8, 125.5, 125.1, 124.6, 124.4, 67.5, 67.2, 58.8, 58.2, 44.7, 44.1, 42.9, 42.7, 35.3, 34.9, 34.7, 24.7, 24.5; IR (thin film) 2942, 1705, 1417, 1279 cm−1; HRMS (ESI) m/z calcd for C20H20ClNNaO2 (M+Na)+, 364.1080; found, 364.1084.
(±)-9-Chloro-2-(cyclohexylmethyl)-1,2,3,4,5,6-hexahydro-1,6-methanobenzo[c]azocine (MFG-1046)
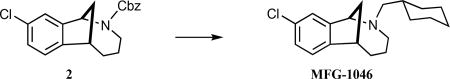
Iodotrimethylsilane (384 mg, 0.27 mL 1.92 mmol) was added in one portion to a solution of carbamate 2 (220 mg, 0.64 mmol) in CH2Cl2 (10.0 mL) at 0 °C in flask wrapped in foil. The cooling bath was removed after 2 h, and the reaction was stirred for an additional 3.25 h, whereupon the mixture was cooled to 0 °C, and methanolic HCl (2.0 M, 15 mL) was added. After stirring for 5 min, the volatiles were removed under reduced pressure, 2 N HCl(aq) (30 mL) was added, and the aqueous mixture was extracted with Et2O (2 × 15 mL). The pH of the aqueous layer was adjusted to ~9 by the slow addition of 2.2 M NaOH(aq) and then extracted with CH2Cl2 (3 × 20 mL). The combined organic extracts were dried (Na2SO4), filtered, and concentrated under reduced pressure to afford 120 mg (90%) of the crude secondary amine which was of sufficient purity (>95% LC and 1H NMR) to be used in the next step without purification. Cyclohexanecarboxaldehyde (S8, 47 mg, 50 μL, 0.42 mmol) was added to a solution of secondary amine (30 mg, 0.14 mmol) in DCE (1 mL), followed by the addition of sodium triacetoxyborohydride (88 mg, 0.42 mmol). The mixture was stirred for 24 h and then quenched by the addition of saturated sodium bicarbonate solution(aq). After stirring for 5 min, the layers were separated, and H2O (~1 mL) was added and the resultant mixture extracted with CH2Cl2 (3 × 5 mL). Brine was added to the aqueous mixture, which was then extracted with CH2Cl2 (1 × 5 mL). The combined organic extracts were dried (MgSO4), filtered, and concentrated under reduced pressure to give the crude product, which was purified via flash column chromatography (SiO2), eluting with 100% hexanes → 10% EtOAc/hexanes to afford 13 mg (30%) MFG-1046 as a colorless oil: 1H NMR (500 MHz, CDCl3) δ 7.36 (d, J = 10.0 Hz, 1 H), 7.26 (d, J = 10.0 Hz, 1 H), 7.20–7.16 (m, 1 H), 4.76–4.68 (m, 1 H), 3.39–3.34 (m, 1 H), 3.22–3.08 (m, 1 H), 2.83–2.58 (comp, 4 H), 2.41–2.30 (m, 1 H), 2.23–1.89 (comp, 5 H), 1.79–1.69 (comp, 2 H), 1.67–1.49 (comp, 4 H), 1.36–0.99 (comp, 4 H); 13C NMR (125 MHz, CDCl3) δ 151.9, 153.3, 133.1, 131.5, 126.4, 126.1, 67.1, 61.1, 51.8, 40.7, 34.4, 34.1, 32.3, 32.0, 30.0, 25.8, 25.6; IR (thin film) 2935, 2860, 1458 cm−1; HRMS (ESI) m/z calcd for C19H27ClN (M+H)+, 304.18265; found, 304.18300.
(±)-Benzyl-9-(4-(trifluoromethyl)phenyl)-3,4,5,6-tetrahydro-1,6-methanobenzo[c]azocine-2(1H)-carboxylate (S10)
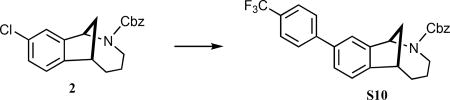
A solution of carbamate 2 (258 mg, 0.75 mmol), 4-(trifluoromethyl)phenylboronic acid (286 mg, 1.5 mmol), Cs2CO3 (489 mg, 1.5 mmol), and palladium(bis)(tert-butyl)3 phosphine (19 mg, 37 μmol) in degassed 1,4-dioxane (3.2 mL) was stirred for 1 min at room temperature and then 4 h at 98 °C. The reaction was cooled to room temperature and filtered through a pad of Celite, and the filter pad was rinsed with EtOAc (~30 mL). The combined filtrates were concentrated under reduced pressure, and the crude residue was purified via radial plc (SiO2), eluting with 100% hexanes → 5% EtOAc/hexanes → 10% EtOAc/hexanes to give 299 mg S10 (88%) as a white foam: 1H NMR (500 MHz, CDCl3, as a mixture of rotamers) δ 7.70–7.64 (m, 1 H), 7.61 (d, J = 10.0 Hz, 1 H), 7.53 (br s, 1 H), 7.52–7.42 (comp, 2 H), 7.39–7.31 (comp, 4 H), 7.23–7.20 (comp, 3), 5.66 (dd, J = 7.5 Hz, 0.4 H), 5.50 (dd, J = 7.5 Hz, 0.6 H), 5.37–5.29 (m, 1 H), 5.21–5.08 (m, 1 H), 4.21 (dd, J = 15.0, 5.0 Hz, 0.6 H), 3.93 (dd, J = 15.0, 5.0 Hz, 0.4 H), 3.50–3.45 (m, 1 H), 3.34–3.22 (m, 1 H), 2.46–2.34 (m, 1 H), 2.25–2.16 (m, 1 H), 1.95–1.85 (comp, 2 H), 1.65–1.55 (m, 1 H), 1.14–1.00 (m, 1 H); 13C NMR (125 MHz, CDCl3, mixture of rotamers) δ 155.7, 155.6, 147.1, 146.8, 146.7, 146.5, 144.7, 138.9, 138.7, 136.9, 136.7, 128.6, 128.5, 128.4, 128.2, 127.9, 127.8, 127.4, 127.2, 125.6 (q, JC-F = 3.75 Hz), 124.2, 124.0, 123.8, 123.3, 67.5, 67.2, 59.0, 58.3, 44.8, 44.2, 43.2, 43.0, 35.4, 35.3, 35.0, 34.9, 24.9, 24.7; IR (thin film) 2928, 1698, 1334 cm−1; HRMS (ESI) m/z calcd for C27H24F3NNa2O2 (M+Na)+, 474.1651; found, 474.1657.
(±)-2-Cyclopentyl-9-(4-(trifluoromethyl)phenyl)-1,2,3,4,5,6-hexa-hydro-1,6-methanobenzo[c]azocine (DKR-1051)
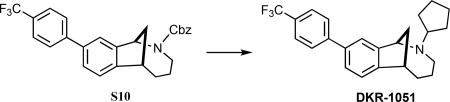
A mixture of S10 (297 mg, 0.65 mmol) and 10% Pd/C (27 mg) in ethanol (10 mL) was stirred under an atmosphere of H2 for five h. More 10% Pd/C (44 mg) was added, and the reaction continued for an additional 3 h. The catalyst was removed by filtration through a cotton plug, and the combined filtrates were concentrated under reduced pressure to provide 208 mg (99%) of crude product, which was of sufficient purity (1H NMR) to use in the subsequent reaction. Cyclopentanone (S11, 36 mg, 40 μL, 0.43 mmol) was added to a solution of secondary amine (69 mg, 0.21 mmol) in DCE (2.0 mL), and sodium triacetoxyborohydride (91 mg, 0.43 mmol) was added. The mixture was stirred for 24 h, and then saturated sodium bicarbonate solution(aq) (3 mL) was added. After stirring for 5 min, the layers were separated, H2O was added (~1 mL), and the aqueous mixture was extracted with CH2Cl2 (3 × 5 mL). The combined organic extracts were dried (MgSO4), filtered, and concentrated under reduced pressure to give the crude product, which was purified via radial plc (SiO2), eluting with 10% EtOAc/hexanes → 15% EtOAc/hexanes to give 66 mg (79%; 78% over two steps) of DKR-1051 as a colorless oil: 1H NMR (500 MHz, CDCl3) δ 7.72–7.65 (comp, 4 H), 7.54–7.45 (m, 1 H), 7.48 (d, J = 7.5 Hz, 1 H), 7.29 (d, J = 7.5 Hz, 1 H), 4.55–4.46 (m, 1 H), 3.37 (app t, J = 5.0 Hz, 1 H), 3.08–3.00 (m, 1 H), 2.82–2.73 (m, 1 H), 2.43–2.23 (comp, 2 H), 2.19–2.06 (comp, 2 H), 2.02–1.90 (comp, 2 H), 1.83–1.71 (comp, 4 H), 1.65–1.57 (comp, 4 H), 1.38–1.27 (m, 1 H); 13C NMR (125 MHz, CDCl3) δ 149.7, 145.1, 138.6, 129.1, 128.9, 127.4, 125.6 (q, JC-F = 3.75 Hz), 124.4, 123.9, 123.7, 123.3, 65.0, 63.5, 50.2, 42.3, 34.3, 32.9, 31.9, 30.7, 24.0; IR (thin film) 2963, 2866, 1623, 1327, 1128 cm−1; HRMS (ESI) m/z calcd for C24H27F3N (M+H)+, 386.2090; found 386.2092.
(±)-9-(4-(Trifluoromethyl)phenyl)-3,4,5,6-tetrahydro-1,6-methanobenzo[c]azocin-2(1H)-yl)propan-1-ol (UKH-1114)
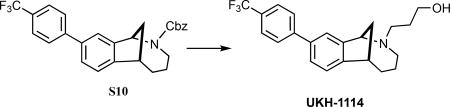
The benzyl carbamate of S10 was removed with hydrogenolysis conditions as reported for the synthesis of DKR-1051. The secondary amine was of sufficient purity (1H NMR) to use in the subsequent reaction. 3-Bromo-1-propanol (S12, 42 mg, 30 μL, 0.30 mmol) and K2CO3 (55 mg, 0.40 mmol) were added to a solution of secondary amine (32 mg, 0.10 mmol) in CH3CN (2.0 mL), and the mixture was heated at 55 °C for 17 h. After cooling to room temperature, the solids were removed by filtration, and the filter cake was rinsed with CH2Cl2. The combined filtrates were concentrated under reduced pressure to give the crude product, which was purified via radial plc (SiO2), eluting with 50% EtOAc/hexanes → 100% EtOAc → 3% MeOH/EtOAc to give 24 mg (64%; 63% over two steps) of UKH-1114 as a colorless oil: 1H NMR (400 MHz, CD2Cl2) δ 7.78–7.68 (comp, 4 H), 7.57 (s, 1 H), 7.53 (dd, J = 7.9, 1.8 Hz, 1 H), 7.33 (d, J = 7.9 Hz, 1 H), 4.41 (d, J = 5.6 Hz, 1 H), 3.93–3.82 (comp, 2 H), 3.38 (t, J = 6.2 Hz, 1 H), 3.01–2.92 (m, 1 H), 2.86–2.78 (m, 1 H), 2.63 (dd, J = 12.0, 8.4 Hz, 1 H), 2.24–2.11 (comp, 3 H), 2.04–1.94 (m, 1 H), 1.94–1.82 (m, 1 H), 1.78–1.65 (comp, 3 H), 1.42–1.30 (m, 1 H); 13C NMR (150 MHz, CDCl3) δ 144.6, 138.8, 129.4, 129.1, 128.3, 127.5, 125.7 (q, JC-F = 3.3 Hz), 125.2, 124.2, 124.1, 123.4, 66.5, 66.4, 50.3, 45.3, 41.5, 33.3, 27.9, 23.5, 22.3; IR (thin film) 3361, 2935, 2853, 1609, 1334, 1121 cm−1; HRMS (ESI) m/z calcd for C22H25F3NO (M+H)+, 376.1883; found 376.1892.
Sigma Receptor Binding Assay Protocol
Receptor binding assays were performed by the Psychoactive Drug Screening Program (PDSP) at Chapel Hill, North Carolina. The assay protocol book can be accessed free of charge at: https://pdspdb.unc.edu/pdspWeb/content/PDSP%20Protocols%20II%202013-03-28.pdf. Briefly, σ1R were sourced from guinea pig brain and Sig1R binding affinity (Ki) was determined through competition binding assays with [3H]-(+)-pentazocine. σ2R were obtained from rat PC12 cells, and the Sig2R ligand binding affinity (Ki) was determined through competition binding assays using the radioligand [3H]-ditolylguanidine in the presence of (+)-pentazocine to block σ1R binding sites. Ki values are calculated from best-fit IC50 determinations and are the average of two or more independent runs performed in triplicate.
Sample Preparation for in Vivo Use
Dry compounds were reconstituted in dimethyl sulfoxide (DMSO) to a stock concentration between 25 and 100 mM and then diluted down to 10 μg total drug in a volume of 5 μL for IT injections. The total amount of DMSO for IT injections was always less than 5%, and the vehicle injections always contained the same amount of DMSO. Siramesine (Tocris) sample was reconstituted and diluted in the same way. Samples were diluted in the same manner for IV injections.
Gene Expression Analysis
Transcripts per million (TPM) was used to quantify relative gene abundance. Fragments per kilobase of transcript per million mapped reads (FPKMs) for coding genes were obtained by running the Tophat/Cuffdiff57 pipeline or from published data. These were then normalized to sum to 1 million in order to generate TPMs. For mouse single cell RNA-seq data sets, low sequencing depth makes TPMs inaccurate due to high sampling variation, and the fraction of cells with presence of sequenced reads from genes of interest for different cell types are presented instead.
Statistics
Data are shown as mean ± standard error of the mean, and the number of animals or samples used in each analysis are shown in figure legends. GraphPad Prism 7 for Mac OSX was used to analyze data. Two-way ANOVAs were used to analyze von Frey data with Bonferroni’s multiple comparison post hoc test. Two tailed t tests were used to analyze rotorod data. Significance level was at α < 0.05.
Acknowledgments
Funding
This work was supported by NIH grants R01NS065926 (T.J.P.) and R56NS098826 (T.J.P.), The University of Texas STARS program (T.J.P.), The Robert A. Welch Foundation (F-0652) (S.F.M.), and the Dell Medical School’s Texas Health Catalyst program (S.F.M. and J.J.S.).
ABBREVIATIONS
- σ1R
sigma 1 receptor
- σ2R
sigma 2 receptor
- AD
Alzheimer’s disease
- DRG
dorsal root ganglion
- IT
intrathecal
- IV
intravenous
- MCI
mild cognitive impairment
- SNI
spared nerve injury
- TMEM97
transmembrane protein 97
- TPM
transcripts per million mapped reads
Footnotes
Author Contributions
J.J.S. prepared compounds; J.J.S., G.L.M., and P.R.R. performed experiments; J.J.S., P.R.R, S.F.M., and T.J.P. designed experiments; J.J.S., P.R.R., S.F.M., and T.J.P. wrote the paper.
The authors declare no competing financial interest.
References
- 1.Finnerup NB, Attal N, Haroutounian S, McNicol E, Baron R, Dworkin RH, Gilron I, Haanpaa M, Hansson P, Jensen TS, Kamerman PR, Lund K, Moore A, Raja SN, Rice AS, Rowbotham M, Sena E, Siddall P, Smith BH, Wallace M. Pharmacotherapy for neuropathic pain in adults: a systematic review and meta-analysis. Lancet Neurol. 2015;14:162–173. doi: 10.1016/S1474-4422(14)70251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsumoto RR, Bown WD, Su TP. Sigma receptors: Chemistry, cell biology and clinical implications. Springer; New York: 2007. [Google Scholar]
- 3.Schmidt HR, Zheng S, Gurpinar E, Koehl A, Manglik A, Kruse AC. Crystal structure of the human sigma1 receptor. Nature. 2016;532:527–530. doi: 10.1038/nature17391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis MP. Sigma-1 receptors and animal studies centered on pain and analgesia. Expert Opin. Drug Discovery. 2015;10:885–900. doi: 10.1517/17460441.2015.1051961. [DOI] [PubMed] [Google Scholar]
- 5.Gris G, Portillo-Salido E, Aubel B, Darbaky Y, Deseure K, Vela JM, Merlos M, Zamanillo D. The selective sigma-1 receptor antagonist E-52862 attenuates neuropathic pain of different aetiology in rats. Sci. Rep. 2016;6:24591. doi: 10.1038/srep24591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alon A, Schmidt HR, Wood MD, Sahn JJ, Martin SF, Kruse AC. Identification of the gene that codes for the sigma 2 receptor. Proc. Natl. Acad. Sci. U.S.A. 2017 doi: 10.1073/pnas.1705154114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartz F, Kern L, Erz D, Zhu M, Gilbert D, Meinhof T, Wirkner U, Erfle H, Muckenthaler M, Pepperkok R, Runz H. Identification of cholesterol-regulating genes by targeted RNAi screening. Cell Metab. 2009;10:63–75. doi: 10.1016/j.cmet.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez-Pulido L, Ponting CP. TM6SF2 and MAC30, new enzyme homologs in sterol metabolism and common metabolic disease. Front. Genet. 2014;5:439. doi: 10.3389/fgene.2014.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebrahimi-Fakhari D, Wahlster L, Bartz F, Werenbeck-Ueding J, Praggastis M, Zhang J, Joggerst-Thomalla B, Theiss S, Grimm D, Ory DS, Runz H. Reduction of TMEM97 increases NPC1 protein levels and restores cholesterol trafficking in Niemann-pick type C1 disease cells. Hum. Mol. Genet. 2016;25:3588–3599. doi: 10.1093/hmg/ddw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi B, Sahn JJ, Ardestani PM, Evans AK, Scott LL, Chan JZ, Iyer S, Crisp A, Zuniga G, Pierce JT, Martin SF, Shamloo M. Small molecule modulator of sigma 2 receptor is neuroprotective and reduces cognitive deficits and neuroinflammation in experimental models of Alzheimer’s disease. J. Neurochem. 2017;140:561–575. doi: 10.1111/jnc.13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cahill MA. Progesterone receptor membrane component 1: an integrative review. J. Steroid Biochem. Mol. Biol. 2007;105:16–36. doi: 10.1016/j.jsbmb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Kabe Y, Nakane T, Koike I, Yamamoto T, Sugiura Y, Harada E, Sugase K, Shimamura T, Ohmura M, Muraoka K, Yamamoto A, Uchida T, Iwata S, Yamaguchi Y, Krayukhina E, Noda M, Handa H, Ishimori K, Uchiyama S, Kobayashi T, Suematsu M. Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance. Nat. Commun. 2016;7:11030. doi: 10.1038/ncomms11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hand RA, Craven RJ. Hpr6.6 protein mediates cell death from oxidative damage in MCF-7 human breast cancer cells. J. Cell. Biochem. 2003;90:534–547. doi: 10.1002/jcb.10648. [DOI] [PubMed] [Google Scholar]
- 14.Guo L, Zhen X. Sigma-2 receptor ligands: neurobiological effects. Curr. Med. Chem. 2015;22:989–1003. doi: 10.2174/0929867322666150114163607. [DOI] [PubMed] [Google Scholar]
- 15.Cognition Therapeutics. Clinical Trial of CT1812 in Mild to Moderate Alzheimer’s Disease, ClinicalTrials.gov Identifier: NCT02907567 2016
- 16.Minerva Neurosciences. Study to Evaluate the Efficacy, Safety, and Tolerability of MIN-101 in Patients With Negative Symptoms of Schizophrenia, EudraCT number: 2014-004878-42. 2014. [Google Scholar]
- 17.Sahn JJ, Hodges TR, Chan JZ, Martin SF. Norbenzomorphan Scaffold: Chemical Tool for Modulating Sigma Receptor-Subtype Selectivity. ACS Med. Chem. Lett. 2017;8:455. doi: 10.1021/acsmedchemlett.7b00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 19.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 20.Izzo NJ, Xu J, Zeng C, Kirk MJ, Mozzoni K, Silky C, Rehak C, Yurko R, Look G, Rishton G, Safferstein H, Cruchaga C, Goate A, Cahill MA, Arancio O, Mach RH, Craven R, Head E, LeVine H, Spires-Jones TL, 3rd, Catalano SM. Alzheimer’s therapeutics targeting amyloid beta 1–42 oligomers II: Sigma-2/PGRMC1 receptors mediate Abeta 42 oligomer binding and synaptotoxicity. PLoS One. 2014;9:e111899. doi: 10.1371/journal.pone.0111899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roh DH, Kim HW, Yoon SY, Seo HS, Kwon YB, Kim KW, Han HJ, Beitz AJ, Na HS, Lee JH. Intrathecal injection of the sigma(1) receptor antagonist BD1047 blocks both mechanical allodynia and increases in spinal NR1 expression during the induction phase of rodent neuropathic pain. Anesthesiology. 2008;109:879–889. doi: 10.1097/ALN.0b013e3181895a83. [DOI] [PubMed] [Google Scholar]
- 22.de la Puente B, Nadal X, Portillo-Salido E, Sanchez-Arroyos R, Ovalle S, Palacios G, Muro A, Romero L, Entrena JM, Baeyens JM, Lopez-Garcia JA, Maldonado R, Zamanillo D, Vela JM. Sigma-1 receptors regulate activity-induced spinal sensitization and neuropathic pain after peripheral nerve injury. Pain. 2009;145:294–303. doi: 10.1016/j.pain.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Merlos M, Romero L, Zamanillo D, Plata-Salaman C, Vela JM. Sigma-1 Receptor and Pain. Handb. Exp. Pharmacol. 2017 doi: 10.1007/164_2017_9. [DOI] [PubMed] [Google Scholar]
- 24.Zhu S, Wang C, Han Y, Song C, Hu X, Liu Y. Sigma-1 Receptor Antagonist BD1047 Reduces Mechanical Allodynia in a Rat Model of Bone Cancer Pain through the Inhibition of Spinal NR1 Phosphorylation and Microglia Activation. Mediators Inflammation. 2015;2015:265056. doi: 10.1155/2015/265056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perregaard J, Moltzen EK, Meier E, Sanchez C. Sigma ligands with subnanomolar affinity and preference for the sigma 2 binding site. 1. 3-(omega-aminoalkyl)-1H–indoles. J. Med. Chem. 1995;38:1998–2008. doi: 10.1021/jm00011a019. [DOI] [PubMed] [Google Scholar]
- 26.Abate C, Perrone R, Berardi F. Classes of sigma2 (sigma2) receptor ligands: structure affinity relationship (SAfiR) studies and antiproliferative activity. Curr. Pharm. Des. 2012;18:938–949. doi: 10.2174/138161212799436485. [DOI] [PubMed] [Google Scholar]
- 27.Zeng C, Rothfuss JM, Zhang J, Vangveravong S, Chu W, Li S, Tu Z, Xu J, Mach RH. Functional assays to define agonists and antagonists of the sigma-2 receptor. Anal. Biochem. 2014;448:68–74. doi: 10.1016/j.ab.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.GTEx Consortium. Lonsdale J, Thomas J, Salvatore M, et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The ENCODE Project Consortium. Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjostedt E, Asplund A, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 31.Duff MO, Olson S, Wei X, Garrett SC, Osman A, Bolisetty M, Plocik A, Celniker SE, Graveley BR. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature. 2015;521:376–379. doi: 10.1038/nature14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson S, Golden JP, Copits BA, Ray PR, Vogt SK, Valtcheva MV, Schmidt RE, Ghetti A, Price TJ, Gereau RW., IV Group II mGluRs suppress hyperexcitability in mouse and human nociceptors. Pain. 2016;157:2081–2088. doi: 10.1097/j.pain.0000000000000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilad Y, Mizrahi-Man O. A reanalysis of mouse ENCODE comparative gene expression data. F1000Research. 2015;4:121. doi: 10.12688/f1000research.6536.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benoit J, Ayoub AE, Rakic P. Transcriptomics of critical period of visual cortical plasticity in mice. Proc. Natl. Acad. Sci. U. S. A. 2015;112:8094–8099. doi: 10.1073/pnas.1509323112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerhold KA, Pellegrino M, Tsunozaki M, Morita T, Leitch DB, Tsuruda PR, Brem RB, Catania KC, Bautista DM. The star-nosed mole reveals clues to the molecular basis of mammalian touch. PLoS One. 2013;8:e55001. doi: 10.1371/journal.pone.0055001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eipper-Mains JE, Kiraly DD, Duff MO, Horowitz MJ, McManus CJ, Eipper BA, Graveley BR, Mains RE. Effects of cocaine and withdrawal on the mouse nucleus accumbens transcriptome. Genes, Brain and behavior. 2013;12:21–33. doi: 10.1111/j.1601-183X.2012.00873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huan T, Meng Q, Saleh MA, Norlander AE, Joehanes R, Zhu J, Chen BH, Zhang B, Johnson AD, Ying S, et al. Integrative network analysis reveals molecular mechanisms of blood pressure regulation. Mol. Syst. Biol. 2015;11:799. doi: 10.15252/msb.20145399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romero L, Zamanillo D, Nadal X, Sanchez-Arroyos R, Rivera-Arconada I, Dordal A, Montero A, Muro A, Bura A, Segales C, Laloya M, Hernandez E, Portillo-Salido E, Escriche M, Codony X, Encina G, Burgueno J, Merlos M, Baeyens JM, Giraldo J, Lopez-Garcia JA, Maldonado R, Plata-Salaman CR, Vela JM. Pharmacological properties of S1RA, a new sigma-1 receptor antagonist that inhibits neuropathic pain and activity-induced spinal sensitization. Br. J. Pharmacol. 2012;166:2289–2306. doi: 10.1111/j.1476-5381.2012.01942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji RR, Chamessian A, Zhang YQ. Pain regulation by non-neuronal cells and inflammation. Science. 2016;354:572–577. doi: 10.1126/science.aaf8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nieto FR, Cendan CM, Sanchez-Fernandez C, Cobos EJ, Entrena JM, Tejada MA, Zamanillo D, Vela JM, Baeyens JM. Role of sigma-1 receptors in paclitaxel-induced neuropathic pain in mice. J. Pain. 2012;13:1107–1121. doi: 10.1016/j.jpain.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 43.Diaz JL, Cuberes R, Berrocal J, Contijoch M, Christmann U, Fernandez A, Port A, Holenz J, Buschmann H, Laggner C, Serafini MT, Burgueno J, Zamanillo D, Merlos M, Vela JM, Almansa C. Synthesis and biological evaluation of the 1-arylpyrazole class of sigma(1) receptor antagonists: identification of 4-{2-[5-methyl-1-(naphthalen-2-yl)-1H–pyrazol-3-yloxy]ethyl}-morpholine (S1RA, E-52862) J. Med. Chem. 2012;55:8211–8224. doi: 10.1021/jm3007323. [DOI] [PubMed] [Google Scholar]
- 44.Mavlyutov TA, Duellman T, Kim HT, Epstein ML, Leese C, Davletov BA, Yang J. Sigma-1 receptor expression in the dorsal root ganglion: Reexamination using a highly specific antibody. Neuroscience. 2016;331:148–157. doi: 10.1016/j.neuroscience.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu G, Sun W, Zou Y, Cai Z, Wang P, Lin X, Huang J, Jiang L, Ding X, Hu G. RNA interference against TMEM97 inhibits cell proliferation, migration, and invasion in glioma cells. Tumor Biol. 2015;36:8231–8238. doi: 10.1007/s13277-015-3552-6. [DOI] [PubMed] [Google Scholar]
- 46.Asiedu MN, Tillu DV, Melemedjian OK, Shy A, Sanoja R, Bodell B, Ghosh S, Porreca F, Price TJ. Spinal protein kinase M zeta underlies the maintenance mechanism of persistent nociceptive sensitization. J. Neurosci. 2011;31:6646–6653. doi: 10.1523/JNEUROSCI.6286-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melemedjian OK, Asiedu MN, Tillu DV, Sanoja R, Yan J, Lark A, Khoutorsky A, Johnson J, Peebles KA, Lepow T, Sonenberg N, Dussor G, Price TJ. Targeting adenosine monophosphate-activated protein kinase (AMPK) in preclinical models reveals a potential mechanism for the treatment of neuropathic pain. Mol. Pain. 2011;7:70. doi: 10.1186/1744-8069-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melemedjian OK, Khoutorsky A, Sorge RE, Yan J, Asiedu MN, Valdez A, Ghosh S, Dussor G, Mogil JS, Sonenberg N, Price TJ. mTORC1 inhibition induces pain via IRS-1-dependent feedback activation of ERK. Pain. 2013;154:1080–1091. doi: 10.1016/j.pain.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 50.Decosterd I, Woolf CJ. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain. 2000;87:149–158. doi: 10.1016/S0304-3959(00)00276-1. [DOI] [PubMed] [Google Scholar]
- 51.Hylden JL, Wilcox GL. Intrathecal morphine in mice: a new technique. Eur. J. Pharmacol. 1980;67:313–316. doi: 10.1016/0014-2999(80)90515-4. [DOI] [PubMed] [Google Scholar]
- 52.Pangborn AB, Giardello MA, Grubbs RH, Rosen RK, Timmers FJ. Safe and convenient procedure for solvent purification. Organometallics. 1996;15:1518–1520. [Google Scholar]
- 53.Still WC, Kahn M, Mitra A. Rapid Chromatographic Technique for Preparative Separations with Moderate Resolution. J. Org. Chem. 1978;43:2923–2925. [Google Scholar]
- 54.Sahn JJ, Martin SF. Expedient synthesis of norbenzomorphan library via multicomponent assembly process coupled with ring-closing reactions. ACS Comb. Sci. 2012;14:496–502. doi: 10.1021/co300068a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sahn JJ, Hodges TR, Chan JZ, Martin SF. Norbenzomorphan Framework as a Novel Scaffold for Generating Sigma 2 Receptor/PGRMC1 Subtype-Selective Ligands. ChemMed-Chem. 2016;11:556–561. doi: 10.1002/cmdc.201500551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahn JJ, Martin SF. Facile Syntheses of Substituted, Conformationally-Constrained Benzoxazocines and Benzazocines via Sequential Multicomponent Assembly and Cyclization. Tetrahedron Lett. 2011;52:6855–6858. doi: 10.1016/j.tetlet.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]